Contact-dependent growth inhibition (CDI) systems are used in bacterial competition to hinder the growth of neighboring microbes. These systems utilize a two-partner secretion mechanism to display the CdiA exoprotein at the bacterial cell surface.
KEYWORDS: contact-dependent inhibition, Pseudomonas aeruginosa
ABSTRACT
Contact-dependent growth inhibition (CDI) systems are used in bacterial competition to hinder the growth of neighboring microbes. These systems utilize a two-partner secretion mechanism to display the CdiA exoprotein at the bacterial cell surface. CdiA forms a long filamentous stalk that facilitates binding to a target cell and delivery of a C-terminal toxin (CT) domain. This CT domain is processed and delivered into the cytoplasm of a target cell upon contact. CDI systems also encode a cognate immunity protein (CdiI) that protects siblings and resistant targeted cells from intoxication by high-affinity binding to the CT. CdiA CT domains vary among strains within a species, and many alleles encode enzymatic functions that target nucleic acids. This variation is thought to help drive diversity and adaptation within a species. CdiA diversity is well studied in Escherichia coli and several other bacteria, but little is known about the extent of this diversity in Pseudomonas aeruginosa. The purpose of this review is to highlight the variability that exists in CDI systems of P. aeruginosa. We show that this diversity is apparent even among strains isolated from a single geographical region, suggesting that CDI systems play an important role in the ecology of P. aeruginosa.
INTRODUCTION
Bacteria are complex social organisms that live in mixed communities where they must compete for limited space and resources (1). As a result, they have evolved multiple systems to scavenge nutrients and interfere with the growth of other microbes (2). Multiple transport proteins facilitate uptake of crucial nutrients (3), while siderophores scavenge key metals (4). Secreted antibiotics and bacteriocins act to inhibit or kill competitors beyond arm’s reach (5–7). Bacteria can also intoxicate neighboring cells upon direct cell contact in several ways, including by outer membrane exchange (8) and type VI secretion and contact-dependent growth inhibition systems (9, 10). These contact-mediated competition systems are capable of bombarding competitors with an assorted array of antimicrobial effectors. Advances in whole-genome sequencing have rapidly increased our appreciation of the vast effector repertoire found throughout these bactericidal systems (11).
Contact-dependent growth inhibition (CDI) systems consist of an outer membrane Omp85-TpsB superfamily β-barrel protein (CdiB) that facilitates secretion of a large exoprotein (CdiA) from the periplasm onto the bacterial cell surface in a type Vb (i.e., two-partner) secretion process (12, 13). CdiA then facilitates delivery of its own C-terminal toxin domain into a neighboring bacterium to kill or prevent the growth of susceptible target strains (14). Siblings are protected from intoxication because they produce an immunity protein (CdiI) to neutralize the toxin domain. CdiA is a polymorphic toxin, meaning that different bacterial strains within a species contain CdiA proteins with different toxin domains (15). Many of these target nucleic acids for degradation (16, 17), while others have been shown to form pores in the inner membrane (18). This diversity is thought to drive important interactions that help shape bacterial populations (19).
A considerable amount of effort has been devoted to understanding variation in the CDI system of Escherichia coli and several other bacterial species (15, 20, 21), but this system has been less thoroughly examined in Pseudomonas aeruginosa. Here, we review what can be inferred about the P. aeruginosa CDI system by comparing it to the CDI system of E. coli, and we describe its genetic organization and diversity.
CURRENT MODEL OF CdiA-DEPENDENT INTOXICATION
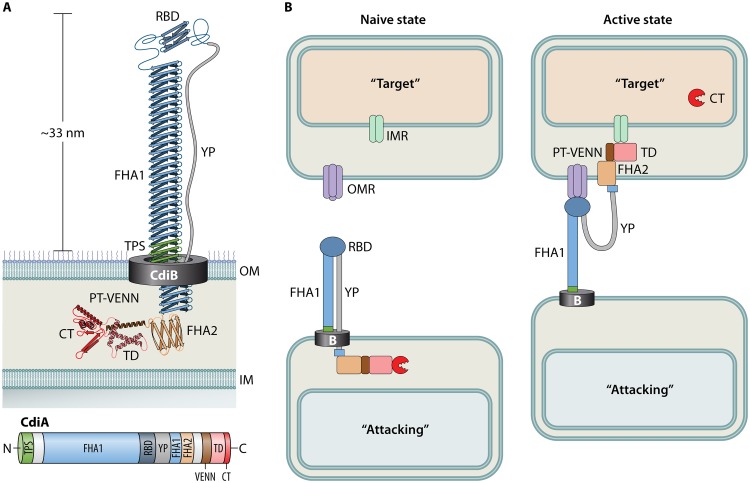
As with other bacteria, the key component of the P. aeruginosa CDI system is the CdiA exoprotein. Little experimental work has been done on P. aeruginosa CdiA, but its mechanism can be inferred from what is known about CdiA of E. coli. CdiA is a large modular protein containing several domains with distinct functions. Some domains are highly conserved within a species, while others appear to rearrange in various combinations to generate functional diversity (22, 23). All CdiA proteins contain a conserved two-partner secretion (TPS) domain (Pfam identifier [ID] PF05860) and an FHA1 domain (Pfam ID PF05594) of variable length (Fig. 1A). The FHA1 domain is composed of dozens of β-strands that presumably stack to form a rigid extracellular filament called a β-helix, as described in the filamentous hemagglutinin protein of Bordetella pertussis (24). In E. coli, this long extracellular filament is anchored at the cell surface and presents a receptor binding domain (RBD) at its distal end (Fig. 1A) (25). The RBD is required for adherence to neighboring bacteria during CDI (Fig. 1B) (22, 25). Adjacent to the RBD is a stretch of amino acids with an abundance of tyrosine and proline residues that is referred to as the YP domain. This region is thought to act as an extended linker between the RBD and the following FHA2 domain (Pfam ID PF13332). In its native conformation, the FHA2 and remaining C-terminal domains are sequestered in the periplasm of the attacking cell until contact of the RBD with its cognate receptor on a target bacterium (25). The signal of receptor binding is then likely transduced through the YP domain to release the sequestered domains and continue the intoxication process (Fig. 1B).
FIG 1.
Current model of CDI intoxication. (A) A cartoon diagram of CdiA in its native state at the cell surface of an attacking bacterium, as modeled by Ruhe et al. (25). The corresponding CdiA protein domains are illustrated to scale below the cartoon. See the text for details. (B) CdiA-dependent intoxication of a target bacterium. Upon binding of the CdiA-RBD to an outer membrane receptor (OMR) on a target bacterium, the periplasmic domains are released from the attacking bacterium to deliver the CT into the cytosol of the target bacterium. This is thought to occur through a stepwise process whereby the FHA2 domain delivers the TD and CT into the periplasm of a target cell and the TD binds an inner membrane receptor (IMR) that facilitates translocation of the CT into the cytosol through an unknown mechanism.
In E. coli, the FHA2 domain was shown to localize with the outer membrane of target cells following receptor binding, and labeling experiments suggest that this domain facilitates translocation of the remaining C-terminal domains across the target outer membrane into the periplasm (Fig. 1B) (25). The final destination of a C-terminal toxin (CT) domain depends upon its mechanism of action. CTs that disrupt membrane potential by forming pores can function from within the periplasm by attacking the periplasmic face of the inner membrane (18). In contrast, nucleic acid-degrading CTs must be further translocated to the cytosol to reach their target. This is thought to be accomplished by specific translocation domains (TD), sandwiched between the FHA2 and CT domains, which interact with specific inner membrane receptors (IMRs) to facilitate translocation of the CT across the inner membrane into the target cell cytosol (Fig. 1B) (23). IMRs that have been identified in E. coli function as various metabolite permeases, but the inherent transport function of these proteins is not required for CDI intoxication. Rather, it was proposed that the inner membrane-associated AAA+ superfamily protein transporter FtsH may be responsible for the actual translocation of the CT across the inner membrane and that TD binding to a cognate IMR facilitates an interaction with FtsH. This type of interaction has since been modeled with the secreted bacteriocin colicin D (26). Residues 410 to 437 of the colicin D central domain constitute a binding site for interaction with the inner membrane type I signal peptidase LepB. This interaction is required for FtsH-dependent import of the colicin D C-terminal RNase domain (CRD) and release of the CRD into the target cell cytoplasm (27). Likewise, it is thought that the CdiA CT separates from the TD during translocation into the target cell (28). Some have proposed the E. coli PT-VENN domain (Pfam ID PF04829) immediately preceding the TD domain may facilitate cleavage of the CT from the filament for proper delivery to the target bacterium (29), but it is also possible that FtsH-dependent cleavage of the CT occurs during import, as occurs with the nuclease colicins (27, 30).
GENETIC ORGANIZATION OF cdi LOCI IN P. AERUGINOSA
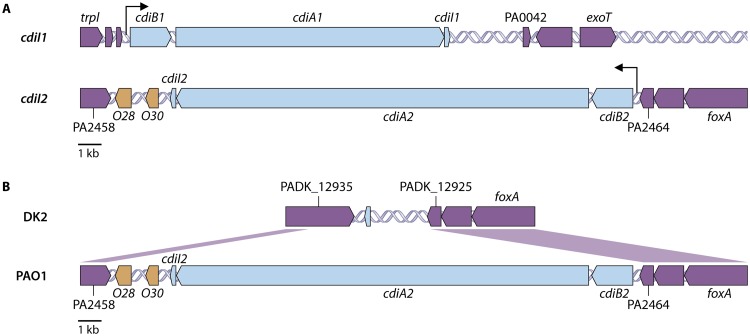
CDI systems are widespread among proteobacteria (15), and individual strains within these different groups may possess multiple cdi loci (31). An analysis of 104 completed P. aeruginosa genomes representing strains from four different continents (NCBI strains [see Table S1 in the supplemental material]) revealed that all strains contain a cdi homolog in the same genetic location upstream of the gene encoding the type III secretion system effector ExoT (Fig. 2A). In addition, 81% of strains were found to contain a second cdi locus downstream of the ferrioxamine receptor gene (foxA) (Fig. 2A). In P. aeruginosa strain PAO1, the cdiA genes located in proximity to exoT and foxA were previously designated cdiA1 (PA0041) and cdiA2 (PA2464), respectively (Fig. 2A) (32, 33). A single promoter upstream of cdiB is thought to drive expression of the entire cdiBAI operon at each locus (Fig. 2A). This genetic arrangement is similar to that observed in E. coli and many other Gram-negative bacteria (14, 16). In contrast to the high proportion of P. aeruginosa strains containing multiple cdi loci, only approximately 15% of sequenced E. coli strains contain any cdi genes (19), suggesting an important role for CDI systems in P. aeruginosa biology.
FIG 2.
Organization of P. aeruginosa CDI systems. (A) The cdi1 and cdi2 loci are displayed as blue arrows in their genetic orientation within P. aeruginosa strain PAO1. Core genes outside the cdi loci are colored purple. O28 and O30, colored orange, represent orphan immunity genes located in the intergenic region downstream of the functional cdiBAI cluster. Black arrows indicate the predicted promoter sites. (B) Alignment of the P. aeruginosa strain PAO1 cdi2 locus and flanking regions to the same genetic region in strain DK2, which does not contain a cdi2 locus.
Variations in both cdiA1 and cdiA2 suggest that the 3′ end of cdiA undergoes diversification through homologous recombination (Fig. S3), but how such genetic material would spread by horizontal gene transfer in different P. aeruginosa populations is unclear. Immediately downstream of the cdi2 locus on the complementary strand is another polymorphic gene encoding a predicted outer membrane protein with numerous Rhs repeats (Pfam ID PF05593) (Fig. 2B). This gene also contains 3′ termini that differ between strains (data not shown). Polymorphic genes encoding Rhs repeat proteins have been associated with type VI secretion systems (34); however, the function of these specific genes in P. aeruginosa is unknown. It is intriguing that two polymorphic genes with variations in their respective 3′ ends share an intergenic region, and the question remains as to whether there is a shared mechanism of 3′ diversification through some common mobile genetic element. Of note, this Rhs repeat protein is truncated in P. aeruginosa strain PAO1 (PA2458) (Fig. 2B), but this not the case for most cdiA2-containing strains. All other cdi2 flanking regions appear intact, with no obvious genetic signatures to suggest a mechanism for horizontal transfer. P. aeruginosa CdiA1UCBPP-PA14 was found on an isolated contig from Enterobacter cloacae strain e403 (contig ERS380557SCcontig000071), suggesting that horizontal transfer of cdiA may occur; however, no additional P. aeruginosa cdiA alleles have been identified outside the genus Pseudomonas. CDI systems have been observed within mobile genetic elements in other organisms. E. coli and Burkholderia pseudomallei carry cdi gene clusters on different pathogenicity islands (35), and Xenorhabdus doucetiae contains a cdi locus on an integrative conjugative element (36). Burkholderia thailandensis E264 carries a cdi locus on a novel 210-kb composite transposon that can exist as an extrachromosomal circular double-stranded DNA (dsDNA) intermediate referred to as a “megacircle” (37). Since P. aeruginosa is an environmental bacterium, it is possible that the reservoirs of mobile elements that facilitate horizontal transfer of cdiA genes in P. aeruginosa exist in this niche and not in the clinical setting, where most strains are obtained for study (38). Additional sampling from natural environments where this type of diversification likely occurs may help answer some of these questions.
THE AMINO-TERMINAL HALF OF CdiA IS CONSERVED
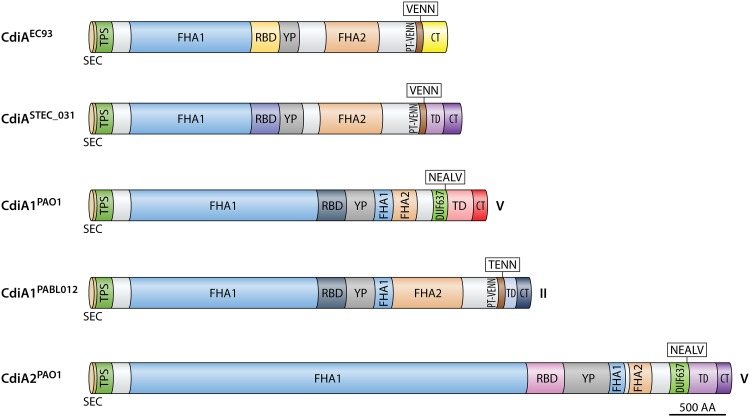
There is substantial similarity in the overall domain structure between the CdiA proteins of E. coli and P. aeruginosa, suggesting that they likely adopt the same general architecture (Fig. 3). P. aeruginosa CdiA proteins contain longer FHA1 domains than those of E. coli, with the FHA1 domain of CdiA2 having over twice the length (Fig. 3). In E. coli strain EC93, the CdiA filament was shown by cryo-electron tomography to be approximately 33 nm in length (25). If the model of CdiA in its native conformation is correct, the RBD would be the most distal domain on the filament (Fig. 1A). The length of the primary amino acid sequence to the end of the RBD in E. coli EC93 is approximately 1,635 residues. Based upon the ratio of this number of residues over the measured length by tomography in E. coli, the P. aeruginosa CdiA1 and CdiA2 filaments are predicted to extend approximately 45 nm and 84 nm, respectively. Consequently, the YP domain of CdiA2 is larger than that of CdiA1, which may accommodate the longer β-helical filament of CdiA2. The consequences of these size differences are unknown, as both systems promote bacterial competition (32, 33).
FIG 3.
CdiA protein diversity. CdiA protein domain organization of E. coli CdiAEC93 and CdiASTEC_031 based upon the work of Ruhe et al. (24) and of CdiA1PAO1, CdiA1BL012, and CdiA2PAO1 from this work. Roman numerals indicate the P. aeruginosa CdiA class type (20). Colors reflect the different predicted protein domains from sequence alignments, domain searches, and domain descriptions in E. coli. Sec, general secretory signal.
The only region of sequence divergence in the amino half of CdiA occurs in the receptor-binding domain (RBD). Four separate CdiA-RBDs have been identified in E. coli (22, 39, 40), and 5 have been identified in P. aeruginosa (denoted rbdA through rbdE) based upon sequence alignments (see Fig. S1 in the supplemental material). The primary amino acid sequences of the respective RBDs from P. aeruginosa and E. coli do not share any similarity and therefore do not provide direct insight into the specific cellular receptors. Interestingly, all P. aeruginosa CdiA1 proteins contain the same RBD, which was not observed in any of the CdiA2 proteins. This suggests that recombination between this portion of the two cdi loci does not occur or is extremely rare. Of the 4 identified RBDs from E. coli, 3 have respective target cell receptors that have been identified. The RBD from CdiAEC93 binds to BamA, a component of the outer membrane β-barrel protein assembly complex, through specific interactions that target extracellular loops L6 and L7 (39, 41). CdiAEC536 interacts with extracellular loops L4 and L5 of osmoporin OmpC (40), and CdiASTEC031 interacts with the outer membrane nucleoside transporter Tsx (22). Bacteriocins from E. coli are known to hijack these same outer membrane protein transporters to mediate their toxic effects. For example, the E. coli colicin N was shown to use OmpF as a receptor for binding and translocation (42) while colicin 10 uses Tsx (43). Unlike bacteriocins, which typically require energy from the Ton or Tol systems for toxin uptake by the receptor (5), CDI systems do not have this same energy dependency for intoxication, and the interaction of the CdiA-RBD with its cognate receptor is thought only to promote intercellular binding and not outer membrane translocation of CT into the periplasm (44). The receptor overlap between bacteriocins and CdiA proteins in E. coli suggests a similar relationship in P. aeruginosa. The Pseudomonas bacteriocin Pyocin S2 binds the ferripyoverdine receptor FpvA (45, 46), while pyocin S5 uses the ferripyochelin receptor FptA for uptake (47). In addition, the lectin-like bacteriocin LlpA interacts with specific residues on extracellular loop 6 of BamA to facilitate intoxication (48), and the colicin M-like bacteriocin PaeM4 uses the heme receptor HxuC (49). These proteins are possible candidates for CdiA receptors in P. aeruginosa; however, further work is needed to determine the exact receptors used by different P. aeruginosa CdiA proteins.
THE CARBOXY-TERMINAL HALF OF CdiA IS DIVERSE
CDI systems among Pseudomonas species can be divided into 5 classes based upon a set of defined protein motifs immediately preceding the translocation domain (TD) that demarcate the highly variable carboxy terminus of CdiA (32). Similar pretoxin motifs have been described for E. coli (VENN) and Burkholderia species (NXXLYN) (15). In Pseudomonas species, these motifs include WVHN (class I), TENN (class II), LYVT (class III), DAMV (class IV), and NEALV (class V), of which P. aeruginosa CdiA proteins are restricted to the class II and class V groups. Class II CdiA proteins of P. aeruginosa contain a large FHA2 domain followed by a PT-VENN domain (Pfam ID PF04829) and a TENN motif (Fig. 3). Class V CdiA proteins contain a smaller FHA2 domain followed by a DUF637 domain (Pfam ID PF04830) and a NEALV motif (Fig. 3). As stated, the FHA2 domain was proposed to facilitate translocation of the final CdiA C-terminal domains across the target cell outer membrane in E. coli (25). It is unclear whether the smaller class V FHA2 domain could also facilitate outer membrane translocation of the C terminus in P. aeruginosa.
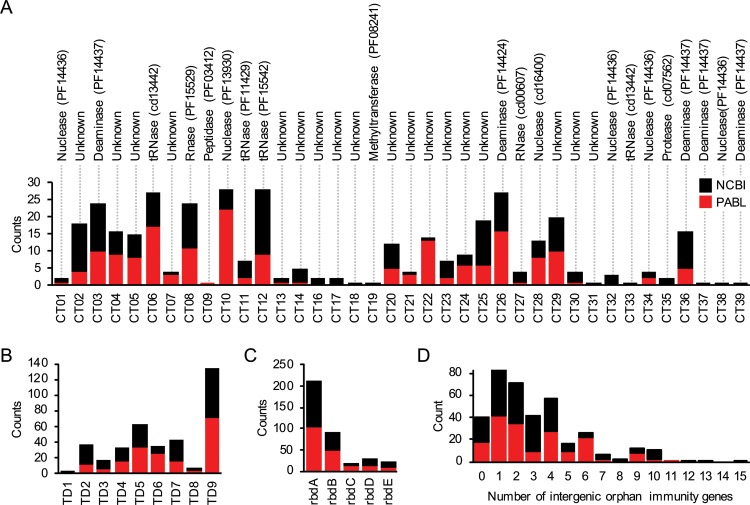
The greatest diversity in CdiA proteins occurs at their carboxy termini (Fig. 3). Studies in E. coli have shown these regions to be highly modular, with various pairings of different TD and CT domains (23). An analysis of CdiA proteins from 104 P. aeruginosa NCBI strains uncovered a total of 9 different TDs and 37 different CTs that were arranged in different combinations (see Fig. S3 in the supplemental material). Eighteen of the 37 CTs had no known function based upon peptide sequence, and most CTs with a predicted function had enzymatic activities that target nucleic acids (Table 1). CTs containing a MafB19-like deaminase domain (Pfam ID PF14437) or an EndoU RNase domain (PF14436) occurred the most (Fig. 4); however, the primary amino acid sequences within these predicted groups were highly diverse (Table S3). This sequence diversity was also mirrored in the respective CdiI proteins. Both MafB19-like deaminase domains and EndoU RNase domains are common among protein systems involved in bacterial competition (11, 50), and such sequence diversity has been shown to dictate substrate specificity for EndoU toxins in other bacterial species (51). Overall, P. aeruginosa CDI systems contain substantial C-terminal diversity on par with that observed in CDIs from other Gram-negative bacteria.
TABLE 1.
Putative activity and distribution of CTs
| Function | No. of unique CTs | Frequency of CTsa |
|---|---|---|
| Unknown | 18 | 39.2 |
| Deaminase | 5 | 16.6 |
| Methyltransferase | 1 | 0.3 |
| Nuclease | 7 | 15.0 |
| Peptidase | 2 | 1.1 |
| RNase | 2 | 7.1 |
| tRNase | 4 | 20.7 |
Among all P. aeruginosa strains analyzed.
FIG 4.
Distribution of CdiA domains in the PABL and NCBI strain collections. (A to C) The occurrence of each CT (A), TD (B), and RBD (C) variant was determined for the P. aeruginosa PABL and NCBI strain collections. Each bar represents the number of strains that contain the indicated domain variant, which are labeled based on designations in Fig. S3. Notations above the CT graph reflect predicted enzymatic functions determined by a conserved domain search (https://www.ncbi.nlm.nih.gov/cdd). (D) Counts represent the number of intergenic orphan cdiI genes at a cdi locus for each strain collection.
REGIONAL AND GLOBAL CdiA DIVERSITY
It is conceivable that CDI diversity in P. aeruginosa occurs on a global scale but that only a few distinct CTs are found in any one geographic location. Alternatively, P. aeruginosa bacteria encoding many different CTs may coexist in the same region. To examine this question, we determined the extent of CdiA diversity among P. aeruginosa isolates within a restricted geographical area. Sequences from a published collection of 100 clinical P. aeruginosa bloodstream (PABL) isolates from Northwestern Memorial Hospital in Chicago, IL, were analyzed for their CdiA diversity (see Table S2 in the supplemental material) (52). As with the NCBI genomes, CdiA homologs of the PABL strains were highly diverse. All 9 TDs and 5 RBDs were observed within this small collection, along with 27 CTs, including 1 new CT. The proportions of each TD and RBD were equivalent between the NCBI and PABL strains, but the proportions of individual CTs varied between the two collections (Fig. 4).
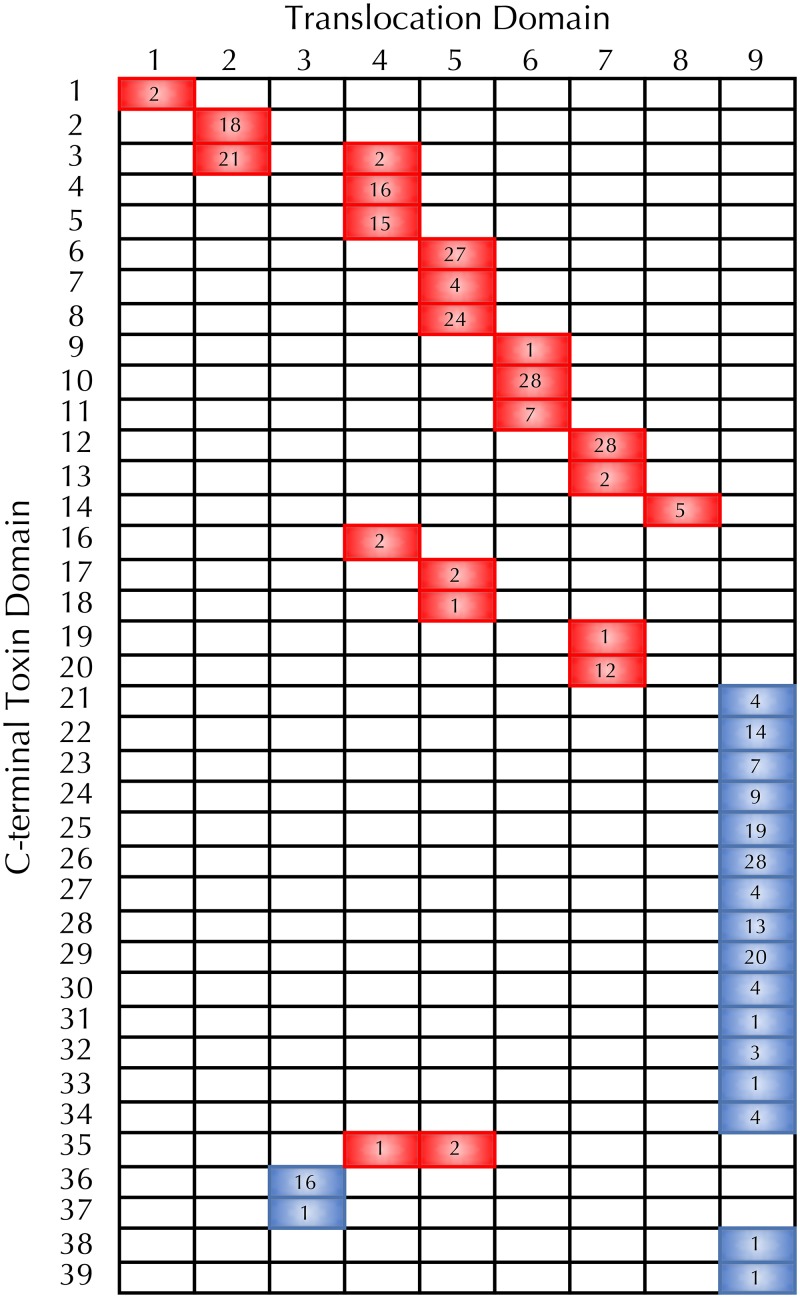
Sequence divergence in the CdiA carboxy terminus suggests that the TD and CT domains recombine as modular units to generate substantial diversity within the species; however, distribution of the different TD and CT domains suggests some biological limitation to the extent of this recombination in P. aeruginosa. Of the 9 TDs observed in P. aeruginosa, 6 were unique to the cdi1 locus, 2 were unique to the cdi2 locus, and only a single TD was shared between the two loci (Fig. 5). Except for two instances, each of the CTs was always associated with a specific TD. Furthermore, the distribution of CT domains was found to be evenly split between the two cdi loci with no overlap. This distribution suggests that there is little recombination between the two cdi loci; however, recombination must help drive cdiA diversity within a given locus. The tight association of CTs with specific TDs also suggests that CTs may have evolved preferred translocation pathways that are the most efficient for a given CT (23).
FIG 5.
P. aeruginosa CdiA C-terminal diversity. Pairwise comparison of CdiA translocation domains (TD) and respective C-terminal toxin (CT) domains were performed for the NCBI and PABL strain collections. Cells colored red indicate that the TD-CT combination was observed at the CDI1 locus, and blue indicates the CDI2 locus. Numbers indicate the number of strains with that observed TD-CT combination.
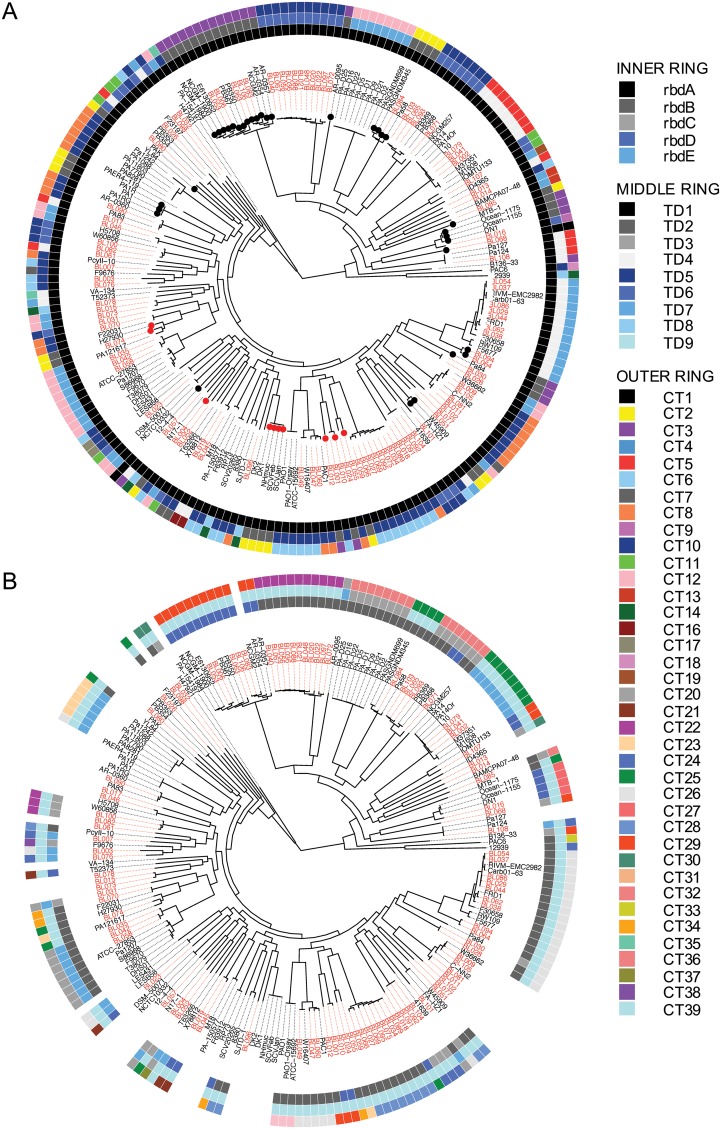
Analysis of the CdiA phylogenetic distribution revealed that strains within the same clonal lineage tended to share the same cdiA allele, but a given cdiA allele could be found in several unrelated clonal lineages. Likewise, different clades typically contained CdiA proteins with different combinations of RBD, TD, and CTs (Fig. 6). This suggests that substantial recombination of CdiA C terminus alleles occurs in P. aeruginosa and, as a consequence, may facilitate the evolution of clonal lineages, even within isolates from a single type of infection at one hospital.
FIG 6.
Phylogenetic distribution of cdiA alleles. The RBDs (inner ring), TDs (middle ring), and CTs (outer ring) for CdiA1 (A) and CdiA2 (B) were mapped onto maximum likelihood core genome trees of the PABL (red) and NCBI (black) strains. Ring colors represent different variants of the respective domains as noted in Fig. S1 and S3. Branches with dots at the tips indicate a class II C terminus. A red circle means the downstream orphan class V CdiA C terminus is the same as the functional class V CdiA C terminus of the closest phylogenetic neighbor.
ORPHAN IMMUNITY GENES
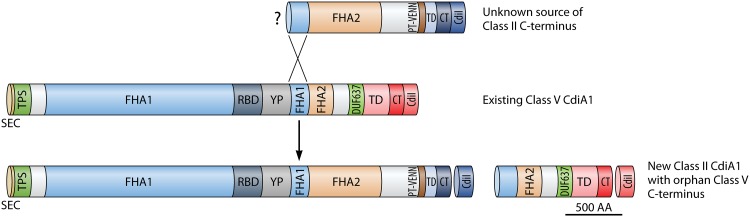
cdi loci often contain orphan 3′ cdiA fragments consisting of partial CT genes and their cognate immunity genes in the intergenic region downstream of the functional cdiA gene (Fig. 1A) (16). While these orphan CT gene fragments no longer facilitate bacterial competition, the orphan immunity genes are functional and may protect bacteria from assault by strains expressing the corresponding CdiA CTs (16). Some investigated P. aeruginosa strains had no orphan genes at either cdi locus, while others had up to 15 orphan immunity genes (Fig. 4D). On average, an individual P. aeruginosa strain was observed to contain 2 and 4 orphan immunity genes at the cdi1 and cdi2 loci, respectively. Of particular interest was a unique orphan that encompassed an entire class V carboxy terminus, including the FHA2 domain, with a unique CT (CT15). The coding sequence for this unusually large orphan motif occurred immediately downstream of a functional class II cdiA1 gene in P. aeruginosa NCGM2 and closely related strains. This orphan CT was predicted to contain a ParB-like nuclease domain (smart00470) and was never observed as a functional CdiA molecule in either the NCBI or PABL collections. Further investigations revealed that all class II cdiA genes encode a class V cdiA C terminus in the immediate downstream intergenic region. Often, this orphan class V C terminus was the actual class V CdiA C terminus of the closest phylogenetic neighbor (Fig. 6). As an example, the cdiA1 class V C-terminal allele of PABL001 can be found just downstream of the cdiA1 class II C-terminal allele of the nearest phylogenetic neighbor, PABL012 (Fig. 6; see Fig. S4 in the supplemental material). Thus, it appears that a class II cdiA C terminus coding sequence can insert into an established class V cdiA1 gene, effectively shifting the original class V C terminus coding sequence into the downstream intergenic region (Fig. 7). This insertion occurs just upstream of the region encoding the FHA2 domain. Neither the mechanism of this insertion nor the source of the class II cdiA C terminus is known. These data suggest that the class V alleles were ancestral at the cdi1 locus and that class II alleles were later horizontally acquired and inserted. Moreover, this phenomenon was only observed at the cdi1 locus, as cdiA genes at the cdi2 locus encoded only class V C termini (Fig. 6). Overall, this may indicate that separate mobile elements encoding CdiA C termini recombine with the cdi1 and cdi2 loci, but more work is needed for a complete understanding of CdiA diversification in P. aeruginosa.
FIG 7.
Illustration of cdiA1 C-terminal class switching. Alignments from Fig. S4 indicate that a cdiA class II C terminus can recombine with a region of conserved sequence immediately upstream of the FHA2 domain, shifting the original class V C terminus into the downstream intergenic region. The source of the class II C terminus is unknown.
CONCLUSIONS
This report reveals the breadth of P. aeruginosa CDI diversity and suggests that these systems play an important role in shaping populations within the species. Studies have shown that the majority of bacterial proteins are modular, created by genetically recombining functional domains in various combinations (53). This is especially true for many proteins involved in bacterial competition (11). Such diversity is thought to drive competition within a particular niche (2) and may lead to coordinated or symbiotic actions that promote survival or niche expansion (54).
Understanding the diverse biology of CDI systems may have implications outside bacterial competition. In some organisms, CDI systems have been shown to mediate behaviors independent from their bacterial growth inhibition function. CDI systems in Burkholderia thailandensis promote biofilm growth in a process termed “contact-dependent signaling” (CDS) (55). Aggregation phenotypes associated with CDI systems in Erwinia chrysanthemi EC16, Xylella fastidiosa, and Xanthomonas axonopodis, are required for bacterial virulence on plant hosts (56–58). In Neisseria meningitidis, mutants of the cdiA homolog hrpA were attenuated for intracellular survival in HeLa cells (59), and Pseudomonas aeruginosa PAO1 cdiA mutants had decreased virulence on lettuce and in Caenorhabditis elegans infection models (33). For many of these examples, it is not known whether these phenotypes can be attributed to the specific CT or if they are a general function of the CdiA protein. Thus, understanding the function and diversity of cdiA alleles in P. aeruginosa and other microbes may help elucidate how these systems aid in colonization and virulence. Ultimately, it is clear that CDI systems play an important role in the biology of many Gram-negative bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health/National Institute of Allergy and Infectious Disease (grants R01 AI118257, R01 AI053674, U19 AI135964, K24 104831, and R21 AI129167, awarded to A.R.H., and grant 1F32 AI108247, awarded to J.P.A.) and the American Heart Association (grant 15POST25830019, awarded to J.P.A.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors of this study have the following competing interests: A.R.H. serves on an advisory board and as a consultant for Microbiotix, Inc., to develop type III secretion inhibitors. This does not alter our adherence to all policies on sharing data and materials.
Biographies

Jonathan P. Allen, Ph.D., is a postdoctoral research fellow in the Department of Microbiology/Immunology at Northwestern University Feinberg School of Medicine in Chicago, IL. He completed his doctoral degree at Wayne State University School of Medicine in Detroit, MI. His current research involves identifying novel Pseudomonas aeruginosa virulence factors and understanding their role in host-microbe interactions.

Alan R. Hauser, M.D., Ph.D., is professor and vice chair of the Department of Microbiology/Immunology and professor of medicine in the Division of Infectious Diseases at Northwestern University in Chicago, IL. He completed his medical diploma, doctoral degree, and medicine residency at the University of Minnesota, and completed his infectious diseases fellowship at the University of California, San Francisco. His laboratory studies the pathogenesis of infections caused by Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter baumannii. He is a fellow of the American Academy of Microbiology and of the Infectious Disease Society of America and currently serves as the director of research for the Infectious Disease Fellowship Program at Northwestern University.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00776-18.
REFERENCES
- 1.Stubbendieck RM, Vargas-Bautista C, Straight PD. 2016. Bacterial communities: interactions to scale. Front Microbiol 7:1234. doi: 10.3389/fmicb.2016.01234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stubbendieck RM, Straight PD. 2016. Multifaceted interfaces of bacterial competition. J Bacteriol 198:2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheepers GH, Lycklama A, Poolman B. 2016. An updated structural classification of substrate-binding proteins. FEBS Lett 590:4393–4401. doi: 10.1002/1873-3468.12445. [DOI] [PubMed] [Google Scholar]
- 4.Khan A, Singh P, Srivastava A. 2018. Synthesis, nature and utility of universal iron chelator—siderophore: a review. Microbiol Res 212-213:103–111. doi: 10.1016/j.micres.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 7.Procopio RE, Silva IR, Martins MK, Azevedo JL, Araujo JM. 2012. Antibiotics produced by Streptomyces. Braz J Infect Dis 16:466–471. doi: 10.1016/j.bjid.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Dey A, Vassallo CN, Conklin AC, Pathak DT, Troselj V, Wall D. 2016. Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. J Bacteriol 198:994–1004. doi: 10.1128/JB.00964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki SK, Poole SJ, Hayes CS, Low DA. 2011. Toxin on a stick: modular CDI toxin delivery systems play roles in bacterial competition. Virulence 2:356–359. doi: 10.4161/viru.2.4.16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cianfanelli FR, Monlezun L, Coulthurst SJ. 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi PS, Bernstein HD. 2010. Sequential translocation of an Escherchia coli two-partner secretion pathway exoprotein across the inner and outer membranes. Mol Microbiol 75:440–451. doi: 10.1111/j.1365-2958.2009.06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazar J, Cotter PA. 2007. New insight into the molecular mechanisms of two-partner secretion. Trends Microbiol 15:508–515. doi: 10.1016/j.tim.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 15.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. 2010. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t’Kint de Roodenbeke C, Low DA, Hayes CS. 2011. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, Goulding CW. 2012. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci U S A 109:21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki SK, Webb JS, Braaten BA, Low DA. 2009. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol 191:1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruhe ZC, Low DA, Hayes CS. 2013. Bacterial contact-dependent growth inhibition. Trends Microbiol 21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson MS, Garcia EC, Cotter PA. 2012. The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet 8:e1002877. doi: 10.1371/journal.pgen.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Gregorio E, Esposito EP, Zarrilli R, Di Nocera PP. 2018. Contact-dependent growth inhibition proteins in Acinetobacter baylyi ADP1. Curr Microbiol 75:1434–1440. doi: 10.1007/s00284-018-1540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruhe ZC, Nguyen JY, Xiong J, Koskiniemi S, Beck CM, Perkins BR, Low DA, Hayes CS. 2017. CdiA effectors use modular receptor-binding domains to recognize target bacteria. mBio 8:e00290-17. doi: 10.1128/mBio.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willett JL, Gucinski GC, Fatherree JP, Low DA, Hayes CS. 2015. Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc Natl Acad Sci U S A 112:11341–11346. doi: 10.1073/pnas.1512124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. 2001. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol 42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- 25.Ruhe ZC, Subramanian P, Song K, Nguyen JY, Stevens TA, Low DA, Jensen GJ, Hayes CS. 2018. Programmed secretion arrest and receptor-triggered toxin export during antibacterial contact-dependent growth inhibition. Cell 175:921–933.e14. doi: 10.1016/j.cell.2018.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JW, Sato Y, Ogawa T, Arakawa T, Fukai S, Fushinobu S, Masaki H. 2018. Crystal structure of the central and the C-terminal RNase domains of colicin D implicated its translocation pathway through inner membrane of target cell. J Biochem 164:329–339. doi: 10.1093/jb/mvy056. [DOI] [PubMed] [Google Scholar]
- 27.Chauleau M, Mora L, Serba J, de Zamaroczy M. 2011. FtsH-dependent processing of RNase colicins D and E3 means that only the cytotoxic domains are imported into the cytoplasm. J Biol Chem 286:29397–29407. doi: 10.1074/jbc.M111.242354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb JS, Nikolakakis KC, Willett JL, Aoki SK, Hayes CS, Low DA. 2013. Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS One 8:e57609. doi: 10.1371/journal.pone.0057609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamet A, Nassif X. 2015. New players in the toxin field: polymorphic toxin systems in bacteria. mBio 6:e00285. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker D, Mosbahi K, Vankemmelbeke M, James R, Kleanthous C. 2007. The role of electrostatics in colicin nuclease domain translocation into bacterial cells. J Biol Chem 282:31389–31397. doi: 10.1074/jbc.M705883200. [DOI] [PubMed] [Google Scholar]
- 31.Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. 2014. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb Perspect Med 4:a010025. doi: 10.1101/cshperspect.a010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercy C, Ize B, Salcedo SP, de Bentzmann S, Bigot S. 2016. Functional characterization of Pseudomonas contact dependent growth inhibition (CDI) systems. PLoS One 11:e0147435. doi: 10.1371/journal.pone.0147435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melvin JA, Gaston JR, Phillips SN, Springer MJ, Marshall CW, Shanks RMQ, Bomberger JM. 2017. Pseudomonas aeruginosa contact-dependent growth inhibition plays dual role in host-pathogen interactions. mSphere 2:e00336-17. doi: 10.1128/mSphere.00336-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willett JL, Ruhe ZC, Goulding CW, Low DA, Hayes CS. 2015. Contact-dependent growth inhibition (CDI) and CdiB/CdiA two-partner secretion proteins. J Mol Biol 427:3754–3765. doi: 10.1016/j.jmb.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogier JC, Duvic B, Lanois A, Givaudan A, Gaudriault S. 2016. A new member of the growing family of contact-dependent growth inhibition systems in Xenorhabdus doucetiae. PLoS One 11:e0167443. doi: 10.1371/journal.pone.0167443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ocasio AB, Cotter PA. 2019. CDI/CDS system-encoding genes of Burkholderia thailandensis are located in a mobile genetic element that defines a new class of transposon. PLoS Genet 15:e1007883. doi: 10.1371/journal.pgen.1007883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu X, Kulasekara BR, Lory S. 2009. Role of horizontal gene transfer in the evolution of Pseudomonas aeruginosa virulence. Genome Dyn 6:126–139. doi: 10.1159/000235767. [DOI] [PubMed] [Google Scholar]
- 39.Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, Low DA. 2008. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol 70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beck CM, Willett JL, Cunningham DA, Kim JJ, Low DA, Hayes CS. 2016. CdiA effectors from uropathogenic Escherichia coli use heterotrimeric osmoporins as receptors to recognize target bacteria. PLoS Pathog 12:e1005925. doi: 10.1371/journal.ppat.1005925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruhe ZC, Wallace AB, Low DA, Hayes CS. 2013. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio 4:e00480-13. doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baboolal TG, Conroy MJ, Gill K, Ridley H, Visudtiphole V, Bullough PA, Lakey JH. 2008. Colicin N binds to the periphery of its receptor and translocator, outer membrane protein F. Structure 16:371–379. doi: 10.1016/j.str.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilsl H, Braun V. 1995. Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol Microbiol 16:57–67. doi: 10.1111/j.1365-2958.1995.tb02391.x. [DOI] [PubMed] [Google Scholar]
- 44.Beck CM, Diner EJ, Kim JJ, Low DA, Hayes CS. 2014. The F pilus mediates a novel pathway of CDI toxin import. Mol Microbiol 93:276–290. doi: 10.1111/mmi.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denayer S, Matthijs S, Cornelis P. 2007. Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J Bacteriol 189:7663–7668. doi: 10.1128/JB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baysse C, Meyer JM, Plesiat P, Geoffroy V, Michel-Briand Y, Cornelis P. 1999. Uptake of pyocin S3 occurs through the outer membrane ferripyoverdine type II receptor of Pseudomonas aeruginosa. J Bacteriol 181:3849–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elfarash A, Dingemans J, Ye L, Hassan AA, Craggs M, Reimmann C, Thomas MS, Cornelis P. 2014. Pore-forming pyocin S5 utilizes the FptA ferripyochelin receptor to kill Pseudomonas aeruginosa. Microbiology 160:261–269. doi: 10.1099/mic.0.070672-0. [DOI] [PubMed] [Google Scholar]
- 48.Ghequire MGK, Swings T, Michiels J, Buchanan SK, De Mot R. 2018. Hitting with a BAM: selective killing by lectin-like bacteriocins. mBio 9:e02138-17. doi: 10.1128/mBio.02138-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghequire MGK, Ozturk B. 2018. A colicin M-type bacteriocin from Pseudomonas aeruginosa targeting the HxuC heme receptor requires a novel immunity partner. Appl Environ Microbiol 84:e00716-18. doi: 10.1128/AEM.00716-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyer LM, Zhang D, Rogozin IB, Aravind L. 2011. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res 39:9473–9497. doi: 10.1093/nar/gkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michalska K, Quan Nhan D, Willett JLE, Stols LM, Eschenfeldt WH, Jones AM, Nguyen JY, Koskiniemi S, Low DA, Goulding CW, Joachimiak A, Hayes CS. 2018. Functional plasticity of antibacterial EndoU toxins. Mol Microbiol 109:509–527. doi: 10.1111/mmi.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheetz MH, Hoffman M, Bolon MK, Schulert G, Estrellado W, Baraboutis IG, Sriram P, Dinh M, Owens LK, Hauser AR. 2009. Morbidity associated with Pseudomonas aeruginosa bloodstream infections. Diagn Microbiol Infect Dis 64:311–319. doi: 10.1016/j.diagmicrobio.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apic G, Gough J, Teichmann SA. 2001. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol 310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 54.Wall D. 2016. Kin recognition in bacteria. Annu Rev Microbiol 70:143–160. doi: 10.1146/annurev-micro-102215-095325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia EC, Perault AI, Marlatt SA, Cotter PA. 2016. Interbacterial signaling via Burkholderia contact-dependent growth inhibition system proteins. Proc Natl Acad Sci U S A 113:8296–8301. doi: 10.1073/pnas.1606323113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guilhabert MR, Kirkpatrick BC. 2005. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute to X. fastidiosa biofilm maturation and colonization and attenuate virulence. MPMI 18:856–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- 57.Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. 2009. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One 4:e4358. doi: 10.1371/journal.pone.0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A 99:13142–13147. doi: 10.1073/pnas.202358699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tala A, Progida C, De Stefano M, Cogli L, Spinosa MR, Bucci C, Alifano P. 2008. The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell Microbiol 10:2461–2482. doi: 10.1111/j.1462-5822.2008.01222.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.