Numerous bacterial species encode proteins predicted to have similarity with Get3- and ArsA-type anion transporters. Our studies provide evidence that these proteins, which we named BagA and BagB, are unlikely to be involved in anion transport. In addition, BagA and BagB are conserved in all mycobacterial species, including the causative agent of leprosy, which has a highly decayed genome. This conservation suggests that BagAB constitutes a part of the core mycobacterial genome and is needed for some yet-to-be-determined part of the life cycle of these organisms.
KEYWORDS: Get3, Mycobacterium, tuberculosis
ABSTRACT
We characterized an operon in Mycobacterium tuberculosis, Rv3679-Rv3680, in which each open reading frame is annotated to encode “anion transporter ATPase” homologues. Using structure prediction modeling, we found that Rv3679 and Rv3680 more closely resemble the guided entry of tail-anchored proteins 3 (Get3) chaperone in eukaryotes. Get3 delivers proteins into the membranes of the endoplasmic reticulum and is essential for the normal growth and physiology of some eukaryotes. We sought to characterize the structures of Rv3679 and Rv3680 and test if they have a role in M. tuberculosis pathogenesis. We solved crystal structures of the nucleotide-bound Rv3679-Rv3680 complex at 2.5 to 3.2 Å and show that while it has some similarities to Get3 and ArsA, there are notable differences, including that these proteins are unlikely to be involved in anion transport. Deletion of both genes did not reveal any conspicuous growth defects in vitro or in mice. Collectively, we identified a new class of proteins in bacteria with similarity to Get3 complexes, the functions of which remain to be determined.
IMPORTANCE Numerous bacterial species encode proteins predicted to have similarity with Get3- and ArsA-type anion transporters. Our studies provide evidence that these proteins, which we named BagA and BagB, are unlikely to be involved in anion transport. In addition, BagA and BagB are conserved in all mycobacterial species, including the causative agent of leprosy, which has a highly decayed genome. This conservation suggests that BagAB constitutes a part of the core mycobacterial genome and is needed for some yet-to-be-determined part of the life cycle of these organisms.
INTRODUCTION
Mycobacterium tuberculosis kills more people than any other bacterial species (1). For this reason, there is a global interest in identifying pathways important for M. tuberculosis pathogenesis that can be targeted for chemotherapy. In an effort to find new pathways that could potentially be targeted for drug development, we searched the M. tuberculosis genome for open reading frames that might have unappreciated activities. We identified an operon, Rv3679-Rv3680, that is predicted to encode proteins with ATPase activity and structural similarity to a eukaryotic chaperone complex called guided entry of tail-anchored proteins 3 (Get3 in fungi or TRC40 in mammalian cells). Rv3679 and Rv3680 also have sequence similarity to bacterial anion transporters, although there are no data to support this function for these proteins. Interestingly, Mycobacterium leprae, which is the causative agent of leprosy and is thought to be evolving toward a minimal, “core” genome (2), has intact Rv3679-Rv3680 homologues, suggesting that these genes are important for a fundamental process in the physiology of mycobacteria.
In this work, we determined that Rv3679 and Rv3680 form a heterodimeric ATPase. While the Rv3679-Rv3680 complex was found to have some structural similarity to the homodimeric Get3 complex, we also observed several notable differences. Furthermore, while Rv3679 and Rv3680 had sequence similarity with anion transporters, our data did not support a model in which these proteins act as anion transporters. Finally, deletion of these genes from M. tuberculosis did not affect pathogenesis in mice, suggesting that this ATPase complex provides a fitness advantage under a condition that remains to be determined.
RESULTS
Rv3679 and Rv3680 form a heterodimeric complex with ATPase activity.
Rv3679 (1,023 bp, 340 amino acids [aa]) and Rv3680 (1,161 bp, 386 aa) encode proteins that share 57% similarity over 157 amino acids (see Fig. S1 in the supplemental material). We used structure prediction analysis (HHPred) to determine if these gene products had similarity to one or more known proteins (3). Both proteins were predicted to have high structural similarity with the fungal protein Get3, which forms soluble ATPase homodimers that target tail-anchored proteins into the membrane of the endoplasmic reticulum (ER). The hydrophobic cleft in a Get3 dimer protects the hydrophobic transmembrane regions of tail-anchored proteins until they reach the Get1/2 membrane complex, which receives cargo-loaded Get3 and uses cycles of ATP hydrolysis to insert tail-anchored polypeptides into the ER membrane (4, 5). Based on the predicted high structural similarity of Rv3679 and Rv3680 with Get3, we named the M. tuberculosis proteins BagA and BagB for bacterial Get3-like proteins A and B.
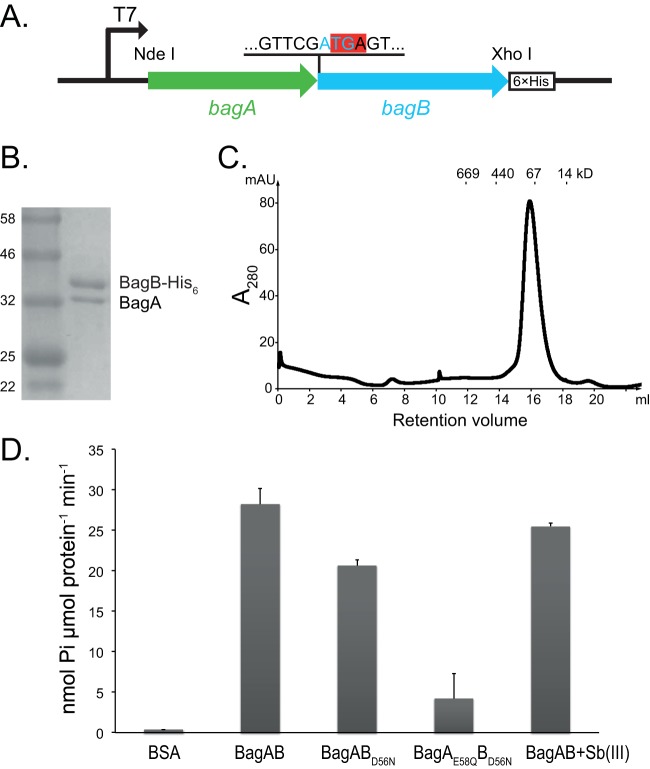
Although Get3 is formed from two molecules of the same protein, we hypothesized that M. tuberculosis BagA and BagB formed a heterodimer. We cloned M. tuberculosis bagAB into an Escherichia coli expression vector (Table 1). The stop codon of bagA overlaps the start codon of bagB; therefore, we cloned the coding sequences in their native form into pET24b(+), which added a hexahistidine (His6) epitope to the carboxyl terminus of BagB (Fig. 1A). After induction of gene expression in E. coli, we copurified BagA and BagB at close to a 1:1 stoichiometry (Fig. 1B), supporting a hypothesis whereby bagA and bagB are cotranscribed and cotranslated. According to the gel filtration profile, the predicted molecular mass of the complex is around 80 kDa, suggesting that these proteins form a heterodimer (Fig. 1C).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant genotype, description, or sequence (5′–3′) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ thi-1 gyrA96 relA1 | Gibco, BRL |
| BL21(DE3) | For inducible T7 expression; F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12 (λs) | New England Biolabs, Inc. |
| M. tuberculosis | ||
| H37Rv | Wild type | ATCCa 25618 |
| MHD1428 | Hygr ΔbagAB::hyg | This work |
| MHD1385 | Kanr H37Rv/pMV306.kan | This work |
| MHD1431 | Hygr Kanr ΔbagAB::hyg ppsE/pMV306.kan | This work |
| MHD1432 | Hygr Kanr ΔbagAB::hyg ppsE/pMV-bagAB | This work |
| Plasmids | ||
| pMV306.kan | Kanr; plasmid that integrates at attB site on the M. tuberculosis chromosome | 33 |
| pMV-bagAB | Kanr; pMV306.kan with 2.3-kb bagAB insert including 200 bp of upstream sequence | This work |
| pET24b(+) | Kanr; for production of C-terminal His6 epitope-tagged protein | Novagen |
| pET24b(+)-bagAB | Kanr; for production of BagA-BagB-His6 | This work |
| pET24b(+)-bagABD56N | Kanr; for production of BagA-BagBD56N-His6 | This work |
| pET24b(+)-bagAE58QBD56N | Kanr; for production of BagAE58Q-BagBD56N-His6 | This work |
| pYUB854 | Hygr; allelic exchange vector. | 34 |
| pYUB-ΔbagAB | Hygr; allelic exchange vector with ΔbagAB::hyg | This work |
| pET24b(+)-bagAB | Kanr; for production of BagAB-His6 | This work |
| pET24b(+)-bagABD56N | Kanr; for production of BagABD56N-His6 | This work |
| pET24b(+)-bagAE58QBD56N | Kanr; for production of BagAE58QBD56N-His6 | This work |
| DNA primers | ||
| For cloning bagAB into pET24b(+) | ||
| Rv3679NdeIf1 | GCCATATGGTGGCAACCACATCTAGCGGCGG | |
| Rv3680XhoIr1 | GGCTCGAGGCGGCCCGCTGCGCGGCCCGCATC | |
| For making the ΔbagAB::hyg mutant | ||
| stuI-bagABup-for | CATGAGGCCTTCAACTCGACTTCCACCGGCGCATC | |
| xbaI-bagABup-rev | CGGTTCTAGACATGGCATGCTAGCCCGATGGCCTG | |
| xhoI-bagABdown-for | CATGCTCGAGGCGCTCGCCGACCAGCTCACCACGG | |
| speI-bagABdown-rev | CGGTACTAGTATACAGGTAACACCGATGTGCATGG | |
| For making the bagAB complementation plasmid | ||
| Xbal_BagA_F | GTGTCTAGAGATCTGTTTGGTGGCGTGCGTCAACAGC | |
| SOE_BagAB_F | GTCACACCGAAGACCCTCGATATGG | |
| SOE_BagAB_R | CCATATCGAGGGTCTTCGGTGTGAC | |
| BagB_EcoRV_R | TAAGATATCTCAGCGGCCCGCTGCGC | |
| For sequencing the bagAB locus | ||
| Rv3679seqf2 | CTGTTAGAAGCGCTGCCGGTG | |
| Rv3680seqr2 | CCGAGCACGGTGGACAACGCC | |
| For making the bagAE58Q mutant | ||
| Rv3679_E58Q_F | AGTCCTACTCGTCGAAGTCCAGGGGCGCCAGGGGATTGC | |
| Rv3679_E58Q_R | GCAATCCCCTGGCGCCCCTGGACTTCGACGAGTAGGACT | |
| For making the bagBD56N mutant | ||
| Rv3680_D56N_F | TGTGGTCGTTTTGACGATTAACCCAGCCAAGCGATTGGC | |
| Rv3680_D56N_R | GCCAATCGCTTGGCTGGGTTAATCGTCAAAACGACCACA | |
| For checking the ppsE sequence | ||
| ppsE seq1 | CTAAACCTGAAGTACAGGGAGGATAAGACC | |
| ppsE seq2 | ATGGGCTTCGGCGATGACATCGGCTTGAGC |
ATTC, American Type Culture Collection.
FIG 1.
BagAB copurify as a heterodimer with ATPase activity. (A) The bagAB operon was cloned into pET24b(+), maintaining the original junction between the genes. The start codon of bagB (cyan) overlaps the stop codon of bagA (highlighted in red). The His6 tag was fused to the C terminus of BagB. (B) Coomassie brilliant blue-stained SDS-PAGE gel of copurified BagAB-His6. The molecular masses (standards are on the left in kilodaltons) of BagA and BagB are 36 and 41 kDa, respectively. (C) Size exclusion profile of BagAB-His6 using a Superose 6 10/300 GL column, showing only one peak with a molecular mass around 80 kDa. Size standards are marked above the chromatographic profile. (D) ATPase activities of BagAB and its variants as determined by malachite green phosphate assay. Bovine serum albumin (BSA) was used as a negative control. Error bars represent standard deviations of three measurements.
BagA and BagB polypeptides have ATPase domains and are predicted to be members of the P-loop nucleoside triphosphatase (NTPase) protein subgroup (6). Additionally, these proteins have predicted similarity with proteins of the arsenite transporter (ArsA) family (reviewed in reference 7), suggesting that BagAB could bind to small molecules. We quantified the ATPase activity of the BagAB complex and found that the recombinant protein had an ATPase activity of 28 nmol min−1 μmol−1 (or 0.35 nmol min−1 mg−1) (Fig. 1D). This activity was comparable to that of E. coli ArsA (0.06 nmol min−1 mg−1) but much lower than that of Saccharomyces cerevisiae Get3 (∼400 nmol min−1 mg−1) (5, 8). We mutated a predicted catalytic residue, aspartate 56 in the BagB switch I region, which is a conserved motif that binds to the γ-phosphate of ATP (Fig. S1), to asparagine (D56N). The mutation caused an ∼30% decrease in ATPase activity, while the addition of a switch I mutation in BagA (E58Q) further reduced ATPase activity of the heterodimer (Fig. 1D). We tried to purify BagAE58QB but could not get any soluble protein when it was produced in E. coli (data not shown).
BagA and BagB have sequence similarity to members of the ArsA protein family; therefore, we tested whether or not antimonite [Sb(III)], an arsenite substitute that potently stimulates ArsA activity (8), could stimulate ATPase activity. Antimonite failed to enhance ATPase activity of the BagAB complex (Fig. 1D, last bar). Together, these data showed that BagAB is a heterodimeric ATPase complex with both subunits actively engaged in ATP hydrolysis and that it is unlikely to be an arsenite or antimonite transporter like ArsA.
The BagAB structure is distinct from those of Get3 and ArsA and has a closed conformation.
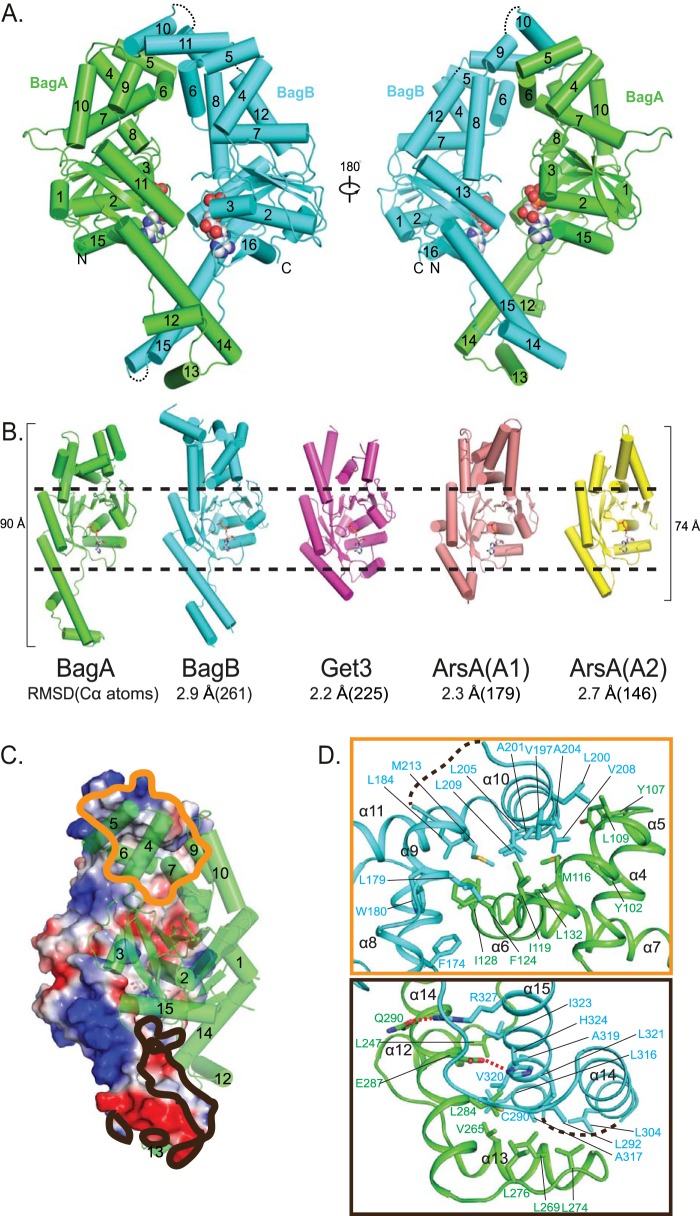
We next used X-ray crystallography to gain an understanding of the BagAB structure. Crystals did not form in the absence of a nucleotide; therefore, we solved structures of the heterodimer in the presence of either ADP, the weakly hydrolyzed ATP analogue ATPγS, or the nonhydrolyzable analogue adenylyl-imidodiphosphate (AMP-PNP) at a resolution of 2.3, 2.5, and 3.2 Å, respectively (Table 2). Like Get3 and ArsA, BagA and BagB each has a core nucleotide-binding α/β subdomain flanked by two α-helical subdomains (Fig. 2A). Superimposition of BagA to Get3 and two ATPase domains of ArsA showed root mean square deviations (RMSD) of 2.2, 2.3, and 2.7 Å for 225, 179, and 146 pairs of C-alpha atoms, respectively (Fig. 2B). The top helical subdomains of BagA and BagB correspond to the substrate transmembrane helix-binding domain in Get3 and the As(III)-binding domain in ArsA, but they contain neither a hydrophobic groove as seen in nucleotide-bound Get3 nor an anion-binding site as seen in ArsA (5, 9). The bottom helical subdomains of BagA and BagB, corresponding to the zinc-coordinated hinge site in Get3, lack a cysteine-(any amino acid)2-cysteine (CXXC) motif and instead contain a long α-helix, making BagAB more elongated (90 Å) than Get3 (73 Å) or ArsA (74 Å) (Fig. 2B). The two helical subdomains of BagA, like two arms, interact with their respective corresponding subdomains of BagB, thereby forming a diamond-shaped central cavity that somewhat resembles the ArsA structure.
TABLE 2.
Data collection and refinement statistics
| Parameter | Valuea
for: |
|||
|---|---|---|---|---|
| Pt2+ derivative | BagAB-ADP | BagAB-ATPγS | BagAB–AMP-PNP | |
| Data collection statistics | ||||
| Wavelength (Å) | 1.05969 | 1.07822 | 1.07822 | 1.05969 |
| Space group | P3221 | P3221 | P3221 | P3221 |
| Cell dimensions (Å) | a = b = 121.47, c = 119.50, α = β = 90°, γ = 120° | a = b = 121.34, c = 120.97, α = β = 90°, γ = 120° | a = b = 121.63, c = 120.06, α = β = 90°, γ = 120° | a = b = 121.13, c = 120.54, α = β = 90°, γ = 120° |
| Resolution range (Å) | 105.19–3.43 | 52.42–2.30 | 48.23–2.50 | 28.28–3.10 |
| Rmerge (%) | 18.6 (62.5) | 6.8 (78.0) | 12.4 (80.2) | 24.3 (95.8) |
| I/σI | 13.4 (5.7) | 13.5 (2.2) | 18.6 (4.4) | 8.2 (3.0) |
| Completeness (%) | 99.9 (100) | 99.4 (100) | 97.0 (100) | 99.5 (100) |
| Unique reflections | 14,042 (2,026) | 45,825 (4,586) | 34,865 (3,552) | 18,830 (1,845) |
| Multiplicity | 19.6 (19.3) | 5.7 (5.9) | 20.4 (21) | 6.7 (6.9) |
| Refinement statistics | ||||
| Rwork/Rfree | 0.198/0.226 | 0.191/0.225 | 0.219/0.248 | |
| Non-H atoms | 5,459 | 5,421 | 5,176 | |
| Protein | 5,146 | 5,154 | 5,144 | |
| Ligand | 63 | 60 | 32 | |
| Average B-factors (Å2) | 63.33 | 54.02 | 25.84 | |
| Macromolecules | 63.29 | 54.30 | 25.87 | |
| Ligands | 51.06 | 31.32 | 20.31 | |
| Root mean square deviations | ||||
| Bond length (Å) | 0.014 | 0.013 | 0.012 | |
| Bond angle (°) | 1.77 | 1.75 | 1.39 | |
| Ramachandran statistics (%) | ||||
| Favored | 98.22 | 98.22 | 98.07 | |
| Allowed | 1.78 | 1.78 | 1.93 | |
| Outlier (%) | 0.00 | 0.00 | 0.00 | |
| Molprobity score | 1.31 | 1.21 | 1.63 | |
| Clashscore | 5.70 | 4.27 | 13.08 | |
Values in parentheses are for the highest-resolution shell.
FIG 2.
Crystal structure of ADP-bound BagAB. (A) Two opposing views of BagAB shown in cartoon form. BagA is in green, and BagB is in cyan. The two ADP molecules are shown as spheres. The three disordered BagB loops connecting α9 and α10, between α11 and α12 as well as between α14 and α15, are labeled by black dashes. (B) A comparison of the structures of BagA, BagB, Get3, and ArsA shown in cartoon form, with BagA in green, BagB in cyan, nucleotide-bound Get3 in purple (PDB 2WOJ), the N-terminal half of ArsA in salmon (PDB 1F48), and the C-terminal half of ArsA in yellow (PDB 1F48). These structures are aligned but shown separately, with the RMSD values and numbers of aligned C-alpha atoms shown below. Superimpositions were carried out using secondary structure matching in the program COOT (28). The black dashes highlight the conserved nucleotide-binding domains. (C) BagA is shown in the green cartoon view and BagB in a surface potential view. The orange contour marks the main interaction between the two upper helical subdomains of BagA and BagB. The black contour marks the interface between the two lower helical subdomains. (D) The upper panel highlights the hydrophobic interactions between BagA and BagB in the region marked by the orange contour in panel C. The lower panel highlights the hydrogen-bonding and hydrophobic interactions between BagA and BagB in the region delineated by the black contour in panel C. Two hydrogen bonds are labeled as red dashes. Disordered loops between α10 and α11 and between α14 and α15 are shown as black dashes.
The overall structures of BagA and BagB are similar, with an RMSD of 2.9 Å for 261 pairs of C-alpha atoms when superimposed. The structure of the nucleotide-binding subdomain is highly conserved between BagA and BagB, but their bottom α-helical subdomains differ; BagA contains three α-helices (α12 to α14), but BagB only has two α-helices (α14 and α15) (see Fig. S2 in the supplemental material). Furthermore, BagB seems to be more flexible, as the three loops connecting α9 and α10, α11 and α12, and α14 and α15 are disordered; the corresponding loops in BagA are well ordered (Fig. 2A). The nucleotide-bound BagAB heterodimer is in a closed conformation, similar to that seen in the nucleotide-bound Get3 structure, with a 2,200-Å2 buried interface. The interface is primarily in the two helical subdomains flanking the nucleotide-bound α/β subdomain (Fig. 2C). At one end, four α-helices of BagB, α8 to α11 of BagB, wrap around α5 to α7 of BagA by extensive hydrophobic interactions (Fig. 2D, upper panel). At the other end, α14 and α15 of BagB interface with α12 to α14 of BagA, also by extensive hydrophobic interactions. There are two hydrogen (H) bonds between the two subunits in this region: Q290 and E287 in BagA form H bonds with R327 and H32 in BagB, respectively (Fig. 2D, lower panel).
BagAB has two nonequivalent nucleotide-binding sites.
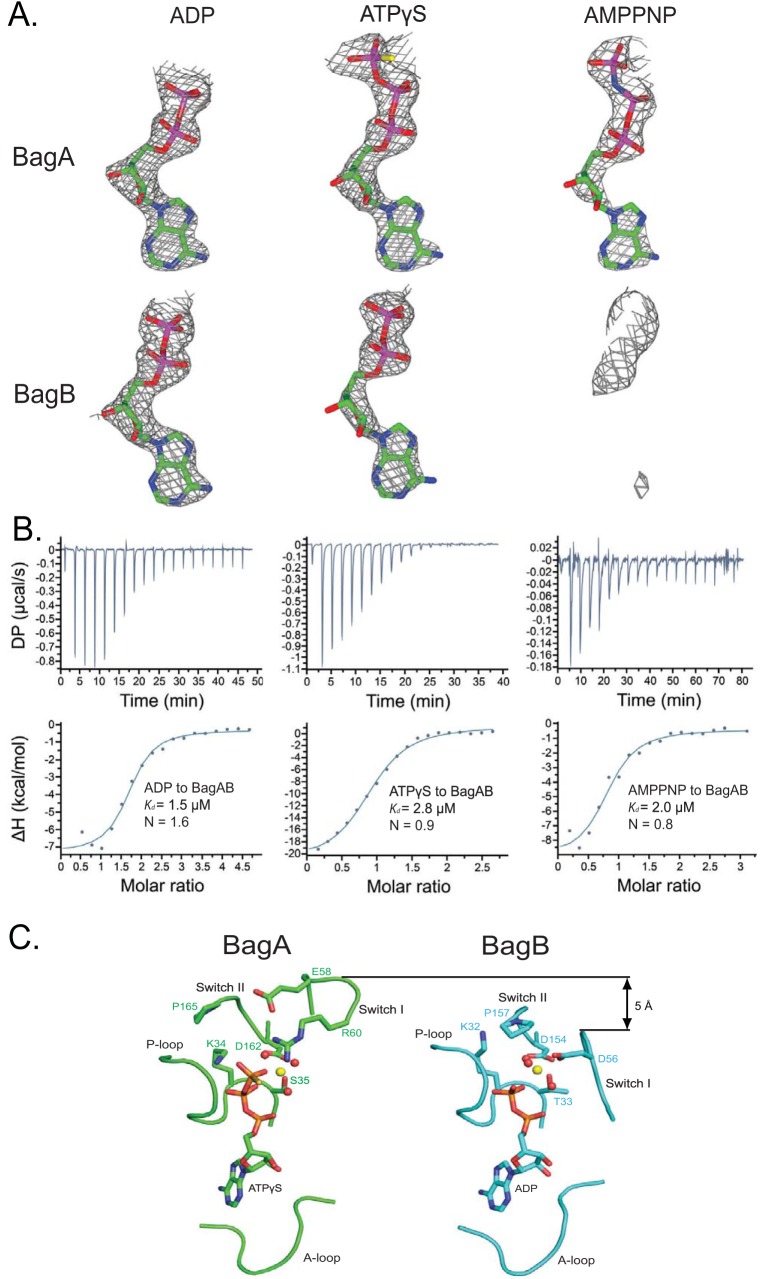
In the cocrystal structures of BagAB complexed with different nucleotides, BagAB adopted nearly identical conformations with an RMSD of 0.11 to 0.24 Å. However, their nucleotide-binding states were different. In the ADP-bound structure, a well-defined electron density of ADP was found in the nucleotide-binding sites of both BagA and BagB (Fig. 3A, left). In contrast to the structure determined in the presence of ATPγS, BagA was occupied with ATPγS, while BagB had ADP in its site (Fig. 3A, middle). This configuration was possible because ATPγS is weakly hydrolyzable. In the AMP-PNP-bound structure, AMP-PNP was found only in BagA and not in BagB (Fig. 3A, right). This heterogeneous ATP analogue-binding mode of BagAB was unlikely due to a crystallization artifact, because the structures were determined by cocrystallization and not by nucleotide soaking. To further assess the physiological relevance of the heterogeneous ATP analogue binding of BagAB, we used isothermal titration calorimetry (ITC) to measure the binding stoichiometry of ADP, ATPγS, and AMP-PNP in solution. The results showed that ADP bound to BagAB with a stoichiometry of 1.6 (Fig. 3B, left), suggesting that one BagAB can bind two ADP molecules. In contrast, the binding stoichiometries of ATPγS and AMP-PNP were only 0.9 and 0.8, respectively (Fig. 3B, middle and right), indicating that one BagAB can bind to only one molecule of ATPγS or AMP-PNP. Therefore, the solution nucleotide-binding behavior of BagAB was consistent with what we observed in the cocrystal structures. We note that the ITC data points for ATPγS were somewhat linear and did not fit well with the classic sigmoidal curve (Fig. 3B). This was likely due to the slow hydrolysis of ATPγS, which is consistent with our cocrystal structure of BagAB in the presence of ATPγS, in which one of the two ATPγS molecules is hydrolyzed to ADP.
FIG 3.
The nucleotide-binding sites in BagA and BagB are not equivalent. (A) Simulated-annealing omit maps of cocrystals with ADP (left), ATPγS (middle), and AMP-PNP (right) in BagA (upper panel) and BagB (lower panel) displayed at a threshold of 2.5σ. Atomic models of the observed nucleotides are shown as sticks. (B) In vitro binding of ADP (left), ATPγS (middle), and AMP-PNP (right) to BagAB as measured by ITC at 25°C. (C) The nucleotide-binding sites of BagA (left) and BagB (right) in the structure determined in the presence of ATPγS. The A-loop, P-loop, switch I region, and switch II region are shown as Cα trace. The nucleotides are shown as sticks. Mg2+ and water are shown as yellow and red spheres, respectively.
To understand the structural basis of the nonequivalent nucleotide binding in BagAB, we compared the two nucleotide-binding sites in the structure determined in the presence of ATPγS, in which BagA bound an ATPγS and BagB bound an ADP. We found that the two sites share similar positioning and configuration of the adenine-binding loop (A-loop), the phosphate-binding loop (P-loop), and the switch II region but that their switch I regions are different (Fig. 3C): the switch I region of BagA is 5 Å higher than that of BagB, making the BagA nucleotide pocket large enough to accommodate the triphosphate of ATPγS (Fig. 3C). Moreover, Arg60 in BagA stabilizes the γ-phosphate via a hydrogen bond (Fig. 3C, left); there is no equivalent residue in BagB. The structural differences suggest that BagA is better adapted to accommodate ATP than BagB and that BagB may have a lower ATPase activity than BagA. However, given our inability to produce the soluble protein of BagAE58QB to test the ATP hydrolysis activity, it remains to be determined if BagA indeed contributes more than BagB to the ATPase activity of the heterodimer and, more importantly, their respective activities in the presence of a substrate.
Global proteomics analysis did not reveal conspicuous changes in the absence of bagAB.
To gain a better understanding of the function of BagAB, we analyzed the proteomes of the wild-type (WT), bagAB mutant, and complemented strains under routine culture conditions in triplicate as described previously (10, 11). PpsE was dramatically reduced in the bagAB strain with or without the integrated complementation plasmid (see Table S1 in the supplemental material); however, we recently noticed a spontaneous mutation in ppsE in a population of our parental WT strain stocks after whole-genome sequencing of several strains made for another study (E. Ballister and K. H. Darwin, unpublished observations). We checked the ppsE sequence in the parental bagAB strain using PCR and sequencing of the products (see Table 1 for primer sequences) and confirmed the presence of a frameshift mutation in ppsE (deletion of a guanine in codon 241 of 1,488) that we had observed in our other studies. Other than the loss of PpsE, we did not observe other substantial changes among the strains.
BagAB is not required for M. tuberculosis pathogenesis in mice.
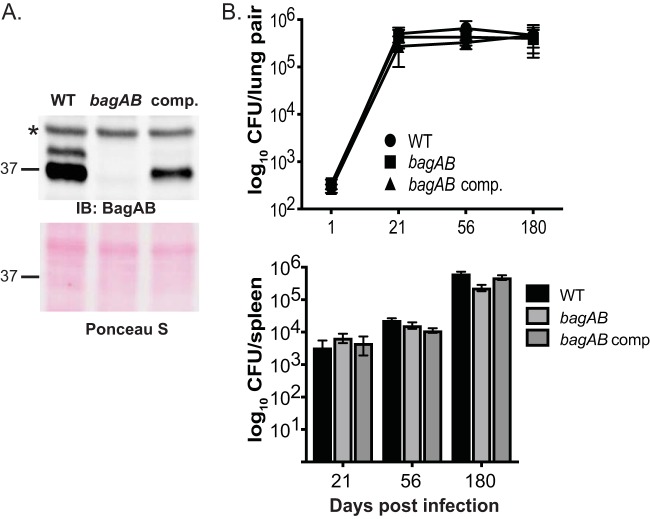
To determine whether or not bagAB is important for the pathogenesis of M. tuberculosis, we deleted and disrupted both genes from the chromosome of strain H37Rv (Table 1; see Materials and Methods). We also raised polyclonal rabbit antibodies to the BagAB-His6 complex to confirm the loss of BagAB from this strain (Fig. 4A). Although there appears to be more BagA than BagB in lysates, we could not make this conclusion based on immunoblotting; detection can be dependent on the sensitivity of the antibodies. However, quantitative mass spectrometry (MS) of total cell lysates of the WT strain showed that BagA was two to three times more abundant than BagB in the lysates (Table S1). It is likely that BagA is more abundant due to bagA transcription and translation before bagB. While it is possible that BagA (or BagB) homodimerizes, we were unable to produce detectable amounts of BagA-His6 or BagB-His6 in the absence of the other protein in E. coli, suggesting that each protein is unstable in the absence of the other (data not shown).
FIG 4.
BagAB is not essential for growth in C57BL6/J mice. (A) Immunoblot (IB) analysis of total cell lysates of WT, ΔbagAB::hyg, and ΔbagAB::hyg-complemented (comp.) strains. The WT and mutant strains carry the empty vector used for making the complementation plasmid. The asterisk indicates a protein that cross-reacts with the polyclonal rabbit antiserum in all strains. Below is the same membrane used for immunoblotting but stained with Ponceau S prior to IB analysis, which shows even loading. (B) All three strains grew equally well in the lungs (top) and spleens (bottom) of C57BL6/J mice. This experiment shows combined data from two independent experiments (day 1, 6 mice; days 21 and 56, 8 mice each; day 180, 10 mice).
Complementation with bagAB expressed from its native promoter in single copy (integrated at the attB site of the M. tuberculosis chromosome) partially restored protein levels (Fig. 4A, right lane). We infected mice with a low to moderate dose (∼200 CFU/mouse) of the WT, mutant, and complemented strains by aerosol and assessed the bacterial burden over time. Curiously, we observed that the bagAB mutant and complemented strains recovered from mouse tissues grew more slowly on agar plates than the parental strain, a phenotype that was inconspicuous when strains were grown in broth and inoculated onto agar (data not shown). The lack of PpsE that we observed in our proteomics analysis may explain the slowed growth of the bagAB mutant and complemented strains that we observed on agar plates after passage through mice. Nonetheless, we did not observe any significant differences in bacterial burden up to 6 months after infection (Fig. 4B), suggesting that this ex vivo small-colony phenotype did not reflect bacterial survival in mice.
We were somewhat surprised that the ppsE mutation had no apparent effect on the growth of the bagAB mutant and complemented strains; ppsE is carried within the complex lipid phthiocerol dimycocerosate (PDIM) biosynthetic pathway gene cluster. It is well established that mutants that do not make PDIM are attenuated in mice (12); however, it has not been determined if disruption of ppsE affects PDIM synthesis or virulence. ppsE mutants in mixed-infection experiments were found to be less fit in cultured macrophages treated with interferon gamma (13), but it is unclear if this phenotype translates into an attenuated phenotype in mice, and this mutant was not complemented. Taken together, our data suggest that neither PpsE nor the BagAB complex is essential for the pathogenesis of M. tuberculosis H37Rv. Alternatively, these proteins may be required under specific conditions during a natural human infection or one that is not present in C57BL6/J mice.
DISCUSSION
In this work, we characterized M. tuberculosis BagAB, which was predicted to have high structural similarity with a eukaryotic complex called Get3. We found that BagA and BagB form a heterodimeric duplex that belongs to the P-loop NTPase protein subgroup and shares a similar topology with ArsA and Get3 (6). Unlike ArsA and Get3, however, BagAB complexes consist of two different but similar subunits, presenting a new dimerization mode in the ArsA protein family. Despite its high conservation among mycobacterial species, we did not identify a function for BagAB for axenic or in vivo growth.
What is the biological function of BagAB in M. tuberculosis? While we do not yet know its purpose in bacterial physiology, our data suggest that BagAB is functionally distinct from both the ArsA and Get3 proteins. First, BagAB ATPase activity was not stimulated by an arsenite analogue, suggesting that it is not a transporter for this family of elements. Second, BagAB does not form a hydrophobic channel, which is characteristic and essential for the function of Get3 in targeting membrane proteins to the ER; the helical domain in BagAB corresponding to the tail-anchored protein-binding domain contains more helices and shows a complicated, asymmetric interaction interface. The nature of the substrate(s) (protein or otherwise) is unknown. Future studies will determine if BagAB has a unique substrate or is able to bind to numerous partners for a function that remains to be determined.
BagAB alone maintains a low ATP hydrolysis activity. The ATPase activity of BagAB may be enhanced by a substrate, in a manner similar to that for the bacterial protein disaggregase ClpB, whose ATPase activity is low in the absence of a substrate but is enhanced by severalfold when bound to a model substrate casein (14). The function of the basal level ATPase activity of BagAB is currently unclear and awaits the identification of the substrate. In the case of the P-glycoprotein, its basal level ATPase activity is suggested to maximize the number of proteins in a substrate-binding-competent form (15).
Interestingly, BagAB binds nucleotide asymmetrically. The nucleotide-based asymmetry has been observed in some dimeric ATPases, such as the DNA mismatch repair protein MutS (16, 17). In MutS, the two ATPase domains are packed tightly against each other, which distorts one of the two P-loops allowing the two DE-loops (corresponding to switch I in BagAB) to interact asymmetrically. In contrast, the two ATPase domains of BagAB are spatially separated by the large central cavity, and they do not physically interact with each other. Therefore, BagAB and MutS may operate under similar mechanisms but likely with some distinction. The asymmetric nucleotide binding of BagAB is also reminiscent of ArsA. In the crystal structure of ArsA complexed with AMP-PNP, AMP-PNP is found only in the nucleotide-binding site of A2 and not in A1 (18). The nucleotide forms fewer hydrogen bonds with the A2 site than with A1 (9). Fluorescence studies revealed that only A1 participates in uni-site catalysis in the absence of antimonite, but both A1 and A2 hydrolyze ATP in the presence of antimonite (6, 19). The relationship between substrate binding and nonequivalent nucleotide binding observed in ArsA may hint at a potential mechanism of action for BagAB. Thus, we propose that the asymmetric ATP-binding mode of BagAB is a functional property of BagAB activity. The identification of a BagAB substrate or substrates may reveal a role for asymmetric nucleotide binding and catalysis by BagAB.
Our studies here expand the membership of the ArsA protein family and provide a foundation to help address the role of BagAB in mycobacterial physiology. Our data support the previous prediction by genome-wide high-density transposon mutagenesis that neither bagA nor bagB is essential in M. tuberculosis H37Rv (20). Interestingly, proteins with high similarity to BagAB are found only in actinomycetes. It is possible that BagAB is required under conditions that are not present in the C57BL6/J mouse model tested in this work or that this complex plays a role in M. tuberculosis pathogenesis that cannot be captured through assessment of bacterial survival alone.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and culture conditions.
Table 1 lists the strains, plasmids and primers used in this study. All DNA oligonucleotides were purchased from Thermo Fisher Scientific/Invitrogen. M. tuberculosis strains were grown in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, 0.5% fraction V bovine serum albumin (BSA), 0.2% dextrose, and 0.085% sodium chloride (the final broth is referred to as 7H9c). M. tuberculosis cultures were grown without shaking in 25- or 75-cm2 vented flasks (Corning) at 37°C. Middlebrook 7H11 agar (Difco) supplemented with 0.5% glycerol and BBL TM Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment (BD) was used for growth on solid medium (referred to as 7H11). M. tuberculosis was transformed as described previously (21). E. coli strains used for cloning and expression were grown in LB-Miller broth (Difco) at 37°C with aeration on a shaker or on LB agar. E. coli strains were chemically transformed as previously described (22). The final concentrations of antibiotics used for growth were as follows: for M. tuberculosis, hygromycin at 50 μg/ml and streptomycin at 25 μg/ml, and for E. coli, hygromycin at 150 μg/ml and streptomycin at 50 μg/ml.
Gene fragments encoding full-length BagA and BagB were amplified and cloned into the pET24b(+) vector (Novagen) using the NdeI and XhoI restriction sites. A His6 tag was added to the C terminus of BagB. Site-directed mutations were introduced into bagA or bagB using the Stratagene QuikChange mutagenesis kit. Plasmids were used to transform E. coli BL21(DE3).
To make the ΔbagAB::hyg strain (MHD1428), we used allelic exchange as described in detail previously (23), using the plasmid and primers indicated in Table 1. Loss of BagA and BagB was confirmed by immunoblotting with rabbit antibodies to BagAB-His6. Polyclonal rabbit antiserum was raised against purified BagAB-His6 at Covance, Inc. (Denver, PA).
For all PCRs, we used Phusion high-fidelity polymerase (New England Biolabs, Inc.). For checking the sequence of ppsE in the bagAB mutant, we used PCR to amplify a specific region of ppsE, since we knew that some of our parental strain bacterial stock had point mutations in this region based on ongoing whole-genome sequencing studies (Ballister and Darwin, unpublished). We sequenced the PCR products from two independent reactions using the same primers and confirmed that the identical ppsE mutation observed in isolates from our parental strain stock was present in the bagAB strain (Table 1). All sequencing was performed by Genewiz, Inc. (South Plainfield, NJ).
Purification of BagAB-His6.
Bacteria were cultured in LB broth at 37°C, and gene expression was induced by isopropyl-β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 0.2 mM. Cells were collected by centrifugation (5,000 × g, 10 min) and lysed in buffer containing 20 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 5 mM imidazole. The cell lysates were passed through a 5-ml Ni2+-nitrilotriacetate acid (Ni-NTA)–agarose column. Target proteins were eluted with a linear gradient concentration of imidazole and further purified in a Superose 6 10/300 GL gel filtration column. Fractions containing target proteins were concentrated to 20 mg/ml in buffer containing 20 mM Tris-HCl (pH 8.0) and 200 mM NaCl. The purity of the protein was examined by SDS-PAGE. The protein concentration was determined using a NanoDrop ND-1000 spectrophotometer (A280).
Crystallization, data collection, and structure determination.
Crystallization of nucleotide-bound BagAB was performed using the hanging-drop vapor diffusion method at 20°C. BagAB at a concentration of 10 mg/ml was first incubated with 5 mM ADP, ATPγS, or AMP-PNP on ice for 30 min. Then, protein solution was mixed with an equal volume of reservoir solution containing 0.1 M imidazole (pH 8.0), 0.2 M calcium acetate, and 10% polyethylene glycol (PEG) 8000. Crystals appeared overnight and were harvested using 30% glycerol as a cryoprotectant. Platinum (Pt) derivatives were prepared by soaking native crystals for 2 h in the mother liquid containing 5 mg/ml K2PtCl4. Pt single-wavelength anomalous dispersion (SAD) and native diffraction data were collected from crystals flash-cooled at 100 K at Advance Photon Source, Argonne National Laboratory, and processed by iMOSFLM (24) and SCALA (25). The Pt positions and initial phases were determined using PHENIX (26). High-resolution native data sets were used for structural refinement. COOT (27, 28) and PHENIX were used for iterative model building and refinement. The quality of the final model was analyzed with Molprobity (27). The statistics for data collection and refinement are shown in Table 2. We note that the average B-factors of the two higher-resolution structures (BagAB-ADP at 2.3 Å and BagAB-ATPγS at 2.5 Å) are relatively high (63 Å2 and 54 Å2, respectively), nearly twice that of the lower-resolution structure (BagAB–AMP-PNP, 3.2 Å), which has a better B-factor of 26 Å2. Two factors likely contributed to the high B-factors in the highest-resolution structures. One factor was the two large flexible regions at the top and bottom of the structures having very high B-factors, approaching 100. These regions significantly raised the averaged B-factors of the whole molecules. The other factor was radiation damage, as we observed a significant loss of high-resolution diffractions during data collection for the highest-resolution crystals of BagAB complexed with ADP or with ATPγS.
ATPase assays.
ATPase activities were determined by measuring the production of inorganic phosphate using the malachite green phosphate assay kit (Sigma-Aldrich, Inc.). The assays were started by adding 2.5 mM ATP to a solution containing 36 μg of BagAB protein, 20 mM Tris-HCl (pH 7.4), 10 mM MgCl2, and 50 mM NaCl. The reactions were performed for 30 min at 37°C. Reaction products were collected every 10 min and added to 20 μl malachite green reagent in a 96-well plate. The amount of the released inorganic phosphate was determined by measuring the absorbance change at 620 nm.
ITC.
Isothermal titration calorimetry (ITC) experiments were performed in 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 5 mM MgCl2 at 25°C using a PEAQ-ITC microcalorimeter (MicroCal). We titrated 600 μM ADP, 400 μM ATPγS, and 400 μM AMP-PNP into 25 μM, 30 μM, and 25 μM BagAB proteins, respectively. Titration curves were analyzed using PEAQ-ITC software.
Sample preparation for mass spectrometry analysis.
M. tuberculosis cell lysates were prepared from three replicate cultures of WT (MHD1385), bagAB (MHD1431), and complemented (MHD1432) strains by harvesting equivalents of an optical density at 580 nm (OD580) of 10 under denaturing conditions. OD-normalized cultures were centrifuged at 3,200 × g and 25°C for 5 min, and cell pellets were washed three times with phosphate-buffered saline (PBS)–0.05% Tween. Cell pellets were resuspended in 500 μl urea lysis buffer (100 mM Tris, 1 mM EDTA, 8 M urea, pH 8.0) and transferred to bead beating tubes with 250 μl zirconia silica beads (BioSpec Products). Cells were lysed by three rounds of 30-s bead beating in a BioSpec Mini Bead Beater with 45 s of icing in between. Samples were then clarified by centrifugation for 2 min and passed through a 0.2-μm nylon filter to sterilize.
One hundred micrograms of each protein lysate was reduced using dithiothreitol (5 μl of a 0.2 M solution) for 1 h at 55°C. The reduced cysteines were subsequently alkylated with iodoacetamide (5 μl of a 0.5 M solution) for 45 min in the dark at room temperature. Next, 100 mM ammonium bicarbonate (pH 8.0) was added to dilute the urea concentration to 2 M, and the protein lysates were digested with trypsin (Promega) at a 100:1 (protein/enzyme) ratio overnight at room temperature. The pHs of the digested protein lysates were lowered to <3 using trifluoroacetic acid (TFA). The digested lysates were desalted using C18 solid-phase extraction (Sep-Pak; Waters), and 40% acetonitrile (ACN) in 0.5% acetic acid followed by 80% ACN in 0.5% acetic acid was used to elute the desalted peptides. The peptide eluate was concentrated in a SpeedVac and stored at −80°C.
Liquid chromatography-MS/MS analysis.
One microgram of each sample was loaded onto a trap column (Acclaim PepMap 100 precolumn [Thermo Scientific] [75 μm by 2 cm, C18, 3 μm, 100 Å]) connected to an analytical column (EASY-Spray column [Thermo Scientific] [50 m by 75 μm inner diameter]; PepMap RSLC [Thermo Scientific] [C18, 2 μm, 100 Å]) using the autosampler of an Easy nLC 1000 (Thermo Scientific) with solvent A consisting of 2% ACN in 0.5% acetic acid and solvent B consisting of 80% acetonitrile in 0.5% acetic acid. The peptide mixture was gradient eluted into a Lumos mass spectrometer (Thermo Scientific) using the following gradient: 5% to 35% solvent B in 180 min, 35% to 45% solvent B in 10 min, and then 45% and 100% solvent B in 20 min. The full scan was acquired with a resolution of 240,000 (at m/z 200), a target value of 1e6 and a maximum ion time of 50 ms. After each full scan, the most intense ions above 2e4 were selected for fragmentation in the linear ion trap using the “Top Speed” method. The tandem MS (MS/MS) spectra were acquired with an isolation window of 0.7 m/z, target value of 2e4, maximum ion time of 18 ms, normalized collision energy (NCE) of 30, and dynamic exclusion of 30 s.
Proteomic data analysis.
The MS/MS spectra were searched against the M. tuberculosis H37Rv UniProt reference proteome database using Andromeda (29). Protein quantitation between samples was performed using summed up eXtracted ion current (XIC) of all isotopic clusters within MaxQuant (30) (version 1.5.2.8). For the comparison of BagA and BagB (Rv3679 and Rv3680) within each sample, intensity-based absolute quantification (iBAQ) (31) was performed. The mass tolerance was set to 10 ppm for MS1 and MS2 searches. False-discovery rate (FDR) filtration was done first on the peptide level and then on the protein level. Both filtrations were done at a 1% FDR using a standard target-decoy database approach. All proteins identified with fewer than two unique peptides were excluded from analysis. Bioinformatics analysis was performed with Perseus. The intensity values were log2 transformed, and missing values were imputed using the normal distribution as implemented by Perseus. A two-sided Welch t test was performed, and the fold changes and P values were represented in a volcano plot.
Mouse infections.
Mouse infections were performed essentially as described previously (32). Six- to 8-week-old female C57BL6/J mice (The Jackson Laboratory, Inc.) were infected by aerosol to deliver ∼200 bacilli per mouse, using a Glas-Col inhalation exposure system (Terre Haute, IN). The strains used were WT/pMV306.kan (MHD1385), ΔbagAB::hyg/pMV306.kan (MHD1431), and ΔbagAB::hyg complemented (MHD1432). This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes for Health. Mice were humanely euthanized according to an approved Institutional Animal Care and Use Committee protocol at New York University School of Medicine. Lungs and spleens were harvested and homogenized in PBS–0.05% Tween 80 using a Bullet blender with 1.6-mm stainless steel beads (Next Advance, Inc.) at the indicated time points to determine bacterial CFU.
Accession number(s).
The coordinate and structure factor files of ADP, ATPγS, and AMP-PNP-bound BagAB were deposited in PDB with accession number 6BS3, 6BS4, and 6BS5, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants AI088075 awarded to K.H.D. and AI070285 awarded to H.L. A.T.J. was supported in part by Public Health Service Institutional Research Training Award T32 AI007180 (principal investigator, Ian Mohr). The mass spectrometric work was partially supported by the NYU School of Medicine and a shared instrumentation grant from the NIH (1S10OD010582-01A1) for the purchase of an Orbitrap Fusion Lumos.
We thank A. Cornelius for assistance with the animal experiments.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00159-19.
REFERENCES
- 1.WHO. 2017. Tuberculosis. http://www.who.int/mediacentre/factsheets/fs104/en/. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Eiglmeier K, Parkhill J, Honore N, Garnier T, Tekaia F, Telenti A, Klatser P, James KD, Thomson NR, Wheeler PR, Churcher C, Harris D, Mungall K, Barrell BG, Cole ST. 2001. The decaying genome of Mycobacterium leprae. Lepr Rev 72:387–398. [PubMed] [Google Scholar]
- 3.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ. 2009. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leipe DD, Wolf YI, Koonin EV, Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 7.Castillo R, Saier MH. 2010. Functional promiscuity of homologues of the bacterial ArsA ATPases. Int J Microbiol 2010:187373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu CM, Rosen BP. 1989. Characterization of the catalytic subunit of an anion pump. J Biol Chem 264:17349–17354. [PubMed] [Google Scholar]
- 9.Zhou T, Radaev S, Rosen BP, Gatti DL. 2000. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J 19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. 2010. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis [corrected]. PLoS One 5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jastrab JB, Wang T, Murphy JP, Bai L, Hu K, Merkx R, Huang J, Chatterjee C, Ovaa H, Gygi SP, Li H, Darwin KH. 2015. An adenosine triphosphate-independent proteasome activator contributes to the virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 112:E1763–E1772. doi: 10.1073/pnas.1423319112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox JS, Chen B, McNeil M, Jacobs WR Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 13.Rengarajan J, Bloom BR, Rubin EJ. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A 102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazal H, Iljina M, Barak Y, Elad N, Rosenzweig R, Goloubinoff P, Riven I, Haran G. 2019. Tunable microsecond dynamics of an allosteric switch regulate the activity of a AAA+ disaggregation machine. Nat Commun 10:1438. doi: 10.1038/s41467-019-09474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Shawi MK, Polar MK, Omote H, Figler RA. 2003. Transition state analysis of the coupling of drug transport to ATP hydrolysis by P-glycoprotein. J Biol Chem 278:52629–52640. doi: 10.1074/jbc.M308175200. [DOI] [PubMed] [Google Scholar]
- 16.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 17.Lamers MH, Winterwerp HH, Sixma TK. 2003. The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J 22:746–756. doi: 10.1093/emboj/cdg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, Radaev S, Rosen BP, Gatti DL. 2001. Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J Biol Chem 276:30414–30422. doi: 10.1074/jbc.M103671200. [DOI] [PubMed] [Google Scholar]
- 19.Zhou T, Shen J, Liu Y, Rosen BP. 2002. Unisite and multisite catalysis in the ArsA ATPase. J Biol Chem 277:23815–23820. doi: 10.1074/jbc.M203432200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. 2012. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog 8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatfull GF, Jacobs WRJ. 2000. Molecular genetics of mycobacteria. ASM Press, Washington, DC. [Google Scholar]
- 22.Sambrook J, Maniatis T, Fritsch E. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Peterson SN, Darwin KH. 2011. A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol Microbiol 79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 30.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 31.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 32.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. 2003. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 33.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR Jr, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 34.Tufariello J, Bardarov S, Hatfull G, Larsen M, Bardarov S, Chan J, Pavelka MS, Sambandamurthy V, Jacobs WR Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.