Abstract
Given the many cell types and molecular components of the human immune system, along with vast variations across individuals, how should we go about developing causal and predictive explanations of immunity? A central strategy in human studies is to leverage natural variation to find relationships among variables, including DNA variants, epigenetic states, immune phenotypes, clinical descriptors, and others. Here, we focus on how natural variation is used to find patterns, infer principles, and develop predictive models for two areas: (a) immune cell activation—how single-cell profiling boosts our ability to discover immune cell types and states—and (b) antigen presentation and recognition—how models can be generated to predict presentation of antigens on MHC molecules and their detection by T cell receptors. These are two examples of a shift in how we find the drivers and targets of immunity, especially in the human system in the context of health and disease.
Keywords: systems immunology, single-cell genomics, single-cell RNA sequencing, immune cell types, antigen presentation, T cell receptor
CHALLENGES FOR SYSTEMS IMMUNOLOGY
A few decades of molecular and genetic analyses of the immune system have made it clear that a healthy immune system eliminates pathogens and maintains tissue homeostasis through extraordinarily complex networks with feedback systems that lead to elimination of pathogens, while avoiding potentially massive tissue destruction. The networks are composed of dozens of immune cell types (as well as hundreds of nonimmune cell types) that deploy hundreds of cytokines, chemokines, and surface proteins to enable communication and thousands of extra- and intracellular factors to regulate and carry out numerous functions. The failure of this system can result in lethal or recurrent infections, autoimmunity, degenerative diseases, and many other disorders.
Given the complexity of this system, our challenge is to determine whether and how it will be possible to address the burning questions that we need to answer to create effective immunotherapies: (a) Which factors should we target to reprogram the immune system and improve human health in diverse diseases? (b) Can we mechanistically explain variations in immunity across the human population as a function of the thousands of components making up the system? We have to address these two questions to succeed in finding the most effective and safe therapies to trigger potent immunity against pathogens or tumors, or to block harmful autoimmunity, allergy/asthma, and other disorders that result from an overactive immune response.
Systems immunology encompasses a set of strategies and methods to measure how components change and interact over time, and across space, in response to environmental perturbations or genetic variations, with the goal of pinpointing the most critical components (molecules, cells, tissues) and interactions that drive observed immune responses. A systems-wide strategy is essential for modeling immune system functions and dysfunctions. While there are many topics that have been investigated with systems immunology strategies, we focus on two: molecular profiling of cells in health and disease, and methods for understanding and predicting antigen presentation by major histocompatibility complex (MHC) protein and detection of peptide-MHC (pMHC) complexes by T cell receptors (TCRs) on T cells. We review a subset of papers that illustrate recent progress in these areas and discuss some new directions for future study.
MOLECULAR PROFILING TO FIND DISEASE SIGNATURES AND UNDERSTAND CELL TYPES
For many years, systems immunology research has focused on monitoring global molecular profiles of cells and tissues in response to perturbations or in the context of disease (1, 2). Such a strategy has been particularly useful in studies of the human immune system (3–5), where the variation across individuals needs to be systematically monitored to define disease versus healthy states, and where many components need to be tracked to translate the findings from animal models (6, 7). A central strategy has been deriving gene expression signatures associated with disease or therapy using gene expression datasets (microarray or next-generation RNA sequencing) collected from blood or tissue, with the goal of identifying associated cells, molecules, and pathways. Examples of key signatures include those associated with effective immune responses to vaccines, or viral and bacterial infections (8−17), progression to active disease in tuberculosis (18, 19), severity of autoimmune diseases such as systemic lupus erythematosus (SLE) (20–23), as well as immune responses as a function of age, genetics, and environment (24–27). While systematic in the monitoring of thousands of human transcripts, these signature-finding strategies have typically profiled whole blood, peripheral blood mononuclear cells, or whole tissue samples, thus aggregating and blurring RNA species from different cell types and states together.
An equally important long-term goal of systems immunology has been to define the identity, function, and origin of each immune cell, as well as the factors regulating cellular properties. After the discovery of red blood cells and leukocytes, between the seventeenth and nineteenth centuries, most of the major immune cell types were discovered in the twentieth century (28) and further defined thanks to developing technologies including microscopy, immunohistochemistry, monoclonal antibodies, and fluorescence-activated cell sorting (FACS). These tools, coupled with functional studies in mice and humans, have continuously refined the definitions of immune cells, identifying new types, subtypes, and states based on function, size, morphology, developmental origin, location and relationship to other cells within tissues, and, of course, molecular components. More recently, genomic approaches have been used extensively to inventory all the known cell types of the immune system. An important effort by the ImmGen consortium of laboratories has already profiled hundreds of known mouse immune cell types across tissues and perturbations. The consortium shared its high-quality datasets with the entire community online (https://www.immgen.org), with new data and analyses being continuously added as it finds more markers, regulators, and functions for each cell type. A similarly scaled effort in human immunology, capturing all known immune cell types, would provide a much-needed reference dataset. This is gathering momentum through projects such as the Human Cell Atlas consortium (29) (https://www.humancellatlas.org) and the Blueprint Epigenome project (http://www.blueprint-epigenome.eu), which has publicly shared more than 50 immune cell type profiles (expression and epigenomic) from healthy and disease states.
DEFINING CELLULAR IDENTITY ONE CELL AT A TIME
Although global population profiling, as described above, is tremendously informative, there is a need to measure changes at single-cell resolution to enable de novo cell classification and analysis of molecular states that do not confound cell types. These measurements can include many layers of single-cell molecular omics, including DNA, RNA, methylation, protein, chromatin modification and accessibility, and many other molecular state readouts that have been reviewed by others (30−42). Single-cell molecular profiling allows data-driven modeling of cellular subtypes that can capture stable and plastic cells for which markers may not yet be known (33, 43, 44). While it is currently possible to profile tens of thousands of cells by single-cell RNA sequencing (scRNA-seq) and millions of cells by protein mass cytometry, it will remain important to keep increasing the scale, sensitivity, resolution, and number of different analytes measured.
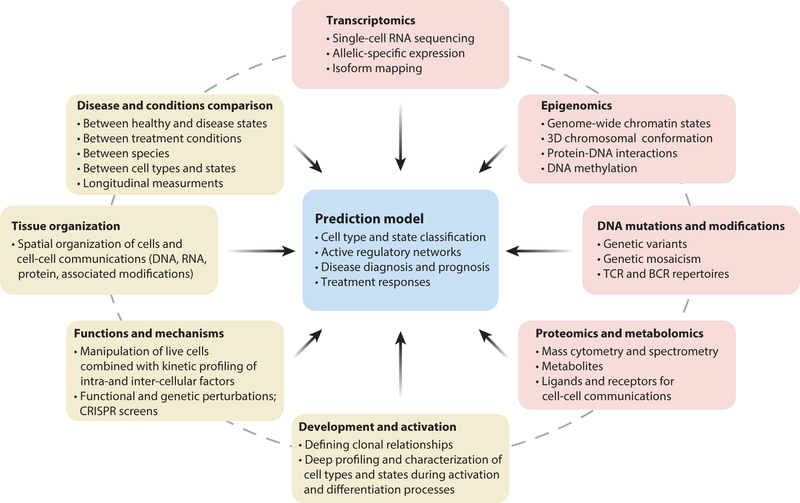
Combining as many different layers of single-cell measurements as possible (45, 46)—such as protein and RNA (47, 48) or RNA and DNA (49) or genotyping together with gene expression and methylation (50)—will ultimately lead to a more sophisticated view of a cell than we have today, not only as an instantiation of a predefined type but also as a sum of historical and dynamic factors that shape its identity (Figure 1). While the field of single-cell measurements is still in its infancy, the last three years have witnessed explosive progress and provide an early framework for applying single-cell systems immunology to addressing open problems. Here we will review initial application of single-cell approaches to define cell types, analyze continuous cell states, and uncover mechanisms in health and disease.
Figure 1.
Single-cell experimental tools and datasets needed for generating models of cell types, states, regulatory networks, and disease/therapy signatures. The boxes in the outer circle represent data types (yellow) as well as experimental and analytical frameworks (pink) needed for describing the properties of a cell and for creating the prediction models described in the center, which include classifying cells and states, learning regulatory networks, and identifying predictive signatures of treatment and disease. Abbreviations: BCR, B cell receptor; CRISPR, clustered regularly interspaced short palindromic repeat; TCR, T cell receptor.
Single-Cell Technologies: High-Dimensional Flow and Mass Cytometry
Our understanding of the human immune system has greatly expanded over the last several decades, due in part to the emergence of single-cell technologies, including modern flow cytometers that enable the simultaneous detection of dozens of proteins per cell. A recent example is a systematic effort by Farber and colleagues to characterize immune cells in lymphoid and mucosal tissues from human organ donors by multicolor flow cytometry (51–55). They provided one of the first quantitative frequencies of immune cell subsets as a function of tissue, age, and other variables in humans, shifting our conception of how T cells change their fates as they move between blood, lymphoid tissues, and mucosal sites. Given the relatively high accuracy, sensitivity, and robustness of this technology, it is not surprising that thousands of studies have and will continue to learn new biology using flow cytometry.
A more recent development is mass cytometry, which combines mass spectrometry with flow cytometry to quantify 30−45 rare earth metal isotope labels on antibodies per cell, with higher numbers expected soon. Spitzer & Nolan (31) published a comprehensive review of the current state of mass cytometry, including both instrumentation and novel analytical methods required for identifying patterns in high-dimensional data, offering solutions beyond what flow cytometry analysis tools can currently offer (56). This method has now been used in numerous studies, including one of the first ones that defined the phenotypic and functional differences across hematopoietic cells (57), as well as subsequent studies of myeloid cells (58), lymphocytes (59), B cells (60), natural killer (NK) cells (61, 62), CD8+ T cells (63), follicular helper T (Tfh) cells (64), T cells (65), dendritic cells (DCs) (66), innate lymphoid cells (67), peripheral and mucosal immune cells (68), the effects of circadian rhythms on immune cell populations across the body (69), and the importance of systemic helper T (Th) cells in controlling melanoma in mice (70). An interesting set of examples came from two groups that used flow cytometry or mass cytometry to quantify markers in twins and identified which markers were more subject to genetic (71) or environmental (72) control, offering a new view on the stability of immune phenotypes in human subjects. Finally, an important application is the modeling of developmental trajectories, which becomes feasible because of the high number of cells that can be profiled in high-dimensional space, allowing reconstruction of differentiation paths (60, 73).
Mass cytometry, together with high-dimensional flow cytometry, allows a more complete interrogation of cellular phenotypes based on protein levels as well as their posttranslational modifications. While the limited number of markers will often restrict the potential for new discoveries (especially when analyzing one-of-a-kind human samples), the ability to quantify protein abundance with specific antibodies in millions of cells will remain a central tool in immunology for the foreseeable future.
Single-Cell Genomics to Quantify, Discover, and Characterize Immune Cell Types
The emergence of scRNA-seq has enabled de novo discovery of immune cell types and states, and the development of new mechanistic hypotheses (74, 75). We review a small number of examples illustrating the use of this technology in immunology. Jaitin et al. (76) were among the first groups to use scRNA-seq approaches to decipher the cellular components of a tissue without using known markers. An analysis of several thousand mouse splenic cells identified the known cell types in proportions similar to the expected frequencies and discovered novel heterogeneity in DCs before and after lipopolysaccharide (LPS; a component of gram-negative bacteria) activation. Other studies have used similar approaches to identify cell types in mice, such as DC progenitors (77) and differentiating tissue-resident macrophages during organogenesis (78). Several studies added epigenomic profiling to better define the regulatory circuitry underlying transcription and cell identity, such as in blood murine monocytes (79), thymic natural killer T (NKT) cells (80), CD8+ T cells (81, 82), intestinal innate lymphoid cells (ILCs) (83), and murine microglia (84). In one case, a more complete analysis of immune cell types was performed using ATAC-seq for open chromatin profiling (85, 86), though this was done in predefined populations. These studies represent a new paradigm for de novo immune cell type discovery and elucidation of regulatory circuits, with potential applications to any problem in immunology.
An elegant example illustrates the potential for rapid application of single-cell profiling results to test a new model of immune cell ontogeny (87). In this study, single-cell profiling addressed whether distinct myeloid differentiation pathways arise from common myeloid progenitors (CMPs) versus lymphoid-primed multi-potent progenitors by scRNA-seq analysis. Upon identifying Gata1 as a discriminating marker across potential myeloid progenitors, a mouse expressing a Gata1-GFP reporter was engineered, which allowed prospective sorting, profiling, and analysis of lineage potential. Gata1-GFP+ cells were found to give rise to one set of cells (erythrocytes, megakaryocytes, eosinophils, and mast cells) and Gata1-GFP− cells to a distinct set (neutrophils, monocytes, lymphocytes), which challenged the current conception of an early blood-lineage fate decision model.
Discovery of genes that mark a particular immune cell population by scRNA-seq can generate new opportunities for therapeutic targeting and manipulation of a particular cell type, as demonstrated by Yu et al.’s (88) discovery and characterization of innate lymphoid cell progenitors (ILCPs). Based on 10 clusters initially identified by performing scRNA-seq of cells isolated from a particular cell gate in the bone marrow, the authors focused follow-up studies on one cluster identified as the putative ILCP population based on the expression of some known markers. This led them to identify PD-1 as a discriminating surface marker for these cells, allowing efficient prospective isolation of ILCPs without using genetic reporters and complex gating schemes, and functional experiments validating the developmental potential of Lin−PD-1hi cells. Upon expanding the scope of their study and validating PD-1 as a marker also expressed in tissue-resident ILCs, the authors demonstrated that administrating anti-PD-1 antibody (which is normally used to induce cytotoxic cells that target cancer cells) depleted Lin−PD-1hi cells and led to changes in immunity to influenza and acute lung damage. This study highlights how better cell markers can be discovered de novo through a systematic scRNA-seq approach and be used to deplete and study an important cell type that had been challenging to manipulate hitherto.
Another important application of scRNA-seq is to remap the cells of the human immune system (89−94). For example, Björklund (89) found that ILCs from tonsil tissue segregated into the known 3 ILC subsets (ILC1, ILC2, ILC3), with each subset expressing a unique set of markers, receptors, and signaling pathway components. While there were no new ILC subsets, ILC3 further split into 3 subpopulations defined by novel markers. On a technical note, this study also linked protein and RNA levels in single cells by measuring protein levels through index sorting of single cells into plates prior to generating full-length cDNA and sequencing libraries with the Smart-seq2 protocol (developed by this group; 95, 96), which helps link known markers to discovered cell types. A second example in humans, from our group, also used deep single-cell profiling by Smart-seq2 to identify 11 clusters corresponding to putative subtypes of DCs, monocytes, and DC progenitors (90). Based on surface markers identified by scRNA-seq, we developed strategies for prospective isolation of several clusters (DC2, DC3, 2 subsets of DC5, and a putative cDC progenitor). We validated the purification strategy by performing additional scRNA-seq on sorted cells and showing high enrichment or purity of these prospectively sorted cells through projection of profiles on top of the originally discovered cells. Upon validating enrichment strategies, newly sorted cells were then used to do functional studies, showing for example that the putative DC progenitor differentiated into both types of conventional DCs in vitro and that plasmacytoid dendritic cells (pDCs) isolated using standard methods are contaminated with a new DC subtype, DC5, that accounts for much of the T cell stimulatory potential previously assigned to pDCs. We note that a cell that has overlapping properties with DC5 was independently discovered by See and colleagues (91) using CyTOF and scRNA-seq; they found it to be rare and heterogeneous, with the ability to differentiate into conventional DCs, to stimulate naive T cells, and to also be contained within the traditionally defined pDC flow gate. Our study thus provides a framework for using scRNA-seq data to identify markers that can be used to isolate newly discovered immune cell populations for deeper functional characterization. The strategies used in these two studies will be useful in developing a more complete atlas of the human immune system, which should now be possible using emerging high-throughput scRNA-seq methods (93, 97–99).
An important goal is to tackle immune populations across different organs to understand the sites of disease. While not focused on immune cells, some recent studies illustrated the power of single-cell profiling in human tissues (100–102). Baron et al.’s (100) profiling of 2,000 pancreatic cells from mice and 10,000 from humans allowed a parallel comparison of these mouse and human organs, showing 14 and 13 distinct clusters (including immune cells such as tissue-resident macrophages, mast cells, B cells, and cytotoxic T cells) in human and mouse, respectively. While almost all the cell types were conserved, the expression profiles of the 4 endocrine cell types (α, β, γ, and δ) were the most conserved across species. A similar study, only in humans, from Murano et al. (101) identified 9 distinct clusters (excluding immune cells), most of which overlapped with the ones reported by Baron et al. Both studies reported lists of de novo uncovered cell-specific markers for each cell cluster observed, a rich resource for the community, and validated some of the newly predicted markers by immunohistochemistry.
An important use of any tissue atlas is to find the cell types that are most likely to contribute to disease. By mapping the expression of disease susceptibility genes from genome-wide association studies (GWAS) of type 1 and type 2 diabetes to their pancreatic single-cell dataset, Baron et al. (100) and others (103) showed that DLK1 is the only GWAS risk gene specifically expressed in β cells. Despite mouse models of type 2 diabetes (104) showing that leptin and its receptor are important in β cells (105), the leptin receptor appears to be δ cell–specific in humans, suggesting an important species difference and emphasizing the value of accurately mapping genes to cell types in humans and mice.
To overcome the loss of spatial information in dissociated cells used for scRNA-seq, Halpern and colleagues (106) combined single-molecule fluorescence in situ hybridization (smFISH) of six landmark markers with scRNA-seq to map mouse liver cells. Briefly, using smFISH they segmented liver cells into different zonation patterns and inferred the mapping of 1,500 single cells to different liver segments based on their scRNA-seq profiles. The accuracy of the map was validated by smFISH of an additional 20 genes. Their strategy led to a new classification of mouse liver cells. This elegant approach can be easily generalized to any structured organ across healthy and disease states, as was shown by other groups who combined scRNA-seq with in situ expression measurements of landmark genes to map cells in space (107, 108). Spatial sequencing continues to be improved with NICHE-seq (109), fluorescent in situ RNA sequencing (FISSEQ) (110), multiplexed error-robust fluorescence in situ hybridization (MERFISH) (111), and RNA sequential probing of targets (SPOTs) (112).
Many challenges remain ahead, including finding solutions for increasing the throughput at a scale that would enable efficiently capturing even the rarest events in a cost-effective manner. This could be done, for example, by using individual-specific natural genetic variation (113). For the immunology community, generating a detailed census and spatial map of all immune cell types and states across all healthy tissues is no small endeavor, since immune cells are present in all tissues. An international community has organized around the goal of creating a human cell atlas (29), an evolving project that will keep adding new layers of information, such as cell activity states, dynamic transitions, physical locations, lineage relationships, and developmental origins. Importantly, while we mostly discussed RNA single-cell measurements as a means of classifying cells, ultimately many additional layers of information—such as epigenetic landscape, protein localization, gene functions, spatial localization, and ontogeny—in steady-state and activated contexts will need to be integrated to generate a more complete model of the immune system (45–50) (Figure 1).
A Continuous Spectrum of Phenotypes in Cells
Categorization of cells into discrete types has been the focus of multiple single-cell genomic studies. However, cells of the same type can also exhibit substantial heterogeneity, reflecting finer subtypes, functional variation, or inherent stochasticity. Rather than falling into discrete states, cells sometimes take on states along multiple continuous dimensions. For the immune system, this may be an important mechanism to generate the plasticity needed to face diverse and changing pathogens. Such a continuous spectrum of cells is also observed in development and hematopoiesis, which we do not review comprehensively though it is a field where scRNA-seq is having a major impact. While flow and mass cytometry can be used to monitor continuous states, such as T cell polarization, they are less likely to discover new states (if there are unknown ones) or identify candidate regulators for which the markers remain unknown. In addition, while profiling millions of cells by mass cytometry does help in reconstructing trajectories of differentiation and activation (60, 73), a more definitive trajectory and molecular model should be possible to infer in most cases once single-cell transcriptomes can be profiled in larger numbers. Of course, the required sampling density ultimately depends on the number and complexity of paths and intersections to be modeled, and different experimental strategies can be implemented that can iteratively enrich for rare, transient populations to reconstruct a trajectory. These dynamic processes may occur as part of development or in response to an environmental trigger or physiological cue. Below, we provide some examples of principles and strategies to study continuous states.
Following an initial study (114) showing heterogeneity in mouse bone marrow–derived DCs (BMDCs) stimulated with LPS, a second study of several hundred cells (115) identified a small subset (<1%) of LPS-induced BMDCs that expressed IFN-β at early time points and appeared to coordinate the subsequent response of other cells through paracrine signaling. To demonstrate a role for cell-cell communication in the activation of these cells, they profiled the cells—interacting together, isolated from each other (in sealed chambers), or deficient in regulators of interferon—and showed how cell-to-cell communication contributes to the induction of cellular heterogeneity during immune responses. These were some of the first examples of studying dynamic activation processes using scRNA-seq and determining the contribution of intercellular communication to single-cell decisions.
Activated T cells represent another example of a continuous spectrum of states. T cells often become polarized toward one fate or a combination of fates but do not necessarily form well-separated single-cell clusters by gene expression. A study of Th17 cells by Gaublomme et al. (116) used scRNA-seq to show that pathogenic Th17 cells, which appeared during induction of autoimmunity in a model of experimental autoimmune encephalomyelitis (EAE) or could be generated in vitro under certain conditions, were not distinct subtypes from nonpathogenic Th17 cells but rather represent states along a spectrum. This type of analysis was still able to nominate candidate regulators for pathogenicity (i.e., Gpr65, Plzp, Toso, and Cd5l), some of which were subsequently validated in knockout mouse models (117), suggesting the possibility of suppressing pathogenic Th17 cells but not the nonpathogenic cells that are critical for maintaining tissue homeostasis. This study highlights that a spectrum of cell states can thus underlie differences in function and be analyzed to discover new regulators that drive cell function.
Some of the continuous spectrum of cellular phenotypes captured by single cells can also arise from technical and biological confounders, such as cell cycling. Failing to account for confounders can mask interesting biology. To consider cell cycling confounders, Buettner et al. (118) developed and used a single-cell latent variable model (scLVM) to remove cell cycle effects and discover structure in scRNA-seq data from CD4+ Th2 cells, revealing genes that distinguish two differentiation states—neither of which was detectable when the cell cycle covariates were not accounted for. While reviewing all single-cell analytical computational challenges and solutions is beyond the scope of this review (33, 119, 120), this particular example illustrates the need to consider confounders to unmask interesting biology.
Nonetheless, correcting for confounders such as cell cycling should be done carefully. Proserpio et al. (121) assessed the heterogeneity of CD4+ T cells from infected mice and found three CD4+ T cell states during the differentiation process from naive to effector Th2 cells: activated yet nondividing cells, proliferating cells that are not differentiated, and Th2-like cells that are cycling and differentiated. They validated their model through subsequent in vitro differentiation and polarization of Th2 cells, live imaging, and additional profiling. The authors concluded not only that cell cycle entry is an essential condition for differentiation but also that these faster cycling cytokine-secreting mature cells may be critical for infection clearance. Although not surprising, this model illustrates that cell cycle gene expression modules can also be associated with important biology.
scRNA-seq analysis can also highlight new biological principles when comparing different contexts or environmental perturbations. Gury-BenAri et al. (83) asked how administration of antibiotics affects ILCs of the small intestinal lamina propria. By integrating population transcriptomics, epigenetics (ChIP-seq, ATAC-seq), and single-cell transcriptomics, they revealed 15 transcriptional states, including 2 novel states, and found that absence of microbiota (e.g., after antibiotics or in germ-free housing) blocks ILC3 plasticity toward ILC1 fate, demonstrating how microbiota maintain certain ILCs in the intestinal microenvironment. Another variable that may alter immune cell behavior is aging, as one group observed upon profiling CD4+ T cells from mice. They discovered increased cell-to-cell transcriptional variability in aged mice compared to young mice (122). This property of Th cells may turn out to be a feature of aging that underlies the decline in immunity, although the cause remains a mystery.
Dynamic processes in vivo can also be analyzed, and two particular studies demonstrated the power of profiling single cells along a time course of differentiation to generate a model of a biological process and cell fate trajectory. Kakaradov et al. (81) analyzed CD8+ T cell fate specification by scRNA-seq during lymphocytic choriomeningitis virus infection in mice. Based on trajectory analysis and epigenetic profiling, cells undergoing their first division appeared to be composed of two subpopulations that reflected much later terminally differentiating effector and memory cells. By classifying cells at each intermediate time point after infection according to their predicted future fates, they inferred differentiation pathways and validated a central role of a key regulator, Ezh2, which was initially identified as a differentially expressed gene between effector and memory cells. Their data led to a new model in which differentiation of CD8+ terminal effector T cells begins with an early transcriptional burst that is subsequently refined by epigenetic silencing of memory-associated transcripts, whereas the induction of memory is a slower process marked by a gradual increase in expression of a few genes.
Using a similar experimental strategy, Lönnberg et al. (123) profiled single CD4+ T cells after Plasmodium infection in mice and resolved the time of bifurcation of CD4+ T cells toward Th1 and Tfh fates. By using endogenous TCR sequences captured by scRNA-seq as cell barcodes using a method reported by Stubbington et al. (124) (see other studies that have also looked at single-cell TCR analysis, in some cases jointly with transcriptional analysis; 94, 125–128), they were able to find Th1/Tfh cells that arise from the same clones, supporting the idea that a single naive CD4+ T cell gives rise to more than one cell fate. Through leveraging their scRNA-seq temporal data, they also identified transcriptional signatures associated with bifurcation of Th1 and Tfh fate trajectories (e.g., including transcription factor Tcf7f for Tfh fate and Id2 for Th1 fate), through which both subsets arise by day 7. By profiling myeloid cells at single-cell resolution, they were able to hypothesize cell-cell interactions between chemokine receptor–expressing T cells and chemokine-expressing myeloid cells. Additionally, the authors provided a new modeling framework that can characterize cell differentiation toward multiple cell fates (i.e., GPfates), and made their single-cell datasets available for analysis by the community (http://www.plasmoth.org)—a type of resource that would benefit any single-cell studies generating such rich datasets. Noteworthy, Ner-Gaon et al. (129) have created the JingleBells data repository for immune-related scRNA-seq datasets (http://jinglebells.bgu.ac.il).
Molecular Mechanisms of Intracellular and Intercellular Circuits in Health and Disease
To build models of regulatory circuits, one typically correlates the levels of particular regulators with cellular phenotypes and then tests the hypothesized connection through genetic perturbation (130). However, to directly discover regulator-target relationships, one can also perturb each gene using CRISPR/Cas9 short guide RNAs (sgRNAs) and monitor the phenotypic effects using RNA-seq or other profiling methods. Although studies have used this approach with success (131), a shift toward scRNA-seq readouts would increase the scale of perturbations while removing confounding results caused by different cells responding differently to perturbations. Several groups recently implemented such a method and showed that scRNA-seq could be used both to monitor the impact of sgRNAs on transcriptomic phenotype and simultaneously to detect the guide that is present in each cell (132–135), thus combining the power of single-cell studies with systematic genetic perturbations. These methods are still in their infancy and promise to enable very powerful genetic-phenotype association studies in cells in vitro and in vivo. Other emerging approaches for mapping intracellular circuitries include microfluidic systems enabling dynamic manipulations of single cells coupled with time-lapse microscopy for live cell measurements and readout by scRNA-seq, as recently demonstrated by Lane et al. (136) and Junkin et al. (137), who both studied myeloid cell signaling.
There is always cross talk between host and pathogen, allowing host cells to integrate bacterial signals and bacteria to regulate virulence in response to the host. In several studies, scRNA-seq analysis identified cell-to-cell variability in host cells infected with pathogens, leading to distinct host cell innate immune responses and control of pathogen infection and replication (99, 138–140). The results of such studies will be critical for understanding the basis for incomplete protection against specific pathogens as a result of host heterogeneity or defects, as well as mechanisms of resistance and tolerance by the pathogen.
The power of single-cell analysis of disease is evident from decades of flow cytometry and histopathological studies. The emergence of single-cell genomics readouts is now enabling extensive characterization of the disease cellular ecosystem in tissues, resulting in richer molecular signatures and deeper understanding of the cells involved in driving the pathogenesis. Comparing immune cell residents’ and infiltrates’ profiles by scRNA-seq across wild-type and disease conditions in animal models can lead to new disease pathogenesis hypotheses (141–144). For example, Keren-Shaul et al. (141) identified a novel type of microglia associated with neurodegenerative disease (DAM), along with specific markers, spatial localization, and pathways associated with these pathogenic cells, leading the authors to propose a new disease model. In another study, population-level and scRNA-seq analyses of CD8+ tumor-infiltrating lymphocytes, followed by genetic perturbation modeling, allowed Singer et al. (142) to find a regulatory program that contributes to dysfunctional CD8+ T cells in the tumor.
Generating single-cell, genome-wide information from many patients will allow us to link the cellular ecosystem in cancer lesions with disease course, treatment response, and immunity. scRNA-seq studies of freshly resected tumor samples have already been published for many cancers: melanoma (145), glioblastoma (146), low-grade glioma (147), isocitrate dehydrogenase–mutant glioma (148), colorectal tumors (149), breast cancer (150), liver cancer (151), renal cell carcinoma (152), ovarian cancer (153), chronic lymphocytic leukemia (154), acute lymphoblastic leukemia (155), and myeloproliferative neoplasms (156). Unsupervised analysis distinguishes malignant, stromal, endothelial cells and several immune cell types, even inferring defective pathways and cell states, such as those associated with dysfunctional T cell states (142, 145). Additionally, in breast cancer tissues, spatial analysis of multiplex protein expression by imaging mass cytometry (157, 158) has allowed classification of infiltrating immune cells and malignant cells based on the neighborhood of surrounding cells. Furthermore, cell type and state signatures that are identified in these single-cell genomic studies can also be found in bulk tissue or blood samples (145, 146, 159) and could be studied for their association with clinical outcomes. In addition, to analyze the communication between tumor and immune cells, scRNA-seq could also associate regulatory programs in cancer cells with immune infiltrate cell types and states. Furthermore, the immune cell landscape in tumor lesions can be studied by mass cytometry, as shown recently in stage I lung adenocarcinoma (160) and clear cell renal cell carcinoma (161); specific immune cell states were found in tumor but not healthy tissue. Litzenburger et al. (162) reported how single-cell chromatin accessibility could uncover cancer heterogeneity, with potential implications for clonal dynamics, drug sensitivity, and immune responses. We expect that increasingly detailed maps of tumor cells along with immune and other cells in the microenvironment will improve our understanding and approach to cancer immunotherapy.
Similar strategies for analyzing immune cellular ecosystems at single-cell resolution are also being applied to the field of autoimmunity and inflammatory conditions, identifying cell types, states, and pathways that may drive pathogenesis. Initial findings have been reported for peanut allergy (163), immune response to surgical trauma (164), type 1 diabetes (102, 165), type 2 diabetes (102, 166–168), lupus nephritis (113, 169, 170), and rheumatoid arthritis (171, 172).
We expect future studies to apply single-cell profiling to thousands of patients to find how genetic variation affects gene transcription and cell phenotype, and to pinpoint the cellular context in which disease susceptibility genes drive disease (Figure 1). Thinking toward the more distant future, if we can uncover the drivers of each individual’s immune response, we will be able to customize therapy to address the specific configuration of each person’s immune system and thus advance precision medicine.
PATTERNS AND PREDICTORS OF ANTIGEN PRESENTATION AND T CELL IMMUNITY
The adaptive immune system is critical for clearance and long-term protection against pathogens and tumors, but it also underlies autoimmunity and allergy. The complex repertoire of antigen receptors expressed in T and B cells is created at the chromosomal level through recombination and somatic mutation, leading to receptors that bind highly molecularly diverse antigens, including self and nonself antigens. In contrast to antibodies that bind antigens in their native form outside the cell, TCRs only recognize protein fragments bound to major histocompatibility complex (MHC) proteins on the surface of cells. These fragments are generated inside the cell, transported to the endoplasmic reticulum (ER), and loaded onto MHC molecules for display on the cell surface. Almost all cells of the body have the capability to present antigens on MHC class I molecules, thereby revealing the contents of any cells to T cells for recognition. In this way, viruses and bacteria, as well as tumor antigens and self-antigens, within cells can be detected by T cells and their host cells selectively killed by cytotoxic CD8+ T cells. In addition, specialized antigen-presenting cells and other cells can also present antigens from internalized materials on MHC class II molecules, leading to activation of CD4+ T cells, which help orchestrate immune responses, including activation of cytotoxic T cells and B cells.
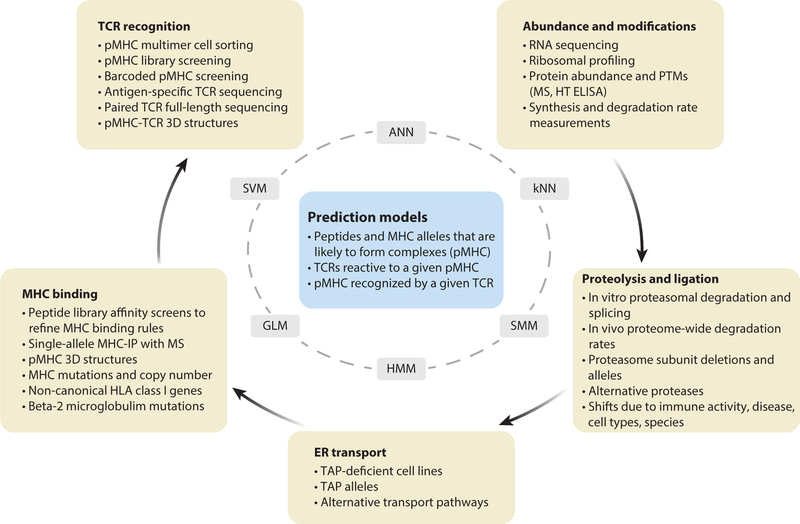
A long-term goal in computational immunology is to predict which antigens are immunogenic and which antigen receptors interact with particular antigens. If we can do this with high accuracy, we will be able to more effectively identify the critical antigens underlying control of infections and cancer as well as destruction of tissue by autoreactive lymphocytes. TCR-antigen interactions are constrained by the universe of antigens that are presented by MHC, and we focus our discussion on studies aimed at predicting the presentation of peptide antigens on MHC molecules and the subsequent recognition of the formed peptide-MHC (pMHC) complexes by TCRs. We review progress in generating and using large-scale datasets to derive rules of MHC-I antigen presentation and recognition (which are better developed than for MHC-II), proteolytic processing of proteins into peptides, transport to the ER, loading onto diverse alleles of MHC proteins, and recognition of pMHC by TCRs (Figure 2). Ultimate success for computational systems immunology would be to predict whether a given antigen and MHC allele will form a pMHC complex and how likely it is to induce a T cell response in a particular individual or, conversely, to predict how likely a given TCR is to recognize any particular pMHC. These predictive algorithms could then be applied to design individual-specific vaccines to target evolving pathogens and highly heterogeneous tumors, as exemplified in three studies demonstrating personalized cancer vaccines in humans (173–175).
Figure 2.
Experimental tools and datasets needed to develop predictive models of antigen presentation and TCR recognition. The boxes in the outer circle represent steps in the process of MHC-I antigen presentation and recognition, and the experimental strategies and types of datasets that need to be collected to train predictive models. The inner circle lists some of the analytical methods for learning the underlying rules that govern each step or the integrated process. The box in the center shows the output (or goals) of the predictive models. Abbreviations: ANN, artificial neural networks; GLM, generalized linear models; HMM, hidden Markov models; HT, high throughput; kNN, k-nearest neighbors; MHC-IP, immunoprecipitation of MHC proteins from cells; MS, mass spectrometry; pMHC, peptide-MHC complex; PTMs, posttranslational modifications; SMM, stabilized matrix method; SVM, support vector machines; TAP, transporter associated with antigen processing.
PEPTIDE GENERATION THROUGH PROTEIN PROTEOLYSIS
The Role of Proteasomal Subunits in Generating Peptides for Antigen Presentation
MHC-I-presented antigens originate from degraded intracellular proteins, either self or non-self, typically broken down by the proteasome. The proteasome is a multiunit protein complex that exists in different flavors depending on the proteolytic enzymes that make up its catalytic core subunits. At homeostasis, most cells express the constitutive proteasome with characteristic units β1, β2, and β5, while IFN-γ promotes one or more subunits of the immunoproteasome (β1i, β2i, β5i). Whether alternative proteasome variants have characteristic cleavage patterns and what their impact is on the MHC ligandome are pertinent questions for antigen prediction and have been researched extensively (176–178). We highlight a study by de Verteuil and coworkers (179), who addressed these questions in an in vivo system by comparing eluted pMHC from wild-type DCs, which naturally express both the constitutive proteasome and the immunoproteasome, and LMP7 (β5i) MEC1 (β2i) double knockout DCs. They found that half of the peptides detected in wild-type DCs were at lower levels and 14% undetectable in cells lacking the immunoproteasome subunits. Changes in gene expression were also observed in immunoproteasome-deficient cells; however, their role and mechanism are not understood and they did not explain the differences in observed peptides. In a more recent study with mice lacking all three immunoproteasome subunits, Kincaid et al. (180) found that MHC expression was significantly reduced in the immunodeficient animals and that antigen repertoires were ~50% different from those of wild-type mice. More work remains to be done in human and mouse cells (immune, nonimmune, and tumor cells) to better define the role of the different proteasomes, as it varies across cell types, immune activity, disease, and species (181). In addition, proteolysis for MHC-II presentation, which is not covered here, also requires further research.
Other Proteolytic Enzymes
As complex as the proteasome is, it is also important to consider the plethora of other proteases that shape the antigen repertoire, albeit to a lesser extent. An overview of ~20 proteases that act in the MHC-I pathway and can alter presented epitopes is available from Lazaro and colleagues (182; also see 183). Mouse models deficient for most have been studied and, in general, do not suffer global defects in antigen presentation; however, certain peptides are known to require particular enzymes to be generated (182).
Cleavage Prediction Tools
As an integral part of the MHC-I presentation pathway, rules of proteolytic degradation have been captured into cleavage site prediction models. Two sources of data are available for the training of such models: in vitro proteasomal digestions and in vivo epitopes presented on MHC molecules, both read out with mass spectrometry (MS). It is important to note that in vivo MHC-eluted peptides are a surrogate, rather than direct representation, of proteasome activity due to the confounding effects of auxiliary intracellular proteases and MHC selectivity. Different subsets of these types of data and different techniques have been used to develop predictors [as reviewed by Lundegaard et al. (184)] with NetChop (185), an artificial neural network–based method trained on in vivo data representative of both constitutive proteasomes and immunoproteasomes often shown to outperform other predictors based on the AUC (area under the receiver operating curve) metric, especially on in vivo data (186, 187). Unlike NetChop and other similar methods that predict the likelihood of cleavage at particular residues, a few algorithms predict the likelihood of the full peptide being formed, for instance, FragPredict (188, 189) and PepCleave_II (190). Toward a similar goal, we recently took advantage of the vastly increased set of available MS-sequenced epitopes to train a neural network predictor on more than 100,000 epitopes (191), and expand processing rules to include cleavage preferences at both termini as well as within the peptide. We observed that arginine and lysine were enriched at both upstream and downstream positions, while proline was depleted, likely due to its rigid conformation. Correspondingly, proline was enriched within the epitope sequence, preventing internal cleavage. A strong preference for peptides originating from the protein C terminus, such that a single cut is required to make the peptide, was also noted. These rules are likely to be refined significantly in the near future, given the dramatic increase in the number of MHC-associated peptides identified by MS, and, hopefully, the additional profiling of different cell types with varying configurations of proteasome subunits.
Peptide Splicing Expands the Repertoire of Presented Antigens
Apart from canonical proteasome-degraded peptides, spliced peptides can also be formed by the proteasome acting in reverse to create peptide bonds. While specific spliced peptides were discovered with low-throughput methods as early as 2004 (192–195), systematic identification of splicing rules was explored in 2015 by Berkers and colleagues (196). The group used a small pool of short peptide precursors and in vitro ligation assays to screen for sequences that are favorable for transpeptidation. The combinations of hydrophobic amino acid at P1 with a basic or small residue at P2, or negatively charged or polar residues at P1 followed by a small or polar residue at P2, were found to be most conducive to splicing at the N terminus, while the presence of C-terminal ligation partners, rather than the particular sequence, emerged as the most important determinant of spliced peptide formation—with the caveat that a single HLA allele and a limited set of antigen precursors were examined. Additionally, both cis (ligation between precursors from the same degradation fragment) and trans (ligation partners from two different fragments) events were observed in vitro, but experiments with heavy-labeled precursors confirmed that cis splicing is predominant for longer protein fragments, as expected.
A high-throughput, unbiased approach for identifying spliced peptide epitopes was recently adopted by Liepe et al. (197). Combining powerful MS techniques and custom database search strategies, they interrogated MS spectra of HLA class I–eluted peptides from multiple cell lines for spliced peptides. As much as 30% of unique sequenced epitopes were found to be spliced. The finding of spliced peptides by MS is likely to spur many groups to search for these peptides in their existing MS data, which is important for validating as well as expanding the list of spliced antigens. An important point is that this study did not identify the same biases as Berkers et al. (196), which may be consistent with cell line–specific peptide splicing preferences and in part due to the presence of distinct HLA alleles. Furthermore, predicted HLA binding scores for spliced peptides were lower than for nonspliced peptides. The authors argue that this could be because prediction algorithms are trained on data that do not contain spliced peptides, but it could also indicate that some of the identified spliced peptides are false-positives. The diagnostic and therapeutic potential of spliced peptides needs to be explored further, for example, by looking for spliced peptides that are validated with synthetic peptides detected by MS, that are found in multiple patients with a disease, and that can induce T cells that recognize cells naturally expressing the spliced antigen (as in 198, for example).
Proteolysis prediction is made difficult by the existence of multiple proteasome subtypes and catalytic units, the proteasome’s ability to not only degrade but also ligate proteins, the lack of direct measurement methods, the activity of additional proteases, and differences between cell types and species. By using MS to identify and quantify MHC-associated peptides in cells with or without different proteases (deleted using CRISPR/Cas9 in cells or mice) as a function of cell type, it should be possible to reach a more predictive model of peptide generation.
PEPTIDE TRANSPORT PREFERENCES
Fragments of endogenous proteins degraded by the proteasome are translocated into the ER for further processing, typically via the transporter associated with antigen processing (TAP). TAP-mediated transport is the predominant mechanism of antigen transport used across all cell types, although alternative ER transport pathways have been reported (199). As such, whether TAP is permissive of all peptides or exhibits any binding preferences that may restrict MHC-I display is of interest. Several groups inferred rules from systematic studies with increasingly larger combinatorial libraries of peptides screened for TAP affinity showing a minimal peptide length requirement and constraints on amino acid properties in the first three N-terminal positions and the C terminus, while the middle of the peptide, involved in TCR recognition, was found to be unconstrained (200–202). Different computational approaches have been utilized to train TAP affinity predictors—including consensus scoring matrix (203), support vector machines (TAPPred; 204, 205), neural networks, and hidden Markov models (PREDTAP; 206)—and have confirmed the experimentally observed importance of peptide positions 1, 2, 3, and 9. To evaluate the contribution of TAP to the prediction of presented antigens, Peters et al. (203) considered a two-step strategy. Of 87 HLA-A0201-restricted peptides, those with predicted TAP affinity below a specified threshold were filtered out before MHC binding was predicted, which resulted in a significant AUC increase, from 0.919 to 0.932. A more systematic evaluation of the predictive value of TAP awaits.
PEPTIDES THAT BIND DIVERSE ALLELES OF THE MHC PROTEINS
High-Throughput Detection of MHC-Bound Peptides
Peptides are loaded onto MHC molecules in the ER and then shuttled to the cell surface. With more than 10,000 HLA alleles identified to date (207), the rules of MHC peptide selection play a central role in shaping the antigen presentation landscape. Various assays have been developed to gather data on peptide-MHC binding (reviewed in 208). Historically, synthetic pools of peptides were assessed for binding against specific HLA alleles in competition binding assays. While in vitro assay data are a valuable resource, synthetic peptide libraries can be very biased, are relatively low throughput, and do not reflect the biological processing and presentation processes of the cell. Alternatively, many groups have developed pipelines for isolating MHC-associated peptides and identifying them by MS (see 209 for one of the first examples), which also allows for the detection of posttranslational modifications. Most recently, Bassani-Sternberg and colleagues, who applied a high-throughput workflow for identifying in vivo HLA-presented peptides by MS (210, 211), found >100,000 naturally presented epitopes from cell lines and patient samples across two studies, constituting the largest endogenous epitope dataset to date. A significant positive correlation between protein abundance and antigen presentation was observed (210), and patient-specific neoantigens were detected (211). Although MS methods allow for the detection of naturally presented epitopes, one caveat of using this approach to gather data for the development of predictive algorithms is the fact that cells express up to six different HLA alleles, which necessitates preexisting knowledge of binding motifs to assign peptides to alleles and thus prevents de novo unbiased motif learning. To this end, our group recently applied a streamlined mono-allelic MS strategy for finding HLA allele–specific peptides and developed improved predictors of HLA binding, especially for alleles with fewer known epitopes (191).
Building Predictors of MHC Class I–Peptide Binding
The selectivity of the MHC binding step of the presentation pathway is stronger than preferences in degradation or ER transport. Thus, experimental peptide-MHC binding data form the basis for the development of in silico peptide screening methods. Highly accurate computational algorithms that predict whether a certain epitope and MHC allele pair are compatible have been created by exploiting the information content of peptide sequences already known to bind (reviewed in 184, 212). Naturally, studies have been carried out to compare the performance of different predictors side-by-side, and careful considerations are taken to ensure fair evaluation, such as the size of the dataset, its allele composition, and its overlap with any training datasets (213–215). NetMHC (216, 217) and NetMHCpan (218, 219) repeatedly perform well and are the most widely used (152, 184, 214). Both tools are neural networks with a single hidden layer utilizing two different amino acid–encoding schemes: binary encoding and similarity encoding. NetMHCpan stands out for its ability to make binding predictions even for alleles for which there are few or no training data. This is achieved by featurizing the sequence of the allele itself in addition to the sequence of the peptide and borrowing information from other alleles with similar binding properties. Similarly to most MHC binding predictors, NetMHC and NetMHCpan, except for the most recent version, have been trained on in vitro binding affinity data and predict IC50 binding affinity; more recent versions also predict the rank of a given peptide-MHC pair affinity score amongst 400,000 randomly selected genomic peptides. The rank score is valuable because it helps address biases when comparing alleles with different distributions of binding affinities. The training of the latest version of NetMHCpan, NetMHCpan 4.0, now incorporates MS-derived data, and benchmarking confirmed the increased predictive power for naturally presented peptides (220).
The metric that is most commonly used to evaluate and compare the performance of different predictors is the AUC. In fact, it is near saturated with results consistently exceeding 0.9 (AUC is measured on a [0,1] scale). While this speaks to the maturity of prediction algorithms, it also hinders progress, due to the lack of guiding optimization goals. Furthermore, the AUC integrates over all false-positive rate thresholds and does not reward for higher true-positive rates achieved at lower false-positive rates, which is highly desirable for applications such as selecting epitopes for personalized vaccine development (191). To this end, we have proposed positive predictive value (PPV), the percentage of true-positives among all positive calls, as an alternative evaluation metric for its ability to distinguish better-performing algorithms among predictors with the same AUC results (191).
Endogenous Peptide Presentation Prediction
As discussed above, in vitro binding affinity data do not reflect intracellular antigen processing and presentation properties. Hence, in order to predict whether a certain peptide is likely to be presented on HLA in vivo, systems that combine predictors from the different stages of the pathway have been developed (221–223). For example, NetCTL and NetCTLpan integrate proteasomal cleavage, TAP transport, and MHC binding predictors (221, 223). Alternatively, since MS data can capture peptides eluted from MHC on the surface of cells, MHC binding predictors that are trained on such data implicitly model endogenous antigen presentation. This distinction is important to consider in different application domains—if the task is to predict the propensity of a peptide to bind MHC in isolation, pure MHC binding tools will be better suited; however, if the task is to predict HLA-presented epitopes in a patient, then integrative methods should be used (191, 221–223). In addition to modeling specific pathway components, systems for antigen presentation prediction often take into consideration general molecular features such as the localization or abundance of source proteins. One way to account for the effect of protein availability is to filter antigens whose precursors do not meet a predefined abundance threshold. Alternatively, transcript expression level from RNA sequencing data, which is more feasible to obtain in a clinical setting, can be used as a proxy for protein abundance and integrated within the prediction model (191). Features pertaining to the translation efficiency of source transcripts, such as number of exons, number of upstream open reading frames, and mRNA length, and protein properties such as length and density of ubiquitination sites were also recently proposed to influence epitope presentation (224). Therefore, constructing a rich feature space by capturing these and other predictive variables into integrative modes, along with optimizing the quality of each independent component, should yield increasingly powerful tools for endogenous antigen prediction.
TCR LIGAND RECOGNITION AND TCR REPERTOIRES
Predicting Which Antigens Are More Likely to Be Recognized by T Cell Receptors
The rules of TCR ligand recognition are less understood than the rules of MHC binding. While the peptide-MHC binding prediction problem is complicated by the polymorphism of HLA genes, pMHC recognition by TCR is further exacerbated by the extreme diversity of TCRs per organism. Furthermore, truly large-scale experimental assays that allow for TCR immunogenicity measurements along each of the three main degrees of freedom—MHC allele, peptide, and TCR—are still lacking. Nevertheless, evidence from pMHC-TCR crystal structures and experimentally verified immunogenic interactions has shown that the most highly variable region of the TCR (the CDR3 region) interacts with the peptide, especially at positions P4–P6 (225). In a systematic study of immunogenic peptide properties, analysis of a curated collection of ~800 immunogenic and nonimmunogenic peptides recapitulated the importance of positions 4–6 and revealed an overrepresentation of large and/or aromatic residues (226). The importance of every position and the enrichment score of each amino acid in immunogenic epitopes were then used to develop the first online immunogenicity predictor. More recently, Chowell and coworkers (227) considered positional amino acid and physicochemical differences in TCR contact residues of immunogenic antigens within a larger dataset of ~10,000 peptides. They found a strong overall preference for hydrophobic amino acids and developed neural network models that predict the immunogenicity potential of a given peptide for two MHC alleles. When applied in conjunction with a consensus MHC binding tool, the models improved the ranking of CTL epitopes, albeit in a relatively limited dataset. Two collaborating groups, Łuksza et al. (228) and Balachandran et al. (229), developed a new approach for assessing whether a tumor is immunogenic based in part on the estimated likelihood of TCR recognition for each predicted neoantigen. These estimates were computed from the sequence similarities between the predicted neoantigens and a database of immunogenic epitopes, under the assumption that neoantigens resembling infectious-disease-associated antigens that are known to stimulate T cells are more likely to be immunogenic. The proposed model was validated by demonstrating improved separation of responders from nonresponders in three patient cohorts under checkpoint blockade therapy. Taken together, these studies establish the feasibility of creating computational models for predicting the immunogenicity of a given peptide as well as their broad utility.
High-Throughput Experimental Systems for Finding MHC-Bound Ligands of T Cell Receptors
Advances in next-generation sequencing technologies, TCR sequence reconstruction methods, and multimer-based detection of antigen-specific T cells have made it possible to characterize the TCR repertoires of subsets of cell or whole organisms and led to studies comparing TCR sequences between individuals or T cell types (reviewed in 230–233), among others. A study by Birnbaum et al. (234) coupled deep sequencing with pMHC yeast-display libraries to assess the extent to which specific TCRs recognize multiple ligands. Multiple-round selection of cognate peptides for three distinct TCRs targeting the same antigens was performed. Hundreds of unique peptides were identified per TCR after the third round of enrichment; however, only a handful of peptides remained after the fourth round and they matched the motif of the a priori known antigen. The identified peptides were used to evaluate amino acid preference at each position, resulting in similar recognition motifs across the three TCRs. Additionally, synergistic effects of amino acids within peptides were demonstrated by cooperativity analysis, while clustering confirmed the shared motif, as every ligand was found to differ by at most three amino acids compared to another ligand. Although the yeast-display approach is likely to undersample the cross-reactivity space due to the limited number of peptides present in the library, the consistency of emerging rules led the authors to attribute TCR cross-reactivity to increased flexibility in noninterface residues and constrained variability in contact positions, rather than degeneracy. These findings allow for the development of an algorithm that identifies the cognate epitopes for a given TCR, with the caveat that the binding characteristics of each TCR first need to be characterized by a cognate peptide screen. An epitope discovery method based on substitution matrices was assessed by computationally predicting novel ligands (encoded in the human or microbial genomes) of an established autoimmune TCR (targeting myelin), which included many genes unrelated to the known antigen, and then experimentally verifying these antigens with a 94% success rate, revealing novel homologous self and environmental agonists. The same screening strategy was applied to TCRs isolated from tumor infiltrating cells from colorectal cancer patients with the goal of finding their unknown peptide ligands (235). Of the 20 TCRs screened, cognate antigens were identified and validated for 4 TCRs. As the authors point out, one explanation for this low success rate is that the peptide libraries were HLA-A*:02 restricted. While these results demonstrate the utility of peptide screens in defining the space of targets for a given TCR and then developing predictive rules that allow discovery of novel cognate human or nonhuman antigens, they also pinpoint the limitations of current techniques to fully and unbiasedly explore the pMHC space at scale.
TCR Binding Patterns Revealed by High-Throughput Sequencing of TCR Repertoires
In order to discover the general characteristics of self-reactive TCRs, Stadinski and colleagues (236) analyzed 53 CDR3β structures of human and mouse TCRs and found that either P6 or P7 of the CDR3β is in contact with the peptides in all structures, and both were utilized for binding in 43 of 53 cases (reviewed in 237). To understand the mechanisms acting at these positions, the authors went on to sequence TCRs from preselection and activated thymocytes and showed a positive correlation between hydrophobicity at residues P6–P7 and activation, suggesting that hydrophobic residues at CDR3β P6–P7 are more important for contacting antigens. Consistent with these results, TCRs from CD4+ and CD8+ T cells of MHC-deficient mice were more likely to have strongly interacting amino acids in P6–P7, which would have otherwise been screened out by central tolerance. Apart from extending our knowledge of TCR binding mechanisms, this work also relates the findings to biases in thymic selection for the different T cell subsets. Using similar techniques, Chen et al. (238) characterized the TCR repertoires of multiple individuals in response to two viral antigens, finding preferential V-J pairings and CDR3 lengths.
In back-to-back papers, single-cell TCR sequencing was coupled with pMHC tetramer staining to derive features of TCRs that target the same ligand/antigen (239, 240). Glanville and coworkers (239) profiled TCR repertoires from up to 10 donors against a set of 8 pMHC tetramers, building a set of ~2,100 reactive TCRs. They searched for motifs of length 2, 3, or 4 that were enriched at CDR3 contact residues and clustered TCRs by global and local similarity to reveal that most TCRs reactive to the same cognate epitope fell into the same or related groups. Based on these observations, the authors developed an algorithm, GLIPH, to cluster TCRs into specificity groups. This approach grouped 14% of TCRs, with 95% of clustered members grouped with other TCRs of the same specificity. Unobserved but predicted TCRs of high specificity for a previously profiled antigen were also validated. In a similar manner, Dash et al. (240) set out to characterize epitope specificity by profiling mouse and human TCR repertoires for 10 specific epitopes (~4,600 sequences), identifying enriched CD3 sequences, and developing a nearest-neighbor TCR classifier according to sequence similarity. The models correctly assigned ~80% of TCRs to their corresponding antigen. Taken together, these studies demonstrate that TCR protein sequences can be arranged in specificity groups that tend to coincide with their associated antigen(s). This organization is informative in various applications—including predicting matches of sequenced TCRs to antigens (based on prior observed matches) and optimizing TCR recognition of antigens for vaccines and immunotherapies—and ultimately allows us to better comprehend the apparently vast complexity of TCR repertoires.
Challenges and Future Directions in Predicting Presented Antigens and Their Cognate T Cell Receptors
The ability to accurately predict pMHC for a given TCR and, conversely, TCRs reactive to a given pMHC is a long-standing goal in systems immunology. Recent developments in high-throughput experimental assays, such as peptide library screens and TCR sequencing, coupled with analytical methods, such as mathematical modeling and clustering techniques, have brought us that much closer to this goal. Unquestionably, however, key challenges remain to be addressed. The first large-scale datasets characterizing TCR diversity provide novel mechanistic insights into antigen recognition, but they rely on a reduced complexity of the space by fixing the MHC, the peptide, or the receptor dimension such that truly de novo pMHC-TCR pairing prediction remains elusive. While we know that contacts between the CDR1 and CDR2 regions with the MHC molecule are important for binding, we have yet to understand how much the particular allele affects the interaction. Another obstacle faced by peptide library screens is the need to engineer and optimize MHC constructs, which proves to be especially challenging for class I alleles for their closed conformation and variable topology with respect to the light chain (β2 microglobulin) (230). Library profiling for class II poses its own challenges because the molecules are heterodimers, which expands diversity combinatorially. One way to address this is to barcode multiple HLA alleles in a pooled screen.
Although antigen presentation is much better understood than TCR recognition, we have more to learn here as well. Monoallelic MS epitope sequencing is well positioned to catalogue a collection of alleles that covers over 90% of the population; however, the relatively low sensitivity and lack of negative observations (i.e., peptides that do not bind) pose challenges in creating predictive models. Furthermore, the characterization of peptide binding preferences for class II is complicated by the expanded diversity of alleles, a more permissive peptide length register, and a shifting binding core. In terms of processing, we have yet to grasp the full repertoire of proteasomal flavors and how they manifest in different cell types or under different conditions. Finally, it is important to keep in mind that the pMHC-TCR interaction naturally occurs in the close proximity of other important immune receptors, such as CD8, that can exert structural influence on the docking geometry and alter downstream signaling (241), which is ideally addressed by in vivo assays.
CONCLUSIONS
The examples described above represent exciting directions in systems immunology. We expect that single-cell global profiling will continue to define new cell types and states associated with healthy immunity and disease, leading to new mechanistic hypotheses and therapeutic directions. It will also eventually be feasible to use these tools in the clinic to monitor and predict immune health and disease. The problem of antigen presentation and recognition by TCRs is a more restricted one than the problem of defining cell types and states, as demonstrated by the emergence of increasingly powerful predictive models that promise to transform our ability to analyze both antigens and antigen receptors that contribute to immunity in each patient. Ultimately, more predictive modeling of recognition (i.e., detection of antigen) and cognition (i.e., integration of signals for cells to make decisions) by immune cells should lead to better understanding of immunity in individual patients and more appropriate personalized therapies.
ACKNOWLEDGMENTS
A.-C.V. was supported by the Canadian Institute of Health Research Banting Postdoctoral Fellow-ship. S.S. was supported by grant T32 HG002295 from the National Human Genome Research Institute, NIH. N.H. was supported by the David P. Ryan, MD, Endowed Chair in Cancer Research, NIH/NHGRI CEGS P50 HG006193, NIH/NIAID U24 AI118668, NIH/NCI R01 CA208756, Cancer Research Institute, and the Blavatnik Family Foundation.
Footnotes
DISCLOSURE STATEMENT
N.H. is a founder, shareholder, and scientific advisor for Neon Therapeutics.
LITERATURE CITED
- 1.Brodin P, Davis MM. 2017. Human immune system variation. Nat. Rev. Immunol 17:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis MM, Tato CM, Furman D. 2017. Systems immunology: just getting started. Nat. Immunol 18:725–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayday AC, Peakman M. 2008. The habitual, diverse and surmountable obstacles to human immunology research. Nat. Immunol 9:575–80 [DOI] [PubMed] [Google Scholar]
- 4.Davis MM. 2008. A prescription for human immunology. Immunity 29:835–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germain RN, Schwartzberg PL. 2011. The human condition: an immunological perspective. Nat. Immunol 12:369–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinman RM, Mellman I. 2004. Immunotherapy: bewitched, bothered, and bewildered no more. Science 305:197–200 [DOI] [PubMed] [Google Scholar]
- 7.von Herrath MG, Nepom GT. 2005. Lost in translation: barriers to implementing clinical immunother-apeutics for autoimmunity. J. Exp. Med 202:1159–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, et al. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med 205:3119–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulendran B 2009. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat. Rev. Immunol 9:741–47 [DOI] [PubMed] [Google Scholar]
- 10.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol 10:116–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, et al. 2011. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol 12:786–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, et al. 2014. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 157:499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, et al. 2015. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity 43:1186–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andres-Terre M, McGuire HM, Pouliot Y, Bongen E, Sweeney TE, et al. 2015. Integrated, multicohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity 43:1199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haralambieva IH, Ovsyannikova IG, Kennedy RB, Zimmermann MT, Grill DE, et al. 2016. Transcriptional signatures of influenza A/H1N1-specific IgG memory-like B cell response in older individuals. Vaccine 34:3993–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ovsyannikova IG, Salk HM, Kennedy RB, Haralambieva IH, Zimmermann MT, et al. 2016. Gene signatures associated with adaptive humoral immunity following seasonal influenza A/H1N1 vaccination. Genes Immun 17:371–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobolev O, Binda E, O’Farrell S, Lorenc A, Pradines J, et al. 2016. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat. Immunol 17:204–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, et al. 2016. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387:2312–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweeney TE, Braviak L, Tato CM, Khatri P. 2016. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir. Med 4:213–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, et al. 2003. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. PNAS 100:2610–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, et al. 2016. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 165:1548–50 [DOI] [PubMed] [Google Scholar]
- 22.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, et al. 2008. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 29:150–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. 2015. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523:612–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carr EJ, Dooley J, Garcia-Perez JE, Lagou V, Lee JC, et al. 2016. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol 17:461–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Jong S, Neeleman M, Luykx JJ, ten Berg MJ, Strengman E, et al. 2014. Seasonal changes in gene expression represent cell-type composition in whole blood. Hum. Mol. Genet 23:2721–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dopico XC, Evangelou M, Ferreira RC, Guo H, Pekalski ML, et al. 2015. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun 6:7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furman D, Chang J, Lartigue L, Bolen CR, Haddad F, et al. 2017. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat. Med 23:174–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajdu SI. 2003. A note from history: the discovery of blood cells. Ann. Clin. Lab. Sci 33:237–38 [PubMed] [Google Scholar]
- 29.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, et al. ; Hum. Cell Atlas Meet. Particip. 2017. The Human Cell Atlas. eLife 6:e27041 10.1101/121202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svensson V, Natarajan KN, Ly LH, Miragaia RJ, Labalette C, et al. 2017. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14:381–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spitzer MH, Nolan GP. 2016. Mass cytometry: single cells, many features. Cell 165:780–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crosetto N, Bienko M, van Oudenaarden A. 2015. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet 16:57–66 [DOI] [PubMed] [Google Scholar]
- 33.Wagner A, Regev A, Yosef N. 2016. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol 34:1145–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanay A, Regev A. 2017. Scaling single-cell genomics from phenomenology to mechanism. Nature 541:331–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakadan SM, Shalek AK, Weitz DA. 2017. Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. Nat. Rev. Genet 18:345–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W. 2016. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol 17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartzman O, Tanay A. 2015. Single-cell epigenomics: techniques and emerging applications. Nat. Rev. Genet 16:716–26 [DOI] [PubMed] [Google Scholar]
- 38.Cuvier O, Fierz B. 2017. Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells. Nat. Rev. Genet 18:457–72 [DOI] [PubMed] [Google Scholar]
- 39.Gawad C, Koh W, Quake SR. 2016. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet 17:175–88 [DOI] [PubMed] [Google Scholar]
- 40.Woodworth MB, Girskis KM, Walsh CA. 2017. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet 18:230–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Navin NE. 2015. Advances and applications of single-cell sequencing technologies. Mol. Cell 58:598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, et al. 2017. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65:631–43.e4 [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C 2015. Defining cell types and states with single-cell genomics. Genome Res 25:1491–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grun D, van Oudenaarden A. 2015. Design and analysis of single-cell sequencing experiments. Cell 163:799–810 [DOI] [PubMed] [Google Scholar]
- 45.Macaulay IC, Ponting CP, Voet T. 2017. Single-cell multiomics: multiple measurements from single cells. Trends Genet 33:155–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock C, Farlik M, Sheffield NC. 2016. Multi-omics of single cells: strategies and applications. Trends Biotechnol 34:605–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, et al. 2017. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol 35(10):936–39 [DOI] [PubMed] [Google Scholar]
- 48.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, et al. 2017. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14(9):865–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. 2015. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol 33(3):285–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheow LF, Courtois ET, Tan Y, Viswanathan R, Xing Q, et al. 2016. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods 13(10):833–36 [DOI] [PubMed] [Google Scholar]
- 51.Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, et al. 2013. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 38:187–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, et al. 2014. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159:814–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thome JJ, Farber DL. 2015. Emerging concepts in tissue-resident T cells: lessons from humans. Trends Immunol 36:428–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thome JJ, Bickham KL, Ohmura Y, Kubota M, Matsuoka N, et al. 2016. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat. Med 22:72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Granot T, Senda T, Carpenter DJ, Matsuoka N, Weiner J, et al. 2017. Dendritic cells display subset and tissue-specific maturation dynamics over human life. Immunity 46:504–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saeys Y, Gassen SV, Lambrecht BN. 2016. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol 16:449–62 [DOI] [PubMed] [Google Scholar]
- 57.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, et al. 2011. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332:687–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, et al. 2014. High-dimensional analysis of the murine myeloid cell system. Nat. Immunol 15:1181–89 [DOI] [PubMed] [Google Scholar]
- 59.Sen N, Mukherjee G, Sen A, Bendall SC, Sung P, et al. 2014. Single-cell mass cytometry analysis of human tonsil T cell remodeling by varicella zoster virus. Cell Rep 8:633–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bendall SC, Davis KL, Amir ED, Tadmor MD, Simonds EF, et al. 2014. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157:714–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, et al. 2013. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med 5:208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, et al. 2015. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci. Transl. Med 7:297ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. 2012. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36:142–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong MT, Chen J, Narayanan S, Lin W, Anicete R, et al. 2015. Mapping the diversity of follicular helper T cells in human blood and tonsils using high-dimensional mass cytometry analysis. Cell Rep 11:1822–33 [DOI] [PubMed] [Google Scholar]
- 65.Wong MT, Ong DE, Lim FS, Teng KW, McGovern N, et al. 2016. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 45:442–56 [DOI] [PubMed] [Google Scholar]
- 66.Guilliams M, Dutertre CA, Scott CL, McGovern N, Sichien D, et al. 2016. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45:669–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, et al. 2017. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity 46:148–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Unen V, Li N, Molendijk I, Temurhan M, Hollt T, et al. 2016. Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity 44:1227–39 [DOI] [PubMed] [Google Scholar]
- 69.Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, et al. 2015. An interactive reference framework for modeling a dynamic immune system. Science 349:1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, et al. 2017. Systemic immunity is required for effective cancer immunotherapy. Cell 168:487–502.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roederer M, Quaye L, Mangino M, Beddall MH, Mahnke Y, et al. 2015. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell 161:387–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, et al. 2015. Variation in the human immune system is largely driven by non-heritable influences. Cell 160:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, et al. 2016. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol 34:637–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Proserpio V, Mahata B. 2016. Single-cell technologies to study the immune system. Immunology 147:133–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neu KE, Tang Q, Wilson PC, Khan AA. 2017. Single-cell genomics: approaches and utility in immunology. Trends Immunol 38:140–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, et al. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343:776–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, et al. 2015. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol 16:718–28 [DOI] [PubMed] [Google Scholar]
- 78.Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, et al. 2016. Specification of tissue-resident macrophages during organogenesis. Science 353:aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mildner A, Schonheit J, Giladi A, David E, Lara-Astiaso D, et al. 2017. Genomic characterization of murine monocytes reveals C/EBPβ transcription factor dependence of Ly6C− cells. Immunity 46:849–62.e7 [DOI] [PubMed] [Google Scholar]
- 80.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, et al. 2016. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol 17:728–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, et al. 2017. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol 18:422–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pace L, Goudot C, Zueva E, Gueguen P, Burgdorf N, et al. 2018. The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359(6372):177–86. [DOI] [PubMed] [Google Scholar]
- 83.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, et al. 2016. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166:1231–46.e13 [DOI] [PubMed] [Google Scholar]
- 84.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, et al. 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353:aad8670. [DOI] [PubMed] [Google Scholar]
- 85.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, et al. 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48:1193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buenrostro JD, Corces R, Wu B, Schep AN, Lareau C, et al. 2017. Single-cell epigenomics maps the continuous regulatory landscape of human hematopoietic differentiation. bioRxiv 109843. 10.1101/109843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Drissen R, Buza-Vidas N, Woll P, Thongjuea S, Gambardella A, et al. 2016. Distinct myeloid progenitor- differentiation pathways identified through single-cell RNA sequencing. Nat. Immunol 17:666–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu Y, Tsang JC, Wang C, Clare S, Wang J, et al. 2016. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature 539:102–6 [DOI] [PubMed] [Google Scholar]
- 89.Björklund ÅK , Forkel M, Picelli S, Konya V, Theorell J, et al. 2016. The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol 17:451–60 [DOI] [PubMed] [Google Scholar]
- 90.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, et al. 2017. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356:eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.See P, Dutertre CA, Chen J, Gunther P, McGovern N, et al. 2017. Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356:eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Breton G, Zheng S, Valieris R, Tojal da Silva I, Satija R, Nussenzweig MC. 2016. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med 213:2861–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O’Rourke P, et al. 2018. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol 3:eaan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Picelli S, Björklund ÅK , Faridani OR, Sagasser S, Winberg G, Sandberg R. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10:1096–98 [DOI] [PubMed] [Google Scholar]
- 96.Picelli S, Faridani OR, Björklund ÅK , Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9:171–81 [DOI] [PubMed] [Google Scholar]
- 97.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, et al. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161:1187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161:1202–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gierahn TM, Wadsworth MH 2nd, Hughes TK, Bryson BD, Butler A, et al. 2017. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14:395–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, et al. 2016. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst 3:346–60.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Muraro MJ, Dharmadhikari G, Grun D, Groen N, Dielen T, et al. 2016. A single-cell transcriptome atlas of the human pancreas. Cell Syst 3:385–94.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang YJ, Schug J, Won KJ, Liu C, Naji A, et al. 2016. Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65:3028–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li J, Klughammer J, Farlik M, Penz T, Spittler A, et al. 2016. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep 17:178–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Drel VR, Mashtalir N, Ilnytska O, Shin J, Li F, et al. 2006. The leptin-deficient (ob/ob) mouse: a new animal model of peripheral neuropathy of type 2 diabetes and obesity. Diabetes 55:3335–43 [DOI] [PubMed] [Google Scholar]
- 105.Morioka T, Asilmaz E, Hu J, Dishinger JF, Kurpad AJ, et al. 2007. Disruption of leptin receptor expression in the pancreas directly affects β cell growth and function in mice. J. Clin. Investig 117:2860–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, et al. 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542:352–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Achim K, Pettit JB, Saraiva LR, Gavriouchkina D, Larsson T, et al. 2015. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat. Biotechnol 33:503–9 [DOI] [PubMed] [Google Scholar]
- 109.Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, et al. 2017. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 358(6370):1622–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, et al. 2014. Highly multiplexed subcellular RNA sequencing in situ. Science 343(6177):1360–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. 2015. RNA imaging: spatially resolved, highly multiplexed RNA profiling in single cells. Science 348(6233):aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eng CL, Shah S, Thomassie J, Cai L. 2017. Profiling the transcriptome with RNA SPOTs. Nat. Methods 14(12):1153–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, et al. 2018. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol 36(1):89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, et al. 2013. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498:236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, et al. 2014. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510:363–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, et al. 2015. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163:1400–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, et al. 2015. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell 163:1413–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, et al. 2015. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol 33:155–60 [DOI] [PubMed] [Google Scholar]
- 119.Stegle O, Teichmann SA, Marioni JC. 2015. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet 16:133–45 [DOI] [PubMed] [Google Scholar]
- 120.Yuan GC, Cai L, Elowitz M, Enver T, Fan G, et al. 2017. Challenges and emerging directions in single-cell analysis. Genome Biol 18:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Proserpio V, Piccolo A, Haim-Vilmovsky L, Kar G, Lönnberg T, et al. 2016. Single-cell analysis of CD4+ T-cell differentiation reveals three major cell states and progressive acceleration of proliferation. Genome Biol 17:103 Erratum. 2016. Genome Biol. 17:133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martinez-Jimenez CP, Eling N, Chen HC, Vallejos CA, Kolodziejczyk AA, et al. 2017. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 355:1433–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lönnberg T, Svensson V, James KR, Fernandez-Ruiz D, Sebina I, et al. 2017. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol 2:eaal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, et al. 2016. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 13:329–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Han A, Glanville J, Hansmann L, Davis MM. 2014. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol 32:684–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Afik S, Yates KB, Bi K, Darko S, Godec J, et al. 2017. Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res 45:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McDaniel JR, DeKosky BJ, Tanno H, Ellington AD, Georgiou G. 2016. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat. Protoc 11:429–42 [DOI] [PubMed] [Google Scholar]
- 128.Redmond D, Poran A, Elemento O. 2016. Single-cell TCRseq: paired recovery of entire T-cell alpha and beta chain transcripts in T-cell receptors from single-cell RNAseq. Genome Med 8(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ner-Gaon H, Melchior A, Golan N, Ben-Haim Y, Shay T. 2017. JingleBells: a repository of immune-related single-cell RNA–sequencing datasets. J. Immunol 198:3375–79 [DOI] [PubMed] [Google Scholar]
- 130.Yosef N, Regev A. 2016. Writ large: genomic dissection of the effect of cellular environment on immune response. Science 354:64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, et al. 2015. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162:675–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dixit A, Parnas O, Li B, Chen J, Fulco CP, et al. 2016. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167:1853–66.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, et al. 2016. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell 167:1883–96.e15 [DOI] [PubMed] [Google Scholar]
- 134.Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, et al. 2016. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167:1867–82.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, et al. 2017. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lane K, Van Valen D, DeFelice MM, Macklin DN, Kudo T, et al. 2017. Measuring signaling and RNA-Seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst 4:458–69.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Junkin M, Kaestli AJ, Cheng Z, Jordi C, Albayrak C, et al. 2016. High-content quantification of single-cell immune dynamics. Cell Rep 15:411–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Avraham R, Haseley N, Brown D, Penaranda C, Jijon HB, et al. 2015. Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell 162:1309–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saliba AE, Li L, Westermann AJ, Appenzeller S, Stapels DA, et al. 2016. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat. Microbiol 2:16206. [DOI] [PubMed] [Google Scholar]
- 140.Wills QF, Mellado-Gomez E, Nolan R, Warner D, Sharma E, et al. 2017. The nature and nurture of cell heterogeneity: accounting for macrophage gene-environment interactions with single-cell RNA-Seq. BMC Genom 18:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, et al. 2017. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169:1276–90.e17 [DOI] [PubMed] [Google Scholar]
- 142.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, et al. 2016. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell 166:1500–11.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ikebuchi R, Teraguchi S, Vandenbon A, Honda T, Shand FH, et al. 2016. A rare subset of skin-tropic regulatory T cells expressing Il10/Gzmb inhibits the cutaneous immune response. Sci. Rep 6:35002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rahman K, Vengrenyuk Y, Ramsey SA, Vila NR, Girgis NM, et al. 2017. Inflammatory Ly6Chi monocytes and their conversion to M2 macrophages drive atherosclerosis regression. J. Clin. Investig 127:2904–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352:189–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, et al. 2016. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539:309–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, et al. 2017. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355:eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, et al. 2017. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet 49:708–18 [DOI] [PubMed] [Google Scholar]
- 150.Chung W, Eum HH, Lee HO, Lee KM, Lee HB, et al. 2017. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun 8:15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, et al. 2017. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169:1342–56.e16 [DOI] [PubMed] [Google Scholar]
- 152.Kim KT, Lee HW, Lee HO, Song HJ, Jeong DE, et al. 2016. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol 17:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Winterhoff BJ, Maile M, Mitra AK, Sebe A, Bazzaro M, et al. 2017. Single cell sequencing reveals heterogeneity within ovarian cancer epithelium and cancer associated stromal cells. Gynecol. Oncol 144:598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang L, Fan J, Francis JM, Georghiou G, Hergert S, et al. 2017. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res 27:1300–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ebinger S, Ozdemir EZ, Ziegenhain C, Tiedt S, Castro Alves C, et al. 2016. Characterization of rare, dormant, and therapy-resistant cells in acute lymphoblastic leukemia. Cancer Cell 30:849–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, et al. 2017. SC3: consensus clustering of single-cell RNA-seq data. Nat. Methods 14:483–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, et al. 2014. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11:417–22 [DOI] [PubMed] [Google Scholar]
- 158.Schapiro D, Jackson HW, Raghuraman S, Zanotelli VRT, Fischer JRR, et al. 2017. Systematic analysis of cell phenotypes and cellular social networks in tissues using the multiplexed image cytometry analysis toolbox (miCAT). bioRxiv 109207 10.1101/109207 [DOI] [Google Scholar]
- 159.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir ED, et al. 2015. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162:184–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, et al. 2017. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169:750–65.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, et al. 2017. An immune atlas of clear cell renal cell carcinoma. Cell 169:736–49.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Litzenburger UM, Buenrostro JD, Wu B, Shen Y, Sheffield NC, et al. 2017. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol 18:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ryan JF, Hovde R, Glanville J, Lyu SC, Ji X, et al. 2016. Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets. PNAS 113:E1286–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gaudilliere B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, et al. 2014. Clinical recovery from surgery correlates with single-cell immune signatures. Sci. Transl. Med 6:255ra131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cerosaletti K, Barahmand-Pour-Whitman F, Yang J, DeBerg HA, Dufort MJ, et al. 2017. Single-cell RNA sequencing reveals expanded clones of islet antigen-reactive CD4+ T cells in peripheral blood of subjects with type 1 diabetes. J. Immunol 199:323–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xin Y, Kim J, Okamoto H, Ni M, Wei Y, et al. 2016. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab 24:608–15 [DOI] [PubMed] [Google Scholar]
- 167.Segerstolpe A, Palasantza A, Eliasson P, Andersson EM, Andreasson AC, et al. 2016. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab 24:593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lawlor N, George J, Bolisetty M, Kursawe R, Sun L, et al. 2017. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res 27:208–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, et al. 2017. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2:93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.O’Gorman WE, Kong DS, Balboni IM, Rudra P, Bolen CR, et al. 2017. Mass cytometry identifies a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric SLE patients. J. Autoimmun 81:74–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mizoguchi F, Slowikowski K, Wei K, Marshall JL, Rao DA, et al. 2018. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun 9:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stephenson W, Donlin LT, Butler A, Rozo C, Rashidfarrokhi A, et al. 2017. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low cost microfluidic instrumentation. bioRxiv 140848. 10.1101/140848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, et al. 2017. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547:217–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, et al. 2017. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547:222–26 [DOI] [PubMed] [Google Scholar]
- 175.Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, et al. 2015. Cancer immunotherapy: A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348:803–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rock KL, Farfan-Arribas DJ, Shen L. 2010. Proteases in MHC class I presentation and cross-presentation. J. Immunol 184:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Van den Eynde BJ, Morel S. 2001. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr. Opin. Immunol 13:147–53 [DOI] [PubMed] [Google Scholar]
- 178.Toes RE, Nussbaum AK, Degermann S, Schirle M, Emmerich NP, et al. 2001. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med 194:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.de Verteuil D, Muratore-Schroeder TL, Granados DP, Fortier MH, Hardy MP, et al. 2010. Deletion of immunoproteasome subunits imprints on the transcriptome and has a broad impact on peptides presented by major histocompatibility complex I molecules. Mol. Cell Proteom 9:2034–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, et al. 2011. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol 13:129–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sesma L, Alvarez I, Marcilla M, Paradela A, Lopez de Castro JA. 2003. Species-specific differences in proteasomal processing and tapasin-mediated loading influence peptide presentation by HLA-B27 in murine cells. J. Biol. Chem 278:46461–72 [DOI] [PubMed] [Google Scholar]
- 182.Lazaro S, Gamarra D, Del Val M. 2015. Proteolytic enzymes involved in MHC class I antigen processing: a guerrilla army that partners with the proteasome. Mol. Immunol 68:72–76 [DOI] [PubMed] [Google Scholar]
- 183.Neefjes J, Jongsma ML, Paul P, Bakke O. 2011. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol 11:823–36 [DOI] [PubMed] [Google Scholar]
- 184.Lundegaard C, Hoof I, Lund O, Nielsen M. 2010. State of the art and challenges in sequence based T-cell epitope prediction. Immunome Res 6(Suppl. 2):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nielsen M, Lundegaard C, Lund O, Kesmir C. 2005. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33–41 [DOI] [PubMed] [Google Scholar]
- 186.Saxova P, Buus S, Brunak S, Kesmir C. 2003. Predicting proteasomal cleavage sites: a comparison of available methods. Int. Immunol 15:781–87 [DOI] [PubMed] [Google Scholar]
- 187.Calis JJ, Reinink P, Keller C, Kloetzel PM, Kesmir C. 2015. Role of peptide processing predictions in T cell epitope identification: contribution of different prediction programs. Immunogenetics 67:85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Holzhutter HG, Frommel C, Kloetzel PM. 1999. A theoretical approach towards the identification of cleavage-determining amino acid motifs of the 20 S proteasome. J. Mol. Biol 286:1251–65 [DOI] [PubMed] [Google Scholar]
- 189.Holzhutter HG, Kloetzel PM. 2000. A kinetic model of vertebrate 20S proteasome accounting for the generation of major proteolytic fragments from oligomeric peptide substrates. Biophys. J 79:1196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ginodi I, Vider-Shalit T, Tsaban L, Louzoun Y. 2008. Precise score for the prediction of peptides cleaved by the proteasome. Bioinformatics 24:477–83 [DOI] [PubMed] [Google Scholar]
- 191.Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, et al. 2017. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46:315–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hanada K, Yewdell JW, Yang JC. 2004. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427:252–56 [DOI] [PubMed] [Google Scholar]
- 193.Vigneron N, Stroobant V, Chapiro J, Ooms A, Degiovanni G, et al. 2004. An antigenic peptide produced by peptide splicing in the proteasome. Science 304:587–90 [DOI] [PubMed] [Google Scholar]
- 194.Warren EH, Vigneron NJ, Gavin MA, Coulie PG, Stroobant V, et al. 2006. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 313:1444–47 [DOI] [PubMed] [Google Scholar]
- 195.Dalet A, Vigneron N, Stroobant V, Hanada K, Van den Eynde BJ. 2010. Splicing of distant peptide fragments occurs in the proteasome by transpeptidation and produces the spliced antigenic peptide derived from fibroblast growth factor-5. J. Immunol 184:3016–24 [DOI] [PubMed] [Google Scholar]
- 196.Berkers CR, de Jong A, Schuurman KG, Linnemann C, Meiring HD, et al. 2015. Definition of proteasomal peptide splicing rules for high-efficiency spliced peptide presentation by MHC class I molecules.J. Immunol 195:4085–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Liepe J, Marino F, Sidney J, Jeko A, Bunting DE, et al. 2016. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354:354–58 [DOI] [PubMed] [Google Scholar]
- 198.Platteel AC, Liepe J, Textoris-Taube K, Keller C, Henklein P, et al. 2017. Multi-level strategy for identifying proteasome-catalyzed spliced epitopes targeted by CD8+ T cells during bacterial infection. Cell Rep 20(5):1242–53 [DOI] [PubMed] [Google Scholar]
- 199.Oliveira CC, van Hall T. 2015. Alternative antigen processing for MHC class I: Multiple roads lead to Rome. Front. Immunol 6:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Neefjes JJ, Momburg F, Hammerling GJ. 1993. Selective and ATP-dependent translocation of peptides by the MHC-encoded transporter. Science 261:769–71 [DOI] [PubMed] [Google Scholar]
- 201.van Endert PM, Riganelli D, Greco G, Fleischhauer K, Sidney J, et al. 1995. The peptide-binding motif for the human transporter associated with antigen processing. J. Exp. Med 182:1883–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Uebel S, Kraas W, Kienle S, Wiesmuller KH, Jung G, Tampe R. 1997. Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. PNAS 94:8976–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Peters B, Bulik S, Tampe R, van Endert PM, Holzhutter HG. 2003. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J. Immunol 171:1741–49 [DOI] [PubMed] [Google Scholar]
- 204.Bhasin M, Raghava GP. 2004. Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci 13:596–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lam TH, Mamitsuka H, Ren EC, Tong JC. 2010. TAP Hunter: a SVM-based system for predicting TAP ligands using local description of amino acid sequence. Immunome Res 6(Suppl. 1):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang GL, Petrovsky N, Kwoh CK, August JT, Brusic V. 2006. PRED(TAP): a system for prediction of peptide binding to the human transporter associated with antigen processing. Immunome Res 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. 2015. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43:D423–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gfeller D, Bassani-Sternberg M, Schmidt J, Luescher IF. 2016. Current tools for predicting cancer-specific T cell immunity. Oncoimmunology 5:e1177691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, et al. 1992. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255:1261–63 [DOI] [PubMed] [Google Scholar]
- 210.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. 2015. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell Proteom 14:658–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bassani-Sternberg M, Braunlein E, Klar R, Engleitner T, Sinitcyn P, et al. 2016. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun 7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kessler JH, Melief CJ. 2007. Identification of T-cell epitopes for cancer immunotherapy. Leukemia 21:1859–74 [DOI] [PubMed] [Google Scholar]
- 213.Kim Y, Sidney J, Buus S, Sette A, Nielsen M, Peters B. 2014. Dataset size and composition impact the reliability of performance benchmarks for peptide-MHC binding predictions. BMC Bioinform 15:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Trolle T, Metushi IG, Greenbaum JA, Kim Y, Sidney J, et al. 2015. Automated benchmarking of peptide-MHC class I binding predictions. Bioinformatics 31:2174–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. 2008. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol 9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, et al. 2003. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12:1007–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Andreatta M, Nielsen M. 2016. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32:511–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, et al. 2009. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nielsen M, Andreatta M. 2016. NetMHCpan-3.0: improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. 2017. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 199(9):3360–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, et al. 2005. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur. J. Immunol 35:2295–303 [DOI] [PubMed] [Google Scholar]
- 222.Doytchinova IA, Guan P, Flower DR. 2006. EpiJen: a server for multistep T cell epitope prediction. BMC Bioinform 7:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Stranzl T, Larsen MV, Lundegaard C, Nielsen M. 2010. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics 62:357–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pearson H, Daouda T, Granados DP, Durette C, Bonneil E, et al. 2016. MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Investig 126:4690–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rudolph MG, Stanfield RL, Wilson IA. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu.Rev. Immunol 24:419–66 [DOI] [PubMed] [Google Scholar]
- 226.Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, et al. 2013. Properties of MHC class I presented peptides that enhance immunogenicity. PLOS Comput. Biol 9:e1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chowell D, Krishna S, Becker PD, Cocita C, Shu J, et al. 2015. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. PNAS 112:E1754–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, et al. 2017. A neoantigens fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551(7681):517–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, et al. 2017. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551(7681):512–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Birnbaum ME, Dong S, Garcia KC. 2012. Diversity-oriented approaches for interrogating T-cell receptor repertoire, ligand recognition, and function. Immunol. Rev 250:82–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Newell EW, Davis MM. 2014. Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nat. Biotechnol 32:149–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bentzen AK, Hadrup SR. 2017. Evolution of MHC-based technologies used for detection of antigen-responsive T cells. Cancer Immunol. Immunother 66:657–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. 2017. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol 17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, et al. 2014. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157:1073–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gee MH, Han A, Lofgren SM, Beausang JF, Mendoza JL, et al. 2018. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172:549–63.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Stadinski BD, Shekhar K, Gomez-Tourino I, Jung J, Sasaki K, et al. 2016. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat. Immunol 17:946–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chakraborty AK. 2017. A perspective on the role of computational models in immunology. Annu. Rev.Immunol 35:403–39 [DOI] [PubMed] [Google Scholar]
- 238.Chen G, Yang X, Ko A, Sun X, Gao M, et al. 2017. Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens. Cell Rep 19:569–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, et al. 2017. Identifying specificity groups in the T cell receptor repertoire. Nature 547:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, et al. 2017. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, et al. 2011. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 35:681–93 [DOI] [PMC free article] [PubMed] [Google Scholar]