Abstract
Organismal responses to environmental stresses are a determinant of the effect of climate change. These can occur through the regulation of gene expression, involving genetic adaptation and plastic changes as evolutionary strategy. Heat shock protein (hsp) family genes are extensively expanded and play important roles in thermal adaptation in oysters. We investigated expression of all heat-responsive hsps in two allopatric congeneric oyster species, Crassostrea gigas and C. angulata, which are respectively distributed along the northern and southern coasts of China, using common garden and reciprocal transplant experiments. Our results showed that hsps in C. gigas have evolved higher basal levels of expression under ambient conditions at each field site, with lower expression plasticity in response to heat stress in comparison to C. angulata, which exhibited lower baseline expression but higher expression plasticity. This pattern was fixed regardless of environmental disturbance, potentially implying genetic assimilation. Our findings indicate divergent adaptive strategies with underlying evolutionary trade-offs between genetic adaptation and plasticity at the molecular level in two oyster congeners in the face of rapid climate change.
Keywords: oyster, expression pattern, genetic adaptation, plasticity, evolutionary trade-off, genetic assimilation
1. Introduction
Understanding individual responses to environmental stresses, a central goal of evolutionary ecology, is an urgent task in the face of rapid climate change. Genetic adaptation and phenotypic plasticity represent two forms of evolutionary strategy to respond to environmental variability [1,2]. The choice between these strategies is species specific and often involves evolutionary trade-offs between genetic variation and plasticity, which depend on interactions between genes and the environment [3,4]. The regulation of gene expression is a common factor reflecting both genetic and environmental variations. Many constitutive divergences have a heritable genetic basis, indicating evolutionary adaptation under normal conditions [5], whereas environmentally induced regulatory patterns indicate plastic change. In most cases, expression patterns of gene regulation after stressful stimuli indicate divergence among related species with different thermotolerances [4].
Oysters inhabiting highly variable intertidal zones provide an ideal model to investigate such adaptive strategies, and the destruction of oyster reefs worldwide necessitates assessment of their adaptive potential [6,7]. Oysters have evolved a series of genetic and physiological adaptations that confer resilience against harsh environmental conditions, especially in heat shock protein (hsp) genes. Hsp family genes are extensively expanded in oysters and are strongly upregulated as a stress response to changes in ambient conditions [8,9], which is a common feature in other marine invertebrates such as amphipods and corals [4,10]. Meanwhile, the Bcl-2 associated athanogene (bag) genes are highly conserved across wide evolutionary distances and have been found to be involved in stress tolerance by positively or negatively regulating the function of hsp genes [11]. Analysing these genes should provide insights into adaptive mechanisms and enable assessment of evolutionary potential.
Two allopatric congeneric oyster species, Crassostrea gigas and C. angulata, are the dominant oyster species, respectively, distributed along the northern and southern coasts of China, which are separated by the Yangtze River [12]. Phylogenetic analysis has shown that they diverged approximately 2.7 Ma [13]. Comparative laboratory and field studies have demonstrated their adaptive divergence in detail at morphological, physiological, molecular and genomic levels [3,14,15]. However, the evolutionary relationship between the two oysters is controversial and some authors have considered them as a single species [16], while other authors concluded that they are genetically closely related species [12,14,17]. Few studies have been reported regarding the evolutionary responses of these two oyster congeners in response to climate change. To investigate these strategies, we collected wild oysters of both species, conducted common garden and reciprocal transplant experiments and analysed the patterns of baseline and plastic expression between northern and southern oysters to reveal their adaptive mechanisms in response to climate change.
2. Material and methods
(a). Common garden and reciprocal transplant experiments
The experimental design is described in detail in a previous study by Li et al. [3]. Briefly, two oyster congeners, C. gigas and C. angulata, were collected from their respective northern and southern coastline habitats in Qingdao (QD, 35°44′ N) and Xiamen (XM, 24°33′ N). Mean air and sea surface temperatures at the southern site were 8.7°C and 6.6°C higher than those at the northern site during the past 2 years, respectively [3]. We conducted a one-generation common garden experiment under identical conditions for both oyster congeners to alleviate maternal effects on gene expression [18]. For each species, eggs from 30 mature females were mixed and divided among 30 beakers. Sperm from 30 mature males were crossed individually with each beaker of eggs. Culture of zygotes and larvae was conducted under farm conditions, and spat and juvenile oysters were cultured in the sea [3]. Eight-month-old F1 progenies were transported to their origin sampling sites as a reciprocal transplant experiment to test the effects of the local environment. After two months of acclimatization, adult progeny oysters were collected for gene expression measurements at each site. These were cleaned to remove epifauna and reared in aquaria for 15 days with aerated and sand-filtered seawater at 20°C, changed daily. Commercial spirulina powder was added once per day as a food source. No mortality was detected during laboratory acclimation.
(b). Gene expression measurements
Oysters reared as described above were exposed to 35°C, the sublethal temperature of C. gigas and the maximum air temperature of the southern site [3], for 48 h. Gills were sampled at 0, 3, 6, 12, 24 and 48 h and immediately frozen in liquid nitrogen and stored at −80°C for subsequent RNA extraction (n = 15 per time point). RNA extraction, cDNA synthesis using 1 µg mixed RNA from each of five oysters, and qRT-PCR using two technical replicates for each cDNA were performed as described in previous studies, with six replicates for each time point [3,7]. All strongly thermally induced hsp family genes (including the hsp-regulating gene bag4 and collectively called hsps) were selected as candidate genes [8,19]. Primers are shown with their amplification efficiency for the hsps and the reference gene elongation factor alpha (ef1α) in electronic supplementary material, table S1. Relative expression under heat shock was determined by the method of [20]. Adult oysters of both species were collected from ambient conditions of each field site, and basal expression of hsps in the gills was determined in the same manner (n = 15 for each site). To compare basal levels of hsps between C. gigas and C. angulata under ambient conditions, we first calculated the mean basal level in C. angulate (). Relative basal level of each replicate hsp in C. gigas () was individually contrasted with , using the following formula:
where indicates the value of ith replicate, c indicates each candidate gene, r indicates reference gene (ef1α), a indicates C. angulata, g indicates C. gigas.
(c). Statistical analyses
All statistical analyses were carried out using R v. 3.5.0. The comparison of basal expression between C. angulata () and the relative basal level for C. gigas () was made using a one-sample Wilcoxon single-rank test. The comparison for relative expression under heat shock for each hsp between two oyster congeners was conducted using a t-test after confirming normality of distribution using a Kolmogorov–Smirnov test and homogeneity of variance using Bartlett's test. Species, hsp genes, time of sampling and their interactions were modelled as fixed factors. The significance of fixed factors was evaluated using likelihood ratio tests. If an effect of species, gene or time was found to be significant at p < 0.05 following false discovery rate correction using the Bonferroni method, a post hoc Tukey test using the ghlt function of the multcomp package [21] was used to evaluate the significance of each pairwise comparison.
3. Results
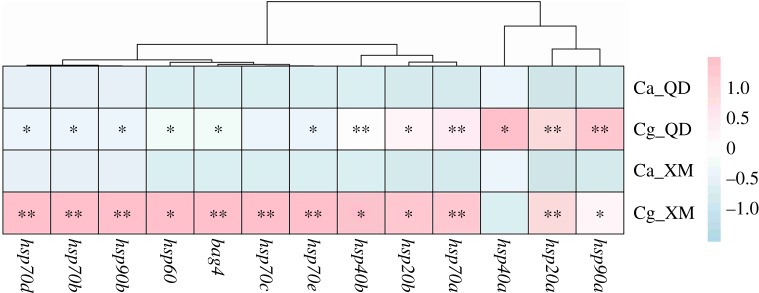
Both basal expression of hsps under ambient conditions and relative expression under heat shock were strongly affected by species, gene, time point and their interactions at both sites (p < 0.05, electronic supplementary material, table S2). The relative basal expression of 12 of the 13 hsps in C. gigas was significantly higher than in C. angulata at both sampling sites (1.3–4.2-fold higher at Qingdao and 1.6–8.6-fold higher at Xiamen; p < 0.05; figure 1).
Figure 1.
Heatmap for the basal level of hsps in two congeneric oyster species under ambient field conditions. Asterisks indicate significant difference between species at each habitat (Ca_QD versus Cg_QD, Ca_XM versus Cg_XM; *0.05 < p < 0.01, **p < 0.01). Ca, C. angulata; Cg, C. gigas; QD, Qingdao; XM, Xiamen. (Online version in colour.)
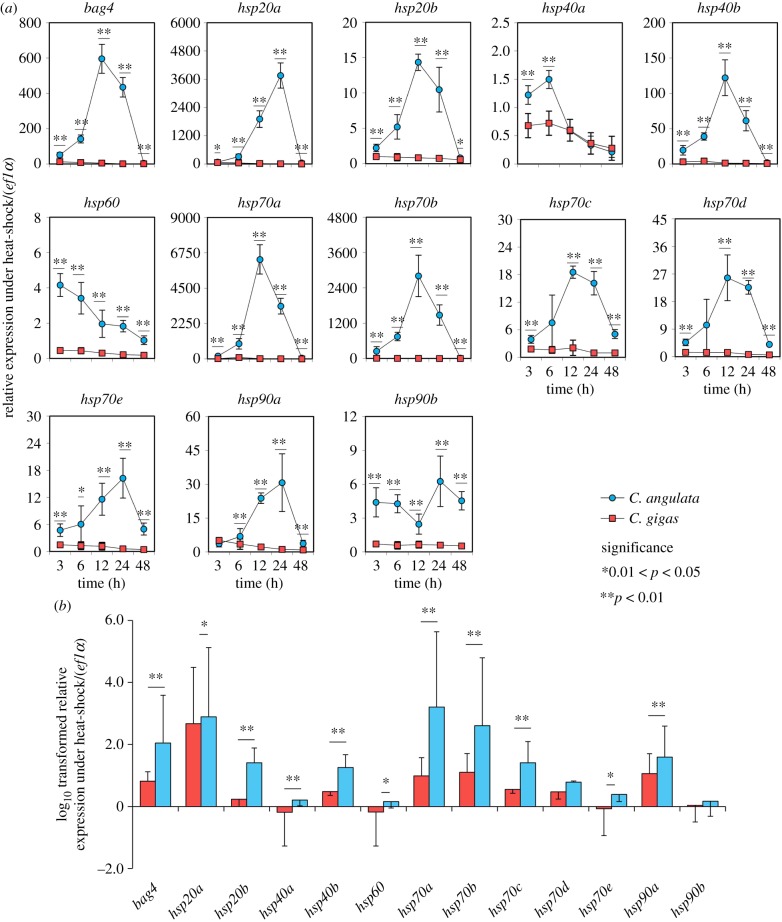
All genes in C. angulata showed dramatic fluctuations in expression from 1.2-fold (hsp40a at 3 h) to 6.3 × 103-fold (hsp70a at 12 h) at the northern site, as well as a 1.6 × 103-fold (hsp70a at 6 h) change at the southern site when the oysters were exposed to high temperature (figure 2a). However, 11 of 13 genes in C. gigas showed no more than 10-fold changes, and the greatest thermally induced change was only 85-fold (hsp70a at 6 h) at the northern site (electronic supplementary material, figure S1). Similarly, all hsps in C. gigas showed more moderate expression patterns than in C. angulata at the southern site (figure 2b). Furthermore, highly increased expression of most hsps in C. angulata occurred at later stages (12–24 h) during heat shock, but occurred at earlier stages (3–6 h) in C. gigas.
Figure 2.
Relative expression of hsps in C. gigas in comparison to C. angulata under heat shock at 35°C. Oysters were sampled at (a) Qingdao and (b) Xiamen (log10 transformed). Asterisks indicate significant differences between species at each sampling time, and error bars represent 1 s.d.
Expression of most genes (11 of 13) in C. angulata obviously increased at 6 h and sharply decreased at 24 h, while those in C. gigas continuously decreased after 3 h during heat stress at QD. We chose 6 h as a time point to show the magnitude of divergence between the two species cultured at XM.
4. Discussion
The basal levels of hsp expression under ambient conditions at each field site and relative expression under heat shock showed divergent expression patterns between the two species. Southern oysters (C. angulata) showed greater expression plasticity than the northern species (C. gigas) in response to thermal stress. Other marine invertebrates, such as corals, show a similar pattern, where inshore warm-adapted populations require a greater degree of plastic changes to be resistant to a thermally stressful environment [22]. Considering previous findings [3,6,7,23], we propose that oysters inhabiting thermally stressful environments may preferentially evolve high expression plasticity. However, a concern should be raised that this sublethal temperature may have led to cellular damage in the northern oysters, resulting in lower expression plasticity. Furthermore, warm-adapted C. angulata exhibited higher expression plasticity of hsps at a later stage of heat shock, while this occurred in northern C. gigas at an early stage. This delay is consistent with our previous findings that a greater extent of transcriptional plasticity in thermotolerant oyster populations is delayed in comparison to their thermosensitive counterparts [7]. Whereas there is high species specificity in transcriptional responses to temperature, the fact that cold-adapted species may exhibit a long lag time in upregulating hsp synthesis—as seen in this study and in a study of congeneric marine snails [24]—suggests that the speed with which an adaptive response can be elicited may vary with species' adaptation temperatures.
However, C. gigas showed higher basal expression under ambient conditions at both field sites. This finding confirms that there are evolutionary trade-offs between the basal and plastic expression levels of hsps in these species, implying evolutionary trade-offs between the acquisition of extreme tolerance limits and the retention of highly plastic tolerance limits [25]. Identical expression patterns have been similarly detected in a common fish Fundulus heteroclitus, in which hsc70 mRNA in southern individuals showed higher thermally induced expression plasticity than that in northern individuals [26]. This expression trade-off is species specific, while in contrast, the thermotolerant amphipod Eulimnogammarus cyaneus dwelling in the upper littoral zone exhibited higher constitutive expression of HSP70 proteins under normal physiological conditions and lower plastic expression after heat stress than those of its more thermosensitive congener E. verrucosus in the sublittoral zone [4]. However, further studies are required to quantify relationships between hsp gene and HSP protein expression under these conditions. In addition to the molecular level, evolutionary trade-offs were also demonstrated at the organismal level: oysters inhabiting environments with greater thermal fluctuation evolve higher thermotolerance but slower growth than their counterparts inhabiting moderate environments [7], as well as oysters dwelling in intertidal or subtidal zones [6].
Interestingly, the decreased gene expression plasticity in northern oysters was consistently exhibited at both sites, regardless of the differences in the environment. This potentially indicates that expression plasticity between the two oyster species may be fixed with a genetic basis. This same pattern of high baseline expression plus lower expression plasticity in the heat-sensitive species is in line with the evolutionary mechanism of genetic assimilation, in which derived populations lose plasticity in new environments [27–30]. Our results provide further evidence of evolutionary divergence of the two oysters and indicate that genetic assimilation is likely to have occurred, mediating the adaptive evolution of these two species.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to Professors Zhinan Zeng and Yue Ning for collecting oysters at Xiamen. We thank Mr Runshan Du, Xuegang Wang and Dr Xueying Tang for breeding the F1 progeny and culturing the oysters, and we thank Mr Hailong Liu for helping to extract RNA.
Ethics
There are no conflicts with animal-ethics or conservation policies, and no licence was required for the experimental treatments. The treatment described in the methods is a standard approach for oysters, and permission for oyster collection was granted by IOCAS.
Data accessibility
Data are provided as electronic supplementary material.
Authors' contributions
L.L. and G.Z. conceived of the study and helped draft the manuscript; A.L. carried out the molecular laboratory work, participated in data analysis and drafted the manuscript; W.W. edited the manuscript; and L.L. and W.W. collected oyster samples and carried out breeding of the F1 progeny. All authors give final approval for publication and agree to be held accountable for the content.
Competing interests
We declare we have no competing interests.
Funding
L.L. is supported by the National Natural Science Foundation of China (grant no. 31572620), National Key R&D Program of China (grant no. 2018YFD0900304) and the Modern Agro-industry Technology Research System (grant no. CARS-49).
References
- 1.Gienapp P, Teplitsky C, Alho JS, Mills JA, Merila J. 2008. Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol. 17, 167–178. ( 10.1111/j.1365-294X.2007.03413.x) [DOI] [PubMed] [Google Scholar]
- 2.Ho WC, Zhang J. 2018. Evolutionary adaptations to new environments generally reverse plastic phenotypic changes. Nat. Commun. 9, 350 ( 10.1038/s41467-017-02724-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li A, Li L, Song K, Wang W, Zhang G. 2017. Temperature, energy metabolism, and adaptive divergence in two oyster subspecies. Ecol. Evol. 7, 6151–6162. ( 10.1002/ece3.3085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedulina DS, et al. 2013. Expression patterns and organization of the hsp70 genes correlate with thermotolerance in two congener endemic amphipod species (Eulimnogammarus cyaneus and E. verrucosus) from Lake Baikal. Mol. Ecol. 22, 1416–1430. ( 10.1111/mec.12136) [DOI] [PubMed] [Google Scholar]
- 5.Gibson G, Weir B. 2005. The quantitative genetics of transcription. Trends Genet. T.I.G. 21, 616–623. ( 10.1016/j.tig.2005.08.010) [DOI] [PubMed] [Google Scholar]
- 6.Li A, Li L, Wang W, Song K, Zhang G. 2018. Transcriptomics and fitness data reveal adaptive plasticity of thermal tolerance in oysters inhabiting different tidal zones. Front. Physiol. 9, 825 ( 10.3389/fphys.2018.00825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, et al. 2018. Divergence and plasticity shape adaptive potential of the Pacific oyster. Nat. Ecol. Evol. 2, 1751–1760. ( 10.1038/s41559-018-0668-2) [DOI] [PubMed] [Google Scholar]
- 8.Zhang G, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. ( 10.1038/nature11413) [DOI] [PubMed] [Google Scholar]
- 9.Guo X, He Y, Zhang L, Lelong C, Jouaux A. 2015. Immune and stress responses in oysters with insights on adaptation. Fish Shellfish Immunol. 46, 107–119. ( 10.1016/j.fsi.2015.05.018) [DOI] [PubMed] [Google Scholar]
- 10.Kenkel CD, Meyer E, Matz MV. 2013. Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 22, 4322–4334. ( 10.1111/mec.12390) [DOI] [PubMed] [Google Scholar]
- 11.Kabbage M, Dickman MB. 2008. The BAG proteins: a ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 65, 1390–1402. ( 10.1007/s00018-008-7535-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Qian L, Liu X, Zhang G, Guo X. 2010. Classification of a common cupped oyster from southern China. J. Shellfish Res. 29, 857–866. ( 10.2983/035.029.0420) [DOI] [Google Scholar]
- 13.Ren J, Liu X, Jiang F, Guo X, Liu B. 2010. Unusual conservation of mitochondrial gene order in Crassostrea oysters: evidence for recent speciation in Asia. B.M.C. Evol. Biol. 10, 394 ( 10.1186/1471-2148-10-394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haure J, Huvet A, Palvadeau H, Nourry M, Penisson C, Martin JLY, Boudry P. 2003. Feeding and respiratory time activities in the cupped oysters Crassostrea gigas, Crassostrea angulata and their hybrids. Aquaculture 218, 539–551. ( 10.1016/s0044-8486(02)00493-3) [DOI] [Google Scholar]
- 15.Wang J, Li L, Zhang G. 2016. A high-density SNP genetic linkage map and QTL analysis of growth-related traits in a hybrid family of oysters (Crassostrea gigas×Crassostrea angulata) using genotyping-by-sequencing. G3 Genes Genome. Genet. 6, 1417–1426. ( 10.1534/g3.116.026971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reece KS, Cordes JF, Stubbs JB, Hudson KL, Francis EA. 2007. Molecular phylogenies help resolve taxonomic confusion with Asian Crassostrea oyster species. Mar. Biol. 153, 709–721. ( 10.1007/s00227-007-0846-2) [DOI] [Google Scholar]
- 17.Gagnaire PA, Lamy JB, Cornette F, Heurtebise S, Degremont L, Flahauw E, Boudry P, Bierne N, Lapegue S. 2018. Analysis of genome-wide differentiation between native and introduced populations of the cupped oysters Crassostrea gigas and Crassostrea angulata. Genome Biol. Evol. 10, 2518–2534. ( 10.1093/gbe/evy194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanford E, Kelly MW. 2011. Local adaptation in marine invertebrates. Annu. Rev. Mar. Sci. 3, 509–535. ( 10.1146/annurev-marine-120709) [DOI] [PubMed] [Google Scholar]
- 19.Zhu Q, Zhang L, Li L, Que H, Zhang G. 2016. Expression characterization of stress genes under high and low temperature stresses in the Pacific oyster, Crassostrea gigas. Mar. Biotechnol. 18, 176–188. ( 10.1007/s10126-015-9678-0) [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 21.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biometrical J. 50, 346–363. ( 10.1002/bimj.200810425) [DOI] [PubMed] [Google Scholar]
- 22.Kenkel CD, Matz MV. 2016. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 1, 0014 ( 10.1038/s41559-016-0014) [DOI] [PubMed] [Google Scholar]
- 23.Somero GN. 2012. The physiology of global change: linking patterns to mechanisms. Annu. Rev. Mar. Sci. 4, 39–61. ( 10.1146/annurev-marine-120710-100935) [DOI] [PubMed] [Google Scholar]
- 24.Tomanek L, Somero GN. 2000. Time course and magnitude of synthesis of heat-shock proteins in congeneric marine snails (Genus Tegula) from different tidal heights. Physiol. Biochem. Zool. 73, 249–256. ( 10.1086/316740) [DOI] [PubMed] [Google Scholar]
- 25.Bozinovic F, Calosi P, Spicer JI. 2011. Physiological correlates of geographic range in animals. Annu. Rev. Ecol. Evol. Syst. 42, 155–179. ( 10.1146/annurev-ecolsys-102710-145055) [DOI] [Google Scholar]
- 26.Fangue NA, Hofmeister M, Schulte PM. 2006. Intraspecific variation in thermal tolerance and heat shock protein gene expression in common killifish, Fundulus heteroclitus. J. Exp. Biol. 209, 2859–2872. ( 10.1242/jeb.02260) [DOI] [PubMed] [Google Scholar]
- 27.Crispo E. 2007. The Baldwin effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution 61, 2469–2479. ( 10.1111/j.1558-5646.2007.00203.x) [DOI] [PubMed] [Google Scholar]
- 28.Healy TM, Schulte PM. 2015. Phenotypic plasticity and divergence in gene expression. Mol. Ecol. 24, 3220–3222. ( 10.1111/mec.13246) [DOI] [PubMed] [Google Scholar]
- 29.Waddington CH. 1952. Selection of the genetic basis for an acquired character. Nature 169, 278 ( 10.1038/169278a0) [DOI] [PubMed] [Google Scholar]
- 30.Kelly M. 2019. Adaptation to climate change through genetic accommodation and assimilation of plastic phenotypes. Phil. Trans. R. Soc. B 374, 20180176 ( 10.1098/rstb.2018.0176) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are provided as electronic supplementary material.