Abstract
Nucleus accumbens–associated protein-1 (NAC1) is a transcriptional repressor encoded by the NACC1 gene, which is amplified and overexpressed in various human cancers and plays critical roles in tumor development, progression, and drug resistance. NAC1 has therefore been explored as a potential therapeutic target for managing malignant tumors. However, effective approaches for effective targeting of this nuclear protein remain elusive. In this study, we identified a core unit consisting of Met7 and Leu90 in NAC1's N-terminal domain (amino acids 1–130), which is critical for its homodimerization and stability. Furthermore, using a combination of computational analysis of the NAC1 dimerization interface and high-throughput screening (HTS) for small molecules that inhibit NAC1 homodimerization, we identified a compound (NIC3) that selectively binds to the conserved Leu-90 of NAC1 and prevents its homodimerization, leading to proteasomal NAC1 degradation. Moreover, we demonstrate that NIC3-mediated down-regulation of NAC1 protein sensitizes drug-resistant tumor cells to conventional chemotherapy and enhances the antimetastatic effect of the antiangiogenic agent bevacizumab both in vitro and in vivo. These results suggest that small-molecule inhibitors of NAC1 homodimerization may effectively sensitize cancer cells to some anticancer agents and that NAC1 homodimerization could be further explored as a potential therapeutic target in the development of antineoplastic agents.
Keywords: cancer therapy, transcription target gene, protein targeting, inhibitor, cancer biology, combination therapy, dimerization, inhibitor, NAC1
Introduction
Overexpression or mutation of oncogenes and loss of tumor-suppressor genes are important events in cancer development and progression (1, 2). Because transcription is a critical control point in the production of many proteins, alterations of oncogenic or tumor suppressor proteins often involve dysregulated transcription (3). Indeed, certain transcription factors have been found to be in excess amount or overactive in various types of cancer and contribute to survival, unrestrained replication, and apoptosis escape in cancers (4). Therefore, inhibiting the activity of oncogenic transcription factors is considered as a promising strategy in anticancer treatment. However, many of those transcription factors seen in human cancers are considered “undruggable” due to their large protein–protein interaction interfaces and their lack of deep protein pockets and intracellular localization. For drugging oncogenic transcription factors, several new approaches have been developed to overcome the undruggable nature of these transcription factors (5). For instance, the BRD4-based small-molecule bromodomain inhibitor (JQ1) can directly block MYC transcription and Myc-dependent transcriptional target genes; the conformation-disrupting Aurora A inhibitors can destabilize the complex of Aurora A with Myc and promote the degradation of Myc protein (6, 7). Two compounds, IS3-295 and S3I-201, were reported to exert their antitumor activity partially through blocking the Stat3-mediated DNA-binding activity (8, 9). Molecular docking and synthesis of novel quinazoline analogues were shown to inhibit NF-κB–dependent gene transcription (10).
Nucleus accumbens–associated protein-1 (NAC1)4 is a transcription cofactor belonging to the bric-à-brac, tramtrack, broad complex/pox virus and Zn finger (BTB/POZ) family (11, 12). The conserved BTB protein–protein interaction domain is required for NAC1 homodimerization, which plays important roles in various biological processes (11, 13). NAC1 is overexpressed in various types of cancer, including ovarian cancer, cervical cancer, and breast cancer (14, 15), and has been shown to contribute to tumor-suppressor inactivation (16), autophagic survival response (17), cellular senescence escape (18), cancer cell cytokinesis (19), anaerobic glycolysis (20), and fatty-acid synthase expression (21). Also, high expression of NAC1 was closely associated with chemotherapy resistance and tumor recurrence (11, 22), and inhibition of NAC1 by siRNA or dominant-negative mutant increased apoptosis induced by anticancer agents (17, 20, 23). These observations indicate that NAC1 may represent a potential molecular target for cancer treatment; however, approaches to effectively targeting this nucleic oncogenic protein remain elusive.
It is known that exposure of the hydrophobic interface of a dimeric protein may lead to conformational change, causing destabilization and degradation of a protein via proteasomal or autophagic pathways (24–26). In this study, we identified a core unit consisting of Met7 and Leu90 in the N-terminal domain (amino acids 1–130) of NAC1, which is critical for its dimerization and stability. To test our hypothesis that inhibiting the dimerization of NAC1 can destabilize NAC1 protein and promote its degradation, we searched for chemicals able to inhibit the homodimerization of NAC1 using a combination approach of computational analysis of the dimerization interface and high-throughput screening. Here, we report a compound, NIC3, that has the ability to selectively bind with the conserved Leu90 of NAC1 and to inhibit NAC1 dimerization, resulting in proteasomal degradation of the NAC1 protein. We further assessed the therapeutic potential of NIC3 in combination therapy. We showed both in vitro and in vivo that down-regulation of NAC1 protein by NIC3 significantly overcame tumor cell resistance to conventional chemotherapy and enhanced antimetastatic efficacy of the antiangiogenic agent bevacizumab. The results of this study not only underscore the potential of NAC1 as an anticancer target but also demonstrate the therapeutic benefits of the small-molecule inhibitors of NAC1 dimerization in cancer treatment.
Results
Analysis of dimerization domain and residues of NAC1
The conserved BTB/POZ domain is essential for NAC1 dimerization, which plays important roles in tumor development (11); however, no evidence has been provided about the mechanisms and biologic consequences of NAC1 dimerization. To study this, we first assessed the association of the two forms of NAC1 proteins tagged with either V5 or Myc epitopes by co-IP after their coexpression in HEK-293T cells. Fig. 1A demonstrates the association of V5-NAC1 with Myc-NAC1 in the cells. To further assess the interaction between two tagged NAC1 proteins, we used bacterially expressed GST-NAC1 protein in an in vitro dimerization assay. Fig. 1B shows that with increasing concentrations of disuccinimidyl suberate (DSS), a noncleavable bivalent chemical cross-linker that is commonly used to detect direct protein–protein interactions, the intensity of NAC1 monomers was reduced gradually accompanied by the appearance of the expected NAC1 homodimers. NAC1 homodimers could not be detected in the bacterially expressed NAC1(ΔN130) protein (BTB/POZ domain deletion) (Fig. 1C).
Figure 1.
Homodimerization of NAC1 via the BTB domain and binding residues Met7 and Leu90. A, V5-NAC1 and Myc-NAC1 were cotransfected into HEK-293T cells. The lysates of these cells were immunoprecipitated with an anti-V5 antibody. The immunoprecipitates and input were examined by immunoblotting with the respective antibodies. Cells cotransfected withV5-NAC1 and empty Myc vector (−) or Myc-NAC1 and empty V5 vector (−) served as negative controls. B, NAC1 homodimerization in vitro. GST-NAC1 fusion protein was incubated with increasing concentrations of the bivalent chemical cross-linker DSS. The cross-linked complexes were resolved by nondenaturing PAGE and analyzed by Western blotting using an anti-GST antibody. C, GST-NAC1 and GST-ΔN130 fusion proteins were incubated with increasing concentrations of DSS, and the cross-linked proteins were analyzed by nondenaturing PAGE analysis. Nonspecific bands are indicated with asterisks. D, HEK-293T cells were transfected with a V5-NAC1 plasmid, Myc-NAC1 plasmid, V5-NAC1(L90A) plasmid, or Myc-NAC1(L90A) plasmid for 24 h and then treated with 5 μm MG132 for another 6 h. Cell lysates were immunoprecipitated with an anti-Myc antibody and then immunoblotted (IB) for V5. E, HEK-293T cells were transfected with a V5-NAC1 plasmid or V5-NAC1(L90A) plasmid for 24 h and then treated with 5 μm MG132 for another 6 h. NAC1 protein was examined by Western blotting. β-Actin was used as a loading control. F, HEK-293T cells were transfected with a V5-NAC1 plasmid or V5-NAC1(L90A) plasmid for 24 h and then subjected to pulse-chase analysis in the presence of cycloheximide (20 μg/ml) for the indicated time periods. Error bars represent S.D. Data are presented as mean ± S.D. (n = 3). G, proposed model for the role of binding residues Met7 and Leu90 in NAC1 homodimerization.
To identify the sites in BTB/POZ domain required for association between the two tagged NAC1 proteins, contacts between atoms of the protein were examined using the Protein Contacts module in Molecular Operating Environment (MOE). Among them, one dimerization core unit consisting of four residues, Met7 and Leu90 from one chain and the same residues from another chain, appears to be crucial for NAC1 dimerization (Fig. S1, A and B). To further delineate the role of binding residues (Met7 and Leu90) in dimerization, using a site-directed mutagenesis method we generated two different tagged NAC1 mutants in which Leu90 sites were altered to Arg. We found that single Leu90 mutation decreased NAC1 dimer formation, and double Leu90 mutations further decreased NAC1 dimerization (Figs. 1D and S2A). Fig. S2B shows that with increasing concentrations of DSS the intensity of NAC1 monomers was reduced gradually and was accompanied by the appearance of the expected NAC1 homodimers; NAC1 homodimers could not be detected in the NAC1(S91A) protein. Introduction of Leu90 mutation led to a significant reduction of NAC1 protein amount in HEK-293T cells, and reduction of NAC1 protein caused by Leu90 mutation could be rescued by the proteasome inhibitor MG132 (Fig. 1E). The pulse-chase experiments demonstrated that NAC1 mutant (L90A) had a shorter half-life than WT NAC1 (4.6 h versus 20 min) (Fig. 1F). Notably, ubiquitination of the NAC1 mutant (L90A) was significantly increased as compared with WT NAC1 (Fig. S2C). These results suggest that the dimerization core unit of NAC1 consisting of Leu90 and Met7 is critical for the formation of stable NAC1 dimers; disrupting this core unit would suppress NAC1 dimerization and destabilize this protein.
Identification of NIC3 as an inhibitor of NAC1 dimerization
To search for small-molecule compounds able to target the dimeric interface of NAC1, we screened ∼200,000 compounds using an in silico docking approach (Fig. 2A). Of the 50 hits, one compound, NIC3 (Fig. 2B), was chosen for the current study based on the molecular docking showing that the nitrogen atoms in NIC3 form hydrogen bonds with the critical dimerization core residue Leu90, and this confers this compound druglike properties (Figs. 2, C–E, and S3). Validation of NIC3 as an inhibitor of NAC1 dimerization was carried out using co-IP assay. We showed that NIC3 could abolish co-IP of ectopic NAC1 protein tagged with V5 or Myc epitopes (Fig. 2F). Using the purified NAC1 protein (GST-NAC1) and nondenaturing PAGE analysis, we also showed the inhibition of formation of NAC1 dimerization by NIC3 in vitro (Fig. 2G). In these experiments, we incubated the bacterially expressed proteins with increasing concentrations of NIC3 and demonstrated that NAC1 dimerization was suppressed in a dose-dependent manner.
Figure 2.
Identification and validation of NIC3 as an inhibitor of NAC1 homodimerization. A, flowchart of the number of candidate compounds identified at each step, resulting in 50 candidate compounds after virtual screening. B, the structure of NIC3. C, 3D binding mode of the receptor–ligand complex with ligand colored in cyan, receptor atoms in green, and backbone in magenta. D, the tentative surfaces of the binding sites on NAC1 of which exposed, polar, and hydrophobic areas are colored in red, magenta, and green, respectively. E, 2D binding mode of the same complex. F, HEK-293T cells were transfected with a V5-NAC1 plasmid and Myc-NAC1 plasmid for 12 h and then treated with increasing concentrations of NIC3 for another 24 h. Cell lysates were immunoprecipitated with an anti-Myc antibody followed by immunoblotting (IB) for V5. G, purified GST-NAC1 protein was incubated with increasing concentrations of NIC3. Cell lysates were resolved by nondenaturing PAGE and analyzed by Western blotting using an anti-GST antibody.
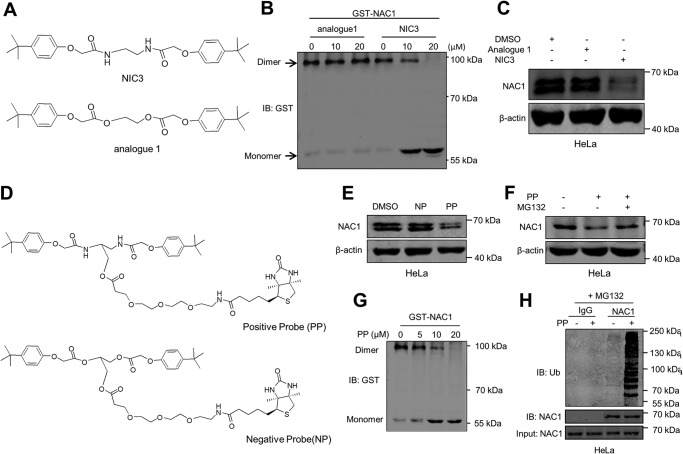
To determine how NIC3 docks into NAC1, we prepared several chemical probes (Fig. S4). Structure–activity relationship studies revealed that nitrogen atoms are essential for inhibition of the NAC1 dimerization as the NIC3 treatment was able to dissociate the dimeric NAC1 into monomers, whereas analogue 1 could not inhibit the formation of NAC1 dimerization and decrease NAC1 level (Fig. 3, A–C). Based on this information, we synthesized a positive probe (PP) and a negative probe (NP) with biotin and an ester bond linker (Fig. 3D). We found that the biotin-tagged positive probe possessed the ability to effectively inhibit NAC1 expression, whereas the biotin-tagged negative probe did not have this ability at the same concentration (Fig. 3E), and reduction of NAC1 protein caused by PP could be rescued by the proteasome inhibitor MG132 (Fig. 3F). Furthermore, NAC1 dimerization was inhibited, and ubiquitination of the NAC1 protein was significantly increased by PP (Fig. 3, G and H). In co-IP experiments, we detected the association of NAC1 with PP but not with NP (Fig. 4A). To determine whether NIC3 directly binds to NAC1, we generated a bacterially expressed GST-NAC1 protein. The purified GST-NAC1 was incubated with increasing concentrations of PP bound to biotin-GSH particles followed by immunoblotting with biotin antibody. As shown in Fig. 4B, the intensity of GST-NAC1 increased with increasing concentrations of PP. However, NIC3 lost its inhibitory effect on NAC1 dimerization when the nitrogen atoms were replaced with oxygen atoms (Fig. 3B). Molecular docking demonstrated that Leu90, Ser91, and Met92 form hydrogen bonds with the nitrogen atoms of NIC3 (Fig. 2, C–E). To determine which of these residues are critical for the binding with NIC3, we mutated these leucine, serine, and methionine residues of NAC1 to alanine residues (Fig. 4, C and D). Among these mutants, we found that L90A mutant completely lost the ability to bind to NIC3, whereas S91A and M92A retained binding to NIC3 in a manner similar to WT NAC1 (Fig. 4, C and D). To determine the effects of those mutations on NAC1 structure, we calculated and compared the backbone root-mean-square deviations between the WT NAC1 and each of the mutants. None of the mutants showed significant structural changes, suggesting that neither L90A nor other NAC1 mutants tested in the studies have significant effects on the NAC1 monomer structure as compared with the WT (Fig. S1, C and D). These results imply that NIC3 can efficiently and specifically dock into NAC1 and that Leu90 of NAC1 is essential for its interaction with NIC3.
Figure 3.
Synthesis and biological validation of chemical probes. A, the chemical structure of an analogue of NIC3. B, GST-NAC1 fusion protein was incubated with increasing concentrations of analogue 1 and NIC3. Cell lysates were resolved by nondenaturing PAGE and analyzed by Western blotting using an anti-GST antibody. C, HeLa cells were incubated with 20 μm NIC3 or 20 μm analogue 1 for 48 h. NAC1 protein was determined by immunoblotting (IB). β-Actin was used as a loading control. D, chemical structures of biotin-labeled NIC3 (PP) and biotin-labeled analogue 1 (NP). E, HeLa cells were incubated with 20 μm PP or NP for 48 h. NAC1 protein was determined by immunoblotting. β-Actin was used as a loading control. F, HeLa cells were incubated with 20 μm PP for 42 h and then treated with 5 μm MG132 for an additional 6 h. NAC1 protein was examined by Western blotting. β-Actin was used as a loading control. G, purified GST-NAC1 protein was incubated with increasing concentrations of PP. Cell lysates were resolved by nondenaturing PAGE and analyzed by Western blotting using an anti-GST antibody. H, HeLa cells were treated with 20 μm PP for 36 h and then treated with vehicle or 5 μm MG132 for an additional 6 h. Cell lysates were immunoprecipitated with a control IgG or anti-NAC1 antibody. The immunoprecipitates and input were probed for ubiquitin (Ub) and NAC1 by immunoblotting.
Figure 4.
Leu90 of NAC1 is essential for its binding to NIC3. A, HeLa cells were incubated with 20 μm PP or NP for 24 h. The lysates of these cells were immunoprecipitated with a biotin antibody, and the immunoprecipitates were examined by immunoblotting (IB) with an NAC1 antibody. B, GST-NAC1 fusion protein was incubated with increasing concentrations of PP for 12 h at 4 °C followed by immunoblotting with biotin or GST. C and D, HEK-293T cells were transfected with V5-NAC1, V5-NAC1(L90A), V5-NAC1(S91A), or V5-NAC1(M92A) plasmids or Myc-NAC1, Myc-NAC1(L90A), Myc-NAC1(S91A), or Myc-NAC1(M92A) plasmids for 12 h followed by treatment with PP (20 μm) or NP (20 μm) for 24 h and with 5 μm MG132 for an additional 6 h. Cell lysates were immunoprecipitated with a biotin antibody followed by immunoblotting for V5 or Myc.
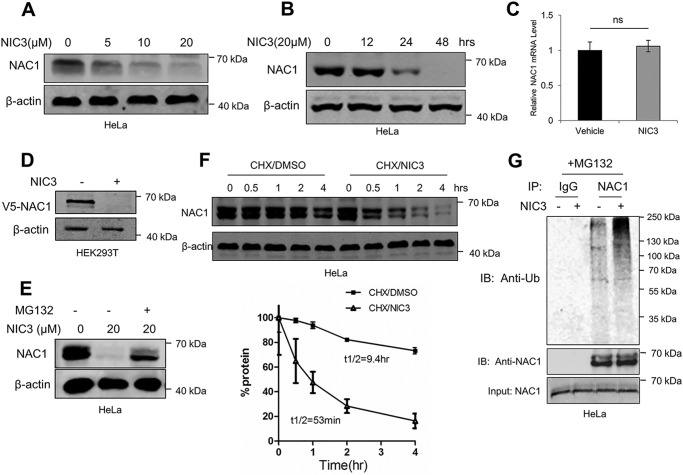
NIC3 accelerates the proteasomal degradation of NAC1 protein
As the dimerization of NAC1 contributed to its stability (Fig. 1), we next wanted to know whether inhibiting NAC1 dimerization by NIC3 would facilitate proteasomal degradation of NAC1 protein. In these experiments, we first treated HeLa cells with a series of concentrations of NIC3 for different periods of time and then analyzed the level of endogenous NAC1 protein using immunoblotting. Fig. 5, A and B, show that NIC3 treatment caused a reduction of NAC1 protein in a dose- and time-dependent manner. Similar effects of NIC3 on the NAC1 level were observed in SKOV3 cells (Fig. S2D). The mRNA level of NAC1 was not affected by NIC3 treatment, as measured by quantitative RT-PCR (Fig. 5C). NIC3 treatment also reduced the level of ectopic V5-tagged NAC1 in HEK-293T cells (Fig. 5D). The reduction of NAC1 protein in the NIC3-treated cells could be rescued by the proteasome inhibitor MG132 (Fig. 5E). Fig. 5F shows that the turnover of NAC1 protein was greatly facilitated in the cells treated with NIC3 as compared with that in the control cells (t½, 53 versus 560 min), as measured by pulse-chase experiments. Additionally, NIC3 treatment enhanced the ubiquitination of NAC1 protein (Figs. 5G and S2C). To confirm these findings, we examined the effects of NIC3 on the NAC1-regulated proteins HIF-1α and Gadd45-interacting protein 1 (16, 20, 23). We found that NIC3 treatment indeed affected HIF-1α and Gadd45-interacting protein 1 (Fig. S9), further supporting the role of NIC3 in accelerating the ubiquitin-proteasomal degradation of NAC1 protein via prohibiting its dimerization.
Figure 5.
NIC3 induces proteasome degradation of NAC1 protein. A, HeLa cells were treated with increasing concentrations of NIC3 as indicated for 48 h. NAC1 protein was determined by immunoblotting. β-Actin was used as a loading control. B, HeLa cells were treated with 20 μm NIC3 for the indicated periods of time. NAC1 protein was determined by immunoblotting. β-Actin was used as a loading control. Error bars represent S.D. C, HeLa cells were treated with 20 μm NIC3 for 24 h. NAC1 mRNA was measured by quantitative RT-PCR and plotted after normalization. Bars are mean ± S.D. (n = 3). D, HEK-293T cells were transfected with a V5-NAC1 plasmid and then treated with 20 μm NIC3 for 48 h. Ectopic V5-tagged NAC1 was determined by immunoblotting. β-Actin was used as a loading control. E, HeLa cells were treated with 20 μm NIC3 for 40 h and then treated with 5 μm MG132 for another 6 h. NAC1 protein was determined by immunoblotting. β-Actin was used as a loading control. F, HeLa cells were treated with 20 μm NIC3 for 40 h and then subjected to pulse-chase analysis in the presence of cycloheximide (20 μg/ml) for the indicated periods of time. Error bars represent S.D. Data are presented as mean ± S.D. (n = 3). G, HeLa cells were treated with 20 μm NIC3 for 36 h and then treated with vehicle or 5 μm MG132 for another 6 h. Cell lysates were immunoprecipitated with a control IgG or an anti-NAC1 antibody. The immunoprecipitates and input were probed for ubiquitin (Ub) and NAC1 by immunoblotting. ns, not significant.
NIC3 overcomes chemoresistance in vitro and in vivo
Conventional chemotherapy is an important part of treatment for many cancers, but its effectiveness is often limited by acquired drug resistance (27). Previous studies showed that high expression of NAC1 was closely associated with chemotherapy resistance and tumor recurrence (11, 22). Therefore, we determined the effects of NIC3 on tumor-cell sensitivity to cisplatin and Adriamycin, two commonly used anticancer drugs, in drug-resistant cancer cell lines. NIC3 itself did not show any cytotoxicity in the cancer cell lines until the concentration reached 100–1000 μm (Fig. S5). To test whether NIC3 can sensitize drug-resistant tumor cells to cisplatin or Adriamycin, we used the sublethal concentration of 20 μm, which can inhibit NAC1 expression significantly. The human MCF-7/ADR (Adriamycin-resistant) and HeLa/DDP (cisplatin-resistant) cell lines were used in these experiments. We observed that combination treatment of NIC3 with Adriamycin or cisplatin significantly augmented the cytotoxic effect in the resistant cancer cell lines (Fig. 6, A–D). The increased cytotoxicity seen in the cells treated with the combination of cisplatin or Adriamycin with NIC3 was associated with enhanced apoptosis, as evidenced by elevated levels of cleaved PARP (Fig. 6, E and F). We also showed that the combination of cisplatin or Adriamycin with NIC3 resulted in decreased cell proliferation and increased apoptotic cell death in the human ovarian cancer cell line SKOV3 (Fig. S6, A–C). By contrast, NIC3 did not show a sensitizing effect in ES-2, an ovarian cancer cell line that has low intrinsic NAC1 expression (Fig. S7, A and B). Furthermore, NIC3 can also reverse drug resistance in an animal xenograft model. In mice bearing MCF-7/ADR cells (inoculated subcutaneously), NIC3 treatment alone had no effect on tumor growth; Adriamycin treatment alone showed a moderate inhibitory effect on the growth of the xenografted tumors (Fig. 6G). In contrast, the combination of NIC3 with Adriamycin significantly suppressed the growth of the xenografted tumors (Figs. 6G and S8A). Concomitantly, as compared with either vehicle control, NIC3, or Adriamycin alone, the combination treatment of NIC3 with Adriamycin caused a significant increase in the terminal deoxynucleotidyltransferase dUTP nick end labeling (TUNEL) staining–positive cells and a decrease in Ki67, a proliferation marker (Fig. 6H), and in NAC1 protein expression (Figs. 6H and S8B). No significant changes in body weight were observed in the mice treated with the combination therapy (Fig. 6G). These results suggest that NIC3 is synergistic with cisplatin or Adriamycin in suppressing cancer cell growth, which is associated with induction of apoptotic cell death in drug-resistant tumor cells both in vitro and in vivo.
Figure 6.
NIC3 significantly sensitizes drug-resistant tumor cells to cisplatin and Adriamycin in vitro and in vivo. A and B, HeLa/DDP cells and MCF-7/ADR cells were treated with the indicated concentrations of cisplatin (DDP) or Adriamycin (ADM) or with or without NIC3. 48 h later, cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. C and D, HeLa/DDP cells and MCF-7/ADR cells treated as described earlier were plated and incubated for 10 days at 37 °C. Cells were stained, and colonies were counted under a light microscope. Error bars represent S.D. Bars are mean ± S.D. (n = 3). E and F, HeLa/DDP cells and MCF-7/ADR cells were subjected to treatment as described earlier. Apoptosis was determined by Western blot analysis of cleaved PARP. G, top, experimental schedule/regimen. Bottom left, the mice inoculated with MCF-7/ADR cells were divided into four treatment groups: 1) control (Ctrl), 2) NIC3 (25 mg/kg), 3) Adriamycin (5 mg/kg), and 4) NIC3 (25 mg/kg) + Adriamycin (5 mg/kg). Tumor size was measured every 3 days. Error bars represent S.D. Each point represents mean ± S.D. (n = 5). Bottom right, mouse body weight. H, the tumor specimens were stained for NAC1 protein and Ki67 protein and subjected to TUNEL. Bars are mean ± S.D. (n = 5). Scale bar, 50 μm. ns, not significant; *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
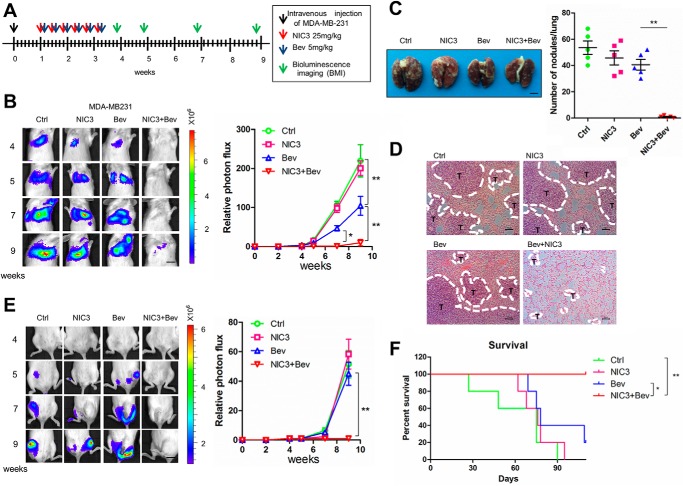
NIC3 potentiates the antimetastatic effect of bevacizumab in a metastatic breast cancer model
Bevacizumab is an antiangiogenic agent used in treating metastatic breast cancer (28). However, hypoxia environment or cancer stem cell enrichment induced by bevacizumab may limit its antimetastatic efficacy (29–31). We and other groups previously showed that silencing NAC1 could mitigate the hypoxic microenvironment (20) and regulate stem cell differentiation (32, 33). Therefore, we next determined whether NIC3 could reinforce the antimetastatic effect of bevacizumab. An experimental metastatic breast cancer model established by intravenous injection of luciferase-labeled MDA-MB-231 cells was used. We observed that the combination treatment of bevacizumab with NIC3 significantly reduced the lung and bone disseminations of the tumor-bearing mice as compared with either control, NIC3, or bevacizumab alone, as evidenced by the luciferase expression in the thoracic cavity and in the hind limbs (Fig. 7, B and E). H&E staining of the sections corresponding to this region confirmed the decrease of lung metastasis (Fig. 7, C and D). Additionally, the enhanced antimetastatic effect of this combination treatment significantly extended the survival of the tumor-bearing mice (Fig. 7F). These observations suggest that NIC3 could be used as a sensitizer in antimetastatic therapy.
Figure 7.
NIC3 significantly strengthens the therapeutic efficacy of bevacizumab in a metastatic breast cancer mouse model. Luciferase-labeled MDA-MB-231 human breast tumor cells (1 × 106) were injected into the tail vein of SCID mice. A, experimental schedule/regimen. B, the mice inoculated with MDA-MB-231 cells were divided into four treatment groups: 1) control (Ctrl), 2) NIC3 (25 mg/kg), 3) bevacizumab (Bev) (5 mg/kg), and 4) NIC3 (25 mg/kg) + bevacizumab (5 mg/kg). Error bars represent S.D. *, p < 0.05; **, p < 0.01. Left panel, at the indicated time after xenografting, representative bioluminescence images were acquired; right panel, normalized photon flux of lung metastasis. Scale bar, 10 mm. C, lungs were dissected, fixed, and inspected for metastatic nodules on the surface. The graph represents numbers of observable metastatic nodules in the lung surface of each mouse. Error bars represent S.D. **, p < 0.01. Scale bar, 3 mm. Error bars represent S.D. **, p < 0.01. D, lungs showing in C were fixed, embedded in paraffin, sectioned, and stained by H&E for histological analyses. Scale bar, 50 μm. E, left panel, at the indicated week after xenografting, representative bioluminescence images were acquired; right panel, normalized photon flux of bone metastasis. Scale bar, 10 mm. F, mice were monitored for survival. Kaplan–Meier plots illustrate survival of the animals (n = 5/group; *, p < 0.05; **, p < 0.01, log-rank test), combination treatment versus bevacizumab alone.
Discussion
Because of the critical role of NAC1 in oncogenesis, therapeutic targeting of this protein has been explored as a potential strategy for cancer treatment. For example, to inactivate NAC1, Wu et al. (34) developed a series of self-inhibitory peptides that block NAC1 dimerization by rebinding at the dimerization interface. Although this method might be an effective approach to inhibiting NAC1, the biological effects of these peptides and physicochemical challenges in the application of those peptides in vivo remain obscure. Thus, nonpeptidic molecules capable of targeting NAC1 appear to be better candidates. It is known that NAC1 homodimerization is critical for tumor cell proliferation and survival; however, the mechanism by which NAC1 dimerization occurs and its biologic consequences are unclear (11). In the current study, we first focused on the molecular basis of NAC1 protein dimerization (Figs. 1 and S1). We show that this type of homodimerization is important for the stability of NAC1 protein (Figs. 1 and S2C). Furthermore, using computational analysis of the dimerization interface and high-throughput screening approach, we identified a small-molecule compound, NIC3, that is able to inhibit NAC1 dimerization via targeting the critical dimerization core residues and can promote proteasome-dependent degradation of NAC1 protein (Figs. 2, F and G; 5, F and G; and S2, D and E). These results are consistent with the concept that exposure of the hydrophobic interface of a dimeric protein often leads to its conformational change and destabilization (24, 25). We demonstrate that degradation of NAC1 protein caused by NIC3 is specifically based on its properties as an inhibitor of NAC1 homodimerization but is not a consequence of an off-target effect. Using biotin-labeled NIC3 as a probe, we show that NIC3 directly targets NAC1 protein, and through both computational and experimental studies we further show that NIC3 inhibits NAC1 homodimerization through docking into critical residues such as Leu90, thereby preventing the interaction between the two NAC1 molecules (Fig. 4).
A number of oncogenes such as kinases are valuable druggable targets. However, there are oncogenic proteins that are regarded as undruggable, and most of these are transcriptional factors, including MYB, MYC, and NF-κB (35, 36). Targeting nuclear proteins has been considered challenging, mainly due to their relatively larger interaction surfaces and lack of a druggable binding pocket (5). Nevertheless, based on new concepts in drug development and better understanding of the interaction surfaces, a number of so-called undruggable molecules have now been successfully targeted (37–42). The results reported here provide another example of targeting a nuclear factor through its hydrophobic pockets and demonstrate that computational analysis of the hydrophobic dimerization interface to identify the core units is a feasible approach to searching for small-molecule compounds for inhibiting homodimerization of the target proteins.
Cancer metastasis and therapeutic resistance are the major clinical challenges that account for cancer-related deaths. We show that NIC3 can sensitize drug-resistant tumor cells to chemotherapeutic drugs such as Adriamycin and cisplatin and enhance the antimetastatic efficacy of the antiangiogenic agent bevacizumab. We and others have previously shown that NAC1 contributes to chemoresistance through tumor-suppressor Gadd45 inactivation (16), autophagic survival response (17), cellular senescence escape (18), and cancer cell cytokinesis (19). Thus, targeting NAC1 may weaken those survival advantages in tumor cells treated with anticancer drugs. Indeed, the combination treatment of NIC3 with Adriamycin or bevacizumab shows more potent antitumor efficacy than Adriamycin or bevacizumab alone in drug-resistant tumors both in vitro and in vivo. NAC1 has previously been shown to promote a hypoxic microenvironment through hypoxia-induced up-regulation of HIF-1α (20) and to maintain stemness of stem cells (32, 33). Antiangiogenic agents such as bevacizumab may induce hypoxia in the tumor microenvironment and increase the population of cancer stem cells that favors metastatic spread (29–31). Down-regulation of NAC1 through preventing its dimerization by small-molecule inhibitors such as NIC3 may mitigate the hypoxic tumor environment and reduce the population of cancer stem cells.
Whether or not NIC3 can be further developed as a therapeutic agent for treatment of human cancer remain to be investigated. For instance, the pharmacokinetic properties of this compound are currently unknown. Bioisosteric replacements and scaffold hopping are useful approaches in drug design as they facilitate optimization of pharmacokinetic properties and activity (43). Whether bioisosterism and scaffold hopping such as substitution of a lipophilic group with a hydrophilic group or ethylenediamine with piperazine or homopiperazine may improve pharmacokinetics, solubility, metabolic stability, binding ability, or simplification of the synthetic route of NIC3 requires further studies.
Collectively, the findings reported here uncover not only a biologically critical but also a therapeutically exploitable role for NAC1 homodimerization. The inhibitor of NAC1 homodimerization that we identified, NIC3, shows potent effects on sensitizing drug-resistant tumor cells to chemotherapy and reinforcing the antimetastatic efficacy of the antiangiogenic agent bevacizumab. Thus, small-molecule inhibitors of NAC1 homodimerization may have great potential to be used as therapeutic agents in cancer treatment and may warrant further investigation.
Experimental procedures
Cell lines and culture
The human ovarian cancer cell lines SKOV3 and ES-2; the human cervical cancer cell lines HeLa and HeLa/DDP (the cisplatin-resistant cell line); the human breast cancer cell lines MDA-MB-231, MCF-7, and MCF-7/ADM (the Adriamycin-resistant cell line); and the human embryonic kidney HEK-293T cells were purchased from ATCC (Manassas, VA). The identities of these cell lines were verified by short tandem repeat analysis. HeLa, HeLa/DDP, ES-2, MDA-MB-231, MCF-7, and MCF-7/ADM cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. The SKOV3 cell line was cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. Cells were cultured at 37 °C in a humidified atmosphere of 20% O2, 5% CO2 (normoxia) or 1% O2, 5% CO2 (hypoxia). All cultures were monitored routinely and found to be free of contamination by Mycoplasma or fungi. Cell lines did not surpass 10 passages between thawing and use.
Reagents and antibodies
The following antibodies were used in Western blotting, coimmunoprecipitation, or immunohistochemistry: anti-NAC1 (Novus Biologicals, Littleton, CO, catalogue number NB110-77345), anti-PARP (Cell Signaling Technology, Danvers, MA, catalogue number 5625S), anti-V5 (Cell Signaling Technology, catalogue number 13202), anti-GST (Cell Signaling Technology, catalogue number 2622s), anti-Myc (Cell Signaling Technology, catalogue number 2278p), anti-β-actin (Santa Cruz Biotechnology, Dallas, TX, catalogue number sc-47778), anti-ubiquitin (Santa Cruz Biotechnology, catalogue number sc-8017), anti-Ki67 (Proteintech, China, catalogue number 19972-1-AP), streptavidin–horseradish peroxidase conjugate (GE Healthcare, catalogue number RPN1231), and mouse IgG and rabbit IgG (Beyotime, Shanghai, China, catalogue numbers A0216 and A0208). Cisplatin and Adriamycin were purchased from Sigma-Aldrich (catalogue numbers P4394 and 1225758). MG132 and cycloheximide were purchased from Sigma-Aldrich (catalogue numbers 5087390001 and M8699–1). In Situ Cell Death Detection kit (TUNEL staining) was purchased from Roche Applied Science (catalogue number 11684817910).
Molecular docking
The 2D structures of the ligands were drawn in ChemBioDraw 2013 and converted to 3D in MOE 2014 through energy minimization. Protein structures were downloaded from the Protein Data Bank (PDB code 3GA1) and prepared with the Structure Preparation workflow in MOE to correct structural errors such as missing atoms or nonstandard atom names. Then, the protonation state of the proteins and the orientation of the hydrogens were optimized by LigX at a pH of 7 and temperature of 300 K. Prior to docking, the force field of AMBER10: EHT and the implicit solvation model of Reaction Field (R-field) were selected. The docking workflow followed the “induced fit” protocol of MOE-Dock, in which the side chains of the receptor pocket were allowed to move according to ligand conformations, with a constraint on their positions. The weight used for tethering side-chain atoms to their original positions was 10. For each ligand, all docked poses were ranked by London dG scoring first, and then a force field refinement was carried out on the top 30 poses followed by a rescoring of GBVI/WSA dG, respectively.
Chemical synthesis
Syntheses of the PP and NP were carried out according to Fig. S4.
Plasmids and plasmid transfection
V5-NAC1 plasmid was described previously (20). The Myc-NAC1 plasmid was generated by replacing V5 tag with Myc tag. To generate the mutants of NAC1, we replaced the leucine site Leu90, serine site Ser91 with arginine, and methionine site Met92 with leucine. Site-directed mutagenesis was performed using the QuikChange kit (Stratagene). Transfection of the plasmids was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Quantitative real-time PCR
Total RNA was prepared using TRIzol reagent (Roche Applied Science). First-strand cDNA was synthesized using an Omniscript reverse transcription kit (Qiagen) with random primers. Quantitative RT-PCR was performed on ABI 7500 using Brilliant II SYBR Green QPCR Master Mix (Stratagene) and primers. After 40 cycles, data were collected and analyzed using the ABI 7500 software.
Immunoblotting and coimmunoprecipitation
Proteins (10–20 μg) were resolved by SDS-PAGE and then transferred to PVDF membrane (Bio-Rad). Membranes were incubated with primary antibodies in 3% BSA at 4 °C overnight followed by incubation with secondary antibodies at room temperature for 1 h. The protein signals were detected by ECL method. For co-IP, appropriate antibodies were first incubated with protein A/G beads (Santa Cruz Biotechnology) at 4 °C overnight, and then cell lysates were incubated with the protein A/G beads at 4 °C for 6 h. At the end of incubation, the beads were washed three times with radioimmune precipitation assay buffer, and the immunoprecipitates were eluted with SDS buffer and then subjected to immunoblotting.
Nondenaturing PAGE
Purified NAC1 was incubated with increasing amounts of DSS (Thermo Scientific) or different concentrations of NIC3 before mixing with an equal volume of sample buffer (100 mmol/liter Tris, pH 8.0, 20% glycerol, 0.005% bromphenol blue, 2% Triton X-100, and 100 mmol/liter DTT) followed by incubation at room temperature for 30 min. The cross-linking reaction was stopped by addition of 1 μl of 200 mm Tris-HCl, pH 7.4, and incubated for 15 min at room temperature to quench the cross-linker. After centrifugation at 11,000 × g for 10 min, the supernatants were separated by electrophoresis on a 12% Tris/glycine polyacrylamide gel followed by transfer to PVDF membrane for Western blot analysis as described previously (44).
Cell viability assay
Cellular viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Briefly, cells were plated at a density of 5 × 103 cells/well in 96-well tissue culture plates and subjected to different treatments. Following 48-h incubation at 37 °C in a humidified atmosphere containing 5% CO2, 95% air, the cells were incubated for another 4 h with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reagent. The formazan product was dissolved in DMSO and read at 570 nm on a Victor3 Multi Label plate reader (PerkinElmer Life Sciences).
Clonogenic assay
Cells subjected to different treatments were plated in 35-mm tissue culture dishes (numbers of cells varied with different cell lines to generate single colonies). Following incubation at 37 °C in a humidified atmosphere containing 5% CO2, 95% air for 12 days, cells were stained with 1% methylene blue in 50% methanol, and colonies (>30 cells) were counted.
Histology
For immunohistochemistry analysis of xenograft tumors, tumor specimens were fixed in 4% paraformaldehyde and stained with H&E. For in situ determination of cell proliferation or apoptosis, sample slides were incubated with anti-Ki67 antibody (1:200 dilution) and subjected to TUNEL.
Animal experiments
Animal maintenance and experimental procedures were approved by the Institutional Animal Care and Use Committee of Soochow University. Female severe combined immunodeficient (SCID) mice (5 weeks old, female) were inoculated subcutaneously with MCF-7/ADM cells (2 × 106 cells/mouse, mixed (1:1 volume) with BD Matrigel). Two weeks after inoculation, the tumor-bearing mice were divided into four groups (five mice per group): 1) control, 2) Adriamycin, 3) NIC3, and 4) Adriamycin + NIC3. Adriamycin (5 mg/kg) was given intraperitoneally q3d six times. NIC3 (25 mg/kg) was given via tail vein q3d six times. Tumor volumes were determined by measuring the length (L) and the width (W) of the tumors and calculating using the formula V = L × W2/2. At the end of the experiment (on day 27), the mice were euthanized, and tumors were surgically dissected. The tumor specimens were fixed in 4% paraformaldehyde for histopathologic examination. In the experimental metastasis mouse model, luciferase-labeled MDA-MB-231 human breast tumor cells (1 × 106) were injected into the tail vein of SCID mice. One week after injection, the mice were divided into four groups (five mice per group): 1) control, 2) bevacizumab, 3) NIC3, and 4) bevacizumab + NIC3. Bevacizumab (5 mg/kg) was given intraperitoneally q3d six times. NIC3 (25 mg/kg) was given via tail vein q3d for 2 weeks. Mice were imaged for luciferase expression on a weekly basis for 7 weeks via a Xenogen IVIS bioluminescence imager. Mice were injected with 5 μl/g of body weight of 30 mg/ml Luciferin-D (Gold Biotechnology, catalogue number LUCK-1G) in PBS 5 min prior to imaging. Photon flux was calculated using region of interest measurements of the ventral thoracic area for lung and bone metastases. At the experiment end point, mice were euthanized, and lung tissue was harvested for ex vivo analysis and subsequent histology.
Statistical analysis
Statistical analyses were performed using Microsoft Excel software and GraphPad Prism. The results are presented as mean ± S.D. from at least three independent experiments. The p values for comparisons between experimental groups were obtained by Student's t test. All statistical tests were two-sided. *, p < 0.05; **, p < 0.01.
Author contributions
X. W., H. Z., Y. X., Z. M., and Y. Z. formal analysis; X. W., C. J., Y. R., B. L., Z. R., B. W., and Y. Z. investigation; X. W. visualization; X. W., H. Z., Y. R., L. S., Y. X., B. L., Z. R., and B. W. methodology; C. J. and Y. Z. funding acquisition; C. J. and J. Y. writing-review and editing; Y. X. data curation; Z. M., X. R., and J. S. resources; J. Y. and Y. Z. supervision; J. Y. and Y. Z. writing-original draft; Y. S. and Y. H. chemical synthesis; L. G. and W. Z. pathology slides reviewing.
Supplementary Material
Acknowledgments
We thank Dr. Zhihong Liu from Sun Yat-sen University for providing aforementioned modeling software and Wecomput Technology for providing computation consulting.
This work was supported by National Natural Sciences Foundation of China Grants 81473240 and 81773749 (to Y. Z.) and Natural Science Foundation of Jiangsu Province of China Grant BK20151209 (to C. J.) and sponsored by the Qing Lan Project (to Y. Z.) and by a project funded by the Priority Academic Program Development of the Jiangsu Higher Education Institutes (PAPD). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9.
- NAC1
- nucleus accumbens–associated protein-1
- BTB/POZ
- bric-à-brac, tramtrack, broad complex/pox virus and Zn finger
- co-IP
- coimmunoprecipitation
- GST
- glutathione S-transferase
- DSS
- disuccinimidyl suberate
- MOE
- Molecular Operating Environment
- PP
- positive probe
- NP
- negative probe
- PARP
- poly(ADP-ribose) polymerase
- TUNEL
- terminal deoxynucleotidyltransferase dUTP nick end labeling
- H&E
- hematoxylin and eosin
- PVDF
- polyvinylidene difluoride
- q3d
- every 3 days
- ADR
- Adriamycin-resistant
- DDP
- cis-diamminedichloroplatinum(II) (cisplatin)
- SCID
- severe combined immunodeficient.
References
- 1. Futreal P. A., Kasprzyk A., Birney E., Mullikin J. C., Wooster R., and Stratton M. R. (2001) Cancer and genomics. Nature 409, 850–852 10.1038/35057046 [DOI] [PubMed] [Google Scholar]
- 2. Brivanlou A. H., and Darnell J. E. Jr. (2002) Signal transduction and the control of gene expression. Science 295, 813–818 10.1126/science.1066355 [DOI] [PubMed] [Google Scholar]
- 3. Darnell J. E., Jr. (2002) Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2, 740–749 10.1038/nrc906 [DOI] [PubMed] [Google Scholar]
- 4. Denhardt D. T. (1996) Oncogene-initiated aberrant signaling engenders the metastatic phenotype: synergistic transcription factor interactions are targets for cancer therapy. Crit. Rev. Oncog. 7, 261–291 10.1615/CritRevOncog.v7.i3-4.70 [DOI] [PubMed] [Google Scholar]
- 5. Dang C. V., Reddy E. P., Shokat K. M., and Soucek L. (2017) Drugging the 'undruggable' cancer targets. Nat. Rev. Cancer 17, 502–508 10.1038/nrc.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Otto T., Horn S., Brockmann M., Eilers U., Schüttrumpf L., Popov N., Kenney A. M., Schulte J. H., Beijersbergen R., Christiansen H., Berwanger B., and Eilers M. (2009) Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell 15, 67–78 10.1016/j.ccr.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 7. Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W. B., Fedorov O., Morse E. M., Keates T., Hickman T. T., Felletar I., Philpott M., Munro S., McKeown M. R., Wang Y., Christie A. L., et al. (2010) Selective inhibition of BET bromodomains. Nature 468, 1067–1073 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Turkson J., Zhang S., Mora L. B., Burns A., Sebti S., and Jove R. (2005) A novel platinum compound inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells. J. Biol. Chem. 280, 32979–32988 10.1074/jbc.M502694200 [DOI] [PubMed] [Google Scholar]
- 9. Siddiquee K., Zhang S., Guida W. C., Blaskovich M. A., Greedy B., Lawrence H. R., Yip M. L., Jove R., McLaughlin M. M., Lawrence N. J., Sebti S. M., and Turkson J. (2007) Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc. Natl. Acad. Sci. U.S.A. 104, 7391–7396 10.1073/pnas.0609757104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu L., and Russu W. A. (2013) Molecular docking and synthesis of novel quinazoline analogues as inhibitors of transcription factors NF-κB activation and their anti-cancer activities. Bioorg. Med. Chem. 21, 540–546 10.1016/j.bmc.2012.10.051 [DOI] [PubMed] [Google Scholar]
- 11. Nakayama K., Nakayama N., Davidson B., Sheu J. J., Jinawath N., Santillan A., Salani R., Bristow R. E., Morin P. J., Kurman R. J., Wang T. L., and Shih I.-M. (2006) A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proc. Natl. Acad. Sci. U.S.A. 103, 18739–18744 10.1073/pnas.0604083103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perez-Torrado R., Yamada D., and Defossez P. A. (2006) Born to bind: the BTB protein-protein interaction domain. BioEssays 28, 1194–1202 10.1002/bies.20500 [DOI] [PubMed] [Google Scholar]
- 13. Yeasmin S., Nakayama K., Rahman M. T., Rahman M., Ishikawa M., Katagiri A., Iida K., Nakayama N., Otuski Y., Kobayashi H., Nakayama S., and Miyazaki K. (2012) Biological and clinical significance of NAC1 expression in cervical carcinomas: a comparative study between squamous cell carcinomas and adenocarcinomas/adenosquamous carcinomas. Hum. Pathol. 43, 506–519 10.1016/j.humpath.2011.05.021 [DOI] [PubMed] [Google Scholar]
- 14. Nakayama K., Rahman M. T., Rahman M., Yeasmin S., Ishikawa M., Katagiri A., Iida K., Nakayama N., and Miyazaki K. (2010) Biological role and prognostic significance of NAC1 in ovarian cancer. Gynecol. Oncol. 119, 469–478 10.1016/j.ygyno.2010.08.031 [DOI] [PubMed] [Google Scholar]
- 15. Shih I.-M., Nakayama K., Wu G., Nakayama N., Zhang J., and Wang T. L. (2011) Amplification of the ch19p13.2 NACC1 locus in ovarian high-grade serous carcinoma. Mod. Pathol. 24, 638–645 10.1038/modpathol.2010.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakayama K., Nakayama N., Wang T. L., and Shih I.-M. (2007) NAC-1 controls cell growth and survival by repressing transcription of Gadd45GIP1, a candidate tumor suppressor. Cancer Res. 67, 8058–8064 10.1158/0008-5472.CAN-07-1357 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Cheng Y., Ren X., Zhang L., Yap K. L., Wu H., Patel R., Liu D., Qin Z. H., Shih I. M., and Yang J. M. (2012) NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene 31, 1055–1064 10.1038/onc.2011.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y., Cheng Y., Ren X., Hori T., Huber-Keener K. J., Zhang L., Yap K. L., Liu D., Shantz L., Qin Z. H., Zhang S., Wang J., Wang H. G., Shih I.-M., and Yang J. M. (2012) Dysfunction of nucleus accumbens-1 activates cellular senescence and inhibits tumor cell proliferation and oncogenesis. Cancer Res. 72, 4262–4275 10.1158/0008-5472.CAN-12-0139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yap K. L., Fraley S. I., Thiaville M. M., Jinawath N., Nakayama K., Wang J., Wang T. L., Wirtz D., and Shih I.-M. (2012) NAC1 is an actin-binding protein that is essential for effective cytokinesis in cancer cells. Cancer Res. 72, 4085–4096 10.1158/0008-5472.CAN-12-0302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y., Ren Y. J., Guo L. C., Ji C., Hu J., Zhang H. H., Xu Q. H., Zhu W. D., Ming Z. J., Yuan Y. S., Ren X., Song J., and Yang J. M. (2017) Nucleus accumbens-associated protein-1 promotes glycolysis and survival of hypoxic tumor cells via the HDAC4-HIF-1α axis. Oncogene 36, 4171–4181 10.1038/onc.2017.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ueda S. M., Yap K. L., Davidson B., Tian Y., Murthy V., Wang T. L., Visvanathan K., Kuhajda F. P., Bristow R. E., Zhang H., and Shih I.-M. (2010) Expression of fatty acid synthase depends on NAC1 and is associated with recurrent ovarian serous carcinomas. J. Oncol. 2010, 285191 10.1155/2010/285191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishibashi M., Nakayama K., Yeasmin S., Katagiri A., Iida K., Nakayama N., Fukumoto M., and Miyazaki K. (2008) A BTB/POZ gene, NAC-1, a tumor recurrence-associated gene, as a potential target for Taxol resistance in ovarian cancer. Clin. Cancer Res. 14, 3149–3155 10.1158/1078-0432.CCR-07-4358 [DOI] [PubMed] [Google Scholar]
- 23. Jinawath N., Vasoontara C., Yap K. L., Thiaville M. M., Nakayama K., Wang T. L., and Shih I. M. (2009) NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene 28, 1941–1948 10.1038/onc.2009.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agashe V. R., Shastry M. C., and Udgaonkar J. B. (1995) Initial hydrophobic collapse in the folding of barstar. Nature 377, 754–757 10.1038/377754a0 [DOI] [PubMed] [Google Scholar]
- 25. Lins L., and Brasseur R. (1995) The hydrophobic effect in protein folding. FASEB J. 9, 535–540 10.1096/fasebj.9.7.7737462 [DOI] [PubMed] [Google Scholar]
- 26. Kubota H. (2009) Quality control against misfolded proteins in the cytosol: a network for cell survival. J. Biochem. 146, 609–616 10.1093/jb/mvp139 [DOI] [PubMed] [Google Scholar]
- 27. Szakács G., Paterson J. K., Ludwig J. A., Booth-Genthe C., and Gottesman M. M. (2006) Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 5, 219–234 10.1038/nrd1984 [DOI] [PubMed] [Google Scholar]
- 28. Miller K., Wang M., Gralow J., Dickler M., Cobleigh M., Perez E. A., Shenkier T., Cella D., and Davidson N. E. (2007) Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N. Engl. J. Med. 357, 2666–2676 10.1056/NEJMoa072113 [DOI] [PubMed] [Google Scholar]
- 29. Ebos J. M., Lee C. R., Cruz-Munoz W., Bjarnason G. A., Christensen J. G., and Kerbel R. S. (2009) Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell 15, 232–239 10.1016/j.ccr.2009.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pàez-Ribes M., Allen E., Hudock J., Takeda T., Okuyama H., Viñals F., Inoue M., Bergers G., Hanahan D., and Casanovas O. (2009) Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell 15, 220–231 10.1016/j.ccr.2009.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Conley S. J., Gheordunescu E., Kakarala P., Newman B., Korkaya H., Heath A. N., Clouthier S. G., and Wicha M. S. (2012) Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc. Natl. Acad. Sci. U.S.A. 109, 2784–2789 10.1073/pnas.1018866109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim J., Chu J., Shen X., Wang J., and Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 10.1016/j.cell.2008.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malleshaiah M., Padi M., Rué P., Quackenbush J., Martinez-Arias A., and Gunawardena J. (2016) Nac1 coordinates a sub-network of pluripotency factors to regulate embryonic stem cell differentiation. Cell Rep. 14, 1181–1194 10.1016/j.celrep.2015.12.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu T., He P., Wu W., Chen Y., and Lv F. (2018) Targeting oncogenic transcriptional corepressor Nac1 POZ domain with conformationally constrained peptides by cyclization and stapling. Bioorg. Chem. 80, 1–10 10.1016/j.bioorg.2018.05.024 [DOI] [PubMed] [Google Scholar]
- 35. Baud V., and Karin M. (2009) Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 8, 33–40 10.1038/nrd2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dang C. V. (2012) MYC on the path to cancer. Cell 149, 22–35 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Piunti A., Hashizume R., Morgan M. A., Bartom E. T., Horbinski C. M., Marshall S. A., Rendleman E. J., Ma Q., Takahashi Y. H., Woodfin A. R., Misharin A. V., Abshiru N. A., Lulla R. R., Saratsis A. M., Kelleher N. L., et al. (2017) Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 23, 493–500 10.1038/nm.4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Overman J., Fontaine F., Moustaqil M., Mittal D., Sierecki E., Sacilotto N., Zuegg J., Robertson A. A., Holmes K., Salim A. A., Mamidyala S., Butler M. S., Robinson A. S., Lesieur E., Johnston W., et al. (2017) Pharmacological targeting of the transcription factor SOX18 delays breast cancer in mice. eLife 6, e21221 10.7554/eLife.21221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grimley E., Liao C., Ranghini E. J., Nikolovska-Coleska Z., and Dressler G. R. (2017) Inhibition of Pax2 transcription activation with a small molecule that targets the DNA binding domain. ACS Chem. Biol. 12, 724–734 10.1021/acschembio.6b00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rath K. S., Naidu S. K., Lata P., Bid H. K., Rivera B. K., McCann G. A., Tierney B. J., Elnaggar A. C., Bravo V., Leone G., Houghton P., Hideg K., Kuppusamy P., Cohn D. E., and Selvendiran K. (2014) HO-3867, a safe STAT3 inhibitor, is selectively cytotoxic to ovarian cancer. Cancer Res. 74, 2316–2327 10.1158/0008-5472.CAN-13-2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qi J., Dong Z., Liu J., Peery R. C., Zhang S., Liu J. Y., and Zhang J. T. (2016) Effective targeting of the survivin dimerization interface with small-molecule inhibitors. Cancer Res. 76, 453–462 10.1158/0008-5472.CAN-15-1874 [DOI] [PubMed] [Google Scholar]
- 42. Vilaboa N., Boré A., Martin-Saavedra F., Bayford M., Winfield N., Firth-Clark S., Kirton S. B., and Voellmy R. (2017) New inhibitor targeting human transcription factor HSF1: effects on the heat shock response and tumor cell survival. Nucleic Acids Res. 45, 5797–5817 10.1093/nar/gkx194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown N. (2014) Bioisosteres and scaffold hopping in medicinal chemistry. Mol. Inform. 33, 458–462 10.1002/minf.201400037 [DOI] [PubMed] [Google Scholar]
- 44. Majerciak V., Pripuzova N., Chan C., Temkin N., Specht S. I., and Zheng Z. M. (2015) Stability of structured Kaposi's sarcoma-associated herpesvirus ORF57 protein is regulated by protein phosphorylation and homodimerization. J. Virol. 89, 3256–3274 10.1128/JVI.03721-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.