Abstract
Caldicellulosiruptor bescii is an extremely thermophilic, cellulolytic bacterium with a growth optimum at 78 °C and is the most thermophilic cellulose degrader known. It is an attractive target for biotechnological applications, but metabolic engineering will require an in-depth understanding of its primary pathways. A previous analysis of its genome uncovered evidence that C. bescii may have a completely uncharacterized aspect to its redox metabolism, involving a tungsten-containing oxidoreductase of unknown function. Herein, we purified and characterized this new member of the aldehyde ferredoxin oxidoreductase family of tungstoenzymes. We show that it is a heterodimeric glyceraldehyde-3-phosphate (GAP) ferredoxin oxidoreductase (GOR) present not only in all known Caldicellulosiruptor species, but also in 44 mostly anaerobic bacterial genera. GOR is phylogenetically distinct from the monomeric GAP-oxidizing enzyme found previously in several Archaea. We found that its large subunit (GOR-L) contains a single tungstopterin site and one iron-sulfur [4Fe-4S] cluster, that the small subunit (GOR-S) contains four [4Fe-4S] clusters, and that GOR uses ferredoxin as an electron acceptor. Deletion of either subunit resulted in a distinct growth phenotype on both C5 and C6 sugars, with an increased lag phase, but higher cell densities. Using metabolomics and kinetic analyses, we show that GOR functions in parallel with the conventional GAP dehydrogenase, providing an alternative ferredoxin-dependent glycolytic pathway. These two pathways likely facilitate the recycling of reduced redox carriers (NADH and ferredoxin) in response to environmental H2 concentrations. This metabolic flexibility has important implications for the future engineering of this and related species.
Keywords: glycolysis, cellulose, metabolism, metabolomics, metalloenzyme, oxidation-reduction (redox), anaerobic glycolysis, iron-sulfur protein, pterin, tungsten
Introduction
Caldicellulosiruptor bescii is an extremely thermophilic bacterium with an optimal growth temperature of 78 °C, making it the most thermophilic cellulose degrader identified to date (1–3). This organism is an attractive target for use in biotechnological applications due to its ability to ferment high concentrations of unpretreated plant biomass, thereby potentially mitigating the costly thermochemical pretreatment steps that are typically required for lignocellulose conversion (4–6). C. bescii and other species of the Caldicellulosiruptor genus do not contain the well-characterized cellulosomes described for other cellulolytic bacteria, such as Clostridium thermocellum, but rather secrete a suite of multidomain noncellulosomal glycosyl hydrolases that work synergistically to convert plant biomass into fermentable sugars (5, 7). C. bescii ferments C6 and C5 sugars derived from cellulose and hemicellulose primarily to hydrogen gas, lactate, acetate, and carbon dioxide (8, 9). The development of a genetic system for this organism has created the potential to metabolically-engineer it for consolidated biomass processing to produce liquid fuels and other bio-based products from renewable biomass. Engineered strains of C. bescii that can produce moderate amounts of ethanol directly from the conversion of plant biomass have already been achieved (10, 11). The recent developments of a high temperature kanamycin-resistance gene and of an inducible promoter have further increased the potential use of this organism as a platform for biotechnological applications (12–15). The further development of efficient metabolic engineering strategies will require an in-depth understanding of its primary metabolism.
We recently reported that C. bescii has a completely uncharacterized aspect to its primary redox metabolism involving the metal tungsten (16). This was very surprising; tungsten is an element that is rarely utilized in biological systems, as virtually all forms of life utilize the chemically analogous element molybdenum (17). Molybdoenzymes are ubiquitous, serving roles in carbon, nitrogen, and sulfur metabolism (18). In contrast, the number of microorganisms known to require tungsten is relatively few (17). Tungsten is chemically very similar to molybdenum with comparable coordination chemistries and physical properties. They are both coordinated to enzymes via pyranopterin cofactors and they both stabilize three redox states (IV, V, and VI) in the biological range, although for tungsten these are at much lower reduction potentials than those of molybdenum (19). Despite this, there is a large bias toward molybdoenzymes in all three domains of life, whereas tungstoenzymes appear to be more abundant in thermophilic organisms (20).
Tungsten metabolism is best characterized in hyperthermophilic Archaea, represented by Pyrococcus furiosus, which grows optimally at 100 °C. It contains five members of the aldehyde oxidoreductase (AOR)3 family of tungstoenzymes, which are phylogenetically unrelated to any of the three families of molybdoenzymes (21). All five have been characterized and they are referred to as aldehyde ferredoxin oxidoreductase (AOR) (22), glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) (23), and formaldehyde ferredoxin oxidoreductase (FOR) (24), together with two tungstoenzymes of unknown function, WOR4 (25) and WOR5 (26). All of these enzymes oxidize aldehydes of various types. The prototypical AOR has a broad substrate specificity and is thought to be involved in peptide catabolism wherein it oxidizes amino acid-derived aldehydes (17). However, of all of these archaeal tungstoenzymes only the function of GAPOR is definitively understood (23). GAPOR replaces the conventional NAD-dependent glyceraldehyde-3-phosphate (GAP) dehydrogenase (GAPDH) and is part of a modified glycolytic pathway in Pyrococcus furiosus and several other archaeal species (23, 27, 28). In addition to using ferredoxin rather than NAD as an electron acceptor, the GAPOR reaction oxidizes GAP directly to 3-phosphoglycerate rather than to the 1,3-bisphosphoglycerate generated by GAPDH (23). Hence, this unusual GAPOR-containing glycolytic pathway found in these Archaea does not contain phosphoglycerate kinase and consequently GAP oxidation does not result in net ATP synthesis by substrate level phosphorylation.
The notion of C. bescii utilizing tungsten came from the identification of a 12-gene cluster of highly expressed genes (Athe_0820-Athe_0831) (Fig. S1) that are completely conserved across all 14 members of the Caldicellulosiruptor genus that have been characterized to date (29). These genes encode proteins involved in pyranopterin synthesis and tungstate transport, and a putative tungstopterin-containing enzyme (Athe_0821) belonging to the AOR family of tungstoenzymes that we termed XOR to signify its uncharacterized status. To demonstrate that C. bescii could utilize tungsten, the known tungstoenzyme AOR of P. furiosus was heterologously produced in an active form (16). Additionally, we showed that during growth on simple and complex C6 and C5 sugars, Athe_0821 is constitutively expressed and its protein product represents ∼2% of the cytoplasmic protein in C. bescii. XOR is phylogenetically distinct from the characterized AOR enzymes and was proposed to represent a new member of this family of tungstoenzymes (16). XOR is also unusual in that the gene encoding Athe_0821 is adjacent to another highly-expressed gene (Athe_0820) encoding a polyferredoxin-like protein predicted to contain four [4Fe-4S] clusters. It was not known if this functioned as an electron carrier for XOR or was an additional subunit.
It was therefore of importance to elucidate the function of this tungstoenzyme in the Caldicellulosiruptor genus, both in terms of understanding its primary metabolism and in utilizing this information to engineer these organisms for biotechnological applications. In contrast to the archaeon P. furiosus, C. bescii does not grow on peptides and its genome encodes the standard Embden-Meyerhof (EM) pathway, including a classical phosphorylating GAPDH (Athe_1406). Moreover, cell-free extracts of C. bescii did not catalyze the oxidation of any of the aldehydes, including GAP, used by the AOR-type enzymes of P. furiosus (16). Herein we show that XOR is indeed a new member of the AOR family of tungstoenzymes and is a novel bacterial-type of glyceraldehyde-3-phosphate ferredoxin oxidoreductase. We will refer to it as GOR to differentiate it from the phylogenetically-distinct GAPOR found primarily in Archaea. Affinity-tagged GOR was purified and kinetically characterized. Utilizing a deletion mutant strain lacking GOR, metabolomics was used to show that GOR provides C. bescii with an alternative glycolytic pathway, generating reduced ferredoxin rather than NADH. This is the first report of a functional “GAPOR” in a bacterium.
Results
Purification of affinity-tagged GOR
The genes encoding XOR (Athe_0821) and the putative polyferredoxin (PFD, Athe_0820) were modified by the inclusion of a polyhistidine tag at the N terminus of XOR and the expression of both genes was placed under control of the high level promoter of the S-layer protein (slp) to give the overexpression strain (OE-XOR/PFD, Fig. S1). The slp gene is expressed at a level that is ∼10-fold higher than that of Athe_0821 and of Athe_0820 in WT C. bescii (16). XOR was purified from the cytoplasmic extract of the OE-XOR/PFD strain by ion exchange, affinity, and size exclusion chromatography and its properties are described below. The first priority was to determine a catalytic activity for the purified enzyme. XOR was assayed for its ability to oxidize a variety of aldehydes and reduce the artificial electron acceptor benzyl viologen (BV). These included the following compounds: 2-ethylhexanal, 2-methylbutyraldehyde, 2-methylpentanal, 2-methoxybenzaldehyde, 2-naphthaldehyde, 2-phenylpropionaldehyde, 3-phenylbutyraldehyde, 3,4-dihydroxybenzaldehyde, acetaldehyde, acetoin, arabinose, butyraldehyde, cinnamaldehyde, coniferaldehyde, dihydroxyacetone, galactose, glucose, glucose 6-phosphate, glyceraldehyde, glycoaldehyde, glycolic acid, glyoxal, glyoxylic acid, hexanal, isobutyraldehyde, mannose, methyl glyoxal, phenylpropionaldehyde, ribose, sinapaldehyde, syringaldehyde, vanillin, and xylose. However, none were utilized as a substrate by XOR. We had previously shown that a cytoplasmic extract of WT C. bescii was unable to oxidize several other aldehydes that are known substrates of the AOR family members in the Archaea (16). These included formaldehyde, propionaldehyde, crotonaldehyde, glutaraldehyde, isovaleraldehyde, benzaldehyde, and glyceraldehyde 3-phosphate (GAP). Similarly, purified XOR was unable to use these substrates, with the notable exception of GAP. Surprisingly, GAP (1 mm) was readily oxidized by purified XOR with a specific activity (at 70 °C) of 48.3 units/mg and the activity increased with increasing concentrations of XOR in the assay (Table S1). However, contrary to expectation, only a very low level of GAP oxidation activity was detected in the cytoplasmic extract of the OE-XOR/PFD strain (0.01 units/mg). As previously reported, the same was also observed with extracts from the WT strain (< 0.01 unit/mg). Because the purified XOR enzyme clearly utilizes GAP as a substrate, we will now refer to XOR as a GAP oxidoreductase or GOR.
The lack of significant benzyl viologen (BV)-linked GAP-oxidation activity in the cytoplasmic extract of the WT strain appeared to be due to the presence of an inhibitor of GOR activity. For example, as shown in Table 1, the total GAP-oxidizing units increased ∼4-fold following the affinity purification of GOR. This was accompanied by a 76-fold purification yielding 2.1 mg of purified protein from 50 g (wet weight) of cells. A similar response to the partial purification from the cytoplasmic fraction was observed with GAPDH, where a greater than 100-fold increase in the NAD-dependent oxidation of GAP was observed after an anion exchange chromatography step (Fig. S2). The addition of the cytoplamic extract to purified GOR resulted in a ∼90% reduction in its GAP-oxidizing activity (Fig. 1), indicating that the cytosol contains an inhibitory factor. To investigate the nature of this inhibitory factor, the cytoplasmic extract from the OE-XOR/PFD strain was fractionated using a 30-kDa filter (and the protein fraction <30 kDa was washed with buffer). The washed extract (nominally >30 kDa) and the resulting filtrate (<30 kDa) were each tested for their capacity to inhibit the GAP-oxidizing activity of purified GOR (Fig. 1). The inhibitor appeared only in the high molecular mass fraction (>30 kDa) as the low molecular mass fraction (<30 kDa) did not have any inhibitory effect on GOR (Fig. 1). Moreover, heat treatment (95 °C for 60 min) of the washed cytoplasmic extract eliminated the inhibitory effect, indicating that the inhibitor is likely to be a protein.
Table 1.
Affinity purification of His-tagged GOR
| Step | Protein | Volume | Activity | Specific activity | Purification | Yield |
|---|---|---|---|---|---|---|
| mg | ml | units | units/mg | -fold | % | |
| S100 | 3361.9 | 235 | 33.6 | 0.01 | ||
| Q-HP | 518.1 | 370 | 339 | 0.7 | 1 | 100 |
| Nickel-nitrilotriacetic acid | 2.9 | 2.4 | 144 | 50.1 | 76 | 43 |
| Sephadex 200 | 2.1 | 2.1 | 101 | 48.3 | 74 | 30 |
Figure 1.
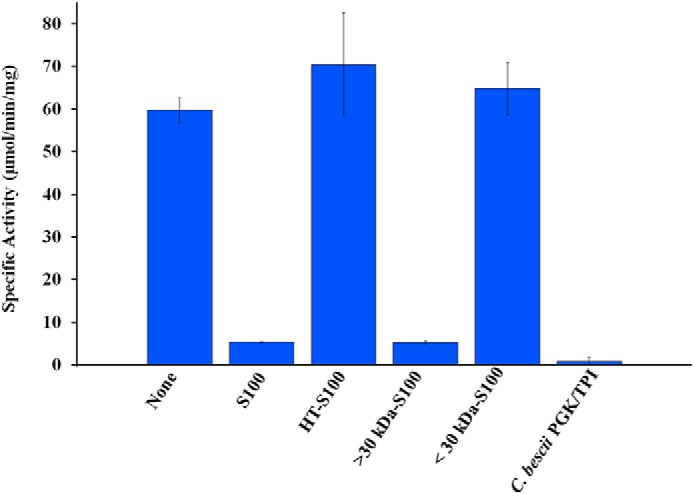
The activity of purified GOR (0.006 mg/ml) was measured in the presence of cytoplasmic extract (S100, 0.174 mg/ml), heat-treated (95 °C for 60 min) cytoplasmic extract (HT-S100, 0.036 mg/ml), cytoplasmic extract washed using a 30-kDa membrane (> 30 kDa-S100, 0.325 mg/ml), and the flow-through material that passed through the 30-kDa membrane (<30 kDa-S100, protein was not measured), or partially purified PGK/TPI (C. bescii PGK/TPI, 0.002 mg/ml). The concentrations given in parentheses are the final concentration of each component in the assay cuvette.
Based on its ability to inhibit purified GOR activity, the inhibiting factor was purified from the flow-through fraction of the GOR affinity purification step by hydrophobic interaction chromatography. Analysis of the purified inhibitor fraction by LC-tandem MS (LC-MS/MS) (Proteomics and Mass Spectrometry Facility, University of Georgia) identified a 70-kDa bifunctional phosphoglycerate kinase/triose-phosphate isomerase (PGK/TPI) fusion protein (Athe_1405). Addition of the purified PGK/TPI protein to the GOR assay virtually abolished GAP-oxidizing activity (Fig. 1).
TPI catalyzes the interconversion of GAP and dihydroxyacetone phosphate (DHAP) with a strong bias in favor of DHAP (∼95:5) (30). Hence, TPI activity is responsible for the inhibition of GOR and GAPDH in cytoplasmic extracts by a “substrate-stealing” mechanism and thus its separation from these enzymes during their purification results in a dramatic increase in their GAP-dependent activities. The presence of the PGK/TPI protein in the standard GOR assay enabled reduction of BV to be measured in the presence of DHAP (which generates GAP) in the absence of added GAP (Table S2). Likewise, the presence of PGK/TPI in the standard GAPDH assay enabled the reduction of NAD if DHAP (rather than GAP) was added (Table S2). Interestingly, DHAP itself partially inhibits GOR activity in the absence of PGK/TPI and, conversely, increases GAPDH activity in the absence of PGK/TPI (Table S2). GOR activity is reduced by 70% in the presence of 10 mm DHAP in the absence of TPI, but only decreases by 22% in the presence of 1 mm DHAP. In contrast, GAPDH activity increased 4-fold in the presence of 10 mm DHAP. Notably, TPI inhibition of both can be overcome by sufficiently high concentrations of GAP (5 and 15 mm) as measured by the initial activities (Table S2). However, in all cases where PGK/TPI was present, activity diminished within 15 s, compared with >30 s in the standard assays.
A kinetic analysis of purified GOR with varying concentrations of GAP revealed a linear increase in activity, with no saturation observed up to 20 mm GAP with BV (1 mm) as the electron acceptor. Although these data precludes the determination of Km and kcat values, kcat/Km was determined as the slope of the plot of activity versus substrate concentration to be 4.7 × 104 s−1 m−1 at 70 °C (Fig. S3A). However, with C. bescii ferredoxin as the electron carrier, saturation kinetics were observed, with an estimated kcat value of 5.57 s−1 and a Km value for oxidized Fd of 38.9 ± 10.7 μm at 70 °C (using 5 mm GAP) (Fig. S4). Like GOR, partially purified GAPDH also showed a linear increase in activity with increasing GAP concentrations utilizing NAD (1 mm) as the electron acceptor, with no saturation observed at GAP concentrations up to 20 mm (Fig. S3B).
Size exclusion chromatography of affinity purified GOR showed that GOR eluted as two distinct symmetrical peaks that had the same specific activities with approximate molecular masses of 315 and 73 kDa (Fig. S5). SDS-PAGE revealed identical profiles for the peaks with two major bands near 60 and 15 kDa. These corresponded to the expected gene products of Athe_0821 (65,916 Da) and Athe_0820 (14,618 Da), which will now be referred to as the large (GOR-L) and small (GOR-S) subunits of GOR. The identity of the two subunits was confirmed by peptide analysis using LC-MS (Fig. S6). We propose that the two peaks from the size exclusion chromatography column represent the holoenzyme (L4S4), which is a tetramer of heterodimers, and the dissociated heterodimer (LS, Fig. S5). GOR-L has 29% sequence identity (47% similarity) to P. furiosus GAPOR (which is 67 amino acids larger) and is 31% identical (48% similar) to the structurally-characterized AOR of P. furiosus, which contains one tungstopterin site and one [4Fe-4S] cluster. The cluster coordinating cysteine residues and the tungstopterin-binding residues of AOR are conserved in GOR-L and GAPOR (although they lack the residues that coordinate the monomeric iron atom in AOR, see Fig. S7). GOR-S, annotated as a putative polyferredoxin, contains 16 cysteinyl residues in four motifs and these are predicted to coordinate four [4Fe-4S] clusters (Fig. S7) (31). Metal analysis of purified GOR yielded an iron to tungsten ratio (Fe/W) of 20.4:1, consistent with one and four [4Fe-4S] clusters in GOR-L and GOR-S, respectively, together with a monomeric tungstopterin site in GOR-L.
Deletion of the genes encoding GOR
To assess the functional importance of GOR, mutants were generated by individually disrupting gorL (Athe_0821) and gorS (Athe_0820) by replacing each with the gene encoding kanamycin resistance (Cbhtk) (Fig. S1) to generate the mutant strains ΔgorL and ΔgorS (Table S3). As shown in Fig. 2, compared with the parent, growth on xylose (C5) of both mutant strains exhibited a longer lag phase. Although the exponential growth rates were comparable (Figs. 2 and Fig. S8), both mutant strains reached a significantly higher cell density in stationary phase, up to 50% higher than that of the parent. Similar results were obtained during growth on other sugars such as: glucose, fructose, mannose, galactose, rhamnose, arabinose, and lactose (Fig. S8).
Figure 2.
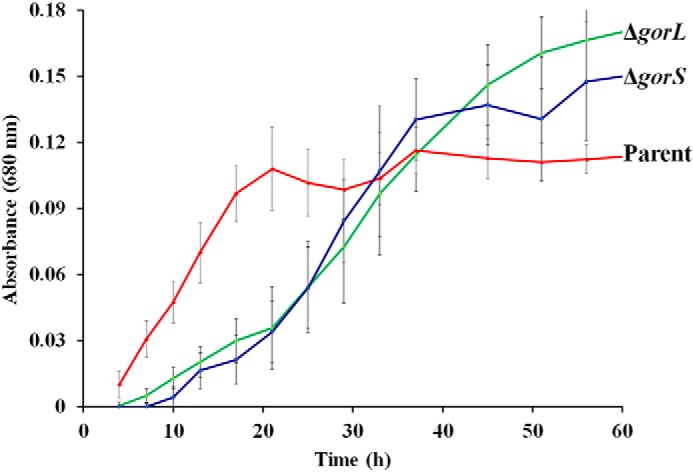
Growth of parent strain (red) and the ΔgorL (green) and ΔgorS (blue) strains using xylose as the carbon source at 75 °C (n = 3).
Metabolomic analyses of the ΔgorL strain
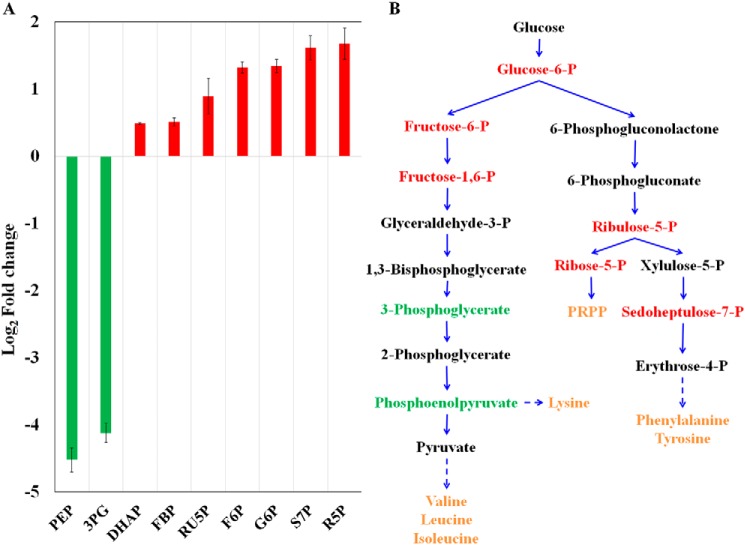
To provide further insight into the metabolic role of GOR, MS-based metabolomics were employed to evaluate changes in the concentrations of key metabolites that result from the deletion of gorL. A total of 19 intermediary metabolites were found to have significant changes in concentration in the ΔgorL strain relative to the parent strain (Table S4), the most notable of which were intermediates of the glycolytic and pentose phosphate pathways. As shown in Fig. 3, concentrations of glucose 6-phosphate, fructose 6-phosphate, and fructose 1,6-bisphosphate were higher in the in the ΔgorL strain, whereas those of 3-phosphoglycerate and phosphoenolpyruvate were lower, consistent with a bottleneck in the glycolytic pathway at the GAP oxidation step.
Figure 3.
A, relative concentrations of intracellular metabolites in the ΔgorL strain relative to the parent strain as indicated by the log2 fold-change. Error bars represent the mean ± S.D. (n = 3 replicates). B, glycolytic and pentose phosphate pathways in C. bescii with relative abundance of key metabolites. Those with higher abundance in the mutant strain are shown in red, whereas those with lower abundance are shown in green. The further metabolism of intermediates of the pathways to amino acids and nucleotide precursors are indicated in orange. Abbreviations: PEP, phosphoenolpyruvate; 3PG, 3-phosphoglycerate; FBP, fructose-1,6-phosphate; RU5P, ribulose 5-phosphate; F6P, fructose 6-phosphate; G6P, glucose 6-phosphate; S7P, sedoheptulose 7-phosphate; R5P, ribose 5-phosphate; PPRP, phosphoribosyl pyrophosphate.
Phylogenetic analyses of GOR
A phylogenetic tree of the AOR family of enzymes was constructed using 4,063 homologs of the GOR-L sequence based on the InterPro domains (IPR013983 and IPR001203). Although the five previously characterized types of tungsten-containing oxidoreductases, AOR, FOR, GAPOR, WOR4, and WOR5, are all from Archaea, such as P. furiosus, the AOR family is also well-represented in the bacterial domain with 64% of the sequences from Bacteria and 30% from Archaea, with the remaining (6%) from uncharacterized metagenomes or environmental samples (Fig. S9). The five types of previously characterized members of the AOR family from P. furiosus form distinct clades that are well-separated from each other and C. bescii GOR is clearly more closely related to GAPOR than it is to any of the other enzymes, sharing a more recent common ancestor. Nevertheless, the GOR and GAPOR branches also subdivide into bacterial and archaeal subclades (Fig. S9B). Hence, the phylogeny supports distinctive terms for GOR and GAPOR, especially as they are found predominantly in Bacteria and Archaea, respectively.
Discussion
Herein we describe the first glyceraldehyde-3-phosphate ferredoxin oxidoreductase from a bacterium and designate it GOR to show that is it is distinct from the previously characterized GAPOR from Archaea (16). We also demonstrate that GOR is directly involved in the glycolytic pathway of C. bescii. This result was unexpected because the genome of C. bescii encodes the standard pathway that includes the classical phosphorylating GAPDH. This catalyzes the conversion of GAP to 1,3-bisphosphoglycerate, which is then used to conserve energy in the form of ATP by PGK generating 3-phosphoglycerate. Note that GOR oxidizes GAP directly to 3-phosphoglycerate and so ATP is not synthesized in the GOR-dependent glycolytic pathway. Both GAPDH and PGK/TPI enzymes were partially purified in this study, indicating that the enzymes are present at substantial concentrations in the cell. However, the results of our genetic and metabolomics-based studies clearly indicate an important glycolytic role for GOR under the usual growth conditions. Moreover, deletion of either of the genes encoding GOR affects the efficient oxidation of GAP, as evident from the accumulation of metabolic intermediates upstream of this step in the glycolytic pathway and depletion of intermediates downstream (Fig. 3).
Lack of saturation kinetics for both GOR and GAPDH is a highly unusual feature for enzymes of this type. Although there are other examples of nonsaturable enzymes (e.g. AhpC from Helicobacter pylori) this phenomenon is rare (32). In the case of GOR, the order of kcat/Km (4.7 × 104 s−1 m−1) indicates that the lack of saturation is not because the enzyme is diffusion limited. Instead, it likely has an extremely low affinity for GAP but with a high turnover rate, such that formation of the [ES] complex is slow, but turnover occurs rapidly upon complex formation. Nonsaturation kinetics is even more surprising for GAPDH. This enzyme has been extremely well-studied from many organisms with Km values for GAP ranging from 20 μm to 1.4 mm (33, 34). However, in an organism like C. bescii with two competing primary metabolic pathways for GAP oxidation, it is not unreasonable that the two GAP-oxidizing enzymes have low affinities for the substrate as a means of regulating substrate flux. It is also possible that another component is necessary in the assay for these enzymes to be saturable with GAP, as in the case with human ADH5 (class III alcohol dehydrogenase), which cannot be saturated with ethanol (up to 3 m) in the absence of activation by fatty acids (35).
We show here that both GAPDH and GOR activities are masked by the presence of a highly active TPI, which is biased toward DHAP formation thereby maintaining only a relatively low concentration of GAP that is available to both enzymes. The high TPI activity might play a unique thermoprotective role by minimizing the pool of the thermally-labile glycolytic intermediate GAP by reversibly converting it to the more thermostable DHAP, thus regulating the glycolytic flux through GOR and GAPDH (36). This is in contrast to the situation in some Archaea where GAPOR is the sole GAP-oxidizing enzyme in a modified EM pathway generating 3-phosphoglycerate in a single irreversible step, where GAPDH serves solely in a gluconeogenic role (23, 37, 38). The concurrent presence of GOR and GAPDH activity suggest that in C. bescii both enzymes operate in parallel. The operation of parallel pathways in glucose degradation is unusual, although not without precedent in other thermophilic microorganisms. For example, the archaeon Thermoproteus tenax has been demonstrated to utilize modified versions of both EMP and ED pathways (39–42).
The inhibition of the activity of GOR by DHAP, although GAPDH appears to be activated by DHAP (Table S2), was unanticipated. However, it does explain our inability to measure significant GAP-dependent oxidoreductase activity (BV-dependent) in cytoplasmic extracts previously. That DHAP is a regulator of both GOR and GAPDH cannot be ruled out as it could potentially activate GAPDH, whereas inhibiting GOR, but further study will be required to clarify this, especially because high and likely nonphysiological concentrations of DHAP (10 mm) were required to observe significant effects.
If the inhibition of GAPDH activity in cytosolic extracts results from PGK/TPI activity, it would be expected that this effect should have been observed in the purification of GAPDH from other organisms where TPI is present. This situation in C. bescii is extremely unusual as shown by comparison with a related anaerobic fermentative thermophile, Thermotoga maritima (Topt 80 °C). Like C. bescii, T. maritima also contains a PGK/TPI fusion protein but its GAPDH behaves like a conventional enzyme. It shows no inhibition in cytoplasmic extracts and has been purified by conventional chromatography with no significant increases in activity (43). Notably, T. maritima lacks GOR. Thus, we hypothesize that the PGK/TPI effect is related to the presence of GOR in C. bescii. Similarly, this effect may be related to the lack of saturation kinetics discussed above as a means of regulating the dynamic pathways for GOR oxidation present in this organism.
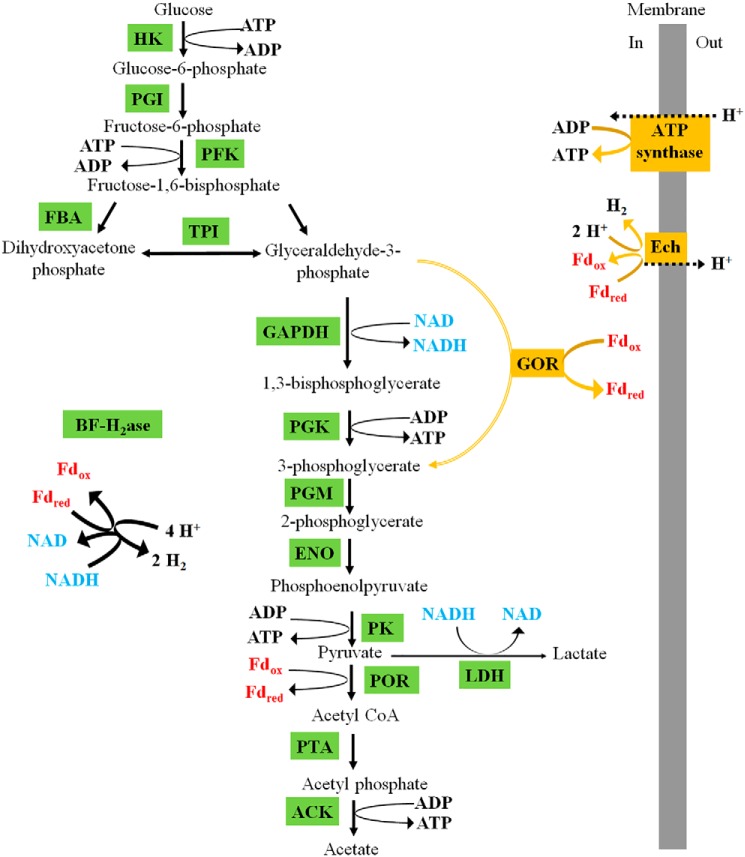
We speculate that the role of GOR in C. bescii and other Caldicellulosiruptor species is to serve as an alternative glycolytic pathway (Fig. 4) that is used by the cell to regulate carbon and electron flux during growth under conditions in which C. bescii is unable to efficiently recycle its electron carriers. This might be particularly important when C. bescii is exposed to excess carbon loading, such as during growth on unprocessed biomass (9). For example, the genome of C. bescii encodes two hydrogenases, one using only reduced ferredoxin as the electron donor, whereas the other simultaneously uses both reduced ferredoxin and NADH. The latter is a cytoplasmic ferredoxin/NADH-dependent bifurcating [FeFe] hydrogenase that is the primary source of H2 production during growth on glucose-based substrates (44). The former is a membrane-bound ferredoxin-dependent [NiFe] hydrogenase (Ech) that can generate a proton gradient across the cell membrane (45). The standard EM pathway utilizing the classic phosphorylating GAPDH is likely the primary glycolytic route used by C. bescii as this pathway produces ATP. Although ferredoxin-dependent conversion of GAP to 3-phosphoglycerate by GOR yields less ATP than the standard EM pathway, it allows glycolysis to proceed with only ferredoxin as the redox carrier. This can be recycled via H2 production by Ech, avoiding the potential generation of excess NADH. This hypothesis is supported by the finding that genes necessary for the biosynthesis of GOR are up-regulated in the closely-related Caldicellulosiruptor saccharolyticus in the presence of high H2 concentrations (46), along with the recent discovery that GOR (XOR) is regulated by Rex, a redox-sensing transcription factor that responds to the intracellular NADH/NAD ratio (47). These findings suggest that high concentrations of NADH or H2, either of which could interfere with the regeneration of oxidized cofactors via the bifurcating hydrogenase (13, 48), steer carbon flux toward GOR to utilize the more thermodynamically favorable Ech pathway, thereby maintaining a high rate of H2 production (13, 48). However, it is not possible at present to reconcile such an analysis with the observed growth phenotype of the Δgor mutant strains, which exhibit a longer lag phase but higher cell densities compared with the parent strain (Fig. 2 and Fig. S8).
Figure 4.
Proposed dual glycolytic pathways in C. bescii. The conventional pathway is shown in green where oxidation of NADH and reduced ferredoxin leads to H2 production via the bifurcating hydrogenase. The alternative GOR-dependent pathway and oxidation of the resulting reduced ferredoxin by the Ech hydrogenase and the associated ATP synthase is shown in dark yellow. Abbreviations: HK, hexokinase; PGI, phosphoglucose isomerase; PFK, phosphofructokinase; FBA, fructose-bisphosphate aldolase; BFH2ase, bifurcating [FeFe] hydrogenase; PGM, phosphoglycerate mutase; ENO, enolase; PK, pyruvate kinase; Fd, ferredoxin; POR, pyruvate ferredoxin oxidoreductase; LDH, lactate dehydrogenase; PTA, phosphotransacetylase; ACK, acetate kinase.
Phylogenetic analyses of the AOR family of enzymes (Fig. S9) show that each of the five characterized members of the AOR family of enzymes from P. furiosus can be assigned to distinct clusters, which is in accordance with their function in terms of both their substrate specificities and oligomeric structures. Hence AOR from P. furiosus and also from Pyrobaculum aerophilum are homodimeric enzymes and catalyze the oxidation of a wide range of both aliphatic and aromatic aldehydes (31, 49). FOR from Thermococcus litoralis and P. furiosus are homotetrameric enzymes but with a more limited substrate range than AOR, oxidizing short chain aliphatic aldehydes (24, 50). WOR4 and WOR5, both from P. furiosus, are also homodimeric enzymes. WOR5, like AOR, can oxidize a wide range of aldehydes but the substrate for WOR4 is not known and this enzyme was isolated purely based on its tungsten content (26, 51). GAPORs from Methanococcus maripauldis, P. furiosus, and Pyrobaculum aerophilum are monomeric enzymes and are absolutely specific for glyceraldhyde 3-phosphate as they are not known to oxidize any other aldehyde (23, 27, 28). We show here that C. bescii GOR-L is part of a distinct clade, separate from other members of the AOR family, including GAPOR (Fig. S9), its closest characterized homolog. The GOR-L clade contains all of the Caldicellulosiruptor species plus 44 other anaerobic genera, representing 9 taxonomic classes: Archaeoglobi, Bacilli, Clostridia, Deltaproteobacteria, Fusobacteria, Methanomicrobia, Nitrospira, Thermodesulfobacteria, and Thermoprotei. Although many of the genera are thermophilic or hyperthermophilic some are mesophilic; however, almost all species with GOR-L also have a co-located GOR-S homolog (SynTax web server) (52). Moreover, this analysis shows that, unlike GAPOR, which is found primarily in Archaea, GOR is found primarily in Bacteria. For example, the GOR-L clade could be divided into three subclades representing a bacterial-only clade, making up 54% of all GOR-L clade sequences, an archaeal-only making up 26%, and a mixed clade (split 50/50) making up 20% (Fig. S9).
We also show here that GOR is part of a small minority of AOR family members that are heteromeric. These include a homolog of AOR (termed Aa AorB) that is a heterotrimer containing the equivalent of the GOR-S subunit with a string of iron-sulfur clusters (53). In addition, a second heteromeric member of the AOR family described recently is part of an enzyme complex that reduces benzoyl-CoA (54). Termed BamB, the AOR homolog also has a partner subunit (BamC) with multiple iron-sulfur clusters. Furthermore, of the characterized AOR family members from hyperthermophilic Archaea, the gene encoding WOR5 has an adjacent gene homologous to that encoding GOR-S, suggesting that WOR5, like GOR, has an additional iron-sulfur containing subunit that was likely not detected during standard SDS analysis. Finally, the heteromeric AOR family group also includes an AOR from Escherichia coli termed EcYdhV that contains a polyferredoxin-like subunit similar to GOR-S. Interestingly, YdhV contains molybdenum rather than tungsten as E. coli does not metabolize tungsten (55). It is not clear why GOR and these other AOR family members require an additional subunit with four [4Fe-4S] clusters, which in GOR are proposed to transfer electrons between between the catalytic subunit (GOR-L) and ferredoxin. GOR has a high affinity for C. bescii ferredoxin (Km ∼ 40 μm), consistent with this being the physiological electron acceptor, although it is possible that GOR could provide reductant to the Ech hydrogenase directly, whereas ferredoxin is required to interact with the bifurcating hydrogenase.
In any event, the results presented here lay the foundation for a more comprehensive understanding of the two glycolytic pathways that are present in the cellulolytic genus Caldicellulosiruptor and for more effective metabolic engineering projects in the future. Moreover, this work suggests that alternative glycolytic pathways using either GOR or GAPOR may be more widespread than previously thought in both the bacterial and archaeal worlds.
Experimental procedures
Growth of C. bescii
C. bescii strains used or constructed in the present study are listed in Table S3. Strains were transformed as previously described (16, 56). For growth of uracil auxotrophic strains, uracil was added at a concentration of 20 μm. When required, kanamycin and 5-fluoroorotic acid were used at concentrations of 50 μg/ml and 4 mm, respectively, unless otherwise noted. Growth experiments were performed with 5-ml cultures in screw cap hungate tubes sealed with butyl rubber stoppers in modified DSMZ 516 medium with the following composition per liter: 1× salt solution, 1× vitamin solution, 1× trace element solution, 0.16 μm sodium tungstate, 0.25 mg of resazurin, 5 g of xylose (unless otherwise noted), 1 g of cysteine hydrochloride, 1 g of sodium bicarbonate, and 1 mm potassium phosphate buffer (pH 7.2). The ×50 stock salt solution contained the following per liter: 16.5 g of NH4Cl, 16.5 g of KCl, 16.5 g of MgCl2·6H2O, and 7 g of CaCl2×2H2O. Stock solutions of ×200 vitamins and ×1,000 trace elements were prepared as previously described (1). Cultures were also grown in a 15-liter fermenter at 75 °C with 20 g/liter of xylose as the growth substrate. The culture was sparged at 2 liters/h with 80% (v/v) nitrogen and 20% (v/v) carbon dioxide and stirred at 150 rpm with a flat blade impeller. The pH was maintained at 7.0 (25 °C) by the addition of 10% (w/v) sodium bicarbonate. Cells were harvested in the late-exponential phase by centrifugation at 6,000 × g for 10 min (Beckman Avanti J-30I JLA 10.500 rotor) and the resulting cell pellets were flash frozen in liquid nitrogen and stored at −80 °C. The cell yield from the fermenter was ∼12 g (wet weight).
Vector construction
Plasmids were constructed from PCR products using NEBuilder® HiFi DNA Assembly Master Mix (New England BioLabs). PCR was performed using Takara PrimeSTAR Max DNA polymerase (Takara R045A). Genomic DNA, plasmid DNA, and PCR products were purified with kits from Zymo Research and Stratagene. The templates for PCR were either previously sequenced plasmids or genomic DNA from C. bescii. All primers used in this study can be found in Table S5 and plasmids constructed for this study can be found in Fig. S10. Plasmids were cloned into New England Biolabs 10-β competent E. coli (New England BioLabs C3019I), followed by Sanger sequencing through GeneWiz.
Metabolomics
For the MS-based metabolomics, parent and ΔgorL strains of C. bescii were grown in duplicate in 16-mm sealed hungate tubes in modified DSMZ 516 medium as described above with glucose as the carbon source. Cultures were grown to an OD600 of ∼0.15 and each culture was deposited by vacuum filtration onto a 0.2-μm nylon membrane (47 mm diameter). The membrane was then placed (cells down) into 1.5-ml cold (−20 °C) dry ice extraction solvent (20:20:10 (v/v/v), acetonitrile:methanol:water) in a small Petri dish and swirled. After a few moments the filter was inverted (cells up) and solvent was passed over the surface of the membrane several times to maximize extraction. Finally, the cell extract was stored at −80 °C. Cell extracts from E. coli (K-12 substrain MG1655 rph+ ilvG+) grown in [13C]glucose were used as internal standards for quantitation of metabolites in C. bescii strains. E. coli cells were grown aerobically to an OD600 of ∼0.45 in M9 minimal medium containing 0.4% universally labeled [13C]glucose as the sole carbon source. To assure complete labeling of metabolites in E. coli, inoculation was performed using 1:50 dilution from an overnight culture that was also grown on 13C-labeled glucose. E. coli metabolites were extracted in a similar manner to the procedure used for C. bescii.
C. bescii and 13C-labeled E. coli extracts were thawed, centrifuged at 14,000 rpm for 10 min to remove cell debris, and the C. bescii supernatants were mixed with 13C-labeled E. coli supernatant in ratios of 1:1, 5:1, and 1:0 (no 13C-labeled internal standards). These mixed samples were then fully dried under a stream of nitrogen and subsequently re-suspended with HPLC running solvent (97:3, water:methanol containing 10 mm tributylamine and 9.8 mm acetate, pH 8.2) in half the initial volume. Samples were then centrifuged to remove particulates, transferred to HPLC vials, and analyzed by HPLC/MS for quantification as previously described (57).
Enzyme assays
All assays were carried out in serum-stoppered cuvettes under anaerobic conditions at 70 °C, unless noted otherwise. To remove trace amounts of O2, sodium dithionite was added to the assay mixture to give an A600 of ∼0.2. The extract was added and after a 1-min incubation period the reaction was initiated by addition of the substrate. Aldehyde-oxidizing activity of cytoplasmic extracts of C. bescii strains and purified GOR were determined by following the reduction of ferredoxin or BV (1 mm) in 50 mm EPPS buffer (pH 8.0), at 425 or 600 nm, respectively, using various aldehydes (1 mm) as the substrate. With GAP as the substrate the assay temperature was 70 °C. GAPDH activity was measured by following GAP-dependent reduction of NAD (1 mm) at 320 nm. Extinction coefficients of 8.2, 6.2, and 8.1 mm−1 cm−1 were used for reduced benzyl viologen, NADH, and reduced ferredoxin, respectively. Specific enzyme activities are expressed as units/mg of protein, where 1 unit represents 1 μmol of substrate oxidized per min.
Purification of affinity-tagged GOR
All purification steps were performed under strictly anaerobic conditions. Frozen cells (50 g, wet weight) of C. bescii strain OE XOR/PFD were thawed and suspended in 25 mm Tris-HCl (pH 8.0), containing 1 mg ml−1 lysozyme (Sigma) in a ratio of 3 ml/g of cells. The suspended cells were incubated at room temperature for 15 min followed by four 15-s intervals of sonication (amplitude 40; Qsonica Q55) interspaced by at least 30 s. Cell lysates were clarified by ultracentrifugation at 100,000 × g for 1 h (Beckman L90K ultracentrifuge 70.1Ti rotor) yielding clarified cell lysate (235 ml, 14.3 mg of protein ml−1). This was loaded onto a 250-ml QHP column equilibrated with buffer A (25 mm Tris, pH 8.0, containing 2 mm cysteine), at a rate of 10 ml min−1 diluted 1:5 with buffer A. Protein was eluted with a linear gradient (2500 ml) from 0 to 500 mm NaCl at 10 ml min−1 and collected as 75-ml fractions. Fractions were assayed for GOR and GAPDH activity. Fractions containing GAPDH activity were reserved for further purification, whereas fractions containing GOR activity were pooled and applied to a 5-ml HisTrap Excel column (GE Healthcare) equilibrated in buffer B (50 mm phosphate, pH 7.2, containing 300 mm NaCl). The column was washed with 5 volumes of buffer B, and GOR was eluted with a gradient of 0 to 500 mm imidazole in buffer C (buffer B containing 500 mm imidazole) over 20 column volumes. Fractions were collected in sealed serum bottles. Fractions containing GOR activity were pooled and concentrated (15-kDa Amicon Ultra centrifugal filter) to yield 2.9 ml of GOR (1.2 mg/ml). This was loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated with 50 mm Tris-HCl (pH 8.2), containing 200 mm NaCl at a rate of 1.25 ml min−1 and collected in 4-ml fractions in sealed serum bottles. Protein-containing fractions were analyzed for purity using denaturing SDS-PAGE gradient electrophoresis using precast 4–12% BisTris gels; Novex NuPAGE gels (Invitrogen) and stained with Imperial protein stain (Novex). Protein bands were identified via a Mascot MS/MS Ions Search of a custom C. bescii database (version 2.6) using LC-tandem MS (LC-MS/MS) data (Proteomics and Mass Spectrometry Facility, University of Georgia).
Partial purification of GAPDH
To further purify GAPDH, the fraction containing the highest GAPDH activity from the QHP column was concentrated (10-kDa Amicon Ultra centrifugal filter) to yield 4.1 ml (14.2 mg/ml) and loaded onto a HiLoad 16/60 Superdex 200 and eluted in the same manner as above. Fractions containing GAPDH activity were pooled and applied to a 1-ml HiTrap Blue HP column (GE Healthcare) equilibrated in 20 mm EPPS (pH 8). The column was washed with 5 column volumes of the equilibration buffer. Proteins were eluted with 2 ml of the equilibration buffer containing 20 mm NAD. The eluate was concentrated and buffer exchanged (10-kDa Amicon Ultra centrifugal filter) into 50 mm EPPS (pH 8) to remove NAD, yielding 0.4 ml of partially purified GAPDH (1.6 mg/ml).
Estimation of molecular mass and metal analysis
For size determination of the purified GOR, the elution volumes (Ve) were determined by measuring the absorbance at 280 nm and the total void volume (Vo) was determined by measuring the elution of blue dextran. The known molecular mass standards thyroglobulin (669 kDa), apoferritin (443 kDa), amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), and carbonic anhydrase (29 kDa) were used as references. The molecular mass of the purified protein in each peak was estimated from the plot of Ve/Vo against the logarithm of the molecular mass (kDa). Metal concentrations of the final chromatography fractions were measured using an Agilent 7900 inductively coupled plasma mass spectrometer (ICP-MS) fitted with MicroMist nebulizer, UHMI-spray chamber, Pt cones, and an Octopole Reaction System collision cell (Agilent Technologies, Santa Clara, CA) as described previously (58).
Purification of PGK/TPI
To isolate the GOR inhibiting factor, the flow-through from the affinity purification of GOR described above was applied to a 60-ml Phenyl-Sepharose column equilibrated with buffer A containing 1 m (NH4)2SO4. Protein was eluted with a linear gradient (800 ml) from 1.0 to 0 m (NH4)2SO4 at 7.5 ml min−1. The resulting fractions were buffer exchanged with 50 mm EPPS (pH 8.0), using ultrafiltration with a 30-kDa filter and tested for the ability to inhibit GAP (1 mm) oxidation by purified GOR. Fractions exhibiting the ability to inhibit GOR activity were analyzed by SDS-PAGE using precast 4–12% BisTris Novex NuPAGE gels (Invitrogen) and protein bands were identified via MS/MS as described above.
Phylogenetic analyses
In InterPro (version 63.0), GOR-L (Athe_0821; UniProt ID: B9MQI2_CALBD) is annotated to have one aldehyde ferredoxin oxidoreductase N-terminal (IPR013983) domain and one aldehyde ferredoxin oxidoreductase C-terminal (IPR001203) domain (59). One of each domain was entered into the InterPro Domain Architecture search tool, yielding 4,107 GOR-L sequence homologs. The 4,107 sequences were aligned using Clustal Omega, version 1.2.1, with the default parameters (60). TrimAl was used to remove multiple alignment positions with greater than 99.7% gaps (i.e. −gt 0.003) (61), and the resulting alignment was used to construct a maximum likelihood phylogenetic tree using IQ-Tree (version 1.5.5) (62). The IQ-Tree standard model selection test was used to automatically determine the best-fit model (i.e. LG + F + R10) (63). This model tree was refined using ultrafast bootstrap approximation (UFBoot) and Shimodaira-Hasegawa–like approximate likelihood ratio tests with 3,000 bootstrap replicates for each. Only sequences that were not marked deleted by Uniprot were used to display the tree, leaving 4063 sequences. iTOL (version 4.2.1) was used for analysis and display of the phylogenetic tree (64). For visualization the tree was re-rooted and branches with ultrafast bootstrap confidence values less than 94% were removed. Uniprot Retrieve was used to lookup the kingdom of the organism for each of the 4,063 sequences and label them in iTOL (65).
Author contributions
I. M. S., G. M. R., F. L. P., D. M. S., and D. A.-N. data curation; I. M. S., G. M. R., F. L. P., G. L. L., G. J. S., A. M. W.-R., D. M. S., and D. A.-N. formal analysis; I. M. S., G. M. R., F. L. P., G. J. S., A. M. W.-R., D. M. S., and D. A.-N. investigation; I. M. S., G. M. R., F. L. P., G. L. L., G. J. S., A. M. W.-R., D. M. S., D. A.-N., and M. W. W. A. methodology; I. M. S. and G. M. R. writing-original draft; G. L. L. and M. W. W. A. resources; G. L. L., G. J. S., D. M. S., D. A.-N., R. M. K., and M. W. W. A. writing-review and editing; G. J. S. and M. W. W. A. supervision; R. M. K. and M. W. W. A. conceptualization; R. M. K. and M. W. W. A. funding acquisition; R. M. K. and M. W. W. A. project administration.
Supplementary Material
Acknowledgments
We thank Jeffrey Zurawski, Piyum Khatibi, and Laura Lee for many helpful discussions and Daehwan Chung and Janet Westpheling for providing strains and plasmids of C. bescii. We also acknowledge the University of Georgia Proteomics and Mass Spectrometry Facility for performing MS analysis and computational support. The phylogenetic analysis was made possible by the resources and technical expertise of the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia's Office of the Vice President for Research and Office of the Vice President for Information Technology.
This work was supported by United States Department of Energy, Office of Science, Office of Biological and Environmental Research Grant DE-SC0019391, Bioenergy Science Center at Oak Ridge National Laboratory Grant DE-PS02–06ER64304, and United States National Science Foundation Grant CBET1264053. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S10 and Tables S1–S5.
- AOR
- aldehyde oxidoreductase
- PFD
- polyferredoxin
- GAPOR
- glyceraldehyde-3-phosphate ferredoxin oxidoreductase
- FOR
- formaldehyde ferredoxin oxidoreductase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- BV
- benzyl viologen
- DHAP
- dihydroxyacetone phosphate
- PGK
- phosphoglycerate kinase
- TPI
- triose-phosphate isomerase
- EPPS
- 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GOR
- glyceraldehyde-3-phosphate ferredoxin oxidoreductase
- Ech
- [NiFe] hydrogenase.
References
- 1. Yang S. J., Kataeva I., Wiegel J., Yin Y., Dam P., Xu Y., Westpheling J., and Adams M. W. (2010) Classification of “Anaerocellum thermophilum” strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int. J. Syst. Evol. Microbiol. 60, 2011–2015 10.1099/ijs.0.017731-0 [DOI] [PubMed] [Google Scholar]
- 2. Straub C. T., Zeldes B. M., Schut G. J., Adams M. W., and Kelly R. M. (2017) Extremely thermophilic energy metabolisms: biotechnological prospects. Curr. Opin. Biotechnol. 45, 104–112 10.1016/j.copbio.2017.02.016 [DOI] [PubMed] [Google Scholar]
- 3. Blumer-Schuette S. E., Brown S. D., Sander K. B., Bayer E. A., Kataeva I., Zurawski J. V., Conway J. M., Adams M. W., and Kelly R. M. (2014) Thermophilic lignocellulose deconstruction. FEMS Microbiol. Rev. 38, 393–448 10.1111/1574-6976.12044 [DOI] [PubMed] [Google Scholar]
- 4. Yang S. J., Kataeva I., Hamilton-Brehm S. D., Engle N. L., Tschaplinski T. J., Doeppke C., Davis M., Westpheling J., and Adams M. W. W. (2009) Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl. Environ. Microb. 75, 4762–4769 10.1128/AEM.00236-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blumer-Schuette S. E., Giannone R. J., Zurawski J. V., Ozdemir I., Ma Q., Yin Y., Xu Y., Kataeva I., Poole F. L. 2nd, Adams M. W., Hamilton-Brehm S. D., Elkins J. G., Larimer F. W., Land M. L., Hauser L. J., et al. (2012) Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J. Bacteriol. 194, 4015–4028 10.1128/JB.00266-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olson D. G., McBride J. E., Shaw A. J., and Lynd L. R. (2012) Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 23, 396–405 10.1016/j.copbio.2011.11.026 [DOI] [PubMed] [Google Scholar]
- 7. Brunecky R., Alahuhta M., Xu Q., Donohoe B. S., Crowley M. F., Kataeva I. A., Yang S.-J., Resch M. G., Adams M. W., Lunin V. V., Himmel M. E., and Bomble Y. J. (2013) Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342, 1513–1516 10.1126/science.1244273 [DOI] [PubMed] [Google Scholar]
- 8. Kataeva I., Foston M. B., Yang S. J., Pattathil S., Biswal A., Poole F. L., Basen M., Rhaesa A. M., Thomas T. P., Azadi P., Olman V., Saffold T. D., Mohler K. E., Lewis D. L., Doeppke C., et al. (2013) Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energ. Environ. Sci. 6, 2186–2195 10.1039/c3ee40932e [DOI] [Google Scholar]
- 9. Basen M., Rhaesa A. M., Kataeva I., Prybol C. J., Scott I. M., Poole F. L., and Adams M. W. W. (2014) Degradation of high loads of crystalline cellulose and of unpretreated plant biomass by the thermophilic bacterium Caldicellulosiruptor bescii. Bioresource Technol. 152, 384–392 10.1016/j.biortech.2013.11.024 [DOI] [PubMed] [Google Scholar]
- 10. Williams-Rhaesa A. M., Rubinstein G. M., Scott I. M., Lipscomb G. L., Poole Ii F. L., Kelly R. M., and Adams M. W. W. (2018) Engineering redox-balanced ethanol production in the cellulolytic and extremely thermophilic bacterium, Caldicellulosiruptor bescii. Metab. Eng. Commun. 7, e00073 10.1016/j.mec.2018.e00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung D., Cha M., Guss A. M., and Westpheling J. (2014) Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc. Natl. Acad. Sci. U.S.A. 111, 8931–8936 10.1073/pnas.1402210111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Groom J., Chung D., Kim S.-K., Guss A., and Westpheling J. (2018) Deletion of the Clostridium thermocellum recA gene reveals that it is required for thermophilic plasmid replication but not plasmid integration at homologous DNA sequences. J. Ind. Microbiol. Biot. 45, 753–763 10.1007/s10295-018-2049-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sander K., Chung D., Hyatt D., Westpheling J., Klingeman D. M., Rodriguez M. Jr, Engle N. L., Tschaplinski T. J., Davison B. H., and Brown S. D. (2019) Rex in Caldicellulosiruptor bescii: novel regulon members and its effect on the production of ethanol and overflow metabolites. Microbiologyopen 8, e00639 10.1002/mbo3.639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lipscomb G. L., Conway J. M., Blumer-Schuette S. E., Kelly R. M., and Adams M. W. (2016) A highly thermostable kanamycin resistance marker expands the toolkit for genetic manipulation of Caldicellulosiruptor bescii. Appl. Environ. Microbiol. 82, 4421–4428 10.1128/AEM.00570-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams-Rhaesa A. M., Awuku N. K., Lipscomb G. L., Poole F. L., Rubinstein G. M., Conway J. M., Kelly R. M., and Adams M. W. (2018) Native xylose-inducible promoter expands the genetic tools for the biomass-degrading, extremely thermophilic bacterium Caldicellulosiruptor bescii. Extremophiles 22, 629–638 10.1007/s00792-018-1023-x [DOI] [PubMed] [Google Scholar]
- 16. Scott I. M., Rubinstein G. M., Lipscomb G. L., Basen M., Schut G. J., Rhaesa A. M., Lancaster W. A., Poole F. L. 2nd, Kelly R. M., and Adams M. W. (2015) A new class of tungsten-containing oxidoreductase in the genus of the plant biomass-degrading, thermophilic bacteria Caldicellulosiruptor. Appl. Environ. Microbiol. 81, 7339–7347 10.1128/AEM.01634-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kletzin A., and Adams M. W. (1996) Tungsten in biological systems. FEMS Microbiol. Rev. 18, 5–63 10.1111/j.1574-6976.1996.tb00226.x [DOI] [PubMed] [Google Scholar]
- 18. Iobbi-Nivol C., and Leimkuhler S. (2013) Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1827, 1086–1101 10.1016/j.bbabio.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 19. Bevers L. E., Hagedoorn P.-L., and Hagen W. R. (2009) The bioinorganic chemistry of tungsten. Coord. Chem. Rev. 253, 269–290 10.1016/j.ccr.2008.01.017 [DOI] [Google Scholar]
- 20. Havarushka N., Fischer-Schrader K., Lamkemeyer T., and Schwarz G. (2014) Structural basis of thermal stability of the tungsten cofactor synthesis protein MoaB from Pyrococcus furiosus. PLOS One 9, e86030 10.1371/journal.pone.0086030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hille R., Hall J., and Basu P. (2014) The mononuclear molybdenum enzymes. Chem. Rev. 114, 3963–4038 10.1021/cr400443z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mukund S., and Adams M. W. (1991) The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase, evidence for its participation in a unique glycolytic pathway. J. Biol. Chem. 266, 14208–14216 [PubMed] [Google Scholar]
- 23. Mukund S., and Adams M. W. (1995) Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270, 8389–8392 10.1074/jbc.270.15.8389 [DOI] [PubMed] [Google Scholar]
- 24. Roy R., Mukund S., Schut G. J., Dunn D. M., Weiss R., and Adams M. W. (1999) Purification and molecular characterization of the tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus: the third of a putative five-member tungstoenzyme family. J. Bacteriol. 181, 1171–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy R., and Adams M. W. (2002) Characterization of a fourth tungsten-containing enzyme from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 184, 6952–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bevers L. E., Bol E., Hagedoorn P.-L., and Hagen W. R. (2005) WOR5, a novel tungsten-containing aldehyde oxidoreductase from Pyrococcus furiosus with a broad substrate specificity. J. Bacteriol. 187, 7056–7061 10.1128/JB.187.20.7056-7061.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reher M., Gebhard S., and Schönheit P. (2007) Glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) and nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN), key enzymes of the respective modified Embden–Meyerhof pathways in the hyperthermophilic crenarchaeota Pyrobaculum aerophilum and Aeropyrum pernix. FEMS Microbiol. Lett. 273, 196–205 10.1111/j.1574-6968.2007.00787.x [DOI] [PubMed] [Google Scholar]
- 28. Park M.-O., Mizutani T., and Jones P. R. (2007) Glyceraldehyde-3-phosphate ferredoxin oxidoreductase from Methanococcus maripaludis. J. Bacteriol. 189, 7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee L. L., Blumer-Schuette S. E., Izquierdo J. A., Zurawski J. V., Loder A. J., Conway J. M., Elkins J. G., Podar M., Clum A., Jones P. C., Piatek M. J., Weighill D. A., Jacobson D. A., Adams M. W. W., and Kelly R. M. (2018) Genus-wide assessment of lignocellulose utilization in the extremely thermophilic genus Caldicellulosiruptor by genomic, pangenomic, and metagenomic analyses. Appl. Environ. Microbiol. 84, e02694–e02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rozovsky S., and McDermott A. E. (2007) Substrate product equilibrium on a reversible enzyme, triosephosphate isomerase. Proc. Natl. Acad. Sci. U.S.A. 104, 2080–2085 10.1073/pnas.0608876104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan M. K., Mukund S., Kletzin A., Adams M. W., and Rees D. C. (1995) Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science 267, 1463–1469 10.1126/science.7878465 [DOI] [PubMed] [Google Scholar]
- 32. Baker L. M., Raudonikiene A., Hoffman P. S., and Poole L. B. (2001) Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183, 1961–1973 10.1128/JB.183.6.1961-1973.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bell R. A., Smith J. C., and Storey K. B. (2014) Purification and properties of glyceraldehyde-3-phosphate dehydrogenase from the skeletal muscle of the hibernating ground squirrel, Ictidomys tridecemlineatus. Peer J. 2, e634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seidler N. W. (2012) GAPDH: biological properties and diversity, Springer Science & Business Media, Netherlands [Google Scholar]
- 35. Engeland K., Höög J. O, Holmquist B., Estonius M., Jörnvall H., and Vallee B. L. (1993) Mutation of Arg-115 of human class III alcohol dehydrogenase: a binding site required for formaldehyde dehydrogenase activity and fatty acid activation. Proc. Natl. Acad. Sci. U.S.A. 90, 2491–2494 10.1073/pnas.90.6.2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahmed H., Tjaden B., Hensel R., and Siebers B. (2004) Embden–Meyerhof–Parnas and Entner–Doudoroff pathways in Thermoproteus tenax: metabolic parallelism or specific adaptation? Biochem. Soc. Trans. 32, 303–304 10.1042/bst0320303 [DOI] [PubMed] [Google Scholar]
- 37. van der Oost J., Schut G., Kengen S. W., Hagen W. R., Thomm M., and de Vos W. M. (1998) The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. J. Biol. Chem. 273, 28149–28154 10.1074/jbc.273.43.28149 [DOI] [PubMed] [Google Scholar]
- 38. Schäfer T., and Schönheit P. (1993) Gluconeogenesis from pyruvate in the hyperthermophilic archaeon Pyrococcus furiosus: involvement of reactions of the Embden–Meyerhof pathway. Arch. Microbiol. 159, 354–363 10.1007/BF00290918 [DOI] [Google Scholar]
- 39. Selig M., Xavier K. B., Santos H., and Schönheit P. (1997) Comparative analysis of Embden-Meyerhof and Entner–Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch. Microbiol. 167, 217–232 10.1007/BF03356097 [DOI] [PubMed] [Google Scholar]
- 40. Siebers B., and Hensel R. (1993) Glucose catabolism of the hyperthermophilic archaeum Thermoproteus tenax. FEMS Microbiol. Lett. 111, 1–7 10.1111/j.1574-6968.1993.tb06353.x [DOI] [Google Scholar]
- 41. Siebers B., Tjaden B., Michalke K., Dörr C., Ahmed H., Zaparty M., Gordon P., Sensen C. W., Zibat A., Klenk H.-P., Schuyster S. C., and Hensel R. (2004) Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data. J. Bacteriol. 186, 2179–2194 10.1128/JB.186.7.2179-2194.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bräsen C., Esser D., Rauch B., and Siebers B. (2014) Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175 10.1128/MMBR.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wrba A., Schweiger A., Schultes V., Jaenicke R., and Závodszky P. (1990) Extremely thermostable d-glyceraldehyde-3-phosphate dehydrogenase from the eubacterium Thermotoga maritima. Biochemistry 29, 7584–7592 10.1021/bi00485a007 [DOI] [PubMed] [Google Scholar]
- 44. Cha M., Chung D., and Westpheling J. (2016) Deletion of a gene cluster for [Ni-Fe] hydrogenase maturation in the anaerobic hyperthermophilic bacterium Caldicellulosiruptor bescii identifies its role in hydrogen metabolism. Appl. Microbiol. Biotechnol. 100, 1823–1831 10.1007/s00253-015-7025-z [DOI] [PubMed] [Google Scholar]
- 45. Schut G. J., and Adams M. W. (2009) The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457 10.1128/JB.01582-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bielen A. A., Verhaart M. R., VanFossen A. L., Blumer-Schuette S. E., Stams A. J., van der Oost J., Kelly R. M., and Kengen S. W. (2013) A thermophile under pressure: Transcriptional analysis of the response of Caldicellulosiruptor saccharolyticus to different H2 partial pressures. Int. J. Hydrogen Energy 38, 1837–1849 10.1016/j.ijhydene.2012.11.082 [DOI] [Google Scholar]
- 47. McLaughlin K. J., Strain-Damerell C. M., Xie K., Brekasis D., Soares A. S., Paget M. S., and Kielkopf C. L. (2010) Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol. Cell 38, 563–575 10.1016/j.molcel.2010.05.006 [DOI] [PubMed] [Google Scholar]
- 48. Bielen A. A., Verhaart M. R., van der Oost J., and Kengen S. W. (2013) Biohydrogen production by the thermophilic bacterium Caldicellulosiruptor saccharolyticus: current status and perspectives. Life 3, 52–85 10.3390/life3010052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hagedoorn P. L., Chen T., Schröder I., Piersma S. R., de Vries S., and Hagen W. R. (2005) Purification and characterization of the tungsten enzyme aldehyde: ferredoxin oxidoreductase from the hyperthermophilic denitrifier Pyrobaculum aerophilum. J. Biol. Inorg. Chem. 10, 259–269 10.1007/s00775-005-0637-5 [DOI] [PubMed] [Google Scholar]
- 50. Mukund S., and Adams M. (1993) Characterization of a novel tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon, Thermococcus litoralis: a role for tungsten in peptide catabolism. J. Biol. Chem. 268, 13592–13600 [PubMed] [Google Scholar]
- 51. Roy R., and Adams M. W. (2002) Tungsten-dependent aldehyde oxidoreductase: a new family of enzymes containing the pterin cofactor. Met. Ions Biol. Syst. 39, 673–697 [PubMed] [Google Scholar]
- 52. Oberto J. (2013) SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics 14, 4 10.1186/1471-2105-14-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arndt F., Schmitt G., Winiarska A., Saft M., Seubert A., Kahnt J., and Heider J. (2019) Characterisation of an aldehyde oxidoreductase from the mesophilic bacterium Aromatoleum aromaticum EbN1, a member of a new subfamily of tungsten-containing enzymes. Front. Microbiol. 10, 71 10.3389/fmicb.2019.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weinert T., Huwiler S. G., Kung J. W., Weidenweber S., Hellwig P., Stärk H.-J., Biskup T., Weber S., Cotelesage J. J., George G. N., Ermler U., and Boll M. (2015) Structural basis of enzymatic benzene ring reduction. Nat. Chem. Biol. 11, 586 10.1038/nchembio.1849 [DOI] [PubMed] [Google Scholar]
- 55. Reschke S., Duffus B. R., Schrapers P., Mebs S., Teutloff C., Dau H., Haumann M., and Leimkühler S. (2019) Identification of YdhV as first molybdoenzyme binding a bis-Mo-MPT cofactor in Escherichia coli. Biochemistry 58, 2228–2242 10.1021/acs.biochem.9b00078 [DOI] [PubMed] [Google Scholar]
- 56. Chung D., Cha M., Farkas J., and Westpheling J. (2013) Construction of a stable replicating shuttle vector for Caldicellulosiruptor species: use for extending genetic methodologies to other members of this genus. PLOS One 8, e62881 10.1371/journal.pone.0062881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rydzak T., Garcia D., Stevenson D. M., Sladek M., Klingeman D. M., Holwerda E. K., Amador-Noguez D., Brown S. D., and Guss A. M. (2017) Deletion of type I glutamine synthetase deregulates nitrogen metabolism and increases ethanol production in Clostridium thermocellum. Metab. Eng. 41, 182–191 10.1016/j.ymben.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 58. Vaccaro B. J., Menon A. L., Lancaster W. A., and Adams M. W. (2012) Metallomics using inductively coupled plasma mass spectrometry. Curr. Protoc. Chem. Biol. 4, 249–274 10.1002/9780470559277.ch120031 [DOI] [Google Scholar]
- 59. Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., Gwad M., Hurwitz D. I., Lu F., Marchler G. H., Song J. S., et al. (2016) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Capella-Gutiérrez S., Silla-Martínez J. M., and Gabaldón T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nguyen L.-T., Schmidt H. A., von Haeseler A., and Minh B. Q. (2014) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., and Jermiin L. S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Letunic I., and Bork P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.