Abstract
Prolyl hydroxylase 3 (PHD3) has initially been reported to hydroxylase hypoxia-inducible factor α (HIFα) and mediate HIFα degradation. More recent studies have shown that, in addition to HIFα, PHD3 has also other substrates. Moreover, pHD3 is believed to act as a tumor suppressor, but the underlying mechanism remains to be elucidated. Here, we demonstrate that PHD3 stabilizes p53 in a hydroxylase-independent manner. We found that PHD3 overexpression increases and PHD3 knockdown decreases p53 levels. Mechanistically, PHD3 bound MDM2 proto-oncogene (MDM2) and prevented MDM2 from interacting with p53, thereby inhibiting MDM2-mediated p53 degradation. Interestingly, we found that PHD3 overexpression could enhance p53 in the presence of the prolyl hydroxylase inhibitor dimethyloxalylglycine, and the prolyl hydroxylase activity-deficient variant PHD3-H196A also inhibited the p53-MDM2 interaction and stabilized p53. Genetic ablation of PHD3 decreased p53 protein levels in mice intestinal epithelial cells, but a genetic knockin of PHD3-H196A did not affect p53 protein levels in vivo. These results suggest that the prolyl hydroxylase activity of PHD3 is dispensable for its ability to stabilize p53. We found that both PHD3 and PHD3-H196A suppress the expression of the stem cell-associated gene NANOG and inhibited the properties of colon cancer stem cells through p53. Our results reveal an additional critical mechanism underlying the regulation of p53 expression and highlight that PHD3 plays a role in the suppression of colon cancer cell stemness in a hydroxylase-independent manner.
Keywords: p53, protein stability, mouse double minute 2 homolog (MDM2), cancer stem cells, colon cancer, prolyl hydroxylase 3
Introduction
Prolyl hydroxylases (PHD1, 2, and 3)2 are dioxygenases that use oxygen and 2-oxoglutarate (2-OG) as co-substrates. PHDs are involved in the cellular response to oxygen by hydroxylating conserved prolyl residues of hypoxia-inducible factor α (HIFα) (1–3). The hydroxylated HIFα is recognized by von Hippel–Lindau protein and then subjected to ubiquitination and proteasomal degradation. In hypoxia, the prolyl hydroxylase activity is suppressed, resulting in accumulation of HIFα. HIFα is a master transcription factor. It dimerizes with HIF-1β and translocates into the nucleus, where it initiates the transcription of many genes. The hydroxylase activity of PHD is strictly controlled by oxygen availability over the entire physiologic range (2). These properties make PHDs well-suited to act as oxygen sensors.
Recent studies have shown that, in addition to HIFα, PHDs have other substrates. For example, PHD3 was found to mediate oxygen-dependent stability of the activating transcriptional factor 4 (ATF4) (4) and the β2-adrenergic receptor (5) and to prevent degradation of myogenin (6). PHD3 regulates DNA damage response through hydroxylating the human homolog of the Caenorhabditis elegans biological clock protein CLK-2 (7). A recent study demonstrates that PHD3 hydroxylates and stabilizes MAPK6 (8). We found that PHD3 repressed IKK/NF-κB signaling (9).
A few studies have demonstrated that PHD3 acts as a tumor suppressor. Down-regulation of PHD3 was found in a few cancers (9–11). PHD3 up-regulation was linked to cell apoptosis (12), and its activation suppressed xenograft growth of melanoma cells (13). PHD3 caused apoptosis of cervical cancer HeLa cells (14) and inhibited proliferation of gastric cancer cells (15) and renal carcinoma cells (16). Epidemiology studies showed that expression of PHD3 was correlated with good prognostic factors in breast cancers (17), and it was a favorable prognosticator for gastric cancer (18). PHD3 was shown to inhibit tumor growth via EGF receptor signaling (19).
Although studies have indicated that PHD3 functions as a tumor suppressor, the underlying mechanism remains unclear. In this manuscript we demonstrate that PHD3 blocks the interaction of p53 and MDM2, thereby inhibiting the MDM2-mediated p53 destruction, in a hydroxylase-independent mechanism. The PHD3-induced p53 stabilization inhibits NANOG expression, leading to inhibition of colon cancer stem cells. Our findings reveal a new mechanism underlying the regulation of p53 stability through PHD3 and highlight the role of PHD3 in suppression of cancer cell stemness independent of its hydroxylase activity.
Results
PHD3 stabilizes p53
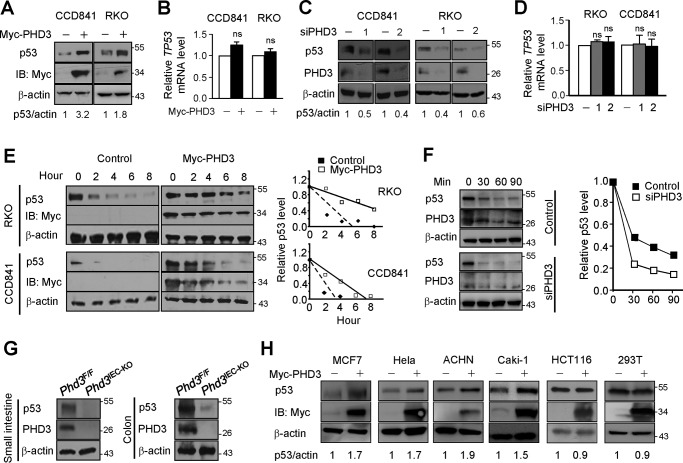
This study was kindled by an accidental discovery that PHD3 influenced the expression of p53. We found that overexpression of PHD3 enhanced the protein levels of p53 in colon cancer RKO and normal colon epithelial CCD841 cells (Fig. 1A). Overexpression of PHD3 did not affect the transcript level of TP53 (Fig. 1B). In agreement with the above results, knockdown of PHD3 reduced p53 protein (Fig. 1C) with little effect on TP53 transcript levels (Fig. 1D). The data imply that PHD3 regulates the expression of p53 in a post-transcriptional manner. We found that overexpression of PHD3 retarded degradation of p53 and increased the half-life of p53 (Fig. 1E). We determined the effect of PHD3 knockdown on the half-life of p53, and the results show that knockdown of PHD3 increased the rate of p53 degradation (Fig. 1F). These results suggest that PHD3 stabilizes p53. We next did experiments to verify that PHD3 could modulate the expression of p53 in vivo. We employed Phd3IEC-KO mice in which Phd3 was deleted in intestinal epithelial cells. Generation of Phd3IEC-KO mice is as described under “Experimental procedures.” The results show that genetic ablation of Phd3 led to a dramatic decrease of p53 in both small intestine and colon epithelial cells in mice (Fig. 1G). We examined other cells and found that overexpression of PHD3 increased p53 in cervical cancer HeLa, breast cancer MCF-7, and renal cancer ACHN and Caki-1 cells but not in colon cancer HCT116 cells (Fig. 1H). We also examined human embryonic kidney 293T cells, and overexpression of PHD3 had no effect on expression of p53 in the cells (Fig. 1H). This is probably because 293T cells express adenoviral oncoproteins e1a/e1b55k, which prevents p53 degradation (20, 21). All these cells we examined have WT p53.
Figure 1.
PHD3 stabilizes p53. A, overexpression of PHD3 increased p53. CCD841 and RKO cells were transfected with control or myc-PHD3 plasmid. After 24 h, the cells were harvested for Western blotting. B, overexpression of PHD3 had little effect on mRNA levels of TP53. The cells were treated as in A. C, knockdown of PHD3 decreased p53. The cells were transfected with control or PHD3 siRNA oligonucleotides. After 48 h, the cells were harvested. D, RKO and CCD841 cells were transfected with control or siPHD3 oligonucleotides as indicated. After 48 h, the cells were harvested for determination of TP53 mRNA level by qPCR. E, CCD841 and RKO cells were transfected as indicated. After 24 h, cycloheximide (10 μg/ml) was added to inhibit new protein synthesis, and the cells were harvested at different time intervals. The right panel shows the relative p53 level at different time point. F, CCD841 cells were transfected with control or siPHD3 oligonucleotides. After 48 h, the cells were treated with cycloheximide (10 μg/ml) for different time intervals. The right panel shows the relative p53 level. G, proteins extracted from small intestinal and colonic epithelial cells of Phd3F/F and Phd3IEC-KO mice were subjected to Western blotting. The p53(DO-7) antibody was used to detect mouse p53. H, the cells were transfected with control or myc-PHD3 plasmid as indicated. After 24 h, the cells were harvested. The Western blots were quantified using ImageJ, and the results are from one experiment. IB, immunoblotting; ns, no significance.
PHD3 stabilizes p53 through MDM2
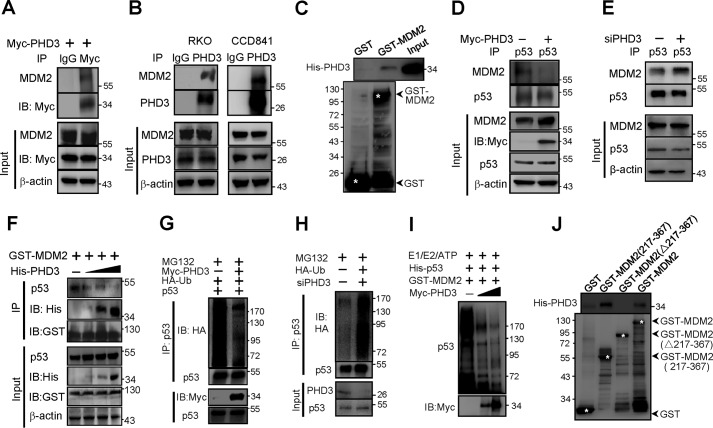
Because MDM2 is the primary E3 ubiquitin ligase of p53 (22), we presumed that PHD3 might stabilize p53 through MDM2. To this end, we first determined whether PHD3 associated with MDM2. We found that both exogenous (Fig. 2A) and endogenous (Fig. 2B) PHD3 interacted with MDM2. The bacterial produced His-PHD3 and GST-MDM2 bound each other, suggesting that PHD3 binds MDM2 directly (Fig. 2C). Thus, PHD3 might bind MDM2 to interfere with p53-MDM2 interaction. As expected, overexpression of PHD3 blocked the interaction of p53 and MDM2 (Fig. 2D). Consistent with the results, knockdown of PHD3 enhanced the interaction of p53 and MDM2 (Fig. 2E). Furthermore, we found that PHD3 inhibited p53–MDM2 interaction in a dose-dependent manner (Fig. 2F).
Figure 2.
PHD3 stabilizes p53 via MDM2. A, RKO cells were transfected with myc-PHD3 vector. After 24 h the cells were harvested, and cellular proteins (350 μg) were used for immunoprecipitation (IP) assay. 50 μg of proteins were used in input (14%). B, the endogenous PHD3 and MDM2 bound each other. Cellular proteins (500 μg) of RKO and CCD841 cells were used to do the immunoprecipitation assay with 50 μg of the proteins as Input (10%). C, PHD3 bound MDM2 directly. Equal amounts of bacterial lysates (containing His-PHD3) were incubated with the GSH-Sepharose beads that had already captured GST-MDM2 or GST proteins. The beads were washed, and His-PHD3 retained on beads was determined by immunoblotting (IB). D, PHD3 inhibited p53-MDM2 interaction. RKO cells were transfected as indicated. After 24 h, the cells were treated with MG132 (10 μm) for 3 h, followed by immunoprecipitation experiments. E, RKO cells were transfected as indicated. After 48 h, the cells were treated with MG132 (10 μm) for 2 h. The cells were then harvested for immunoprecipitation experiment. F, the E. coli supernatant containing GST-MDM2 protein was incubated with beads at 4 °C for 2 h. The beads were washed and incubated at 4 °C with RKO cell lysates containing p53 and different amounts of His-PHD3 protein. After 3 h, the beads were washed and subjected to immunoblotting. G, overexpression of PHD3 reduced ubiquitination of p53. RKO cells were transfected with myc-PHD3, HA-Ub, and p53 vectors as indicated. After 24 h, MG132 (10 μm) was added, and the cells were incubated for another 3 h, followed by immunoprecipitation. H, knockdown of PHD3 enhanced p53 ubiquitination. RKO cells were transfected as indicated. After 48 h, MG132 (10 μm) was added, and the cells were incubated for another 3 h. The cells were harvested for immunoprecipitation. I, in vitro p53 ubiquitination was performed as described under “Experimental procedures.” J, equal amounts of bacterial lysates containing His-PHD3 were incubated with the beads that had already captured GST, GST-MDM2, or mutated GST-MDM2. The beads were washed, and His-PHD3 retained on the beads was determined by immunoblotting. The asterisk indicates the band that the arrow pointed.
We determined the effect of PHD3 on ubiquitination of p53. The results show that overexpression of PHD3 decreased (Fig. 2G), and knockdown of PHD3 increased p53 ubiquitination (Fig. 2H). We did in vitro p53 ubiquitination assay, and the results show that PHD3 decreased the MDM2-mediated ubiquitination of p53 (Fig. 2I).
It is known that the central acid domain of MDM2 binds p53, which is essential for MDM2 to mediate p53 ubiquitination (23–25). We asked whether PHD3 bound this central domain of MDM2 to impede p53–MDM2 interaction. To determine this, we constructed a vector encoding GST-MDM2(Δ217–367) that did not have the central domain. Our results show that PHD3 did not bind GST-MDM2(Δ217–367) (Fig. 2J). We also constructed a vector encoding GST-MDM2(217–367) and found that PHD3 bound GST-MDM2(217–367) (Fig. 2J). These results suggest that PHD3 binds the central domain of MDM2, which prevents MDM2 from mediating p53 destruction.
PHD3 stabilizes p53 independent of its hydroxylase activity
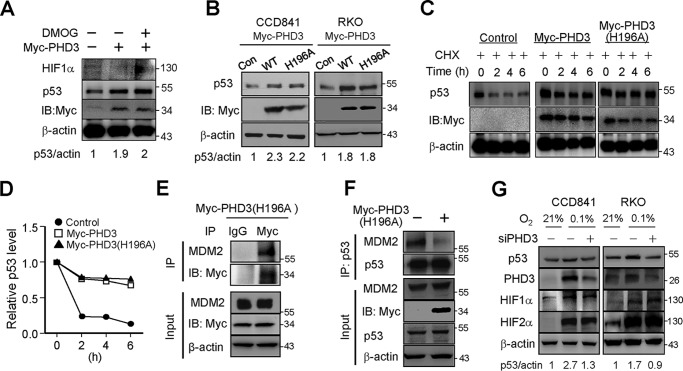
PHD3 has prolyl hydroxylase-independent functions (9, 26). We wanted to know whether the prolyl hydroxylase activity is required for PHD3 to stabilize p53. To this end, we employed the prolyl hydroxylase inhibitor DMOG in our work. Treatment of the cells with DMOG increased HIF1α dramatically (Fig. 3A), indicating that the prolyl hydroxylase activity is suppressed. Interestingly, overexpression of PHD3 still enhanced p53 protein in the presence of DMOG (Fig. 3A). We next employed a vector coding prolyl hydroxylase-deficient PHD3(H196A) in our work (27). We found that overexpression of PHD3(H196A) also enhanced p53 protein (Fig. 3B). Moreover, we found that PHD3(H196A) increased the half-life of p53 as PHD3 did (Fig. 3, C and D). These results suggest that PHD3 stabilizes p53 in a hydroxylase-independent manner. We found that PHD3(H196A) associated with MDM2 (Fig. 3E), and overexpression of PHD3(H196A) suppressed the interaction between p53 and MDM2 (Fig. 3F). Thus, PHD3(H196A) may enhance p53 in a same way as PHD3 does.
Figure 3.
PHD3(H196A) induced p53. A, RKO cells were transfected with control or Myc-PHD3 vector as indicated. After 24 h, DMOG (2 mm) was added, and the cells were incubated for another 4 h, followed by immunoblotting (IB). B, RKO and CCD841 cells were transfected with control, Myc-PHD3, or Myc-PHD3(H196A) vector. After 24 h, the cells were harvested for immunoblotting (IB). C and D, RKO cells were transfected with control, Myc-PHD3, or Myc-PHD3(H196A) vector. After 24 h, cycloheximide (CHX) (10 μg/ml) was added to inhibit new protein synthesis, and the cells were harvested at different time intervals. The right panel shows the relative p53 level at different time point. Relative p53 was shown in D. E, RKO cells were transfected with myc-PHD3(H196A) vector. After 24 h, the cells were harvested, and immunoprecipitation (IP) was performed to determine the interaction between PHD3(H196A) and MDM2. F, RKO cells were transfected with control or PHD3(H196A) vector. After 24 h, MG132 (10 μm) was added, and the cells were incubated for another 3 h before harvesting the cells. Immunoprecipitation was performed to determine the effect of Myc-PHD3(H196A) on the interaction between p53 and MDM2. G, RKO and CCD841 cells were transfected with control or PHD3 siRNA oligonucleotides as indicated. After 48 h, the cells were incubated in normal or 0.1% O2 for 10 h, followed by Western blotting. The Western blots were quantified using ImageJ, and the results of quantification are from one experiment.
In hypoxia, the expression of PHD3 is transcriptionally activated by HIFα (28). A few studies have shown that hypoxia also induces expression of p53 (29–32), so we asked whether PHD3 was involved in hypoxic induction of p53. To know this, we determined the effect of knockdown of PHD3 on expression of p53 in hypoxia. We found that hypoxia enhanced p53, and knockdown of PHD3 attenuated the hypoxia-induced p53 in CCD841 and RKO cells (Fig. 3G). These results suggest that PHD3 is required for hypoxic induction of p53. Further, we found that knockdown of PHD3 had little effect on HIF-1 and -2α protein levels, suggesting that PHD3 does not regulate hypoxic induction of p53 via HIFα.
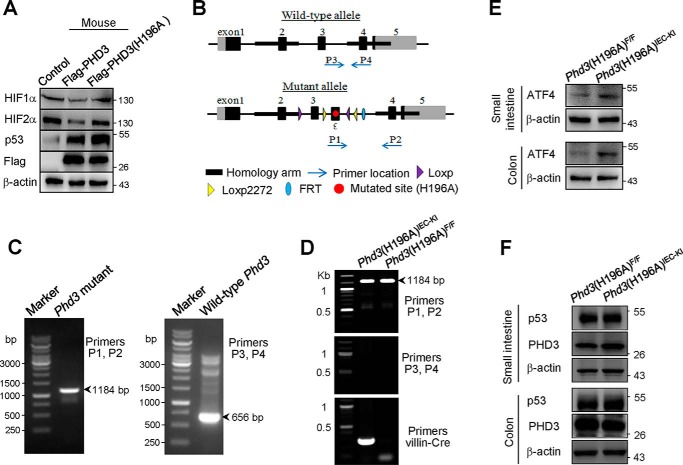
To confirm that PHD3 regulates p53 expression in a hydroxylase-independent manner, we employed intestinal epithelial PHD3(H196A) knockin (Phd3(H196A)IEC-KI) mice in our work. PHD3 of mouse is quite similar to that of human (97% identities). Both human PHD3 and mouse PHD3 consist of 239 amino acids, and they both have residue His196. We constructed mouse PHD3 and PHD3(H196A) vectors and found that overexpression of mouse PHD3, but not PHD3(H196A), decreased HIF-1α and HIF-2α proteins (Table 1 and Fig. 4A), implying that the mouse PHD3(H196A) is deficient of prolyl hydroxylase activity. We found that overexpression of mouse PHD3(H196A) enhanced p53 protein as WT PHD3 did (Fig. 4A).
Table 1.
Primers for determining wildtype (P3 and P4) and mutated (P1 and P2) allele of Phd3 and Villin-Cre
| Primer | Sequence (5′ → 3′) |
|---|---|
| P1 | GTTCGCCTATCAGCTTTGGG |
| P2 | GCATAAAGCCCCAGCATCAGAAA |
| P3 | AGGATTCACTAGCATACAAA |
| P4 | GCATAAAGCCCCAGCATCAGAAA |
| Villin-Cre | ATTTGCCTGCATTACCGGTCGCAGCATTGCTGTCACTTGGTC |
Figure 4.
PHD3(H196A) knockin did not affect the expression of p53. A, RKO cells were transfected with FLAG-tagged mouse PHD3 or PHD3(H196A). After 24 h, the cells were harvested for immunoblotting. B, structure of WT and mutant allele of Phd3 of mice. C, determination of the mutated Phd3 (left panel) and WT Phd3 (right panel) by PCR. Primers 1 and 2 are used for determining the mutated Phd3. Primers 3 and 4 are used for determining the WT Phd3. The homozygous Phd3(H196A) had a band of 1184 bp, and WT Phd3 had a band of 656 bp. The primers for determining WT and mutated Phd3 were shown in Table 1. D, the genotyping of Phd3(H196A)F/F and Phd3(H196A)IEC-KI mice. The primers for determining Villin-Cre was shown in Table 1. E and F, The proteins extracted from small intestine and colon epithelial cells of Phd3(H196A)F/F and Phd3(H196A)IEC-KI mice were subjected to immunoblotting with ATF4 (E) and p53 (F) antibodies. The p53(DO-7) antibody was used to detect mouse p53.
Generation of Phd3(H196A)IEC-KI mice is as described under “Experimental procedures.” The structures of WT and mutant allele of Phd3 of mice were shown (Fig. 4B). PCR results show that the homozygous Phd3(H196A) had a band of 1184 bp, and the WT Phd3 had a band of 656 bp (Fig. 4C). Genotyping of Phd3(H196A)F/F and Phd3(H196A)IEC-KI are shown in Fig. 4D. The right lane that had a mutated Phd3 band indicates Phd3(H196A)F/F mice. The left lane having mutated Phd3 and Villin-Cre bands indicates the Phd3(H196A)IEC-KI mice. It is known that PHD3 hydroxylates ATF4, leading to ATF4 destruction (4). Therefore, we determined the effect of PHD3(H196A) knockin on expression of ATF4 in small intestine and colon epithelial cells. The results show that ATF4 protein level is enhanced in intestinal epithelial cells from Phd3(H196A)IEC-KI mice (Fig. 4E), implying that the mutated PHD3 is deficient of hydroxylase activity. Finally, we determined the protein level of p53 in these cells and found that PHD3(H196A) knockin had no effect p53 protein levels (Fig. 4F).
PHD3 inhibits the expression of NANOG through p53
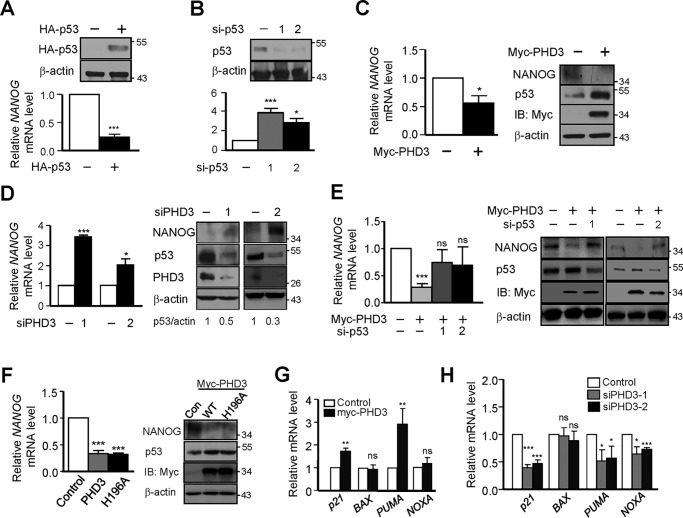
Several studies have implicated a critical role of p53 in stem cells through regulating expression of genes related to these cells (33). Among these genes, NANOG is a major one (34). The expression of NANOG was demonstrated to be regulated negatively by p53 (35). Therefore, we asked whether PHD3 influenced the expression of NANOG through p53. In agreement with previous results, overexpression of p53 decreased (Fig. 5A), and knockdown of p53 enhanced the expression of NANOG (Fig. 5B). As expected, overexpression of PHD3 inhibited (Fig. 5C) and knockdown of PHD3 stimulated (Fig. 5D) the expression of NANOG. To verify that PHD3 regulates the expression of NANOG through p53, we introduced p53 siRNA oligonucleotides into the cells. We found that knockdown of p53 reversed the expression of NANOG suppressed by overexpression of PHD3 (Fig. 5E). These results imply that PHD3 inhibits NANOG expression through p53. In addition, we found that overexpression of PHD3(H196A) also restrained the expression of NANOG as PHD3 did (Fig. 5F). The results suggest that PHD3 attenuates the expression of NANOG in a hydroxylase-independent manner.
Figure 5.
PHD3 induces NANOG. A, RKO cells were transfected with control or HA-p53 vector. After 48 h, the expression of NANOG was determined by Western blotting and quantitative PCR. B, RKO cells were transfected with control or p53 siRNA oligonucleotides as indicated. After 48 h, the expression of NANOG was determined. C, RKO cells were transfected with control or myc-PHD3 vector. After 24 h, the mRNA and protein levels of NANOG were determined. D, RKO cells were transfected with control or PHD3 siRNA oligonucleotides. After 48 h, the cells were harvested for determination of expression of NANOG. E, RKO cells were transfected as indicated. After 48 h, the expression of NANOG was determined. F, RKO cells were transfected as indicated. After 24 h, the cells were harvested for determination of mRNA and protein levels of NANOG. G, RKO cells were transfected as indicated. After 24 h, the cells were harvested for qPCR. H, RKO cells were transfected as indicated. After 48 h, the cells were harvested for qPCR. The Western blots were quantified using ImageJ. The quantification of Western blotting is from one experiment. Con, control; IB, immunoblotting; ns, no significance. *, p < 0.05; ***, p < 0.001.
We also determined the effect of PHD3 on other p53 downstream genes including p21, BAX, PUMA, and NOXA in RKO cells. The results show that overexpression of PHD3 induced the expression of p21 and PUMA (Fig. 5G), and knockdown of PHD3 decreased the expression of p21, PUMA, and NOXA (Fig. 5H).
PHD3 inhibits properties of colon cancer stem cells (CSCs)
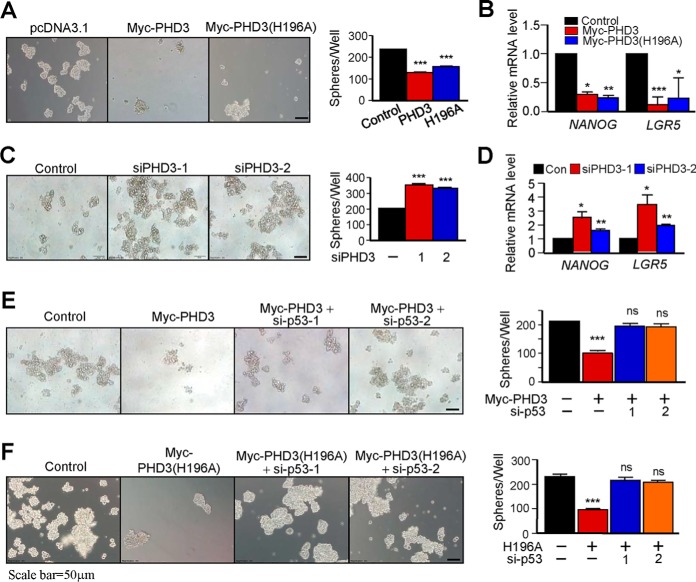
Because PHD3 induced the expression of p53 and inhibited the expression of NANOG (Fig. 5), we presumed that PHD3 was capable of suppressing the properties of colon CSCs. To know this, we determined the effects of PHD3 on sphere formation of RKO cells. A sphere-forming assay has been widely used to identify stem cells (36). Our results show that overexpression of PHD3 suppressed sphere formation of the cells (Fig. 6A). As expected, overexpression of PHD3 inhibited the expression of colon CSC genes NANOG and LGR5 of the cells (Fig. 6B). Overexpression of PHD3(H196A) also repressed sphere formation (Fig. 6A) and attenuated the expression of NANOG and LGR5 (Fig. 6B). On the contrary, knockdown of PHD3 promoted sphere formation (Fig. 6C) and enhanced the expression of NANOG and LGR5 (Fig. 6D). Next, we determined whether PHD3 affects sphere formation through p53. We found that knockdown of p53 reversed the sphere formation inhibited by overexpression of PHD3 (Fig. 6E). Similarly, knockdown of p53 also prevented PHD3(H196A) from suppressing sphere formation of the cells (Fig. 6F). Together, these results suggest that PHD3 functions to inhibit the properties of colon CSCs through p53.
Figure 6.
PHD3 inhibits CSC properties of colon cancer cells. A, RKO cells were transfected with control, Myc-PHD3 or Myc-PHD3(H196A) vector. The sphere formation was determined as described under “Experimental procedures.” The right panel shows that number of spheres/well. B, RKO cells were transfected with control, myc-PHD3, or myc-PHD3(H196A) vector. The mRNA levels of NANOG and LGR5 were determined as described under “Experimental procedures.” C, RKO cells were transfected with control or PHD3 siRNA oligonucleotides. Sphere formation was determined. The right panel shows that number of spheres/well. D, RKO cells were transfected with control or PHD3 siRNA oligonucleotides as indicated. The mRNA levels of NANOG and LGR5 were determined. E, RKO cells were transfected with Myc-PHD3 in the absence or presence of p53 siRNA oligonucleotides. The right panel shows that number of spheres/well. F, RKO cells were transfected with Myc-PHD3(H196A) vector with or without p53 siRNA oligonucleotides. The right panel shows that number of spheres/well. Con, control; ns, no significance. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
We have demonstrated in this manuscript that PHD3 stabilizes p53 by inhibiting the interaction between p53 and MDM2, independent of its hydroxylase activity. The PHD3-induced stabilization of p53 leads to attenuation of the expression of NANOG and suppresses the properties of colon CSCs. Our results reveal a new mechanism of regulation of p53 expression through PHD3 and highlight a role of PHD3 in suppressing colon CSCs.
Stability of p53 is regulated by the ubiquitin-proteasome system, and MDM2 acts as the E3 ubiquitin ligase that mediates p53 degradation (37). Binding to the central domain of MDM2 is essential for p53 ubiquitination and destruction (23–25, 38). We found that PHD3 also bound the central domain of MDM2 (Fig. 2) as p53 did. Thus, PHD3 may block the accessing of p53 to the central domain of MDM2, which prevents MDM2 from mediating p53 destruction. Interestingly, our results showed that the hydroxylase-deficient PHD3(H196A) also stabilized p53 (Fig. 3). It bound MDM2 and blocked the interaction of p53 and MDM2 as the WT PHD3 did (Fig. 3, E and F). We generated PHD3(H196A) knockin mice and found that loss of prolyl hydroxylase activity of PHD3 had little influence on protein level of p53 in vivo (Fig. 4). Considering the fact that PHD3 knockout decreased p53 in mice intestinal epithelial cells (Fig. 1G), our data suggest that PHD3 induces p53 independent of its hydroxylase activity. In hypoxia, HIFα is accumulated and initiates the expression of PHD3 (28). Hypoxia also induces expression of p53 (29–32). The hypoxia-induced PHD3 may reduce MDM2–p53 interaction and stabilize p53. Indeed, we found that knockdown of PHD3 attenuated the hypoxic induction of p53 (Fig. 3G). Thus, the hypoxia-induced PHD3 may contribute the hypoxic induction of p53.
Rodriguez et al. (39) reported recently that PHD3 hydroxylates p53, leading to stabilization of p53. They found that PHD3 hydroxylated p53 to promote the interaction between p53 and deubiquitinases, leading to p53 stabilization. They showed that hypoxia resulted in a reduction of p53 in HeLa and HepG2 cells, implying that the hydroxylase activity is required for p53 expression. We found that the prolyl hydroxylase-deficient PHD3(H196A) also enhanced p53 protein in colon epithelial RKO and CCD841 cells as the WT PHD3 did. The cause for the discrepancy is not clear. One possible reason is that different cells were examined in these two works. Hypoxic induction of p53 is cell type–specific. For instance, Rodriguez et al. (39) demonstrated that hypoxia did not enhance p53 in HeLa and HepG2 cells. Some other reports showed that hypoxia induced accumulation of p53 in lymphoblastoid GM02184B cells (29) and RKO cells (30). Hypoxic induction of p53 is complex. Pan et al. (31) found that hypoxia alone could not induce p53. An et al. (32) reported that HIF-1α is required for stabilization of p53 in hypoxia. It is known that the regulation of expression of p53 is complicated and may be at multiple levels, including transcription, translation, and post-translation. Therefore, it is not surprising in the discrepancies of these works. PHD3 may stabilize p53 in different mechanisms. It is noted that PHD3 had little effect on the expression of p53 in HCT116 cells (Fig. 1G), suggesting that PHD3 stabilizes p53 in a cell type–specific manner.
p53 suppresses self-renewal during the oncogenic process, and it is a barrier to the formation of CSCs (40–42). p53 mediates the stem cell function and suppresses self-renewal via regulating the expression of NANOG (33). NANOG is a crucial factor that confers certain CSCs properties (34), including colon CSCs (43, 44). PHD3 stabilizes p53 and suppresses the expression of NANOG (Fig. 5). Thus, PHD3 may function to inhibit colon CSCs. In fact, our data show that PHD3 functions to suppress the properties of colon CSCs.
PHDs initially are recognized as hydroxylases that catalyze the proline hydroxylation of proteins. Recent studies have indicated that PHDs may also have hydroxylase-independent functions. For instance, Chan et al. (45) demonstrated that PHD2 regulated angiogenesis independent of its hydroxylase activity. Fu and Taubman (26) reported that PHD3 competed with cIAP1 for IKKγ binding, leading to inhibition of cIAP1-IKKγ interaction, IKKγ ubiquitination, and IKK/NF-κB signaling. We found that PHD3 stabilized the tight junction protein occludin (46) in a hydroxylase-independent mechanism. These results suggest that PHD3 works through hydroxylase-dependent and -independent mechanisms.
In this study, we have shown that PHD3 stabilizes p53 and inhibits colon CSC properties independent of its hydroxylase activity. These results broaden the functional scope of PHD3 and expand our understanding of PHD3 as a suppressor of cancer cells.
Experimental procedures
Animals
The C57BL/6J mice were purchased from the Shanghai Experimental Animal Center. The intestinal epithelia-specific Phd3 knockout mice (Phd3IEC-KO) were generated by intercrossing the Phd3flox/flox (Phd3F/F) mice with Villin-Cre ones as described (46). The Phd3(H196A)flox/flox (Phd3(H196A)F/F) mice were created at Shanghai Research Center for Model Organisms. Intestinal epithelial Phd3(H196A) knockin (Phd3(H196A)IEC-KI) mice were generated by intercrossing the Phd3(H196A)F/F mice with Villin-Cre mice. The animals were housed in specified pathogen-free conditions. All experimental procedures were approved by the Institutional Animal Care and Use Committee at the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Cell culture
Human colon cancer RKO, human embryonic kidney 293T, human breast cancer MCF7, human renal cancer ACHN, and human cervical cancer HeLa cells were grown in Dulbecco's modified Eagle's medium. Human normal colon epithelial CCD841 cells were grown in RPMI 1640 medium. Human colon cancer HCT116 cells and human renal cancer Caki cells were grown in M5A medium. All medium were supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 g/ml streptomycin. The cells were cultured at 37 °C in an incubator with 5% CO2.
Antibodies and reagents
The antibodies against PHD3(NB100–303) and HIF-2α(NB100–122) were from Novus Biologicals. The p53(DO-1), p53(DO-7), MDM2(SMP14), ATF4, GST, His, HA, and Myc antibodies were from Santa Cruz Biotechnology. The HIF-1α(610959) and NANOG(Ab80892) antibodies were from BD and Abcam, respectively. β-Actin antibody was from Sigma. EGF was purchased from Life Technologies. B27 and basic fibroblast growth factor was purchased from Invitrogen and Peprotech, respectively.
Construction of vectors
The vectors encoding myc-tagged human PHD3 and PHD3(H196A) were as described (9). The lentivirus encoding FLAG-tagged human PHD3 was purchased from Genechem (Shanghai, China). The HA-p53 vector was from Dr. Zhixue Liu (Institute for Nutritional Sciences, Chinese Academy of Sciences), and p53 vector was from Dr. Hai Jiang (Shanghai Institute for Biochemistry and Cell Biology, Chinese Academy of Sciences). The vector encoding glutathione S-transferase (GST)–MDM2 fusion protein was constructed by inserting PCR-generated DNA fragments encoding regions of MDM2 into pGEX-4T-1. The His-PHD3 vector was generated as described (9). The BL21-Gold(DE3)pLysS Escherichia coli cells were transformed with pGEX-4T-1 or pET28a vectors and treated with isopropyl-d-thiogalactoside (0.1 mm) for 4 h.
siRNA and short hairpin RNA
siRNA duplexes were synthesized by GenePharma (Shanghai, China). The cells were transfected with siRNA oligonucleotides using Lipofectamine-2000: siP53–1, 5′-GUACCACCAUCCACUACAATT-3′; siP53–2, 5′-GUAAUCUACUGGGACGGAATT-3′; siPHD3–1, 5′-GUGAUGGUCGCUGCAUCATT-3′; siPHD3–2, 5′-GGAGAGGUCUAAGGCAAUGTT-3′; and control, 5′-UUCUCCGAACGUGUCACGUTT-3′.
Quantitative real-time PCR (qPCR)
qPCR was performed as described (9). β-Actin was used as the internal control. The primers are as follows: TP53 forward, 5′-GAGAGCTGAATGAGGCCTTG-3′; TP53 reverse, 5′-TTATGGCGGGAGGTAGACTG-3′; NANOG forward, 5′-GGTGGCAGAAAAACAACTGG-3′; NANOG reverse, 5′-CATCCCTGGTGGTAGGAAGA-3′; LGR5 forward, 5′-GGTCGCTCTCATCTTGCTCA-3′; LGR5 reverse, 5′-GCCACAGGGCAGTTTAGGAT-3′; p21 forward, 5′-TGGGGATGTCCGTCAGAACC-3′; p21 reverse, 5′-TCACCCTCCAGTGGTGTCTC-3′; PUMA forward, 5′-CGGAGCAGCACCTGGAGTCG-3′; PUMA reverse, 5′-TTGAGGTCGTCCGCCATCCG-3′; NOXA forward, 5′-CGCAAGAACGCTCAACCGAG-3′; NOXA reverse, 5′-CTGCCGGAAGTTCAGTTTGTCTC-3′; BAX forward, 5′-ACCAAGAAGCTGAGCGAGTGT-3′; BAX reverse, 5′- CCAGTTGAAGTTGCCGTCAG-3′; β-actin forward, 5′-GATCATTGCTCCTCCTGAGC-3′; and β-actin reverse, 5′-ACTCCTGCTTGCTGATCCAC-3′.
In vitro p53 ubiquitination assay
The MDM2/p53 ubiquitination kit (catalog no. K-200B) from BostonBiochem was used in the experiment. The in vitro ubiquitination assay of p53 was performed as described (47). In brief, 293T cells were transfected to express myc-PHD3. 100 or 400 μg of cellular proteins were immunoprecipitated with 0.25 or 1 μg of Myc antibody at 4 °C for 3 h. 40 μl of protein A/G Plus-agarose beads were added, and the incubation continued at 4 °C overnight. The beads were washed, spun, and resuspended in a reaction volume of 30 μl. The mixture was incubated at 37 °C for 1 h. After the reaction completed, the supernatant was used for p53 ubiquitination analysis in a Western blot. The beads were boiled in loading buffer, resolved in SDS-PAGE, and immunoblotted with Myc antibody.
Isolation of intestinal epithelial cells
The intestine was removed and washed free of fecal material with solution A (96 mm NaCl, 27 mm sodium citrate, 1.5 mm KCl, 0.8 mm KH2PO4, 5.6 mm Na2HPO4, 5,000 units/liter penicillin, 5 mg/liter streptomycin, 0.5 mm DTT, and 2 mm phenylmethylsulfonyl fluoride, pH 7.4). Square pieces of tissue were placed in solution A (10 ml) at 37 °C for 10 min with gentle shaking. This removed the mucus, bacteria, and other lumen contents. The tissue fragments were then incubated in solution B (0.1 mm EDTA, 115 mm NaCl, 25 mm NaHCO3, 2.4 mm K2HPO4, 0.4 mm KH2PO4, 0.5 mm DTT, 5 mg/liter streptomycin, 2.5 mm glutamine, 5,000 U/liter penicillin, and 2 mm phenylmethylsulfonyl fluoride, pH 7.4) at 37 °C for 30 min. The disruption of the mucosa and elution of cells was stopped by adjusting to 1 mm CaCl2. The cells recovered in the suspension were collected by centrifugation and lysed in radioimmune precipitation assay buffer for immunoblotting.
Tumor sphere formation assay
The transfected cells were seeded in 96-well ultra-low attachment plates at a density of 2 × 103 cells/well in 200 μl of serum-free Dulbecco's modified Eagle's medium/F-12 medium containing 2% B27, 20 ng/ml EGF, 10 ng/ml basic fibroblast growth factor, and 5 μg/ml insulin. The cells were incubated in a humidified atmosphere at 37 °C with 5% CO2. After 120 h, spheres (>50 μm in diameter) were counted under a microscope.
Statistical analysis
Statistical analysis was made using the unpaired two-tailed Student's t test or two-way analysis of variance with GraphPad Prism 5.0. The data represent means ± S.E. from three independent experiments except where indicated. p < 0.05 is considered statistically significant.
Author contributions
Y. Xu, Q. G., Y. Xue, X. L., L. X., and Y. Q. investigation; C. L. visualization; J. F. conceptualization; J. F. supervision; J. F. funding acquisition; J. F. writing-review and editing.
This work was supported by Ministry of Science and Technology of China Grant 2017YFD0400206, National Natural Science Foundation of China Grants 31470769 and 31670785, Chinese Academy of Sciences Grant KFZD-SW-213, and Shanghai Ministry of Science and Technology Grant 16JC1406100. The authors declare that they have no conflicts of interest with the contents of this article.
- PHD
- prolyl hydroxylase
- HIF
- hypoxia-inducible factor
- DMOG
- dimethyloxalylglycine
- ATF
- activating transcriptional factor
- CSC
- cancer stem cell
- EGF
- epidermal growth factor
- GST
- glutathione S-transferase
- qPCR
- quantitative real-time PCR.
References
- 1. Fong G. H., and Takeda K. (2008) Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 15, 635–641 10.1038/cdd.2008.10 [DOI] [PubMed] [Google Scholar]
- 2. Kaelin W. G. Jr., and Ratcliffe P. J. (2008) Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 10.1016/j.molcel.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 3. Schofield C. J., and Ratcliffe P. J. (2004) Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354 10.1038/nrm1366 [DOI] [PubMed] [Google Scholar]
- 4. Köditz J., Nesper J., Wottawa M., Stiehl D. P., Camenisch G., Franke C., Myllyharju J., Wenger R. H., and Katschinski D. M. (2007) Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood 110, 3610–3617 10.1182/blood-2007-06-094441 [DOI] [PubMed] [Google Scholar]
- 5. Xie L., Xiao K., Whalen E. J., Forrester M. T., Freeman R. S., Fong G., Gygi S. P., Lefkowitz R. J., and Stamler J. S. (2009) Oxygen-regulated β2-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci. Signal. 2, ra33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fu J., Menzies K., Freeman R. S., and Taubman M. B. (2007) EGLN3 Prolyl hydroxylase regulates skeletal muscle differentiation and myogenin protein stability. J. Biol. Chem. 282, 12410–12418 10.1074/jbc.M608748200 [DOI] [PubMed] [Google Scholar]
- 7. Xie L., Pi X., Mishra A., Fong G., Peng J., and Patterson C. (2012) PHD3-dependent hydroxylation of HCLK2 promotes the DNA damage response. J. Clin. Invest. 122, 2827–2836 10.1172/JCI62374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodriguez J., Pilkington R., Garcia Munoz A., Nguyen L. K., Rauch N., Kennedy S., Monsefi N., Herrero A., Taylor C. T., and von Kriegsheim A. (2016) Substrate-trapped interactors of PHD3 and FIH cluster in distinct signaling pathways. Cell Rep. 14, 2745–2760 10.1016/j.celrep.2016.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xue J., Li X., Jiao S., Wei Y., Wu G., and Fang J. (2010) Prolyl hydroxylase-3 is down-regulated in colorectal cancer cells and inhibits IKKβ independent of hydroxylase activity. Gastroenterology 138, 606–615 10.1053/j.gastro.2009.09.049 [DOI] [PubMed] [Google Scholar]
- 10. Place T. L., Fitzgerald M. P., Venkataraman S., Vorrink S. U., Case A. J., Teoh M. L., and Domann F. E. (2011) Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS One 6, e14617 10.1371/journal.pone.0014617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hatzimichael E., Dasoula A., Shah R., Syed N., Papoudou-Bai A., Coley H. M., Dranitsaris G., Bourantas K. L., Stebbing J., and Crook T. (2010) The prolyl-hydroxylase EGLN3 and not EGLN1 is inactivated by methylation in plasma cell neoplasia. Eur. J. Haematol. 84, 47–51 10.1111/j.1600-0609.2009.01344.x [DOI] [PubMed] [Google Scholar]
- 12. Lee S., Nakamura E., Yang H., Wei W., Linggi M. S., Sajan M. P., Farese R. V., Freeman R. S., Carter B. D., Kaelin W. G. Jr., and Schlisio S. (2005) Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell 8, 155–167 10.1016/j.ccr.2005.06.015 [DOI] [PubMed] [Google Scholar]
- 13. Tennant D. A., and Gottlieb E. (2010) HIF prolyl hydroxylase-3 mediates α-ketoglutarate-induced apoptosis and tumor suppression. J. Mol. Med. 88, 839–849 10.1007/s00109-010-0627-0 [DOI] [PubMed] [Google Scholar]
- 14. Rantanen K., Pursiheimo J., Högel H., Himanen V., Metzen E., and Jaakkola P. M. (2008) Prolyl hydroxylase PHD3 activates oxygen-dependent protein aggregation. Mol. Biol. Cell 19, 2231–2240 10.1091/mbc.e07-11-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui L., Qu J., Dang S., Mao Z., Wang X., Fan X., Sun K., and Zhang J. (2014) Prolyl hydroxylase 3 inhibited the tumorigenecity of gastric cancer cells. Mol. Carcinog. 53, 736–743 10.1002/mc.22025 [DOI] [PubMed] [Google Scholar]
- 16. Tanaka T., Torigoe T., Hirohashi Y., Sato E., Honma I., Kitamura H., Masumori N., Tsukamoto T., and Sato N. (2014) Hypoxia-inducible factor (HIF)-independent expression mechanism and novel function of HIF prolyl hydroxylase-3 in renal cell carcinoma. J. Cancer Res. Clin. Oncol. 140, 503–513 10.1007/s00432-014-1593-7 [DOI] [PubMed] [Google Scholar]
- 17. Peurala E., Koivunen P., Bloigu R., Haapasaari K. M., and Jukkola-Vuorinen A. (2012) Expressions of individual PHDs associate with good prognostic factors and increased proliferation in breast cancer patients. Breast Cancer Res. Treat. 133, 179–188 10.1007/s10549-011-1750-5 [DOI] [PubMed] [Google Scholar]
- 18. Su C., Huang K., Sun L., Yang D., Zheng H., Gao C., Tong J., and Zhang Q. (2012) Overexpression of the HIF hydroxylase PHD3 is a favorable prognosticator for gastric cancer. Med. Oncol. 29, 2710–2715 10.1007/s12032-012-0171-6 [DOI] [PubMed] [Google Scholar]
- 19. Henze A.-T., Garvalov B. K., Seidel S., Cuesta A. M., Ritter M., Filatova A., Foss F., Dopeso H., Essmann C. L., Maxwell P. H., Reifenberger G., Carmeliet P., Acker-Palmer A., and Acker T. (2014) Loss of PHD3 allows tumours to overcome hypoxic growth inhibition and sustain proliferation through EGFR. Nat. Commun. 5, 5582 10.1038/ncomms6582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grand R. J., Turnell A. S., Mason G. G., Wang W., Milner A. E., Mymryk J. S., Rookes S. M., Rivett A. J., and Gallimore P. H. (1999) Adenovirus early region 1A protein binds to mammalian SUG1: a regulatory component of the proteasome. Oncogene 18, 449–458 10.1038/sj.onc.1202304 [DOI] [PubMed] [Google Scholar]
- 21. Lowe S. W., and Ruley H. E. (1993) Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7, 535–545 10.1101/gad.7.4.535 [DOI] [PubMed] [Google Scholar]
- 22. Moll U. M., and Petrenko O. (2003) The MDM2-p53 interaction. Mol. Cancer Res. 1, 1001–1008 [PubMed] [Google Scholar]
- 23. Meulmeester E., Frenk R., Stad R., de Graaf P., Marine J. C., Vousden K. H., and Jochemsen A. G. (2003) Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol. Cell. Biol. 23, 4929–4938 10.1128/MCB.23.14.4929-4938.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu G. W., Rudiger S., Veprintsev D., Freund S., Fernandez-Fernandez M. R., and Fersht A. R. (2006) The central region of HDM2 provides a second binding site for p53. Proc. Natl. Acad. Sci. U.S.A. 103, 1227–1232 10.1073/pnas.0510343103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma J., Martin J. D., Zhang H., Auger K. R., Ho T. F., Kirkpatrick R. B., Grooms M. H., Johanson K. O., Tummino P. J., Copeland R. A., and Lai Z. (2006) A second p53 binding site in the central domain of Mdm2 is essential for p53 ubiquitination. Biochemistry 45, 9238–9245 10.1021/bi060661u [DOI] [PubMed] [Google Scholar]
- 26. Fu J., and Taubman M. B. (2013) EGLN3 inhibition of NF-κB is mediated by prolyl hydroxylase-independent inhibition of IκB kinase γ ubiquitination. Mol. Cell. Biol. 33, 3050–3061 10.1128/MCB.00273-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bruick R. K., and McKnight S. L. (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 10.1126/science.1066373 [DOI] [PubMed] [Google Scholar]
- 28. Pescador N., Cuevas Y., Naranjo S., Alcaide M., Villar D., Landázuri M. O., and Del Peso L. (2005) Identification of a functional hypoxia-responsive element that regulates the expression of the egl 9 homologue 3 (egln3/phd3) gene. Biochem. J. 390, 189–197 10.1042/BJ20042121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Graeber T. G., Peterson J. F., Tsai M., Monica K., Fornace A. J. Jr., and Giaccia A. J. (1994) Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol. Cell. Biol. 14, 6264–6277 10.1128/MCB.14.9.6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koumenis C., Alarcon R., Hammond E., Sutphin P., Hoffman W., Murphy M., Derr J., Taya Y., Lowe S. W., Kastan M., and Giaccia A. (2001) Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell. Biol. 21, 1297–1310 10.1128/MCB.21.4.1297-1310.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan Y., Oprysko P. R., Asham A. M., Koch C. J., and Simon M. C. (2004) p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene 23, 4975–4983 10.1038/sj.onc.1207657 [DOI] [PubMed] [Google Scholar]
- 32. An W. G., Kanekal M., Simon M. C., Maltepe E., Blagosklonny M. V., and Neckers L. M. (1998) Stabilization of wild-type p53 by hypoxia-inducible factor 1α. Nature 392, 405–408 10.1038/32925 [DOI] [PubMed] [Google Scholar]
- 33. Bonizzi G., Cicalese A., Insinga A., and Pelicci P. G. (2012) The emerging role of p53 in stem cells. Trends Mol. Med. 18, 6–12 10.1016/j.molmed.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 34. Gong S., Li Q., Jeter C. R., Fan Q., Tang D. G., and Liu B. (2015) Regulation of NANOG in cancer cells. Mol. Carcinog. 54, 679–687 10.1002/mc.22340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin T., Chao C., Saito S., Mazur S. J., Murphy M. E., Appella E., and Xu Y. (2005) p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 7, 165–171 10.1038/ncb1211 [DOI] [PubMed] [Google Scholar]
- 36. Pastrana E., Silva-Vargas V., and Doetsch F. (2011) Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 8, 486–498 10.1016/j.stem.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hock A. K., and Vousden K. H. (2014) The role of ubiquitin modification in the regulation of p53. Biochim. Biophys. Acta 1843, 137–149 10.1016/j.bbamcr.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 38. Karni-Schmidt O., Lokshin M., and Prives C. (2016) The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11, 617–644 10.1146/annurev-pathol-012414-040349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodriguez J., Herrero A., Li S., Rauch N., Quintanilla A., Wynne K., Krstic A., Acosta J. C., Taylor C., Schlisio S., and von Kriegsheim A. (2018) PHD3 regulates p53 protein stability by hydroxylating proline 359. Cell Rep. 24, 1316–1329 10.1016/j.celrep.2018.06.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu D., Ou L., Clemenson G. D. Jr., Chao C., Lutske M. E., Zambetti G. P., Gage F. H., and Xu Y. (2010) Puma is required for p53-induced depletion of adult stem cells. Nat. Cell Biol. 12, 993–998 10.1038/ncb2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao T., and Xu Y. (2010) p53 and stem cells: new developments and new concerns. Trends Cell Biol. 20, 170–175 10.1016/j.tcb.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 42. Aloni-Grinstein R., Shetzer Y., Kaufman T., and Rotter V. (2014) p53: the barrier to cancer stem cell formation. FEBS Lett. 588, 2580–2589 10.1016/j.febslet.2014.02.011 [DOI] [PubMed] [Google Scholar]
- 43. Zhang J., Espinoza L. A., Kinders R. J., Lawrence S. M., Pfister T. D., Zhou M., Veenstra T. D., Thorgeirsson S. S., and Jessup J. M. (2013) NANOG modulates stemness in human colorectal cancer. Oncogene 32, 4397–4405 10.1038/onc.2012.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ibrahim E. E., Babaei-Jadidi R., Saadeddin A., Spencer-Dene B., Hossaini S., Abuzinadah M., Li N., Fadhil W., Ilyas M., Bonnet D., and Nateri A. S. (2012) Embryonic NANOG activity defines colorectal cancer stem cells and modulates through AP1- and TCF-dependent mechanisms. Stem Cells 30, 2076–2087 10.1002/stem.1182 [DOI] [PubMed] [Google Scholar]
- 45. Chan D. A., Kawahara T. L., Sutphin P. D., Chang H. Y., Chi J. T., and Giaccia A. J. (2009) Tumor vasculature is regulated by PHD2-mediated angiogenesis and bone marrow-derived cell recruitment. Cancer Cell 15, 527–538 10.1016/j.ccr.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y., Zhang H. S., Fong G. H., Xi Q. L., Wu G. H., Bai C. G., Ling Z. Q., Fan L., Xu Y. M., Qin Y. Q., Yuan T. L., Sun H., and Fang J. (2015) PHD3 stabilizes the tight junction protein occludin and protects intestinal epithelial barrier function. J. Biol. Chem. 290, 20580–20589 10.1074/jbc.M115.653584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., Zhao W., Gao Q., Fan L., Qin Y., Zhou H., Li M., and Fang J. (2016) pVHL mediates K63-linked ubiquitination of IKKβ, leading to IKKβ inactivation. Cancer Lett. 383, 1–8 10.1016/j.canlet.2016.09.009 [DOI] [PubMed] [Google Scholar]