Abstract
Jasmonoyl-isoleucine (JA-Ile) is a phytohormone that orchestrates plant defenses in response to wounding, feeding insects, or necrotrophic pathogens. JA-Ile metabolism has been studied intensively, but its catabolism as a potentially important mechanism for the regulation of JA-Ile–mediated signaling is not well-understood. Especially the enzyme(s) responsible for specifically glycosylating 12-hydroxy-jasmonic acid (12-OH-JA) and thereby producing 12-O-glucopyranosyl-jasmonic acid (12-O-Glc-JA) is still elusive. Here, we used co-expression analyses of available Arabidopsis thaliana transcriptomic data, identifying four UDP-dependent glycosyltransferase (UGT) genes as wound-induced and 12-OH-JA–related, namely, UGT76E1, UGT76E2, UGT76E11, and UGT76E12. We heterologously expressed and purified the corresponding proteins to determine their individual substrate specificities and kinetic parameters. We then used an ex vivo metabolite-fingerprinting approach to investigate these proteins in conditions as close as possible to their natural environment, with an emphasis on greatly extending the range of potential substrates. As expected, we found that UGT76E1 and UGT76E2 are 12-OH-JA-UGTs, with UGT76E1 contributing a major in vivo UGT activity, as deduced from Arabidopsis mutants with abolished or increased UGT gene expression. In contrast, recombinant UGT76E11 acted on an unidentified compound and also glycosylated two other oxylipins, 11-hydroxy-7,9,13-hexadecatrienoic acid (11-HHT) and 13-hydroxy-9,11,15-octadecatrienoic acid (13-HOT), which were also accepted by recombinant UGT76E1, UGT76E2, and UGT76E12 enzymes. UGT76E12 glycosylated 12-OH-JA only to a low extent, but also accepted an artificial hydroxylated fatty acid and low amounts of kaempferol. In conclusion, our findings have elucidated the missing step in the wound-induced synthesis of 12-O-glucopyranosyl-jasmonic acid in A. thaliana.
Keywords: glycosylation, glycosyltransferase, phytohormone, plant defense, CRISPR/Cas, oxylipin
Introduction
Necrotrophic pathogens, feeding insects, and wounding in general are serious threats for all plants and cause severe losses in agricultural yields (1). The phytohormone jasmonoyl-isoleucine (JA-Ile)2 is a ubiquitous key player in the organization of the response to those threats (2). Its precursor jasmonic acid (JA) and respective derivatives are collectively known as jasmonates and belong to the large group of oxidized polyunsaturated fatty acids, so-called oxylipins (3).
In Arabidopsis thaliana, JA biosynthesis starts with the oxygenation of α-linolenic acid (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid) to 13S-hydroperoxy-octadecatrienoic acid (13-HPOT) by four specific 13-linoleate lipoxygenase enzymes (13-LOX). Next, the allene oxide synthase produces an unstable allene oxide, which is subsequently converted to cis-(+)-12-oxo-phytodienoic acid (12-OPDA) by the allene oxide cyclase. 12-OPDA is transported from the plastid to the peroxisome, reduced by 12-OPDA reductase 3 (OPR3), and subjected to three rounds of β-oxidation yielding the core molecule JA (2). An alternative JA pathway lacks the OPR3-mediated reduction of 12-OPDA, which undergoes β-oxidation directly, thus yielding didehydro-JA (ddh-JA). ddh-JA is finally reduced to JA by OPR2 (4). In contrast to other phytohormones, JA gains its activity through conjugation to the amino acid isoleucine by the JA-amido synthetase, named JA resistance 1 (JAR1, GH3.11) (5).
JA-Ile reprograms affected plants toward a defense mode, however, signal termination is highly important for a timely restoration of efficient growth (6, 7). For this, P450 enzymes oxidize JA-Ile at the ω-terminus of the pentenyl side chain to yield the first inactive form, 12-hydroxy-JA-Ile (12-OH-JA-Ile) (8–12). Further oxidation yields 12-carboxy-JA-Ile (12-COOH-JA-Ile) (10). Alternatively, all three oxidation states can be de-conjugated (13, 14), hereby yielding JA, 12-hydroxy-JA (12-OH-JA), or 12-carboxy-JA (12-COOH-JA). JA levels are also depleted through a direct oxidation of JA to 12-OH-JA by jasmonate-induced oxygenases 1 to 4 (JOX1–4), thereby down-regulating the JA-Ile–dependent responses (15–17).
12-OH-JA does not exhibit JA-typical responses like inhibition of root growth, germination, or expression of JA-responsive genes (18, 19). It is not only synthesized, but can also be modified further in two different ways. The specific sulfotransferase 2a (ST2a) forms 12-hydroxy-JA sulfate (12-HSO4-JA) (18), however, the 12-hydroxy group can also be glycosylated to form 12-O-glucopyranosyl-JA (12-O-Glc-JA) (2), for which the responsible enzymes are still elusive (2, 7). Both compounds have been identified in different plant species in an organ-specific manner (19, 20). Although their synthesis is induced by JA-Ile/COI, they do not stimulate the defense reaction themselves (19, 20). Similar to 12-OH-JA and 12-HSO4-JA, 12-O-Glc-JA may be part of the inactivation process of the JA-Ile–mediated defense (18, 19) and the balance of 12-HSO4-JA and 12-O-Glc-JA seems to be tightly regulated by competition for the joint substrate 12-OH-JA (21, 22). 12-OH-JA and/or 12-O-Glc-JA have been reported as mobile compounds in Solanum tuberosum and Nicotiana tabacum (20). In mimosa, 12-O-Glc-JA may induce leaf closure (23). However, without the responsible enzymes known, specific and individual functions of 12-OH-JA and its respective glycoside cannot be addressed.
Generally, glycosylated forms are well-known in A. thaliana for most phytohormones except for ethylene (24). Glycosylation increases the solubility of compounds and promotes their metabolic transport, thereby regulating the bioactivity and the homeostasis of specialized metabolites and phytohormones, for example (24–26). 2-O-Glucosyl-salicylic acid (2-O-Glc-SA) for instance, was found in the vacuoles of tobacco and soybean cells (27, 28). With respect to jasmonates, even more glucose derivatives like JA-glucosyl ester (JA-GE) (29) and 12-O-glucopyranosyl-JA-Ile (12-O-Glc-JA-Ile) (30) have been identified, yet again, the responsible enzymes are still missing.
Glycosylation is mostly mediated by UDP-dependent glycosyltransferases (UGTs), which are ubiquitous glycosyltransferases that transfer sugar moieties from UDP-activated sugars to small acceptor molecules, such as hormones, lipids, and secondary metabolites (25, 31). To identify the genes responsible for the so far uncharacterized reaction in JA-Ile catabolism, we characterized the enzyme specificities and function of four UGTs from Arabidopsis, (originally selected based on their wound-induced expression profile) and established the role of UGT76E1 as the predominant 12-O-Glc-JA forming in vivo activity in response to mechanical wounding in A. thaliana.
Results
UGT76E1, UGT76E2, UGT7E11, and UGT76E12 are related to JA-Ile metabolism
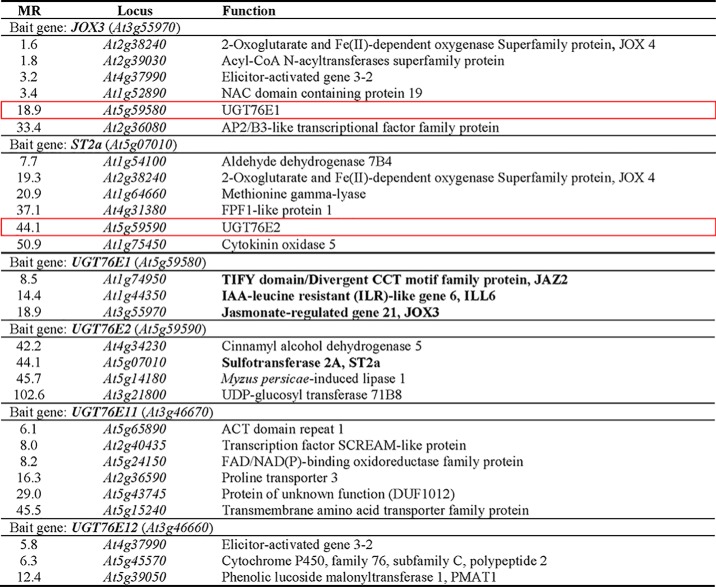
The JA-Ile–mediated defense response in plants is achieved through quick and rapid modifications of jasmonates (2, 15). Therefore, the gene(s) encoding the UGTs acting on 12-OH-JA are most likely transcribed within the same time period in which the corresponding 12-O-Glc-JA accumulates after wounding. We hence performed RNA-seq co-expression analyses on publically available Arabidopsis data sets (32), using JOX1–4 as bait genes, which encode oxidases catalyzing the last step of 12-OH-JA synthesis (16). JOX3 showed co-expression with UGT76E1 (At5g59580), a so far uncharacterized UGT (Table 1), whereas JOX1, JOX2, and JOX4 gave no hits with UGTs. Vice versa, UGT76E1 itself correlated with JAZ2, ILL6, and JOX3, which are connected to the JA-Ile–mediated wound-response in A. thaliana (Table 1) (2). Next, we used ST2a as bait gene, of which the gene product forms 12-HSO4-JA from 12-OH-JA. ST2a showed co-expression with UGT76E2 (At5g59590) (Table 1), a second uncharacterized UGT that lies right next to UGT76E1 on chromosome 5 and probably originates from a gene duplication. UGT76E2 mirrors this co-expression but did not show further correlation to any of the other JA-metabolizing gene products.
Table 1.
Co-expression analyses for identification of putative UDP-glucose: 12-OH-JA glucosyltransferase genes
RNA-seq co-expression analysis of genes involved in 12-OH-JA metabolism. Jasmonic acid oxidase 3 (JOX3) and sulfotransferase 2a (ST2a) were used as bait genes to search for UGT genes. Two genes encoding for uncharacterized glycosyltransferases, UGT76E1 and UGT76E2, were identified (red frames). RNA-seq co-expression analysis of genes correlating with UGT76E1, UGT76E2, UGT76E11, and UGT76E12. Identified gene products related to JA metabolism are given in bold. JAZ2, jasmonate-ZIM-domain protein 2; JOX3, jasmonic acid oxidase 3. Given are the complete database outputs for every bait. Gene products are ranked by Mutual Rank (MR), giving an average correlation of two genes indicating stronger correlation by smaller values. Co-expression data are publically available from (32).
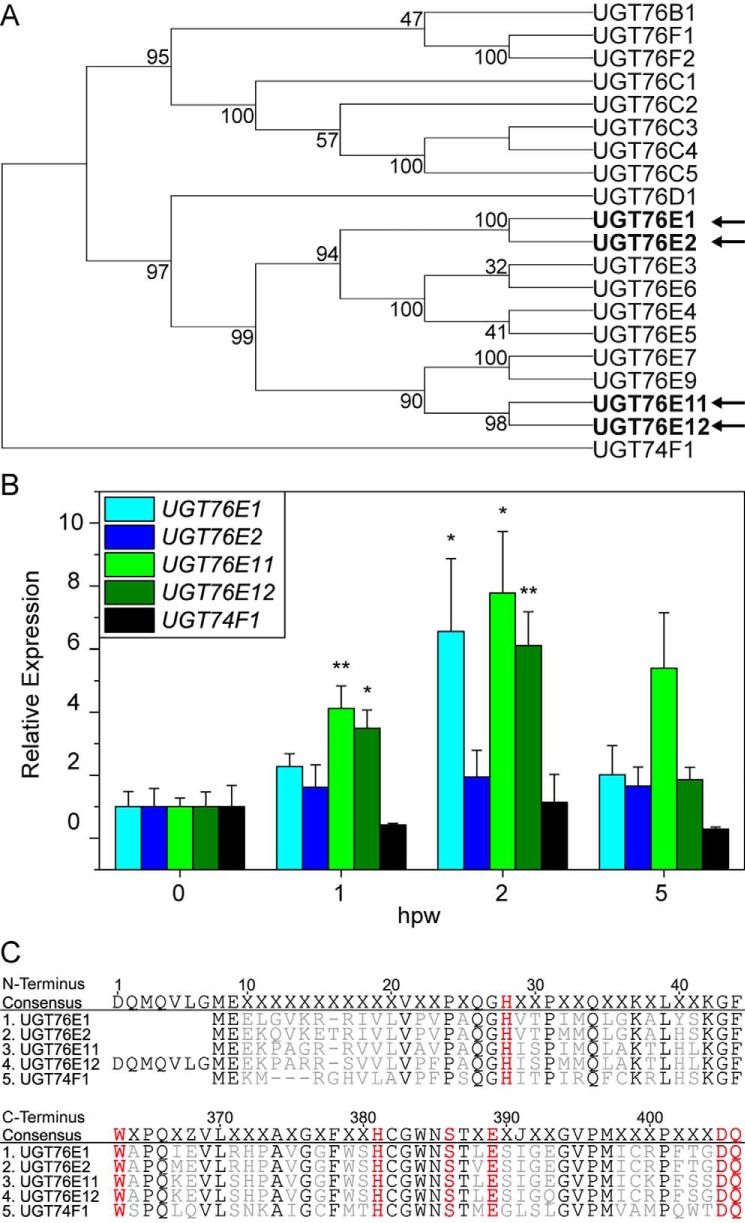
The phylogeny of the A. thaliana UGT superfamily has been studied before (26, 33). UGT76E1 and UGT76E2 show a high amino acid sequence identity (88%) and are part of group H, which comprises 19 Arabidopsis members of the UGT76 family, but of which only three have been described in more detail so far (Fig. 1A) (34). The same group harbors two more UGT genes, UGT76E11 (At3g46660) and UGT76E12 (At3g46670), also with elevated transcript levels in shoots after wounding (35). In addition, UGT76E11 and UGT76E12 also cluster as homologues with 84% amino acid sequence identity. For all subsequent experiments, we included UGT74F1 as a reference glycosyltransferase, which glycosylates salicylic acid (SA), another plant hormone active in a different defense pathway (33, 36) (Fig. 1A), and which groups into the well-studied UGT74 family.
Figure 1.
Sequence and expression analysis of tentative UGTs for 12-OH-JA glycosylation. A, molecular phylogenetic analysis for 20 of 107 UGT genes of A. thaliana by Maximum Likelihood method. The selected branch shows the group H of a complete analysis (26) and the outgroup UGT74F1. The tree of the highest likelihood is shown with the percentage of associated taxa clustering given next to the branches. The four UGT genes, which are induced after wounding are indicated in bold with arrows. Evolutionary analyses were conducted in MEGA X (63). B, quantitative real-time PCR of UGT76E1, UGT76E2, UGT76E11, UGT76E12, and UGT74F1 after wounding. Plants were grown for 6 weeks under short day conditions and wounded. Damaged rosette leaves were harvested at the indicated time points (hpw) and RNA was isolated. All expression values are normalized to Actin 8 as reference and to their respective unwounded levels (0 hpw). Each data point represents the mean ± S.E. of three independent experiments. Asterisks indicate significance by one-sided t test with: *, p < 0.05; **, p < 0.01. C, amino acid alignment of the four UGT76Es (1–4) and the control UGT74F1 (5). Amino acids, which are conserved in the six indicated UGTs, are highlighted in black. The catalytic histidine at the N terminus and the UDP-Glc binding motif at the C terminus are shown in red. Alignments were created in Geneious version 8.1.
Transcripts of UGT76E1, UGT7E11, and UGT76E12 accumulate after wounding
First of all, quantitative real-time PCR was performed to prove the findings of the co-expression analyses (Fig. 1B). The data confirmed the accumulation of UGT76E1, UGT76E11, and UGT76E12 transcripts in shoots at 1 and/or 2 h post wounding (hpw). Remarkably, UGT76E2 was not expressed significantly higher at these time points in wounded leaves, but was nevertheless included in all further experiments.
Sequence analysis confirmed the presence of both for UGTs mandatory catalytic motifs (37): all four enzymes carry a catalytic histidine at position 28 of the N-terminal consensus sequence as well as six conserved amino acids of the UDP-Glc–binding motif at the C terminus, which were determined by co-alignment with the UGT74F1 as reference (Fig. 1C).
UGT76E1, UGT7E2, and UGT76E12 catalyze the glycosylation of 12-OH-JA to 12-O-Glc-JA
For biochemical characterization, all five proteins were heterologously expressed and purified from recombinant Escherichia coli by affinity chromatography and size exclusion chromatography (Fig. S1). Although the four UGT76Es are very similar with respect to their properties (Mr, extinction coefficient, pI, localization, and the optimal reaction temperature; Table S1), the purification protocols required individual optimization. The optimal pH values differed between pH 7.5 and 8.0 for UGT76E1, UGT76E2 and UGT76E11, UGT76E12, respectively (Table S1). To test their catalytic activity toward 12-OH-JA, UDP-Glc-dependent glycosylation of 12-OH-JA was monitored by high resolution LC-MS (Fig. 2). The extracted ion chromatograms for the enzymatic product 12-O-Glc-JA ([M-H]− 387.116, 3.3 min) depicted the highest activity for UGT76E2 (100%), UGT76E1 and UGT76E12 both showed 65%, whereas only trace amounts could be detected for UGT76E11. UGT74F1 showed no product signal. The identity of the tentatively assigned product 12-O-Glc-JA was confirmed by comparison with authentic 12-O-Glc-JA (kindly provided by Dr. Otto Miersch, IPB Halle, Germany). The signal of [M-H]− 387.116 (from the analysis of the UGT-catalyzed reactions with 12-OH-JA) had an identical retention time as well as the same fragmentation pattern by high resolution MS/MS analysis as the authentic standard (Fig. 3, A and B, Stp and P). The analytical fragment of m/z 207.101 derived from the neutral loss of the Glc moiety and m/z 59.013 represented the acetyl moiety of JA (Fig. 3C). In conclusion, UGT76E1, UGT76E2, and UGT76E12 form 12-O-Glc-JA in vitro and can be thus classified as 12-OH-JA-UGTs.
Figure 2.
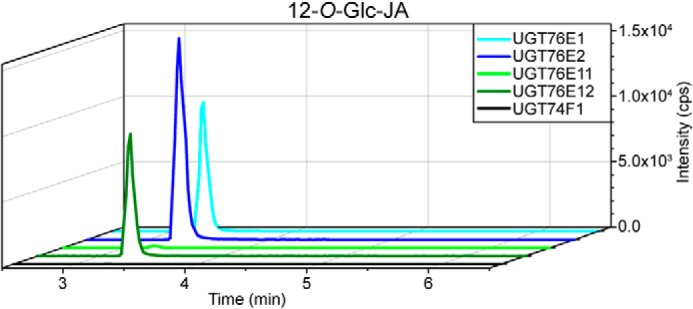
LC-MS–based activity assays of UGT76E1, UGT76E2, UGT7E11, UGT76E12, and UGT74F1 with 12-OH-JA. Activity tests of 10 μg of purified UGT76E1, UGT76E2, UGT76E11, UGT76E12, and UGT74F1 were performed with 0.1 mm 12-OH-JA and 0.5 mm UDP-Glc for 1 h at 25 °C. The reactions were stopped by adding 25 μl of acetonitrile and analyzed by LC-MS. Extracted ion chromatograms are shown for the product 12-O-Glc-JA ([M-H]− 387.116). Signal intensity is given in counts per second (cps). Data represent a single experiment.
Figure 3.
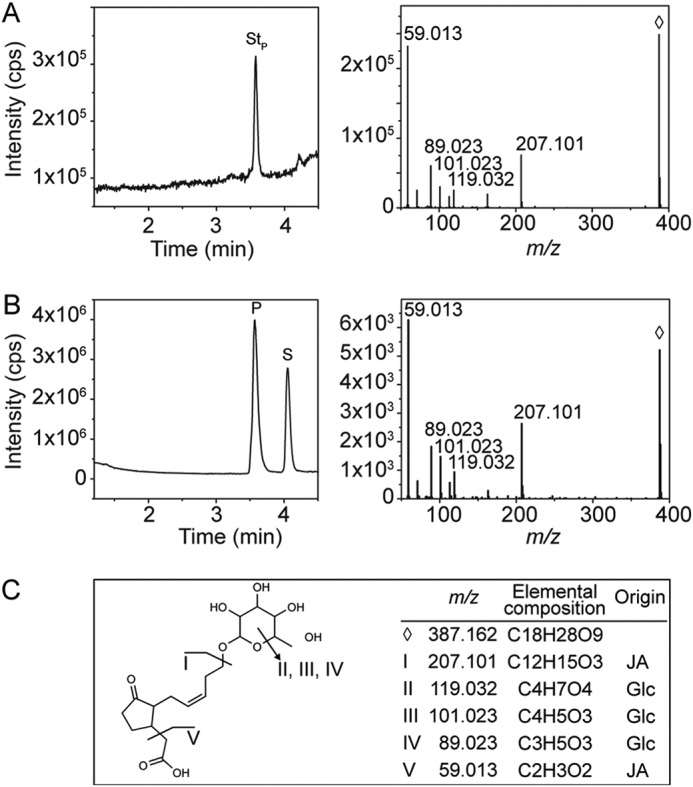
Structure confirmation of 12-O-Glc-JA. An authentic chemical standard of 12-O-Glc-JA ([M-H]− 387.116, ♢) was compared with the reaction product of UGT76E2 with 12-OH-JA by LC-MS/MS. A, total ion chromatogram and the corresponding MS/MS spectrum of the authentic 12-O-Glc-JA (StP) standard. B, total ion chromatogram of the in vitro reaction of UGT76E2 with 12-OH-JA showing the signals for the product (P) and substrate (S) and the MS/MS spectrum of the respective product. C, assignment of the MS/MS fragments to the structure of 12-O-Glc-JA.
UGT76E1 and UGT76E2 show substrate preference for 12-OH-JA
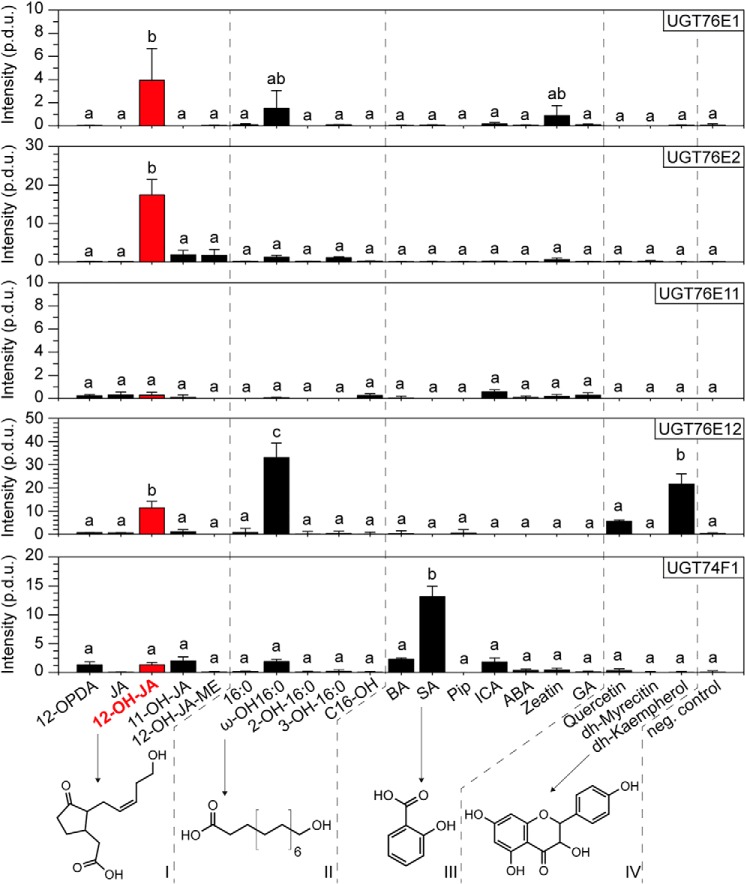
UGT enzymes are known to have rather broad substrate specificities (24). Hence, we investigated the substrate preferences of UGT76E1, UGT76E2, UGT76E11, and UGT76E12 with different approaches. First, four different groups of possible substrates for glycosylation were tested in a radioactive assay using UDP-Glc with a uniformly 14C-labeled Glc moiety ([U-14C]Glc) as co-substrate: (i) JA, its precursor (12-OPDA) as well as 12-OH-JA, 11-hydroxy-JA (11-OH-JA), and 12-hydroxy-JA-methyl ester (12-OH-JA-ME), (ii) palmitic acid (a saturated, aliphatic fatty acid of 16 carbons), its hydroxylated derivatives as well as the corresponding fatty alcohol, (iii) other plant hormones and stress-related molecules carrying free hydroxy and/or carboxyl groups on aromatic or nonaromatic structures, as well as (iv) several flavonoids (Fig. 4) (Table S2). Sets (iii) and (iv) hold known substrates of already characterized UGTs (24, 33, 38), including SA, which is the in vivo substrate for the reference protein UGT74F1 (36, 38). UGT76E1 displayed the strongest activity toward 12-OH-JA and ω-hydroxy-hexadecanoic acid (ω-OH-16:0). These substrates contain, to varying extents, aliphatic chains with a ω-hydroxy group and a free α-carboxyl group. Surprisingly, zeatin, a phytohormone that includes a purine, was accepted as substrate to a minor extent too. UGT76E2 also glycosylates preferentially 12-OH-JA and to very low amounts of 11-OH-JA and 12-OH-JA-ME. For UGT76E11, the signal intensities were too low to consider them as the result of true enzymatic activities. The recombinant protein was either not active or the right substrate was not present in the set of substrates. UGT76E12 demonstrated the highest activities toward ω-OH-16:0, whereas the flavonoid dihydro-kaempferol (dh-kaempferol), as well as 12-OH-JA, were also accepted as substrate, but to lower amounts. UGT76E12 thus seemed to recognize particularly the terminal hydroxy group of ω-OH-16:0 and 12-OH-JA as well as the polyphenolic structure of dh-kaempferol (Fig. 4). As expected, the reference UGT74F1 showed an exclusive activity toward its native substrate SA. In analogy to this, UGT76E1 and UGT76E2 exhibited the highest activity toward 12-OH-JA. UGT76E12 accepted the broadest range of substrates, also including 12-OH-JA to lesser extent. However, none of the candidates were able to glycosylate JA or 12-OPDA. Hence, substrates with a (terminal) hydroxy group and a free carboxyl function seemed to be preferred for catalysis.
Figure 4.
Substrate specificity of UGT7E1, UGT7E2, UGT76E11, UGT76E12, and UGT74F1. The substrate specificities were analyzed by an activity assay with [U-14C]Glc-labeled UDP-Glc (0.03 mm, 0.02 μCi), the indicated substrates (0.06 mm), and 5 μg of purified UGT each for 1 h at 25 °C. After metabolite extraction and TLC, the plates were exposed to phosphor screens and radioactive signals were detected. Given are relative product intensities (procedure defined units, p.d.u.). UGT76E1, UGT76E2, UGT76E11, UGT76E12, and UGT74F1 were each incubated with four sets of different aglycones (I–IV): 12-oxo-phytodienoic acid (12-OPDA), jasmonic acid (JA), 12-hydroxy-JA (12-OH-JA), 11-hydroxy-JA (11-OH-JA), 12-hydroxy-JA-methyl ester (12-OH-JA-ME), hexadecanoic acid (16:0), ω-hydroxy-16:0 (ω-OH-16:0), 2-hydroxy-16:0 (2-OH-16:0), 3-hydroxy-16:0 (3-OH-16:0), hexadecanol (OH-C16), benzoic acid (BA), SA, pipecolic acid (Pip), indole-3-caboxylic acid (ICA), abscisic acid (ABA), zeatin, gibberellic acid (GA), quercetin, dihydro-myricetin (dh-myricetin), dh-kaempferol, and a negative control with inactivated enzyme and ω-OH-16:0 (neg. control). The data are three independent experiments, mean ± S.E. and one-sided ANOVA test (p = 0.05, Turkey test) for every enzyme.
Analysis of quasi-native metabolite extracts identified 11-HHT and 13-HOT as substrates of UGT76E2, UGT76E11, and UGT76E12
In the second, so-called ex vivo approach we searched for further possible substrates in a nontargeted way. The purified proteins were incubated in an extract obtained from wounded A. thaliana leaves (2 hpw) mimicking an almost native, metabolite-rich environment, and subsequently analyzed by metabolite fingerprinting (39). Besides unknown substrates, this approach also provides suspected substrates, like the previously mentioned 12-OH-JA-Ile, which represents the major form of inactivated JA-Ile. 12-OH-JA-Ile is strongly enriched in extracts of wounded leaves, but commercially not available. We decided to use plant extracts obtained from A. thaliana rosettes at 2 hpw, when both 12-OH-JA and 12-OH-JA-Ile are already present, but the internal abundance of 12-O-Glc-JA is still low (15).
This nontargeted metabolome approach allows the identification of tentative UGT products as well as corresponding substrates and is based on the assumption that a glycosylated product increases in an enzyme-dependent matter, ideally at the expense of the nonglycosylated substrate. Such substrate-product pairs are thus characterized by an inverse accumulation pattern, a mass shift of 162.053 Da (which corresponds to the neutral loss of the glucose moiety), and a characteristic shift in retention time due to higher hydrophilicity of the glycoside.
Data analysis delivered four substrate-product pairs that unambiguously passed the above criteria (Fig. 5), including the expected reaction pair 12-OH-JA and 12-O-Glc-JA. Interestingly, two oxylipins namely 13-hydroxy-9,11,15-octadecatrienoic acid (13-HOT, 276.210 Da, 6.59 min) and 11-hydroxy-7,9,13-hexadecatrienoic acid (11-HHT, 266.186 Da, 6.08 min) were identified as new substrates for all four UGTs in question (40). This resulted in the accumulation of the corresponding products 13-glucosyl-O-octadecatrienoic acid (13-O-Glc-HOT, 465.274 Da, 5.65 min) and 11-O-glycosyl-hexadecatrienoic acid (11-O-Glc-HHT, 428.240 Da, 5.25 min). 13-HOT was almost completely consumed in the extracts by UGT76E11 and UGT76E12, whereas the minor levels of 11-HHT were strongly depleted by all four UGTs (Fig. 5, A and B). Furthermore, we detected a substrate that appeared specific for UGT76E11 (Fig. 5C). It has a deduced sum formula of C11H18O3 (198.126 Da, 5.47 min), and the corresponding product (360.179 Da, retention time 4.65 min) showed the inverse intensity pattern as well as the indicative neutral loss of 162.053 Da (Table S3). However, the chemical structure of this new compound remained to be elucidated. As proof-of-concept, the box plots for the reaction pair 12-OH-JA and 12-O-Glc-JA are shown (Fig. 5D). The signal intensity for endogenous 12-OH-JA (as substrate) was slightly reduced for UGT76E11 and UGT76E12 as compared with the control assays (assay with inactive enzymes, Fig. 5D). For UGT76E1, however, it was decreased by a third, and nearly abolished for UGT76E2. This already suggested that the substrate was completely used for the synthesis of 12-O-Glc-JA, as confirmed by the strong increase in intensity of the corresponding product peak. Again, recombinant UGT76E2 showed the highest activity (Fig. 5D), as observed in the previous experiment. Last, the glycosylation of 12-OH-JA-Ile to 12-O-Glc-JA-Ile was of special interest in this study. The tentative substrate 12-OH-JA-Ile (323.205 Da, 4.9 min) was abundant in the 2 hpw extracts. However, we did not detect the corresponding product of 501.257 Da, suggesting that none of the four candidate UGTs accepts 12-OH-JA-Ile for catalysis (Fig. S2).
Figure 5.
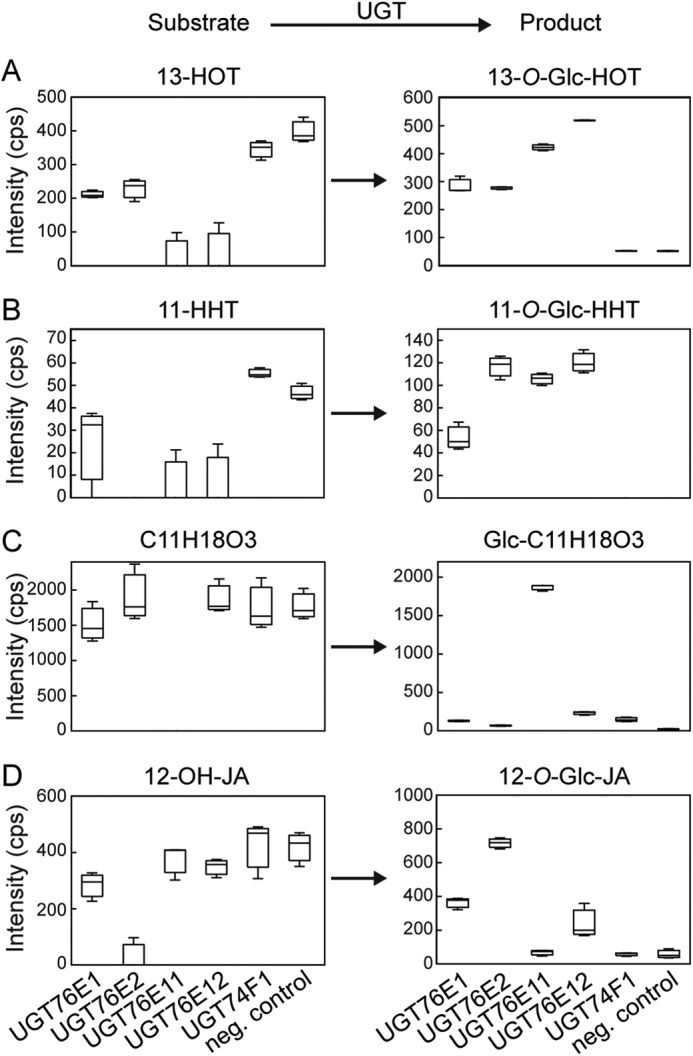
Substrates of UGT76E1, UGT7E2, UGT76E11, UGT76E12, and UGT74F1 identified in quasi-native metabolite extracts by metabolite fingerprinting. For the ex vivo approach, recombinant proteins were incubated with total metabolite extracts of wounded A. thaliana plants (2 hpw). The extracts were resolved in protein buffer and the assay was performed with 0.1 mm UDP-Glc and 100 μg of the indicated active UGT-enzyme or inactive enzyme (neg. control) for 1 h at 25 °C. The reactions were stopped by adding acetonitrile and analyzed by LC-MS–based metabolite fingerprinting approach. The particular features for: A, 13-HOT and 13-O-Glc-HOT; B, 11-HHT and 11-O-Glc-HHT; C, 12-OH-JA and 12-O-Glc-JA; and D, C11H18O3 and C11H18O3-Glc, are shown as Box-Whisker plots with intensities given as counts per second (cps). The data represent three technical replicates of one experiment. Data were analyzed using the MarVis tool (70).
UGT76E1 and UGT76E2 prefer 12-OH-JA as substrate, whereas UGT76E11 and UGT76E12 show preferences toward 9- and 13-HOT
Having a set of substrates available, we compared the substrate specificities of all four UGTs and the control UGT74F1 in an LC-MS–based activity assay (Table 2). The necessary amounts of 11-HHT could not be provided for the enzymatic assays, it was therefore exchanged for 9-hydroxy-(10E,12Z,15Z)-10,12,15-octadecatrienoic acid (9-HOT), which originates from the co-product of the LOX reaction also forming 13-HOT. Thus, relative conversion rates were determined for ω-OH-16:0, 12-OH-JA, 9-HOT, 13-HOT, and SA. UGT76E12 showed the highest conversion rate (set to 100%) of all five UGTs for ω-OH-16:0, 13-HOT, and 9-HOT. 13-HOT was also glycosylated by UGT76E2 and UGT76E11 with comparable conversion rates. 9-HOT was a much poorer substrate for both (with 22 and 5%, respectively). 12-OH-JA is preferably glycosylated by UGT76E2 (100%) and by about one-third of that by UGT76E1 and UGT76E12. UGT74F1 shows high activity to its native substrate SA, as well as some side activity toward ω-OH-16:0 (4% of the UGT76E12 activity) (Table 2).
Table 2.
Relative substrate preferences of UGT76E1, UGT76E2, UGT76E11, UGT76E12, and UGT74F1
Substrates so far identified for UGT76E1, UGT76E2, UGT76E11, UGT76E12, and UGT74F1 were evaluated by an in vitro assay. The relative conversion rate (%) was analyzed by LC-MS: ω-OH-16:0, SA, 12-OH-JA, 9-HOT, and 13-HOT. 10 μg of homogenous enzyme were incubated with 0.05 mm substrate and 0.25 mm UDP-Glc at 25 °C for 30 min. The reactions were stopped by adding acetonitrile. The data are representative for one experiment.
| Substrate | UGT76E1 | UGT76E2 | UGT76E11 | UGT76E12 | UGT74F1 |
|---|---|---|---|---|---|
| ω-OH-16:0 | 9 | 36 | 2 | 100 | 4 |
| SA | NDa | ND | ND | ND | 100 |
| 12-OH-JA | 29 | 100 | ND | 28 | ND |
| 9-HOT | ND | 22 | 5 | 100 | ND |
| 13-HOT | 6 | 94 | 98 | 100 | ND |
a ND, not detected.
UGT76E1 is highly specific toward 12-OH-JA, but UGT76E2 has a higher turnover rate
Subsequently, we resolved the kinetic constants of UGT76E1, UGT76E2, UGT76E11, and UGT76E12 for their main substrates (12-OH-JA, 13-HOT, and 9-HOT) by a continuous photospectrometric assay (Fig. S3). For this, the UGT reaction was coupled to a NADPH consumptive reaction (41) to record the initial velocity and calculate their Km value, maximal turnover rate (kcat), as well as catalytic efficiency (kcat/Km). The catalytic efficiency of UGT76E1 for 12-OH-JA was 202 s−1 m−1 with a reasonable affinity toward 12-OH-JA of 61 μm, but low turnover rate of 0.012 s−1. UGT76E2 showed a much higher catalytic efficiency of 6744 s−1 m−1 with a Km of 219 μm and a kcat of 1.477 s−1. In conclusion, the affinity of UGT76E2 for 12-OH-JA was 3-fold lower than for UGT76E1, but, at the same time, the turnover rate was 100-fold faster (Table 3). Furthermore, the kinetic parameters of the joint substrate 13-HOT (Fig. 5A) were determined for UGT76E2, UGT76E11, and UGT76E12. UGT76E1 did not show detectable activities with 13-HOT in this assay. UGT76E11 exhibited a low and UGT76E12 a substantial affinity toward 13-HOT (Km of 156 and 23 μm, respectively), but both with very low turnover rates, resulting in poor catalytic efficiencies. UGT76E2 glycosylated 13-HOT with a high catalytic efficiency, because it acted fast, although with low affinity. In case of 9-HOT, UGT76E12 exclusively showed a similar catalytic efficiency of 33 s−1 m−1 as compared with 13-HOT (22 s−1 m−1, Table 3). For comparison, the kinetic parameter of UGT74F1 with SA were determined (catalytic efficiency of 13 s−1 m−1) and confirmed that the here obtained kinetic values of UGT76E1 and UGT76E2 for 12-OH-JA were in a reasonable range, because the 12-OH-JA concentration was found to increase to 6 nmol/g fresh weight at 5 hpw in leaves (Fig. S4). This was consistent with former studies measuring up to 4 nmol/g fresh weight at 6 hpw (19).
Table 3.
Kinetic parameters of the UGT76E1, UGT76E2, UGT76E11, UGT76E12, and UGT74F1 towards their best substrates
Steady-state kinetics were recorded for all four UGTs with the joint substrates 9-HOT and 13-HOT. UGT76E1 and UGT76E2 were also tested towards 12-OH-JA. UGT74F1 was measured with SA. A coupled spectrophotometric assay with the pyruvate kinase and the lactate dehydrogenase was used to measure the reactions of the UGTs in a 1:1:1 stoichiometry by the decrease of NADH at 340 nm (41). The data were fitted with the hyperbolic function and the correlations are given. The kinetic parameters were calculated as Km, the turnover rate (kcat), and the catalytic efficiency (kcat/Km). The data are representative for three biological replicates of one experiment.
| Enzyme | Substrate | Correlation | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| μm | s−1 | s−1 m−1 | |||
| UGT76E1 | 12-OH-JA | 0.982 | 61 | 0.0123 | 202 |
| 9-HOT | NDa | ND | |||
| 13-HOT | ND | ND | |||
| UGT76E2 | 12-OH-JA | 0.998 | 219 | 1.4770 | 6744 |
| 9-HOT | ND | ND | |||
| 13-HOT | 0.951 | 4000 | 0.5000 | 125 | |
| UGT76E11 | 9-HOT | ND | ND | ||
| 13-HOT | 0.993 | 156 | 0.0007 | 4 | |
| UGT76E12 | 9-HOT | 0.983 | 21 | 0.0007 | 33 |
| 13-HOT | 0.990 | 23 | 0.0005 | 22 | |
| UGT74F1 | SA | 0.998 | 212 | 0.0027 | 13 |
a ND, not detected.
A loss-of-function mutation in UGT76E1 reduces the amount of 12-O-Glc-JA in planta
To investigate the functional activities of UGT76E1 and UGT76E2 in vivo, we performed jasmonate analyses of different A. thaliana mutant plants. First of all, we generated and isolated a homozygous ugt76e1 loss-of-function mutant using the CRISPR/Cas9 approach. This mutant allele carries a thymine insertion at the Cas9-cutting site within the first exon, resulting in a frameshift and an early stop-codon (position 46 of the amino acid sequence). Furthermore, we obtained a homozygous T-DNA insertion line in UGT76E1 from the European Arabidopsis Stock Centre (SAIL_738_A03C, A. thaliana Columbia 0 background), in which the T-DNA was confirmed to lie within the promotor region as annotated. Surprisingly this resulted in a 10-fold increased expression rate of UGT76E1 as determined by both semi-quantitative PCR and real-time analysis (Fig. 6A, Fig. S5). Next, a transposon-tagged UGT76E2 line was isolated from the RIKEN collection (A. thaliana, Nossen background (42, 43)). This line carries an insertion in the first exon that totally abolishes accumulation of the UGT76E2 transcript (Fig. 6A). The mutants and their respective controls were then used in a targeted metabolomics study to determine jasmonate levels with a focus on 12-OH-JA, 12-HSO4-JA, and 12-O-Glc-JA in wounded A. thaliana leaves (5 hpw). As mentioned before, the levels of 12-OH-JA and its metabolic successors have been reported to influence each other (21, 22). Hence, we calculated the relative amount of 12-O-Glc-JA of all three metabolites. Disruption of a functional UGT76E1 reduced the amount of 12-O-Glc-JA significantly (Fig. 6B), whereas overexpression led to an increased accumulation (Fig. 6B, Fig. S4). On the other hand, deletion of a functional UGT76E2 did not show any significant effects on either 12-O-Glc-JA or related metabolites (Fig. 6B, Fig. S4). We further investigated the impact of the observed changes in 12-O-Glc-JA levels on the expression of JA-Ile–responsive genes in wounded leaves (5 hpw) of both the overexpression and loss-of-function mutant of UGT76E1. These included LOX2, JOX2, ST2a, VSP1 (VEGETATIVE STORAGE PROTEIN1), PDF1.5 (PLANT DEFENSIN 1.5), and ORA59 (OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF gene 59) (Fig. S6). RNS1 (RIBONUCLEASEI) was included as a JA-independent control for wounding (44). For all genes investigated, the observed fold-changes ranged merely between 0.5 and 1.5 of the WT, suggesting that jasmonate signaling remained largely unaffected in these mutants upon mechanical wounding.
Figure 6.
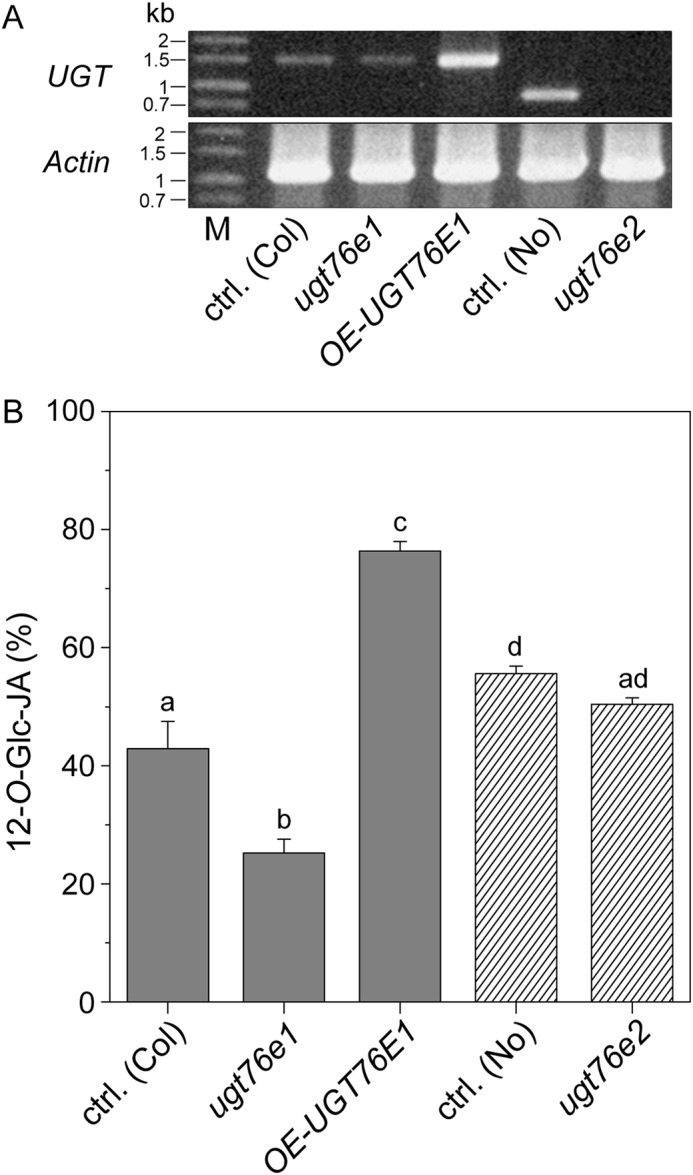
UGT76E1 forms 12-O-Glc-JA in wounded plants. A, detection of the UGT76E1 transcript in WT Columbia 0 plants (ctrl. (Col)), a loss-of-function UGT76E1-CRISPR/Cas-mutant allele (ugt76e1), a T-DNA insertion mutant of UGT76E1 (OE-UGT76E1) as well as UGT76E2 transcript in a parent (ctrl. (No)) and the transposon-tagged ugt76e2 mutant allele, both of the Nossen ecotype. RT-PCR was performed on RNA from wounded leaf samples (2 hpw). UGT76E1 was amplified as full-length transcript, UGT76E2 was amplified using an internal reverse primer (as listed in Table S5). Actin 8 was used as a positive control. B, relative levels of 12-O-Glc-JA (% of 12-OH-JA + 12-SO4-JA + 12-O-Glc-JA) in wounded A. thaliana leaves (5 hpw). Metabolite levels of ctrl. (Col), ugt76e1, OE-UGT76E1 plants as well as ctrl. (No) and the respective ugt76e2 plants were determined by targeted LC-MS/MS. Signals were identified and quantified to internal standards. Data show the 3–6 biological replicates, mean ± S.E. and one-sided ANOVA test (p = 0.05, Turkey test).
Discussion
JA catabolism is a highly important process for the timely removal of the biologically active compound(s) to limit further expenditure of resources and ensure smooth plant growth and development. In addition to the well-described depletion of JA-Ile via oxidation (8–12), this may also be accomplished by glycosylation of selected JA-derivatives, such as 12-OH-JA. It is known that plant UGTs are mainly controlled on the transcriptional level to regulate their time of action (25). By linking JA metabolism to co-expression studies of wounded plants, we identified four sequence-related UGTs in A. thaliana as candidates for the synthesis of 12-O-Glc-JA (Table 1, Fig. 1). LC-MS analyses confirmed that recombinant UGT76E1, UGT76E2, and UGT76E12 effectively glycosylated 12-OH-JA to 12-O-Glc-JA (Fig. 7).
Figure 7.
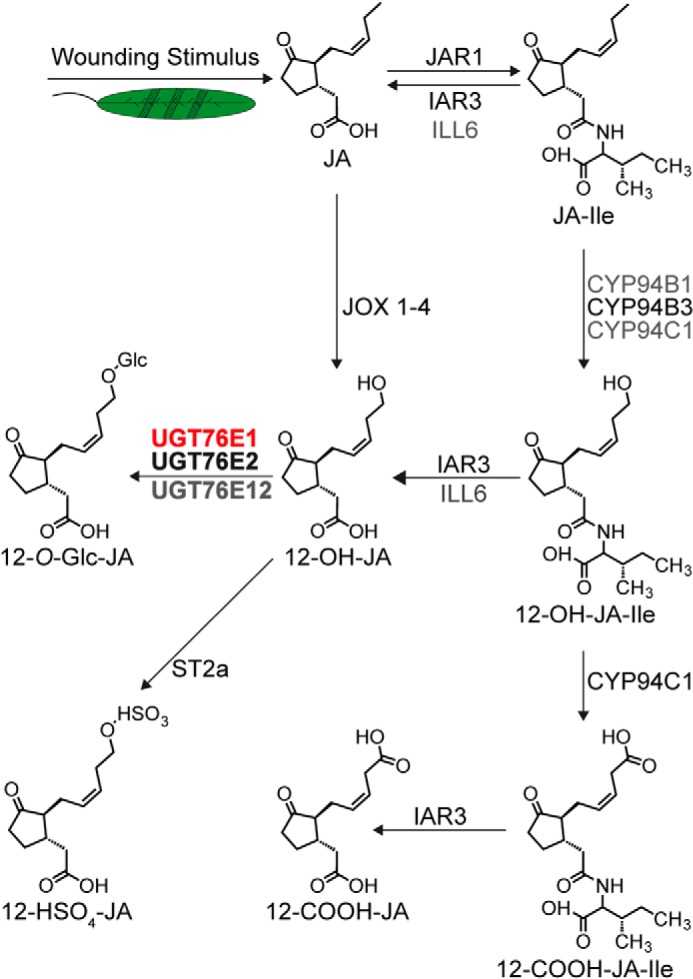
Wound-induced metabolic fate of jasmonates. Wounding stimuli lead to the activation of JA biosynthesis. The hormone JA is activated through conjugation to isoleucine (JA-Ile). CYP94B1, CYP94B3, and CYP94C1 oxidize JA-Ile to 12-OH-JA-Ile and CYP94C1 to 12-carboxy-JA-Ile (12-COOH-JA-Ile). JA-Ile, 12-OH-JA-Ile, and 12-COOH-JA-Ile are de-conjugated by indole-3-acetic acid (IAA) alanine-resistant 3 (I) and IAA-leucine-resistant-like 6 (ILL6) forming JA, 12-OH-JA, and 12-carboxy-JA (12-COOH-JA), respectively. JA is oxidized to 12-OH-JA by jasmonate-induced oxidases 1 to 4 (JOX1–4). 12-OH-JA is modified to 12-HSO4-JA by sulfotransferase 2a (ST2a). This work characterized UGT76E1 and UGT76E2 (bold) to specifically glycosylate 12-OH-JA and UGT76E12 to have minor activity forming 12-O-Glc-JA in vitro. In addition, UGT76E1 was shown to synthesize 12-O-Glc-JA levels in vivo (red). The scheme was modified from Ref. 15 with additions from Refs. 14 and 16.
UGTs tend to be rather unspecific enzymes and were therefore also exposed to different substrates to identify their true, native reaction partners (24, 25). Indeed, when different substrate classes were tested using the radiolabeled co-substrate (UDP-[U-14C]Glc), the same 3 enzymes displayed distinct activities and specificities, which contrasted with those of the A. thaliana UGT74F1 control, which showed a specific activity toward its native substrate SA in all experiments (Fig. 4).
UGT76E1, and also UGT76E2 and UGT76E12 require a hydroxy group and mostly a free carboxyl function, although only the hydroxy group is glycosylated. Interestingly, a shift of the hydroxy group from position 12 (12-OH-JA) to position 11 (11-OH-JA) was not accepted by UGT76E1 in cyclopentane substrates, whereas aliphatic fatty acid derivatives with a hydroxyl group in the middle of the molecule (11-HHT, 13-HOT) were tolerated (Fig. 5). The kinetic analyses confirmed that UGT76E1 glycosylates 12-OH-JA with high affinity (Table 3). Measurements of 12-O-Glc-JA in A. thaliana WT, a knock-out mutant (ugt76e1), and an over-expresser (OE-UGT76E1) clearly illustrate a role of UGT76E1 in glycosylation of 12-OH-JA in vivo (Fig. 6).
UGT76E2 was not significantly induced after wounding, but showed co-expression with the gene encoding for ST2a, which catalyzes the concurrent reaction from 12-OH-JA to 12-HSO4-JA (Table 1, Fig. 1). Recombinant UGT76E2 showed the highest activities toward 12-OH-JA and the best catalytic efficiency of all analyzed UGTs (Table 3). However, the specificity for the ω-hydroxy group seems less pronounced, because side activities toward 11-OH-JA and 3-OH-16:0 were observed and oxylipins with mid-chain groups (11-HHT, 9-HOT, and 13-HOT) were also accepted as substrates (Figs. 4 and 5, Tables 2 and 3). The kinetic parameters of UGT76E2 indicated a high specificity toward 12-OH-JA. We did not observe a significant reduction of 12-O-Glc-JA in the ugt76e2 loss-of-function mutant, which fits well with its lower expression level as compared with UGT76E1. However, it should be noted that this particular mutant originates from a Nossen ecotype, which may differ from Columbia 0 with respect to its JA-Ile-mediated defense.
Despite the high sequence homology between UGT76E11 and UGT76E12, recombinant UGT76E11 did not show any convincing activity in the first screens for possible substrates (Fig. 2). However, when exposed to a quasi-native substrate environment in the ex vivo experiment, it glycosylated 11-HHT and 13-HOT, which appear to serve as a general substrate for all wound-inducible UGT76Es (Fig. 5). Furthermore, this approach, which supplies the enzymes with a large range of putative, but otherwise not available or especially unexpected substrates, revealed a so far unknown compound of C11H18O3 as exclusive substrate for UGT76E11. This unknown metabolite neither gave any data bank hit nor could it be structurally identified by MS/MS fragmentation analysis. The elucidation of its chemical structure might enlighten the function of UGT76E11.
Expression of UGT76E12 is highly induced after biotic and abiotic stresses in A. thaliana (45). It was found to be up-regulated in abscisic acid-sensitive seedlings and after application of 12-OPDA (46). Compared with the other three UGTs, UGT76E12 shows high product formations with oxylipins like 11-HHT, 9-HOT, 13-HOT, and ω-OH-16:0 (Figs. 4 and 5, Table 2). In addition, UGT76E12 also accepts 12-OH-JA as substrate, but to a lower extent and with lower catalytic efficiency than the more specific enzymes UGT76E1 and UGT76E2. UGT76E12 tolerates the hydroxy group both at the ω-position (ω-OH-16:0, 12-OH-JA) and in the middle of the molecule (11-HHT, 9-HOT, and 13-HOT). For the glycosylation of the latter two substrates UGT76E12 displays a high affinity, but rather low turnover rates (Table 3). The occurrence of such glycosylated oxylipins has been described before in Lemna paucicostata (47) and 13-HOT, which is produced by the 13-LOX pathway and is sufficient to induce pathogenesis-related genes in Hordeum vulgare (48). Altogether, UGT76E12 shows high and broad preference toward oxylipin substrates, but their occurrence and tentative function in planta have to be evaluated in more detail the future.
The free carboxyl group of 12-OH-JA seems to be crucial for substrate recognition. In line with this, none of the UGTs in question glycosylated JA itself to its corresponding JA-GE. Recently, this was also documented by Yang and co-workers, who systematically tested a total of 54 A. thaliana UGTs (including the ones analyzed here) for activities toward plant secondary metabolites. Instead, they reported that UGT84A1, UGT84A2, and UGT75B1 glycosylate JA (49). So far, very low amounts of a glycosylated version of 12-OH-JA-Ile were identified in A. thaliana, Glycine max, and N. tabacum by targeted LC-MS/MS analysis of wounded plant material (30). Although 12-OH-JA-Ile was present as substrate in our ex vivo assay, its glycoside could not be detected as the enzymatic product. It should be emphasized that the metabolite-rich extracts of the ex vivo approach provided enough potency to unambiguously identify 12-COOH-JA in wounded leaves of A. thaliana for the first time (Fig. S7). This finding supports the mutant analysis of Smirnova and co-workers (17), and puts 12-COOH-JA not only in the context of flower development (15) but also in that of the late wound-response of A. thaliana.
So far, it was not possible to assign an individual biological function to 12-OH-JA or its glycoside. We generated a ugt76e1-mutant through genome editing, which already showed the expected decrease in 12-O-Glc-JA (Fig. 6B, Fig. S4C) and led to an increase of the concurrent product 12-SO4-JA (Fig. S4D). Furthermore, an increased expression of UGT76E1 was sufficient to turn this pattern around, resulting in elevated 12-O-Glc-JA, but reduced 12-SO4-JA and 12-OH-JA levels as compared with the respective WT. However, jasmonate signaling seems not affected in either of these mutants because gene expression of several JA-responsive candidates (LOX2, JOX2, ST2a, VSP1, and ORA59) was unchanged, which may be explained by the residual amounts of 12-O-Glc-JA and the concomitant increase in 12-HSO4-JA levels (or vice versa).
It is tempting to speculate why two different metabolic modifications of 12-OH-JA are needed. A specific hydrolase for 12-O-Glc-JA in Oryza sativa suggests a role as a reversible storage compound at least in rice (50). Therefore, to further investigate the physiological role of both derivatives in vivo, more single and especially higher order mutants up to a ugt76e1 ugt76e2 st2a triple mutant should be characterized in more detail with respect to possible metabolic functions in signaling, inactivation, degradation, storage, or recycling (24, 25).
Altogether, A. thaliana UGT76E1 and UGT76E2 were characterized as specific 12-OH-JA-UGTs in vitro. Additionally, UGT76E1 influenced the formation of 12-O-Glc-JA in vivo, thus adding to the set of known enzymes acting on jasmonates after wounding.
Experimental procedures
Plant material and generation of plant mutants
If not stated otherwise, A. thaliana ecotype Columbia 0 was used in this study and grown as described in Ref. 51. A T-DNA insertion mutant of UGT76E1 (SAIL_738_A03C, A. thaliana; ecotype Columbia 0), was obtained from the European Arabidopsis Stock Centre. A transposon-tagged UGT76E2 line (PST00878) and its respective parents (PST91000 and PST99999, used as controls) were obtained from the RIKEN collection (A. thaliana; ecotype Nossen). The Arabidopsis transposon-tagged line was developed by the plant genome project of RIKEN Genomic Sciences Center 8 (42, 43).
The CRISPR/Cas9 method aimed at targeting all four genes, UGT76E1, UGT76E2, UGT76E11, and UGT76E12, simultaneously, according to Ref. 52. Putative target sequences were identified with three different online tools: CRISPRdirect (53), CRISPR-P (54–56), and The Genetic Perturbation Platform (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design).3 Selected candidate sequences (Table S6) were used to replace the dummy nucleotides in the CRISPR/Cas9 construct within the pHEE401E vector (Addgene) (57). Correct plasmids were transformed into Agrobacterium tumefaciens for plant transformation into A. thaliana WT Columbia 0 by floral dip (58). Positive T1 transformants were selected on hygromycin. Mutant screening was performed in the T2 generation and led to the identification of a homozygous ugt76e1 knockout mutant, in which a thymine was inserted at position 98 after the transcription start at the UGT76E1 target sequence (Table S6), and which resulted in an early stop-codon (position 46 of the amino acid sequence). Sequence analyses confirmed that none of the other target genes were mutated in this mutant line. Jasmonate measurements and expression analyses were conducted in the following generation.
Plant wounding conditions
Six-week-old plants grown under short-light conditions were wounded according to the procedure described in Ref. 59 and harvested before (0 hpw) and at different time points after wounding. Subsequently, tissue samples were frozen in liquid nitrogen and homogenized for analysis.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from 100 mg of A. thaliana rosette leaves using the protocol of Onate-Sanchez and Vicente-Carbajosa (60). cDNA was produced using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). RT-quantitative PCR assay was performed using Takyo No Rox SYBR® MasterMix dTTP Blue (Eurogentec) following the manufacturer's instructions. Special quantitative PCR primers were designed with the online tools primer3prefold and primer3plus (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi)3 and are listed in Table S5. The 2-ΔΔCT method (61) was used to estimate relative gene expression, which was normalized to the expression of the reference gene actin 8 (At1g49240, GenBankTM accession number AY063089.1).
Phylogenetic analysis
The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (62). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. Evolutionary analyses were conducted in MEGA X (63).
Heterologous protein expression and purification
The coding sequences of UGT76E1 (At5g59580, GenBankTM accession number OAO90200.1), UGT76E2 (At5g59590, GenBankTM accession number OAO92843.1), UGT76E11 (At3g46670, GenBankTM accession number OAP05192.1), UGT76E12 (At3g46660, GenBankTM accession number AEE78190.1), and UGT74F1 (At2g43840.2, GenBankTM accession number, AEC10332.1) were cloned into pET28a (Novagen). The respective oligonucleotides are listed in Table S5. Correct constructs were transformed into E. coli BL21 Star(DE3) cells (Thermo Fisher Scientific). Expression cultures were grown in autoinduction medium following the protocol of Studier (64) at 16 °C for 3 days. Cell pellets of 2-liter cultures were resuspended in purification buffers (Table S4) and lysed by high pressure in a Microfluidizer M-110L device (Microfluidic). The protein purification was performed with a two-step protocol of immobilized metal affinity chromatography and size exclusion chromatography. UGT76E1 was captured with cobalt-affinity (Takara Bio Europe), and the remaining UGTs by nickel-affinity (GE Healthcare). All UGTs were purified by HiLoad 16/600 Superdex 75 prep grade (GE Healthcare). Individual buffer requirements were optimized for each of the UGTs and are listed in Table S5.
Enzyme assays
LC-MS–based activity assay
For reaction optima and general substrate assays, 30 μg of UGT were incubated with 0.5 mm UDP-Glc (Merck) and 0.1 mm substrate in their individual buffer conditions (Table S4) for 1 h at 25 °C or the indicated temperature. The reactions were stopped by adding ¼ volume of acetonitrile, centrifuged at 20,000 × g for 5 min, and analyzed by LC-MS. For the relative conversion rate, 10 μg of UGT, 0.25 mm UDP-Glc, and 0.05 mm substrate were mixed in their individual buffer conditions (Table S4) and incubated for 30 min at 25 °C. The assays were stopped by adding 1 volume of acetonitrile. The precipitated protein was removed by centrifugation and the supernatant was analyzed by ultra performance liquid chromatography (UPLC, Waters Inc.) coupled to an orthogonal TOF-MS (LCT Premier, Waters Inc.). For LC, an ACQUITY UPLC HSS T3 column (1 × 100 mm, 1.8 μm particle size, Waters Inc.) was used at a temperature of 40 °C and a flow rate of 0.2 ml min−1. A binary gradient of solvent A (water:formic acid (100:0.1, v/v)) and solvent B (acetonitrile:formic acid (100:0.1, v/v)) was applied as follows: for solvent B, 0–0.5 min 40% B, 0.5–3 min from 40 to 100% B, 3–5 min 100% B, 5–7 min 40% B. The TOF-MS was operated in W in negative electrospray ionization (ESI) mode. Data were acquired by MassLynx 4.1 software in centroided format over a mass range of m/z 50–700 with a scan duration of 0.5 s and an inter-scan delay of 0.1 s. The following source parameters were used: capillary and cone voltage at 2700 and 30 V, respectively, desolvation and source temperature at 350 and 80 °C, respectively, nitrogen flow as cone and desolvation gas at 30 and 800 liter h−1, respectively). Dynamic range enhancement mode and leucine-enkephaline as reference spray ([M-H]− 554.2615 or [M+H]+ 556.2771 as well as its 13C isotopologue ([M-H]− 555.2615 or its double 13C isotopologue [M+H]+ 558.2836) was used for data recording.
Radioactive activity assay
The radioactive substrate assay was performed with 60 μm substrate, 3.3 μm labeled UDP-[U-14C]Glc (0.02 μCi), 28 μm UDP-Glc, and 5 μg of enzyme in their individual buffer conditions (Table S4) at 25 °C for 1 h. A negative control was performed with inactivated enzyme. 30 μl of n-butyl alcohol were added to stop the reactions and extract the metabolites (65, 66). The organic phases of two extractions were evaporated and resolved in 20 μl of ethanol (70%, v/v) for separation by TLC. The TLC plates (Silica Gel 60 plate, Merck KGaA) were run in dichloromethane:methanol:water (80: 20: 2, v/v/v) (67). Radioactive signals were transferred to phosphor screens for 3 days and scanned with a FLA-3000 fluorescent image analyzer (Fujifilm Corporation) with appropriate software (Datinf GmbH). Signal intensities were calculated as product signal minus background over area.
Enzymatic synthesis and purification of 9-HOT and 13-HOT
Both 9-HOT and 13-HOT were synthesized enzymatically as follows. For 9-HOT, a bacterial cell pellet of 10 ml of recombinant E. coli expressing the LOX from S. tuberosum (68) culture was resuspended in 1 ml of lysis buffer (50 mm Tris/HCl, pH 7.5, 300 mm NaCl, 10% glycerol, 1 μl Tween 20). After cell lysis, the crude lysate was mixed with 6 ml of reaction buffer (100 mm Tris/HCl, pH 7.5) and 60 μl of linolenic acid (250 mg ml−1). 13-HOT was produced by mixing 100 μl of LOX from G. max (0.250 mg ml−1, Merck KGaA) with 2 ml of borate buffer (200 mm borate, pH 9) and 60 μl of linolenic acid (250 mg ml−1) in an open vessel at room temperature for 30 min. The respective reactions were stirred on ice for 30 min, terminated by adding 10 ml of SnCl2 (0.375%, w/v) in methanol) and incubated at room temperature for an additional 10 min. After adding 100 μl of concentrated acetic acid and 10 ml of chloroform, the solutions were vigorously mixed and centrifuged at 3220 × g for 10 min. The lower phases were harvested and the remaining fractions re-extracted with chloroform. After combining and evaporating the lower phases, the respective residues containing either 9-HOT or 13-HOT were dissolved in 500 μl of ethanol each. Subsequently, both HOTs were purified by straight-phase HPLC in isocratic conditions in hexane:isopropyl alcohol:TFA (100:1:0.02, v/v/v) on a ZORBAX RX-SIL (4.6 × 50 mm, 1.8-μm particle size, Agilent) at 0.8 ml min−1. The elution of 9-HOT and 13-HOT was monitored at 234 nm and their identity confirmed by co-elution with authentic standards. The final concentrations of 9-HOT and 13-HOT were determined at 234 nm with the shared molar extinction coefficient of 23,000 m−1 cm−1.
Determination of kinetic parameters
To record steady-state kinetics, a coupled spectrophotometric assay was modified as described in Ref. 41. The UGT reaction was coupled in a 1:1:1 stoichiometry via the co-product UDP to the pyruvate kinase and lactate dehydrogenase reactions, and measured by the decrease of NADPH at 340 nm. All measurements were performed in biological triplicates in Quartz SUPERSIL cuvettes (Hellma Analytics) in a V-630 UV-visible Spectrophotometer (JASCO Corporation). UGT76E1 was stabilized with BSA (0.13%, w/v). For the assay, 10 μg of UGT were incubated with 5 mm UDP-Glc (Merck) and 0–1200 mm of the respective substrate in the individual buffer conditions (Table S4) for 5–10 min at 25 °C. The linear slopes of the reactions were determined with Spectra Manager II Software (JASCO Corporation). For evaluation, the velocities were calculated to kcat values by a modified Lambert-Beer equation and plotted against the given substrate concentration by SIGMA Plot software.
| (Eq. 1) |
For Equation 1, kcat, maximal turnover number; Vmax, maximal reaction velocity; [E], concentration of the enzyme; ϵ(NADH), specific extinction coefficient of NADH, 6220 m−1 cm−1; path length (for the used cuvettes) = 1 cm; and t, reaction time.
Two-phase metabolite extraction of A. thaliana rosette leaves
Metabolites were extracted from plant tissue as described previously (69). Briefly, 100 mg of ground leaves was used for each analysis. Frozen plant material was mixed with 750 μl of methanol and 2.5 ml of methyl-tert-butyl ether (MTBE). After 60 min shaking at 4 °C in darkness, 600 μl of water were added for phase separation. The upper MTBE phase was collected and the lower phase re-extracted. This time, both the upper and the lower phase (but without any debris or material from the interphase) were taken, combined with the upper phase of the first extraction step, evaporated under a stream of nitrogen and re-dissolved in methanol (LC-MS grade). After evaporation, the samples were finally dissolved either in protein buffer (for the ex vivo experiment) or in chromatographic starting conditions (for the LC-MS–based jasmonate analysis). For quantification of jasmonates in the latter analysis, 10 ng of D6-JA and 10 ng of D3-JA-Leu (kindly provided by Dr. Otto Miersch, IPB, Halle/Saale, Germany) were added as internal standards.
LC-MS–based jasmonate analysis
Plant metabolite extracts were analyzed by the UPLC nanoESI MS/MS method. Here, an Acquity UPLC system equipped with an ACQUITY UPLC HSS T3 column (100 × 1 mm, 1.8-μm particle size; both from Waters Corp.) was coupled to a nanoESI chip ion source (TriVersa NanoMate). The MS analysis was performed with an Applied Biosystems 4000 quadrupole/linear ion trap mass spectrometer (MDS Sciex). For analysis, 10 μl of the metabolite extracts were injected. The binary gradient system consisted of solvent A (0.3 mm NH4COOH) and solvent B (acetonitrile:water, 90:10, v/v, 0.3 mm NH4COOH), both adjusted to pH 3.5 with formic acid, and the following gradient program: 10% solvent B for 0.5 min, followed by a linear increase of solvent B up to 40% within 1.5 min, held for 2 min, and followed by an increase of solvent B up to 95% within 1 min. An isocratic run at 95% solvent B was then held for 2.5 min. To recover the starting conditions, a linear decrease to 10% B within 3 min was performed. The flow rate was 0.16 ml min−1. For stable nanoESI conditions, 70 μl min−1 of 2-propanol:acetonitrile:water (70:20:10, v/v/v, 0.3 mm NH4COOH), adjusted to pH 3.5 with formic acid and delivered by a Pharmacia 2248 HPLC pump (GE Healthcare), were added just after the column via a mixing tee valve. By using a second post column splitter, 502 nl min−1 of the eluent were directed to the nanoESI chip with 5-μm internal diameter nozzles. Ionization voltage was set to −1.7 kV, phytohormones were negatively ionized and detected in a scheduled multiple reaction monitoring mode with AB Sciex 4000 QTRAP tandem mass spectrometer (AB Sciex). For the scheduled mode, the multiple reaction monitoring detection window was set to 72 s and a target scan time of 1.2 s. The mass analyzers were adjusted to a resolution of 0.7 atomic mass units full-width at half-height. The ion source temperature was 40 °C and the curtain gas was set at 10, given in arbitrary units. Mass transitions were as follows: 214/62 (declustering potential (DP) −35 V, entrance potential (EP) 8.5 V, collision energy (CE) −24 V) for D6-JA, 209/59 (DP −30 V, EP −4.5 V, CE −214 V) for JA, 225/59 (DP −35 V, EP −9.0 V, CE 28 V) for 11/12-OH-JA, 305/97 (DP −30 V, EP-4.0 V, CE 32 V) for 12-HSO4-JA, and 387/59 (DP −85 V, EP −9.0 V, CE −52 V) for 12-O-Glc-JA. Quantification of JA and 12-OH-JA was carried out using a calibration curve of m/z. The ratios were calculated of [unlabeled]/[deuterium labeled] versus molar amounts of unlabeled (0.3–1000 pmol). Relative quantification of 12-SO4-JA and 12-O-Glc-JA was performed using D5-JA as internal standard.
Ex vivo metabolite activity assay
Plant metabolite extracts of A. thaliana rosette leaves at 2 hpw, obtained by the two-phase metabolite extraction (see above), were re-dissolved in buffer consisting of 50 mm Tris, pH 8, 100 mm NaCl. Extracts were used as the quasi-native substrate mixture, incubated with 100 μg of purified enzyme and 0.1 mm UDP-Glc for 1 h, at 25 °C. A negative control was performed with heat-inactivated enzymes. The reaction was stopped with ¼ volume of acetonitrile, centrifuged at 20,000 × g for 5 min, and analyzed by the metabolite fingerprinting platform (see below).
Nontargeted metabolite fingerprinting
Samples of the ex vivo metabolite activity assay were analyzed by UPLC-TOF-MS in positive and negative ESI mode as described in Ref. 15. For data processing and data mining the toolbox MarVis was used as described (15, 70). This tool includes the subroutines MarVis Filter and MarVis Cluster. An analysis of variance (ANOVA) test combined with a multiple testing (Benjamini-Hochberg) was performed to filter and extract 638 features with a false discovery rate <5 × 10−4. Data of one experiment were analyzed. Tentative substrates and products pairs were extracted by applying three criteria: (i) a mass difference of 162.016 Da (neutral loss of the Glc moiety), (ii) a retention time shift of the more hydrophilic product to earlier retention times compared with the substrate, and (iii) an inverse intensity pattern to each other. The identity of the UGT-substrates and products was confirmed by UHPLC ESI-QTOF-MS/MS analysis and in vitro reaction assays with pure 9-HOT and 13-HOT.
Structure determination of marker metabolites by UHPLC ESI-QTOF-MS/MS
To confirm the identity of 12-O-Glc-JA as well as the substrates and products of the ex vivo experiment, MS/MS fragment analyses were performed by using a LC 1290 Infinity (Agilent Technologies) coupled with an Applied Biosystems 6540 UHD Accurate Mass QTOF-MS (Agilent Technologies) as described (15). The respective settings and fragments are listed in Table S3.
Statistical analysis
Data show mean ± S.E. and were statistically analyzed by Student's test and one-way ANOVA using the Tukey post hoc test (p < 0.05). We employed for these methods the Microsoft Office Excel 2016 and Origin Pro 8.5 software.
Author contributions
S. H., K. F., A. A. K., and I. F. conceptualization; S. H., K. F., A. A. K., and I. F. data curation; S. H., K. F., A. A. K., K. Z., C. H., and I. F. formal analysis; S. H., K. F., A. A. K., C. H., and I. F. validation; S. H., K. F., A. A. K., K. Z., A. S., C. H., and I. F. investigation; S. H. visualization; S. H., K. F., A. A. K., K. Z., C. H., and I. F. methodology; S. H., K. F., and A. A. K. writing-original draft; K. Z., C. H., and I. F. writing-review and editing; I. F. supervision; I. F. funding acquisition; I. F. project administration.
Supplementary Material
Acknowledgments
We are grateful to Dr. Otto Miersch (IPB, Halle/Saale, Germany) for providing labeled jasmonate and 12-O-Glc-JA standards and Dr. Cornelia Herrfurth for establishing the LC-MS-based jasmonate analysis. We acknowledge Milena Lewandowska, Dr. Corinna Thurow, and Dr. Guido Kriete for supporting the RT-PCR measurements and Dr. Ellen Hornung for assisting with the cloning of the UGTs. We thank Sabine Freitag for enzymatic syntheses and purification of 9-HOT and 13-HOT and Julia Sondermann for mutant analyses.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants IRTG 2172 “PRoTECT”, ZUK 45/2010, and INST 186/822-1 and the “PRoTECT” program of the Göttingen Graduate School of Neuroscience and Molecular Biology (GGNB) (to S. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S6 and Figs. S1–S7.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- JA-Ile
- jasmonoyl-isoleucine
- 11-HHT
- 11-hydroxy-hexadecatrienoic acid
- 11-O-Glc-HHT
- 11-O-glycosyl-hexadecatrienoic acid
- 11-OH-JA
- 11-hydroxy-jasmonic acid
- 12-COOH-JA-Ile
- 12-carboxy-jasmonoyl-isoleucine
- 12-HSO4-JA
- 12-hydroxy-jasmonic acid sulfate
- 12-O-Glc-JA
- 12-O-glucopyranosyl-jasmonic acid
- 12-O-Glc-JA-Ile
- 12-O-glucopyranosyl-jasmonoyl-isoleucine
- 12-OH-JA
- 12-hydroxy-jasmonic acid
- 12-OH-JA-Ile
- 12-hydroxy-jasmonoyl-isoleucine
- 12-OH-JA-ME
- 12-hydroxy-jasmonic acid methyl ester
- 12-OPDA
- cis-(+)-12-oxo-phytodienoic acid
- 13-HOT
- 13-hydroxy-octadecatrienoic acid
- 13-HPOT
- 13-hydroperoxy-octadecatrienoic acid
- 13-LOXs
- specific 13-linoleate lipoxygenase enzymes
- 13-O-Glc-HOT
- 13-glucosyl-O-octadecatrienoic acid
- [U-14C]Glc
- uniformly 14C-labeled Glc moiety
- 2-O-Glc-SA
- 2-O-glucosyl-salicylic acid
- 9-HOT
- 9-hydroxy-octadecatrienoic acid
- CE
- collision energy
- COI1
- coronatine insensitive 1
- ddh-JA
- didehydro-jasmonic acid
- dh-kaempferol
- dehydro-kaempferol
- DP
- declustering potential
- EP
- entrance potential
- ESI
- electrospray ionization
- hpw
- hours post wounding
- IAA
- indole-3-acetic acid
- ILL6
- indole-3-acetic acid-leucine resistant-like 6
- JA-GE
- jasmonic acid-glucosyl ester
- JAZ
- jasmonate-ZIM domain
- JOX1–4
- jasmonate-induced oxygenase 1 to 4
- LOX
- lipoxygenase
- MTBE
- methyl-tert-butyl ether
- OE
- over expresser
- OPR2
- oxo-phytodienoate reductase isoform 2
- OPR3
- 12-oxo-phytodienoate reductase isoform 3
- SA
- salicylic acid
- ST2a
- sulfotransferase 2a
- UGT
- UDP-dependent glycosyltransferases
- ω-OH-16:0
- ω-hydroxy-16:0 fatty acid
- ANOVA
- analysis of variance
- UHPLC
- ultra high performance liquid chromatography.
References
- 1. Howe G. A., and Jander G. (2008) Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66 10.1146/annurev.arplant.59.032607.092825 [DOI] [PubMed] [Google Scholar]
- 2. Wasternack C., and Feussner I. (2018) The oxylipin pathways: biochemistry and function. Annu. Rev. Plant Biol. 69, 363–386 10.1146/annurev-arplant-042817-040440 [DOI] [PubMed] [Google Scholar]
- 3. Gerwick W. H., Moghaddam M., and Hamberg M. (1991) Oxylipin metabolism in the red alga Gracilariopsis lemaneiformis: mechanism of formation of vicinal dihydroxy fatty acids. Arch. Biochem. Biophys. 290, 436–444 10.1016/0003-9861(91)90563-X [DOI] [PubMed] [Google Scholar]
- 4. Chini A., Monte I., Zamarreño A. M., Hamberg M., Lassueur S., Reymond P., Weiss S., Stintzi A., Schaller A., Porzel A., García-Mina J. M., and Solano R. (2018) An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 14, 171–178 10.1038/nchembio.2540 [DOI] [PubMed] [Google Scholar]
- 5. Westfall C. S., Zubieta C., Herrmann J., Kapp U., Nanao M. H., and Jez J. M. (2012) Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 336, 1708–1711 10.1126/science.1221863 [DOI] [PubMed] [Google Scholar]
- 6. Guo Q., Major I. T., and Howe G. A. (2018) Resolution of growth–defense conflict: mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 44, 72–81 10.1016/j.pbi.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 7. Koo A. J. (2018) Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytochem. Rev. 17, 51–80 10.1007/s11101-017-9510-8 [DOI] [Google Scholar]
- 8. Koo A. J., Cooke T. F., and Howe G. A. (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc. Natl. Acad. Sci. U.S.A. 108, 9298–9303 10.1073/pnas.1103542108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kitaoka N., Matsubara T., Sato M., Takahashi K., Wakuta S., Kawaide H., Matsui H., Nabeta K., and Matsuura H. (2011) Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 52, 1757–1765 10.1093/pcp/pcr110 [DOI] [PubMed] [Google Scholar]
- 10. Heitz T., Widemann E., Lugan R., Miesch L., Ullmann P., Désaubry L., Holder E., Grausem B., Kandel S., Miesch M., Werck-Reichhart D., and Pinot F. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287, 6296–6306 10.1074/jbc.M111.316364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koo A. J., Thireault C., Zemelis S., Poudel A. N., Zhang T., Kitaoka N., Brandizzi F., Matsuura H., and Howe G. A. (2014) Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-l-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J. Biol. Chem. 289, 29728–29738 10.1074/jbc.M114.603084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Widemann E., Smirnova E., Aubert Y., Miesch L., and Heitz T. (2016) Dynamics of jasmonate metabolism upon flowering and across leaf stress responses in Arabidopsis thaliana. Plants 5, e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Widemann E., Miesch L., Lugan R., Holder E., Heinrich C., Aubert Y., Miesch M., Pinot F., and Heitz T. (2013) The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J. Biol. Chem. 288, 31701–31714 10.1074/jbc.M113.499228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang T., Poudel A. N., Jewell J. B., Kitaoka N., Staswick P., Matsuura H., and Koo A. J. (2016) Hormone crosstalk in wound stress response: wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 67, 2107–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bruckhoff V., Haroth S., Feussner K., König S., Brodhun F., and Feussner I. (2016) Functional characterization of CYP94-genes and identification of a novel jasmonate catabolite in flowers. PLoS ONE 11, e0159875 10.1371/journal.pone.0159875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caarls L., Elberse J., Awwanah M., Ludwig N. R., de Vries M., Zeilmaker T., Van Wees S. C. M., Schuurink R. C., and Van den Ackerveken G. (2017) Arabidopsis jasmonate-induced oxygenases down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc. Natl. Acad. Sci. U.S.A. 114, 6388–6393 10.1073/pnas.1701101114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smirnova E., Marquis V., Poirier L., Aubert Y., Zumsteg J., Ménard R., Miesch L., and Heitz T. (2017) Jasmonic acid oxidase 2 (JAO2) hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea infection. Mol. Plant 10, 1159–1173 10.1016/j.molp.2017.07.010 [DOI] [PubMed] [Google Scholar]
- 18. Gidda S. K., Miersch O., Levitin A., Schmidt J., Wasternack C., and Varin L. (2003) Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278, 17895–17900 10.1074/jbc.M211943200 [DOI] [PubMed] [Google Scholar]
- 19. Miersch O., Neumerkel J., Dippe M., Stenzel I., and Wasternack C. (2008) Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177, 114–127 [DOI] [PubMed] [Google Scholar]
- 20. Seto Y., Hamada S., Matsuura H., Matsushige M., Satou C., Takahashi K., Masuta C., Ito H., Matsui H., and Nabeta K. (2009) Purification and cDNA cloning of a wound inducible glucosyltransferase active toward 12-hydroxy jasmonic acid. Phytochemistry 70, 370–379 10.1016/j.phytochem.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 21. Wasternack C., and Hause B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development: an update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mugford S. G., Yoshimoto N., Reichelt M., Wirtz M., Hill L., Mugford S. T., Nakazato Y., Noji M., Takahashi H., Kramell R., Gigolashvili T., Flügge U. I., Wasternack C., Gershenzon J., Hell R., Saito K., and Kopriva S. (2009) Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21, 910–927 10.1105/tpc.109.065581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura Y., Mithofer A., Kombrink E., Boland W., Hamamoto S., Uozumi N., Tohma K., and Ueda M. (2011) 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol. 155, 1226–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bowles D., Lim E.-K., Poppenberger B., and Vaistij F. E. (2006) Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57, 567–597 10.1146/annurev.arplant.57.032905.105429 [DOI] [PubMed] [Google Scholar]
- 25. Vogt T., and Jones P. (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5, 380–386 10.1016/S1360-1385(00)01720-9 [DOI] [PubMed] [Google Scholar]
- 26. Ross J., Li Y., Lim E.-K., and Bowles D. J. (2001) Higher plant glycosyltransferases. Genome Biol. 2, reviews3004.3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dean J. V., Mohammed L. A., and Fitzpatrick T. (2005) The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221, 287–296 10.1007/s00425-004-1430-3 [DOI] [PubMed] [Google Scholar]
- 28. Dean J. V., Shah R. P., and Mohammed L. A. (2003) Formation and vacuolar localization of salicylic acid glucose conjugates in soybean cell suspension cultures. Physiol. Plant. 118, 328–336 10.1034/j.1399-3054.2003.00117.x [DOI] [PubMed] [Google Scholar]
- 29. Swiatek A., Dongen W. V., Esmans E. L., and Onckelen H. V. (2004) Metabolic fate of jasmonates in tobacco bright yellow-2 cells. Plant Physiol. 135, 161–172 10.1104/pp.104.040501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kitaoka N., Kawaide H., Amano N., Matsubara T., Nabeta K., Takahashi K., and Matsuura H. (2014) CYP94B3 activity against jasmonic acid amino acid conjugates and the elucidation of 12-O-β-glucopyranosyl-jasmonoyl-l-isoleucine as an additional metabolite. Phytochemistry 99, 6–13 10.1016/j.phytochem.2013.12.019 [DOI] [PubMed] [Google Scholar]
- 31. Mackenzie P. I., Owens I. S., Burchell B., Bock K. W., Bairoch A., Belanger A., Fournel-Gigleux S., Green M., Hum D. W., and Iyanagi T. (1997) The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7, 255–269 10.1097/00008571-199708000-00001 [DOI] [PubMed] [Google Scholar]
- 32. Obayashi T., Aoki Y., Tadaka S., Kagaya Y., and Kinoshita K. (2018) ATTED-II in 2018: a plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol. 59, e3–e3 10.1093/pcp/pcx191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim E. K., Doucet C. J., Li Y., Elias L., Worrall D., Spencer S. P., Ross J., and Bowles D. J. (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 277, 586–592 10.1074/jbc.M109287200 [DOI] [PubMed] [Google Scholar]
- 34. Lim E. K., Li Y., Parr A., Jackson R., Ashford D. A., and Bowles D. J. (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 276, 4344–4349 [DOI] [PubMed] [Google Scholar]
- 35. Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., and Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363 10.1111/j.1365-313X.2007.03052.x [DOI] [PubMed] [Google Scholar]
- 36. Thompson A. M. G., Iancu C. V., Neet K. E., Dean J. V., and Choe J.-y. (2017) Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci. Rep. 7, 46629 10.1038/srep46629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B. G., Tarling C. A., Ford C. M., Bowles D. J., and Davies G. J. (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405 10.1038/sj.emboj.7600970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lim E. K., Ashford D. A., Hou B., Jackson R. G., and Bowles D. J. (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 87, 623–631 10.1002/bit.20154 [DOI] [PubMed] [Google Scholar]
- 39. Kaever A., Landesfeind M., Feussner K., Mosblech A., Heilmann I., Morgenstern B., Feussner I., and Meinicke P. (2015) MarVis-pathway: integrative and exploratory pathway analysis of non-targeted metabolomics data. Metabolomics 11, 764–777 10.1007/s11306-014-0734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montillet J.-L., Cacas J.-L., Garnier L., Montane M.-H., Douki T., Bessoule J.-J., Polkowska-Kowalczyk L., Maciejewska U., Agnel J.-P., Vial A., and Triantaphylides C. (2004) The upstream oxylipin profile of Arabidopsis thaliana: a tool to scan for oxidative stresses. Plant J. 40, 439–451 10.1111/j.1365-313X.2004.02223.x [DOI] [PubMed] [Google Scholar]
- 41. Brown C., Leijon F., and Bulone V. (2012) Radiometric and spectrophotometric in vitro assays of glycosyltransferases involved in plant cell wall carbohydrate biosynthesis. Nat. Protocols 7, 1634–1650 10.1038/nprot.2012.089 [DOI] [PubMed] [Google Scholar]
- 42. Ito T., Motohashi R., Kuromori T., Mizukado S., Sakurai T., Kanahara H., Seki M., and Shinozaki K. (2002) A new resource of locally transposed dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 129, 1695–1699 10.1104/pp.002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuromori T., Hirayama T., Kiyosue Y., Takabe H., Mizukado S., Sakurai T., Akiyama K., Kamiya A., Ito T., and Shinozaki K. (2004) A collection of 11800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37, 897–905 [DOI] [PubMed] [Google Scholar]
- 44. Hillwig M. S., Lebrasseur N. D., Green P. J., and Macintosh G. C. (2008) Impact of transcriptional, ABA-dependent, and ABA-independent pathways on wounding regulation of RNS1 expression. Mol. Genet. Genomics 280, 249–261 10.1007/s00438-008-0360-3 [DOI] [PubMed] [Google Scholar]
- 45. von Saint Paul V., Zhang W., Kanawati B., Geist B., Faus-Kessler T., Schmitt-Kopplin P., and Schäffner A. R. (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23, 4124–4145 10.1105/tpc.111.088443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rehman H. M., Nawaz M. A., Shah Z. H., Ludwig-Müller J., Chung G., Ahmad M. Q., Yang S. H., and Lee S. I. (2018) Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 8, 1875 10.1038/s41598-018-19535-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kai K., Akaike R., Iida K., Yokoyama M., and Watanabe N. (2010) C14-Oxylipin glucosides isolated from Lemna paucicostata. Phytochemistry 71, 1168–1173 10.1016/j.phytochem.2010.04.015 [DOI] [PubMed] [Google Scholar]
- 48. Weichert H., Stenzel I., Berndt E., Wasternack C., and Feussner I. (1999) Metabolic profiling of oxylipins upon salicylate treatment in barley leaves: preferential induction of the reductase pathway by salicylate. FEBS Lett. 464, 133–137 10.1016/S0014-5793(99)01697-X [DOI] [PubMed] [Google Scholar]
- 49. Yang M., Fehl C., Lees K. V., Lim E.-K., Offen W. A., Davies G. J., Bowles D. J., Davidson M. G., Roberts S. J., and Davis B. G. (2018) Functional and informatics analysis enables glycosyltransferase activity prediction. Nat. Chem. Biol. 14, 1109–1117 10.1038/s41589-018-0154-9 [DOI] [PubMed] [Google Scholar]
- 50. Wakuta S., Hamada S., Ito H., Matsuura H., Nabeta K., and Matsui H. (2010) Identification of a β-glucosidase hydrolyzing tuberonic acid glucoside in rice (Oryza sativa L.). Phytochemistry 71, 1280–1288 10.1016/j.phytochem.2010.04.025 [DOI] [PubMed] [Google Scholar]
- 51. König S., Feussner K., Schwarz M., Kaever A., Iven T., Landesfeind M., Ternes P., Karlovsky P., Lipka V., and Feussner I. (2012) Arabidopsis mutants of sphingolipid fatty acid α-hydroxylases accumulate ceramides and salicylates. New Phytol. 196, 1086–1097 10.1111/j.1469-8137.2012.04351.x [DOI] [PubMed] [Google Scholar]
- 52. Xing H.-L., Dong L., Wang Z.-P., Zhang H.-Y., Han C.-Y., Liu B., Wang X., and Chen Q.-J. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Naito Y., Hino K., Bono H., and Ui-Tei K. (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lei Y., Lu L., Liu H.-Y., Li S., Xing F., and Chen L.-L. (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496 10.1093/mp/ssu044 [DOI] [PubMed] [Google Scholar]
- 55. Liu H., Ding Y., Zhou Y., Jin W., Xie K., and Chen L.-L. (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532 10.1016/j.molp.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 56. Ding Y., Li H., Chen L.-L., and Xie K. (2016) Recent advances in genome editing using CRISPR/Cas9. Front. Plant Sci. 7, 703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Z.-P., Xing H.-L., Dong L., Zhang H.-Y., Han C.-Y., Wang X.-C., and Chen Q.-J. (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Clough S. J., and Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 59. Mosblech A., König S., Stenzel I., Grzeganek P., Feussner I., and Heilmann I. (2008) Phosphoinositide and inositolpolyphosphate signalling in defense responses of Arabidopsis thaliana challenged by mechanical wounding. Mol. Plant 1, 249–261 10.1093/mp/ssm028 [DOI] [PubMed] [Google Scholar]
- 60. Oñate-Sanchez L., and Vicente-Carbajosa J. (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1, 93 10.1186/1756-0500-1-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 62. Jones D. T., Taylor W. R., and Thornton J. M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 63. Kumar S., Stecher G., Li M., Knyaz C., and Tamura K. (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Studier F. W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 65. Huang F. C., Hinkelmann J., and Schwab W. (2015) Glucosylation of aroma chemicals and hydroxy fatty acids. J. Biotechnol. 216, 100–109 10.1016/j.jbiotec.2015.10.011 [DOI] [PubMed] [Google Scholar]
- 66. Suzuki H., Hayase H., Nakayama A., Yamaguchi I., Asami T., and Nakajima M. (2007) Identification and characterization of an Ipomoea nil glucosyltransferase which metabolizes some phytohormones. Biochem. Biophys. Res. Commun. 361, 980–986 10.1016/j.bbrc.2007.07.147 [DOI] [PubMed] [Google Scholar]
- 67. Augustin J. M., Drok S., Shinoda T., Sanmiya K., Nielsen J. K., Khakimov B., Olsen C. E., Hansen E. H., Kuzina V., Ekstrøm C. T., Hauser T., and Bak S. (2012) UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol. 160, 1881–1895 10.1104/pp.112.202747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Andreou A.-Z., Hornung E., Kunze S., Rosahl S., and Feussner I. (2009) On the substrate binding of linoleate 9-lipoxygenases. Lipids 44, 207–215 10.1007/s11745-008-3264-4 [DOI] [PubMed] [Google Scholar]
- 69. Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., and Schwudke D. (2008) Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 10.1194/jlr.D700041-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kaever A., Landesfeind M., Feussner K., Morgenstern B., Feussner I., and Meinicke P. (2014) Meta-analysis of pathway enrichment: combining independent and dependent omics data sets. PLoS ONE 9, e89297 10.1371/journal.pone.0089297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.