Abstract
Porcine deltacoronavirus (PDCoV) is a pathogen belonging to the genus Deltacoronavirus that in 2014 caused outbreaks of piglet diarrhea in the United States. To identify suitable therapeutic targets, a more comprehensive understanding of the viral entry pathway is required, particularly of the role of proteases. Here, we identified the proteases that activate the viral spike (S) glycoprotein to initiate cell entry and also pinpointed the host-cellular pathways that PDCoV uses for entry. Our results revealed that cathepsin L (CTSL) and cathepsin B (CTSB) in lysosomes and extracellular trypsin in cell cultures independently activate the S protein for membrane fusion. Pretreating the cells with the lysosomal acidification inhibitor bafilomycin-A1 (Baf-A1) completely inhibited PDCoV entry, and siRNA-mediated ablation of CTSL or CTSB expression significantly reduced viral infection, indicating that PDCoV uses an endosomal pathway for entry. Of note, trypsin treatment of cell cultures also activated PDCoV entry, even when the endosomal pathway was inhibited. This observation indicated that trypsin-induced S protein cleavage and activation in cell cultures enables viral entry directly from the cell surface. Our results provide critical insights into the PDCoV infection mechanism, uncovering two distinct viral entry pathways: one through cathepsin L and cathepsin B in the endosome and another via a protease at the cell surface. Because PDCoV infection sites represent a proteases-rich environment, these findings suggest that endosome inhibitor treatment alone is insufficient to block PDCoV entry into intestinal epithelial cells in vivo. Therefore, approaches that inhibit viral entry from the cell membrane should also be considered.
Keywords: virology, virus, virus entry, protease, proteolytic enzyme, porcine deltacoronavirus, cell entry, cathepsins, trypsin, membrane fusion, endosome, viral pathogenesis, spike glycoprotein, porcine diarrhea
Introduction
The causative agent of porcine diarrhea that first caused outbreaks in 2014 in the United States is a newly identified porcine deltacoronavirus (PDCoV),3 which causes severe diarrhea and vomiting followed by high morbidity and mortality in piglets (1–5). The virus belongs to the genus Deltacoronavirus, and family Coronaviridae (6). Like porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV) in the genus Alphacoronavirus, the jejunum and ileum are the primary sites of PDCoV replication (3). PDCoV was first reported in Hong Kong in 2012 (6) and has been subsequently reported in the United States (4), Canada (7), South Korea (8), China (9), Thailand (10), and Vietnam (11).
PDCoV is an enveloped virus that expresses spike (S) proteins with a high molecular weight. The coronavirus S protein has been recognized as a type I transmembrane glycoprotein with heavy glycosylation (12, 13). The cryo-EM structure of the PDCoV S protein has been determined, and indicates that the structures of the S1 domain are more similar to those of alphacoronaviruses (e.g. PEDV and TGEV) than those of betacoronaviruses (e.g. severe acute respiratory syndrome coronavirus (SARS-CoV), mouse hepatitis virus, and Middle East respiratory syndrome coronavirus (MERS-CoV)) (14). In addition, the S protein of mouse hepatitis virus or MERS-CoV is often post-translationally cleaved into S1 and S2 by endogenous cellular proteases (e.g. furin) (15, 16). A number of features of the PEDV or TGEV S proteins in alphacoronaviruses and PDCoV in deltacoronavirus are conserved; however, they have a low amino acid identity, similar to the S protein between SARS-CoV and those of the other coronaviruses (17). The protein covers the virion surface in a trimeric, integral, and uncleaved format (13). The S trimer is generated in a locked conformation to prevent proteolytic activation triggering membrane fusion (18, 19), which is similar to studies of other coronaviruses (e.g. SARS-CoV (20, 21), PEDV (18), TGEV (22), and human coronavirus 229E (23, 24)). The S1 subunit contains receptor-binding sites, which are responsible for the recognition and binding of its cellular receptor (14, 25–27). After binding to the receptor, conformational changes occur between S1 and S2, which expose the cleavage site to proteases (28). The spike protein is separated into a surface unit, S1, and a transmembrane unit, S2 after cleavage by protease. The cleavage of S protein is the key step for the membrane fusion. The cleaved S2 subunit contains an N-terminal fusion peptide, which can be inserted into the cell membrane and induce virus-cell membrane fusion, leading to viral entry (29, 30).
Host proteases play a crucial role in virus infection and the different proteases used by viruses determine the virus entry pathway to some extent. Four proteases participate in the process of viral infection: 1) membrane-binding proteases, like transmembrane serine protease, which appear to mediate viral entry following virus attachment to cell receptors (31, 32); 2) lysosomal proteases, cathepsin L or cathepsin B activated virus entry after virus endocytosis in virus-targeted cells; 3) extracellular proteases (e.g. intestinal proteases), which are essential for PEDV entry (33); and 4) proprotein convertases (e.g. furin). The S protein is cleaved by furin after production in virus-infected cells.
Although the mechanism of PDCoV entry remains unclear, the functional virus receptor (porcine aminopeptidase N) has been identified as an important factor critical for PDCoV entry into cells (34, 35). However, another crucial factor required for viral entry, in which proteases function as activators of the viral S glycoprotein to activate cell entry, has not been determined. Moreover, which pathway was used by PDCoV for its entry remains unknown. In the present study, we examined the pathways used by PDCoV for cell entry, and the data suggest that cathepsins (i.e. CTSL or CTSB) activate the S protein for fusion activity. The results indicate that PDCoV used an endosomal pathway for its entry and cell infection. Moreover, the function of trypsin in virus infection was also evaluated. We found that trypsin was not necessary for the continuous passage of PDCoV in ST cells, which differed from PEDV. The propagation of PEDV in cell culture requires exogenous trypsin (18, 33). Therefore, whether trypsin plays a role in the entry of PDCoV remains unknown. Our results suggest that treatment with trypsin allowed PDCoV to bypass the endosomal pathway and directly enter the cells from the cell surface. Together, these results suggest that PDCoV used two pathways for its entry. Furthermore, cathepsins (e.g. CTSL and CTSB) and trypsin activated the S protein for viral entry in the endosomal and cell surface pathways, respectively.
Results
PDCoV uses two pathways for its entry
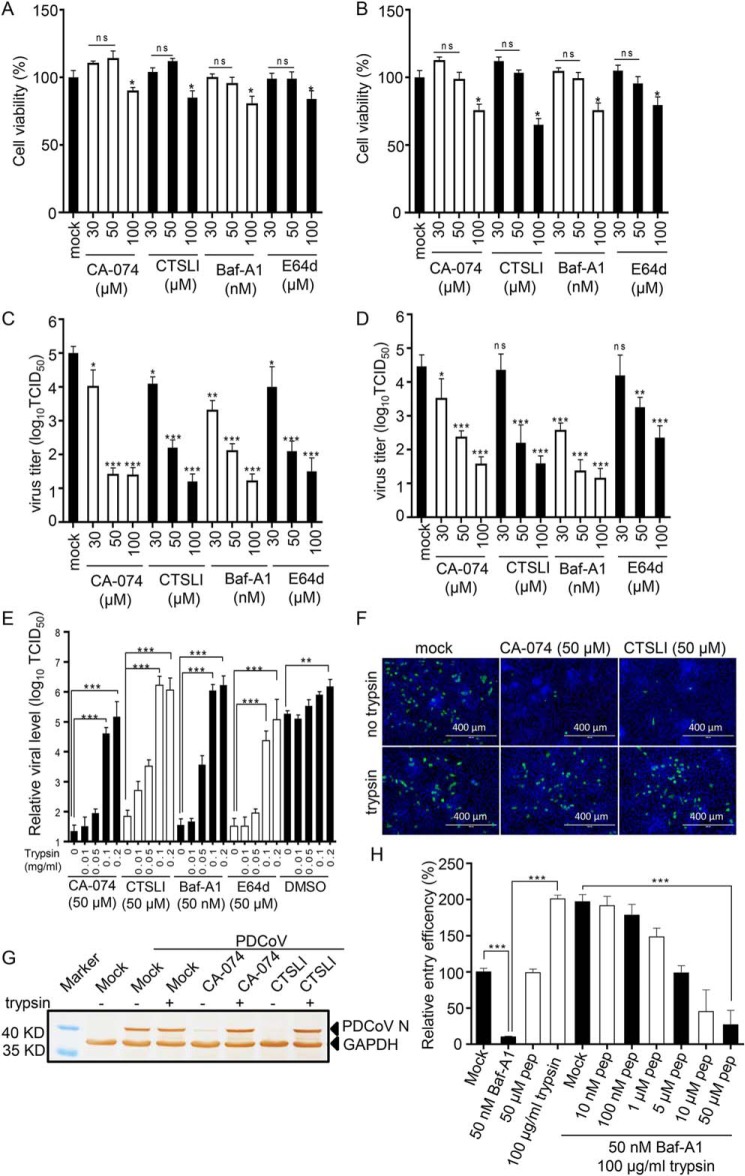
A previous study confirmed that MERS-CoV and SARS-CoV infected cells through two different routes: 1) the endosomal pathway relying on a low pH environment and lysosomal cysteine proteases or 2) directly from the cell surface via fusion of the virus envelope and the cell membrane when the S protein binds to receptors on the cell surface (36, 37). To determine the role of lysosomal cysteine proteases in PDCoV entry, various inhibitors, including bafilomycin A1 (Baf-A1, lysosomal acidification inhibitor), E64d (lysosomal cysteine protease inhibitor), CA-074 (cathepsin B–specific inhibitor), and Z-FY-CHO (CTSLI, cathepsin L–specific inhibitor) were used. The optimal inhibitor concentrations were determined in ST and IPI-2I cells using a CCK-8–based cell viability assay (Fig. 1, A and B, respectively) and virus titer assay by measuring the TCID50 (Fig. 1, C and D, respectively). The results indicate that the inhibitors exhibited cellular toxicity following an increase in the drug concentration. The optimal functional concentrations were 50 μm for E64d, CA-074, and CTSLI and 50 nm for Baf-A1. Furthermore, when ST and IPI-2I cells were treated with inhibitors, PDCoV infection was dramatically inhibited. The results indicate that PDCoV cell entry was activated by lysosomal proteases.
Figure 1.
PDCoV uses two pathways for its entry. Determination of optimum concentrations of inhibitors. A and B, ST cells (A) and IPI-2I cells (B) were pretreated with various concentrations of protease inhibitors at 37 °C for 1 h, and fresh DMEM with 10% FBS was added for 24 h. The optimum concentration of inhibitors for ST and IPI-2I cell viability was determined using a CCK-8 kit. C and D, ST cells (C) and IPI-2I cells (D) were pretreated with various concentrations of protease inhibitors and the cells were infected with PDCoV (m.o.i. = 0.1) at room temperature for 30 min and washed three times with DMEM, followed by an incubation for 48 h at 37 °C. The virus yield was determined by measuring the TCID50. E, trypsin reversed the effect of inhibitors (CA-074, CTSLI, Baf-A1, and E64d) to enhance virus entry. IPI-2I cells were treated with inhibitors (50 μm for E64d, CA-074, and CTSLI, and 50 nm for Baf-A1) for 1 h at 37 °C, and infected with PDCoV at an m.o.i. of 1 for 30 min at 4 °C. Subsequently, the cells were treated with various concentrations of trypsin at room temperature for 10 min and cultured with inhibitors for 6 h. PDCoV entry was detected with RT-qPCR for sgNS7a. F, virus entry was inhibited by inhibitors, and trypsin treatment extensively facilitated virus entry. IPI-2I cells were pretreated with inhibitors (50 μm for CA-074 and CTSLI) and infected with PDCoV at an m.o.i. of 1 for 30 min at 4 °C, followed by treatment with 100 μg/ml trypsin. The cells were fixed and the virus was detected with IFA by anti-PDCoV N mAb at 6 h post-infection. G, alternatively, the expression of the N protein was detected by Western blotting. H, PDCoV directly entered IPI-2I cells from the cell surface. IPI-2I cells were treated with 50 nm Baf-A1 and various concentrations of HR2 peptides for 1 h at 37 °C, infected with PDCoV for 30 min at 4 °C, and treated with 100 μg/ml trypsin. Virus entry was detected with RT-qPCR for sgNS7a after 6 h infection. Data are expressed as the mean ± S.D. for triplicate samples. Level of significance was determined by Student's t test. ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To determine whether PDCoV directly enters from the cell surface, IPI-2I cells were treated with inhibitors (50 μm for E64d, CA-074, CTSLI and 50 nm for Baf-A1) for 1 h at 37 °C to inhibit the endosomal pathway and infected with PDCoV at an m.o.i. of 1 for 30 min at 4 °C (absorption to the cell surface did not allow viral entry into the cells). The cells were subsequently treated with various concentrations of trypsin at room temperature for 10 min and cultured with inhibitors for 6 h. The entry of PDCoV was detected with RT-qPCR for sgNS7a. As shown in Fig. 1E, viral entry was dramatically inhibited following treatment with inhibitors compared with the DMSO-treated group, whereas treatment with 100 μg/ml trypsin extensively facilitated viral entry, showing no differences compared with the mock-treated group. Virus entry in the presence of cathepsin L or cathepsin B inhibitors was also detected using IFA (Fig. 1F) and Western blotting (Fig. 1G). The results suggest that treatment with trypsin could reverse the inhibitory effect of the inhibitors and allow PDCoV entry to bypass the endosomal pathway.
A previous study confirmed that the peptides derived from the heptad repeat (HRP) region of the SARS-CoV S protein can strongly inhibit virus entry from the cell surface induced by proteases but has little effect on the endosomal pathway (38, 39), which has also been found in PEDV as previously described (40). Therefore, to confirm that PDCoV can directly enter cells from the cell surface, the HR2 peptides (GIYNNTILNLTVEINDLQERSKNLSQIADRLQNYIDNLNNTLVDLEWL) of the PDCoV S protein were synthesized. IPI-2I cells were treated with 50 nm Baf-A1 for 1 h at 37 °C, infected with PDCoV for 30 min at 4 °C, and treated with 100 μg/ml trypsin in DMEM containing various concentrations of HR2 peptides. Viral entry was detected using RT-qPCR for sgNS7a at 6 h post-infection. As shown in Fig. 1H, 50 nm Baf-A1 dramatically inhibited virus entry via the endosomal pathway, whereas trypsin treatment reversed this effect, as in the above-mentioned results. However, when the endosomal pathway was inhibited, 10 μm HR2 peptide completely inhibited viral entry, even if trypsin was added. These results indicate that PDCoV bypassed the endosomal pathway and entered the cells directly from the cell surface following trypsin treatment.
Cathepsin L and cathepsin B activate PDCoV entry from the endosome pathway
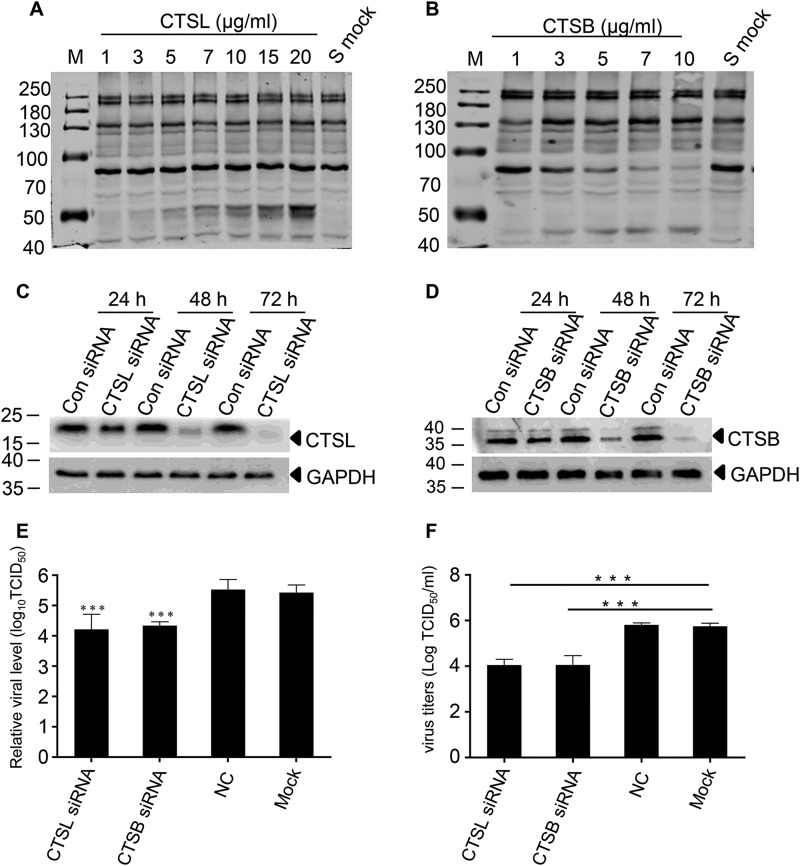
To determine whether cathepsin L or cathepsin B activated the entry of PDCoV by cleaving the S protein, we expressed the S protein in 293T cells. An enzyme cleavage assay was performed with recombinant cathepsin L and cathepsin B in Dulbecco's phosphate-buffered saline (pH 5.6), respectively. The cleavage of the S protein by these two proteases was detected by Western blotting. As shown in Fig. 2, A and B, the extracellular domain of the S protein was ∼180 kDa, as revealed by an anti-FLAG mAb reacting with the C-terminal fraction of the S protein. Cathepsin L cleaved the S protein into two S2 bands of ∼50 kDa, whereas only one S2 band (∼45 kDa) was detected following cathepsin B cleavage.
Figure 2.
PDCoV entry is activated by CTSL and CTSB. A and B, CTSL (A) and CTSB (B) cleaved S protein. The S protein was expressed in 293T cells and the enzyme cleavage assay was performed with recombinant cathepsin L and cathepsin B in Dulbecco's phosphate-buffered saline (pH 5.6), respectively. C and D, the expression of CTSL (C) or CTSB (D) was significantly inhibited by gene-specific siRNA. CTSL- and CTSB-specific siRNA were designed and transfected into IPI-2I cells to knock down endogenous CTSL or CTSB expression. The protein expression at different times was detected by Western blotting. E, siRNA for CTSL or CTSB dramatically decreased virus entry. IPI-2I cells were transfected with siRNAs of CTSL or CTSB; 48 h later, the cells were infected with PDCoV (m.o.i. = 1), then NS7a sgRNA was detected with real-time PCR at 6 h post-infection. F, siRNA for CTSL or CTSB dramatically decreased the virus yield. Following transfection with siRNAs, IPI-2I cells were infected with PDCoV (m.o.i. = 0.01). The viral dose was measured by calculating TCID50 at 48 h post-infection. Data are expressed as the mean ± S.D. for triplicate samples. Level of significance was determined by Student's t test. ns, p > 0.05; ***, p < 0.001.
To confirm that the cathepsins (CTSL and CTSB) activated PDCoV entry, specific siRNAs were designed and transfected into IPI-2I cells to knock down endogenous CTSL or CTSB expression. Western blotting results imply that endogenous CTSL or CTSB expression was successfully knocked down (Fig. 2, C and D). As shown in Fig. 2E, viral entry was inhibited by ∼10-fold lower compared with that of the mock-treated group. The virus yield was also dramatically decreased, as shown by measuring the TCID50 (Fig. 2F). These results also suggest that CTSL or CTSB activated the S protein of PDCoV, which facilitated viral infection from the endosomal pathway.
PDCoV infection up-regulates the expression of CTSL and CTSB
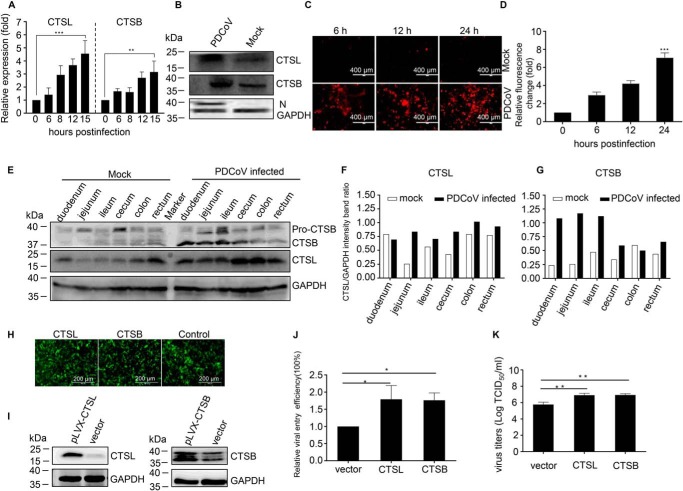
To further understand the relationship between cathepsins (CTSL and CTSB) and PDCoV infection, we examined the changes in protein expression and enzyme activity following viral infection. As determined by RT-qPCR, the levels of CTSL or CTSB mRNA were significantly up-regulated in PDCoV-infected IPI-2I cells at 15 h post-infection (Fig. 3A). Moreover, with regard to the protein levels, virus infection increased the expression of CTSL and CTSB (Fig. 3B). We also examined the changes in cathepsin enzyme activity following PDCoV infection, as described in Fig. 3, C and D. Although viral infection altered the enzyme activity of CTSB, it had no effect on CTSL enzyme activity (data not shown). To examine the changes of CTSL and CTSB in pigs following PDCoV infection in vivo, intestinal tissues from five specific pathogen-free pigs (three infected pigs and two control pigs) used to study the pathogenicity of PDCoV were prepared and subjected to Western blot analyses. As shown in Fig. 3E, PDCoV infection promoted CTSL and CTSB expression in the pig intestinal tissues. The intensity band ratio of CTSL/GAPDH and CTSB/GAPDH is shown in Fig. 3, F and G. To investigate whether these changes contributed to PDCoV infection, we overexpressed CTSL and CTSB in IPI-2I cells with the pLVX-IRES-EGFP plasmid, a lentiviral vector expressing EGFP proteins. As observed in Fig. 3H, more than 90% of the cells displayed green fluorescence, suggesting that the CTSL or CTSB genes had been successfully transduced into the ST cells with high efficiency. Western blot analyses also indicated that CTSL and CTSB, respectively, were overexpressed in IPI-2I cells (Fig. 3I). The relative efficiency of PDCoV entry into CTSL- or CTSB-overexpressing IPI-2I cells was ∼2-fold higher than that of the vector-only transduced cells (Fig. 3J). We also examined the viral yield in the overexpressed cells. The results indicate that the viral yield was higher in CTSL (6.92 ± 0.38 log10 TCID50/ml) or CTSB (6.93 ± 0.28 log10 TCID50/ml)–overexpressing IPI-2I cells than in the mock cells (5.77 ± 0.5 log10 TCID50/ml) (Fig. 3K). These results indicate that PDCoV infection could up-regulate the expression of CTSL and CTSB both in vitro and in vivo, and these changes promoted viral infection.
Figure 3.
PDCoV infection up-regulates the expression of CTSL and CTSB in vitro and in vivo to promote virus infection. A, PDCoV infection increased the level of CTSL and CTSB mRNA. IPI-2I cells were infected with PDCoV (m.o.i. = 1), and the cellular RNA was extracted and determined by RT-qPCR at different time points. B, PDCoV infection increased the level of CTSL and CTSB protein expression in vitro. IPI-2I cells were infected with PDCoV (m.o.i. = 1), and cells were collected and subjected to Western blotting with anti-CTSL– or anti-CTSB–specific antibodies. C, PDCoV infection increased the enzymatic activity of CTSB. IPI-2I cells were infected with PDCoV at an m.o.i. of 1, and the enzymatic activity of CTSL or CTSB was measured with a Magic Red® Cathepsin L or Cathepsin B Detection Kit at different time points. D, the fluorescence intensity of the red fluorescence could be detected at an optimal excitation and emission wavelength of 592 nm and 628 nm, respectively. E, PDCoV infection increased the level of CTSL and CTSB protein expression in vivo. Intestinal tissues from five specific pathogen-free pigs (three infected pigs and two control pigs) were prepared and subjected to Western blot analysis. F and G, the intensity band ratios of CTSL/GAPDH (F) and CTSB/GAPDH (G) were calculated and compared. The overexpression of CTSL or CTSB facilitates PDCoV entry. H, CTSL or CTSB was overexpressed in IPI-2I cells. IPI-2I cells were transfected with CTSL- or CTSB-expressing plasmids and the transfection efficiency was detected by EGFP expression. I, the protein expression was detected by Western blotting. J, overexpression of CTSL or CTSB promoted viral entry. K, the virus yield as determined by measuring the TCID50 at 36 h post-infection. Data are expressed as the mean ± S.D. for triplicate samples. Level of significance was determined by Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Entry of PDCoV from the cell surface is facilitated by trypsin
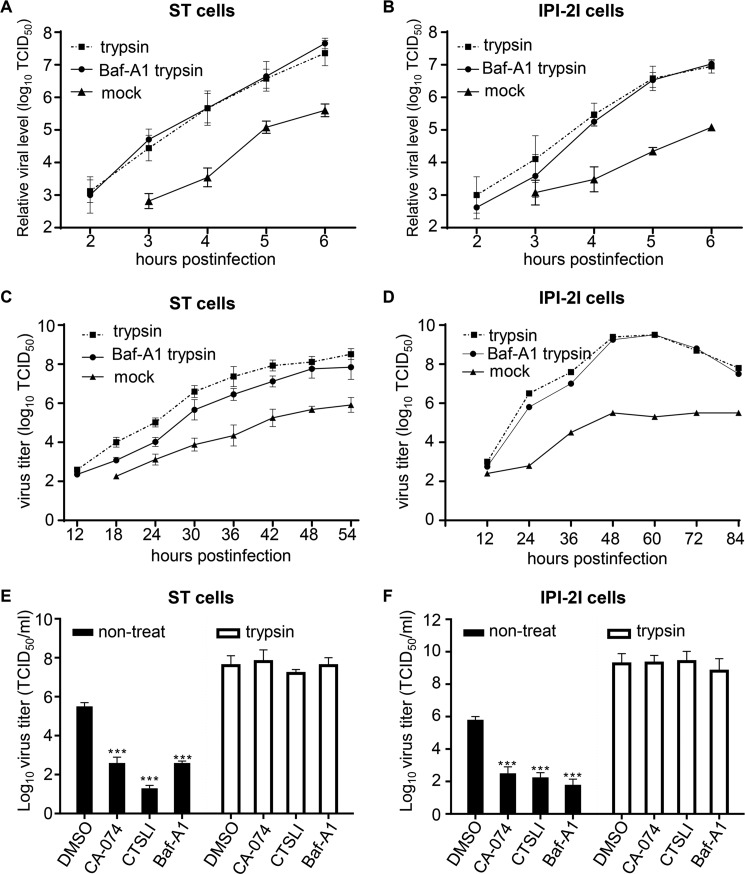
To compare the efficiency of the two pathways or cell types with regard to virus entry, the infection kinetics of PDCoV in cells pretreated with Baf-A1 and subsequently treated with 100 μg/ml trypsin were conducted, in which the virus only enters via the cell surface pathway. The ST and IPI-2I cells treated with trypsin (viral entry from the cell surface) or untreated (viral entry only via the endosome pathway) were used as controls. The time of viral entry from the cell surface was ∼1 h earlier via only the endosomal pathway in ST and IPI-2I cells (Fig. 4, A and B, respectively). The results also indicated that viral entry at any given time was always about 10–100-fold higher in the trypsin-treated cells than in the untreated cells. The viral growth curve was also determined to assess the efficiency of virus spread via the two entry pathways in the presence of trypsin or Baf-A1. The virus titer could be determined in the trypsin or mock-treated cells at 12 h post-infection, whereas the viral dose in the cell cultures containing trypsin and Baf-A1 could be examined at 18 h post-infection. Moreover, the viral titer in the cultures treated with only trypsin was 10- to 100-fold higher than that in the ST cells treated with trypsin and Baf-A1 (Fig. 4C). In IPI-2I cells, the effect of trypsin on PDCoV infection was more obvious than that in the ST cells, especially 24 h post-infection, and the virus yield in trypsin-treated IPI-2I cells (6.5 log10 TCID50/ml) was ∼10,000-fold higher than that in the untreated IPI-2I cells (2.8 log10 TCID50/ml) (Fig. 4D). We also assessed the function of trypsin in the presence of other inhibitors by determining the viral titer. As shown in Fig. 4, E and F, trypsin treatment increased the viral load by about 100- to 1000-fold when compared with that of the untreated cells, and could reverse the inhibitory effect. These results suggest that trypsin treatment enhanced PDCoV infection.
Figure 4.
Trypsin treatment enhances virus entry and dramatically increased the virus yield. A and B, trypsin promoted virus entry into ST cells (A) or IPI-2I cells (B). ST or IPI-2I cells were pretreated with Baf-A1 and infected with PDCoV at an m.o.i. of 1, followed by treatment with 100 μg/ml trypsin. ST and IPI-2I cells treated only with trypsin or left untreated were used as controls. PDCoV entry at the given time was detected with RT-qPCR for sgNS7a. C and D, trypsin increased the virus infection in ST cells (C) and IPI-2I cells (D). ST or IPI-2I cells were pretreated with Baf-A1 and infected with PDCoV at an m.o.i. of 0.001, followed by treatment with 100 μg/ml trypsin. ST and IPI-2I cells treated only with trypsin or left untreated were used as controls. The virus dose at the indicated time points was detected by measuring the TCID50. E and F, trypsin promoted viral infection in the presence of cathepsin L and cathepsin B inhibitors in ST cells (E) and IPI-2I cells (F). ST or IPI-2I cells were pretreated with Baf-A1, CA-074, or CTSLI inhibitors and infected with PDCoV at an m.o.i. of 0.001, followed by treatment with 100 μg/ml trypsin. The virus yield was determined by measuring the TCID50 at 36 h post-infection. Data are expressed as the mean ± S.D. for triplicate samples. Level of significance was determined by Student's t test. ns, p > 0.05; ***, p < 0.001.
Activation of cell fusion and S1–S2 cleavage by trypsin treatment
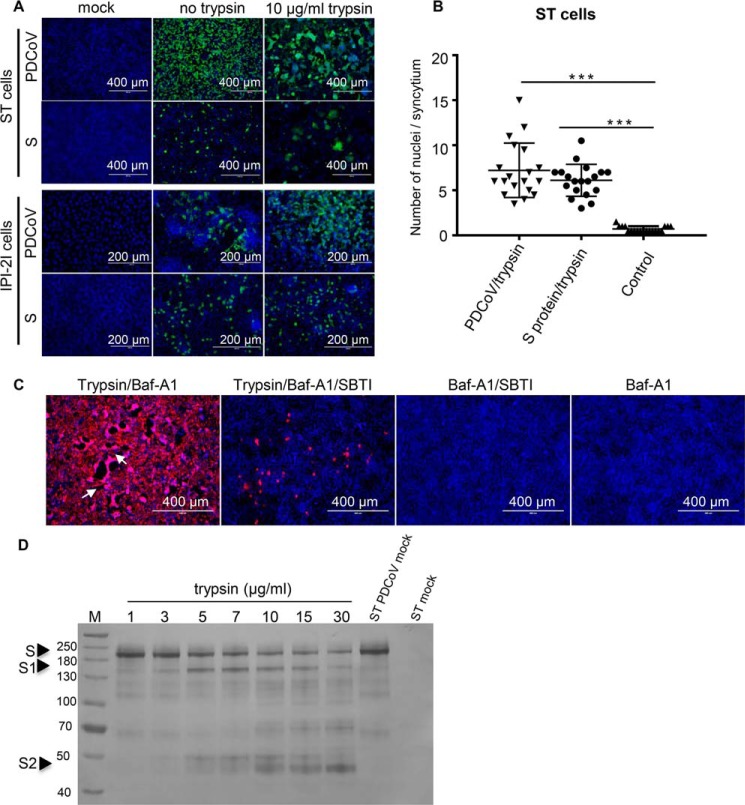
ST cells were infected with PDCoV at an m.o.i. of 1, and the infected cells were treated with 20 μg/ml trypsin or left untreated at room temperature for 20 min. The uninfected cells were used as a control. Cell fusion was detected by IFA and the CPE was observed. The results indicated that PDCoV infection with trypsin treatment induced cell fusion, and the cells displayed several syncytia and obvious CPE. In contrast, no syncytia were observed in the control or untreated ST cells (Fig. 5A). To confirm that cell fusion was induced by the PDCoV S protein after trypsin treatment, the plasmid encoding the S gene (pAAV-PDCoV opti-S) was transfected into ST cells, which were treated with trypsin as described above. At 48 h post-transfection, large cell fusion was observed with trypsin treatment (Fig. 5A), whereas the untreated or mock cells remained largely mononucleated. However, no obvious cell fusion was detected in the PDCoV-infected or transfected IPI-2I cells with the pAAV-PDCoV opti-S plasmid, although the IPI-2I cells were treated with trypsin. The number of nuclei in the syncytia was counted to semi-quantify the extent of cell fusion (Fig. 5B). To further verify the role of trypsin in PDCoV infection, the SBTI was added to the cell culture in the presence of Baf-A1 to block trypsin activity and as shown in Fig. 5C, virus infection was significantly inhibited compared with the trypsin treatment group in the presence of Baf-A1 (Fig. 5C). Large syncytial foci were observed after trypsin treatment in the presence of Baf-A1 (Fig. 5C, white arrow), whereas no syncytium was observed after SBTI treatment, even though in the presence of trypsin. To provide direct biochemical evidence that trypsin cleaved PDCoV S, ST cells were infected with PDCoV to express S protein in the cells. The cells were collected and lysed by sonication and then incubated with various concentrations of trypsin followed by Western blot analysis with S polyclonal antibody. As shown in Fig. 5D, two extra bands (150 and 50 kDa) were detected after trypsin treatment. The results suggested that trypsin could activate PDCoV spike protein to induce cell to cell fusion.
Figure 5.
Cell fusion induced by trypsin treatment. A, trypsin activated cell fusion in ST cells but not in IPI-2I cells. ST and IPI-2I cells were infected with PDCoV or transfected with a PDCoV S-expressing plasmid with or without trypsin, and virus infection was detected by IFA with a mouse anti-PDCoV N protein mAb. Untreated or uninfected cells were used as controls. B, cell fusion was semi-quantified by counting the number of nuclei in the syncytia in ST cells following infection or transfection with trypsin treatment. C, ST cells were pretreated with Baf-A1, then infected with PDCoV (m.o.i. = 0.1) in the presence 10 μg/ml trypsin or trypsin and 40 μg/ml SBTI (Sigma) for 24 h. PDCoV-positive cells were detected by IFA. D, ST cells were infected with PDCoV to express S protein in the cells. The cells were collected and lysed by sonication, and then incubated with various concentrations of trypsin followed by Western blot analysis with S polyclonal antibody. Two extra bands (150 and 50 kDa) were detected after trypsin treatment. Data are expressed as the mean ± S.D. for triplicate samples. Level of significance was determined by Student's t test. ns, p > 0.05; ***, p < 0.001.
Arg-672 in the S protein is critical for trypsin-induced cell fusion
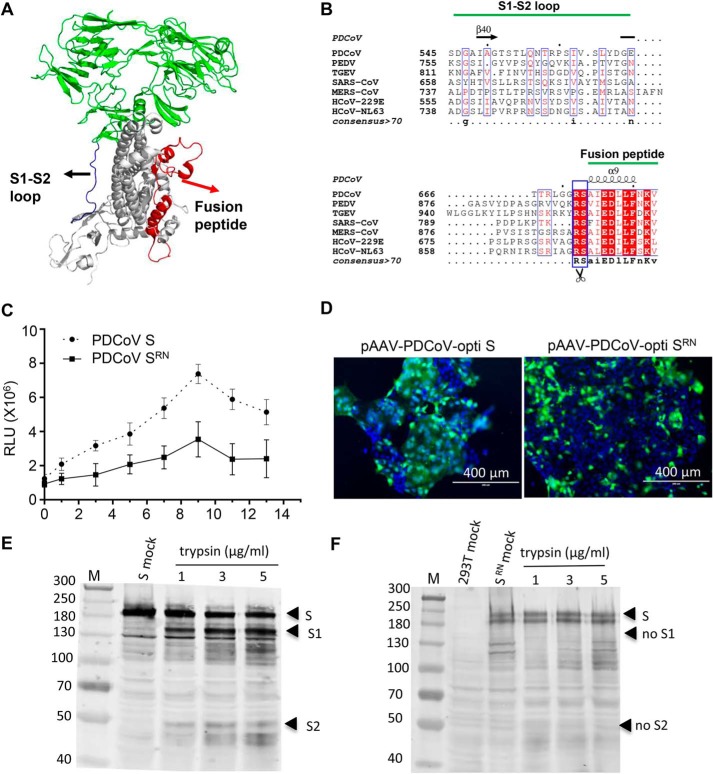
The structure of the PDCoV S protein was determined by cryo-EM (PDB ID: 6B7N). The monomer structure of the S protein is shown in Fig. 6A. A loop structure was observed between the S1 and S2 domains which contains a variety of proteases cleavage sites (e.g. furin protease (16, 41, 42), trypsin (12), and cathepsin L (44)). Whether the loop structure of the PDCoV S protein could be cleaved by proteases remains unknown. A multiple sequence alignment of the S gene was conducted among coronaviruses, which indicated that the S2′ (Arg-672 in PDCoV S protein) cleavage site was conserved (Fig. 6B). This amino acid site was confirmed to be cleaved by trypsin in SARS-CoV and PEDV. We also conducted a cell to cell fusion assay to analyze the effect of Arg-672 in the S-meditated cell fusion induced by trypsin. The results indicate that the extent of cell fusion was dramatically decreased with the mutated S compared with the WT S protein (Fig. 6C). The results were confirmed by S- or SRN-meditated cell fusion, as shown in Fig. 6D; a large amount of cell fusion was detected in the S-transfected ST cells when the cells were treated with trypsin, whereas little or no obvious cell fusion was detected in SRN-transfected ST cells. To determine whether this position (Arg-672) was cleaved by trypsin, we mutated residue Arg-672 to N (SRN) and no detectable cleaved S2 bands were observed (Fig. 6F) with S polyclonal antibody. The cleavage of WT S protein was used as a control and S2 bands could be detected, as shown in Fig. 6E.
Figure 6.
Arg-672 in the S protein is crucial for trypsin-induced cell to cell fusion. A, the monomer structure of the PDCoV spike protein was determined by cryo-EM (PDB ID: 6B7N). Loop regions between S1 and S2 domains and fusion peptide are indicated with blue and red, respectively. B, sequence conservation among the coronavirus glycoproteins. Multiple sequence alignment of the coronavirus S protein sequences. PDCoV strain NH (GenBankTM: ANA78450.1), PEDV CV777 (GenBankTM: AAK38656.1), TGEV Purder strain (GenBankTM: ABG89335.1), SARS coronavirus Urbani (GenBankTM: AAP13441.1), MERS-CoV (GenBankTM: AVN89453.1), HcoV-229E (GenBankTM: ARB07392.1), and HcoV-NL63 (GenBankTM: AFV53148.1). The secondary structure of the PDCoV S protein is indicated above the sequence. C, the R672N mutant inhibited trypsin-induced cell fusion. Effector cells (293T) were co-transfected with WT (pCAGGS-opti S) or mutant S-expressing plasmid (pCAGGS-PDCoV opti-SRN) and pCDNA3.1-T7 vectors. The target cells (ST) were transfected with the pET-32a-IRES-luc plasmid, and then the ST cells were quickly trypsinized and overlaid with 293T cells followed by various concentrations of trypsin treatment to induce cell fusion. Luciferase activity was measured using a luciferase assay kit (Promega) with an EnSpire® multifunctional microplate reader (PerkinElmer). D, cell fusion was significantly inhibited by the R672N mutant. ST cells were transfected with pAAV-PDCoV-opti S and pAAV-PDCoV opti-SRN; 48 h post-transfection, the cells were treated with 100 μg/ml trypsin in DMEM for 10 min at room temperature and cell fusion was detected with a fluorescence microscope. The introduction of a furin site at Arg-672 induced PDCoV S-mediated cell fusion. E and F, 293T cells were transfected with pAAV-PDCoV-opti S (E) and pAAV-PDCoV opti-SRN (F) plasmid. At 48 h post-transfection, cells were collected to perform trypsin cleavage assay.
Discussion
The S protein of coronavirus is a type I membrane glycoprotein with high molecular weight which determines the viral tropism, host range, and pathogenicity by receptor specificity and activation meditated by host proteases during virus entry into cells (13, 16, 27). Because of the essential role of S cleavage for coronavirus infection, targeting of relevant host cell proteases may be a promising therapeutic strategy for coronavirus infection (41, 45, 47). However, the viral entry pathway of PDCoV, particularly of the role of proteases in viral entry has not been determined. In this study, we found that PDCoV used two pathways (CTSL and CTSB in the endosome and trypsin at the cell surface) for its entry.
The roles of CTSL and CTSB have been extensively studied in virus entry, such as SARS-CoV, MERS-CoV, and PEDV (48–50). Here, we found that purified CTSL and CTSB can also cleave S proteins into a fusion-ready formation for membrane fusion under a low pH environment. Furthermore, PDCoV infection could increase CTSL and CTSB expression in vivo and in vitro, and the enzyme activity of CTSB was increased following PDCoV infection. These findings are consistent with those of a previous study showing that porcine reproductive and respiratory syndrome virus (PRRSV) infection up-regulates CTSL expression, but PDCoV infection increased the proteolytic activity of intracellular CTSB instead of the CTSL following PRRSV infection (51). The mechanisms of the up-regulation of CTSL and CTSB to promote PDCoV infection should be further investigated. The enzyme activity was increased by PDCoV infection, and this may represent the best method of efficiently activating viral entry.
PDCoV is enteropathogenic and the primary sites of PDCoV replication are the jejunum and ileum (3). The jejunum and ileum belong to a portion of the small intestine that contains several digestive enzymes (53, 54). Therefore, the small intestine fluid, including trypsin or other proteases, may contribute to viral infection during the viral entry step (18). In fact, the findings of a previous study suggest that the isolation or repeated passage of PEDV and PDCoV in vitro, and the addition of trypsin or small intestinal contents is necessary during cell culture (33, 55). However, whether trypsin plays an important role in virus entry has not been determined. In this study, we found that trypsin activated the membrane fusion induced by the PDCoV S protein in ST cells. This indicated that treatment with trypsin meditated the cell–membrane fusion induced by the S protein in ST cells but not in IPI-2I cells. Moreover, it may meditate fusion of the viral envelope to the cell membrane, which contributes to viral infection. Therefore, we formulated the hypothesis that PDCoV could bypass the endosomal pathway and directly enter cells from the cell surface. The addition of trypsin rendered viral entry by bypassing the endosomal pathway with high efficiency. To further verify this assumption, we compounded the HR2 regions of the PDCoV S protein and found that this peptide could efficiently inhibit viral entry from the cell surface but had little effect on entry from the endosomal pathway as described in a previous study of SARS-CoV (38). Moreover, PDCoV entry was significantly inhibited by the HR2 peptide even though the cells were treated with trypsin. These results confirm the hypothesis that PDCoV could enter the cells via a trypsin-mediated cell surface pathway. We compared the entry efficiency of PDCoV from 2 to 6 h post-infection. The direct entry of PDCoV from the cell surface meditated by trypsin was considered the most effective method of viral infection, and was about 1 h in advance of that via the endosomal pathway. Abundant proteases in the environment of the small intestine provide powerful cleavage for the S proteins of PDCoV, and entry from the cell surface is the most appropriate choice for virus infection. Considering several Toll-like receptors locate in intracellular compartments such as endosomes, bypassing the endosomal pathway for cell entry may minimally induce the body's immune response, which may also be a method of viral immune escape (56, 57).
As described above, PDCoV entry from the cell surface directly meditated by proteases in small intestine is the most efficient method of viral infection. Thus, the question remains as to why the virus retains an inefficient method (endosomal pathway) of entry. Recent reports have shown that an alternative pathway for PEDV infection exists from the nasal cavity to the intestinal mucosa in swine (58). This study provides evidence of airborne transmission for a gastrointestinal coronavirus in which entry via the endosomal pathway may be an important route for viral infection from nasal epithelial cells. A study of PDCoV pathogenicity also found that infection could occur indirectly by contact in piglets. The control gnotobiotic piglets were infected by indirect contact and displayed high levels of viral RNA similar to those of the infected piglets (59). We have sufficient reason to believe that PDCoV may use a pathway similar to PEDV for its infection, although the molecular mechanisms remain to be elucidated.
In summary, our study found that PDCoV uses two pathways for cell entry. Moreover, PDCoV entry from the cell surface was found to be more efficient than that from the endosome. Therefore, treatment with trypsin dramatically increased the virus yield, which may explain why the addition of trypsin contributes to viral isolation. These results suggest that treatment with inhibitors of the cell entry step may be insufficient for blocking only PDCoV entry into intestinal epithelial cells in vivo from endosomes, given that PDCoV can directly enter cells from the cell surface following protease treatment. Therefore, targets designed to inhibit viral entry from the cell membrane also should be considered.
Experimental procedures
Cells and virus
Swine testis cells (ST cells), 293T cells (human embryonic kidney cells), IPI-2I cells (porcine intestinal epithelial cells), and human hepatocellular carcinoma Huh-7 cells were grown in Dulbecco's modified Eagle medium (DMEM) (Gibco), supplemented with 10% FBS (Gibco) and a 1% penicillin-streptomycin solution (Gibco). The cells were grown at 37 °C in a 5% CO2 incubator. The PDCoV strain NH was grown and titrated in ST cells.
Plasmids
The codon-optimized PDCoV S gene (PDCoV strain NH, GenBankTM: KU981062.1) was cloned into the pCAGGS and pAAV-IRES-hrGFP vector. The constructs had a FLAG tag at the C terminus and were designated pCAGGS-PDCoV opti-S and pAAV-PDCoV opti-S. Two pairs of specific primers were used for PCR amplification of the gene Sus scrofa cathepsin L (CTSL) (NCBI Reference Sequence: NM_213892.1) and Sus scrofa cathepsin B (CTSB) (NCBI Reference Sequence: NM_001097458.1) from the cDNA of ST cells, respectively. The PCR products were each cloned into the pLVX-IRES-ZsGreen1 vector with an HA tag at the C terminus to generate the plasmids pLVX-CTSL/pLVX-CTSB. Luciferase plasmids (pcDNA3.1-T7 and pET-32a-IRES-luc) were constructed as described previously to detect cell– fusion (60). The pCDNA3.1-T7 vector, which encodes the T7 polymerase, was PCR amplified from Escherichia coli BL21 (DE3) competent cells. The pET-32a-IRES-luc expressing luciferase was controlled by a T7 promoter, in which the internal ribosome entry site (IRES) sequence and the luciferase sequence were cloned into the BamH I/EcoR I and EcoR I/NotI sites of the pET-32a vector, respectively.
RT-qPCR for sgNS7a and calibration line
The NS7a subgenome (sgNS7a) was amplified using RT-qPCR to quantify the efficiency of viral entry (61). This method was also used with other coronaviruses (e.g. SARS-CoV and MERS-CoV) as described previously (36, 37). Detection of the level of CTSL or CTSB mRNA in IPI-2I cells or pig small intestinal tissue was performed by SYBR Green–based RT-qPCR with GAPDH as the internal control. Relative quantification was performed using the cycle threshold (ΔΔCT) method. The One Step PrimeScript™ RT-PCR Kit (Perfect Real Time) or One Step SYBR® PrimeScript™ RT-PCR Kit II (Perfect Real Time) was used to perform RT-qPCR analysis according to the manufacturer's instructions under the following conditions: reverse transcription (42 °C for 10 min; 95 °C for 10 s), one cycle; PCR (95 °C for 5 s; 60 °C for 20 s), and 45 cycles with a LightCycler 480 instrument (Roche Diagnostics). All of the primers and probes are listed in Table 1. To measure the level of viral entry, IPI-2I cells in 12-well culture plates were infected with 10-fold stepwise diluted PDCoV, from 102 to 106, at a 50% tissue culture infectious dose (TCID50). 6 h later, the total cellular RNA was isolated using 1 ml TRIzol® reagent according to the established protocol and dissolved in 40 μl DNase/RNase-free ddH2O. The amounts of sgNS7a were determined by RT-qPCR. The calibration line was established according to the cycle value and virus titer. The relationship between the viral titer (x axis) and real-time PCR cycles (y axis) was demonstrated to reach a positive correlation (data not shown).
Table 1.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| sgNS7a-leader-Fa | ATCTCCCTAGCTTCGCTAGTTCTCTAC |
| sgNS7a-Ra | GAAACCTTGAGCTGGGCCA |
| sgNS7a-probea | FAM-ACCCCAACAATCCT-MGB |
| CTSL-Fa | GGCAAGCTTGTTTCACTGAG |
| CTSL-Ra | CCTCCATTGTCCTTCACGTA |
| GAPDH-Fa | ACACTCACTCTTCTACCTTTG |
| GAPDH-Ra | CAAATTCATTGTCGTACCAG |
| CTSB-Fa | AACTGCCCGACCATCAAAG |
| CTSB-Ra | CACTCGTCGCCACAACAG |
| CTSL-Fa | AATATGGGATCCATGAAACCTTCACTCTTCCTG |
| CTSL-Rb | AAAATAATACTCGAGCACGGTGGGATAGCTGGCT |
| CTSB-Fb | AAAATAATGGATCCATGTGGCGGCTCTTGGCCAC |
| CTSB-Rb | AAATAACTCGAGGAAATGGGGAGTACATGGGA |
a Primers used for RT-qPCR.
b Primers used for the amplication of CTSL and CTSB genes. The products were cloned into EcoR I (bold) and Xho I (underlined) sites of the pAAV-IRES-hrGFP vector.
Indirect immunofluorescence assay (IFA)
ST and IPI-2I cells in 24-well plates were infected with PDCoV at a multiplicity of infection (m.o.i.) of 0.1 in DMEM. After adsorption at 37 °C for 30 min, the virus was removed, and the cells were treated with 100 μg/ml trypsin in DMEM for 10 min at room temperature, or left untreated. After the trypsin had been removed, the cells were washed three times in DMEM. The cells were cultured in DMEM with or without 10 μg/ml trypsin for 24 h. When the cytopathic effect (CPE) was obvious, the cells were fixed with 4% paraformaldehyde and blocked (5% nonfat dry milk in PBS) overnight. The cells were subsequently incubated for 60 min with a mouse anti-PDCoV N protein mAb, followed by Alexa Fluor® 488-conjugated sheep anti-mouse IgG for 60 min. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and the results were observed using a fluorescence microscope.
ST and IPI-2I cells grown to 90% confluence in 12-well plates were transfected with a pAAV-IRES-hrGFP-PDCoV NH opti-S plasmid using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. At 48 h post-transfection, the cells were treated with 100 μg/ml trypsin in DMEM for 10 min at room temperature, or left untreated. After removing the trypsin, the cells were washed three times in DMEM. The cells were cultured in DMEM with or without 10 μg/ml trypsin for 24 h. The cells were fixed and stained with DAPI. The number of nuclei in the syncytia was counted.
ST cells were pretreated with 50 nm Baf-A1 at 37 °C for 1 h, then inoculated with PDCoV (m.o.i. = 0.1) in the presence 10 μg/ml trypsin or trypsin and 40 μg/ml soybean trypsin inhibitor type I (SBTI, Sigma) for 24 h (18). The cells were fixed and staining with mouse anti-PDCoV N mAb, following with Alexa Fluor® 633–conjugated goat anti-mouse IgG (H+L) antibody to detect PDCoV-positive cells.
Cell to cell fusion assay
To quantify the fusion induced by trypsin, a cell– fusion assay based on luciferase expression was performed as described previously (52). Briefly, effector cells (293T) were co-transfected with WT (pCAGGS-opti S) and pCDNA3.1-T7 vectors. The target cells (ST) were transfected with a pET-32a-IRES-luc plasmid. At 6 h post-transfection, the ST cells were quickly trypsinized and overlaid with 293T cells; 24 h later, the co-cultured cells were treated with various concentrations of trypsin in serum-free DMEM at room temperature for 30 min to induce cell fusion. The cells were incubated for another 6 h with fresh DMEM with 10% FBS and then lysed. Luciferase activity was measured using a luciferase assay kit (Promega) with an EnSpire® multifunctional microplate reader (PerkinElmer).
Inhibitors
Various inhibitors were used as described in previous studies (43, 46, 49). To determine the optimal inhibitor concentration, ST and IPI-2I cells were pretreated with various concentrations of lysosomal acidification inhibitor (bafilomycin-A1, Abcam) and protease inhibitors (CA-074, Sigma-Aldrich; cathepsin L inhibitor Z-FY-CHO, (CTSLI), Santa Cruz Biotechnology; and E64d, Sigma-Aldrich) for 1 h at 37 °C, after which fresh DMEM with 10% FBS was added for 24 h. The effect of inhibitors on the viability of ST and IPI-2I cells was determined with a CCK-8 kit (Dojindo Molecular Technologies, Gaithersburg, MD). The inhibitor-treated ST and IPI-2I cells were infected with PDCoV at an m.o.i. of 1 for 6 h because a previous study showed that one cycle of replication for most coronaviruses takes 6 h. The total cellular RNA was isolated, and RT-qPCR was performed to measure viral entry.
Western blotting
IPI-2I cells were pretreated with 50 μm CA-074 or 50 μm CTSLI for 1 h at 37 °C, and the cells were infected with PDCoV at an m.o.i. of 1 for 30 min at 4 °C, followed by treatment with 100 μg/ml trypsin at room temperature for 20 min and washed three times with DMEM. At 6 h post-infection, the cells were prepared for Western blotting. The proteins were transferred to a nitrocellulose membrane and incubated with mouse anti-pig CTSL (Abcam) or rabbit anti-pig CTSB (Cell Signaling Technology) antibodies, a mouse anti-PDCoV N protein mAb, and mouse anti-GAPDH antibody, followed by horseradish peroxidase (HRP)–conjugated sheep anti-mouse IgG (Sigma-Aldrich) or HRP-conjugated sheep anti-rabbit IgG (Thermo Fisher Science). The expression of proteins was detected using the enhanced chemiluminescence Western blotting detection system (GE Healthcare Life Sciences).
Knockdown
The siRNAs were designed by GenePharma (Shanghai, China) (Table 2). CTSL- or CTSB-specific siRNA pools or control siRNA were used to transfect IPI-2I cells. After 24 h, the cells were prepared for the detection of gene silencing using Western blotting. In some experiments, after 24 h of transfection, IPI-2I cells were infected with PDCoV at an m.o.i. of 1 and the cells were subjected to RT-qPCR analysis 6 h later.
Table 2.
Sequences of siRNA used to ablate CTSL and CTSB protein expression in IPI-2I cells
| Target siRNAs sense and antisense strand sequence (5′–3′) | |
|---|---|
| CTSL | |
| siRNA1 | GCAUGGCCAUGAAUGCCUUTTAAGGCAUUCAUGGCCAUGCTT |
| siRNA2 | CCCUCGAAGGACAGAUGUUTTAACAUCUGUCCUUCGAGGGTT |
| siRNA3 | GCUGCAAUGGUGGCCUAAUTTAUUAGGCCACCAUUGCAGCTT |
| CTSB | |
| siRNA1 | CCGGACACAAUUUCUACAATTUUGUAGAAAUUGUGUCCGGTT |
| siRNA2 | GCCCGACCAUCAAAGAGAUTTAUCUCUUUGAUGGUCGGGCTT |
| siRNA3 | GGAACUUCUGGACAAAGAATTUUCUUUGUCCAGAAGUUCCTT |
| Control | |
| siRNA | UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT |
Enzymatic activity assay
CTSL or CTSB enzymatic activity after PDCoV infection was measured with a Magic Red® Cathepsin L or a Cathepsin B Detection Kit (ImmunoChemistry Technologies, Bloomington, MN) (51). Briefly, IPI-2I cells in 12-well plates were infected with PDCoV at an m.o.i. of 1. At different times (6, 12, and 24 h), the cells were treated with Magic Red® staining solution, which could be cleaved by CTSL or CTSB. The products were observed using a fluorescence microscope equipped with an excitation filter of 550 nm and a long pass >610 nm emission filter pair. The fluorescence intensity of the red fluorescence could be detected at an optimal excitation and emission wavelength of 592 nm and 628 nm, respectively. The experiment was repeated three times.
Growth curves
IPI-2I cells were pretreated with inhibitors for 1 h at 37 °C, and infected with PDCoV at an m.o.i. of 0.01 for 1 h at 37 °C. The cells were treated with 100 μg/ml trypsin at room temperature for 20 min and washed three times with DMEM, followed by incubation at 37 °C. Supernatant samples were collected at different time points and stored at −70 °C. The TCID50 of the virus was quantified after three freeze-thaw cycles (62).
Western blot analysis of spike cleavage by trypsin
ST cells were infected with PDCoV at an m.o.i. of 1, and 24 h later, the cells were collected and lysed by sonication as described previously with some modifications (49). Cell lysates were then incubated with various concentrations of trypsin at 37 °C for 30 min and subjected to Western blot analysis. For RN mutation assay, plasmids encoding S or SRN gene were transfected to 293T cells. 48 h after transfection, the cells were collected, washed with PBS, and lysed by sonication. Trypsin cleavage was performed as described above and Western blot analysis was conducted with S polyclonal antibody.
Statistical analysis
All data are representative of at least three independent experiments. Data were analyzed using a Student's t test, and values are reported with mean ± S.D. A threshold of p < 0.05 was considered significant.
Author contributions
J. Z. and L. F. formal analysis; J. Z. writing-original draft; J. C., H. S., X. Zhang, J. L., X. Zhu, Y. L., and X. W. data curation; J. C. writing-review and editing; D. S. and Z. J. investigation; J. L. project administration; L. C. software; L. F. funding acquisition.
Acknowledgments
We thank Dr. Pinghuang Liu, Xiaojun Wang, and Yulong Gao for reviewing the manuscript.
This work was supported by the National Key Technology R&D Program of China Grant 2016YFD0500103, National Natural Science Foundation of China Grants 31602072 and 31572541, Natural Science Foundation of Heilongjiang Province of China Grant C2017079, and the State Key Laboratory of Veterinary Biotechnology Foundation Grant SKLVBP2018002. The authors declare that they have no conflicts of interest with the contents of this article.
- PDCoV
- porcine deltacoronavirus
- PEDV
- porcine epidemic diarrhea virus
- CTSL
- cathepsin L
- CTSB
- cathepsin B
- TGEV
- transmissible gastroenteritis virus
- SARS-CoV
- severe acute respiratory syndrome coronavirus
- MERS-CoV
- Middle East respiratory syndrome coronavirus
- Baf-A1
- bafilomycin A1
- TCID50
- tissue culture infectious dose 50% ml
- m.o.i.
- multiplicity of infection
- IFA
- immunofluorescence assay
- CPE
- cytopathic effect
- SBTI
- soybean trypsin inhibitor
- ST cells
- swine testis cells
- RT-qPCR
- real-time quantitative reverse transcription PCR.
References
- 1. Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., and Zhang J. (2015) Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology 482, 51–59 10.1016/j.virol.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li G., Chen Q., Harmon K. M., Yoon K. J., Schwartz K. J., Hoogland M. J., Gauger P. C., Main R. G., and Zhang J. (2014) Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2, e00278–14 10.1128/genomeA.00278-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., and Saif L. J. (2015) Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 21, 650–654 10.3201/eid2104.141859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang L., Byrum B., and Zhang Y. (2014) Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 20, 1227–1230 10.3201/eid2007.140296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., and Rovira A. (2014) Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 20, 1347–1350 10.3201/eid2008.140526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woo P. C., Lau S. K., Lam C. S., Lau C. C., Tsang A. K., Lau J. H., Bai R., Teng J. L., Tsang C. C., Wang M., Zheng B. J., Chan K. H., and Yuen K. Y. (2012) Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008 10.1128/JVI.06540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ojkic D., Hazlett M., Fairles J., Marom A., Slavic D., Maxie G., Alexandersen S., Pasick J., Alsop J., and Burlatschenko S. (2015) The first case of porcine epidemic diarrhea in Canada. Can. Vet. J. 56, 149–152 [PMC free article] [PubMed] [Google Scholar]
- 8. Lee S., and Lee C. (2014) Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14–04/2014. Genome Announc. 2, e01191–14 10.1128/genomeA.01191-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong N., Fang L., Zeng S., Sun Q., Chen H., and Xiao S. (2015) Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 21, 2254–2255 10.3201/eid2112.150283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., and Amonsin A. (2016) Porcine deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 22, 757–759 10.3201/eid2204.151852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Vui D., Stott C., Madapong A., Tripipat T., Wegner M., Intrakamhaeng M., and Chongcharoen W. (2017) Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound. Emerg. Dis. 64, 3–10 10.1111/tbed.12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belouzard S., Chu V. C., and Whittaker G. R. (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 106, 5871–5876 10.1073/pnas.0809524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bosch B. J., van der Zee R., de Haan C. A., and Rottier P. J. (2003) The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77, 8801–8811 10.1128/JVI.77.16.8801-8811.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Tai W., Du L., Zhou Y., Zhang W., and Li F. (2018) Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J. Virol. 92, e01556–17 10.1128/JVI.01556-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Haan C. A., Stadler K., Godeke G.-J., Bosch B. J., and Rottier P. J. (2004) Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 78, 6048–6054 10.1128/JVI.78.11.6048-6054.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Millet J. K., and Whittaker G. R. (2014) Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15214–15219 10.1073/pnas.1407087111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rota P. A., Oberste M. S., Monroe S. S., Nix W. A., Campagnoli R., Icenogle J. P., Penaranda S., Bankamp B., Maher K., Chen M-h., Tong S., Tamin A., Lowe L., Frace M., DeRisi J. L., et al. (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300, 1394–1399 10.1126/science.1085952 [DOI] [PubMed] [Google Scholar]
- 18. Wicht O., Li W., Willems L., Meuleman T. J., Wubbolts R. W., van Kuppeveld F. J., Rottier P. J., and Bosch B. J. (2014) Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 88, 7952–7961 10.1128/JVI.00297-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White J. M., Delos S. E., Brecher M., and Schornberg K. (2008) Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 10.1080/10409230802058320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watanabe R., Matsuyama S., Shirato K., Maejima M., Fukushi S., Morikawa S., and Taguchi F. (2008) Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J. Virol. 82, 11985–11991 10.1128/JVI.01412-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao X., Chakraborti S., Dimitrov A. S., Gramatikoff K., and Dimitrov D. S. (2003) The SARS-CoV S glycoprotein: Expression and functional characterization. Biochem. Biophys. Res. Commun. 312, 1159–1164 10.1016/j.bbrc.2003.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garwes D. J., and Pocock D. H. (1975) The polypeptide structure of transmissible gastroenteritis virus. J. Gen. Virol. 29, 25–34 10.1099/0022-1317-29-1-25 [DOI] [PubMed] [Google Scholar]
- 23. Bonavia A., Zelus B. D., Wentworth D. E., Talbot P. J., and Holmes K. V. (2003) Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77, 2530–2538 10.1128/JVI.77.4.2530-2538.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hofmann H., Simmons G., Rennekamp A. J., Chaipan C., Gramberg T., Heck E., Geier M., Wegele A., Marzi A., Bates P., and Pöhlmann S. (2006) Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80, 8639–8652 10.1128/JVI.00560-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sui J., Li W., Murakami A., Tamin A., Matthews L. J., Wong S. K., Moore M. J., Tallarico A. S. C., Olurinde M., Choe H., Anderson L. J., Bellini W. J., Farzan M., and Marasco W. A. (2004) Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 101, 2536–2541 10.1073/pnas.0307140101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K. C., Chen Y. H., Zhang L., and Wang X. (2013) Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 23, 986–993 10.1038/cr.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gallagher T. M., and Buchmeier M. J. (2001) Coronavirus spike proteins in viral entry and pathogenesis. Virology 279, 371–374 10.1006/viro.2000.0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsuyama S., and Taguchi F. (2009) Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 83, 11133–11141 10.1128/JVI.00959-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sainz B. Jr., Rausch J. M., Gallaher W. R., Garry R. F., and Wimley W. C. (2005) Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J. Virol. 79, 7195–7206 10.1128/JVI.79.11.7195-7206.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Madu I. G., Roth S. L., Belouzard S., and Whittaker G. R. (2009) Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 83, 7411–7421 10.1128/JVI.00079-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glowacka I., Bertram S., Müller M. A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T. S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., and Pöhlmann S. (2011) Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85, 4122–4134 10.1128/JVI.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phillips J. M., Gallagher T., and Weiss S. R. (2017) Neurovirulent murine coronavirus JHM.SD uses cellular zinc metalloproteases for virus entry and cell-cell fusion. J. Virol. 91, e01564–16 10.1128/JVI.01564-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hofmann M., and Wyler R. (1988) Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 26, 2235–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang B., Liu Y., Ji C.-M., Yang Y.-L., Liang Q.-Z., Zhao P., Xu L.-D., Lei X.-M., Luo W.-T., and Qin P. (2018) Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J. Virol. 92, e00318–18 10.1128/JVI.00318-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li W., Hulswit R. J. G., Kenney S. P., Widjaja I., Jung K., Alhamo M. A., van Dieren B., van Kuppeveld F. J. M., Saif L. J., and Bosch B.-J. (2018) Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. U.S.A. 115, E5135–E5143 10.1073/pnas.1802879115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsuyama S., Ujike M., Morikawa S., Tashiro M., and Taguchi F. (2005) Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U.S.A. 102, 12543–12547 10.1073/pnas.0503203102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shirato K., Kawase M., and Matsuyama S. (2013) Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 87, 12552–12561 10.1128/JVI.01890-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ujike M., Nishikawa H., Otaka A., Yamamoto N., Yamamoto N., Matsuoka M., Kodama E., Fujii N., and Taguchi F. (2008) Heptad repeat-derived peptides block protease-mediated direct entry from the cell surface of severe acute respiratory syndrome coronavirus but not entry via the endosomal pathway. J. Virol. 82, 588–592 10.1128/JVI.01697-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bosch B. J., Martina B. E., Van Der Zee R., Lepault J., Haijema B. J., Versluis C., Heck A. J., De Groot R., Osterhaus A. D., and Rottier P. J. (2004) Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 101, 8455–8460 10.1073/pnas.0400576101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao P., Wang B., Ji C. M., Cong X., Wang M., and Huang Y. W. (2018) Identification of a peptide derived from the heptad repeat 2 region of the porcine epidemic diarrhea virus (PEDV) spike glycoprotein that is capable of suppressing PEDV entry and inducing neutralizing antibodies. Antiviral Res. 150, 1–8 10.1016/j.antiviral.2017.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gierer S., Müller M. A., Heurich A., Ritz D., Springstein B. L., Karsten C. B., Schendzielorz A., Gnirß K., Drosten C., and Pöhlmann S. (2015) Inhibition of proprotein convertases abrogates processing of the middle eastern respiratory syndrome coronavirus spike protein in infected cells but does not reduce viral infectivity. J. Infect. Dis. 211, 889–897 10.1093/infdis/jiu407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamada Y., and Liu D. X. (2009) Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 83, 8744–8758 10.1128/JVI.00613-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simmons G., Gosalia D. N., Rennekamp A. J., Reeves J. D., Diamond S. L., and Bates P. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881 10.1073/pnas.0505577102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bosch B. J., Bartelink W., and Rottier P. J. (2008) Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82, 8887–8890 10.1128/JVI.00415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bergeron E., Vincent M. J., Wickham L., Hamelin J., Basak A., Nichol S. T., Chrétien M., and Seidah N. G. (2005) Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem. Biophys. Res. Commun. 326, 554–563 10.1016/j.bbrc.2004.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simmons G., Rennekamp A. J., and Bates P. (2006) Proteolysis of SARS-associated coronavirus spike glycoprotein. in The Nidoviruses (Perlman S., and Holmes K. V., eds) pp. 235–240, Springer, Boston: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simmons G., Zmora P., Gierer S., Heurich A., and Pöhlmann S. (2013) Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 100, 605–614 10.1016/j.antiviral.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang J., Liu C., Tao L., and Zhang H. (2016) Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biol. Chem. 291, 9218–9232 10.1074/jbc.M116.716100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu C., Ma Y., Yang Y., Zheng Y., Shang J., Zhou Y., Jiang S., Du L., Li J., and Li F. (2016) Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J. Biol. Chem. 291, 24779–24786 10.1074/jbc.M116.740746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang I. C., Bosch B. J., Li F., Li W., Lee K. H., Ghiran S., Vasilieva N., Dermody T. S., Harrison S. C., Dormitzer P. R., Farzan M., Rottier P. J., and Choe H. (2006) SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 281, 3198–3203 10.1074/jbc.M508381200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo C., Zhu Z., Guo Y., Wang X., Yu P., Xiao S., Chen Y., Cao Y., and Liu X. (2017) Heparanase upregulation contributes to porcine reproductive and respiratory syndrome virus release. J. Virol. 91, e00625–17 10.1128/JVI.00625-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., and Gallagher T. (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85, 873–882 10.1128/JVI.02062-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hampson D. J., and Kidder D. E. (1986) Influence of creep feeding and weaning on brush border enzyme activities in the piglet small intestine. Res. Vet. Sci. 40, 24–31 10.1016/S0034-5288(18)30481-8 [DOI] [PubMed] [Google Scholar]
- 54. Fan M. Z., Stoll B., Jiang R., and Burrin D. G. (2001) Enterocyte digestive enzyme activity along the crypt-villus and longitudinal axes in the neonatal pig small intestine. J. Anim. Sci. 79, 371–381 10.2527/2001.792371x [DOI] [PubMed] [Google Scholar]
- 55. Hu H., Jung K., Vlasova A. N., Chepngeno J., Lu Z., Wang Q., and Saif L. J. (2015) Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 53, 1537–1548 10.1128/JCM.00031-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kawai T., and Akira S. (2007) TLR signaling. in Seminars in Immunology, Vol. 19, pp. 24–32, Elsevier, Cambridge, MA: [DOI] [PubMed] [Google Scholar]
- 57. Pichlmair A., and Reis e Sousa C. (2007) Innate recognition of viruses. Immunity 27, 370–383 10.1016/j.immuni.2007.08.012 [DOI] [PubMed] [Google Scholar]
- 58. Li Y., Wu Q., Huang L., Yuan C., Wang J., and Yang Q. (2018) An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 9, 3811 10.1038/s41467-018-06056-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., and Li J. (2015) Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 6, e00064 10.1128/mBio.00064-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okuma K., Nakamura M., Nakano S., Niho Y., and Matsuura Y. (1999) Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254, 235–244 10.1006/viro.1998.9530 [DOI] [PubMed] [Google Scholar]
- 61. Fang P., Fang L., Hong Y., Liu X., Dong N., Ma P., Bi J., Wang D., and Xiao S. (2017) Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 98, 173–178 10.1099/jgv.0.000690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reed L. J., and Muench H. (1938) A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]