Abstract
The secreted metalloprotease ADAMTS9 has dual roles in extracellular matrix (ECM) turnover and biogenesis of the primary cilium during mouse embryogenesis. Its gene locus is associated with several human traits and disorders, but ADAMTS9 has few known interacting partners or confirmed substrates. Here, using a yeast two-hybrid screen for proteins interacting with its C-terminal Gon1 domain, we identified three putative ADAMTS9-binding regions in the ECM glycoprotein fibronectin. Using solid-phase binding assays and surface plasmon resonance experiments with purified proteins, we demonstrate that ADAMTS9 and fibronectin interact. ADAMTS9 constructs, including those lacking Gon1, co-localized with fibronectin fibrils formed by cultured fibroblasts lacking fibrillin-1, which co-localizes with fibronectin and binds several ADAMTSs. We observed no fibrillar ADAMTS9 staining after blockade of fibroblast fibronectin fibrillogenesis with a peptide based on the functional upstream domain of a Staphylococcus aureus adhesin. These findings indicate that ADAMTS9 binds fibronectin dimers and fibrils directly through multiple sites in both molecules. Proteolytically active ADAMTS9, but not a catalytically inactive variant, disrupted fibronectin fibril networks formed by fibroblasts in vitro, and ADAMTS9-deficient RPE1 cells assembled a robust fibronectin fibril network, unlike WT cells. Targeted LC-MS analysis of fibronectin digested by ADAMTS9-expressing cells identified a semitryptic peptide arising from cleavage at Gly2196–Leu2197. We noted that this scissile bond is in the linker between fibronectin modules III17 and I10, a region targeted also by other proteases. These findings, along with stronger fibronectin staining previously observed in Adamts9 mutant embryos, suggest that ADAMTS9 contributes to fibronectin turnover during ECM remodeling.
Keywords: extracellular matrix, fibronectin, metalloprotease, proteolysis, ADAMTS, fibril, mass spectrometry (MS), primary cilium, ADAM metallopeptidase with thrombospondin type 1 motif (ADAMTS), ECM, fibrillogenesis, proteolytic enzyme
Introduction
An extracellular matrix (ECM)3 is a complex network of proteins, glycoproteins, proteoglycans, and glycosaminoglycans that ensures tissue structural integrity. It also provides positional information and mechanical signals to cells and sequesters growth factors and cytokines (1, 2). Because nearly all signals perceived by cells are potentially modifiable by the ECM, a highly regulated ECM composition and organization are necessary for correct cell and organ function. ECM is continuously remodeled by coordinated biosynthesis and proteolysis of its components (3). Among several enzymes participating in ECM proteolysis are ADAMTS proteases, a family of 19 secreted zinc proteases with diverse and essential roles in mammalian development and homeostatic processes (reviewed in Ref. 4–6). Naturally occurring mutations affecting ADAMTS proteases cause a variety of Mendelian disorders in humans and other species (4), and ADAMTS proteolytic activity may drive the pathology of acquired disorders such as osteoarthritis and coronary artery disease (7, 8).
ADAMTS proteases comprise an N-terminal pro-protease domain upstream of a series of canonical modules constituting an ancillary domain, whose hallmark is the presence of one or several thrombospondin type 1 repeats (TSRs) (9). ADAMTS9 and its homolog ADAMTS20 each contain 15 TSRs and a unique C-terminal module named Gon1 after their Caenorhabditis elegans ortholog Gon1 (10, 11). They have high domain and sequence homology with Gon1, which is necessary for gonadal morphogenesis (10), as well as with the Drosophila melanogaster metalloprotease ADAMTS-A, which is essential for tracheal development (12). Thus, these proteases are evolutionarily conserved and likely to have significant functions in mammals. Indeed, genetically engineered mouse mutants have shown that ADAMTS9 is indispensable for several aspects of mammalian embryogenesis. Adamts9-null mouse embryos fail to undergo gastrulation, a crucial early morphogenetic event during which the definitive germ layers are formed, and die by 7 days of gestation (13). This mouse allele, and others, including a hypomorphic Adamts9 allele (Adamts9Gt), chemically induced point mutations, and conditional Adamts9 deletion have identified crucial functions in craniofacial, neural, vascular, cardiac, eye, pigment, and limb development (14–19). In adult mice, ADAMTS9 has a role in parturition via regulation of ECM control of uterine smooth muscle cell differentiation (20). ADAMTS9 and ADAMTS20 were recently found to be essential for biogenesis of the primary cilium, providing an unexpected role for these secreted proteases in formation of a cellular organelle (21, 33). Genome-wide analysis has linked the ADAMTS9 locus to diverse genetic traits, disease susceptibility markers, and common disorders of complex etiology, including type 2 diabetes mellitus, obesity, and age-related macular degeneration (22–26). ADAMTS9 was identified as a tumor-suppressor gene in gastric, nasopharyngeal, and esophageal squamous cell carcinoma, where it may act via suppression of angiogenesis (27–29). Despite this considerable biological and disease relevance, the only known ADAMTS9 substrates to date are the ECM proteoglycans aggrecan and versican (11).
Previous work has demonstrated that ADAMTS9 enters the secretory pathway, where it undergoes obligate glycosylation, binds therein to several chaperones, and arrives at the cell surface, where its propeptide is cleaved by furin (11, 13, 14, 30–32). Recent work shows that furin-processed, catalytically active ADAMTS9 is endocytosed and trafficked to Rab11 vesicles that form a girdle around the basal body of the primary cilium (33). There, through an as-yet unknown mechanism, ADAMTS9 mediates ciliary growth. Here, adding to the growing understanding of this important protease, we report identification of the ECM and circulating glycoprotein fibronectin as a novel ADAMTS9-binding partner and show that its assembly and abundance are reduced in the presence of ADAMTS9. These findings are consistent with stronger fibronectin staining previously observed in mice heterozygous for Adamts9 and mouse embryos homozygous for the loss-of-function Adamts9Gt mutation (14, 19, 33).
Results
Fibronectin is a novel binding partner of the ADAMTS9 Gon1 domain
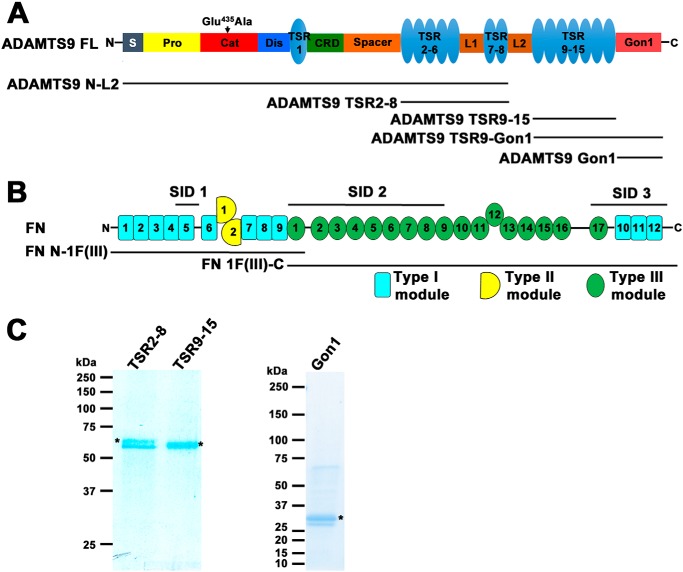
A yeast two-hybrid screen of a human placenta library of Gal4 fusion proteins was undertaken using the human ADAMTS9 Gon1 module (residues 1734–1935, SwissProt accession no. Q9P2N4) (Fig. 1A) as the “bait,” fused in-frame and downstream of the Gal4 DNA-binding domain. In addition to several other secreted or cell-surface proteins, which could be putative binding partners, eight independent ADAMTS9 Gon1-interacting cDNA clones, each encoding part of fibronectin (Fig. 1B), were identified in-frame with the Gal4 DNA-activating domain. The selected interacting domains (SIDs), of fibronectin, which are minimum interacting regions determined by the overlap between these clones (34), are shown in Fig. 1B. The three SIDs, SID1, SID2, and SID3, spanned three nonoverlapping regions of fibronectin comprising both type I and type III repeats (Fig. 1B).
Figure 1.
ADAMTS9 constructs and interaction regions of fibronectin. A, ADAMTS9 domain structure with the various ADAMTS9 constructs indicated. B, domain structure of fibronectin indicating the select interacting domains (SID1, SID2, and SID3) identified in the yeast two-hybrid screen and recombinant FN constructs used in binding assays. C, purified recombinant ADAMTS9 constructs (indicated by asterisks) were analyzed by reducing SDS-PAGE and Coomassie Blue staining. Molecular mass markers (in kDa) are shown on the left.
Yeast two-hybrid interactions occur in a reducing environment between proteins that are synthesized without post-translational modifications typical for secreted mammalian proteins. Such modifications include introduction of disulfide bonds and N-glycosylation. Two disulfide bonds stabilize fibronectin type I modules (35). The human ADAMTS9 Gon1 module contains 10 cysteines (11), which are conserved in other Gon1 modules (http://pfam.xfam.org/family/PF08685)4 and very likely form disulfides, and two sites of potential N-glycosylation. Therefore, experiments were performed in follow-up of the yeast two-hybrid results asking whether the ADAMTS9–fibronectin interactions could be replicated with proteins that had been processed and undergone modification in a eukaryotic secretion pathway. Because we were unable to purify full-length ADAMTS9, myc-His6-tagged ADAMTS9 Gon1 module and longer C-terminal ADAMTS9 constructs with exogenous signal peptides were expressed and purified from the medium of mammalian cells (Fig. 1, A and C). Recombinant ADAMTS9 Gon1 bound to plasma fibronectin, a disulfide-linked dimer of 220-kDa subunits in a solid-phase binding assay (Fig. 2A), as well as to N- and C-terminally truncated fibronectin constructs N-1F(III) and 1F(III)-C, respectively (Fig. 2B). These overlapping constructs were expressed as secreted proteins using recombinant baculoviruses to ensure proper disulfide bonds and represent approximately the N-terminal one-third and the C-terminal two-thirds of fibronectin, respectively (Fig. 1B).
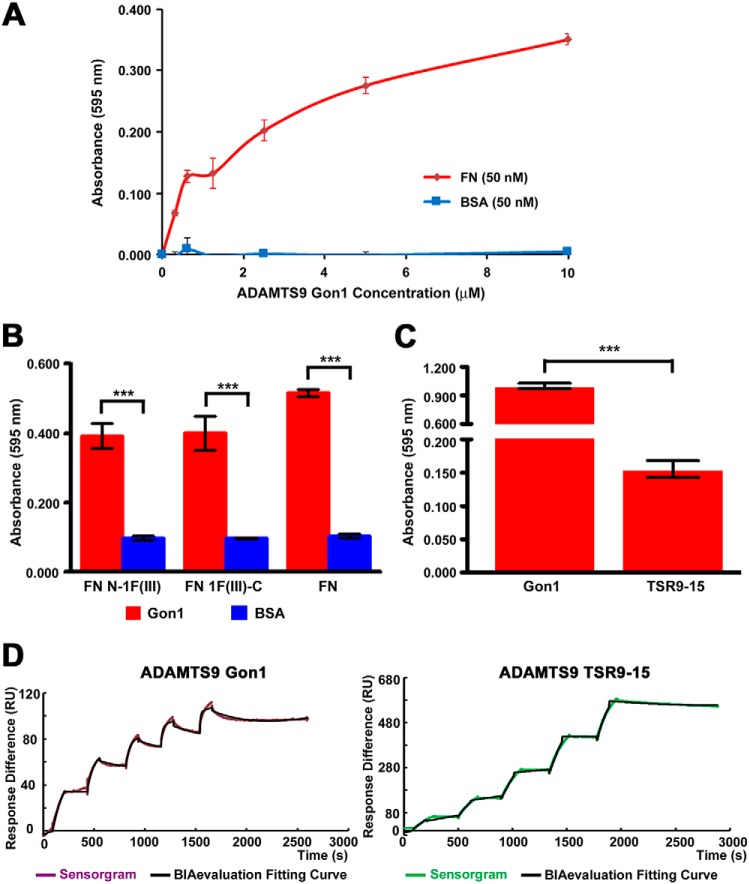
Figure 2.
Fibronectin binding to ADAMTS9 constructs. A, solid-phase binding assay of ADAMTS9 Gon1 module to immobilized fibronectin (FN, red line). BSA (blue line) was used as a control for binding. B, ADAMTS9 Gon1 module was tested for binding to N- and C-terminally truncated fibronectin constructs N-1F(III) and 1F(III)-C, respectively. Binding to these constructs was compared with binding to BSA (negative control). ***, p < 0.0001, n = 4. C, solid-phase binding assay comparing ADAMTS9 Gon1 and ADAMTS9 TSR9–15 binding to fibronectin. ***, p < 0.0001, n = 4. D, surface plasmon resonance analysis of plasma fibronectin (analyte, 16.9–270 mm) binding to ADAMTS9 Gon1 module immobilized on a CM5 chip. The binding KD was 2.8 nm, χ2/Rmax = 4% for ADAMTS9 Gon1, and 104 nm (χ2/Rmax = 6%) for ADAMTS9 TSR9–15. The purple and green lines represent the sensorgram, and the black line is the fitted curve produced by BIAevaluation software.
In solid-phase binding assays, ADAMTS9 Gon1 showed stronger binding to fibronectin than ADAMTS9 TSR9–15 (Fig. 2C), whereas ADAMTS9 TSR2–8 did not bind (data not shown). Purified ADAMTS9 TSR9-Gon1 construct containing both binding regions was unstable and therefore could not be used in the binding assays. ADAMTS9 Gon1 and ADAMTS9 TSR9–15 binding to fibronectin was validated and compared using surface plasmon resonance (Fig. 2D). These constructs bound fibronectin with an estimated KD of 2.8 nm (χ2/Rmax = 4%) and 104 nm (χ2/Rmax = 6%), respectively. Thus, binding of the Gon1 module to different parts of fibronectin as identified in the yeast-two-hybrid screen was replicated using proteins purified after secretion from eukaryotic cells, and the TSR9–15 region of ADAMTS9 was also found to contain one or more binding sites for fibronectin.
ADAMTS9 constructs bind fibronectin fibrils
Many cell types that secrete fibronectin or are provided exogenous fibronectin assemble it into fibrils, starting within a few hours of attachment if plated at sufficiently high density (36). Fibronectin fibrils provide a template for assembly of fibrillin-1 microfibrils, which appear soon after initial fibronectin assembly and co-localize extensively with them (37, 38). Furthermore fibrillin-1, the major fibrillin produced by most cells, is a binding partner for several ADAMTS proteins (39). Therefore, to eliminate fibrillin-1 as a potential confounder in the analysis, we used Fbn1−/− mouse embryo fibroblasts. In the short-term cultures used for these experiments, Fbn1−/− mouse embryo fibroblasts do not assemble fibrillin-2, an ADAMTS ligand and substrate (40–42).
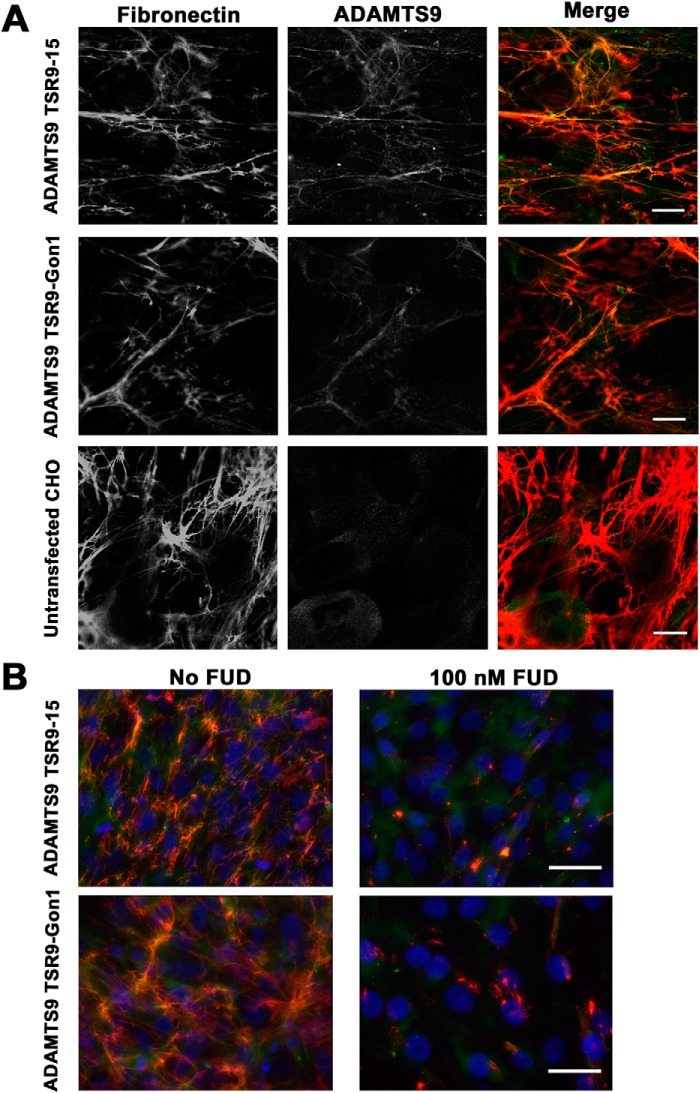
To test whether the ADAMTS9 Gon1 module bound fibronectin fibrils, Fbn1−/− fibroblast monolayer cultures were co-stained with anti-myc (for the exogenous myc-His6-tagged Gon1 domain) and an anti-fibronectin antibody. Despite interacting with full-length fibronectin dimer and fragments of fibronectin monomer (Fig. 2, A, C, and D), recombinant ADAMTS9 Gon1 module did not co-localize with fibronectin fibrils (data not shown). ADAMTS9 constructs containing upstream regions of the ancillary domain were subsequently tested by adding conditioned medium containing them to the fibroblast cultures. ADAMTS9 TSR9-Gon1 bound to fibronectin fibrils, indicated by co-staining of fibrillar structures by anti-myc and fibronectin antibodies (Fig. 3A). We similarly tested binding of an ADAMTS9 TSR9–15 construct to ask whether the TSRs could have a role in fibronectin fibril binding independent of the Gon1 module. This construct co-localized with fibrils in a comparable manner as ADAMTS9 TSR9-Gon1 (Fig. 3A). Because these co-staining experiments were done after addition of exogenous ADAMTS9, i.e. via conditioned medium, they suggest that the ADAMTS9 constructs were incorporated into forming fibrils or bound to the surface of fibronectin fibrils. The experimental approach thus indicated that it was not necessary for ADAMTS9 and fibronectin to undergo initial association in the secretory pathway. Therefore, fibronectin fibrils assembled by one cell or previously deposited in the ECM could bind to ADAMTS9 produced by another cell. Specificity of the ADAMTS9 binding for fibronectin fibrils was demonstrated by the absence of fibrillar myc staining in cultures treated with 100 nm functional upstream domain (FUD) (43) which specifically prevents fibronectin fibril assembly (Fig. 3B). Elimination of the possibility of fibrillin assembly by these MEFs provided additional evidence that ADAMTS9 constructs bound specifically to fibronectin fibrils and not to fibrillin-1 and -2 microfibrils.
Figure 3.
Co-localization of ADAMTS9 ancillary domain constructs with fibronectin fibrils. A, combined immunofluorescence using anti-myc for detection of ADAMTS9 constructs TSR9-Gon1 and TSR9–15 (see Fig. 1A) shows their co-localization with fibronectin fibrils after addition of conditioned medium containing the ADAMTS9 constructs to Fbn1−/− MEFs. In the single fluorescent images, both are shown in gray scale; in the merged image, anti-myc staining is green, and fibronectin is red. B, 100 nm FUD, which abolishes formation of fibronectin fibrils, eliminated anti-myc fibrillar staining in Fbn1−/− mouse embryo fibroblast cultures. The scale bars indicate 10 μm in A and 25 μm in B.
Fibronectin fibrils are fragmented in the presence of catalytically active ADAMTS9
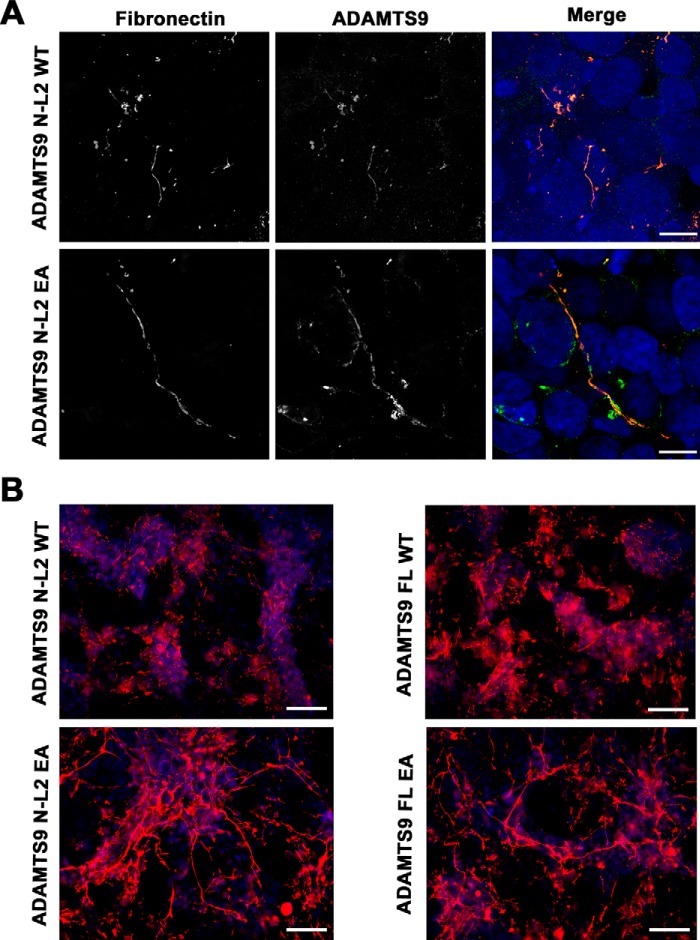
Fibronectin binding by the C-terminal ADAMTS9 constructs suggested that it could be an ADAMTS9 substrate. We therefore investigated the ability of full-length ADAMTS9 and the N-terminal construct ADAMTS9 N-L2, each containing the protease domain, to bind fibronectin fibrils and specifically, to modify fibronectin dimers or fibronectin fibrils proteolytically. Full-length ADAMTS9 is a large and unstable enzyme (>200 kDa), which is expressed at low levels by transfected mammalian cells, and no high-production natural sources are known. Despite multiple attempts, we have been unable to obtain substantially pure recombinant full-length ADAMTS9 or ADAMTS9 N-L2. As an alternative approach, we co-cultured mouse NIH-3T3 cells with HEK293T cells expressing ADAMTS9 or an inactive mutant construct in which the catalytic residue, Glu435 was replaced by Ala. Similar experiments were conducted using the ADAMTS9 N-L2 construct, which excludes the TSR9-Gon1 region (Fig. 1A) and its corresponding inactive mutant (ADAMTS9 N-L2 E435A) and demonstrated that they co-localized with fibronectin fibrils (Fig. 4A).
Figure 4.
Constructs containing the ADAMTS9 metalloproteinase domain co-localize with fibronectin and active ADAMTS9 attenuates fibronectin fibril networks. A, combined immunofluorescence using anti-myc for detection of the constructs ADAMTS9N-L2 and ADAMTS9N-L2 EA (green) expressed by HEK293T cells co-cultured with NIH-3T3 cells shows co-localization with fibronectin fibrils (red). In the single fluorescent images, both are shown in gray scale; in the merged image, ADAMTS9/anti-myc staining is green, and fibronectin is red. B, an interrupted and attenuated fibronectin fibril network (red) was seen in the presence of catalytically active ADAMTS9 constructs ADAMTS9 full-length (FL) WT and ADAMTS9 N-L2 WT compared with the corresponding ADAMTS9 E435A (EA) mutants. The scale bars indicate 10 μm in A and 25 μm in B.
The fibronectin networks showed considerable discontinuity or fragmentation in the presence of catalytically active ADAMTS9 constructs compared with the inactive mutants (Fig. 4, A and B). These observations suggested impaired fibronectin assembly or fibronectin fibril degradation in the presence of catalytically active ADAMTS9. We also studied the human retinal pigment epithelium-derived cell line hTERT RPE-1, which expresses endogenous ADAMTS9, and an RPE-1 clone, D12, in which ADAMTS9 was mutated using CRISPR-Cas9 (33). Fibronectin fibril networks formed by these two lines were compared after 48-h culture in the presence of serum or 24- or 48-h culture subsequent to this period without serum. Under both conditions, a more robust fibril network was observed in D12 cultures (Fig. 5, A and B). Quantitative reverse transcriptase (qRT)–PCR showed no significant difference in FN1 mRNA levels between the two lines (Fig. 5C), strongly suggesting that ADAMTS9 either hindered the assembly of or proteolytically eliminated fibronectin fibrils in hTERT RPE1 cells.
Figure 5.
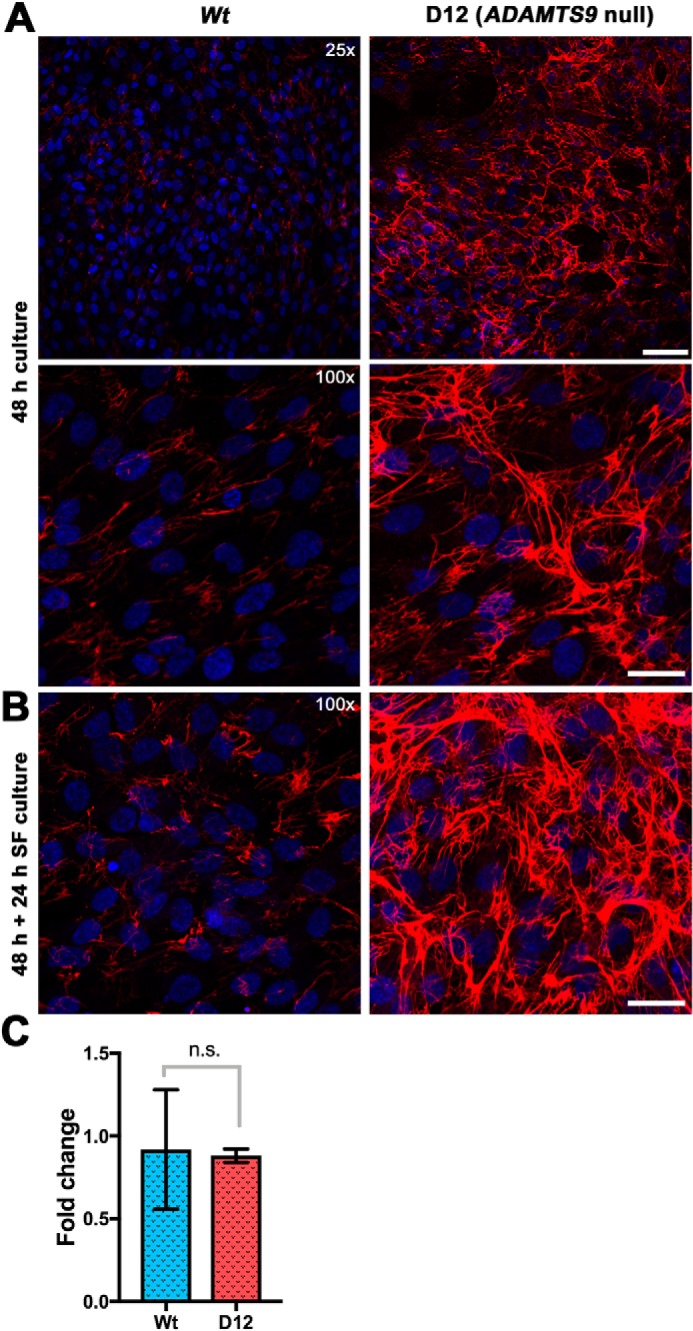
ADAMTS9-deficient RPE-1 cells assemble a more robust fibronectin matrix than WT RPE-1 cells. A, WT RPE-1 cells and RPE-1 D12 cells in which ADAMTS9 was inactivated by CRISPR-Cas9 were cultured for 48 h in the presence of serum. Compared with the WT cells, abundant fibronectin matrix was assembled by the mutant cells. B, The respective cells were cultured for 48 h, followed by 24-h culture in serum-free conditions, with similar observations as in A. C, qRT-PCR for FN1 mRNA showed no significant difference between WT RPE-1 and D12 cells. The scale bars indicate 50 μm in the upper panel of A and 25 μm in the lower panel of A and in B.
ADAMTS9 activity leads to fibronectin cleavage at a specific site
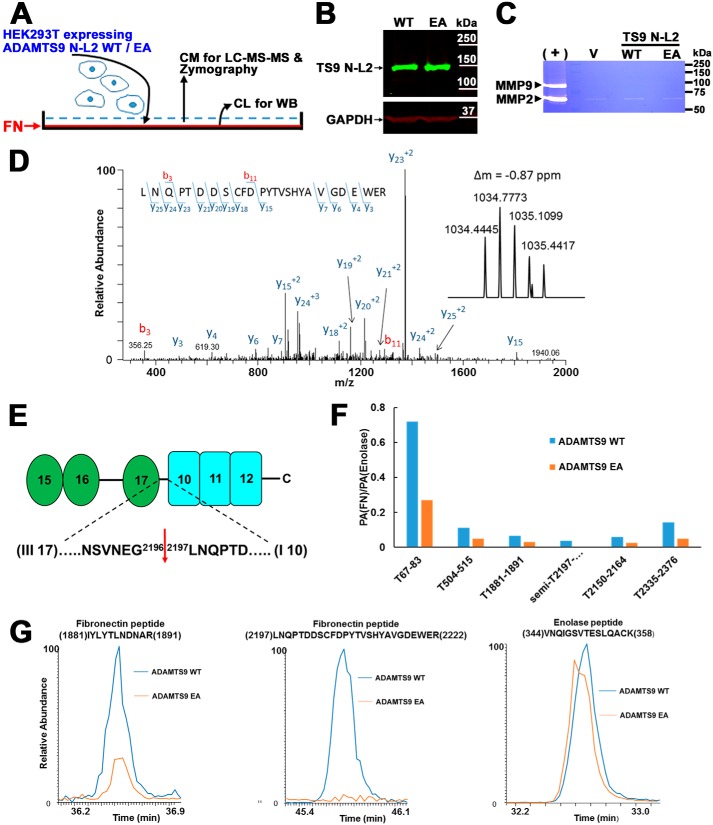
To identify fibronectin cleavages generated in the presence of ADAMTS9, we coated tissue culture wells with human fibronectin on which were cultured ADAMTS9 N-L2- or ADAMTS9 N-L2 EA-expressing HEK293T cells (Fig. 6A). The rationale for this experimental design was that surface-attached and fibrillar fibronectin may have an extended conformation, which would expose cryptic cleavage sites, in contrast to the globular conformation of fibronectin in solution (35, 36). HEK293T cells do not assemble fibronectin fibrils; hence the experiment tested proteolysis of exogenous fibronectin adherent to the plates. The ADAMTS9 N-L2 and ADAMTS9 N-L2 EA constructs were expressed at comparable levels by HEK293T cells (Fig. 6B). Gelatin zymography indicated comparable levels of MMP2 in the medium of the respective HEK293T cells, whereas MMP9 was not detected (Fig. 6C). Conditioned medium from these experiments was obtained after 24 h of culture for targeted analysis of human fibronectin by LC–electrospray ionization–MS/MS. Fibronectin was identified by 44 and 23 tryptic peptides covering 26 and 15% of the fibronectin sequence from the medium of cultures expressing WT and inactive ADAMTS9 N-L2, respectively. The MS1 spectrum identified a unique peptide ion of 1034 Da in digests of fibronectin with active ADAMTS9 N-L2 but not ADAMTS9 N-L2 EA (Fig. 6D, inset). The sequence of this peptide was determined in the MS2 spectrum by assignment of fragment ions (Fig. 6D). The C-terminal Arg residue of this peptide was consistent with tryptic cleavage, but its N terminus was preceded by Gly in full-length fibronectin (Fig. 6E); hence the peptide did not arise by tryptic digestion during sample preparation and represents a “semitryptic peptide” indicative of fibronectin proteolysis at the Gly2196–Leu2197 peptide bond (sequence enumeration after fibronectin isoform with UniProt identifier P02751; Fig. 6E). This peptide bond lies within a short linker connecting fibronectin III17 and I10 modules that may comprise an unstructured region susceptible to proteolytic processing (Fig. 6E). The semitryptic peptide was not observed in analysis of fibronectin exposed to catalytically inactive ADAMTS9 N-L2. We undertook another targeted LC–electrospray ionization–MS/MS analysis, i.e. selective reaction monitoring, for the semitryptic peptide and other fibronectin ions and confirmed its presence exclusively in digests using the active ADAMTS9 N-L2 (Fig. 6F). Indeed, relative quantification of fibronectin peptides detected by LC-MS/MS of the medium of cells expressing WT or inactive ADAMTS9 N-L2 demonstrated that several tryptic fibronectin peptides were more abundant in the medium of cells expressing WT ADAMTS9 N-L2 than ADAMTS9 N-L2 EA (Fig. 6F). The relative abundance of fibronectin was determined in relation to enolase, detected at comparable levels in the samples, which was thus used as a normalization factor (examples of extracted ion chromatograms are shown in Fig. 6G). Thus, in addition to identification of a unique semitryptic peptide, the greater fibronectin sequence coverage and quantitative fibronectin peptide excess upon digestion of adherent fibronectin with active ADAMTS9 N-L2 suggested an overall greater abundance of fibronectin released in these experiments, also supporting fibronectin proteolysis by ADAMTS9. In the absence of an antibody suitable for recognition of the C terminus of fibronectin, we have not been able to obtain additional evidence of cleavage by a Western blotting.
Figure 6.
Fibronectin (FN) is cleaved in the presence of catalytically active ADAMTS9. A, schematic of experimental strategy. CM and CL are conditioned medium and cell lysate, respectively. B, Western blotting with anti-myc mAb of lysates of HEK293T cells expressing catalytically active ADAMTS9 N-L2 or the corresponding inactive mutant showing comparable expression. C, gelatin zymogram showing robust detection of MMP2 and MMP9 in conditioned medium from NIH-3T3 skin fibroblasts (positive control, +) and an evident MMP2 band, but no MMP9 band in the conditioned medium of HEK293T cells expressing ADAMTS9 constructs as indicated. V indicates medium from empty vector-transfected cells. D, MS/MS fragmentation spectrum of a 1034.445-Da semitryptic peptide generated by incubation of fibronectin with cells expressing catalytically active ADAMTS9. The MS/MS spectrum for this peptide is consistent with the semitryptic fibronectin peptide 2197LNQPTDDSCFDPYTVSHYAVGDEWER2222 (using UniProt sequence ID P02751). The parent ion mass distribution obtained by MS is shown in the inset. E, domain and sequence cartoon of FN showing the cleavage site location with flanking sequence and indicating the flanking domains. F, quantitative analysis of fibronectin peptides obtained by analysis of conditioned medium of cells expressing active or catalytically inactive ADAMTS9 grown on a fibronectin substrate. The relative abundance of fibronectin in the medium obtained from cells expressing active or catalytically inactive ADAMTS9 was determined by performing a targeted SRM analysis on six fibronectin peptides and two enolase peptides. Enolase was used as a normalization factor as the amount of this protein appears to be similar in the active and inactive ADAMTS9 samples. These SRM experiments were used to determine the peak area (PA) ratios; PA(FN)/PA(enolase), for six fibronectin peptides. T indicates tryptic peptides, and semi indicates the semitryptic peptide indicative of fibronectin cleavage. The relative abundance of the fibronectin peptides is lower in the mutant sample. G, examples of extracted ion chromatograms for a tryptic fibronectin peptide 1881IYLYTLNDNAR1891, the semitryptic peptide 2197LNQPTDDSCFDPYTVSHYAVGDEWER2222, and the one enolase peptide 344VNQIGSVTESLQACK358. These chromatograms are consistent with similar abundance of enolase in these samples, lower abundance of the 1881IYLYTLNDNAR1891 peptide and the absence of the semitryptic peptide, 2197LNQPTDDSCFDPYTVSHYAVGDEWER2222, in the inactive ADAMTS9 sample compared with active ADAMTS9.
Discussion
Observations from complementary approaches, including the yeast two-hybrid assay, solid-phase binding and surface plasmon resonance using purified proteins collectively support direct interaction of ADAMTS9 with fibronectin. ADAMTS9 localized specifically to fibronectin fibrils, because all ADAMTS9 fibrillar ECM staining was abolished by FUD, which prevents fibronectin polymerization, and these cells could not assemble fibrillin-1, a known ADAMTS ligand. ADAMTS9 produced by HEK293 cells localizes to their cell surface and is released into medium (11), indicative of binding in trans to fibronectin fibrils formed in the co-cultures and demonstrating that the interaction can occur extracellularly. Thus, fibronectin and ADAMTS9 could be co-assembled during fibril assembly, or ADAMTS9 may interact with preformed fibronectin fibrils. Distinguishing between these possibilities is challenging, because fibronectin assembly in cultured cells is a continuous, rapid, and extremely dynamic process involving binding of integrins to the secreted fibronectin dimer, extension of the bound fibronectin by cell movements, and polymerization via exposed binding sites on fibronectin dimers (36). Although the Gon1 module bound to purified fibronectin dimer in solid-phase assays, it did not co-localize with fibronectin fibrils. In contrast, upstream nonoverlapping ADAMTS9 constructs such as TSR9–15 and ADAMTS9 N-L2, both lacking the Gon1 module, consistently bound to fibrils. Taken together with three nonoverlapping fibronectin SIDs identified in the yeast two-hybrid screen, the findings suggest multiple points of contact between the two molecules and distinct modes of interaction of different ADAMTS9 regions with fibronectin dimer or fibrils.
The observed binding of noncatalytic regions of ADAMTS9, i.e. Gon1 and TSR9–15 constructs, logically led to consideration of fibronectin as a potential ADAMTS9 substrate. A semitryptic peptide identified by MS was determined to be specific for immobilized fibronectin digested by active ADAMTS9, because it was not generated in the presence of the catalytically inactive mutant. Detection of higher levels of fibronectin tryptic peptides overall in the presence of active ADAMTS9 suggests additional ADAMTS9 cleavage sites that liberate fibronectin fragments or that cleavage at the Gly2196–Leu2197 bond may promote fibronectin instability and/or depolymerization. One caveat of this study is that it has not been possible to purify ADAMTS9; hence we cannot exclude the possibility that ADAMTS9 is not the terminal protease that degrades fibronectin but instead activates another fibronectinase. Fibronectin was previously identified as a substrate of several proteases, including the metalloproteases ADAM12 (44), MMP2, MMP8, and MMP14 (45, 46), which may cleave it at the Ala1078–Thr1079 bond (47). The ADAMTS9 processing site identified here, Gly2196–Leu2197, was previously identified as a cleavage site for MMP2 and MMP8 (48), but no change in MMP-2 was seen on gelatin zymography in our experimental system. Furthermore, direct binding of ADAMTS9 to both the fibronectin dimer and fibronectin fibrils suggests that it likely cleaves fibronectin directly. The data presented here using either the addition of ADAMTS9 conditioned medium or knockout of endogenous ADAMTS9 each show that fewer fibrils are present in the presence of catalytically active ADAMTS9, supporting its role in turnover of fibronectin fibrils. The observed ADAMTS9 incorporation in trans from conditioned medium or during co-culture experiments suggests that ADAMTS9 is integrated into fibrils as they form, potentially providing an innate mechanism for their physiological turnover, i.e. a “Trojan horse” effect. The cell-surface localization of ADAMTS9 (11, 31, 33) suggests it may access fibronectin fibrils quite early in the fibril assembly process.
Previously, endocytosis of MMP-cleaved fibronectin bound to integrins was implicated as a major degradation pathway operational in cultured cells (46). Like ADAMTS9, MMP14 also attenuates fibronectin matrix assembled by cultured cells. Among ADAMTS proteases, it was recently shown that ADAMTS2, ADAMTS3, and ADAMTS14, which are evolutionarily distinct from ADAMTS9, cleaved the fibronectin Ala292–Val293 bond within a linker between domains I5 and I6 to release an N-terminal 30-kDa fragment that is crucial for fibronectin assembly (49). In addition, this region contains an ADAMTS16 cleavage site just upstream of the ADAMTS2/3/14 cleavage site, i.e. at the Ser285–Phe286 peptide (50). Putative ADAMTS7 cleavage sites, Thr279–Ser280 and Ser281–Gly282 in the linker between I5 and I6, were also recently identified (51). Also like ADAMTS9, ADAMTS16 processing at this site led to reduced fibronectin fibril assembly (50), but the effect of ADAMTS2, ADAMTS3, ADAMTS7, and ADAMTS14 on fibronectin assembly and turnover is yet to be investigated. Like ADAMTS9 processing in the III17-I10 linker, Thr279–Ser280, Ser281–Gly282, Ser285–Phe286, and Ala292–Val293 are present within a linker that may present an exposed, unstructured region susceptible to proteolysis.
Fibronectin is present in the circulation and in many matrices where it is a critical cell adhesion substrate. ADAMTS9 is constitutively produced by vascular endothelium, suggesting a potential physiological role in fibronectin turnover during wound healing or a potential mechanism of its potent anti-angiogenic effect (28). Fibronectin is a foundational matrix molecule that promotes assembly of fibrillin microfibrils, elastic fibers, collagen fibrils, and latent TGF-β binding proteins (36) and is an essential component of embryonic extracellular matrix (52). Thus, fibronectin assembly and turnover have considerable physiological relevance to embryogenesis and adult processes. The present work, together with a recent publication (50), emphasizes that fibronectin proteolysis and not only its transcriptional and assembly mechanisms are crucial for controlling its levels in ECM. Most work on fibronectin proteolysis has utilized cultured cells, and thus few relevant fibronectin-degrading proteases operating at the tissue level are known. ADAMTS9 and MMP14 are among the few for which combined biochemical, in vitro, and in vivo evidence as fibronectinases is available. ADAMTS9 appears to be essential for embryonic fibronectin turnover as evidenced by stronger fibronectin staining in several tissues of Adamts9Gt/Gt embryos. In contrast to Adamts9-null embryos, Adamts9Gt/Gt embryos survive past gastrulation to undertake organogenesis, during which fibronectin-rich ECMs are produced. In previous analysis, Adamts9Gt/Gt umbilical cord was noted to have stronger fibronectin staining than WT littermate umbilical cord, and immunostaining of Adamts9Del/+ eyes demonstrated stronger fibronectin staining (14, 19). More intense fibronectin staining was also seen in the pregnant mouse uterus upon conditional Adamts9 deletion in smooth muscle cells (20). Taken together with these in situ observations, the present data suggest that fibronectin turnover is a widespread morphogenetic role of ADAMTS9.
In contrast, Mmp14 appears not to have as crucial a role in fibronectin turnover during early development, because it is most highly expressed during skeletal development (53), and Mmp14-deficient mice survive to birth without dramatic morphogenetic defects (54, 55). However, Mmp14 appears to be necessary for fibronectin turnover during formation of tendons (47), a tissue where it is highly expressed (53), but Adamts9 is not (56). Thus, diverse proteases may regulate fibronectin content of matrix in specific spatial and temporal contexts, with this abundant, large dimeric molecule and its supramolecular structures being targeted by a plethora of proteases at different sites.
Experimental procedures
Yeast two-hybrid screen
A two-hybrid screen of the human placenta_RP5 library (Hybrigenics, Cambridge, MA) was undertaken using the human ADAMTS9 Gon1 module (residues 1734–1935) fused in-frame and C-terminal of a Gal4 DNA-binding domain in the pB66 vector. The hybrid protein was determined to be neither auto-activating nor toxic in yeast; hence screening was done without inclusion of 3-amino-1,2,4-triazole. 81.4 × 106 yeast clones were screened. A proprietary Predicted Biological Score (34) was computed by Hybrigenics to assess interaction reliability of isolated fibronectin clones and suggested that the probability of the interaction being nonspecific was low. Selected interaction domains representing the minimal region corresponding to all the isolated clones were identified using PIMRiderR (Hybrigenics).
ADAMTS9 expression plasmids and protein purification
The human ADAMTS9 TSR9–15 expression plasmid was previously described (14). Human ADAMTS9 TSR2–8 (residues Phe879–Thr1299, primers 5′-AAGGCGCGCCTTTTACTGGAACAGTCATGGGCCAT-3′, AscI site underlined and 5′-AACTCGAGGGGTCCTTTGAGGGCATGGTGACA-3′, XhoI site underlined) and Gon1 expression plasmids (residues Glu1734-Leu1935 and primers 5′-ACTGGAATTCTTACCCCAGAATTGCAAGGAGG-3′ (EcoRI underlined) and 5′-CAGTCTCGAGATAAAACTCGCACCTCCAGGCCAGTACCAGAGGG-3′ (XhoI underlined) were generated by PCR amplification, sequence-verified in entirety, and cloned into pSecTag2A and pSecTag2C, respectively (Thermo Fisher Scientific) for in-frame expression with an N-terminal signal peptide and C-terminal myc-His6 tag in mammalian cells. Plasmids were transfected into CHO cells (ATCC, Manassas, VA), and stably expressing clones were isolated as previously described. hADAMTS9 TSR2–8, TSR9–15, and Gon1 were purified from the conditioned medium using Ni2+-Sepharose affinity chromatography (Invitrogen) essentially as previously described (14). The purified proteins were analyzed by Coomassie Blue stain (Thermo, Rockford, IL) and Western blotting using anti-myc mouse mAb (clone 9E10, Lerner Research Institute Hybridoma Core). Plasma fibronectin was purified as previously described (57). Purification of fibronectin constructs N-1F(III) and 1F(III)-C was previously described (58). The 1F(III)-C construct lacks alternatively spliced 8F(III) and 13F(III) modules found in tissue fibronectin and absent in plasma fibronectin.
Surface plasmon resonance analysis
Purified ADAMTS9 Gon1 or ADAMTS9 TSR9–15 in 10 mm acetate, pH 4.0 were immobilized on a BIAcore CM5 sensor chip (research grade) with the amine coupling kit following the manufacturer's instructions (GE Healthcare). 900 and 1600 resonance units of ADAMTS9 Gon1 or ADAMTS9 TSR9–15, respectively, were coupled to the chip for analysis in a BIAcore 3000 instrument (GE Healthcare). The single cycle kinetic analysis was performed at 25 °C in 10 mm Hepes buffer, pH 7.4 with 0.15 m NaCl, 1 mm CaCl2, and 0.005% (v/v) surfactant P20 at a flow rate of 30 μl/min. Human plasma fibronectin (57) was diluted in the above buffer at different concentrations and injected through an uncoupled control flow cell in series with a flow cell coupled to ADAMTS9 constructs. The sample injection time was 2 min and was followed by a 1-min pause before the next injection. The dissociation time was 15 min. 50 mm glycine, pH 3.0 was used for chip surface regeneration at a flow rate of 100 μl/min for 30–60 s after Gon1-binding analysis, whereas 50 mm NaOH was required for regeneration of the ADAMTS9 TSR9–15 chip surface. All data were corrected with reference to the background binding in the control flow cell. The kinetic constants were calculated assuming a 1:1 (Langmuir) binding model using the BIAevaluation software (version 4.0.1; GE Healthcare).
Solid-phase binding assay
96-well plates were coated with either fibronectin or BSA (control) at 50 nm/well. After washing and blocking the wells with 2.5% milk, 20 mm Tris-HCl buffer, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 0.05% Tween 20 (TBST Ca2+), 50 μl of purified ADAMTS9 Gon1 in 2.5% milk/TBST Ca2+ at a concentration between 0.3 and 10 μm was added to the coated wells and incubated on a shaker (300 Hz) for 2 h at room temperature. Anti-myc mouse mAb was used to detect bound Gon1. For fibronectin subconstruct binding experiments, 10 μg/ml fibronectin solution was used per well for coating, and 2.5 μm ADAMTS9 Gon1 was used as ligand. To compare binding of different ADAMTS9 constructs, 10 mg/ml of fibronectin was used for coating the wells, and 80 nm of purified ADAMTS9 Gon1, TSR9–15, or TSR2–8 constructs were added to the coated wells. Absorbance was measured at 492 nm.
Cell culture, immunofluorescence, and qRT-PCR
Fbn1−/− mouse embryo fibroblasts were made from Fbn1−/− mice (Fbn1mgN) (59) essentially as previously described (60). hTERT RPE-1 cells were purchased from ATCC (catalog no. CRL-4000). D12, an RPE-1 clone in which ADAMTS9 was mutated using CRISPR-Cas9, was previously described (33). 5 × 104 Fbn1−/− MEFs were seeded on glass coverslips placed in individual wells of a 24-well plate for overnight culture. The medium was replaced with conditioned medium from CHO cells stably expressing ADAMTS9 constructs or untransfected CHO cells, and cells were maintained for a further 48 h. For co-culture experiments of NIH-3T3 cells (ATCC) with transfected HEK293T cells, a ratio of 3:5 was used. The cells were fixed in 4% paraformaldehyde (v/v) containing 4% (w/v) sucrose, and quenched with 1% (w/v) glycine. Anti-fibronectin polyclonal antibody (AB2033, Millipore) and anti-myc mouse mAb 9E10 (LRI Hybridoma Core, Cleveland, OH) were used to detect fibronectin and ADAMTS9, respectively. hTERT RPE-1 and D12 cells were plated at 60% confluence, reaching confluence after 48 h, and were subsequently stained for fibronectin as described above. Imaging was performed on a Leica TCS5 SP11 laser confocal microscope (Figs. 3A and 4A) or a Leica DM RX A2 fluorescence microscope (Fig. 3B and 4B). qRT-PCR was used to compare FN1 mRNA levels in RPE-1 and D12 cells using primers 5′-GTAAACCTGAAGCTGAAGAGAC-3′ and 5′-TCACCAATCTTGTAGGACTG-3′ essentially as previously described (33).
Proteomics determination of fibronectin processing by ADAMTS9
2 × 106 HEK293T cells transiently transfected with ADAMTS9 N-L2 plasmids were washed and suspended in serum-free DMEM and seeded onto wells coated with fibronectin (5 μg/well) in 6-well plates. The conditioned medium was collected 24 h later, and proteins were precipitated using TCA, dried in a SpeedVac, and reconstituted with 50 μl of Tris-HCl buffer containing 6 m urea. The samples were reduced with DTT, alkylated with iodoacetamide, and further diluted in 100 μl of 50 mm ammonium bicarbonate, and trypsin was added at a 1:20 (trypsin:protein) ratio. A first round of digestion was carried out overnight, a second aliquot of trypsin was added, and the sample was incubated for an additional 6 h. The protein digest was desalted using the C18 SPE method and reconstituted in 50 μl of 1% acetic acid for LC-MS analysis. The LC-MS system was a Finnigan LTQ-Orbitrap Elite hybrid mass spectrometer system. The HPLC column was a Dionex 15-cm × 75-μm inner diameter Acclaim Pepmap C18, 2 μm, 100 Å reversed phase capillary chromatography column. 5 μl volumes of the tryptic digest were injected. The peptides were eluted from the column by an acetonitrile, 0.1% formic acid gradient at a flow rate of 0.3 μl/min for introduction into the source of the mass spectrometer online. The nanoelectrospray ion source was operated at 1.9 kV. The digest was analyzed using the data-dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular masses and product ion spectra to determine amino acid sequence in successive instrument scans. The data were analyzed by using all CID spectra collected in the experiment to search the human UniProtKB sequence databases using Mascot (false discovery rate, 0.01%).
Additional samples were analyzed using a targeted selective reaction monitoring (SRM) (61) experiment, which involves the fragmentation of specific ions over the course of the LC experiment, including two peptides from enolase and several fibronectin peptides. The relative abundance of the fibronectin peptides was determined by normalizing the fibronectin (FN) peptide peak areas to the peak areas (PA) of the enolase peptides: PA(FN)/PA(enolase).
Author contributions
L. W. W., D. F. M., B. B. W., and S. S. A. conceptualization; L. W. W., B. B. W., and S. S. A. data curation; L. W. W., B. B. W., and S. S. A. formal analysis; L. W. W., S. N., and B. B. W. investigation; L. W. W., D. S. A., and B. B. W. methodology; L. W. W., D. S. A., J. D., D. F. M., B. B. W., and S. S. A. writing-review and editing; D. S. A., J. D., and D. F. M. resources; B. B. W. validation; B. B. W. and S. S. A. writing-original draft; S. S. A. supervision; S. S. A. funding acquisition.
Acknowledgments
We thank Dr. Francesco Ramirez for providing Fbn1−/− mice for isolation of fibroblasts and Dr. Rahel Schnellmann and Dr. Dirk Hubmacher for helpful discussions.
This work was supported by the Allen Distinguished Investigator Program through support made by the Paul G. Allen Frontiers Group, American Heart Association Grant 17 DIA33820024 (to S. S. A.), National Institutes of Health Grant EY024943 (to S. S. A.), and a Mark Lauer Pediatric Research Grant (to S. N.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ECM
- extracellular matrix
- TSR
- thrombospondin type 1 repeat
- SID
- selected interacting domain
- FUD
- functional upstream domain
- MEF
- mouse embryonic fibroblast
- qRT
- quantitative reverse transcriptase
- CHO
- Chinese hamster ovary
- SRM
- selective reaction monitoring.
References
- 1. Mouw J. K., Ou G., and Weaver V. M. (2014) Extracellular matrix assembly: a multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 15, 771–785 10.1038/nrm3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rozario T., and DeSimone D. W. (2010) The extracellular matrix in development and morphogenesis: a dynamic view. Dev. Biol. 341, 126–140 10.1016/j.ydbio.2009.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonnans C., Chou J., and Werb Z. (2014) Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801 10.1038/nrn3846,10.1038/nrm3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dubail J., and Apte S. S. (2015) Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 44–46, 24–37 10.1016/j.matbio.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 5. Apte S. S. (2016) ADAMTS proteases: mediators of physiological and pathogenic extracellular proteolysis. In Encyclopedia of Cell Biology (Bradshaw R. A., and Stahl P. D., eds) pp. 630–638, Academic Publishers, Waltham, MA [Google Scholar]
- 6. Nandadasa S., Foulcer S., and Apte S. S. (2014) The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol. 35, 34–41 10.1016/j.matbio.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coronary Artery Disease (C4D) Genetics Consortium (2011) A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 43, 339–344 10.1038/ng.782 [DOI] [PubMed] [Google Scholar]
- 8. Stanton H., Melrose J., Little C. B., and Fosang A. J. (2011) Proteoglycan degradation by the ADAMTS family of proteinases. Biochim. Biophys. Acta 1812, 1616–1629 10.1016/j.bbadis.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 9. Hurskainen T. L., Hirohata S., Seldin M. F., and Apte S. S. (1999) ADAM-TS5, ADAM-TS6, and ADAM-TS7, novel members of a new family of zinc metalloproteases. General features and genomic distribution of the ADAM-TS family. J. Biol. Chem. 274, 25555–25563 10.1074/jbc.274.36.25555 [DOI] [PubMed] [Google Scholar]
- 10. Blelloch R., and Kimble J. (1999) Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature 399, 586–590 10.1038/21196 [DOI] [PubMed] [Google Scholar]
- 11. Somerville R. P., Longpre J. M., Jungers K. A., Engle J. M., Ross M., Evanko S., Wight T. N., Leduc R., and Apte S. S. (2003) Characterization of ADAMTS-9 and, ADAMTS-20 as a distinct, ADAMTS subfamily related to Caenorhabditis elegans GON-1. J. Biol. Chem. 278, 9503–9513 10.1074/jbc.M211009200 [DOI] [PubMed] [Google Scholar]
- 12. Ismat A., Cheshire A. M., and Andrew D. J. (2013) The secreted AdamTS-A metalloprotease is required for collective cell migration. Development 140, 1981–1993 10.1242/dev.087908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benz B. A., Nandadasa S., Takeuchi M., Grady R. C., Takeuchi H., LoPilato R. K., Kakuda S., Somerville R. P. T., Apte S. S., Haltiwanger R. S., and Holdener B. C. (2016) Genetic and biochemical evidence that gastrulation defects in Pofut2 mutants result from defects in ADAMTS9 secretion. Dev. Biol. 416, 111–122 10.1016/j.ydbio.2016.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dubail J., Vasudevan D., Wang L. W., Earp S. E., Jenkins M. W., Haltiwanger R. S., and Apte S. S. (2016) Impaired ADAMTS9 secretion: a potential mechanism for eye defects in Peters Plus syndrome. Sci. Rep. 6, 33974 10.1038/srep33974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubail J., Aramaki-Hattori N., Bader H. L., Nelson C. M., Katebi N., Matuska B., Olsen B. R., and Apte S. S. (2014) A new Adamts9 conditional mouse allele identifies its non-redundant role in interdigital web regression. Genesis 52, 702–712 10.1002/dvg.22784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kern C. B., Wessels A., McGarity J., Dixon L. J., Alston E., Argraves W. S., Geeting D., Nelson C. M., Menick D. R., and Apte S. S. (2010) Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol. 29, 304–316 10.1016/j.matbio.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCulloch D. R., Nelson C. M., Dixon L. J., Silver D. L., Wylie J. D., Lindner V., Sasaki T., Cooley M. A., Argraves W. S., and Apte S. S. (2009) ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev Cell. 17, 687–698 10.1016/j.devcel.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tharmarajah G., Eckhard U., Jain F., Marino G., Prudova A., Urtatiz O., Fuchs H., de Angelis M. H., Overall C. M., and Van Raamsdonk C. D. (2018) Melanocyte development in the mouse tail epidermis requires the Adamts9 metalloproteinase. Pigment Cell Melanoma Res. 31, 693–707 10.1111/pcmr.12711 [DOI] [PubMed] [Google Scholar]
- 19. Nandadasa S., Nelson C. M., and Apte S. S. (2015) ADAMTS9-mediated extracellular matrix dynamics regulates umbilical cord vascular smooth muscle differentiation and rotation. Cell Rep. 11, 1519–1528 10.1016/j.celrep.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mead T. J., Du Y., Nelson C. M., Gueye N. A., Drazba J., Dancevic C. M., Vankemmelbeke M., Buttle D. J., and Apte S. S. (2018) ADAMTS9-regulated pericellular matrix dynamics governs focal adhesion-dependent smooth muscle differentiation. Cell Rep. 23, 485–498 10.1016/j.celrep.2018.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choi Y. J., Halbritter J., Braun D. A., Schueler M., Schapiro D., Rim J. H., Nandadasa S., Choi W. I., Widmeier E., Shril S., Körber F., Sethi S. K., Lifton R. P., Beck B. B., Apte S. S., et al. (2019) Mutations of ADAMTS9 cause nephronophthisis-related ciliopathy. Am. J. Hum. Genet. 104, 45–54 10.1016/j.ajhg.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boesgaard T. W., Gjesing A. P., Grarup N., Rutanen J., Jansson P. A., Hribal M. L., Sesti G., Fritsche A., Stefan N., Staiger H., Häring H., Smith U., Laakso M., Pedersen O., Hansen T., et al. (2009) Variant near ADAMTS9 known to associate with type 2 diabetes is related to insulin resistance in offspring of type 2 diabetes patients: EUGENE2 study. PLoS One 4, e7236 10.1371/journal.pone.0007236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fritsche L. G., Chen W., Schu M., Yaspan B. L., Yu Y., Thorleifsson G., Zack D. J., Arakawa S., Cipriani V., Ripke S., Igo R. P. Jr., Buitendijk G. H., Sim X., Weeks D. E., Guymer R. H., et al. (2013) Seven new loci associated with age-related macular degeneration. Nat. Genet. 45, 433–439 10.1038/ng.2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heid I. M., Jackson A. U., Randall J. C., Winkler T. W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M. C., Speliotes E. K., Mägi R., Workalemahu T., White C. C., Bouatia-Naji N., Harris T. B., Berndt S. I., et al. (2010) Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42, 949–960 10.1038/ng.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Helisalmi S., Immonen I., Losonczy G., Resch M. D., Benedek S., Balogh I., Papp A., Berta A., Uusitupa M., Hiltunen M., and Kaarniranta K. (2014) ADAMTS9 locus associates with increased risk of wet AMD. Acta Ophthalmol. 92, e410 10.1111/aos.12341 [DOI] [PubMed] [Google Scholar]
- 26. Zeggini E., Scott L. J., Saxena R., Voight B. F., Marchini J. L., Hu T., de Bakker P. I., Abecasis G. R., Almgren P., Andersen G., Ardlie K., Boström K. B., Bergman R. N., Bonnycastle L. L., Borch-Johnsen K., et al. (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40, 638–645 10.1038/ng.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du W., Wang S., Zhou Q., Li X., Chu J., Chang Z., Tao Q., Ng E. K., Fang J., Sung J. J., and Yu J. (2013) ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene 32, 3319–3328 10.1038/onc.2012.359 [DOI] [PubMed] [Google Scholar]
- 28. Koo B. H., Coe D. M., Dixon L. J., Somerville R. P., Nelson C. M., Wang L. W., Young M. E., Lindner D. J., and Apte S. S. (2010) ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am. J. Pathol. 176, 1494–1504 10.2353/ajpath.2010.090655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lo P. H., Lung H. L., Cheung A. K., Apte S. S., Chan K. W., Kwong F. M., Ko J. M., Cheng Y., Law S., Srivastava G., Zabarovsky E. R., Tsao S. W., Tang J. C., Stanbridge E. J., and Lung M. L. (2010) Extracellular protease ADAMTS9 suppresses esophageal and nasopharyngeal carcinoma tumor formation by inhibiting angiogenesis. Cancer Res. 70, 5567–5576 10.1158/0008-5472.CAN-09-4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koo B. H., and Apte S. S. (2010) Cell-surface processing of the metalloprotease pro-ADAMTS9 is influenced by the chaperone, G. R.P94/gp96. J. Biol. Chem. 285, 197–205 10.1074/jbc.M109.039677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koo B. H., Longpré J. M., Somerville R. P., Alexander J. P., Leduc R., and Apte S. S. (2006) Cell-surface processing of pro-ADAMTS9 by furin. J. Biol. Chem. 281, 12485–12494 10.1074/jbc.M511083200 [DOI] [PubMed] [Google Scholar]
- 32. Koo B. H., Longpré J. M., Somerville R. P., Alexander J. P., Leduc R., and Apte S. S. (2007) Regulation of ADAMTS9 secretion and enzymatic activity by its propeptide. J. Biol. Chem. 282, 16146–16154 10.1074/jbc.M610161200 [DOI] [PubMed] [Google Scholar]
- 33. Nandadasa S., Kraft C. M., Wang L. W., O'Donnell A., Patel R., Gee H. Y., Grobe K., Cox T. C., Hildebrandt F., and Apte S. S. (2019) Secreted metalloproteases ADAMTS9 and ADAMTS20 have a non-canonical role in ciliary vesicle growth during ciliogenesis. Nat. Commun. 10, 953 10.1038/s41467-019-08520-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 10.1101/gr.2659105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maurer L. M., Ma W., and Mosher D. F. (2015) Dynamic structure of plasma fibronectin. Crit. Rev. Biochem. Mol. Biol. 51, 213–227 10.1080/10409238.2016.1184224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh P., Carraher C., and Schwarzbauer J. E. (2010) Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 26, 397–419 10.1146/annurev-cellbio-100109-104020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sabatier L., Chen D., Fagotto-Kaufmann C., Hubmacher D., McKee M. D., Annis D. S., Mosher D. F., and Reinhardt D. P. (2009) Fibrillin assembly requires fibronectin. Mol. Biol. Cell 20, 846–858 10.1091/mbc.e08-08-0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sabatier L., Djokic J., Fagotto-Kaufmann C., Chen M., Annis D. S., Mosher D. F., and Reinhardt D. P. (2013) Complex contributions of fibronectin to initiation and maturation of microfibrils. Biochem. J. 456, 283–295 10.1042/BJ20130699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hubmacher D., and Apte S. S. (2015) ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 47, 34–43 10.1016/j.matbio.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bader H. L., Wang L. W., Ho J. C., Tran T., Holden P., Fitzgerald J., Atit R. P., Reinhardt D. P., and Apte S. S. (2012) A disintegrin-like and metalloprotease domain containing thrombospondin type 1 motif-like 5 (ADAMTSL5) is a novel fibrillin-1-, fibrillin-2-, and heparin-binding member of the ADAMTS superfamily containing a netrin-like module. Matrix Biol. 31, 398–411 10.1016/j.matbio.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hubmacher D., Schneider M., Berardinelli S. J., Takeuchi H., Willard B., Reinhardt D. P., Haltiwanger R. S., and Apte S. S. (2017) Unusual life cycle and impact on microfibril assembly of ADAMTS17, a secreted metalloprotease mutated in genetic eye disease. Sci. Rep. 7, 41871 10.1038/srep41871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hubmacher D., Wang L. W., Mecham R. P., Reinhardt D. P., and Apte S. S. (2015) Adamtsl2 deletion results in bronchial fibrillin microfibril accumulation and bronchial epithelial dysplasia: a novel mouse model providing insights into geleophysic dysplasia. Dis. Model. Mech. 8, 487–499 10.1242/dmm.017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maurer L. M., Tomasini-Johansson B. R., Ma W., Annis D. S., Eickstaedt N. L., Ensenberger M. G., Satyshur K. A., and Mosher D. F. (2010) Extended binding site on fibronectin for the functional upstream domain of protein F1 of Streptococcus pyogenes. J. Biol. Chem. 285, 41087–41099 10.1074/jbc.M110.153692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roy R., Wewer U. M., Zurakowski D., Pories S. E., and Moses M. A. (2004) ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J. Biol. Chem. 279, 51323–51330 10.1074/jbc.M409565200 [DOI] [PubMed] [Google Scholar]
- 45. Doucet A., and Overall C. M. (2011) Broad coverage identification of multiple proteolytic cleavage site sequences in complex high molecular weight proteins using quantitative proteomics as a complement to edman sequencing. Mol. Cell. Proteomics 10, M110.003533 10.1074/mcp.M110.003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi F., and Sottile J. (2011) MT1-MMP regulates the turnover and endocytosis of extracellular matrix fibronectin. J. Cell Sci. 124, 4039–4050 10.1242/jcs.087858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taylor S. H., Yeung C. Y., Kalson N. S., Lu Y., Zigrino P., Starborg T., Warwood S., Holmes D. F., Canty-Laird E. G., Mauch C., and Kadler K. E. (2015) Matrix metalloproteinase 14 is required for fibrous tissue expansion. Elife 4, e09345 10.7554/eLife.09345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Doucet A., and Overall C. M. (2011) Amino-terminal oriented mass spectrometry of substrates (ATOMS) N-terminal sequencing of proteins and proteolytic cleavage sites by quantitative mass spectrometry. Methods Enzymol. 501, 275–293 10.1016/B978-0-12-385950-1.00013-4 [DOI] [PubMed] [Google Scholar]
- 49. Bekhouche M., Leduc C., Dupont L., Janssen L., Delolme F., Vadon-Le Goff S., Smargiasso N., Baiwir D., Mazzucchelli G., Zanella-Cleon I., Dubail J., De Pauw E., Nusgens B., Hulmes D. J., Moali C., et al. (2016) Determination of the substrate repertoire of ADAMTS2, 3, and 14 significantly broadens their functions and identifies extracellular matrix organization and, T. G.F-beta signaling as primary targets. FASEB J. 30, 1741–1756 10.1096/fj.15-279869 [DOI] [PubMed] [Google Scholar]
- 50. Schnellmann R., Sack R., Hess D., Annis D. S., Mosher D. F., Apte S. S., and Chiquet-Ehrismann R. (2018) A selective extracellular matrix proteomics approach identifies fibronectin proteolysis by ADAMTS16 and its impact on spheroid morphogenesis. Mol. Cell. Proteomics 17, 1410–1425 10.1074/mcp.RA118.000676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Colige A., Monseur C., Crawley J. T.B, Santamaria S., and de Groot R. (2019) Proteomic discovery of substrates of the cardiovascular protease ADAMTS7. J. Biol. Chem. 294, 8037–8045 10.1074/jbc.RA119.007492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. George E. L., Georges-Labouesse E. N., Patel-King R. S., Rayburn H., and Hynes R. O. (1993) Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 119, 1079–1091 [DOI] [PubMed] [Google Scholar]
- 53. Apte S. S., Fukai N., Beier D. R., and Olsen B. R. (1997) The matrix metalloproteinase-14 (MMP-14) gene is structurally distinct from other MMP genes and is co-expressed with the, T. I.MP-2 gene during mouse embryogenesis. J. Biol. Chem. 272, 25511–25517 10.1074/jbc.272.41.25511 [DOI] [PubMed] [Google Scholar]
- 54. Holmbeck K., Bianco P., Pidoux I., Inoue S., Billinghurst R. C., Wu W., Chrysovergis K., Yamada S., Birkedal-Hansen H., and Poole A. R. (2005) The metalloproteinase MT1-MMP is required for normal development and maintenance of osteocyte processes in bone. J. Cell Sci. 118, 147–156 10.1242/jcs.01581 [DOI] [PubMed] [Google Scholar]
- 55. Zhou Z., Apte S. S., Soininen R., Cao R., Baaklini G. Y., Rauser R. W., Wang J., Cao Y., and Tryggvason K. (2000) Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl. Acad. Sci. U.S.A. 97, 4052–4057 10.1073/pnas.060037197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jungers K. A., Le Goff C., Somerville R. P., and Apte S. S. (2005) Adamts9 is widely expressed during mouse embryo development. Gene Expr. Patterns 5, 609–617 10.1016/j.modgep.2005.03.004 [DOI] [PubMed] [Google Scholar]
- 57. Mosher D. F., and Johnson R. B. (1983) In vitro formation of disulfide-bonded fibronectin multimers. J. Biol. Chem. 258, 6595–6601 [PubMed] [Google Scholar]
- 58. Xu J., Bae E., Zhang Q., Annis D. S., Erickson H. P., and Mosher D. F. (2009) Display of cell surface sites for fibronectin assembly is modulated by cell adherence to (1)F3 and C-terminal modules of fibronectin. PLoS One 4, e4113 10.1371/journal.pone.0004113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., and Ramirez F. (2006) Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J. Biol. Chem. 281, 8016–8023 10.1074/jbc.M511599200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beene L. C., Wang L. W., Hubmacher D., Keene D. R., Reinhardt D. P., Annis D. S., Mosher D. F., Mecham R. P., Traboulsi E. I., and Apte S. S. (2013) Nonselective assembly of fibrillin 1 and fibrillin 2 in the rodent ocular zonule and in cultured cells: implications for marfan syndrome. Invest. Ophthalmol. Vis. Sci. 54, 8337–8344 10.1167/iovs.13-13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Willard B. B., Ruse C. I., Keightley J. A., Bond M., and Kinter M. (2003) Site-specific quantitation of protein nitration using liquid chromatography/tandem mass spectrometry. Anal. Chem. 75, 2370–2376 10.1021/ac034033j [DOI] [PubMed] [Google Scholar]