Abstract
Leptin receptor (LEPR) signaling may be involved in promoting angiogenesis and proliferation, inhibiting apoptosis and playing a vital role in the progression of carcinogenesis. A number of studies have focused on the association of LEPR rs1137101 variants with susceptibility of cancer, however, the observed results were controversial. We searched literature on the relationship of LEPR rs1137101 G>A polymorphism with cancer risk by using PubMed and Embase databases, covering all publications up to 14 October 2018. In total, 44 case–control studies with 35,936 subjects were included. After combining all eligible studies, we identified null relationship between LEPR gene rs1137101 G>A polymorphism and overall cancer risk [A vs. G: odds ratio (OR ) = 0.97, 95% confidence interval (CI ) = 0.89–1.06, P = 0.547; AA vs. GG: OR = 0.93, 95% CI = 0.78–1.13, P = 0.476; AA/GA vs. GG: OR = 0.99, 95% CI = 0.91–1.09, P= 0.890 and AA vs. GA/GG: OR = 0.92, 95% CI = 0.82–1.04, P= 0.198]. However, in a subgroup analysis, there was an increased susceptibility of oral and oropharyngeal cancer in AA vs. GA/GG genetic model (OR, 1.83; 95% CI, 1.01–3.33; P=0.048). Considering the limited participants were included, the findings might be underpowered. Sensitivity analysis identified that any independent study omitted did not materially influence the pooled ORs and CIs. The results of publication bias detection showed that there was no evidence of bias. In summary, this analysis indicates that no significant association of cancer risk was identified to be correlated with rs1137101 G>A variants, even in stratified analyses.
Keywords: cancer, energy, leptin, polymorphism, receptor, risk
Introduction
Cancer is one of the common health burden worldwide. Due to the prevalence of smoking and drinking, environmental pollution, as well as population aging, the incidence of malignancy is rising. According to the estimation of Global Cancer Statistics 2018, approximately 18.1 million new cancer cases and 9.6 million cancer-related deaths may have occurred in 2018 [1]. In the developing countries, the prognosis of cancer could be poorer than that in the developed countries. The reason of this phenomenon may be due to the diagnosis at an advanced stage combined with limited treatment. Some effective measures can potentially contribute to relieve the global cancer burden, including the application of precise early detection and treatment, tobacco and alcohol control, vaccine injection, sufficient fruits and vegetables intake and appropriate physical exercise. Overweight/obesity influences the health of more than 200 million people [2]. Overweight/obesity may play a vital role in growing morbidity of malignancy [3,4,5]. Thus, it is believed that overweight/obesity-related genes could affect the development of cancer.
Overweight/obesity is attributable to a chronic energy intake and expenditure imbalance. Leptin (LEP) is a common hormone of regulating energy expenditure by inhibiting hunger. LEP receptor (LEPR) is a type I cytokine receptor which is encoded by the LEPR gene and acts as a receptor for the hormone LEP. LEPR is a single transmembrane-domain receptor and composed of extracellular, transmembrane, and intracellular sections. LEP/LEPR signaling may involve in promoting angiogenesis, facilitating cell proliferation, and inhibiting epithelial cell apoptosis [6]. The long isoform in LEPR cytoplasmic domain may be essential for the signal transduction of Janus kinase/signal transducers and activators of transcription pathway [7].
The LEPR gene lies in chromosome 1 (Position_38: 65420652 – 65637493). There are a number of common single nucleotide polymorphisms (SNPs) of the LEPR genes, which have been established. LEPR rs1137101 G>A polymorphism (Arg223Gln) is the most extensively studied association of this SNP with the development of cancer. LEPR rs1137101 locus is a missense variant, which is a substitution of G→A at nucleotide number 668 in exon 6 of LEPR gene [8]. It leads to an Arg→Gln altering in extracellular region [8]. The potential relationships of LEPR rs1137101 variants with susceptibility of cancer have been elucidated in different malignancies; however, the obtained results were inconsistent. Two previous meta-analyses indicated that LEPR rs1137101 G>A polymorphism might not be a risk factor for cancer [9,10]. Recently, more studies concerning the association of LEPR rs1137101 locus with cancer risk were performed [11–26]. Hence, we carried out an updated meta-analysis on such relationship so as to further explore the role of LEPR rs1137101 variants to susceptibility of cancer.
Materials and methods
Searching publications
To obtain the potentially eligible investigations on LEPR rs1137101 G>A polymorphism and cancer susceptibility, we conducted an electronic literature search on PubMed and Embase databases, covering all publications up to 14 October 2018, by using the following searching strategy: (Leptin receptor or LEPR or obese receptor or OBR or CD295) and (carcinoma or cancer or tumor or malignancy or neoplasms) and (polymorphism or SNP or variation). References of the eligible studies and reviews were also screened to identify the additional data. We reported the present study using the Preferred Reporting Items for PRISMA guideline (Supplementary Table S1; PRISMA checklist) [27].
Selection and exclusion criteria
The major selection criteria were: (i) full-text study, (ii) assessing the relationship of LEPR rs1137101 variants with cancer susceptibility, (iii) designed as an unrelated case–control study, (iv) sufficient data could be obtained to calculate the odds ratio (OR) with 95% confidence interval (CI), and (v) genotype distribution conformation to Hardy–Weinberg equilibrium (HWE).
The major exclusion criteria were: (i) genotype data could not be extracted; (ii) not case–control study; (iii) distribution of genotype violated HWE; and (iv) comments, reviews, and letters.
Data extraction
Two authors (G.R. and Y.W.) independently extracted raw data. The following data were collected: the surname of first author, publication year, race, country, number of participants, age, Body Mass Index (BMI), source of control, matching method, genotype frequencies and genotyping method. Ethnicity descents were defined as mixed, Asian, and Caucasian. Cancer types were classified as hepatocellular carcinoma, breast cancer, renal cell carcinoma, esophageal cancer, colorectal cancer, oral and oropharyngeal cancer, non-Hodgkin’s lymphoma and other cancers (lung cancer and bladder cancer). For source of control, the eligible studies were categorized as hospital-based and population-based. When HWE in control group was not available, an online software (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) was harnessed to calculate the P-value of HWE test. If the information extracted was different, two authors reached consensus on each item.
Statistical analysis
The relationship strength of LEPR rs1137101 SNP with susceptibility of cancer was determined by crude ORs with their 95% CIs. The pooled ORs were calculated for allele (A vs. G), dominant (GA/AA vs. GG), recessive (AA versus GA/GG), and homozygote comparison (AA vs. GG) genetic modes. Additionally, stratified analysis was carried out to assess the influence of confounding risk factors: ethnicity (mixed, Asian, and Caucasian) and cancer type (breast cancer, esophageal cancer, hepatocellular carcinoma, renal cell carcinoma, colorectal cancer, oral and oropharyngeal cancer, non-Hodgkin’s lymphoma, and other cancers). We used Chi-square based Q-test and I2 test to assess the potential heterogeneity among the eligible studies. P<0.10 or I2 ≥ 50% was considered as the level of significant heterogeneity. Then a random-effects model (DerSimonian and Laird method) was used to assess the association of LEPR rs1137101 polymorphism with cancer susceptibility [28,29]; otherwise, a fixed-effects model (Mantel–Haenszel method) was harnessed to pool the data [30]. Stratified analysis was conducted to explore the source of heterogeneity. We performed one-way sensitivity analysis to evaluate the effect of an individual study on pooled ORs and CIs. Begg’s funnel plot and the Egger’s test were used to detect the potential bias in included publications, and a P<0.10 was considered significant [31,32]. Statistical analysis of the present study was calculated with STATA 12.0 software (StataCorp, College Station, Texas, U.S.A.).
Results
Study characteristics
In this meta-analysis, 33 publications involving 44 independent case–control studies on the relationship of LEPR rs1137101 G>A polymorphism with cancer risk were recruited [9,11–26,33–48]. In some publications, they contained several subgroups that we treated as independent studies [12,14,40]. Figure 1 shows the eligible study selecting process. In total, 44 independent case–control studies with 35,936 subjects (13,711 cases and 22,225 controls) were included. Among them, 20 were Caucasians [15,16,20–23,34,36,37,39–43,45–48], 14 were Asians [9,11–14,17,24,26,33,44], 1 was African [38], and 9 were mixed populations [18,19,25,35]. Twenty-two case–control studies were designed as hospital-based investigation [9,11–14,17,18,21,22,24,26,33,34,38,44,46,48], seventeen studies were designed as population-based investigation [16,19,25,35–37,40,41,43,45,47], and the source of control in other five studies were unknown [15,20,23,39,42]. Of all the eligible studies, 24 focused on breast cancer [12,14,15,19,20,22,33,35,36,38,41,42,44,47,48], 4 focused on esophageal cancer [13,40], 4 focused on colorectal cancer [34,37,39,46], 3 focused on hepatocellular carcinoma [9,11,26], 2 focused on non-Hodgkin’s lymphoma [43,45], 3 focused on oral and oropharyngeal cancer [18,21,25], 2 focused on renal cell carcinoma [17,24], and 2 focused on other cancers [16,23]. The detailed information of eligible studies is listed in Table 1. The extracted genotypes and HWE are summarized in Table 2.
Figure 1. Flow diagram of the meta–analysis of the association between LEPR rs1137101 G>A polymorphism and cancer risk.
Table 1. Characteristics of the studies in meta-analysis.
| Study | Publication year | Country | Ethnicity | Cancer type | Sample size (case/control) | Case age (years) | Control age (years) | Case BMI (kg/m2) | Control BMI (kg/m2) | Source of control | Match | Genotype method |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang et al. | 2018 | China | Asians | Hepatocellular carcinoma | 584/923 | 53.17 ± 11.76 | 53.72 ± 9.97 | NA | NA | Hospital-based | Age, sex, ethnicity | SNPscan |
| Liu et al. | 2018 | China | Asians | Breast cancer | 488/463 | 43.71 ± 6.13 | 43.35 ± 5.43 | BMI < 24: n=300, BMI ≥ 24, n=148 | BMI < 24: n=289, BMI ≥ 24, n=174 | Hospital-based | Age, sex, ethnicity, region | MS-TOF |
| Liu et al. | 2018 | China | Asians | Breast cancer | 346/342 | 58.55 ± 6.87 | 56.60 ± 6.53 | BMI < 24:n=207, BMI ≥ 24, n=139 | BMI < 24: n=195, BMI ≥ 24, n=147 | Hospital-based | Sex, ethnicity, region | MS-TOF |
| Qiu et al. | 2017 | China | Asians | Esophageal cancer | 507/1,496 | 62.77 ± 8.01 | 62.77 ± 8.84 | 22.27 ± 2.90 | 23.91 ± 3.03 | Hospital-based | Age, ethnicity, sex | SNPscan |
| Yuna et al. | 2018 | China | Asians | Breast cancer | 77/805 | 51.43 ± 11.33 | 48.98 ± 8.83 | Obesity (≥27): n=15, non-obesity, n=62 | Obesity (≥27), n=50, non-obesity, n=751 | Hospital-based | Age, sex, ethnicity, region | MS-TOF |
| Yuna et al. | 2019 | China | Asians | Breast cancer | 79/805 | 49.94 ± 10.10 | 48.98 ± 8.83 | Obesity (≥27), n=12, non-obesity, n=67 | Obesity (≥27), n=50, non-obesity, n=751 | Hospital-based | Age, sex, ethnicity, region | MS-TOF |
| Yuna et al. | 2017 | China | Asians | Breast cancer | 412/805 | 49.73 ± 9.38 | 48.98 ± 8.83 | Obesity (≥27), n=45, non-obesity, n=365 | Obesity (≥27), n=50, non-obesity, n=751 | Hospital-based | Age, sex, ethnicity, region | MS-TOF |
| Yuna et al. | 2017 | China | Asians | Breast cancer | 135/805 | 50.50 ± 9.04 | 48.98 ± 8.83 | Obesity (≥27), n=12, non-obesity, n=123 | Obesity (≥27), n=50, non-obesity, n=751 | Hospital-based | Age, sex, ethnicity, region | MS-TOF |
| El-Hussiny et al. | 2017 | Egypt | Caucasians | Breast cancer | 48/48 | 47.7 ± 7.5 | 43.5 ± 9.2 | 34.37 ± 6.08 | 27.28 ± 3.52 | NA | Sex, ethnicity, region | PCR-RFLP |
| Ali et al. | 2017 | Pakistan | Caucasians | Bladder cancer | 200/200 | 55.5 ± 13.24 | 54.3 ± 9.9 | NA | NA | Population-based | Age, sex, ethnicity | PCR |
| Zhang et al. | 2016 | China | Asians | Renal cell carcinoma | 83/161 | 17–85 (median: 57) | NA | NA | NA | Hospital-based | Ethnicity, age, and sex | PCR-RLFP |
| Rodrigues et al. | 2015 | Brazil | Mixed | Oral and oropharyngeal cancer | 129/186 | 54.9 ± 10.7 | 54.2 ± 11.1 | NA | NA | Hospital-based | Sex, region | PCR-RLFP |
| Slattery et al. | 2015 | America | Mixed | Breast cancer | 239/252 | NA | NA | BMI < 25 | BMI < 25 | Population-based | Sex, region | A multiplexed bead array assay |
| Slattery et al. | 2015 | America | Mixed | Breast cancer | 176/150 | NA | NA | BMI = 25–29 | BMI = 25–29 | Population-based | Sex, region | A multiplexed bead array assay |
| Slattery et al. | 2015 | America | Mixed | Breast cancer | 111/126 | NA | NA | BMI ≥ 30 | BMI ≥ 30 | Population-based | Sex, region | A multiplexed bead array assay |
| Slattery et al. | 2015 | America | Mixed | Breast cancer | 253/239 | NA | NA | BMI < 25 | BMI < 25 | Population-based | Sex, region | A multiplexed bead array assay |
| Slattery et al. | 2015 | America | Mixed | Breast cancer | 205/304 | NA | NA | BMI = 25–29 | BMI = 25–29 | Population-based | Sex, region | A multiplexed bead array assay |
| Slattery et al. | 2015 | America | Mixed | Breast cancer | 148/224 | NA | NA | BMI ≥ 30 | BMI ≥ 30 | Population-based | Sex, region | A multiplexed bead array assay |
| Mahmoudi et al. | 2015 | Iran | Caucasians | Breast cancer | 45/41 | 47.09 ± 11.45 | 48.37 ± 8.80 | NA | NA | NA | Age, sex | PCR-RFLP |
| Hussain et al. | 2015 | India | Caucasians | Oral carcinoma | 306/228 | 33.5 ± 5.79 | 32.7 ± 5.73 | 29.5 ± 5.44 | 23.8 ± 4.88 | Hospital-based | Age, sex, ethnicity and low-risk environment | PCR-RFLP |
| Mohammadzadeh et al. | 2014 | Iran | Caucasians | Breast cancer | 100/100 | 48.16 ± 10.47 | 49.0 ± 7.77 | 27.16 ± 3.96 | 28.31 ± 4.71 | Hospital-based | Age, sex, ethnicity, BMI | PCR-RFLP |
| Unsal et al. | 2014 | Turkey | Caucasians | Lung cancer | 162/130 | 60.96 ± 11.88 | 57.92 ± 14.96 | NA | NA | NA | Age, sex, ethnicity | PCR-RFLP |
| Mu et al. | 2014 | China | Asians | Renal cell carcinoma | 77/161 | 56.22 ± 12.27 | NA | NA | NA | Hospital-based | Ethnicity, age and sex | PCR-RFLP or DNA sequence |
| Domingos et al. | 2014 | Brazil | Mixed | Oral carcinoma | 25/89 | 58.0 ± 13.6 | 55.9 ± 13.9 | NA | NA | Population-based | Age, sex, and smoking habits | RFLP-PCR |
| Li et al. | 2012 | China | Asians | Hepatocellular carcinoma | 417/551 | 52.45 ± 4.6 | 51.95 ± 2.8 | 21.44 ± 3.4 | 22.56 ± 3.2 | Hospital-based | Age, sex, ethnicity | RFLP |
| Kim et al. | 2012 | Korea | Asians | Breast cancer | 400/452 | ≤49: 64.8% | ≤49: 62.4% | ≤25: 68.5% | ≤25: 78.5% | Hospital-based | Age, sex | MS-TOF |
| Karimi et al. | 2011 | Iran | Caucasians | Colorectal cancer | 173/173 | 55.8 ± 12.7 | 44.8 ± 17.2 | 25.1 ± 5.3 | 26.2 ± 7.19 | Hospital-based | BMI, sex and smoking status | RFLP |
| Nyante et al. | 2011 | America | Mixed | Breast cancer | 1972/1776 | 23–74, mean: 50 | 21–74, mean: 51 | BMI < 25, n=712; BMI ≥ 25, n=1219 | BMI < 25, n=545; BMI ≥ 25, n=1194 | Population-based | Age, sex and region | Illumina |
| Cleveland et al. | 2010 | America | Caucasians | Breast cancer | 1065/1108 | NA | NA | BMI < 30, n=291; BMI ≥ 30, n=43 | BMI < 30, n=304; BMI ≥ 30, n=62 | Population-based | Age, sex, region | PCR |
| Pechlivanis et al. | 2009 | Czech Republic | Caucasians | Colorectal cancer | 702/752 | 27–85, mean: 62 | 29–91, mean: 54 | 13.1–44.9, mean: 26.5 | 16.6–44.3, mean: 26.4 | Hospital-based | Sex, region | TaqMan |
| Okobia et al. | 2008 | Nigeria | Africans | Breast cancer | 209/209 | 46.1 ± 12.63 | 47.1 ± 13.50 | NA | NA | Hospital-based | Sex, region | PCR-RFLP |
| Vasků et al. | 2009 | Czech Republic | Caucasians | Colorectal cancer | 102/101 | 68 ± 10.2 | 68.1 ± 5.4 | Male/Female: 26.7 ± 5.1/26.9 ± 5.2 | NA | NA | Age, ethnicity | PCR-RFLP |
| Doecke et al. | 2008 | Australia | Caucasians | Esophageal cancer | 260/1352 | NA | NA | NA | NA | Population-based | Sex, region | Sequenom |
| Doecke et al. | 2008 | Australia | Caucasians | Esophageal cancer | 301/1352 | NA | NA | NA | NA | Population-based | Sex, region | Sequenom |
| Doecke et al. | 2008 | Australian | Caucasians | Esophageal cancer | 213/1352 | NA | NA | NA | NA | Population-based | Sex, region | Sequenom |
| Gallicchio et al. | 2007 | America | Caucasians | Breast cancer | 61/933 | mean: 59 | mean: 59 | NA | BMI < 25, n=479; BMI ≥ 25, n=513 | Population-based | NA | TaqMan |
| Snoussi et al. | 2006 | Tunisia | Caucasians | Breast cancer | 308/222 | 50 ± 24 | 48 ± 14 | NA | NA | NA | Sex, region | PCR-RFLP |
| Willett et al. | 2005 | U.K. | Caucasians | Non-Hodgkin’s lymphoma | 1073/754 | 18–65 | 18–65 | BMI < 25, n=633, BMI ≥ 25, n=603 | BMI < 25, n=524; BMI ≥ 25, n=387 | Population-based | Sex, region | TaqMan |
| Woo et al. | 2006 | Korea | Asians | Breast cancer | 45/45 | NA | NA | BMI < 25, n=27, BMI ≥ 25, n=18 | BMI < 25, n=37; BMI ≥ 25, n=8 | Hospital-based | Age, sex | DNA sequencing |
| Skibola et al. | 2004 | America | Caucasians | Non-Hodgkin’s lymphoma | 376/805 | 21–74 | NA | BMI < 25, n=213; BMI ≥ 25, n=162 | BMI < 25: n=480; BMI ≥ 25, n=321 | Population-based | Age, sex and region | TaqMan |
| Mahmoudi et al. | 2016 | Iran | Caucasians | Colorectal cancer | 261/339 | 56.1 ± 12.6 | 44.3 ± 16.3 | 25.6 ± 4.9 | 25.2 ± 4.2 | Hospital-based | Sex and BMI | PCR-RFLP |
| Dai et al. | 2010 | China | Asians | Hepatocellular carcinoma | 80/102 | 32–65 | 28–60 | NA | NA | Hospital-based | Age, sex, ethnicity | PCR-RFLP |
| Teras et al. | 2009 | America | Caucasians | Breast cancer | 648/659 | mean: 69 | mean: 69 | NA | NA | Population-based | Age, sex | SNPstream |
| Kuptsova et al. | 2008 | Russia | Caucasians | Breast cancer | 110/105 | 56–65 | 50–70 | NA | NA | Hospital-based | Age, sex | PCR |
Abbreviations: MS-TOF, mass spectrometry time of flight; NA, not available; PCR, polymerase chain reaction; PCR-RFLP, PCR-restriction fragment length polymorphism.
Table 2. Distribution of LEPR rs1137101 G>A polymorphism genotype and allele.
| Study | Publication year | Case | Control | Case | Control | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | A [n (%)] | G A [n (%)] | A A [n (%)] | G A [n (%)] | |||
| Zhang et al. | 2018 | 3 | 119 | 453 | 10 | 193 | 717 | 125 (10.87) | 1025 (89.13) | 213 (11.58) | 1627 (88.42) | Yes |
| Liu et al. | 2018 | AA/AG = 101 | NA | 346 | AA/AG = 99 | NA | 363 | NA | NA | NA | NA | Yes |
| Liu et al. | 2018 | AA/AG = 80 | NA | 264 | AA/AG = 79 | NA | 260 | NA | NA | NA | NA | Yes |
| Qiu et al. | 2017 | 6 | 108 | 390 | 21 | 322 | 1146 | 120 (11.90) | 888 (88.10) | 364 (12.22) | 2,614 (87.78) | Yes |
| Yuan et al. | 2017 | AA/AG = 18 | NA | 59 | AA/AG = 178 | NA | 623 | NA | NA | NA | NA | Yes |
| Yuan et al. | 2017 | AA/AG = 17 | NA | 62 | AA/AG = 178 | NA | 623 | NA | NA | NA | NA | Yes |
| Yuan et al. | 2017 | AA/AG = 96 | NA | 314 | AA/AG = 178 | NA | 623 | NA | NA | NA | NA | Yes |
| Yuan et al. | 2017 | AA/AG = 30 | NA | 105 | AA/AG = 178 | NA | 623 | NA | NA | NA | NA | Yes |
| El-Hussiny et al. | 2017 | 9 | 15 | 24 | 2 | 24 | 22 | 33 (34.38) | 63, 65.63) | 28 (29.17) | 68 (70.83) | Yes |
| Ali et al. | 2017 | 51 | 96 | 53 | 67 | 97 | 36 | 198 (49.50) | 202 (50.50) | 231 (57.75) | 169 (42.25) | Yes |
| Zhang et al. | 2016 | 2 | 21 | 61 | 7 | 52 | 102 | 25 (14.88) | 143 (85.12) | 66 (20.50) | 256 (79.50) | Yes |
| Rodrigues et al. | 2015 | 60 | 61 | 8 | 68 | 92 | 26 | 181 (70.16) | 77 (29.84) | 228 (61.29) | 144 (38.71) | Yes |
| Slattery et al. | 2015 | 77 | NA | AG/GG:162 | 69 | NA | AG/GG:183 | NA | NA | NA | NA | Yes |
| Slattery et al. | 2015 | 50 | NA | AG/GG:126 | 50 | NA | AG/GG:100 | NA | NA | NA | NA | Yes |
| Slattery et al. | 2015 | 22 | NA | AG/GG:89 | 38 | NA | AG/GG:88 | NA | NA | NA | NA | Yes |
| Slattery et al. | 2015 | 63 | NA | AG/GG:190 | 80 | NA | AG/GG:159 | NA | NA | NA | NA | Yes |
| Slattery et al. | 2015 | 46 | NA | AG/GG:159 | 83 | NA | AG/GG:221 | NA | NA | NA | NA | Yes |
| Slattery et al. | 2015 | 43 | NA | AG/GG:105 | 52 | NA | AG/GG:172 | NA | NA | NA | NA | Yes |
| Mahmoudi et al. | 2015 | 19 | 25 | 1 | 17 | 18 | 6 | 63 (70.00) | 27 (30.00) | 52 (63.41) | 30 (36.59) | Yes |
| Hussain et al. | 2015 | 48 | 110 | 148 | 12 | 72 | 144 | 206 (33.66) | 406 (66.34) | 96 (21.05) | 360 (78.95) | Yes |
| Mohammadzadeh et al. | 2014 | 25 | 56 | 19 | 54 | 40 | 6 | 106 (53.000) | 94 (47.00) | 148 (74.00) | 52 (26.00) | Yes |
| Unsal et al. | 2014 | 75 | 62 | 25 | 56 | 55 | 19 | 212 (65.43) | 112 (34.57) | 167 (64.23) | 93 (35.77) | Yes |
| Mu et al. | 2014 | 2 | 20 | 55 | 4 | 41 | 116 | 24 (15.58) | 130 (84.42) | 49 (15.22) | 273 (84.78) | Yes |
| Domingos et al. | 2014 | 12 | 7 | 6 | 40 | 38 | 11 | 31 (62.00) | 19 (38.00) | 118 (66.29) | 60 (33.71) | Yes |
| Li et al. | 2012 | 87 | 208 | 122 | 189 | 256 | 106 | 382 (45.80) | 452 (54.20) | 634 (57.53) | 468 (42.47) | Yes |
| Kim et al. | 2012 | 8 | 88 | 294 | 6 | 91 | 350 | 104 (13.33) | 676 (86.67) | 103 (11.52) | 791 (88.48) | Yes |
| Karimi et al. | 2011 | 77 | 75 | 21 | 67 | 80 | 26 | 229 (66.18) | 117 (33.82) | 214 (61.85) | 132 (38.15) | Yes |
| Nyante et al. | 2011 | 494 | 952 | 526 | 416 | 874 | 485 | 1940 (49.19) | 2004 (50.86) | 1706 (48.06) | 1844 (51.94) | Yes |
| Cleveland et al. | 2010 | 173 | 521 | 355 | 187 | 551 | 360 | 867 (41.33) | 1231 (58.67) | 925 (42.12) | 1271 (57.88) | Yes |
| Pechlivanis et al. | 2009 | 179 | 320 | 140 | 202 | 361 | 143 | 678 (53.05) | 600 (46.95) | 765 (54.18) | 647 (45.82) | Yes |
| Okobia et al. | 2008 | 46 | 107 | 56 | 56 | 107 | 46 | 199 (47.61) | 219 (52.39) | 219 (52.39) | 199 (47.61) | Yes |
| Vasků et al. | 2009 | 23 | 56 | 21 | 34 | 45 | 21 | 102 (51.00) | 98 (49.00) | 113 (56.50) | 87 (43.50) | Yes |
| Doecke et al. | 2008 | 73 | 140 | 47 | 419 | 663 | 270 | 286 (55.00) | 234 (45.00) | 1501 (55.51) | 1203 (44.49) | Yes |
| Doecke et al. | 2008 | 84 | 164 | 62 | 419 | 663 | 270 | 332 (53.55) | 288 (46.45) | 1501 (55.51) | 1203 (44.49) | Yes |
| Doecke et al. | 2008 | 64 | 106 | 43 | 419 | 663 | 270 | 234 (54.93) | 192 (45.07) | 1501 (55.51) | 1203 (44.49) | Yes |
| Gallicchio et al. | 2007 | 14 | 24 | 15 | 278 | 443 | 151 | 52 (49.07) | 54 (50.94) | 999 (57.28) | 745 (42.72) | Yes |
| Snoussi et al. | 2006 | 98 | 145 | 65 | 102 | 90 | 30 | 341 (55.36) | 275 (44.64) | 294 (66.22) | 150 (33.78) | Yes |
| Willett et al. | 2005 | 336 | 554 | 183 | 234 | 387 | 133 | 1226 (57.13) | 920 (42.87) | 855 (56.70) | 653 (43.30) | Yes |
| Woo et al. | 2006 | 0 | 12 | 33 | 0 | 8 | 37 | 12 (13.33) | 78 (86.67) | 8 (8.89) | 82 (91.11) | Yes |
| Skibola et al. | 2004 | 115 | 173 | 87 | 226 | 379 | 198 | 403 (53.73) | 347 (46.27) | 831 (51.74) | 775 (48.26) | Yes |
| Mahmoudi et al. | 2016 | 127 | 101 | 33 | 146 | 147 | 46 | 355 (68.01) | 167 (31.99) | 439 (64.75) | 239 (35.25) | Yes |
| Dai et al. | 2010 | 10 | 14 | 58 | 2 | 19 | 81 | 34 (20.73) | 130 (79.27) | 23 (11.27) | 181 (88.73) | Yes |
| Teras et al. | 2009 | AA/AG = 460 | NA | 181 | AA/AG = 439 | NA | 211 | NA | NA | NA | NA | Yes |
| Kuptsova et al. | 2008 | 17 | 69 | 24 | 18 | 51 | 36 | 103 (46.82) | 117 (53.18) | 87 (41.43) | 123 (58.57) | Yes |
Abbreviation: NA, not available.
Results of meta-analysis
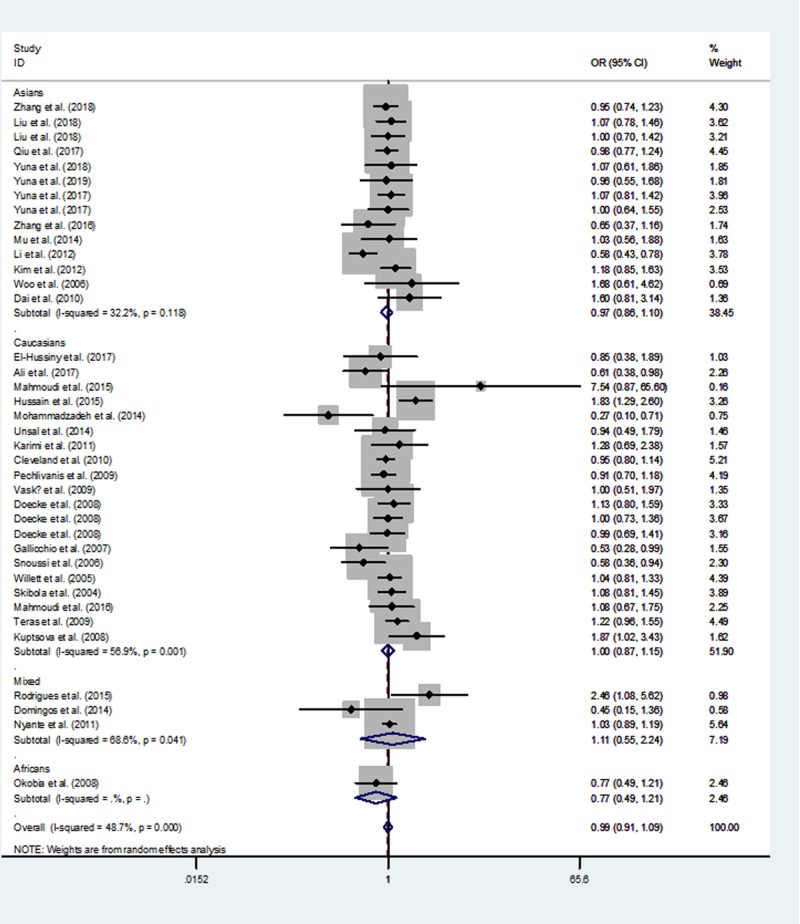
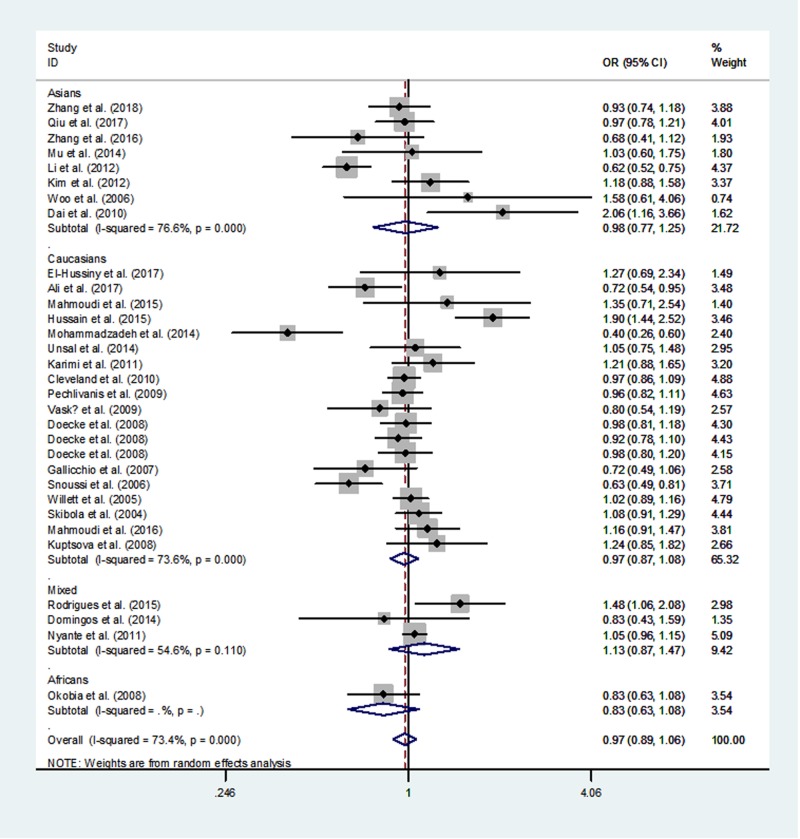
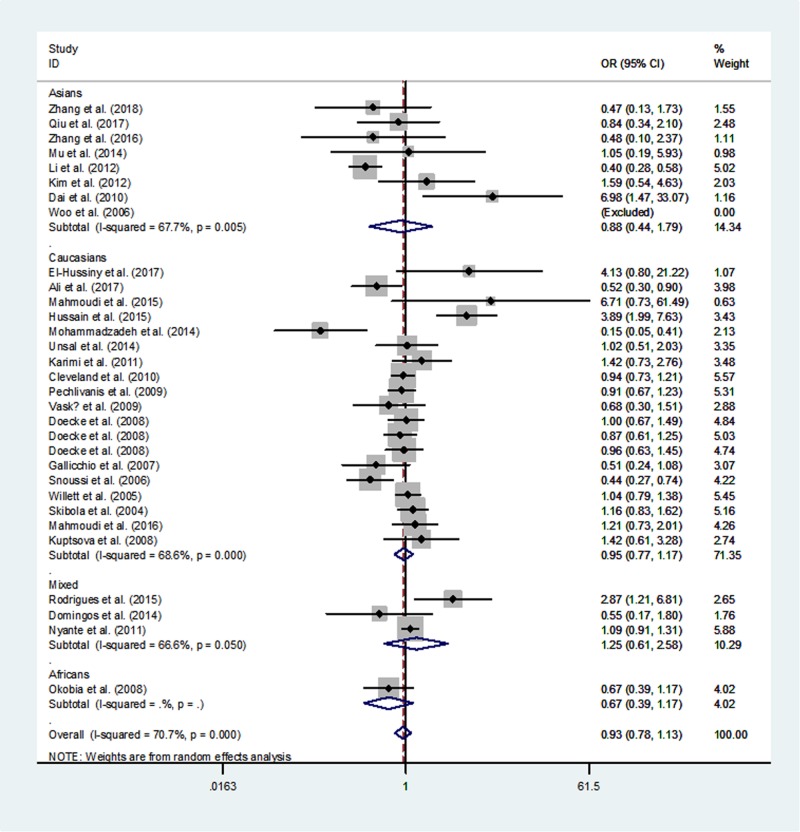
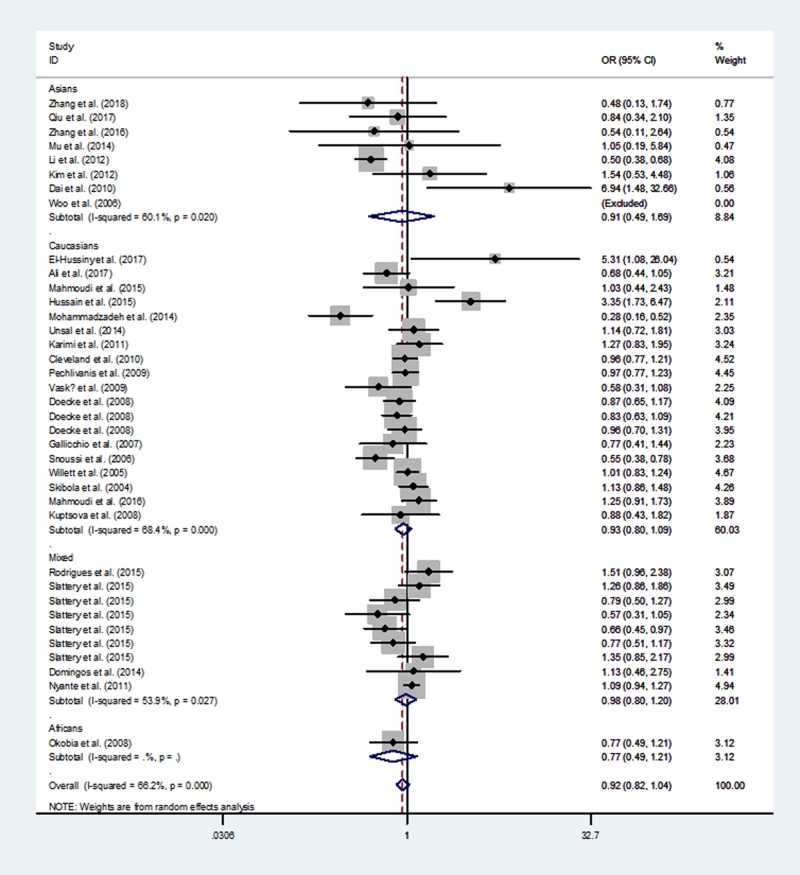
Table 3 lists the overall and subgroup analysis results of the present study. After combining all eligible case–control studies, we identified null relationship between rs1137101 polymorphism in LEPR gene and overall cancer risk under four genetic models (A vs. G: OR = 0.97, 95% CI = 0.89–1.06, P=0.547; AA vs. GG: OR = 0.93, 95% CI = 0.78–1.13, P =0.476; AA/GA vs. GG: OR = 0.99, 95% CI = 0.91–1.09, P=0.890 and AA vs. GA/GG: OR = 0.92, 95% CI = 0.82–1.04, P=0.198, Figures 2–5).
Table 3. Results of the meta-analysis from different genetic models.
| Number of studies | A vs. G | AA vs. GG | AA+GA vs. GG | AA vs.GA+GG | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | I2 | P (Q-test) | OR (95% CI) | P | I2 | P (Q-test) | OR (95% CI) | P | I2 | P (Q-test) | OR (95% CI) | P | I2 | P (Q-test) | ||
| Total | 44 | 0.97 (0.89–1.06) | 0.547 | 73.4% | <0.001 | 0.93 (0.78–1.13) | 0.476 | 70.7% | <0.001 | 0.99 (0.91–1.09) | 0.890 | 48.7% | <0.001 | 0.92 (0.82–1.04) | 0.198 | 66.2% | <0.001 |
| Ethnicity | |||||||||||||||||
| Caucasians | 20 | 0.97 (0.87–1.08) | 0.565 | 73.6% | <0.001 | 0.95 (0.77–1.17) | 0.621 | 68.6% | <0.001 | 1.00 (0.87–1.15) | 0.976 | 56.9% | 0.001 | 0.93 (0.80–1.09) | 0.381 | 68.4% | <0.001 |
| Asians | 14 | 0.98 (0.77–1.25) | 0.864 | 76.6 | <0.001 | 0.88 (0.44–1.79) | 0.733 | 67.7% | 0.005 | 0.96 (0.87–1.06) | 0.456 | 32.2% | 0.118 | 0.91 (0.49–1.69) | 0.765 | 60.1% | 0.020 |
| Mixed | 9 | 1.13 (0.87–1.47) | 0.369 | 54.6% | 0.110 | 1.25 (0.61–2.58) | 0.539 | 60.6% | 0.050 | 1.11 (0.55–2.24) | 0.775 | 68.6% | 0.041 | 0.98 (0.80–1.20) | 0.837 | 53.9% | 0.027 |
| Africans | 1 | 0.83 (0.63–1.08) | 0.167 | - | - | 0.67 (0.39–1.17) | 0.162 | - | - | 0.77 (0.49–1.21) | 0.255 | - | - | 0.77 (0.49–1.21) | 0.255 | - | - |
| Cancer type | |||||||||||||||||
| Breast cancer | 24 | 0.91 (0.77–1.07) | 0.248 | 75.5% | <0.001 | 0.82 (0.57–1.17) | 0.269 | 74.6% | <0.001 | 1.00 (0.88–1.13) | 0.946 | 44.1% | 0.024 | 0.84 (0.69–1.02) | 0.076 | 67.3% | <0.001 |
| Colorectal cancer | 4 | 1.01 (0.90–1.14) | 0.814 | 30.4% | 0.230 | 1.00 (0.79–1.26) | 0.967 | 0.0% | 0.398 | 0.98 (0.80–1.21) | 0.860 | 0.0% | 0.742 | 1.04 (0.88–1.23) | 0.623 | 48.2% | 0.122 |
| Esophageal cancer | 4 | 0.96 (0.87–1.06) | 0.407 | 0.0% | 0.967 | 0.93 (0.75–1.16) | 0.525 | 0.0% | 0.957 | 1.01 (0.87–1.18) | 0.873 | 0.0% | 0.916 | 0.88 (0.74–1.03) | 0.118 | 0.0% | 0.925 |
| Hepatocellular carcinoma | 3 | 0.98 (0.60–1.62) | 0.948 | 89.4% | <0.001 | 0.96 (0.21–4.42) | 0.956 | 84.1% | 0.002 | 0.89 (0.55–1.44) | 0.633 | 80.8% | 0.005 | 1.03 (0.25–4.14) | 0.971 | 81.5% | 0.005 |
| Oral and oropharyngeal cancer | 3 | 1.46 (1.00–2.13) | 0.050 | 64.1% | 0.062 | 2.02 (0.72–5.65) | 0.179 | 75.1% | 0.018 | 1.43 (0.67–3.07) | 0.360 | 69.1% | 0.039 | 1.83 (1.01–3.33) | 0.048 | 60.6% | 0.079 |
| Renal cell carcinoma | 2 | 0.82 (0.57–1.18) | 0.291 | 19.7% | 0.264 | 0.67 (0.21–2.14) | 0.500 | 0.0% | 0.509 | 0.81 (0.53–1.23) | 0.318 | 13.9% | 0.281 | 0.72 (0.22–2.28) | 0.570 | 0.0% | 0.576 |
| Non-Hodgkin’s lymphoma | 2 | 1.04 (0.94–1.16) | 0.451 | 0.0% | 0.577 | 1.09 (0.88–1.35) | 0.438 | 0.0% | 0.641 | 1.06 (0.88–1.28) | 0.547 | 0.0% | 0.839 | 1.05 (0.90–1.24) | 0.528 | 0.0% | 0.526 |
| Others | 2 | 0.86 (0.59–1.25) | 0.426 | 65.9% | 0.087 | 0.68 (0.44–1.04) | 0.076 | 55.3% | 0.135 | 0.71 (0.48–1.04) | 0.079 | 9.8% | 0.292 | 0.86 (0.63–1.18) | 0.600 | 60.7% | 0.111 |
| Sample size | |||||||||||||||||
| <1000 | 32 | 0.98 (0.82–1.17) | 0.816 | 81.1% | <0.001 | 0.98 (0.67–1.43) | 0.913 | 78.6% | <0.001 | 0.97 (0.81–1.15) | 0.694 | 62.4% | <0.001 | 0.93 (0.75–1.14) | 0.466 | 72.9% | <0.001 |
| ≥1000 | 12 | 1.00 (0.95–1.05) | 0.950 | 0.0% | 0.926 | 1.01 (0.91–1.11) | 0.906 | 0.0% | 0.882 | 1.02 (0.95–1.09) | 0.548 | 0.0% | 0.945 | 1.00 (0.92–1.08) | 0.907 | 0.0% | 0.695 |
| Source of control | |||||||||||||||||
| Hospital-based | 22 | 1.03 (0.86–1.23) | 0.744 | 82.7% | <0.001 | 1.04 (0.68–1.57) | 0.869 | 80.2% | <0.001 | 1.03 (0.90–1.19) | 0.651 | 57.4% | <0.001 | 0.99 (0.72–1.36) | 0.965 | 78.6% | <0.001 |
| Population-based | 17 | 0.99 (0.94–1.04) | 0.715 | 20.9% | 0.251 | 0.99 (0.87–1.03) | 0.787 | 25.0% | 0.214 | 1.01 (0.94–1.10) | 0.754 | 29.3% | 0.167 | 0.97 (0.90–1.04) | 0.368 | 29.1% | 0.132 |
| NA | 5 | 0.92 (0.68–1.23) | 0.560 | 61.7% | 0.034 | 1.02 (0.48–2.13) | 0.968 | 68.6% | 0.013 | 0.82 (0.61–1.10) | 0.190 | 38.7% | 0.163 | 0.89 (0.53–1.48) | 0.649 | 69.4% | 0.011 |
I2: ≥50% indicate significant heterogeneity. P (Q-test): <0.10 considered as the criterion of significant heterogeneity.
Abbreviation: NA, not available. Bold values are statistically significant (P<0.05).
Figure 2. Meta-analysis of the association between LEPR rs1137101 G>A polymorphism and cancer risk (AA/GA vs. GG, random–effects model).
Figure 5. Meta-analysis of the association between LEPR rs1137101 G>A polymorphism and cancer risk (A vs. G, random–effects model).
Figure 3. Meta-analysis of the association between LEPR rs1137101 G>A polymorphism and cancer risk (AA vs. GG, random–effects model).
Figure 4. Meta-analysis of the association between LEPR rs1137101 G>A polymorphism and cancer risk (AA vs. GG/GA, random–effects model).
When we conducted a stratified analysis by ethnicity, null association was found in mixed populations, Africans, Asians, and Caucasians. However, when we performed a subgroup analysis by cancer type, there was an increased susceptibility of oral and oropharyngeal cancer in AA vs. GA/GG genetic model (OR, 1.83; 95% CI, 1.01–3.33; P=0.048).
Heterogeneity analysis
Significant heterogeneity among the eligible studies was found in this pooled analysis (Table 3). In the present study, we carried out subgroup analysis to explore the sources of heterogeneity. We found that Caucasians, breast cancer, hepatocellular carcinoma, other cancers, small sample sizes (<1000) and hospital-based case–control studies might contribute the major source to heterogeneity.
Sensitivity analysis
Sensitivity analysis of one-way method identified that any individual study deleted did not materially influence the pooled ORs and CIs under all genetic comparisons. These findings indicated that our observations were stable and reliable (Supplementary Figure S1).
Publication bias
Begg’s funnel plot and Egger’s test were used to evaluate the potential bias. The results of the bias detecting were shown as following: A vs. G: P Begg’s= 0.852, PEgger’s = 0.973; AA vs. GG: P Begg’s= 0.775, PEgger’s = 0.897; AA/GA vs. GG: P Begg’s = 0.950, PEgger’s = 0.869 and AA vs. GA/GG: P Begg’s= 0.703, PEgger’s = 0.897 (Supplementary Figure S2).
Discussion
Recently, variants in LEPR gene and their potential associations with cancer risk have been explored. Rs1137101 G>A polymorphism is one of the important variants in LEPR gene. LEPR rs1137101 G>A polymorphism is located on the exon region of LEPR gene, and it has been thought to be involved in the development of cancer by a number of studies. Several case–control studies reported that LEPR rs1137101 G>A polymorphism might be associated with the decreased risk of cancer [16,22,26,41,42]. However, several primary studies also suggested that LEPR rs1137101 locus could promote the progression of cancers [15,18]. Meanwhile, two meta-analyses were carried out to clarify the correlation of this SNP with susceptibility of overall cancer [9,10]. The results of these meta-analyses indicated that LEPR rs1137101 G>A polymorphism might not be associated with the risk of cancer. Moreover, more epidemiologic data were reported [11–26]. Therefore, an updated meta-analysis is necessary to calrify this issue precisely. In this meta-analysis, data of 44 case–control studies involving 13,711 cases and 22,225 controls, which is higher compared with these previous pooled-analyses mentioned above, were included and analyzed. Therefore, the obtained results may be more convincing.
LEP/LEPR signaling may promote cell proliferation and inhibit epithelial cell apoptosis [6]. In addition, Ben et al. [49] reported that LEPR rs1137101 G>A SNP may affect plasma LEP levels and BMI. A previous study suggested that leptin level was associated with the development of breast cancer [50]. However, null associations of LEPR rs1137101 locus with cancer susceptibility was identified, which was analogous to the results reported in previous meta-analyses [9,10,51,52], but unlike the other four meta-analyses [48,53–55]. Due to lack of sufficient data, the findings of previous systematic reviews and meta-analyses might be conflicting. Ethnicity also may be a vital factor for the difference. The minor allele frequency of LEPR rs1137101 G>A polymorphism was difference among different populations, but in stratified analysis by race, null relationship was found. Additionally, results of stratified analyses by sample size and source of control both found no relationship between LEPR rs1137101 G>A polymorphism with overall cancer susceptibility, highlighting that these variables could not influence the negative findings either.
According to the findings of stratified analysis, we found that LEPR rs1137101 locus might be associated with the susceptibility of oral and oropharyngeal cancer. However, the increased risk of cancer was dubious and hard to explain. Sample size was an important factor to determine the relationship between LEPR rs1137101 G>A polymorphism and cancer risk. In this subgroup, only 460 oral and oropharyngeal cancer cases and 503 controls were included for analysis, the findings might be underpowered.
Significant heterogeneity was found among the eligible studies in multiple genetic models. Thus, we carried out stratified analysis by ethnicity, cancer type, sample size, and source of control. It was obvious that Caucasians, hepatocellular carcinoma, breast cancer, other cancers, small sample sizes (<1000 subjects) and hospital-based case–control studies might contribute to heterogeneity.
In this meta-analysis, as mentioned in results, no publication bias was detected. In addition, findings of sensitivity analysis also indicated that our observation was convincing. Overall, results of this meta-analysis were stable and credible for the studied populations.
However, there were several limitations in the present pooled-analysis. First, some small sample size studies were recruited in this meta-analysis, which could promote the power of study. On the other hand, they led to potential publication bias and significant heterogeneity as well. Second, our findings were based on the crude pooled-results of the eligible case–control studies, the adjustment on characteristics and risk factors (e.g. BMI, physical exercise, age, gender, smoking, alcohol consumption, vegetable and fruit intake, and so on) was not performed. In the future, a more detailed assessment is needed, in which the characteristics and potential risk factors should be considered to adjust the findings. Third, although the cases and controls in eligible studies were fully matched, there was heterogeneity in different cancer types which may influence our results. Fourth, only PubMed and Embase databases were searched to retrieve the eligible. Finally, significant heterogeneity was found in all genetic models, which might influence the findings of our study. Thus, these results should be interpreted with caution.
In summary, this meta-analysis may be the largest sample size so far to assess the potential association of cancer risk with rs1137101 G>A polymorphism in LEPR gene. There is no significant association of cancer risk was identified to be correlated with rs1137101 G>A variants in the overall comparison, and the similar findings was also found in stratified analysis by ethnicity, cancer type, sample size, and source of control. In the future, more large-scale studies are needed to confirm or refute our findings.
Supporting information
Supplementary Figure S1.
Supplementary Figure S2.
Abbreviations
- BMI
body mass index
- CI
confidence interval
- HWE
Hardy–Weinberg equilibrium
- LEP
leptin
- LEPR
leptin receptor
- OR
odds ratio
- SNP
single nucleotide polymorphism
Funding
This work was supported by the Young and Middle-aged Talent Training Project of Health Development Planning Commission in Fujian Province [grant number 2016-ZQN-25]; the Program for New Century Excellent Talents in Fujian Province University [grant number NCETFJ-2017B015]; the Joint Funds for the Innovation of Science and Technology, Fujian province [grant number 2017Y9099]; and the Natural Science Foundation of Fujian Province [grant number 2017J01291].
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
All authors contributed significantly to the present study. Conceived and designed the experiments: G.R. and Y.W. Performed the experiments: G.R., Y.W. and W.T. Analyzed the data: G.R., Y.W., W.T. and H.Q. Contributed reagents/materials/analysis tools: S.C. Wrote the manuscript: G.R., Y.W. and W.T.
References
- 1.Bray F., Ferlay J., Soerjomataram I.. et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Suarez A.L. (2018) Burden of cancer attributable to obesity, type 2 diabetes and associated risk factors. Metabolism 92, 136–146 10.1016/j.metabol.2018.10.013 [DOI] [PubMed] [Google Scholar]
- 3.Spyrou N., Avgerinos K.I., Mantzoros C.S.. et al. (2018) Classic and novel adipocytokines at the intersection of obesity and cancer: diagnostic and therapeutic strategies. Curr. Obes. Rep. 7, 260–275 10.1007/s13679-018-0318-7 [DOI] [PubMed] [Google Scholar]
- 4.Hicks D.F., Bakst R., Doucette J.. et al. (2018) Impact of obesity on outcomes for patients with head and neck cancer. Oral Oncol. 83, 11–17 10.1016/j.oraloncology.2018.05.027 [DOI] [PubMed] [Google Scholar]
- 5.Chadid S., Singer M.R., Kreger B.E.. et al. (2018) Midlife weight gain is a risk factor for obesity-related cancer. Br. J. Cancer 118, 1665–1671, PubMed Central PMCID: PMC6008441 10.1038/s41416-018-0106-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tilg H. and Moschen A.R. (2006) Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 6, 772–783 10.1038/nri1937 [DOI] [PubMed] [Google Scholar]
- 7.Wauman J. and Tavernier J. (2011) Leptin receptor signaling: pathways to leptin resistance. Front. Biosci. 16, 2771–2793 10.2741/3885 [DOI] [PubMed] [Google Scholar]
- 8.Gotoda T., Manning B.S., Goldstone A.P.. et al. (1997) Leptin receptor gene variation and obesity: lack of association in a white British male population. Hum. Mol. Genet. 6, 869–876, PubMed PMID: 9175732 10.1093/hmg/6.6.869 [DOI] [PubMed] [Google Scholar]
- 9.Liu P., Shi H., Liu R.. et al. (2014) Lack of association between LEPR Q223R polymorphisms and cancer susceptibility: evidence from a meta-analysis. J. BUON 19, 855–862 [PubMed] [Google Scholar]
- 10.He J., Xi B., Ruiter R.. et al. (2013) Association of LEP G2548A and LEPR Q223R polymorphisms with cancer susceptibility: evidence from a meta-analysis. PLoS ONE 8, e75135, PubMed Central PMCID: PMC3798550 10.1371/journal.pone.0075135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S., Jiang J., Chen Z.. et al. (2018) Investigation of LEP and LEPR polymorphisms with the risk of hepatocellular carcinoma: a case-control study in Eastern Chinese Han population. Onco. Targets Ther. 11, 2083–2089, PubMed Central PMCID: PMC5905468 10.2147/OTT.S153931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C.R., Li Q., Hou C.. et al. (2018) Changes in body mass index, leptin, and leptin receptor polymorphisms and breast cancer risk. DNA Cell Biol. 37, 182–188 10.1089/dna.2017.4047 [DOI] [PubMed] [Google Scholar]
- 13.Qiu H., Lin X., Tang W.. et al. (2017) Investigation of TCF7L2, LEP and LEPR polymorphisms with esophageal squamous cell carcinomas. Oncotarget 8, 109107–109119, PubMed Central PMCID: PMC5752507 10.18632/oncotarget.22619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan X.L., Xu Z.P., Liu C.R.. et al. (2017) Study of the association between polymorphism of persistent obesity, human leptin gene/leptin receptor gene and molecular subtypes of breast cancer. Zhonghua Yu Fang Yi Xue Za Zhi 51, 533–538 [DOI] [PubMed] [Google Scholar]
- 15.El-Hussiny M.A., Atwa M.A., Rashad W.E.. et al. (2017) Leptin receptor Q223R polymorphism in Egyptian female patients with breast cancer. Contemp. Oncol. 21, 42–47, PubMed Central PMCID: PMC5385477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali S.H.B., Bangash K.S., Rauf A.. et al. (2017) Identification of novel potential genetic predictors of urothelial bladder carcinoma susceptibility in Pakistani population. Fam. Cancer 16, 577–594 10.1007/s10689-017-9991-z [DOI] [PubMed] [Google Scholar]
- 17.Zhang H.P., Zou J., Yin Y.. et al. (2016) High-resolution melting PCR analysis for genotyping Lys109Arg and Gln223Arg in patients with renal cell carcinoma. Ann. Clin. Lab. Sci. 46, 367–373 [PubMed] [Google Scholar]
- 18.Rodrigues P.R., Maia L.L., Santos M.. et al. (2015) Leptin receptor expression and Gln223Arg polymorphism as prognostic markers in oral and oropharyngeal cancer. Genet. Mol. Res. 14, 14979–14988 10.4238/2015.November.24.5 [DOI] [PubMed] [Google Scholar]
- 19.Slattery M.L., Lundgreen A., Hines L.. et al. (2015) Energy homeostasis genes and breast cancer risk: The influence of ancestry, body size, and menopausal status, the breast cancer health disparities study. Cancer Epidemiol. 39, 1113–1122, PubMed Central PMCID: PMC4679560 10.1016/j.canep.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahmoudi R., Noori Alavicheh B., Nazer Mozaffari M.A.. et al. (2015) Polymorphisms of Leptin (-2548 G/A) and Leptin Receptor (Q223R) genes in Iranian women with breast cancer. Int. J. Genom. 2015, 132720, PubMed Central PMCID: PMC4496654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussain S.R., Naqvi H., Gupta S.. et al. (2015) A study on oncogenic role of leptin and leptin receptor in oral squamous cell. Tumour Biol. 36, 6515–6523 10.1007/s13277-015-3342-1 [DOI] [PubMed] [Google Scholar]
- 22.Mohammadzadeh G., Ghaffari M.A., Bafandeh A.. et al. (2014) Effect of leptin receptor Q223R polymorphism on breast cancer risk. Iran. J. Basic Med. Sci. 17, 588–594, PubMed Central PMCID: PMC4240793 [PMC free article] [PubMed] [Google Scholar]
- 23.Unsal M., Kara N., Karakus N.. et al. (2014) Effects of leptin and leptin receptor gene polymorphisms on lung cancer. Tumour Biol. 35, 10231–10236 10.1007/s13277-014-2293-2 [DOI] [PubMed] [Google Scholar]
- 24.Mu H.J., Zou J., Xie P.. et al. (2014) Association of leptin receptor Lys109Arg and Gln223Arg polymorphisms with increased risk of clear cell renal cell carcinoma. Asian Pac. J. Cancer Prev. 15, 4211–4215 10.7314/APJCP.2014.15.10.4211 [DOI] [PubMed] [Google Scholar]
- 25.Domingos P.L., Farias L.C., Pereira C.S.. et al. (2014) Leptin receptor polymorphism Gln223Arg (rs1137101) in oral squamous cell carcinoma and potentially malignant oral lesions. Springer Plus 3, 683, PubMed Central PMCID: PMC4447719 10.1186/2193-1801-3-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z., Yuan W., Ning S.. et al. (2012) Role of leptin receptor (LEPR) gene polymorphisms and haplotypes in susceptibility to hepatocellular carcinoma in subjects with chronic hepatitis B virus infection. Mol. Diagn. Ther. 16, 383–388 [DOI] [PubMed] [Google Scholar]
- 27.Moher D., Liberati A., Tetzlaff J.. et al. (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Intern. Med. 151, 264–269, W64 10.7326/0003-4819-151-4-200908180-00135 [DOI] [PubMed] [Google Scholar]
- 28.Higgins J.P., Thompson S.G., Deeks J.J.. et al. (2003) Measuring inconsistency in meta-analyses. BMJ 327, 557–560, PubMed Central PMCID: PMC192859 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DerSimonian R. and Laird N. (1986) Meta-analysis in clinical trials. Control Clin. Trials 7, 177–188 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 30.Tang W., Qiu H., Ding H.. et al. (2013) Association between the STK15 F31I polymorphism and cancer susceptibility: a meta-analysis involving 43,626 subjects. PLoS ONE 8, e82790, PubMed Central PMCID: PMC3862673 10.1371/journal.pone.0082790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W., Qiu H., Jiang H.. et al. (2014) Aurora-A V57I (rs1047972) polymorphism and cancer susceptibility: a meta-analysis involving 27,269 subjects. PLoS ONE 9, e90328, PubMed Central PMCID: PMC3943872 10.1371/journal.pone.0090328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S., Wang Y., Jiang H.. et al. (2015) Association between the CD28 IVS3 +17T>C (rs3116496) polymorphism and cancer susceptibility: a meta-analysis involving 8,843 subjects. Int. J. Clin. Exp. Med. 8, 17353–17361, PubMed Central PMCID: PMC4694227 [PMC free article] [PubMed] [Google Scholar]
- 33.Kim K.Z., Shin A., Lee Y.S.. et al. (2012) Polymorphisms in adiposity-related genes are associated with age at menarche and menopause in breast cancer patients and healthy women. Hum. Reprod. 27, 2193–2200 10.1093/humrep/des147 [DOI] [PubMed] [Google Scholar]
- 34.Karimi K., Arkani M., Safaei A.. et al. (2011) Association of leptin receptor gene Gln223Arg polymorphism with susceptibility to colorectal cancer. Gastroenterol. Hepatol. 4, 192–198, PubMed Central PMCID: PMC4017430 [PMC free article] [PubMed] [Google Scholar]
- 35.Nyante S.J., Gammon M.D., Kaufman J.S.. et al. (2011) Common genetic variation in adiponectin, leptin, and leptin receptor and association with breast cancer subtypes. Breast Cancer Res. Treat. 129, 593–606, PubMed Central PMCID: PMC3355661 10.1007/s10549-011-1517-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleveland R.J., Gammon M.D., Long C.M.. et al. (2010) Common genetic variations in the LEP and LEPR genes, obesity and breast cancer incidence and survival. Breast Cancer Res. Treat. 120, 745–752, PubMed Central PMCID: PMC3571680 10.1007/s10549-009-0503-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pechlivanis S., Bermejo J.L., Pardini B.. et al. (2009) Genetic variation in adipokine genes and risk of colorectal cancer. Eur. J. Endocrinol. 160, 933–940 10.1530/EJE-09-0039 [DOI] [PubMed] [Google Scholar]
- 38.Okobia M.N., Bunker C.H., Garte S.J.. et al. (2008) Leptin receptor Gln223Arg polymorphism and breast cancer risk in Nigerian women: a case control study. BMC Cancer 8, 338, PubMed Central PMCID: PMC2613914 10.1186/1471-2407-8-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasku A., Vokurka J. and Bienertova-Vasku J. (2009) Obesity-related genes variability in Czech patients with sporadic colorectal cancer: preliminary results. Int. J. Colorectal Dis. 24, 289–294 10.1007/s00384-008-0553-6 [DOI] [PubMed] [Google Scholar]
- 40.Doecke J.D., Zhao Z.Z., Stark M.S.. et al. (2008) Single nucleotide polymorphisms in obesity-related genes and the risk of esophageal cancers. Cancer Epidemiol. Biomark. Prev. 17, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 41.Gallicchio L., McSorley M.A., Newschaffer C.J.. et al. (2007) Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Detect. Prev. 31, 95–101 10.1016/j.cdp.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 42.Snoussi K., Strosberg A.D., Bouaouina N.. et al. (2006) Leptin and leptin receptor polymorphisms are associated with increased risk and poor prognosis of breast carcinoma. BMC Cancer 6, 38, PubMed Central PMCID: PMC1397853 10.1186/1471-2407-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willett E.V., Skibola C.F., Adamson P.. et al. (2005) Non-Hodgkin’s lymphoma, obesity and energy homeostasis polymorphisms. Br. J. Cancer 93, 811–816, PubMed Central PMCID: PMC2361643 10.1038/sj.bjc.6602762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woo H.Y., Park H., Ki C.S.. et al. (2006) Relationships among serum leptin, leptin receptor gene polymorphisms, and breast cancer in Korea. Cancer Lett. 237, 137–142 10.1016/j.canlet.2005.05.041 [DOI] [PubMed] [Google Scholar]
- 45.Skibola C.F., Holly E.A., Forrest M.S.. et al. (2004) Body mass index, leptin and leptin receptor polymorphisms, and non-hodgkin lymphoma. Cancer Epidemiol. Biomark. Prev. 13, 779–786 [PubMed] [Google Scholar]
- 46.Mahmoudi T., Farahani H., Nobakht H.. et al. (2016) Genetic variations in leptin and leptin receptor and susceptibility to colorectal cancer and obesity. Iran. J. Cancer Prev. 9, e7013, PubMed Central PMCID: PMC5038839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teras L.R., Goodman M., Patel A.V.. et al. (2009) No association between polymorphisms in LEP, LEPR, ADIPOQ, ADIPOR1, or ADIPOR2 and postmenopausal breast cancer risk. Cancer Epidemiol. Biomark. Prev. 18, 2553–2557 [DOI] [PubMed] [Google Scholar]
- 48.He B.S., Pan Y.Q., Zhang Y.. et al. (2012) Effect of LEPR Gln223Arg polymorphism on breast cancer risk in different ethnic populations: a meta-analysis. Mol. Biol. Rep. 39, 3117–3122 10.1007/s11033-011-1076-8 [DOI] [PubMed] [Google Scholar]
- 49.Ben Ali S., Kallel A., Sediri Y.. et al. (2009) LEPR p.Q223R Polymorphism influences plasma leptin levels and body mass index in Tunisian obese patients. Arch. Med. Res. 40, 186–190 10.1016/j.arcmed.2009.02.008 [DOI] [PubMed] [Google Scholar]
- 50.Hao J.Q., Zhang Q.K., Zhou Y.X.. et al. (2019) Association between circulating leptin concentration and G-2548A gene polymorphism in patients with breast cancer: a meta-analysis. Arch. Med. Sci. 15, 275–283, PubMed Central PMCID: PMC6425221 10.5114/aoms.2018.75638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin H.Y., Shi H., Li C.Y.. et al. (2015) LEP and LEPR polymorphisms in non-Hodgkin lymphoma risk: a systematic review and pooled analysis. J. BUON 20, 261–268 [PubMed] [Google Scholar]
- 52.Liu C. and Liu L. (2011) Polymorphisms in three obesity-related genes (LEP, LEPR, and PON1) and breast cancer risk: a meta-analysis. Tumour Biol. 32, 1233–1240 10.1007/s13277-011-0227-9 [DOI] [PubMed] [Google Scholar]
- 53.Wang L.Q., Shen W., Xu L.. et al. (2012) The association between polymorphisms in the leptin receptor gene and risk of breast cancer: a systematic review and pooled analysis. Breast Cancer Res. Treat. 136, 231–239 10.1007/s10549-012-2228-9 [DOI] [PubMed] [Google Scholar]
- 54.Luan H., Zhang H., Li Y.. et al. (2017) Association of two obesity-related gene polymorphisms LEPG2548A rs7799039 and LEPRQ223R rs1137101 with the risk of breast cancer. Oncotarget 8, 59333–59344, PubMed Central PMCID: PMC5601736 10.18632/oncotarget.19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu M.B., Xu H., Hu J.M.. et al. (2016) Genetic polymorphisms in leptin, adiponectin and their receptors affect risk and aggressiveness of prostate cancer: evidence from a meta-analysis and pooled-review. Oncotarget 7, 81049–81061, PubMed Central PMCID: PMC5348375 10.18632/oncotarget.12747 [DOI] [PMC free article] [PubMed] [Google Scholar]