Intestinal epithelial cells (IECs) are exposed to the low-oxygen environment present in the lumen of the gut. These hypoxic conditions on one hand are fundamental for the survival of the commensal microbiota and, on the other hand, favor the formation of a selective semipermeable barrier, allowing IECs to transport essential nutrients/water while keeping the sterile internal compartments separated from the lumen containing commensals.
KEYWORDS: HIF-1, barrier function, hypoxia, intestinal epithelial cells, miRNA, tight junctions
ABSTRACT
Intestinal epithelial cells (IECs) are exposed to the low-oxygen environment present in the lumen of the gut. These hypoxic conditions on one hand are fundamental for the survival of the commensal microbiota and, on the other hand, favor the formation of a selective semipermeable barrier, allowing IECs to transport essential nutrients/water while keeping the sterile internal compartments separated from the lumen containing commensals. The hypoxia-inducible factor (HIF) complex, which allows cells to respond and adapt to fluctuations in oxygen levels, has been described as a key regulator in maintaining IEC barrier function by regulating their tight junction integrity. In this study, we sought to better evaluate the mechanisms by which low oxygen conditions impact the barrier function of human IECs. By profiling miRNA expression in IECs under hypoxia, we identified microRNA 320a (miRNA-320a) as a novel barrier formation regulator. Using pharmacological inhibitors and short hairpin RNA-mediated silencing, we could demonstrate that expression of this microRNA (miRNA) was HIF dependent. Importantly, using overexpression and knockdown approaches of miRNA-320a, we could confirm its direct role in the regulation of barrier function in human IECs. These results reveal an important link between miRNA expression and barrier integrity, providing a novel insight into mechanisms of hypoxia-driven epithelial homeostasis.
INTRODUCTION
The human gastrointestinal (GI) tract is the organ forming the largest barrier toward the external environment and a key player in nutrient absorption (1). It consists of a monolayer of intestinal epithelial cells (IECs) separating the lamina propria from the lumen of the gut. This epithelium on the one hand allows for the translocation of nutrients, water, and electrolytes from the lumen to the underlying tissue and, on the other hand, builds up a tight barrier to prevent penetration of commensal bacteria and potential harmful microorganisms (bacterial and viral) to the lamina propria (1). Although these luminal microorganisms have well-characterized beneficial functions for the host, they can represent a risk when epithelial barrier and gut homeostasis are disrupted. Altered barrier functions increase the risk of enteric pathogen infection and can lead to the dysregulation of the mechanisms leading to the tolerance of the commensals which ultimately can lead to inflammation of the GI tract and the development of chronic diseases like inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC) (2, 3). Multiple cellular strategies are utilized to physically separate the content of the gut lumen from the host. First, goblet cells and Paneth cells in the mucosal lining secrete mucus, together with antimicrobial and antiviral peptides, which forms a layer of separation between the intestinal epithelial cells and the luminal content of the digestive tract (4–6). Second, epithelial cells polarize and express tightly juxtaposed adhesive junctional complexes between neighboring cells. These junctional complexes are composed of integral transmembrane proteins that are linked via intracellular scaffolding proteins to the actin cytoskeleton (7). This tight organization of intestinal epithelial cells inhibits paracellular diffusion of ions and other solutes, as well as antigenic material (8). The junctional complex therefore is essential for establishing and maintaining the barrier function of the mucosal layer and is composed of tight and adherens junction proteins such as claudins, occludin, junctional adhesion molecule A (JAM-A), tricellulin, zona occludens 1 (ZO-1), and E-cadherin (8). The interaction between the different tight junction and adherens junction proteins thus creates a tight epithelial barrier and determines selective permeability through the intestinal epithelium.
Within the physiological organization of the GI tract, an important but often overlooked parameter is the low oxygen level present in the lumen of the gut. This environment is fundamental for the survival of many commensals. Within the complex three-dimensional organization of the crypt-villus axis, the tip of the villus protrudes into the almost anoxic environment of the gut. The gut lumen is characterized by an oxygen partial pressure (pO2) below 10 mm Hg (9, 10). This oxygen level increases significantly along the crypt-villus axis where, within the mucosal lining, oxygen-rich blood vessels are located in the subepithelium, providing the stem cell containing crypts with a higher oxygen content of around pO2 of 60 to 85 mm Hg (11, 12). This environment of relatively low oxygen tension is also called physiologic hypoxia and has profound impact on many functions of the gut (10). Besides this oxygen gradient among the intestinal epithelium, the subepithelium of the GI tract is also exposed to daily fluctuations in oxygen content. After food ingestion, the intestinal blood flow and perfusion of the tissue increase and the oxygen content in the subepithelium rises, while under fasting conditions oxygen content decreases (10, 13, 14).
Cells respond to the hypoxic environment by specifically regulating the expression of hundreds of genes through the major hypoxic-induced transcription factor hypoxia-inducible factor (HIF) (15). HIFs are heterodimeric transcription factors that are composed of a constitutively expressed HIF-β subunit and one of the three oxygen-regulated alpha subunits (HIF-1α, HIF-2α, or HIF-3α) (16). Under normoxic conditions, HIF-1α is rapidly hydroxylated at specific proline residues by different prolyl hydroxylases (PHDs), leading to binding to the E3 ubiquitin ligase containing the von Hippel-Lindau (VHL) tumor suppressor protein, polyubiquitination, and subsequent proteasomal degradation of the protein (17). Under hypoxic conditions, lack of substrates such as Fe2+, 2-oxoglutarate, and O2 inhibits hydroxylation (18), therefore stabilizing HIF-1α and leading to dimerization with its constitutively expressed β-subunit (HIF-1β), translocation to the nucleus and binding of the coactivators CBP (CREB-binding protein) and p300 (19). This enables the complex to bind to target genes at the consensus sequence 5′-RCGTG-3′ (where R refers to A or G) and leads to formation of the transcription initiation complex (TIC) with subsequent expression of many genes that promote erythropoiesis, angiogenesis, glucose transport, and metabolism, all of which are needed to adapt to low-oxygen concentrations (20).
Beside the importance of hypoxia for the commensal flora, it has been shown that low-oxygen conditions also impact epithelial cells by inducing secretion of several proteins into the surrounding of the cells, including cytokines and growth factors (21). Precisely, in the context of epithelial barrier function, the intestinal trefoil factors (TFFs) exhibit intestinal cell-specific barrier-protective features and are specifically upregulated under hypoxia in an HIF-1α-dependent manner (22). The molecular mechanisms of TFF function and how they achieve the barrier protection is still not fully understood. Recent publications indicate a stabilizing effect on mucosal mucins (23), induction of cellular signals that modulate cell-cell junctions of epithelia leading to increased levels of claudin-1, impairment of adherens junctions, and facilitation of cell migration in wounded epithelial cell layers (24–26). In addition, it has been shown that pharmacological induction of hypoxia or loss of PHDs result in an increased barrier function due to a decreased amount of cell death by apoptosis, highlighting the importance of hypoxia for regulating the rate of proliferation in the maintenance of barrier function (27–29).
In recent years it has become appreciated that hypoxia additionally regulates the expression of an expanding but specific subset of microRNAs (miRNAs), termed hypoxamiRs (30, 31). miRNAs are endogenous, small noncoding RNAs that consist of 18 to 23 nucleotides. After transcription and subsequent maturation, the functional strand of the mature miRNA is loaded into the RNA-induced silencing complex (RISC), where it silences target mRNAs through mRNA cleavage, translational repression, or deadenylation (32). miRNAs coordinate complex regulatory events relevant to a variety of fundamental cellular processes (33). Although it has been shown that miRNAs can participate in the regulation of barrier function (34), it remains unclear whether the hypoxic environment in the lumen of the gut can induce the expression of a specific subsets of hypoxamiRs, which in turn will influence barrier function of the intestinal epithelium.
In the present study, we sought to investigate how hypoxia impacted the formation of a tight barrier in human intestinal epithelial cells. We found that human intestinal cells grown under hypoxic conditions more rapidly displayed barrier formation compared to cells grown under normoxia. We could correlate this improved barrier function with the faster assembly of the tight junction belt under low oxygen conditions. Through miRNA transcriptome microarray analysis, we identified three hypoxamiRs, miRNA-320a, miRNA-16-5p, and miRNA-34a-5p, known to play a role in barrier formation. Using overexpression and depletion experiments, we could demonstrate that miRNA-320a acts as a key player in promoting barrier formation in human intestinal epithelial cells under hypoxic conditions. Our data demonstrate that the hypoxic condition around intestinal epithelial cells regulates the expression of a specific subset of miRNAs, which in turn participate in the establishment of a fully functional epithelial barrier. Importantly, our work highlights the importance of studying the cellular functions of intestinal epithelial cells under their physiological hypoxic environment.
RESULTS
Low oxygen levels improve barrier function in human intestinal epithelial cells.
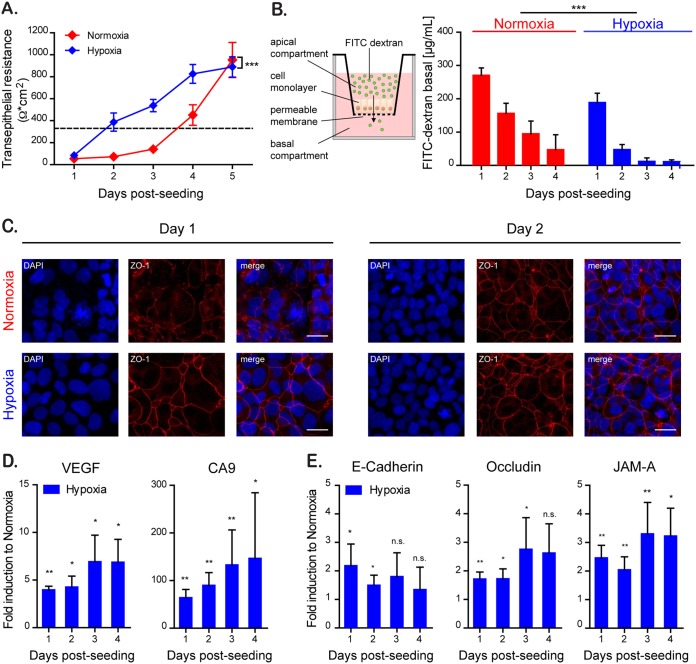
The gastrointestinal tract is characterized by a steep oxygen gradient along the crypt-villus axis with high levels of oxygen at the bottom of the crypts and a low-oxygen environment at the tip of the villi (11). Several studies (22, 28, 35) have shown that low oxygen concentrations can influence the barrier function of epithelial cells in vitro by changing gene expression profiles and inducing secretion of barrier-regulating proteins, i.e., TFFs. To investigate the mechanism by which hypoxic conditions regulate barrier function, the T84 colon adenocarcinoma-derived cell line was seeded onto Transwell inserts (Costar 3415; Corning) and allowed to polarize under normoxic (21% O2) or hypoxic (1% O2) conditions. To determine the effect of hypoxia on the ability of T84 cells to form a tight barrier, transepithelial electrical resistance (TEER) measurements were performed at 24-h intervals for 5 days. TEER is a well-characterized method used to quickly access barrier function characterized by the rise in the electrical resistance over a cell monolayer. Similar to our previous observations (36), normoxic cells reached a polarized state and acquired a fully functional barrier function within 4 to 5 days postseeding (Fig. 1A). However, T84 cells cultured under hypoxic conditions established their barrier function significantly faster compared to cells under normoxic conditions, reaching a polarized state within 2 days postseeding (Fig. 1A). To further assess paracellular permeability and the integrity of the IEC monolayer, the diffusion of fluorescein isothiocyanate (FITC)-labeled dextran across the epithelial monolayer was measured (Fig. 1B). In this assay, when cells are nonpolarized, dextran added to the apical chamber of a Transwell insert is able to rapidly diffuse to the basal compartment. However, upon cellular polarization and creation of a tight barrier, the FITC-dextran is retained in the apical chamber. The results show that, similar to the rapid increase in TEER measurements, T84 cells grown under hypoxic conditions are able to more quickly control FITC-dextran diffusion from the apical into the basal compartment of the Transwell. This indicates that a tight barrier function has been achieved faster under hypoxia compared to normoxia (Fig. 1B). This increase in barrier function was rapid and was already apparent at 1 day postseeding. To determine whether the increase in the rate of polarization and barrier formation was also apparent at the level of the tight junction belt, T84 cells were seeded onto Transwell inserts and the formation of tight junctions was monitored by indirect immunofluorescence of ZO-1 and by quantitative PCR (qPCR) for the tight and adherens junction proteins E-cadherin (CDH1), occludin (OCLN), and junctional adhesion molecule 1 (F11R/JAM-A). The results show that similar to results of the TEER and dextran diffusion assay, cells cultured under hypoxic conditions already showed, within 1 day of seeding, a well-defined tight junction belt characterized by the classical cobblestone pattern. In contrast, cells grown under normoxic conditions did not have well-defined tight junctions 1 day postseeding, and this coincided with the presence of dispersed ZO-1 protein in the cytosol of the cells (Fig. 1C). To address whether hypoxia induces an upregulation of barrier-function protein expression, we performed a quantitative reverse transcription-PCR (qRT-PCR) analysis of cells grown under normoxic and hypoxic conditions. As positive controls, we used the two archetypical hypoxia-driven genes vascular endothelial growth factor (VEGF) and carbonic anhydrase 9 (CA9) genes and confirmed that they were upregulated when cells were cultured under hypoxia (Fig. 1D). Importantly, mRNA expression of the junction proteins E-cadherin, occludin, and JAM-A was increased under hypoxia. E-cadherin showed a higher induction initially after hypoxic culture, while occludin and JAM-A required a prolonged treatment under hypoxia to show increases in their expression (Fig. 1E). Altogether, these results suggest that hypoxia favors the establishment of barrier function in T84 cells.
FIG 1.
Hypoxia improves barrier function in intestinal epithelial cells. T84 cells were seeded onto Transwell inserts and cultured for the indicated time under normoxic (21% O2) (red) or hypoxic conditions (1% O2) (blue). (A) The rate of TEER increase over the cell monolayer was measured every 24 h using the EVOM2 chopstick electrode. A TEER of >330 Ω ⋅ cm2 indicates complete barrier formation and is marked by a dotted line (36). (B) Paracellular permeability of the cell monolayer on Transwell inserts was assessed by adding 4-kDa FITC-dextran to the apical compartment (schematic overview [left panel]). At 3 h postincubation, the basal medium was analyzed for an increase of fluorescence by spectrofluorimetry (right panel). (C) T84 cells cultured for 24 and 48 h under normoxic and hypoxic conditions were evaluated for the expression of the tight junction protein ZO-1 (red). Cell nuclei were stained with DAPI (blue). Scale bar, 25 μm. A representative image is shown. (D) RNA samples of normoxic and hypoxic cultures of T84 cells taken at 24-h intervals for 4 days were analyzed by qPCR for the expression of the hypoxia-induced VEGF and CA9 genes. The results are normalized to normoxic values for each day. (E) RNA samples of normoxic and hypoxic cultures of T84 cells were analyzed over time by qPCR for the expression of the tight junction proteins E-cadherin, occludin, and JAM-A. The results are normalized to normoxic values for each day. (A and B) Values shown represent means ± the standard errors of the mean (SEM) (n = 9) from triplicate experiments. ***, P < 0.0001 (two-way analysis of variance [ANOVA]). (D and E) Experiments were performed in quadruplicate. Error bars indicate the standard deviations (SD). *, P < 0.05; **, < 0.01; n.s., not significant (one-sample t test on log-transformed fold changes).
Chemical stabilization of HIF by DMOG induces a faster barrier formation.
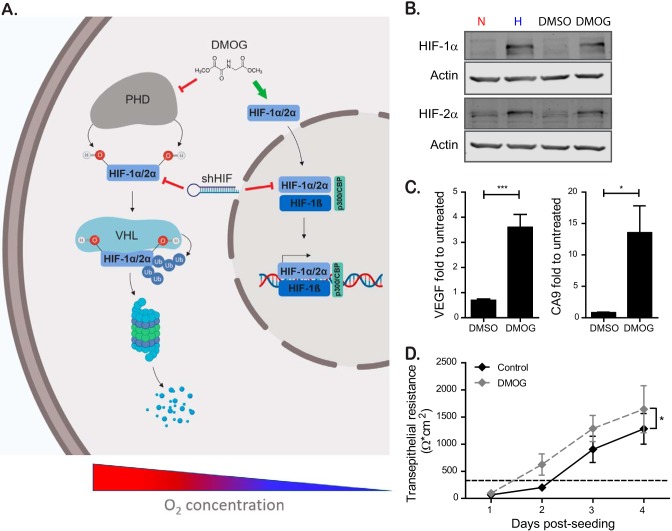
The main transcription factors involved in cellular response following changes in oxygenation are HIF-1α and HIF-2α. To address whether the phenotype of faster barrier establishment under hypoxia was dependent on the activation of HIF1-α or HIF-2α, we aimed at mimicking the hypoxic conditions using the pharmacological HIF-1α/2α activator dimethyloxaloylglycine (DMOG). DMOG exerts its function by inhibiting PHDs, which under normoxic conditions induce the degradation of HIF-1α and HIF-2α (HIF-1α/2α) (28). Therefore, DMOG treatment of normoxic cells stabilizes HIF-1α/2α, allowing for its translocation to the nucleus and induction of HIF-responsive elements (HRE) dependent gene expression (Fig. 2A). To confirm that DMOG was capable of stabilizing HIF-1α/2α, T84 cells were treated with DMOG, and the levels of HIF-1α and HIF-2α were assessed by Western blot analysis. The results show that DMOG treatment induces stabilization of both HIF-1α and HIF-2α to similar levels compared to cells incubated under hypoxic conditions (Fig. 2B). To further confirm that DMOG treatment leads to the upregulation of transcripts known to be induced by HIF-1α/2α, the transcriptional upregulation of the archetypical HIF-1α target proteins VEGF and CA9 were assessed by qPCR. The results show that, similar to hypoxic treatment (Fig. 1D), DMOG treatment results in the significant upregulation of both VEGF and CA9 (Fig. 2C). Finally, to determine whether DMOG treatment can phenocopy hypoxia and can induce an increase in the rate of barrier formation, T84 cells were seeded onto Transwell inserts and incubated under normoxic conditions in the presence or absence of DMOG. The barrier function was assessed by monitoring TEER in 24-h intervals over a 4-day time course. In line with our previous observations, DMOG-treated cells established their barrier function faster than the solvent-treated control cells (Fig. 2D). Together, these results demonstrate that a pharmacological approach to mimic hypoxic conditions promotes barrier function in T84 cells.
FIG 2.
Chemical stabilization of HIF by DMOG to mimic hypoxia induces a faster barrier formation. (A) Schematic showing the regulation of the transcription factor HIF-1α/2α at high and low oxygen concentrations. Under normoxic conditions, HIF-1α/2α is hydroxylated at two specific proline residues by different prolyl hydroxylases (PHDs), leading to binding to the E3 ubiquitin ligase containing the von Hippel-Lindau (VHL) tumor suppressor protein. This mediates the polyubiquitination of HIF-1α/2α and its downstream proteasomal degradation. Under hypoxic conditions, degradation is inhibited due to the lack of substrate for the PHDs, therefore stabilizing HIF-1α/2α, leading to dimerization with its constitutively expressed β-subunit (HIF-1β) and subsequent gene expression. The pharmacological activation of HIF-1-function by DMOG and inhibition by shRNA against HIF mRNA are shown. (B) T84 cells were incubated in normoxic (N) or hypoxic (H) conditions for 6 h or treated with dimethyl sulfoxide (DMSO) or DMOG for 6 h. Protein was harvested, and Western blotting was performed for HIF-1α and HIF-2α. Actin was used as a loading control. Each experiment was performed in triplicate. A representative image is shown. (C) T84 cells were seeded and incubated under normoxic conditions in the presence or absence of DMOG. DMSO was used as a solvent control. RNA was isolated, and the upregulation of VEGF and CA9 was evaluated by qPCR. The results are normalized to untreated cells. (D) T84 cells were seeded on Transwell inserts and incubated under normoxic conditions in the presence or absence of DMOG. DMSO was used as a solvent control. TEER measurements were taken in 24-h intervals for 4 days. A TEER of >330 Ω ⋅ cm2 indicates complete barrier formation and is marked by a dashed line. The values shown represent means ± the SEM (n = 9) from triplicate experiments. (C) *, P < 0.05; ***, P < 0.001 (one-sample t test on log-transformed fold changes). (D) *, P = 0.0417 (two-way ANOVA).
Increased barrier formation induced by hypoxia is HIF-1α dependent.
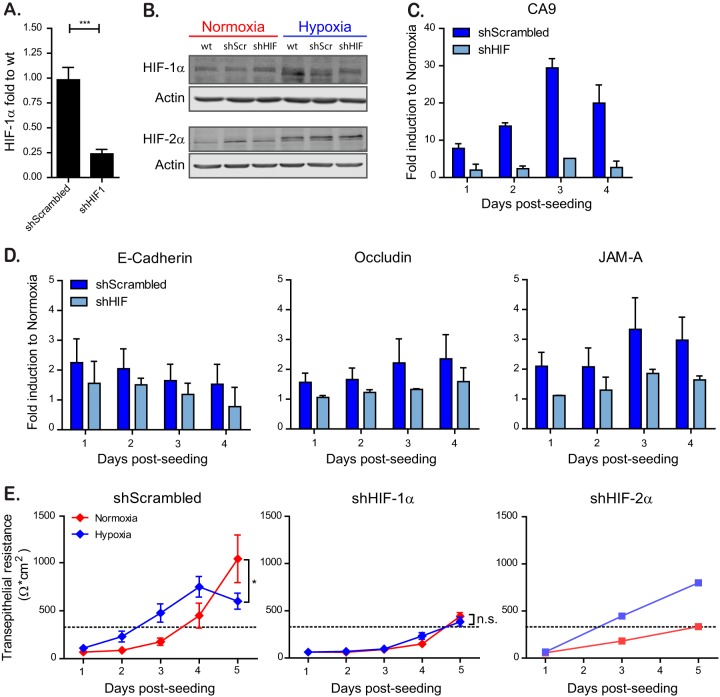
To address whether the observed increase in barrier function was HIF-1α dependent, HIF-1α was knocked down by lentiviral transduction of shRNAs (Fig. 2A). Quantification of HIF-1α knockdown efficiency revealed a 75% reduction of HIF-1α mRNA compared to cells expressing a scrambled shRNA control (shScrambled) (Fig. 3A). Similarly, this downregulation of HIF-1α expression was confirmed by Western blot analysis, where cells expressing an shRNA against HIF-1α grown under hypoxia show a decreased level of HIF-1α compared to WT cells or shScrambled cells (Fig. 3B). Importantly, expression of an shRNA against HIF-1α had no effect on the induction of HIF-2α (Fig. 3B). In addition, downregulation of HIF-1α led to an inhibition of the transcriptional induction of the archetypical HIF-1α-dependent protein CA9 under hypoxic conditions (Fig. 3C). Interestingly, knockdown of HIF-1α resulted in the loss of tight and adherens junction protein upregulation under hypoxia (Fig. 3D). Finally, knockdown of HIF-1α abolished the faster barrier formation under hypoxic conditions, as seen by similar TEER values under normoxic and hypoxic conditions (Fig. 3E). These results were HIF-1α specific since knockdown of HIF-2α did not impact the hypoxia-induced barrier function formation (Fig. 3E). Interestingly, cells expressing HIF-1α shRNA exhibited a slower barrier formation in comparison to scrambled shRNA-expressing cells even under normoxic conditions, revealing a general dependency of barrier formation on HIF-1α even in normal oxygen levels (Fig. 3E). These findings were not due to cytotoxic effects of knocking down HIF-1α since cell proliferation and viability assays did not show any differences between wild type (WT), shScrambled, and cells overexpressing an shRNA against HIF-1α (data not shown). Altogether, these results strongly suggest that faster establishment of barrier function in T84 cells observed under hypoxic conditions is HIF-1α dependent.
FIG 3.
HIF-1α is responsible for faster barrier establishment under hypoxic conditions. T84 cells were transduced with lentiviruses expressing an shRNA targeting HIF-1α or a scrambled control. (A) Knockdown of HIF-1α was controlled by qPCR. (B) T84 cells knocked down for HIF-1α or expressing a scrambled control were incubated under normoxic or hypoxic conditions. At 6 h postincubation, protein samples were harvested and evaluated by Western blotting for HIF-1α or HIF-2α. Each experiment was performed in triplicate; a representative image is shown. (C) T84 cells expressing a scramble shRNA or a shRNA against HIF-1α were evaluated for their induction of the hypoxic marker CA9 over the indicated time course by qPCR. The results are normalized to normoxic values for each day. (D) T84 cells expressing a scramble shRNA or a shRNA against HIF-1α were evaluated for their induction of tight junction proteins over the indicated time course by qPCR. The results are normalized to normoxic values for each day. (E) T84 cells expressing a scramble shRNA or a shRNA against HIF-1α or HIF-2α were seeded on Transwell inserts. Cells were incubated in normoxic or hypoxic conditions, and TEER measurements were taken at 24-h intervals for 5 days. A TEER of >330 Ω ⋅ cm2 indicates complete barrier formation and is marked by a dotted line. Values shown represent means ± the SEM (n = 9) from triplicate experiments for shScrambled and shHIF-1α. For shHIF-2α, one representative experiment is shown of two different anti-HIF-2α shRNAs tested. *, P < 0.05; n.s., not significant (two-way ANOVA). (A, C, and D) Values shown represent the means plus the SD from three independent experiments (***, P < 0.001; one-sample t test on log-transformed fold changes).
Whole-transcriptome miRNA profiling reveals regulation of several miRNAs that are involved in barrier formation.
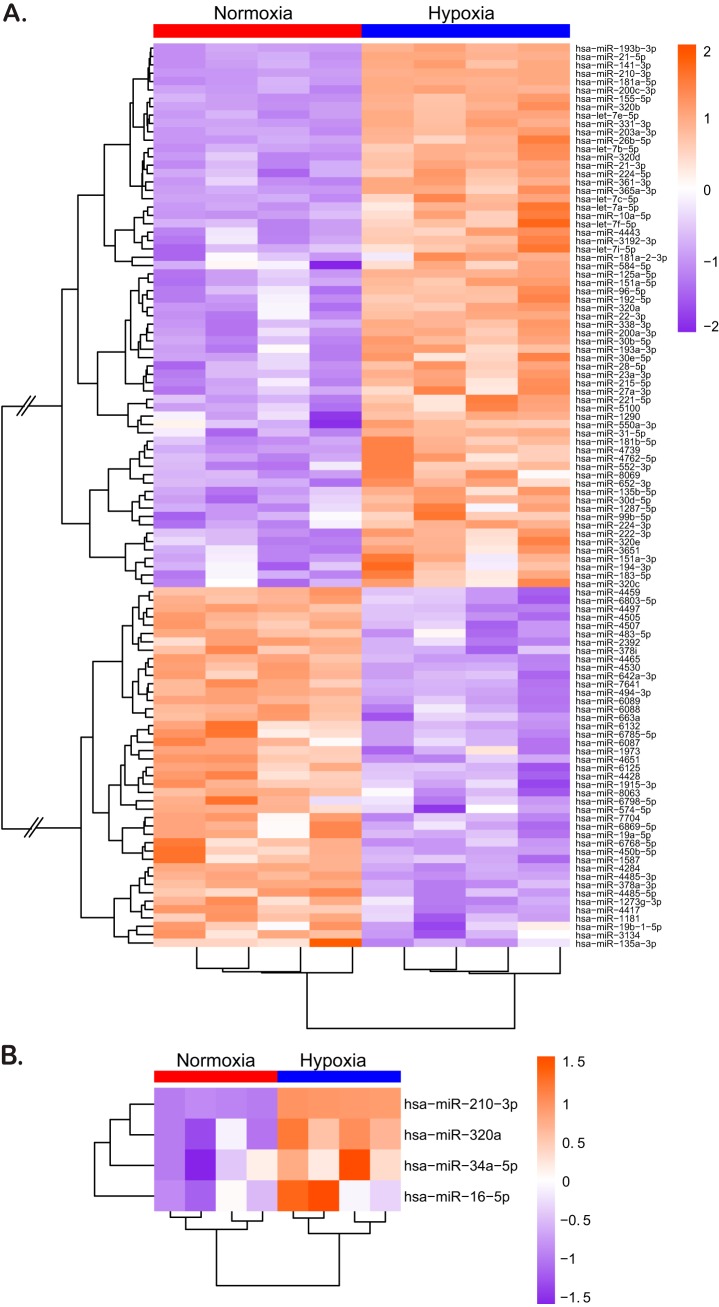
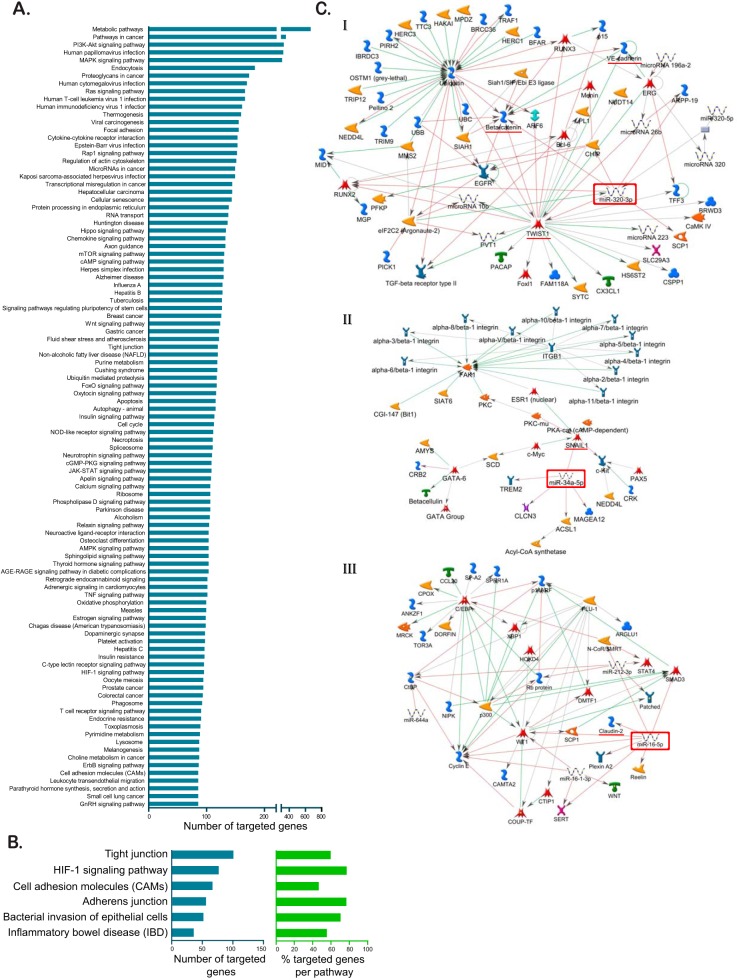
Since significant differences in the barrier state between hypoxic and normoxic conditions could be observed already 24 h after seeding, we hypothesized that the very fast changes in protein expression and barrier establishment must occur within hours after exposure of the cells to hypoxic conditions. Several proteins (22, 24) have been shown to contribute to mucosal repair and barrier formation in intestinal cells, but the role of miRNAs in fine-tuning gene expression involved in barrier formation has recently become appreciated (37). So far, most of these studies have been conducted only under normoxic conditions, hence overlooking the physiological hypoxic conditions of the gut. To directly address the role of miRNAs in regulating barrier functions of IECs under low-oxygen conditions, miRNAome microarray analysis was used for cells incubated under normoxic or hypoxic conditions. This allowed us to broadly screen hypoxia-regulated miRNAs, the so-called hypoxamiRs. By comparing the miRNA expression patterns from normoxic and hypoxic conditions, we could identify a total of 108 differentially regulated hypoxamiRs of which 65 were upregulated and 43 were downregulated under hypoxic conditions (Fig. 4A). Detailed analysis of hypoxamiR expression revealed that upon hypoxic exposure, T84 cells highly upregulate miRNA-210-3p expression (Fig. 4A). This miRNA is a master regulator for adaptation to low oxygen concentration (31) and is a well-characterized hypoxamiR whose expression is strongly linked to hypoxic conditions. This upregulation of miRNA-210-3p strongly suggests that T84 cells have established a hypoxia-specific transcription profile. To probe for miRNAs that could regulate barrier function, we performed KEGG and MetaCore-driven pathway analysis (Fig. 5). This allowed us to identify potential hypoxamiRs involved in barrier function establishment (Fig. 4B and Fig. 5C). To narrow down the list of potential miRNAs involved in regulating barrier function, we decided to focus our attention on three miRNAs for which previous reports described involvement in barrier function. These are miRNA-320a, miRNA-34a-5p, and miRNA-16-5p (Fig. 4B and Fig. 5C). miRNA-320a has been shown to be crucial for intestinal barrier integrity through modulation of the regulatory subunit PPP2R5B of phosphatase PP2A (38). In addition, miRNA-320a was found to both target β-catenin directly (39) and VE-cadherin through inhibition of the transcriptional repressor TWIST1 (40, 41). miRNA-34a-5p has been shown to serve as an inhibitor for the zinc finger transcription factor Snail (42, 43), which in turn functions as a transcriptional repressor of the adherens and tight junction proteins E-cadherin, claudin, and occludin (44–46). Interestingly, we recently (47) determined that miRNA-16-5p acts as a regulator of claudin-2 and cingulin expression and its expression negatively correlated with occurrence of irritable bowel syndrome (IBS) in patients, therefore playing a key role in modulating barrier function.
FIG 4.
Hypoxia leads to changes in expression of several hypoxamiRs known to regulate barrier function. T84 cells were incubated under hypoxic or normoxic conditions for 48 h. miRNA was isolated and evaluated by miRNA microarray. (A and B) Heatmaps of differentially expressed miRNAs in T84 cells cultured under normoxic and hypoxic conditions. The color scale shown on the right illustrates the relative expression levels of differentially expressed miRNAs. Orange indicates upregulated miRNAs (>0), and purple indicates downregulated miRNAs (<0). (A) Heatmap for 108 differentially regulated hypoxamiRs that were significantly up- or downregulated compared to normoxic conditions. Connecting lines in the cluster dendrogram between up- and downregulated miRNAs were shortened to enable visualization (indicated by two skewed lines). (B) Heatmap of miRNA-210-3p (positive control for hypoxic conditions), miRNA-320a, miRNA-34a-5p, and miRNA16-5p, identified by pathway analysis for playing a putative role in barrier formation. For both panels A and B, samples were tested in quadruplicate, and the level of expression of each replicate is shown in the heatmap.
FIG 5.
Pathway analysis by KEGG and MetaCore reveals miRNAs which regulate tight and adherens junction proteins. (A) Target genes of significantly regulated miRNAs were retrieved from miRTarBase database v6.1 and subjected to KEGG pathway analysis. The 100 most targeted pathways by number of targeted genes are shown. (B) Number of targeted genes and percentages of targeted genes per pathway for barrier formation related pathways. (C) MetaCore-driven pathway analysis identified three potential hypoxomiRs involved in barrier function establishment. Interaction maps are shown for miRNA-320a (I), miRNA-34-a-5p (II), and miRNA-16-5p (III). The miRNA of interest is marked by a red square; targeted proteins involved in barrier formation are underlined in red.
To validate the results of the miRNA microarray profiling, we performed qRT-PCR analysis for these specific miRNAs. As observed in our microarray approach, miRNA-210-3p, miRNA-320a, miRNA-34a-5p, and miRNA-16-5p were upregulated under hypoxic conditions in T84 cells 24 or 48 h postseeding (Fig. 6A). On the contrary, qRT-PCR analysis of a miRNA for which expression is not affected by hypoxic conditions (miRNA-1268a) showed no difference between normoxia and hypoxia (Fig. 6A). Since T84 cells are immortalized cells derived from carcinoma, they may show altered gene regulation, protein expression, and signaling pathways. To verify that the observed hypoxia-dependent upregulation of barrier function-related miRNAs was not an artifact of the cancerogenic nature of the T84 cells, stem cell-derived primary intestinal epithelial cells, so called human mini-gut organoids, were used. Organoids are primary cell cultures and thereby retain key features like structural architecture and all major cell lineages present in the inner lining of the gut, hence mimicking the physiological organization of the human gut epithelium in vivo (48). In line with the results found in T84 cells, qRT-PCR confirmed upregulation of all four tested targets under hypoxic conditions by 24 or 48 h postseeding in our human intestinal organoids (Fig. 6B). Our observations made both in immortalized carcinoma-derived cell lines and in primary human IECs therefore confirm the increased expression of the hypoxamiRs miRNA-320a, miRNA-34a-5p, and miRNA-16-5p under hypoxic conditions in the human intestinal epithelial cells.
FIG 6.
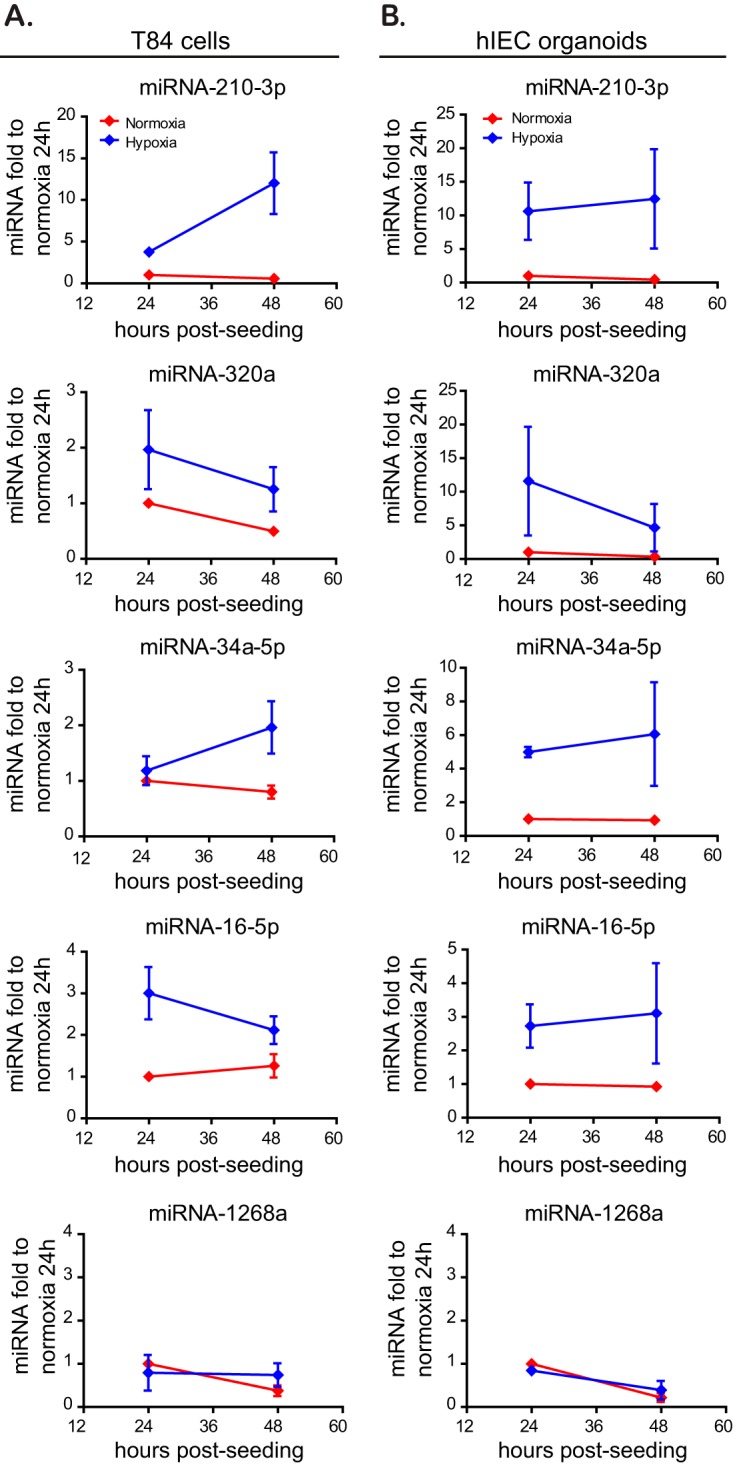
Validation of upregulated hypoxamiRs in T84 cells and primary human mini-gut organoids. T84 (A) and human intestinal epithelial cells (hIEC) cultured as mini-gut organoids (B) were incubated for 24 and 48 h under hypoxic or normoxic conditions. Upregulation of specific miRNAs was evaluated by qRT-PCR. miRNA-210-3p was used as a positive control for a miRNA upregulated under hypoxia. miRNA-1268a was used as a negative control since its expression is not hypoxia dependent. Data were normalized to normoxic cells at 24 h. All experiments were performed in triplicate. Error bars indicate the SEM.
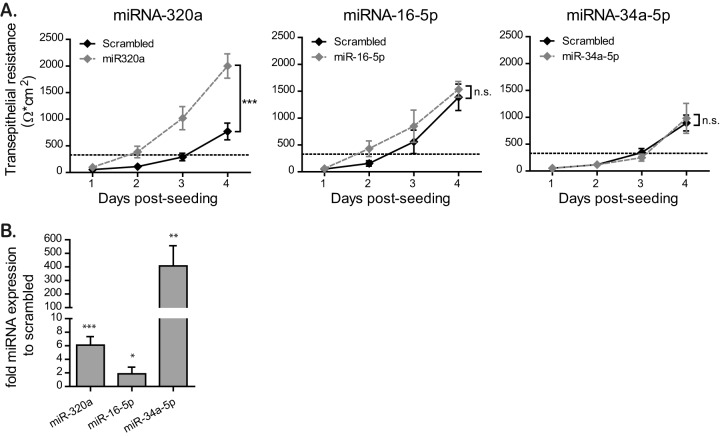
Overexpression of miRNA-320a induces faster barrier formation in T84 cells.
Our above results indicate that miRNA-320a, miRNA-34a-5p and miRNA-16-5p are upregulated under hypoxic conditions. To directly validate that these hypoxamiRs are responsible for the observed improved barrier function under hypoxia, we stably overexpressed these miRNAs in T84 cells by lentiviral transduction. qRT-PCR analysis revealed that all miRNAs were indeed overexpressed in our T84 cells (Fig. 7B). Of note, we achieved only a very low overexpression of miR-16-5p (Fig. 7B). miRNA-overexpressing T84 cells were seeded on Transwell inserts, and their barrier formation was monitored by TEER measurements at 24-h intervals (Fig. 7A). The results show that miRNA-320a-overexpressing cells exhibited a significantly faster barrier formation than scrambled miRNA-expressing cells. miRNA-16-5p- and miRNA-34a-5p-expressing cells showed no significant alteration in barrier formation compared to scrambled miRNA cells (Fig. 7A). Taken together, these data support a model wherein miRNA-320a has a key role in regulating barrier function in intestinal epithelial cells.
FIG 7.
Overexpression of miRNA-320a induces faster barrier formation in T84 cells. (A) T84 cells stably overexpressing miRNA-320a, miRNA-16-5p, and miRNA-34a-5p by lentiviral transduction were seeded onto Transwell inserts, and barrier formation was assessed by TEER measurement in 24-h intervals over 4 days. A TEER of >330 Ω ⋅ cm2 indicates complete barrier formation and is marked by a dotted line. Values shown represent means ± the SEM (n = 6 [miRNA-16-5p and miRNA-34a-5p] or n = 12 [miRNA-320a]) from triplicate or quadruplicate experiments, respectively. ***, P = 0.0002 (two-way ANOVA); n.s., not significant. (B) T84 cells overexpressing miR-320a, miR-16-5p, and miR-34a-5p were harvested, and the overexpression of each miRNA was controlled by qRT-PCR. Values shown represent means ± the SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-sample t test on log-transformed fold changes).
Inhibition of miRNA-320a expression diminishes barrier formation in T84 cells.
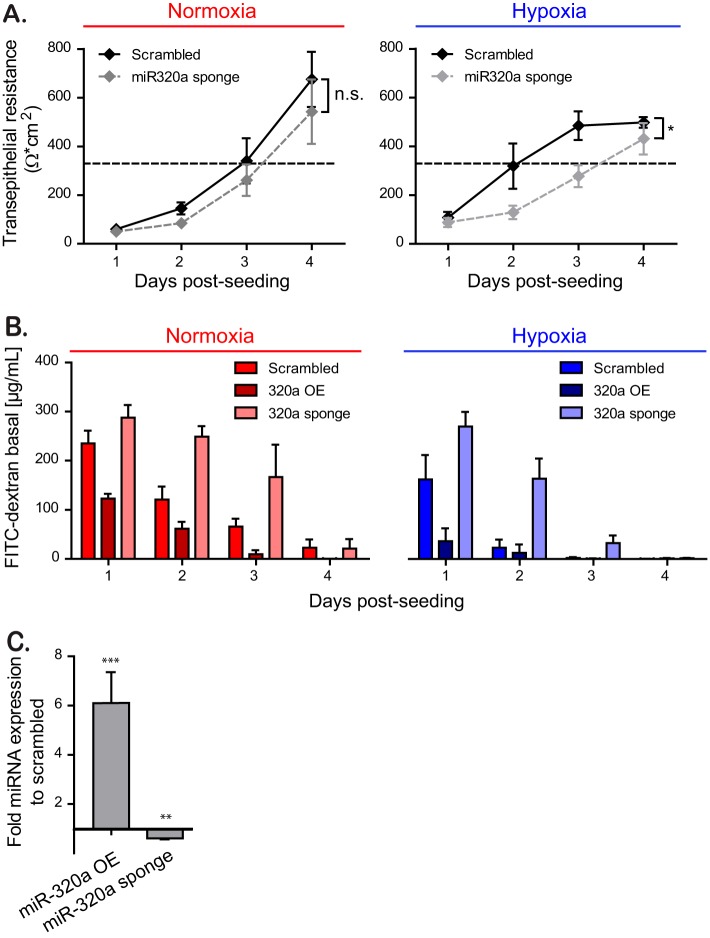
To confirm the role of miRNA-320a in increasing barrier formation under hypoxic conditions, we generated T84 cells expressing a miRNA-320a sponge. We confirmed through qRT-PCR that overexpression of the miRNA sponge leads to a downregulation of miRNA-320a expression (Fig. 8C). In line with our previous results, T84 cells expressing a miRNA-320a sponge displayed a slower establishment of barrier function in comparison to scrambled transduced cells under hypoxic conditions (Fig. 8A). To confirm the role of miRNA-320a in regulating barrier function, T84 cells overexpressing miRNA-320a or depleted of miRNA-320a were seeded on Transwell inserts, and their barrier integrity was monitored using a FITC-dextran diffusion assay. In line with our previous results, miRNA-320a-overexpressing cells show a reduced flux of FITC-dextran to the basal compartment of the Transwell chamber, while cells depleted of miRNA-320a show an increased flux compared to scrambled miRNA cells (Fig. 8B). Taken together, these findings strongly suggest a model where hypoxia-induced expression of miRNA-320a directly regulates the establishment of a functional barrier in the epithelial cells lining the gastrointestinal tract.
FIG 8.
Inhibition of miRNA-320a expression diminishes barrier formation in T84 cells. (A) T84 cells stably expressing miRNA-320a sponge were seeded onto Transwell inserts under normoxic or hypoxic conditions and barrier function was assessed by TEER measurements in 24-h intervals over 4 days. A TEER of >330 Ω ⋅ cm2 indicates complete barrier formation and is marked by a dotted line. Values shown represent means ± the SEM (n = 12) from quadruplicate experiments. *, P = 0.0174 (two-way ANOVA). (B) Paracellular permeability of T84 cells overexpressing the miRNA-320a (overexpression [OE]) or the miRNA-320a sponge. The barrier function on Transwell inserts was assessed by adding 4 kDa FITC-dextran to the apical compartment and measuring the diffusion of fluorescent FITC-dextran to the basal compartment at 3 h posttreatment. A paracellular permeability assay was performed every 24 h for 4 days. Values shown represent the means ± the SEM (n = 3) from triplicate experiments. (C) T84 cells overexpressing (OE) miR-320a or depleted of miR-320a by expression of a sponge were evaluated by qRT-PCR. Values shown represent means ± the SD from three independent experiments. **, P < 0.01; ***, P < 0.001 (one-sample t test on log-transformed fold changes).
miRNA-320a expression is dependent on HIF-1α.
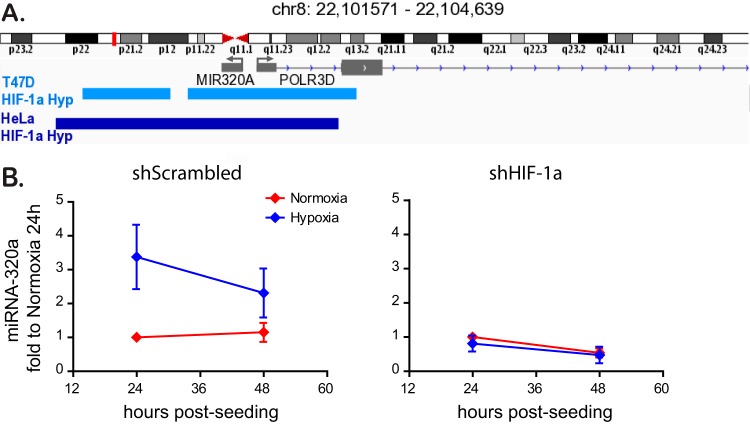
Finally, to investigate whether hypoxia leads to the induction of miRNA-320a in an HIF-1α-dependent manner, we analyzed the promoter region of miRNA-320a. We could identify several HIF-1α binding motives, marked by the HRE consensus sequence 5′-RCGTG-3′, clustered upstream of the promoter of miRNA-320a, strongly indicating a binding of this transcription factor under hypoxic conditions to facilitate miRNA-320a expression (data not shown). In addition, publicly available HIF-1α ChIP-Seq data sets from both breast cancer T47D cells and HeLa cells grown under hypoxia were analyzed. In both cases, binding of HIF-1α was reported over the full length of the genomic region for miRNA-320a (Fig. 9A). These data strongly suggest that HIF-1α directly regulates miRNA-320a expression. To confirm this, HIF-1α knockdown cells cultured under normoxic and hypoxic conditions were analyzed for the expression of miRNA-320a. We could show that knockdown of HIF-1α abrogates the previously observed upregulation of miRNA-320a by hypoxia (Fig. 9B). These data strongly implicate HIF-1α as the transcription factor primarily responsible for the hypoxic induction of miRNA-320a under a low oxygen concentration to enhance barrier formation in intestinal epithelial cells.
FIG 9.
HIF-1α binds to the promoter region of miRNA-320a and controls its expression. (A) HIF-1α binding sites around the miRNA-320a genomic region were analyzed from ChIP-seq data sets of breast cancer T47D (light blue) and HeLa cells (dark blue) cultured under hypoxic conditions. Part of chromosome 8 which contains the coding sequence for miRNA-320a is displayed. (B) T84 cells expressing a scrambled shRNA or a shRNA directed against HIF-1α were incubated under normoxic or hypoxic conditions. At 24 and 48 h postincubation, the levels of miRNA-320a were analyzed by qRT-PCR. Experiments were performed in triplicate; error bars indicate means ± the SEM.
DISCUSSION
In this work, we demonstrate that the physiological hypoxic environment improves intestinal epithelial barrier function of T84 cells as shown by the faster establishment of transepithelial electrical resistance, by the more rapid decrease in barrier permeability to FITC-dextran, as well as by the faster establishment of the tight junction belt compared to normoxic conditions. Using pharmacological inhibitor and knockdown approaches, we could show that this increased barrier function is dependent on the hypoxia regulator HIF-1α. In addition, using a miRNA microarray approach, we identified miRNA-320a as a key miRNA induced under hypoxia being directly responsible for regulating barrier functions in human intestinal epithelial cells. We could demonstrate that its overexpression is sufficient to promote barrier function in epithelial cells while interfering with its expression under hypoxic conditions counteracts the hypoxia-mediated barrier formation establishment. In silico analysis of the genomic region of miRNA-320a, as well as evaluation of miRNA-320a expression in cells expressing a shRNA against HIF-1α, suggests a direct regulation of miRNA-320a by HIF-1α. Taken together, our results show that miRNA-320a is a hypoxia-induced miRNA that plays a key role in regulating barrier function in human intestinal epithelial cells.
The importance of hypoxic conditions in regulating barrier function in intestinal epithelial cells has been previously studied, and several potential mechanisms highlight the central role of the transcription factor HIF. It was shown that specific shRNA-mediated knockdown of HIF-1β in T84 and Caco-2 cells resulted in the decrease of claudin-1 expression on mRNA and protein level accompanied by defects in barrier function and abnormal morphology of tight junctions (49). This is thought to be a direct effect from the HIFs themselves since HIF responsive elements have been identified in the promoter region of claudin-1 (49).
One of the best-characterized means by which hypoxia induces barrier formation involves the HIF-dependent expression of the intestinal trefoil factor (TFFs). The trefoil factor family consists of three peptides: TFF1, TFF2, and TFF3; all three are widely distributed in the gastrointestinal tract and are present in virtually all mucosal membranes (50). Recently, TFFs have been shown to induce a stabilizing effect on mucosal mucins (23). In addition, the Van-Gogh-like protein 1 (Vangl1) was identified as a downstream effector of TFF3 and described to mediate wound healing in IECs, thereby promoting recovery of barrier function under condition of local loss of epithelium integrity (24). Importantly, TFF3 also regulates the expression of tight junctions and adherens junctions in IECs by elevating the levels of claudin-1 and downregulating the expression of E-cadherin (25). This further activates the phosphatidylinositol 3-kinase/Akt signaling pathway, which leads to an increase in barrier function and altered proliferation of cells in the intestinal epithelium (26, 51). Extensions of these studies in vivo revealed the protective role of TFFs on intestinal permeability and barrier function, since both administration of TFFs, as well as administration of a novel PHD inhibitor (FG-4497), were protective and had a beneficial influence on clinical symptoms (weight loss, colon length, tissue tumor necrosis factor alpha [TNF-α]) in a mouse colitis model (29, 52). Correspondingly, HIF-1α was found to be highly expressed in Crohn’s disease and ulcerative colitis patients (53) and seems to play a protective role in inflammatory bowel disorders through improvement of epithelial barrier function (54). It has been suggested that HIF-1α helps to control intestinal inflammation by interacting with the inflammation transcription factor nuclear factor κB (NF-κB) (55).
To date, most of the work aimed at understanding the effect of hypoxia on barrier function in the gut has focused on the transcripts and proteins that are induced under hypoxia. In the emerging field of miRNA, several miRNAs have been identified as potential regulators of barrier function. However, to the best of our knowledge, these miRNAs were not studied under hypoxic conditions but in normal cell culture conditions or in patient samples with inflammatory diseases. For example, McKenna et al. demonstrated that claudin-4 and claudin-7 were not expressed in the apical membrane of intestinal epithelial cells in Dicer1-deficient mice, resulting in impaired intestinal barrier function, thus strongly supporting the importance of miRNA regulation in barrier formation (56). In addition, overexpression of miRNAs has been linked to a regulation of barrier function in intestinal epithelial cells (38, 57). miRNA-31 was found to increase the TEER by decreasing the transepithelial permeability through interaction with tumor necrosis factor superfamily member 15 (TNFSF15) in Caco2-BBE cells (58). Of note, the TNFSF15 gene is a well-known risk gene involved in the pathogenesis of inflammatory bowel syndrome (IBS) and inflammatory bowel disease (59, 60). miRNA-26b was found to regulate the Ste20-like proline/alanine rich kinase (SPAK) involved in epithelial barrier integrity (61), and overexpression of miRNA-21 in patients with ulcerative colitis has been associated with the impaired intestinal epithelial barrier function through targeting the Rho GTPase RhoB (62). We recently identified miRNA-16 and miRNA-125b as being downregulated in patients suffering from IBS with diarrhea and determined that these two miRNAs modulated the tight junction proteins claudin-2 and cingulin (47).
Similar to our work, miRNA-320a was previously reported to play a role in barrier function under normoxic conditions. Cordes et al. could show a functional role of miRNA-320a in stabilizing the intestinal barrier function through reinforcement of barrier integrity in T84 cells and in a murine colitis model (38). These researchers suggest that this is due to a potential modulation of the tight junction complex during intestinal inflammation. However, they did not address how different oxygen concentration could influence expression of this hypoxamiR. Our miRNA expression profiling showed an upregulation in all members of the miRNA-320 family under hypoxic conditions. We further demonstrate that, as a result of the induced expression of miRNA-320, hypoxic conditions favor barrier function of intestinal epithelial cells. As such, we propose that the hypoxic environment present in the lumen of the gut impacts barrier functions not only via direct HIF-mediated regulation of tight junction and adherens proteins expression but also through a miRNA-based regulation of cell-cell contact formation.
To conclude, our work further emphasizes the importance of studying intestinal epithelial cells in their physiological environment. On the one hand, hypoxia directly influences the cell biology of the mucosal layer by regulating cell-to-cell contact, migration, stem cell-ness, and metabolism. On the other hand, a low oxygen concentration is critical for the establishment and maintenance of a stable microbiota. As such, given the growing interests in understanding both host/commensal interactions in health and diseases and the complex interplay between host and pathogens in the gastrointestinal tract, it is critical to integrate the impact of local oxygen concentration and fluctuation in regulating and altering these molecular processes.
MATERIALS AND METHODS
Cell lines.
T84 human colonic adenocarcinoma cells (ATCC CCL-248) were cultured in Gibco Dulbecco modified Eagle medium–F-12 nutrient mixture (1:1), supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco) in collagen-coated T25 cell culture flasks. The cells were kept in a constant humid atmosphere containing 37°C and 5% CO2 and either 21% oxygen (normoxia) or 1% oxygen (hypoxia). HEK293T human embryonic kidney cells (ATCC CRL 3216) were cultured in Iscove modified Dulbecco medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. Human intestinal epithelial organoids were isolated from biopsy tissue provided by the University Hospital Heidelberg, as described previously (63). This study was carried out in accordance with the recommendations of the University Hospital Heidelberg with written informed consent from all subjects in accordance with the Declaration of Helsinki. All samples were received and maintained in an anonymized manner. The protocol was approved by the Ethics Commission of the University Hospital Heidelberg under the protocol S-443/2017. In short, resected intestinal tissue was incubated with 2 mM EDTA in phosphate-buffered saline (PBS) for 1 h at 4°C. Intestinal crypts containing the Lgr5+ stem cell niche were isolated after 2 mM EDTA treatment, washed with ice cold PBS, and resuspended in Matrigel. The Matrigel was then overlaid with basal medium (Advanced DMEM–F-12, supplemented with 1% penicillin-streptomycin, 10 mM HEPES, 50% (vol/vol) L-WRN conditioned medium (ATCC CRL-3276, expressing Wnt3A, R-spondin, and Noggin), 1× B-27 (Life Technology), 1× N-2 (Life Technology), 2 mM GlutaMAX (Gibco), 50 ng/ml epidermal growth factor (Invitrogen), 1 mM N-acetyl-cysteine (Sigma), 10 mM nicotinamide (Sigma), 10 μM SB202190 (Tocris Bioscience), and 500 nM A-83-01 (Tocris) and cultured at 37°C and 5% CO2 and 21 or 1% oxygen.
Antibodies and/or reagents.
Mouse monoclonal antibody against ZO-1 (Invitrogen catalog no. 339100) was used at a 1/100 dilution for immunostaining. Secondary antibodies were conjugated with AF568 (Molecular Probes) and directed against the animal source. For Western blot analysis, mouse antibody against HIF-1α was obtained from BD (catalog no. 610959), the HIF-2α antibody was obtained from Novus Bio (catalog no. NB100-122ss), and the β-actin antibody was obtained from Sigma-Aldrich (catalog no. A5441). ProLong Gold antifade containing DAPI (4′,6′-diamidino-2-phenylindole) was obtained from Thermo Fisher Scientific. FITC-labeled dextran (4 kDa) and DMOG were obtained from Sigma-Aldrich.
Monitoring transepithelial electrical resistance.
To monitor barrier function, 105 T84 cells were grown on Transwell filters (6.5-mm polycarbonate membrane, 3-μm pore size; Corning). The medium was changed 1 day postseeding and subsequently every second day. Transepithelial resistance was measured with an EVOM2 chopstick electrode. T84 cells were considered to have a completely formed barrier when being also fully polarized. Full polarization in our setting was reached with a TEER of 1,000 Ω (36). Taking into account the surface of the membrane, reaching a value of 330 Ω ⋅ cm2 indicated full barrier function (64).
Fluorescent flux assay using FITC-labeled dextran.
A total of 105 T84 cells were grown on collagen-coated Transwell filters under normoxic and hypoxic conditions. Every 24 h, 2 mg/ml FITC-labeled dextran was added to the apical compartment, and medium was collected from the basal compartment at 3 h posttreatment. The increase in the fluorescence in the basal medium was measured using a FLUOstar Omega spectrofluorometer (BMG Labtech) at an excitation wavelength of 495 nm and an emission wavelength of 518 nm. As a positive control, the fluorescence of a 100-μl aliquot of a collagen-coated but cell-free Transwell filter was measured to assess maximum diffusion of FITC-labeled dextran.
Immunofluorescence staining.
A total of 105 T84 cells were grown on Transwell filters. At the indicated times postseeding, the polycarbonate membrane was removed from the Transwell holder, rinsed once in PBS, and fixed in 2% paraformaldehyde (PFA) for 20 min. The PFA was removed, and the cells were washed three times with PBS and permeabilized with 0.5% Triton X-100 (vol/vol) at room temperature for 15 min. After being blocked with 3% bovine serum albumin (BSA)-PBS for 1 h at room temperature, the cells were incubated with primary antibody against ZO-1 in 3% BSA-PBS for 1 h at room temperature. The cells were then washed with 0.1% Tween 20–PBS (vol/vol), followed by incubation with the secondary goat anti-mouse Alexa Fluor 568-conjugated antibody diluted in 1% BSA-PBS at room temperature for 45 min. After 45 min, cells were subjected to three washes with 0.1% Tween 20–PBS. The membrane was then briefly rinsed in Millipore H2O and mounted onto glass slides using ProLong Gold antifade reagent with DAPI. Samples were imaged on a Nikon Eclipse Ti-S inverted microscope using a 40× oil objective.
RNA isolation, cDNA, and qPCR.
Total RNA was purified from lysed T84 colonic adenocarcinoma cells or intestinal organoids using the NucleoSpin RNA extraction kit by Machery-Nagel according to the manufacturer’s instruction. A total of 100 to 250 ng of RNA was reversed transcribed into cDNA using the iScript cDNA synthesis kit according to the manufacturer’s instructions (Bio-Rad Laboratories). qRT-PCR was performed using the Bio-Rad CFX96 real-time PCR detection system and SsoAdvanced Universal SYBR green supermix (Bio-Rad). The data were analyzed with the Bio-Rad CFX Manager 3.0, using the housekeeping HPRT1 gene for normalization. The expression of E-cadherin, occludin, JAM-A, VEGF, and CA9 was analyzed using specific primers for the respective human sequence (Table 1). The expression levels of the investigated genes were calculated as ΔΔCq, where Cq is quantification cycle, normalizing to normoxic control samples and to the normalizing genes.
TABLE 1.
Primer sequences used for qRT-PCR
| Primera | Sequence (5′–3′) |
|---|---|
| CA9 fw | AGGATCTACCTACTGTTGAG |
| CA9 rev | TGGTCATCCCCTTCTTTG |
| E-cadherin fw | CCGAGAGCTACACGTTC |
| E-cadherin rev | TCTTCAAAATTCACTCTGCC |
| JAM-A fw | AAGGGACTTCGAGTAAGAAG |
| JAM-A rev | AAGGCAAATGCAGATGATAG |
| HIF-1a fw | TCCATGTGACCATGAGGAAA |
| HIF-1a rev | CCAAGCAGGTCATAGGTGGT |
| HPRT1 fw | CCTGGCGTCGTGATTAGTGAT |
| HPRT1 rev | AGACGTTCAGTCCTGTCCATAA |
| Occludin fw | GGACTGGATCAGGGAATATC |
| Occludin rev | ATTCTTTATCCAAACGGGAG |
| VEGF fw | CTACCTCCACCATGCCAAGT |
| VEGF rev | AGCTGCGCTGATAGACATCC |
fw, forward; rev, reverse.
Western blotting.
To analyze protein content, cells were rinsed once with 1× PBS and lysed with 1× radioimmunoprecipitation assay buffer (150 mM sodium chloride, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris [pH 8.0]) with phosphatase, and protease inhibitors [Sigma-Aldrich]) for 5 min at room temperature. Lysates were collected, and equal protein amounts determined by a DC protein assay (Bio-Rad) were separated by SDS-PAGE. The separated proteins were transferred to Amersham Hybond ECL nitrocellulose membrane using the Wetblot system (Bio-Rad) at 100 V for 75 min. After blotting, the membrane was washed three times for 10 min each time with 0.1% TBS–Tween and directly blocked by incubation in blocking solution for 2 h at room temperature. The primary antibodies were diluted 1:500 (mouse anti-HIF-1α and rabbit anti-HIF-2α) or 1:5,000 (antiactin) in blocking solution, followed by incubation with the blot at 4°C overnight. After three washing steps in 0.1% TBS–Tween, the blot was incubated with the respective IRDye-conjugated secondary antibody (anti-mouse or anti-rabbit antibody, 1:10,000) in blocking solution for 1 h at room temperature. Blots were washed an additional three times in 0.1% TBS–Tween. Protein bands were detected using the Odyssey CLx imaging system.
miRNA microarray.
The expression of miRNAs under normoxic and hypoxic conditions was analyzed by extracting total RNA, including miRNA, using a Qiagen miRNeasy minikit according to the manufacturer’s instructions. Microarray analysis was performed using an Agilent human miRNA v21 microarray chip. Quantile normalized miRNA expression values were log2 transformed, and differentially expressed miRNAs between experimental conditions were identified using the empirical Bayes approach based on moderated t statistics, as implemented in the Bioconductor package limma. P values were adjusted for multiple testing using the Benjamini-Hochberg correction to control the false discovery rate. Adjusted P values below 5% were considered statistically significant. For heatmap display, miRNAs were scaled across samples, and hierarchical clustering of samples and miRNAs was performed using Euclidean distance and Ward’s linkage. Analyses were carried out using R 3.348, with the add-on package pheatmap. Target genes of significantly regulated miRNAs were retrieved from the miRTarBase database v6.1 using Bioconductor package multiMiR (65). Overrepresentation of KEGG pathways was tested with limma functions kegga and goana. P values were adjusted for multiple testing using the Benjamini-Hochberg correction. Subsequent pathway analysis was performed using MetaCore software.
miRNA validation.
For further validation of miRNA-210-3p, miRNA-320a, miRNA-34a-5p, and miRNA-16-5p, total RNA, including miRNA, was transcribed into cDNA using a miScript II RT kit (Qiagen). After cDNA-synthesis, qRT-PCR was performed using the miScript SYBR green PCR kit (Qiagen) and the respective miScript primer assays (Qiagen) on the Bio-Rad CFX96 real-time PCR detection system, normalizing to RNU6-2 as a housekeeping snRNA. The expression levels (fold values) of the investigated miRNAs were calculated as ΔΔCq, normalizing to normoxic control samples and to the housekeeping snRNA.
Production of lentiviral constructs expressing miRNAs and shRNA against HIF-1α.
Oligonucleotides encoding the sequence for mature miRNA-16-5p, miRNA-34a-5p, and miRNA-320a were designed according to the protocol “Lentiviral Overexpression of miRNAs” (66); oligonucleotides encoding the sequence for HIF-1α knockdown were designed from the TRC library, cloneID TRCN0000003808 (Table 2). The sequences for the oligonucleotides against HIF-2α were kindly provided by the lab of F. Hoppe-Seyler, Heidelberg, Germany. Annealed oligonucleotides were ligated with AgeI-HF- and EcoRI-HF-digested pLKO.1 Puro vector (Addgene, catalog no. 8453) using the T4 DNA ligase (New England Biolabs), and the resulting plasmids were transformed into Escherichia coli DH5α-competent cells. Amplified plasmid DNA was purified using a NucleoBondR PC100 kit (Machery-Nagel) according to the manufacturer’s instructions.
TABLE 2.
Oligonucleotides used for shRNA and miRNA expression
| Oligonucleotidea | Sequenceb |
|---|---|
| shHIF1 fw | CCGGCCGCTGGAGACACAATCATATCTCGAGATATGATTGTGTCTCCAGCGGTTTTTG |
| shHIF1 rev | AATTCAAAAACCGCTGGAGACACAATCATATCTCGAGATATGATTGTGTCTCCAGCGG |
| shHIF2 fw | CCGGGCGACAGCTGGAGTATGAACTCGAGTTCATACTCCAGCTGTCGCTTTTTG |
| shHIF2 rev | AATTCAAAAAGCGACAGCTGGAGTATGAACTCGAGTTCATACTCCAGCTGTCGC |
| MIR-320a fw | CCGGTCGCCCTCTCAACCCAGCTTTTCTCGAGAAAAGCTGGGTTGAGAGGGCGATTTTTG |
| MIR-320a rev | AATTCAAAAATCGCCCTCTCAACCCAGCTTTTCTCGAGAAAAGCTGGGTTGAGAGGGCGA |
| MIR-16-5p fw | CCGGCGCCAATATTTACGTGCTGCTACTCGAGTAGCAGCACGTAAATATTGGCGTTTTTG |
| MIR-16-5p rev | AATTCAAAAACGCCAATATTTACGTGCTGCTACTCGAGTAGCAGCACGTAAATATTGGCG |
| MIR-34a-5p fw | CCGGACAACCAGCTAAGACACTGCCACTCGAGTGGCAGTGTCTTAGCTGGTTGTTTTTTG |
| MIR-34a-5p rev | AATTCAAAAAACAACCAGCTAAGACACTGCCACTCGAGTGGCAGTGTCTTAGCTGGTTGT |
fw, forward; rev, reverse.
Boldfacing indicates the respective target or miRNA sequence.
Lentivirus production and selection of stable cell lines.
HEK293T cells were seeded on 10-cm2 dishes and allowed to adhere for 2 days. When cells reached 70 to 80% confluence, they were transfected with 4 μg of pMD2.G (Addgene, catalog no. 12259), 4 μg of psPAX2 (Addgene, catalog no. 12260), and 8 μg of purified pLKO.1 plasmid containing the shRNA or miRNA constructs. Cell supernatant containing generated lentivirus was harvested at 48 to 72 h posttransfection, filtered through a 0.45-μm Millex-HA filter (Merck Millipore), and purified by ultracentrifugation at 27,000 × g for 3 h. For lentiviral transduction, 3 × 105 T84 cells were seeded onto collagen coated six-well plates. After 24 h, the medium was replaced with 4 ml of medium containing 20 μl of the purified lentivirus or lentivirus encoding the miRNA-320a sponge (Mission Lenti microRNA inhibitor, human; Sigma, catalog no. HLTUD0470). Two to three days after transduction, the medium was supplemented with 10 μg/ml puromycin for the selection of successfully transduced cells.
Bioinformatics.
HIF-1α ChIP-Seq profiles for HeLa and T47D were downloaded from NCBI Gene Expression Omnibus, under accession numbers GSM1634922 and GSM1462475, respectively. The data were visualized with the Integrative Genomics Viewer by the Broad Institute and the Regents of the University of California.
ACKNOWLEDGMENTS
This study was supported by research grants from the Chica and Heinz Schaller Foundation and Deutsche Forschungsgemeinschaft (DFG) in project 240245660 (SFB 1129: project 14 for S.B. and project 20 for M.L.). The project also received funding from the DFG in Project number 278001972 (Project A09 of TRR186) to S.B. This project has received additional funding from the European Union’s Seventh Framework Program under grant agreement 334336 (FP7-PEOPLE-2012-CIG). M.S. was supported by the Brigitte-Schlieben Lange Program from the state of Baden, Württemberg, Germany, and the Dual Career Support from CellNetworks, Heidelberg, Germany. I.J.D.C. is funded by a CellNetworks (Heidelberg, Germany) PostDoc position. This work was additionally supported by German Center for Infection Research (DZIF) Thematic Translational Unit HIV-1 04.704 Infrastructural Measure to M.L.
We thank the lab of Hanno Glimm, NCT, Heidelberg, Germany, for intestinal tissue samples; Himanshu Soni and Björn Tews for providing the HIF-1α shRNA lentivirus construct; and the Genomics and Proteomics core facility of the German Cancer Research Center for preparation and processing of the miRNA microarray samples.
REFERENCES
- 1.König J, Wells J, Cani PD, García-Ródenas CL, MacDonald T, Mercenier A, Whyte J, Troost F, Brummer R-J. 2016. Human intestinal barrier function in health and disease. Clin Transl Gastroenterol 7:e196. doi: 10.1038/ctg.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laukoetter MG, Nava P, Nusrat A. 2008. Role of the intestinal barrier in inflammatory bowel disease. World J Gastroenterol 14:401–407. doi: 10.3748/wjg.14.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groschwitz KR, Hogan SP. 2009. Intestinal barrier function: molecular regulation and disease pathogenesis. J Allergy Clin Immunol 124:3–22. doi: 10.1016/j.jaci.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitman RS, Blumberg RS. 2000. First line of defense: the role of the intestinal epithelium as an active component of the mucosal immune system. J Gastroenterol 35:805–814. doi: 10.1007/s005350070017. [DOI] [PubMed] [Google Scholar]
- 5.Muniz LR, Knosp C, Yeretssian G. 2012. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol 3:310. doi: 10.3389/fimmu.2012.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antoni L, Nuding S, Weller D, Gersemann M, Ott G, Wehkamp J, Stange EF. 2013. Human colonic mucus is a reservoir for antimicrobial peptides. J Crohns Colitis 7:e652–664. doi: 10.1016/j.crohns.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Tsukita S, Furuse M, Itoh M. 2001. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 8.Zihni C, Mills C, Matter K, Balda MS. 2016. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol 17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 9.Fisher EM, Khan M, Salisbury R, Kuppusamy P. 2013. Noninvasive monitoring of small intestinal oxygen in a rat model of chronic mesenteric ischemia. Cell Biochem Biophys 67:451–459. doi: 10.1007/s12013-013-9611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng L, Kelly CJ, Colgan SP. 2015. Physiologic hypoxia and oxygen homeostasis in the healthy intestine: a review in the theme “cellular responses to hypoxia.” Am J Physiol Cell Physiol 309:C350–C360. doi: 10.1152/ajpcell.00191.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeitouni NE, Chotikatum S, von Köckritz-Blickwede M, Naim HY. 2016. The impact of hypoxia on intestinal epithelial cell functions: consequences for invasion by bacterial pathogens. Mol Cell Pediatr 3:14. doi: 10.1186/s40348-016-0041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albenberg L, Esipova T, Judge C, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano R, Lewis J, Li H, Thom S, Bushman F, Vinogradov S, Wu G. 2014. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota in humans and mice. Gastroenterology 147:1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor CT, Colgan SP. 2007. Hypoxia and gastrointestinal disease. J Mol Med (Berl) 85:1295–1300. doi: 10.1007/s00109-007-0277-z. [DOI] [PubMed] [Google Scholar]
- 14.Colgan SP, Taylor CT. 2010. Hypoxia: an alarm signal during intestinal inflammation. Nat Rev Gastroenterol Hepatol 7:281–287. doi: 10.1038/nrgastro.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dengler VL, Galbraith M, Espinosa JM. 2014. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol 49:1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaelin WG, Ratcliffe PJ. 2008. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 18.Schofield CJ, Ratcliffe PJ. 2004. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol 5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL. 2014. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol 9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 20.Greijer AE, van der Groep P, Kemming D, Shvarts A, Semenza GL, Meijer GA, van de Wiel MA, Belien JA, van Diest PJ, van der Wall E. 2005. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J Pathol 206:291–304. doi: 10.1002/path.1778. [DOI] [PubMed] [Google Scholar]
- 21.Fiocchi C. 1996. Cytokines in inflammatory bowel disease. Chapman & Hall, New York, NY. [Google Scholar]
- 22.Furuta GT, Turner JR, Taylor CT, Hershberg RM, Comerford K, Narravula S, Podolsky DK, Colgan SP. 2001. Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J Exp Med 193:1027–1034. doi: 10.1084/jem.193.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomasetto C, Masson R, Linares J, Wendling C, Lefebvre O, Chenard M, Rio M. 2000. pS2/TFF1 interacts directly with the VWFC cysteine-rich domains of mucins. Gastroenterology 118:70–80. doi: 10.1016/S0016-5085(00)70415-X. [DOI] [PubMed] [Google Scholar]
- 24.Kalabis J, Rosenberg I, Podolsky DK. 2006. Vangl1 protein acts as a downstream effector of intestinal trefoil factor (ITF)/TFF3 signaling and regulates wound healing of intestinal epithelium. J Biol Chem 281:6434–6441. doi: 10.1074/jbc.M512905200. [DOI] [PubMed] [Google Scholar]
- 25.Meyer Zum Büschenfelde D, Tauber R, Huber O. 2006. TFF3-peptide increases transepithelial resistance in epithelial cells by modulating claudin-1 and -2 expression. Peptides 27:3383–3390. doi: 10.1016/j.peptides.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 26.Sun Z, Liu H, Yang Z, Shao D, Zhang W, Ren Y, Sun B, Lin J, Xu M, Nie S. 2014. Intestinal trefoil factor activates the PI3K/Akt signaling pathway to protect gastric mucosal epithelium from damage. Int J Oncol 45:1123–1132. doi: 10.3892/ijo.2014.2527. [DOI] [PubMed] [Google Scholar]
- 27.Tambuwala MM, Cummins EP, Lenihan CR, Kiss J, Stauch M, Scholz CC, Fraisl P, Lasitschka F, Mollenhauer M, Saunders SP, Maxwell PH, Carmeliet P, Fallon PG, Schneider M, Taylor CT. 2010. Loss of prolyl hydroxylase-1 protects against colitis through reduced epithelial cell apoptosis and increased barrier function. Gastroenterology 139:2093–2101. doi: 10.1053/j.gastro.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 28.Cummins EP, Seeballuck F, Keely SJ, Mangan NE, Callanan JJ, Fallon PG, Taylor CT. 2008. The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology 134:156–165.e1. doi: 10.1053/j.gastro.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Robinson A, Keely S, Karhausen J, Gerich ME, Furuta GT, Colgan SP. 2008. Mucosal protection by hypoxia-inducible factor prolyl hydroxylase inhibition. Gastroenterology 134:145–155. doi: 10.1053/j.gastro.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loscalzo J. 2010. The cellular response to hypoxia: tuning the system with microRNAs. J Clin Invest 120:3815–3817. doi: 10.1172/JCI45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan YC, Banerjee J, Choi SY, Sen CK. 2012. miR-210: the master hypoxamir. Microcirculation 19:215–223. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 33.He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 34.Tili E, Michaille J-J, Piurowski V, Rigot B, Croce CM. 2017. MicroRNAs in intestinal barrier function, inflammatory bowel disease, and related cancers: their effects and therapeutic potentials. Curr Opin Pharmacol 37:142–150. doi: 10.1016/j.coph.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanai M, Mullen C, Podolsky DK. 1998. Intestinal trefoil factor induces inactivation of extracellular signal-regulated protein kinase in intestinal epithelial cells. Proc Natl Acad Sci U S A 95:178–182. doi: 10.1073/pnas.95.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanifer ML, Rippert A, Kazakov A, Willemsen J, Bucher D, Bender S, Bartenschlager R, Binder M, Boulant S. 2016. Reovirus intermediate subviral particles constitute a strategy to infect intestinal epithelial cells by exploiting TGF-β dependent pro-survival signaling. Cell Microbiol 18:1831–1845. doi: 10.1111/cmi.12626. [DOI] [PubMed] [Google Scholar]
- 37.Cichon C, Sabharwal H, Rüter C, Schmidt MA. 2014. MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers 2:e944446. doi: 10.4161/21688362.2014.944446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordes F, Brückner M, Lenz P, Veltman K, Glauben R, Siegmund B, Hengst K, Schmidt MA, Cichon C, Bettenworth D. 2016. MicroRNA-320a strengthens intestinal barrier function and follows the course of experimental colitis. Inflamm Bowel Dis 22:2341–2355. doi: 10.1097/MIB.0000000000000917. [DOI] [PubMed] [Google Scholar]
- 39.Sun J-Y, Huang Y, Li J-P, Zhang X, Wang L, Meng Y-L, Yan B, Bian Y-Q, Zhao J, Wang W-Z, Yang A-G, Zhang R. 2012. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem Biophys Res Commun 420:787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 40.Li C, Duan P, Wang J, Lu X, Cheng J. 2017. miR-320 inhibited ovarian cancer oncogenicity via targeting TWIST1 expression. Am J Transl Res 9:3705–3713. [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Sun B, Sun R, Zhu D, Zhao X, Zhang Y, Dong X, Che N, Li J, Liu F, Zhao N, Wang Y, Zhang D. 2017. HMGA2 promotes vasculogenic mimicry and tumor aggressiveness by upregulating Twist1 in gastric carcinoma. Sci Rep 7:2229. doi: 10.1038/s41598-017-02494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim NH, Kim HS, Li X-Y, Lee I, Choi H-S, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher CA, Weiss SJ, Yook JI. 2011. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial–mesenchymal transition. J Cell Biol 195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U, Hermeking H. 2011. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle Georget Tex 10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 44.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, García De Herreros A. 2000. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 45.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 46.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. 2003. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci 116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 47.Martínez C, Rodiño-Janeiro BK, Lobo B, Stanifer ML, Klaus B, Granzow M, González-Castro AM, Salvo-Romero E, Alonso-Cotoner C, Pigrau M, Roeth R, Rappold G, Huber W, González-Silos R, Lorenzo J, de Torres I, Azpiroz F, Boulant S, Vicario M, Niesler B, Santos J. 2017. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut 66:1537–1538. doi: 10.1136/gutjnl-2016-311477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato T, Clevers H. 2013. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 49.Saeedi BJ, Kao DJ, Kitzenberg DA, Dobrinskikh E, Schwisow KD, Masterson JC, Kendrick AA, Kelly CJ, Bayless AJ, Kominsky DJ, Campbell EL, Kuhn KA, Furuta GT, Colgan SP, Glover LE. 2015. HIF-dependent regulation of claudin-1 is central to intestinal epithelial tight junction integrity. Mol Biol Cell 26:2252–2262. doi: 10.1091/mbc.E14-07-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aamann L, Vestergaard EM, Grønbæk H. 2014. Trefoil factors in inflammatory bowel disease. World J Gastroenterol 20:3223–3230. doi: 10.3748/wjg.v20.i12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin N, Xu L-F, Sun M. 2013. The protective effect of trefoil factor 3 on the intestinal tight junction barrier is mediated by Toll-like receptor 2 via a PI3K/Akt dependent mechanism. Biochem Biophys Res Commun 440:143–149. doi: 10.1016/j.bbrc.2013.09.049. [DOI] [PubMed] [Google Scholar]
- 52.Kjellev S, Thim L, Pyke C, Poulsen SS. 2007. Cellular localization, binding sites, and pharmacologic effects of TFF3 in experimental colitis in mice. Dig Dis Sci 52:1050–1059. doi: 10.1007/s10620-006-9256-4. [DOI] [PubMed] [Google Scholar]
- 53.Giatromanolaki A, Sivridis E, Maltezos E, Papazoglou D, Simopoulos C, Gatter KC, Harris AL, Koukourakis MI. 2003. Hypoxia inducible factor 1α and 2α overexpression in inflammatory bowel disease. J Clin Pathol 56:209–213. doi: 10.1136/jcp.56.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shah YM. 2016. The role of hypoxia in intestinal inflammation. Mol Cell Pediatr 3:1. doi: 10.1186/s40348-016-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’Ignazio L, Bandarra D, Rocha S. 2016. NF-κB and HIF crosstalk in immune responses. FEBS J 283:413–424. doi: 10.1111/febs.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. 2010. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 139:1654–1664. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu X-Q, Wang J, Chen G-X, Zhang G-Q, Zhang D-Y, Cai Y-Y. 2018. Overexpression of microRNA-495 improves the intestinal mucosal barrier function by targeting STAT3 via inhibition of the JAK/STAT3 signaling pathway in a mouse model of ulcerative colitis. Pathol Res Pract 214:151–162. doi: 10.1016/j.prp.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Nan X, Qin S, Yuan Z, Li Y, Zhang J, Li C, Tan X, Yan Y. 2016. Hsa-miRNA-31 regulates epithelial cell barrier function by inhibiting TNFSF15 expression. Cell Mol Biol (Noisy-le-Grand) 62:104–110. [PubMed] [Google Scholar]
- 59.Gazouli M, Wouters MM, Kapur-Pojskić L, Bengtson M-B, Friedman E, Nikčević G, Demetriou CA, Mulak A, Santos J, Niesler B. 2016. Lessons learned: resolving the enigma of genetic factors in IBS. Nat Rev Gastroenterol Hepatol 13:77–87. doi: 10.1038/nrgastro.2015.206. [DOI] [PubMed] [Google Scholar]
- 60.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar J-P, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan Y, Tan X, Li C, Cao Y, Hong Y, Li W, Xu X, Xu L, Yang L. 2016. Hsa-miRNA-26b regulates SPAK expression during intestinal epithelial cell differentiation. Int J Clin Exp Pathol 9:9821–9836. [Google Scholar]
- 62.Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, Zhang P, Wang F, Yang J, Yang J, Zhu Q, Liang Y, Wu W, Gao R, Yang Z, Zou Y, Qin H. 2013. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun 434:746–752. doi: 10.1016/j.bbrc.2013.03.122. [DOI] [PubMed] [Google Scholar]
- 63.Pervolaraki K, Stanifer ML, Münchau S, Renn LA, Albrecht D, Kurzhals S, Senís E, Grimm D, Schröder-Braunstein J, Rabin RL, Boulant S. 2017. Type I and type III interferons display different dependency on mitogen-activated protein kinases to mount an antiviral state in the human gut. Front Immunol 8:459. doi: 10.3389/fimmu.2017.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corning. 2019. Guidelines for use: Transwell® permeable supports. Corning, Inc, Tewksbury, MA: https://www.corning.com/worldwide/en/products/life-sciences/products/permeable-supports/transwell-guidelines.html. [Google Scholar]
- 65.Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, Mahaffey S, Rossi S, Calin GA, Bemis L, Theodorescu D. 2014. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res 42:e133. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zöllner H, Hahn SA, Maghnouj A. 2014. Lentiviral overexpression of miRNAs. Methods Mol Biol 1095:177–190. doi: 10.1007/978-1-62703-703-7_15. [DOI] [PubMed] [Google Scholar]