Abstract
Auxin-dependent cell expansion is crucial for initiation of fiber cells in cotton (Gossypium hirsutum), which ultimately determines fiber yield and quality. However, the regulation of this process is far from being well understood. In this study, we demonstrate an antagonistic effect between cytokinin (CK) and auxin on cotton fiber initiation. In vitro and in planta experiments indicate that enhanced CK levels can reduce auxin accumulation in the ovule integument, which may account for the defects in the fiberless mutant xu142fl. In turn, supplementation with auxin can recover fiber growth of CK-treated ovules and mutant ovules. We further found that GhPIN3a is a key auxin transporter for fiber-cell initiation and is polarly localized to the plasma membranes of non-fiber cells, but not to those of fiber cells. This polar localization allows auxin to be transported within the ovule integument while specifically accumulating in fiber cells. We show that CKs antagonize the promotive effect of auxin on fiber cell initiation by undermining asymmetric accumulation of auxin in the ovule epidermis through down-regulation of GhPIN3a and disturbance of the polar localization of the protein.
Keywords: Auxin, cotton, cytokinin, fiber initiation, GhPIN3a, polar auxin transport
Cytokinin antagonizes the promotive effect of auxin on cotton fiber initiation by disrupting the localization of GhPIN3a, which mediates asymmetric accumulation of auxin in the ovule epidermis.
Introduction
Cotton (Gossypium hirsutum) is the most prevalent natural fiber crop in the world. Its fibers are single cells that are differentiated from ovule epidermal cells. With a long and unicellular structure, they provide an ideal model for studying cell differentiation and polar growth (Kim and Triplett, 2001). The development of the cotton fiber consists of four distinct phases: initiation, elongation, secondary cell-wall deposition, and maturation (Ryser, 1999; Kim and Triplett, 2001). Fiber initiation, which determines the number and the onset of the subsequent development of fibers, affects both the yield and the quality of cotton (Zhang et al., 2011; Wang et al., 2015).
Phytohormones such as auxin, gibberellins (GAs), jasmonic acid (JA), and brassinosteroids (BRs) are involved in the regulation of cotton fiber initiation (Luo et al., 2007; Xiao et al., 2010; Zhang et al., 2011; Hao et al., 2012; Tan et al., 2012; Zhou et al., 2015; Hu et al., 2016; Xia et al., 2018). Auxin, predominantly represented by indole-3-acetic acid (IAA), plays an essential role in promoting initiation (Beasley, 1973; Seagull and Giavalis, 2004; Zhang et al., 2011). On the day of anthesis, auxin is highly accumulated in the initiating fiber cells, and enhancing its biosynthesis in the ovule epidermal cells can significantly improve fiber yield and fineness (Zhang et al., 2011). Similar to the morphogenesis of many plant lateral organs (Feng and Dickinson, 2007), polar auxin transport is involved in the establishment of a gradient in the cotton ovule epidermis (Zhang et al., 2017), and PIN-formed proteins (PINs) that polarly localize to the plasma membrane and facilitate auxin efflux from cells are key players in this process (Benková et al., 2003; Friml et al., 2003; Petrášek et al., 2006; Petrášek and Friml, 2009; Adamowski and Friml, 2015; Rakusová et al., 2015; Armengot et al., 2016). In a previous study, we found that PIN homologs (GhPINs) in cotton are required for auxin-triggered fiber initiation (Zhang et al., 2017). Of these proteins, GhPIN3a is preferentially expressed in the ovule outer integument on the day of anthesis, and is proposed to be the core regulator for the establishment of the auxin gradient in the epidermis (Zhang et al., 2017). In model plants, PIN undergoes retargeting and localization to the plasma membrane in order to establish a new auxin maximum (Friml et al., 2002; Harrison and Masson, 2008; Kleinevehn et al., 2010). It is known that gravity-induced PIN polar localization participates in the regulation of auxin trafficking in the primary root, lateral root, root hairs, and stem (Petrášek and Friml, 2009). However, since fiber cell initiation occurs across the whole surface of the ovule with any orientation, the distribution pattern of PINs remains unknown as well as how they determine such a scattered distribution of auxin in the ovule epidermal cells.
Interactions between cytokinins (CKs) and auxin regulate many processes of plant growth and development (Francis and Sorrell, 2001; Sakakibara, 2006; Santner and Estelle, 2009; Schaller et al., 2015), including embryogenesis (Müller and Sheen, 2008), meristem development and maintenance (Dello Ioio et al., 2008), lateral root initiation (Li et al., 2006; Laplaze et al., 2007), floral meristem initiation (Zhao et al., 2010), and shoot branching (Tanaka et al., 2006; Shimizu-Sato et al., 2009). Tissue- and context-specific patterns are usually found in their interactions, which occur via various mechanisms including their biosynthesis, degradation, transport and signaling (Sakakibara, 2006; Vanneste and Friml, 2009; Chandler and Werr, 2015; Schaller et al., 2015). It is believed that cytokinin and auxin act synergistically in the formation and maintenance of shoot apical meristem, whilst acting antagonistically in the initiation of lateral root primordia (Chandler and Werr, 2015). In cotton, the study of crosstalk between auxin and CK on fiber development was first reported through use of feeding experiments (Beasley and Ting, 1973, 1974). High concentrations of CK (>5 µM) inhibit fiber growth in vitro, whereas low concentration (<0.5 µM) increase fiber production (Beasley and Ting, 1974). In the fiberless mutant xu142fl, elevated CK levels in the ovules imply a negative effect on fiber production (Liu et al., 1999). Our previous study involving the manipulation of GhCKX in transgenic cotton suggested that CK is indispensable for the initiation of fiber cells (Zeng et al., 2012). Nevertheless, the role of CK in the regulation of fiber development remains unclear, and its interaction with auxin requires investigation.
In this study, we show an antagonistic effect of CK on auxin-triggered fiber initiation. We demonstrate that the auxin efflux carrier GhPIN3a, a pivotal player for cotton fiber initiation, polarly localizes at the plasma membrane in non-fiber cells and delocalizes in fiber cells. We show that an increase of CKs can interfere with the establishment of the auxin gradient in the ovule epidermis through impairment of the localization of GhPIN3a to the membrane, thus inhibiting the initiation of fiber cells.
Materials and methods
Plasmid construction and plant transformation
For the proTCS::iaaM and proDR5::IPT constructs, the cytokinin-inducible promoter proTCS (Müller and Sheen, 2008) and the auxin-inducible promoter proDR5 (Ulmasov et al., 1997) were synthesized with flanking HindIII and XbaI restriction sites. iaaM (Comai and Kosuge, 1982) and IPT sequences (Medford et al., 1989) were amplified by PCR (Novoprotein, China) from Agrobacterium tumefaciens with flanking KpnI and EcoRI restriction sites. After digestion (ThermoFisher Scientific), proTCS linked with iaaM, and proDR5 linked with IPT were separately inserted into the binary vector p5 (Luo et al., 2007). For the proTCS::GUS construct, the proCaMV35S (pro35S) promoter in the binary vector pBI121 was replaced by the proTCS promoter through the HindIII and XbaI restriction sites. For the proGhPIN3a::GUS construct, the proGhPIN3a promoter was amplified from upland cotton (Gossypium hirsutum cultivar ‘Jimian 14’) with flanking HindIII and BamHI restriction sites. After digestion, the proGhPIN3a promoter was placed upstream of the GUS reporter gene in a modified pCambia2300 binary vector. For the proPV::antisenseGhCKX3 construct, an ovule-specific promoter proPV (Goossens et al., 1999; Zhang et al., 2011) was amplified from common bean (Phaseolus vulgaris) with flanking HindIII and BamHI restriction sites, and the partial antisense sequence of GhCKX3 (antisenseGhCKX3) was amplified from the cotton ‘Jimian 14’ with flanking BamHI and EcoRI restriction sites. proPV linked with antisenseGhCKX3 was inserted into the binary vector p5. For the proBAN::GhPIN3a-RNAi construct, 3´-UTR of GhPIN3a was amplified and inserted to a terminal of the first intron of GA20ox1 with SalI and XbaI sites, and inversely inserted to the other terminal with NotI and EcoRI sites in a vector pUCm-T. The RNAi construct was then placed downstream of the ovule epidermis-specific promoter proBAN isolated from Arabidopsis (Debeaujon et al., 2003; Zhang et al., 2011) in a binary vector pLGN modified from pCambia2300. In the T-DNA region of pLGN there is a neomycin phosphotransferase II (NPTII) selection marker fused with a GUS reporter gene under the control of a pro35S promoter. For the pro35S::GhPIN3a::YFP construct, YFP without the start and stop codons was inserted into the GhPIN3a coding sequence (CDS) behind position 1413 bp relative to the start codon through amplifying the fragment with flanking SpeI and SalI restriction sites. The generated GhPIN3a::YFP was placed downstream of a pro35S promoter in the pLGN vector. Information for all constructs including primer sequences and fragment sizes are listed in Supplementary Table S1 at JXB online. These constructs were transformed into cotton ‘Jimian 14’ using A. tumefaciens strain LBA4404 (Luo et al., 2007). Kanamycin-resistant and GUS-positive plantlets were screened out and grown in a greenhouse receiving natural daylight.
Ovule culture
Ovule culture was performed according to the method described previously by Beasley and Ting (1973). Indole-3-acetic acid sodium salt (IAA) and gibberellic acid potassium salt (GA3; both Sigma-Aldrich) were dissolved in distilled water and filter-sterilized to obtain stock solutions of 50 mM and 10 mM, respectively. Trans-zeatin (ZT) was dissolved in DMSO (both Sigma-Aldrich) to obtain a 50-mM stock solution. After autoclaving, plant hormones were added to the growth medium at the various experimental concentrations. The standard concentrations in the medium were 0.5 µM GA3 and 5 µM IAA. Cotton bolls were harvested around 08.00 h at 1 d prior to anthesis and on the day of anthesis [–1 d post anthesis (DPA) and 0 DPA, respectively]. After being surface-sterilized in 75% (v/v) ethanol for 1 min and rinsed in sterile water, bolls were then soaked in 0.1% (w/v) HgCl2 solution for 12 min for full sterilization, followed by rinsing six times with sterile water. For each treatment, 20 ovules in the middle part of each locule were excised and floated on 20 ml of liquid medium in a 100-ml flask, and then incubated in darkness at 32 °C for 3 d (for SEM) or for 15 d (for optical microscopy). Each experiment was repeated three times. An equal volume of DMSO was added to the medium as a negative control.
Quantification of endogenous cytokinins and IAA
Ovules and/or fibers (250 mg FW) harvested around 11.00 h were ground in liquid nitrogen, and then extracted according to the method of Yoshimoto et al. (2009). Then 50 ng of [13C6]IAA plus 10 ng of one the following were added to each extraction buffer as the internal standard: [2H5]ZT (trans-zeatin), [2H5]ZR (ZT riboside), [2H5]Z9G (ZT-9-glucoside), [2H5]ZOG (ZT-O-glucoside), [2H5]ZROG (ZOG riboside), [2H3]DZ (dihydrozeatin), [2H3]DZR (DZ riboside), [2H7]DZOG (DZ-O-glucoside), [2H3]DZ9G (DZ-9-glucoside), [2H7]DZROG (DZOG riboside), [2H6]iP (N6-isopentenyladenine), [2H6]iPR (N6-isopentenyladenosine), and [2H6]iP9G (N6-isopentenyladenosine-9-glucoside) (all OlChemim, Czech Republic). The solutions were then vacuum-evaporated to dryness using Savant ISS110 SpeedVac concentrator (ThermoFisher Scientific) at 40 °C before being redissolved in 100 μl of 10% methanol and filtered through a 0.22-μm pore filter. Portions (20 μl) of the dissolved samples were used for plant hormone quantification using a 4000 Q-Trap LC-ESI-MS/MS system (SCIEX, USA) using a XTerra® MS C18 column (2.1×150 mm, 3.5 μm, Waters) at 30 °C. A series of gradients of methanol/0.01% formic acid (v/v) were applied at a flow-rate of 0.16 ml min–1 as follows: 0~15 min, 10% to 40%; 15~30 min, 40% to 55%; 30~45 minute, 55% to 100%. Cytokinins and IAA were detected in the negative ion mode; other analytical parameters are listed in Supplementary Table S2. Each test was conducted on three biological replicates.
Measurement of total fiber units
Assays for total fiber units (TFUs) were performed according to the method described by Beasley and Ting (1973) with several modifications. In each measurement, 20 ovules, cultured for 15 d, were separated from each other, washed with running water for 15 s, and transferred into a 100-ml conical flask. Then 40 ml of distilled water was added with a drop of hydrochloric acid. After heating in boiling water for 5 min, the ovules were rinsed with distilled water and dried on filter paper (Whatman). They were then dyed in 30 ml of staining solution (0.018% Toluidine Blue O, 16 mM Na2HPO4, 0.01 M citric acid, pH 4.5) for 15 s. After a brief wash in running water for 60 s, ovules were again dried on filter paper, and treated with 25 ml of de-staining solution (acetic acid:ethanol:water, 10:95:5) for 1 h. Absorbance of the de-staining solution was determined at 624 nm and TFU was calculated according to a standard curve. Three biological replicates were used.
RNA extraction and RT-qPCR
RNA was extracted using an EASY spin plant RNA extraction kit (Aidlab, China). After treatment with DNase I (TaKaRa), 1 µg RNA was used to synthesize first-strand cDNA with a PrimeScript™ RT Reagent Kit (TaKaRa). Real-time quantitative PCR (RT-qPCR) was performed on a CFX96TM Real-Time System (Bio-Rad,) with 1×iQ™ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions, and the data were analysed using the software provided. The thermal cycling consisted of a pre-treatment (94 °C, 3 min) followed by 40 amplification cycles (94 °C, 20 s; 56 °C, 20 s; 72 °C, 30 s). The gene-specific primers used for RT-qPCR are shown in Supplementary Table S3. Each test was confirmed by three individual runs (biological replicates) and data from one of them were used to determine expression. GhHIS3 was used as the reference gene.
Ovule microdissection
Ovule microdissection was performed according to the method of Bedon et al. (2012). Fifty fresh ovules harvested from the boll at middle part of each locule were separated. The ovules were then dissected into three types of tissue: outer integument, inner integument, and nucellus. The dissected tissues were ground in liquid nitrogen and then used for RNA extraction and RT-qPCR assays.
Histochemical staining and quantification of GUS activity
Histochemical GUS (β-glucuronidase) staining was performed using the method described by Jefferson et al. (1987). After excision, tissues were immediately immersed in the staining solution [100 mM NaPO4, pH 7.0, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 10 mM EDTA, 0.1% (v/v) Triton X-100, 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid substrate; X-Gluc, Amresco, USA], and then placed in vacuum for 10 min before being kept in the dark at 37 °C for 12 h. The stained samples were then washed and kept in 75% ethanol. Images were captured on an SZX-ILLB2-200 stereo-microscope imaging system (Olympus). The stained ovules were also observed in paraffin-embedded sections on a CKX41 microscope (Olympus). Fluorometric assays of GUS activity in ovules of proTCS::GUS and proDR5::GUS transgenic plants during cotton fiber initiation were performed as described by Hou et al. (2008). Fifty fresh ovules of each sample were ground in liquid nitrogen for the determination. Estimation of proteins was performed using the method described by Bradford (1976). GUS activity was calculated as pmol 4-MU per minute per microgram protein and each test was carried out on three biological replicates.
In situ hybridization
In situ hybridization of GhPIN3a mRNA was carried out using the method previously described by Zhang et al. (2017). Briefly, paraffin sections of ovules sampled at 0 DPA were de-paraffinized and incubated with the Dig-labelled RNA probe of GhPIN3a. The sections were then incubated with anti-Dig-AP conjugate (Roche) and the signal was detected by NBT/BCIP solution (Roche). Sections incubated with the sense RNA probe were used as the negative control. Images were captured using a CKX41 microscope (Olympus).
Microscopic observations
Fiber growth in the feeding experiments and GUS-stained samples were observed using an SZX-ILLB2-200 stereo-microscope imaging system (Olympus). Paraffin-embedded sections were observed on a CKX41 microscope (Olympus). For the observation of fibers during initiation, ovules were frozen in liquid nitrogen, transferred into the cryo-stage of a preparation chamber for platinum coating, and then imaged using an S-3400N SEM (Hitachi) at 2 kV accelerating voltage. For the observation of fluorescence in the pro35S::GhPIN3a::YFP cotton ovule, the ovule was sectioned by hand and then immersed in 2 μM FM4-64 to indicate the plasma membrane. After 30-min incubation in the dark, the sample was mounted between two coverslips with distilled water (pH 7.0) for observation. The fluorescence signal was detected under either a 40× or 63× oil lens by the HyD detector on a Leica SP8 laser-scanning confocal microscope with the following parameters: excitation at 514 nm, laser power at 5%, emission from 523–600 nm (YFP) or from 600–680 nm (FM4-64). The signal intensity was quantified using the Leica Application Suite X software.
Immunohistochemical localization of cytokinins
The procedure for the immunolocalization of CKs was carried out as described by Zhang et al. (2017). In brief, polyclonal antibodies against ZT and ZR (Agrisera) were added to ovule sections at a concentration of 10 μg ml–1. The signal was detected using DyLight549-labeled secondary antibody (Abcam, UK) on a confocal scanning system (FV1000, Olympus). Sections incubated without the primary antibody were used as the negative control.
Results
Contents of cytokinins and auxin exhibit opposite patterns during ovule and fiber development
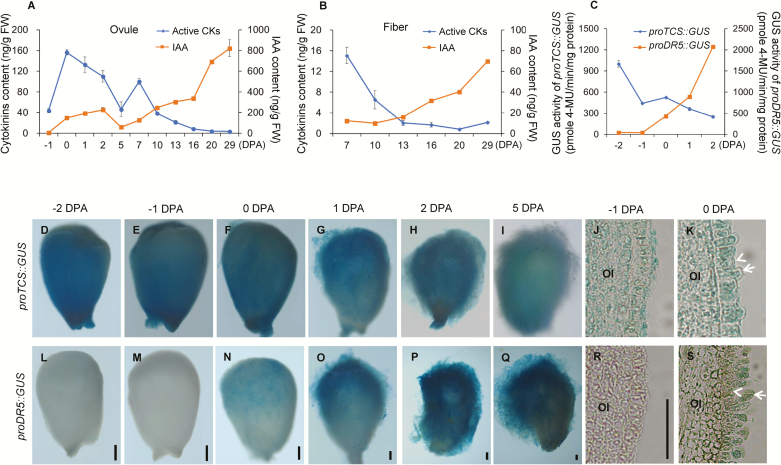
We used LC-MS/MS to examine changes in contents of CKs and IAA in developing ovules and fibers of cotton. The maximum value for content of active CKs, namely ZT, ZR, iP, iPR, plus DZ, which accounted for 83.7% total CKs in the ovule, was recorded on the day of anthesis (0 DPA) (Fig. 1A), after which it gradually declined. The zeatin-type CKs (ZT and ZR) made up the largest portion (89.3%) of the active CKs in 0-DPA ovules, followed by isopentenyladenine-types (iP and iPR, 10.5%), and then the dihydrozeatin-type (DZ, 0.3%) (Supplementary Table S4). A similar decline in CK content was observed in the developing fiber cells from 7–29 DPA (Fig. 1B, Supplementary Table S5). In contrast, IAA exhibited an increase in ovules and fibers during development (Fig. 1A, B, Supplementary Tables S4, S5).
Fig. 1.
Levels of cytokinins (CKs) and IAA in developing cotton ovules and fibers. (A, B) LC-MS/MS determination of contents of CKs and IAA in developing (A) ovules and (B) fibers. Active CKs represent the sum of contents of ZT, ZR, iP, iPR, and DZ. Ovules sampled from bolls at 0– 5 d post anthesis (DPA) contained fibers (because of technical limitations it is difficult to separate fibers from young ovules). Ovules sampled from bolls at –1, and 7–29 DPA were fiber-free. (C) GUS activity assay of proTCS::GUS and proDR5::GUS transgenic ovules. Data are means (±SD) of three biological replicates. (D–S) GUS staining of ovules of transgenic proTCS::GUS (D–I) and proDR5::GUS (L–Q) during cotton fiber initiation, together with paraffin-embedded sections of stained ovules harvested at –1 DPA and 0 DPA (J, K, R, S). Arrows indicate fiber cells and arrowheads indicate non-fiber cells in the ovule epidermis. OI, outer integument. Scale bars are 250 µm in ovule images or 50 µm in section images.
For in situ observation of the changes of CKs and IAA in ovules and fibers, the GUS report gene was fused with either a cytokinin-inducible promoter proTCS (Müller and Sheen, 2008) or an auxin-inducible promoter proDR5 (Ulmasov et al., 1997). Strong staining was observed in the epidermal layer of the ovules of proTCS::GUS plants at –2, –1, and 0 DPA (Fig. 1D–F, J), and the signal appeared in both fiber and non-fiber cells (Fig. 1K). After anthesis, the staining at the ovule surface decreased gradually (Fig. 1G–I). In contrast, in proDR5::GUS ovules, the GUS activity became stronger after flowering (Fig. 1L–R), and the signal appeared mainly in the fiber cells (Fig. 1S). Quantification of GUS activity (Fig. 1C) confirmed the results of the LC-MS/MS analysis. These data indicated opposite changes in the contents of CK and auxin during ovule and fiber development.
Antagonistic effect of cytokinin on auxin promotion of fiber initiation
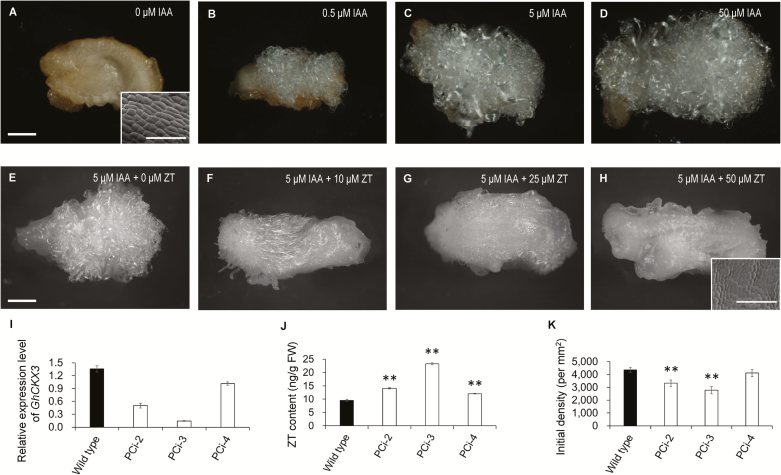
To examine the apparent antagonistic relationship in their regulation of fiber development, we cultured ovules in vitro with various concentrations of IAA or CKs. In the presence of IAA ranging from 0.5 μM to 50.0 μM, ovules harvested at 1 d before anthesis (–1 DPA) were able to produce fibers in a dose-dependent manner (Fig. 2B–D). In contrast, no fibers appeared on cultured ovules without IAA (Fig. 2A). This result supported previous findings that auxin is crucial for fiber initiation (Beasley, 1973; Zhang et al., 2011). In contrast to auxin, CKs displayed an inhibitory effect on initiation (Fig. 2E–H). Fiber growth on the cultured -1-DPA ovules was clearly inhibited when as little as 10 µM ZT was added into the medium (which contained 5 µM IAA+0.5 µM GA3, Fig. 2F). When the concentration of ZT was reached 50 µM, fiber initiation was fully arrested (Fig. 2H). Interestingly, this concentration of ZT was unable to inhibit the fiber growth of 0-DPA ovules, on which fiber cells that have protruded from the surface of the ovule (Supplementary Fig. S1). To increase the endogenous level of CK in the ovule, we down-regulated the expression of cytokinin dehydrogenase (CKX), which catalyses the catabolism of cytokinins to inactive products (Jones and Schreiber, 1997; Zeng et al., 2012) by ovule-specific expression of its antisense gene (proPV::antisenseGhCKX3). Down-regulation of GhCKX3 resulted in a significant increase in ZT content in 0-DPA ovules (Fig. 2I, J). The transgenic lines #2 and #3 showed a significant decrease in fiber density (Fig. 2K), supporting the in vitro results that an excess of CKs inhibited the initiation of cotton fibers.
Fig. 2.
Auxin promotes fiber initiation in cotton, whilst cytokinin (CK) inhibits initiation. (A–D) Fiber growth on ovules cultured with different concentration of IAA. Ovules at –1 d post anthesis (DPA) were cultured in medium containing 0, 0.5, 5, or 50 μM IAA together with 0.5 μM GA3 for 15 d. (E–H) Fiber growth on ovules cultured with different concentration of ZT. Ovules at -1 DPA were cultured in the medium containing 0, 10, 25 or 50 μM ZT together with 0.5 μM GA3 and 5 μM IAA for 15 d. Fiber growth was observed using a stereo-microscope (scale bars are 1 mm) and a SEM (scale bars are 100 μm). (I) Transcription levels of GhCKX3 in proPV::antisenseGhCKX3 (PCi) transformants and the wild-type. Ovules at 0 DPA were used for RT-qPCR assays. Transcription levels (in arbitrary units) are normalized to that of GhHIS3. Data are means (±SD) of three repeats. (J) Content of ZT in ovules at 0 DPA. Data are means (±SD) of three biological replicates. (K) Initial fiber densities on the ovule surface at 0 DPA. Data are means (±SD) of nine ovules. Significant differences compared with the wild-type were determined using Student’s t-test (**P<0.01).
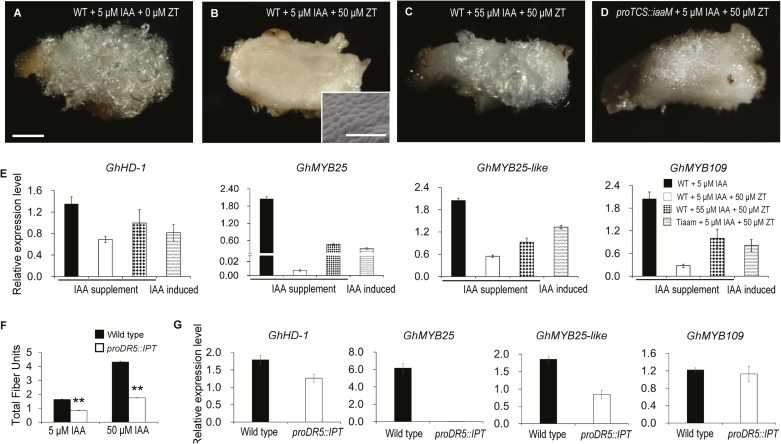
To determine whether auxin could antagonize the inhibitory effect of CKs on fiber initiation, we increased the IAA concentration in the medium. An additional supplement of 50 µM IAA could recover fiber growth of the ovule (Fig. 3C), and four genes associated with fiber initiation, GhHD-1, GhMYB25, GhMYB25-like, and GhMYB109 (Pu et al., 2008; Machado et al., 2009; Walford et al., 2011, 2012), were severely down-regulated in ovules cultured with 50 μM ZT and 5 μM IAA (Fig. 3E). These genes showed restored transcription when the IAA concentration was adjusted to 55 μM (Fig. 3E). These results indicated that auxin can relieve CK repression of fiber initiation.
Fig. 3.
Antagonistic effect of cytokinin (CK) and auxin on fiber initiation in cotton. (A–D) IAA antagonizes the inhibitory effect of ZT on fiber initiation. Ovules at –1 d post anthesis (DPA) were cultured in medium containing different concentrations of IAA and ZT together with 0.5 μM GA3 for 15 d. Fiber growth was observed using a stereo-microscope (scale bars are 1 mm) and a SEM (scale bars are 100 μm). (E) Transcription levels of genes associated with fiber initiation in the ovules shown in (A–D). (F) Cytokinins antagonize the promotive effect of IAA on fiber production. Total fiber units were measured based on 20 ovules of proDR5::IPT transgenic and wild-type ovules cultured with 5 μM or 50 μM IAA together with 0.5 μM GA3 for 15 d. Data are means (±SD) of three biological replicates. Significant differences compared with the wild-type were determined using Student’s t-test (**P<0.01). (G) Transcription levels of genes associated with fiber initiation in proDR5::IPT and wild-type ovules cultured with 50 μM IAA and 0.5 μM GA3 for 3 d and assess using RT-qPCR assays. Transcription levels (in arbitrary units) are normalized to that of GhHIS3. Data are means (±SD) of three repeats. TiaaM, ovules from proTCS:iaaM transgenic cotton.
To further confirm the antagonistic relationship between CK and auxin in fiber initiation, we generated transgenic proTCS::iaaM and proDR5::IPT cotton, in which an IAA or CK biosynthetic gene (iaaM or IPT) was under the control of an cytokinin- or auxin-inducible promoter (proTCS or proDR5). Thus, IAA/CKs could be synthesized in situ where CKs/auxin accumulated. On medium containing 50 μM ZT, proTCS::iaaM ovules produced fibers on the surface, while wild-type ovules did not (Fig. 3A, B, D). At the same time, the transcription levels of genes associated with fiber initiation in proTCS::iaaM ovules on medium containing 50 μM ZT and 5 μM IAA were close to that of wild-type ovules cultured with 50 µM ZT and 55 μM IAA (Fig. 3E). These results indicated that CK-induced IAA production could relieve the inhibition of fiber initiation imposed by CKs. Similarly, IAA-induced CK biosynthesis could attenuate the promotive effect of IAA (Fig. 3F). Instead of promotion, the increased IAA resulted in an inhibitory effect on fiber growth of proDR5::IPT ovules compared with that of the wild-type. The decline in fiber units became even more pronounced as IAA concentration was increased (47.9% lower at 5 μM IAA and 59.4% lower at 50 μM IAA compared with the wild-type, Fig. 3F). At the same time, genes associated with fiber initiation, particularly GhHD-1, GhMYB25, and GhMYB25-like, were down-regulated dramatically in proDR5::IPT ovules (Fig. 3G). These results further demonstrated that CK and auxin antagonize each other in the regulation of fiber initiation.
Cytokinin disrupts auxin accumulation in the ovule epidermis
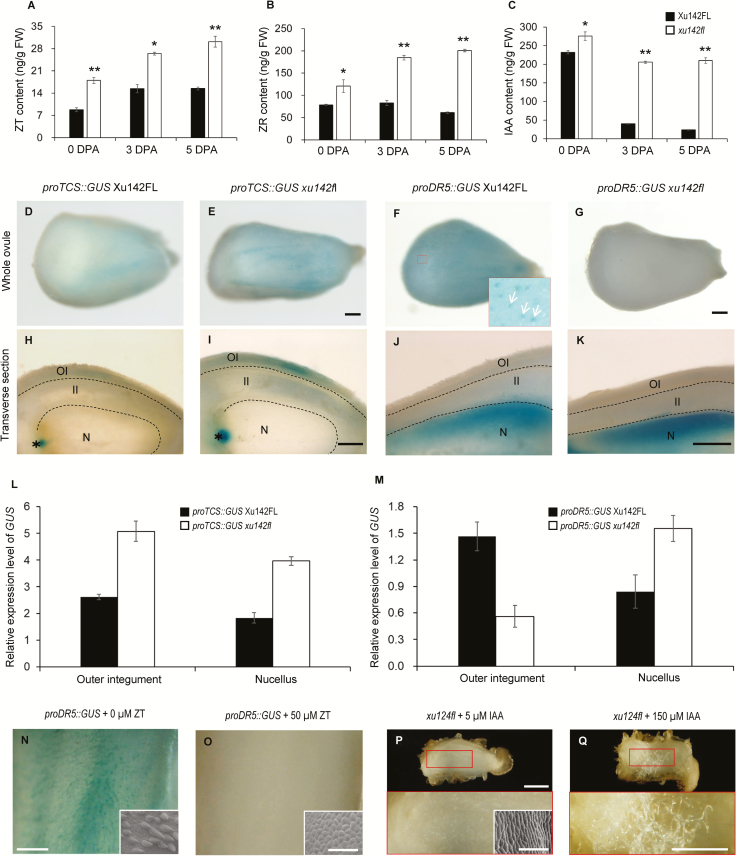
The fiberless mutant xu142fl has been used extensively for studying the molecular mechanism of cotton fiber initiation (Chen et al., 1997; Liu et al., 1999). Our previous study demonstrated that auxin is highly accumulated in initiating fiber cells; however, no noticeable IAA signal was observed in the ovule epidermal layer of xu142fl (Zhang et al., 2011). In the current study, using LC-MS/MS analysis, we compared the contents of IAA and CKs in ovules of the mutant with those in the wild-type (Xu142FL). To our surprise, the IAA content in the mutant ovule at anthesis was significantly higher than that in the wild-type (Fig. 4C). The mutant ovule contained 275.7 ng g–1 IAA, 18.7% higher than that of the wild-type (232.3 ng g–1). For CKs, the concentrations in the ovules of the fiberless mutant were considerably higher than those of the wild-type. In 0-DPA ovules, the concentrations of ZT and ZR of xu142fl were 20.0 ng g–1 and 120.6 ng g–1, respectively, 104.1% and 53.2% higher than that of the wild-type (9.8 ng g–1 and 78.7 ng g–1) (Fig. 4A, B).
Fig. 4.
Cytokinins inhibit cotton fiber initiation through interfering with auxin accumulation in the ovule epidermis. (A–C) Contents of ZT (A), ZR (B), and IAA (C) in ovules of the fiberless mutant xu142fl and wild-type Xu142FL. Ovules at 0, 3, and 5 d post anthesis (DPA) were sampled. Ovules of Xu142FL contained fibers. Data are means (±SD) of three biological replicates. Significant differences between means were determined using Student’s t-test (*P<0.05; **P<0.01). (D, E, H, I) Distribution patterns of proTCS::GUS in ovules of xu142fl and Xu142FL. OI, outer integument; II, inner integument; N, nucellus. Asterisks indicate the chalazal region. Scale bars are 250 μm. (F, G, J, K) Distribution patterns of proDR5::GUS in ovules of xu142fl and Xu142FL. The inset image in (F) is an enlargement of the region indicated by the red square. Arrows indicate fiber cells. Scale are represent 250 μm. (L) Transcription levels of GUS in the outer integument and nucellus of ovules of proTCS::GUS xu142fl and proTCS::GUS Xu142FL. (M) Transcription levels of GUS in the outer integument and nucellus of ovules of proDR5::GUS xu142fl and proDR5::GUS Xu142FL. The outer integument and nucellus were separated from ovules at 0 DPA. Transcription levels (in arbitrary units) are normalized to that of GhHIS3. Data are means (±SD) of three repeats. (N, O) GUS staining of a proDR5::GUS Xu142FL ovule treated with ZT (N) and the control (O). proDR5:GUS Xu142FL ovules at –1 DPA were cultured with 50 μM ZT for 3 d and then used for GUS staining or SEM observation. Scale bars are 500 μm and 100 μm for the insets. (P, Q) Fiber growth of xu142fl ovules was recovered by IAA. Ovules of xu142fl at –1 DPA were cultured with 5 μM IAA (P) or 150 μM IAA (Q) for 15 d. The lower images are enlargements of the regions indicated with red squares in the upper images. The inset in (P) is a SEM image of the ovule surface. Scale bars are 1 mm (upper images), 250 μm (lower images), and 100 μm (SEM image).
In our previous study, we did not observe any noticeable signal of IAA in the epidermal layer of xu142fl (Zhang et al., 2011); however, in the current study a higher concentration of IAA was detected in the ovules of the mutant. To examine this more closely, we investigated the detailed distribution of the two hormones in the ovule by introducing proDR5::GUS and proTCS::GUS reporter gene cassettes into xu142fl by crossing. Plants with the fiberless and wild-type phenotypes that harbored the proDR5::GUS or proTCS::GUS gene cassette were isolated in the F2 generation. On the day of anthesis, a blue signal was hardly detectable on the surface of fiberless proDR5::GUS xu142fl ovules (Fig. 4G), whereas a strong blue dotted signal appeared on the surface of ovules of the wild-type phenotype (proDR5::GUS Xu142FL) (Fig. 4F). We then separated the outer integument and nucellus from ovules and analysed the GUS transcription levels within them. The GUS transcription level in the outer integument of the wild-type ovules was 1.6-fold higher than that of the fiberless ovule (Fig. 4M). Although GUS activity was observed in the nucellus of ovules of both plants, the activity in proDR5::GUS xu142fl was stronger than that in proDR5::GUS Xu142FL (Fig. 4J, K). This indicated that fiberless ovules contained less IAA in the outer integument but more IAA in the nucellus than wild-type ovules.
In contrast to proDR5::GUS xu142fl, GUS staining on the surface of the ovules of the fibreless proTCS::GUS xu142fl was stronger than that on the ovules of the wild-type phenotype (proTCS::GUS Xu142FL) (Fig. 4D, E). Longitudinal sections showed that the blue signal in the outer integument and chalaza region of proTCS::GUS xu142fl ovules was higher than that of proTCS::GUS Xu142FL ovules (Fig. 4H, I). proTCS::GUS expression in the outer integument and nucellus of the mutant was 93.5% and 116.9% higher than that in the wild-type, respectively (Fig. 4L), indicating that the mutant contained more CKs in these tissues. Immunolocalization assays supported this observation: ZT and ZR signals mainly appeared in the outer layer of the ovule integument, and the signal in the mutant ovule was to some degree stronger than that in the wild-type (Supplementary Fig. S2). These results indicated that a higher level of CKs accumulated in the ovule epidermis of the xu142fl mutant as compared with that of the wild-type.
Taking the results for hormone content and hormone-inducible marker gene expression together, we deduced that it was the shortage of auxin in the ovule epidermis rather than that in the whole ovule that accounted for the fiber defects of xu142fl. The higher level of CKs in the outer integument of the mutant might prevent the accumulation of auxin in the ovule epidermis. To test this hypothesis, we observed auxin distribution in the ovules of proDR5::GUS Xu142FL treated with CK. Fiber initiation was entirely inhibited by treatment with 50 μM ZT, whilst no blue signal appeared on the ovule surface (Fig. 4N, O). This confirmed that extra CKs could disrupt the auxin gradient in the ovule epidermal cells, which is a precondition for fiber initiation. As auxin could antagonize the inhibitory effect of CKs on fiber initiation, we were curious as to whether auxin could rescue the fiber initiation of xu142fl ovules. When the IAA concentration of the medium was increased to 150 μM, fiber growth was partially recovered (Fig. 4P, Q). These results supported the idea that cytokinin triggers an obstacle to auxin accumulation and this is associated with the fiber defect of xu142fl.
GhPIN3a is present in the plasma membrane of non-fiber cells but absent in fiber cells, and thus contributes to auxin accumulation in fibers
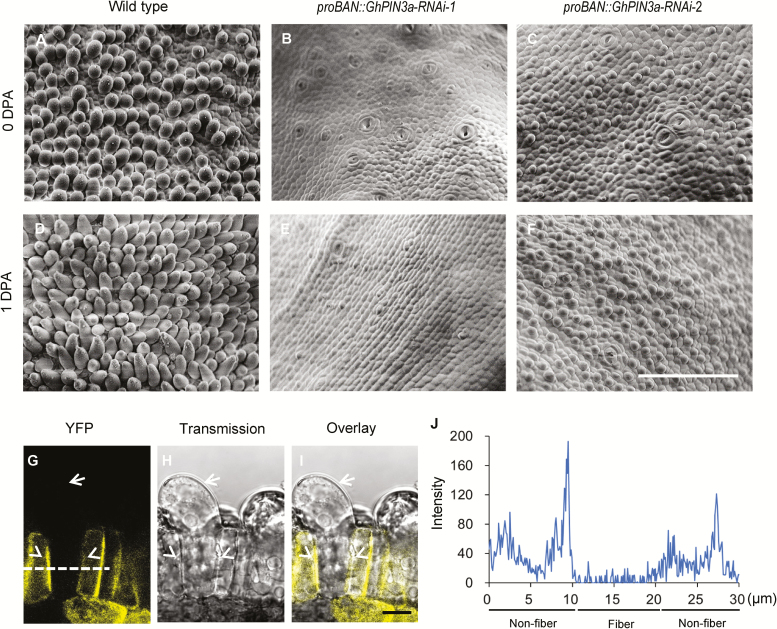
Our previous study indicated that GhPIN3a, which is preferentially expressed in the outer integument of 0-DPA ovules, may be a key regulator in cotton fiber initiation (Zhang et al., 2017). To confirm the role of GhPIN3a in fiber initiation, we expressed an RNAi sequence, generated from the 3´-UTR of GhPIN3a, in the epidermal layer (proBAN::GhPIN3a-RNAi). Down-regulation of this PIN gene (Supplementary Fig. S3) produced severe or even entire inhibition of fiber initiation in transgenic ovules (Fig. 5A–F). We then fused GhPIN3a with yellow fluorescent protein (YFP), and expressed the fused gene. Fluorescence images and quantification of the fluorescence intensity showed that the GhPIN3a::YFP signal mainly appeared at the plasma membrane of non-fiber cells that adjoin the fiber cells (Fig. 5G–J). However, in the neighboring fiber cells, the fluorescent signal was nearly at background level and no localization of GhPIN3a::YFP could be seen at the plasma membrane (Fig. 5G–J, Supplementary Fig. S4). Based on this observation, we suggest that the lack of plasma membrane-located GhPIN3a means that auxin may not be transported out of the fiber cells, and thus it accumulates in the cells.
Fig. 5.
GhPIN3a regulates cotton fiber initiation. (A–F) Fiber initiation on the ovule surface of proBAN::GhPIN3a-RNAi transgenic plants (B, C, E, F) and the wild-type (A, D). Ovules in the middle of each locule were observed using SEM. The scale bar is 100 μm. DPA, d post anthesis. (G–I) GhPIN3a::YFP localization in ovule epidermal cells at 0 DPA. GhPIN3a::YFP is localized to the plasma membrane in non-fiber cells but not in fiber cells. Arrows indicate fiber cells, arrowheads indicate non-fiber cells. The scale bar is 10 μm. (J) Fluorescence intensity along the dashed line in (G).
Cytokinin disturbs the plasma-membrane localization of GhPIN3a in non-fiber cells
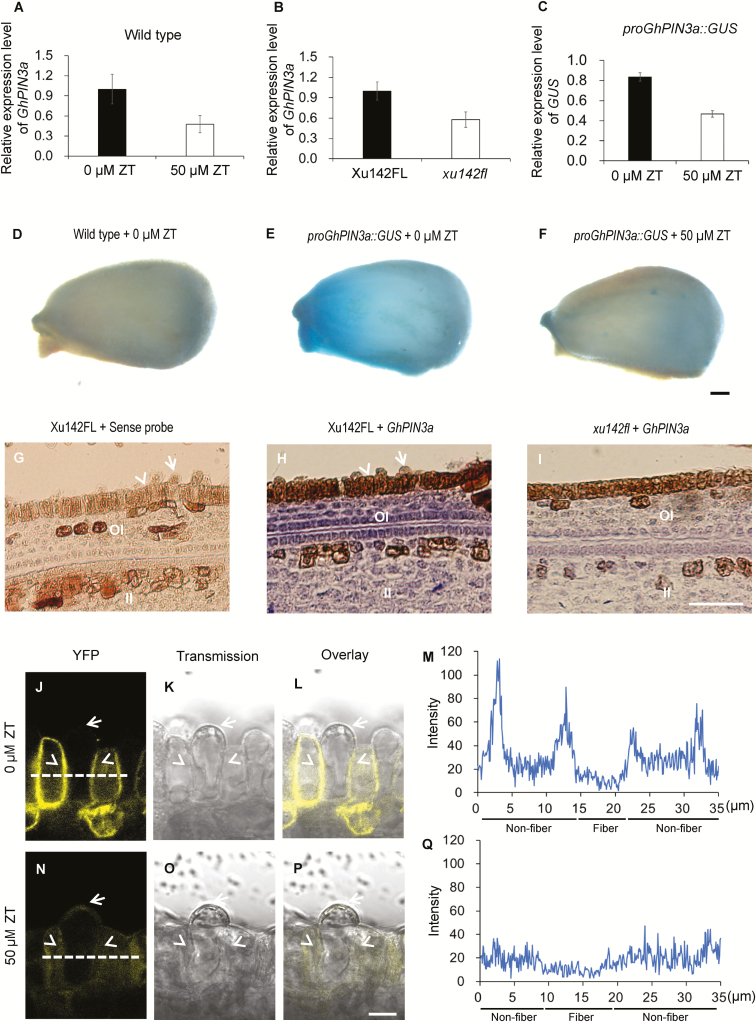
We next investigated the effect of CKs on GhPIN3a expression and protein localization. In ovules treated with ZT the level of transcription of GhPIN3a was considerably decreased (Fig. 6A). Down-regulation of GhPIN3a relative to the wild-type control was also observed in xu142fl ovules (Fig. 6B, G–I). Furthermore, fusing the GhPIN3a promoter sequence with the GUS reporter gene, we found that the GUS signal on the ovule surface of proGhPIN3a::GUS transgenic plants was considerably reduced under treatment with 50 µM ZT (Fig. 6C, D–F), confirming the down-regulation of GhPIN3a by CKs. Interestingly, we found that under ZT treatment, the plasma membrane-located GhPIN3a::YFP in non-fiber cells had almost disappeared, and the signal intensity was dramatically decreased (Fig. 6J–Q), indicating that CKs could induce the delocalization of GhPIN3a in the cells. These results demonstrated that excess CK not only represses the expression of GhPIN3a but also disturbs the polar localization of the protein, which probably in turn undermines the auxin accumulation in cotton fibers.
Fig. 6.
Cytokinins inhibit auxin accumulation in the ovule epidermis of cotton through disturbing GhPIN3a-mediated polar auxin transport. (A) GhPIN3a transcription levels in ZT-treated ovules and the control. Wild-type ovules at 0 d post anthesis (DPA) were treated with 50 μM ZT for 12 h, and then used for RT-qPCR assays. (B) GhPIN3a transcription levels in the outer integument of ovules of the fiberless xu142fl mutant and the Xu142FL wild-type at 0 DPA. (C) GUS transcription levels in ZT-treated proGhPIN3a::GUS ovules and the control. proGhPIN3a::GUS ovules at 0 DPA were treated with 50 μM ZT for 12 h and then used for RT-qPCR assays. Transcription levels (in arbitrary units) are normalized to that of GhHIS3. Data are means (±SD) of three repeats. (D–F) GUS staining of ovules of the wild-type control (D), proGhPIN3a::GUS (E), and ZT-treated proGhPIN3a::GUS (F). Ovules at 0 DPA were treated with or without 50 μM ZT for 12 h. The scale bar is 250 μm. (G–I) GhPIN3a RNA in situ hybridization in the integument of ovules of xu142fl (I) and Xu142FL (H). Transcription of GhPIN3a was repressed in xu142fl as compared with that in Xu142FL. Sections (10 μm) from ovules at 0 DPA were used for in situ hybridization with the GhPIN3a antisense probe. The sense probe was used as the negative control (G). OI, outer integument; II, inner integument. The scale bar is 20 μm. (J–Q) GhPIN3a::YFP localization in the ovule epidermal layer. The polar localization of GhPIN3a::YFP in non-fiber cells was impaired in ovules treated with ZT (N, P, Q) as compared with the control (J, L, M). Ovules at 0 DPA were sectioned by hand and then immersed in 50 μM ZT. After 30 min incubation in the dark, the samples were washed with distilled water and then mounted for observation. Fluorescence intensity along the dashed lines in (J) and (N) was shown in (M) and (Q), respectively. The scale bars is 10 μm. Arrows indicate fiber cells, and arrowheads indicate non-fiber cells in the ovule epidermis.
Discussion
GhPIN3a is the key regulator for fiber initiation
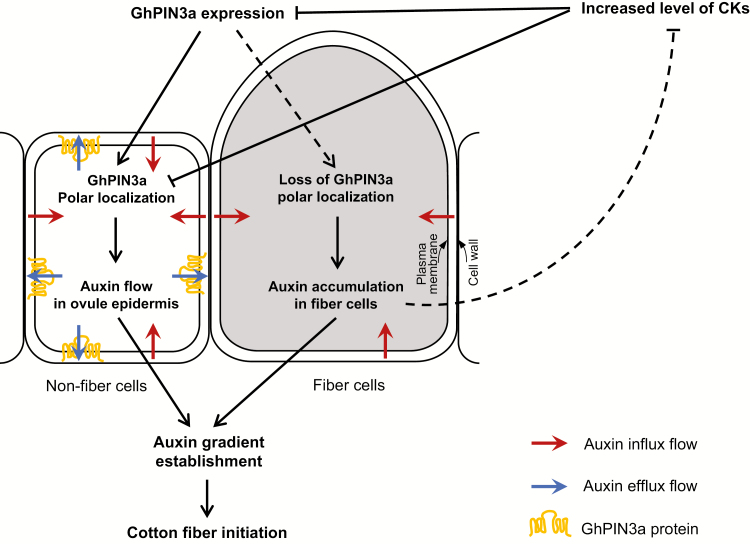
The establishment of an auxin gradient is the precondition for the protrusion of fiber cells (Zhang et al., 2017). Numerous studies have reported that its retargeting to the plasma membrane is the common way for PIN to mediate auxin accumulation (Friml et al., 2002; Harrison and Masson, 2008; Kleinevehn et al., 2010). Recently, we found that GhPIN3a, an auxin efflux carrier, is preferentially expressed in the outer integument of the ovule (Zhang et al., 2017). In this current study, the results from RNA in situ hybridization further showed that the transcription of GhPIN3a in the outer integument is suppressed in the fiberless mutant xu142fl (Fig. 6G–I). Furthermore, seed-coat specific down-regulation of GhPIN3a led to a fiber defect in transgenic ovules (Fig. 5A–F), indicating a crucial role of GhPIN3a in fiber initiation. The question is then how does GhPIN3a direct auxin transport specifically into fiber cells? We demonstrated that GhPIN3a was polarly localized at the plasma membrane of non-fiber cells (Fig. 5G–J). Thus, auxin can be transported within the ovule epidermis through non-fiber cells. At the same, the absence of membrane localization of GhPIN3a in fiber cells would arrest the efflux of auxin out of them, leading to an inevitable accumulation of auxin in the cell for initiation (Fig. 7). This different localization pattern of GhPIN3a in cotton ovule epidermal cells provides an insight as to how auxin can be specifically accumulated in the fiber, a single cell that grows out from a sphere-like surface of the ovule. This also provides evidence that the polar localization of GhPIN3a in cotton ovules may be not associated with gravity, unlike a previous finding regarding PIN proteins (Petrášek and Friml, 2009). Our future work will focus on examining the mechanism of this difference in polar localization between non-fiber and fiber cells.
Fig. 7.
A model showing the antagonistic effect between cytokinins (CKs) and auxin on cotton fiber initiation. An auxin gradient is established in the cotton ovule epidermis by the asymmetric localization of the auxin transporter. GhPIN3a localizes to the plasma membrane of non-fiber cells and facilitates auxin efflux in the ovule integument. However, for reasons yet unknown, this polar localization is absent in neighboring fiber cells, which leads to auxin accumulation in the cell. Increased level of CKs may undermine the establishment of the auxin gradient in the epidermal cells through down-regulating the expression of GhPIN3a and disturbing the polar localization of the protein. The result is that fiber initiation is inhibited. On the other hand, auxin promotes fiber initiation. Augmenting IAA can antagonize the negative effect of CKs and give rise to fiber initiation on the fiberless mutant or on CK-treated ovules.
Cytokinin antagonizes the promotive effect of auxin on fiber initiation through GhPIN3a
What is the relationship between cytokinin and auxin in cotton fiber development? Our data on the contents of CKs and IAA (Fig. 1) and results from genetic manipulation of biosynthesis of cytokinin (proDR5::IPT) or auxin (proTCS::iaaM) (Fig. 3D, F) showed an antagonistic effect between CK and IAA on the initiation of fiber cells. CKs can inhibit IAA accumulation in fiber cells, and thus antagonize the promotive effect of auxin on fiber initiation (Figs 3F, G, 4N, O). On the other hand, auxin can in turn relieve the inhibition of CKs on fiber initiation (Fig. 3A–E). Cytokinin is a crucial hormone for regulating cell proliferation and differentiation in plants (Sakakibara, 2006). Developmentally, cotton fiber is a part of the ovule outer integument. Fiber initiation and elongation is concomitant with enlargement of the seed and seed coat, the process of which requires cell proliferation. Therefore, CK is indispensable for the differentiation and initiation of fiber cells. However, the fine balance of CK homeostasis is crucial. An excessive increase of CK would produce a negative effect on fiber initiation. To increase the yield of cotton fibers and seed, we need to balance the antagonistic relationship between auxin and CK.
In the ovule treated with ZT and ovules of the xu142fl mutant, loss of auxin accumulation in the epidermal layer caused the defects in fiber initiation (Fig. 4G, O). How does CK interfere with fiber-specific accumulation of IAA and thus inhibit the initiation of fiber cells? A series of studies have indicated that CK influences either the transcription of PINs (Laplaze et al., 2007; Dello Ioio et al., 2008; Pernisova et al., 2009; Ruzicka et al., 2009; Moubayidin et al., 2010) or the relocalization and degradation of the protein in order to regulate the establishment of the auxin gradient (Marhavý et al., 2011, 2014). In our study, we found a CK-induced decease of GhPIN3a transcription (Fig. 6A, C, D–F) and a CK-induced reduction of the auxin level in the ovule integument (Fig. 4N, O). At the same time, we observed a CK-induced reduction in plasma membrane localization of GhPIN3a in non-fiber cells (Fig. 6J–Q), which would inevitably block the polar auxin transport (PAT) in epidermal cells, and further result in reduced auxin influx into fiber cells. The lower transcription level of GhPIN3a and lower IAA content in the integument of xu142fl ovules (Figs 6B, G–I, Fig. 4F, G, M) further confirms that GhPIN3a-mediated PAT was disturbed in the mutant, the ovules of which possess higher levels of CKs than the wild-type. In addition, the rapid decrease of CKs after anthesis in wild-type ovules would reduce the negative effect on GhPIN3a-mediated auxin transport, thus allowing more auxin to be imported into the epidermal layer and facilitating cellular accumulation for fiber initiation.
CK mediates the defect of fiber initiation in the xu142fl mutant
The fiberless mutant xu142fl is a useful tool for the study of cotton fiber initiation (Chen et al., 1997; Liu et al., 1999). A genetic assay has indicated that the fiberless phenotype is controlled by two pairs of recessive mutations (Zhang and Pan, 1991). A recent study demonstrated that one of them occurs at the sequence of a MITXA gene, GhMML4-D12, which is responsible for long (lint) fiber production (Wu et al., 2018). In our current study, we demonstrated that by preventing the accumulation of IAA in the outer integument (Fig. 4N, O), the excess of CKs in the ovule may account for the fiber defect of the mutant. LC-MS/MS and auxin-inducible GUS pattern assays confirmed that the mutant ovule had lower levels of auxin in its outer integument than the wild-type control (Fig. 4C, F, G, M). Unexpectedly, the mutant ovule had a significantly higher level of IAA in its nucellus (Fig. 4J, K, M). Our previous work had indicated that auxin in the cotton ovule is mainly from PIN-mediated efflux (Zhang et al., 2017). Accordingly, we suggest that the significantly increased level of CKs in the integument of the mutant prevents auxin from being accumulated there, and this IAA is instead retained in another part of the ovule, particularly in the nucellus. Therefore, it is the attenuation of auxin in the epidermal layer rather than in the whole ovule that accounts for the fiberless phenotype in the xu142fl mutant. The recovered fiber growth of the mutant that results from addition of IAA suggests that the fate of fiber cells still remains in the mutant ovule. That the constraint on fiber initiation can be overcome by auxin implies that the regulation of PAT in epidermal cells may be in the downstream of the MYB transcription factor GhMML4-D12, which is responsible for the production of long fibers. However, the details need further investigation.
Taken together, our results demonstrate an antagonistic effect between CKs and auxin on the initiation of cotton fibers. Different protein localization of GhPIN3a in fiber and non-fiber cells contributes to auxin accumulation in fiber cells. An excess of CKs can disturb GhPIN3a-mediated PAT through both down-regulated gene expression and disabled localization of the protein, and thus antagonize the promotive effect of auxin on fiber cell initiation (Fig. 7). Our study thus provides information that may be valuable for the improvement of cotton yield by the strategy of manipulation of plant hormones.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer and fragment information for plasmid construction.
Table S2. Analysis parameters for CKs and IAA in LC-MS/MS.
Table S3. Primers pairs used for RT-qPCR assays.
Table S4. Contents of CKs and IAA in developing ovules of the cotton cultivar Jimian 14.
Table S5. Contents of CKs and IAA in developing fibers of the cotton cultivar Jimian 14.
Fig. S1. ZT is unable to arrest the growth of initiated fiber cells.
Fig. S2. Immunolocalization of active CKs in ovules on the day of anthesis.
Fig. S3. Transcription levels of GhPIN3a in proBAN::GhPIN3a-RNAi plants compared with the wild-type.
Fig. S4. GhPIN3a::YFP localization in the epidermal layer of ovules on the day of anthesis.
Acknowledgements
We would like to thank Dr Xianlong Zhang, Huazhong Agricultural University, China, for kindly providing the proDR5::GUS cotton. This work was supported by the National Key R&D Program of China (2016YFD0100505-03 to YP), the Fundamental Research Funds for the Central Universities (XDJK2018D021 to JZ), the National Natural Sciences Foundation of China (31130039 to YP, 31871676 to MZ), and the Foundation for the Author of National Excellent Doctoral Dissertation of PR China (201466 to MZ).
Author contributions: YP designed the experiments; all the authors performed the experiments; YP, JZ, and MZ wrote the manuscript; all the authors read and approved the manuscript.
Glossary
Abbreviations:
- CK
cytokinin
- DPA
days post anthesis
- DZ
dihydrozeatin
- iP
N6-isopentenyladenine
- iPR
N6-isopentenyladenosine
- PAT
polar auxin transport
- RT-qPCR
real-time quantitative PCR
- ZR
trans-zeatin riboside
- ZT
trans-zeatin
References
- Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. The Plant Cell 27, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengot L, Marquès-Bueno MM, Jaillais Y. 2016. Regulation of polar auxin transport by protein and lipid kinases. Journal of Experimental Botany 67, 4015–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley CA. 1973. Hormonal regulation of growth in unfertilized cotton ovules. Science 179, 1003–1005. [DOI] [PubMed] [Google Scholar]
- Beasley CA, Ting IP. 1973. The effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. American Journal of Botany 60, 130–139. [Google Scholar]
- Beasley CA, Ting IP. 1974. Effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. American Journal of Botany 61, 188–194. [Google Scholar]
- Bedon F, Ziolkowski L, Osabe K, Venables I, Machado A, Llewellyn D. 2012. Separation of integument and nucellar tissues from cotton ovules (Gossypium hirsutum L.) for both high- and low-throughput molecular applications. Biotechniques 54, 44–46. [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chandler JW, Werr W. 2015. Cytokinin–auxin crosstalk in cell type specification. Trends in Plant Science 20, 291–300. [DOI] [PubMed] [Google Scholar]
- Chen JG, Du XM, Zhou X, Zhao HY. 1997. Levels of cytokinins in the ovules of cotton mutants with altered fiber development. Journal of Plant Growth Regulation 16, 181–185. [Google Scholar]
- Comai L, Kosuge T. 1982. Cloning characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. Journal of Bacteriology 149, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. 2003. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. The Plant Cell 15, 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. [DOI] [PubMed] [Google Scholar]
- Feng X, Dickinson HG. 2007. Packaging the male germline in plants. Trends in Genetics 23, 503–510. [DOI] [PubMed] [Google Scholar]
- Francis D, Sorrell DA. 2001. The interface between the cell cycle and plant growth regulators: a mini review. Plant Growth Regulation 33, 1–12. [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. 2002. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809. [DOI] [PubMed] [Google Scholar]
- Goossens A, Dillen W, De Clercq J, Van Montagu M, Angenon G. 1999. The arcelin-5 gene of Phaseolus vulgaris directs high seed-specific expression in transgenic Phaseolus acutifolius and Arabidopsis plants. Plant Physiology 120, 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Tu L, Hu H, Tan J, Deng F, Tang W, Nie Y, Zhang X. 2012. GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. Journal of Experimental Botany 63, 6267–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison BR, Masson PH. 2008. ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. The Plant Journal 53, 380–392. [DOI] [PubMed] [Google Scholar]
- Hou L, Liu H, Li JB, Yang X, Xiao YH, Luo M, Song SQ, Yang GW, Yan P. 2008. SCFP, a novel fiber-specific promoter in cotton. Science Bulletin 53, 2639–2645. [Google Scholar]
- Hu H, He X, Tu L, Zhu L, Zhu S, Ge Z, Zhang X. 2016. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. The Plant Journal 88, 921–935. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RJ, Schreiber BM. 1997. Role and function of cytokinin oxidase in plants. Plant Growth Regulation 23, 123–134. [Google Scholar]
- Kim HJ, Triplett BA. 2001. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiology 127, 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Kleinevehn J, Ding ZJ, Jones AR, Tasaka M, Morita MT, Friml J. 2010. Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proceedings of the National Academy of Sciences, USA 107, 22344–22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. The Plant Cell 19, 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mo X, Shou H, Wu P. 2006. Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant & Cell Physiology 47, 1112–1123. [DOI] [PubMed] [Google Scholar]
- Liu K, Sun J, Zhang TZ, Pan JJ. 1999. Effects of endogenous and exogenous phytohormones on fiber initiation on ovules from a fuzzless lintless mutant vs. its isogenic wild-type line in upland cotton. Acta Gossypii Sinica 11, 48–56. [Google Scholar]
- Luo M, Xiao Y, Li X, Lu X, Deng W, Li D, Hou L, Hu M, Li Y, Pei Y. 2007. GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. The Plant Journal 51, 419–430. [DOI] [PubMed] [Google Scholar]
- Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES. 2009. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. The Plant Journal 59, 52–62. [DOI] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Developmental Cell 21, 796–804. [DOI] [PubMed] [Google Scholar]
- Marhavý P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benková E. 2014. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Current Biology 24, 1031–1037. [DOI] [PubMed] [Google Scholar]
- Medford JI, Horgan R, El-Sawi Z, Klee HJ. 1989. Alterations of endogenous cytokinins in transgenic plants using a chimeric isopentenyl transferase gene. The Plant Cell 1, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20, 1138–1143. [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisova M, Klima P, Horak J, et al. 2009. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proceedings of the National Academy of Sciences, USA 106, 3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Friml J. 2009. Auxin transport routes in plant development. Development 136, 2675–2688. [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, et al. 2006. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918. [DOI] [PubMed] [Google Scholar]
- Pu L, Li Q, Fan X, Yang W, Xue Y. 2008. The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180, 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusová H, Fendrych M, Friml J. 2015. Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Current Opinion in Plant Biology 23, 116–123. [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Simaskova M, Duclercq J, Petrasek J, Zazimalova E, Simon S, Friml J, Van Montagu MC, Benkova E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proceedings of the National Academy of Sciences, USA 106, 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser U. 1999. Cotton fiber initiation and histodifferentiation. In: Basra AS. ed. Cotton fibers: developmental biology, quality improvement, and textile processing. New York: Haworth Press, 1–46. [Google Scholar]
- Sakakibara H. 2006. Cytokinins: activity, biosynthesis, and translocation. Annual Review of Plant Biology 57, 431–449. [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. 2009. Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin-yang of hormones: cytokinin and auxin interactions in plant development. The Plant Cell 27, 44–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagull R, Giavalis S. 2004. Pre-and post-anthesis application of exogenous hormones alters fiber production in Gossypium hirsutum L. cultivar Maxxa GTO. The Journal of Cotton Science 8, 105–111. [Google Scholar]
- Shimizu-Sato S, Tanaka M, Mori H. 2009. Auxin–cytokinin interactions in the control of shoot branching. Plant Molecular Biology 69, 429–435. [DOI] [PubMed] [Google Scholar]
- Tan JF, Tu LL, Deng FL, Wu R, Zhang XL. 2012. Exogenous jasmonic acid inhibits cotton fiber elongation. Journal of Plant Growth Regulation 31, 599–605. [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. 2006. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. The Plant Journal 45, 1028–1036. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin: a trigger for change in plant development. Cell 136, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Walford SA, Wu Y, Llewellyn DJ, Dennis ES. 2011. GhMYB25-like: a key factor in early cotton fibre development. The Plant Journal 65, 785–797. [DOI] [PubMed] [Google Scholar]
- Walford SA, Wu Y, Llewellyn DJ, Dennis ES. 2012. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. The Plant Journal 71, 464–478. [DOI] [PubMed] [Google Scholar]
- Wang XC, Li Q, Jin X, Xiao GH, Liu GJ, Liu NJ, Qin YM. 2015. Quantitative proteomics and transcriptomics reveal key metabolic processes associated with cotton fiber initiation. Journal of Proteomics 114, 16–27. [DOI] [PubMed] [Google Scholar]
- Wu H, Tian Y, Wan Q, et al. 2018. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytologist 217, 883–895. [DOI] [PubMed] [Google Scholar]
- Xia XC, Hu QQ, Li W, Chen Y, Han LH, Tao M, Wu WY, Li XB, Huang GQ. 2018. Cotton (Gossypium hirsutum) JAZ3 and SLR1 function in jasmonate and gibberellin mediated epidermal cell differentiation and elongation. Plant Cell, Tissue and Organ Culture 133, 249–262. [Google Scholar]
- Xiao YH, Li DM, Yin MH, et al. 2010. Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. Journal of Plant Physiology 167, 829–837. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. 2009. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. The Plant Cell 21, 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng QW, Qin S, Song SQ, Zhang M, Xiao YH, Luo M, Hou L, Pei Y. 2012. Molecular cloning and characterization of a cytokinin dehydrogenase gene from upland cotton (Gossypium hirsutum L.). Plant Molecular Biology Reporter 30, 1–9. [Google Scholar]
- Zhang M, Zeng JY, Long H, Xiao YH, Yan XY, Pei Y. 2017. Auxin regulates cotton fiber initiation via GhPIN-mediated auxin transport. Plant & Cell Physiology 58, 385–397. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zheng X, Song S, et al. 2011. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nature Biotechnology 29, 453–458. [DOI] [PubMed] [Google Scholar]
- Zhang TZ, Pan JJ. 1991. Genetic analysis of fuzzless-lintless mutant in upland cotton. Jiangsu Journal of Agricultural Sciences 7, 13–16. [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. 2010. Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang ZT, Li M, Wei XZ, Li XJ, Li BY, Li XB. 2015. Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnology Journal 13, 269–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.