Silencing expression of SlSWEET1a substantially reduced hexose accumulation in young leaves of tomato plants, revealing the key role of SlSWEET1a in glucose import to sink leaves.
Keywords: Allocation, Micro-tom, phloem, sink and source, Solanum lycopersicum, sugar transport, SWEET, uniporter
Abstract
Sugar allocation from source to sink (young) leaves, critical for plant development, relies on activities of plasma membrane sugar transporters. However, the key sugar unloading mechanism to sink leaves remains elusive. SWEET transporters mediate sugar efflux into reproductive sinks; therefore, they are promising candidates for sugar unloading during leaf growth. Transcripts of SlSWEET1a, belonging to clade I of the SWEET family, were markedly more abundant than those of all other 30 SlSWEET genes in young leaves of tomatoes. High expression of SlSWEET1a was also detected in reproductive sinks, such as flowers. SlSWEET1a was dominantly expressed in leaf unloading veins, and the green fluorescent protein (GFP) fusion protein was localized to the plasma membrane using Arabidopsis protoplasts, further implicating this carrier in sugar unloading. In addition, yeast growth assays and radiotracer uptake analyses further demonstrated that SlSWEET1a acted as a low-affinity (Km ~100 mM) glucose-specific carrier with a passive diffusion manner. Finally, virus-induced gene silencing of SlSWEET1a expression reduced hexose accumulation to ~50% in young leaves, with a parallel 2-fold increase in mature leaves. Thus, we propose a novel function for SlSWEET1a in the uptake of glucose into unloading cells as part of the sugar unloading mechanism in sink leaves of tomato.
Introduction
In plants, sugars are the key carbon source for energy production and macromolecule biosynthesis, and they are important for carbohydrate accumulation in crop organs consumed as food by humans (Braun et al., 2014). Thus, sugar allocation between source leaves (net exporters) and sink organs (net importers), such as young leaves, flowers, and storage organs, is of primary importance for organ development and plant growth, and as a sustainable food supply (Ainsworth and Bush, 2011; Rossi et al., 2015). For long-distance allocation, sugars are primarily synthesized by photosynthetic carbon fixation in source leaves and are loaded, mostly as sucrose, into phloem-specific companion cell (CC)–sieve element (SE) complexes. This loading occurs either symplasmically via plasmodesmata or via the apoplasmic route, with fine-tuned regulation of sugar partitioning (Voitsekhovskaja et al., 2006; Rennie and Turgeon, 2009; Zhang and Turgeon, 2018). Regarding the apoplasmic route, plasma membrane-localized sugar transporters facilitate sucrose efflux and import between adjunct CCs and parenchyma cells of minor veins, promoting mass flow in the phloem for continuous sugar allocation (Sauer, 2007; Chen et al., 2015a; Julius et al., 2017).
During sucrose loading, SWEET (sugar will eventually be exported transporters) proteins, especially clade III SWEET members localized in the plasma membrane of phloem parenchyma cells, are probably the main efflux mechanism to facilitate sucrose export into the apoplast in both dicot (Chen et al., 2012; Eom et al., 2015) and monocot plants (Bezrutczyk et al., 2018). An absence of SWEET exporters significantly suppresses sucrose loading and plant growth, with accumulation of sugars and starch in source leaves (Chen et al., 2012; Le Hir et al., 2015; Bezrutczyk et al., 2018). The final loading step is mediated by SUT1/SUC2-type (sucrose transporter; H+/sucrose symporter) in the plasma membrane of phloem cells, that actively imports apoplasmic sucrose into CC–SE complexes against a concentration gradient (Sauer, 2007; Julius et al., 2017). Plant species with decreased SUT1 uptake ability were stunted, due to lack of sugar loading from source leaves (Gottwald et al., 2000; Srivastava et al., 2008; Slewinski et al., 2009).
Similarly, sucrose unloading from phloem complexes to sink organs can be symplasmic and apoplasmic depending on organ types and developmental stages (Patrick, 1997; Ayre, 2011; Julius et al., 2017). The initial phloem unloading includes sucrose fluxes across SE, CC, and adjacent phloem parenchyma cells. Along the post SE–CC unloading pathway, a fraction of the sucrose could be unloaded from the CCs to the apoplast, probably by simple or facilitated diffusion (Patrick, 1997; Julius et al., 2017). The apoplasmic sucrose can be either directly imported into nearby parenchyma cells by a sucrose transporter, or hydrolyzed by a cell wall invertase into hexoses that are subsequently imported by monosaccharide transporters (Ayre, 2011; Osorio et al., 2014; Julius et al., 2017). Although many sugar transporter families have been characterized in Arabidopsis and other crops, transport mechanisms involved in initial sugar unloading are largely unknown. Some SUT-type sucrose transporters were implicated in sucrose export from CCs (Braun and Slewinski, 2009). Monosaccharide transporters (MSTs) (Sherson et al., 2003), such as STP (sugar transport protein) transporters (Sherson et al., 2000, 2003), may be involved in hexose import; however, experimental evidence for their physiological roles is lacking. SWEET transporters not only mediate sucrose efflux for phloem loading, but are also major sugar unloaders mediating sugar efflux, as well as import, from maternal connecting tissues to reproductive sinks (Lin et al., 2014; Chen et al., 2015b; Yang et al., 2018). SWEETs have been characterized as low-affinity hexose and sucrose bidirectional transporters (exporters or importers), depending on physiological demands. AtSWEET9 specifically localizes in the plasma membrane of inflorescence parenchyma cells to facilitate sucrose efflux for nectar formation (Lin et al., 2014). Clade III SWEETs expressed in maternal cells of developing seeds catalyzed sucrose efflux and import, supporting embryo development in Arabidopsis (Chen et al., 2015b) and rice (Yang et al., 2018). Moreover, the hexose export activity of ZmSWEET4c in corn and OsSWEET4, its ortholog in rice, is of primary importance for seed filling and appears to be linked to seed size (Sosso et al., 2015). These studies highlight the possibility that SWEETs could have a fundamental role in sugar unloading in sink organs as early as the seedling development stage (Eom et al., 2015).
During vegetative stages, young leaves and roots are major sink organs (Wardlaw, 1990), with up to 60% of newly synthesized sucrose exported to sink leaves. Unlike simple rosette leaves in Arabidopsis, tomato (Solanum lycopersicum) has sequentially developed compound leaves, with multiple leaflets that require an extended morphogenetic phase (Shwartz et al., 2016), suggesting that a controlled, fine-gated sugar unloading mechanism is needed through the phloem to developing leaves. Tomato is an important vegetable crop worldwide (United Nations Food and Agriculture Organization; http://www.fao.org/), with many efforts focused on sugar allocation to improve fruit size, sweetness, and quality (Ruan et al., 2012; Gascuel et al., 2017). However, vigorous vegetative growth is the primary determinant of tomato biomass and fruit yield (Vicente et al., 2015). Thus, adequate leaf production could increase flower numbers and fruit weight up to 60% (Jiang et al., 2013; Vicente et al., 2015). However, transport mechanisms regulating initial sugar unloading to sink leaves are largely unknown in tomato. Early studies using either radiotracers, a membrane-impermeable fluorescent carboxyfluorescein, or green fluorescent protein (GFP) fusion proteins have demonstrated that initial phloem unloading in sink leaves is largely via a symplasmic pathway (Schmalstig and Geiger, 1985; Turgeon, 1987; Roberts et al., 1997). However, these data do not exclude an apoplasmic component. For instance, developing maize leaves have been found to switch dynamically from symplasmic to apoplasmic phloem unloading (Baker et al., 2016). These results imply that putative, as yet unidentified, membrane carriers which reside in unloading cells may participate in the apoplasmic sugar unloading into tomato sink leaves.
To date, only two sugar transporters, LeSUT1 and LeSUT2 sucrose transporters, have been implicated in source to sink sugar distribution during vegetative growth of tomato plants (Barker et al., 2000; Weise et al., 2000; Osorio et al., 2014). LeSUT1 proteins are localized in the SEs and appear to have a major role in sucrose loading in source leaves (Barker et al., 2000; Weise et al., 2000), as impaired LeSUT1 expression increased accumulation of soluble sugars and starch in mature leaves (Hackel et al., 2006). Although SE-localized LeSUT2 has been implicated in sucrose unloading to sink leaves, LeSUT2 antisense mutants reduced fruit fertility, with no apparent changes in sugar concentrations in leaves or vegetative tissues (Hackel et al., 2006). Therefore, LeSUT2 is probably not the major sucrose unloader required for sugar unloading in young leaves. Based on the insensitivity of sucrose unloading to a metabolic inhibitor (Schmalstig and Geiger, 1985), and a steep sucrose concentration gradient between SE–CC complexes and the apoplast in most sink organs performing apoplasmic unloading (Patrick, 1997), a passive sucrose facilitator, such as SWEET sucrose carriers, has been suggested to mediate the efflux (Osorio et al., 2014; Julius et al., 2017). Yet, the genetic evidence is still missing.
On the other hand, a hexose facilitator may also be required to act in series to re-load apoplasmically hydrolyzed sucrose into the downstream phloem parenchyma cells (Patrick, 1997). For instance, radiotracer analysis indicated that glucose can be retrieved from the apoplasm of sink leaves by phloem parenchyma cells (Schmalstig and Geiger, 1985). Thus, a glucose facilitator (e.g. SWEETs) on the plasma membrane of parenchyma cells could be a possible candidate to perform this function driven by the relatively low glucose concentrations in the cytoplasm, compared with the apoplasm (Slewinski, 2011; Ruan, 2014).
We deduced that the sink leaf-expressed SlSWEET transporter could be a promising sugar unloader for vegetative growth. Although the complete SWEET gene family has been identified in tomato ‘Heinz 1706’ and its expression pattern characterized by RNA sequencing (RNA-seq) (Feng et al., 2015; Shammai et al., 2018), the role of SWEET proteins in phloem unloading during the vegetative stage in tomato is unknown. The objective in this study was to characterize transport properties of the most highly expressed SlSWEET1a (Solyc04g064610) in young developing leaves of tomato plants. Dominant expression of SlSWEET1a–β-glucuronidase (GUS) fusion proteins in unloading veins and high correlation between SlSWEET1a transcripts and carbon importing activity in sink leaves implicated that it had a specific role in sugar unloading. Furthermore, reduced hexose accumulation in young leaves consistently occurred in SlSWEET1a-silenced tomato plants. Thus, a putative role for SlSWEET1a in apoplasmic sugar unloading to sink leaves is discussed.
Materials and methods
Plant materials and growth conditions
The Micro-tom cultivar of tomato (Solanum lycopersicum) was used, except where indicated. Tomato seeds were surface-sterilized for 8 min using diluted bleach solution (30% CLOROX bleach and 0.1% Triton X-100, v/v) and then washed three times with sterilized water. To propagate tomato plants, sterilized seeds were directly germinated in water at room temperature for 2 d and then transferred to a hydroponic system or pots (diameter, 11.5 cm) containing a mixture of soil and peat moss (1:1). All plants were grown in a controlled chamber (25 °C and 16/8 h light/dark regime with ~100 μmol m−2 s−1 illumination) before harvest or experiments. To collect various organs (except fruits), tomato plants were grown hydroponically for 3 weeks (for vegetative organs) to 5 weeks (for reproductive organs), then roots, stems, mature leaves (the terminal leaflet ~4 cm), young leaves (the terminal leaflet <2 cm), flower buds (developing green buds), and flowers (first day of anthesis) were collected (Supplementary Fig. S1 at JXB online). For leaves, all leaflets from one leaf were pooled together as one sample. Various stages of fruits were collected from soil-grown plants after 14, 21, 35, or 42 d of flowering. To analyze expression in different regions of leaves, the terminal leaflets of young leaves that were 80% of the final size (~3–3.5 cm) were harvested from 5-week-old plants. Then, segments of 1 cm in length from the leaf base to tip were separated for mRNA analysis. For mesophyll protoplast isolation, Arabidopsis thaliana ecotype Col-0 plants were grown in peat moss, vermiculite, and perlite (6:1:1) under short-day conditions (22 °C, 10/14 h light/dark regime, ~80 μmol m−2 s−1 illumination) for 4–5 weeks before isolation.
Analysis of mRNA transcripts
Total RNA was isolated from tomato organs with TRIsure reagent (Bioline, http://www.bioline.com/) with an RNA purification column (GeneMark, http://www.genemarkbio.com/), whereas RNA samples from fruits were prepared using the cetyltrimethylammonium bromide (CTAB) method (Ouyang et al., 2014). Total RNA transcripts were reverse-transcribed, and gene-specific primers for all 31 SlSWEET genes (primer sequences are listed in Supplementary Table S1) were used for real-time quantitative PCR (qRT-PCR) as described (Chen et al., 2015). Expression of the reference gene SlActin7 (Solyc11g005330) was used to determine relative expression levels according to the following equation: 1000×2−(CtSlSWEET−CtSlActin7) (Ct=threshold cycle; Chen et al., 2015).
Localization of GUS fusion proteins
To generate SlSWEET1a–GUS fusion proteins for observation, the entire DNA fragment of SlSWEET1a (1467 bp), including all introns and deletion of the stop codon, was amplified from genomic DNA prepared from Micro-tom leaves with specific primers (forward, 5'-CCGCGGATGGGTGTTGTTCATACTCTG-3'; and reverse, 5'-CTGCAGTCCAGCGCCAACTT GCTCAAGCCTTGACTT-3'). The resulting fragments were first cloned into a pGM-T vector using AT cloning technology and then cloned into pUTKan (including the GUS fragment) binary vector by SacII and PstI restriction sites (pUTKan-gSlSWEET1a). Subsequently, the native promoter of SlSWEET1a (2000 bp) was amplified with specific primers (forward, 5'-TTAAACCTGATCGTCACATAAA-3'; and reverse, 5'-CAGAGTATGAACAACACCCATGTC-3') and cloned into pGM-T, and then cloned into a pUTKan-gSlSWEET1a binary vector, using SacI and KpnI restriction sites. Then, Micro-tom plants were transformed with the resulting pUTKan-PSlSWET1a::gSlSWEET1a–GUS construct in the Academia Sinica Transgenic Plant Core Lab (http://transplant.sinica.edu.tw/en/aboutus/intro/index3.htm). In total, five T1 transgenic tomato plants were obtained. Due to limited numbers of T1 transgenic tomato plant leaves, three young leaves were collected from three independent T1 transgenic lines and three mature leaves were collected from other independent lines. Flowers were collected from T2 lines. All samples were examined by histochemical staining, 16 h for young leaves and mature leaves, and 9 h for flowers, as described (Guo et al., 2014). Stained organs were imaged with a LEICA Z16 APO microscope.
Translocation of carboxyfluorescein
To explore the phloem unloading fluxes in tomato sink leaves, the abaxial surface of the terminal leaflet of mature source leaves from 2-week-old tomato plants was gently abraded with fine sandpaper. A drop of 200 µl of CFDA [5(6)-carboxyfluorescein diacetate, Sigma-Aldrich, USA] solution (300 µg ml−1) was applied to each leaf, which was then covered with plastic wrap to prevent evaporation. After 3 h, fluorescence in the terminal leaflets (<2 cm) of attached sink leaves on intact plants was observed and imaged using a Leica fluorescence microscope equipped with a SOLA light engine (Lumencor, USA) and a Nikon EOS700d digital camera. Images were taken under a GFP filter set and exported by the EOS utility software.
Subcellular localization of GFP fusion proteins
To generate SlSWEET1a–GFP fusion proteins, the cDNA fragment of SlSWEET1a (741 bp) without the stop codon was amplified from the pDRf1-GW-SlSWEET1a plasmid (see ‘Expression of clade I SlSWEET1a in yeast’) with specific primers (forward, 5'-CACCATGGGTGTTGTTCATACT-3'; and reverse, 5'-GGCGCCAACTTGCTCAAGCCTTGACTTGC-3') and cloned into a pENTR-D-TOPO vector using TOPO cloning technology (Invitrogen, USA). Subsequently, the cDNA was transferred into pUBN-GFP and pUBC-GFP via an LR reaction using Gateway technology (Grefen et al., 2010). Arabidopsis and tobacco protoplasts were then transformed with the resulting pUBN-SlSWEET1a and pUBC-SlSWEET1a constructs to express N- and C-terminal translational GFP fusion proteins, respectively.
Arabidopsis mesophyll protoplasts were isolated as described (Wu et al., 2009). A portion of the protoplast preparation (200 µl at a cell density of 2×105 ml−1) was mixed with 10–20 μg of plasmids and 220 μl of polyethylene glycol (PEG) solution prior to incubation for 15 min, at 25 °C in the dark. Protoplasts were then washed and re-suspended in 1 ml of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7) and incubated for 24–48 h in the dark before observation. To determine the position of the plasma membrane, protoplasts were either co-transformed with the membrane marker pRT101-AtPIP2A–red fluorescent protein (RFP) in a 1:1 ratio (Nelson et al., 2007) or stained with 0.5 μl of 10 μM FM4-64 dye just before observation (Chen et al., 2015). Fluorescence was observed on a Carl Zeiss LSM780 confocal microscope (Instrument Development Center, National Cheng Kung University) as described (Chen et al., 2015).
Expression of clade I SlSWEET in yeast
To generate expression constructs for yeast, cDNA sequences of clade I SlSWEET were amplified using Phusion polymerase (New England Biolabs, USA) with gene-specific primers (primer sequences are given in Supplementary Table S2) from leaf cDNA derived from tomato cultivar ‘TS19’. The cDNA sequences were confirmed by sequencing and had 100% identity with those in the Sol Genomics Network (https://solgenomics.net/). Then, cDNA was first cloned into the pDONR221-f1 vector and subsequently transferred to the pDRf1-GW vector using Gateway technology. The yeast strains YSL2-1 (Chen et al., 2015) and EBY4000 were then transformed with the resulting pDRf1-GW-SlSWEET plasmids using the lithium acetate (LiAC) method (Gietz and Schiestl, 2007). Transformants were selected on synthetic deficient medium without uracil (SDM-U medium, containing 1.7 g of yeast nitrogen base without amino acids, 5 g of ammonium sulfate, 2% maltose, 2% agar, and 0.01% histidine, leucine, and tryptophan in 1 liter). The pDRf1 empty vector with a Gateway cassette deleted and a plasmid pDRf1-Hxt5 that harbored a yeast hexose transporter were also transformed as a negative and positive control, respectively, of transport activity (Chen et al., 2015).
Yeast growth assay
Yeast cells from single colonies were grown at 30 °C overnight and then refreshed with new SDM-U medium until they reached early mid-log phase growth (OD600 of 0.8–1.0). Cells were collected and diluted with SD-U medium (SDM-U medium without 2% maltose) to an OD of 0.2. Then, serial dilutions (1, 10–1, 100–1, and 1000–1) of all desired cells were prepared and 5 μl from each dilution was spotted on SDM-U medium or SD-U medium supplemented with 2% glucose, galactose, mannose, fructose, or sucrose. To maintain the media at pH 5 or 7, Tris-MES buffers (6.054 g of Tris and 9.76 g of MES in 1 liter) were included. Plates were incubated at 30 °C for 3–6 d and then scanned.
Radiotracer uptake assay in yeast
One portion of overnight yeast culture was diluted with 50 ml of fresh SDM-U medium to an OD600 of 0.2. After incubation for 4 h at 30 °C, yeast cells (~0.5 OD600) were collected, washed, and re-suspended in 50 mM sodium phosphate buffer (112 mg of Na2HPO4 and 6.8 g of NaH2PO4 in 1 liter, pH 5) to an OD600 of 5 before assay. For a time-dependent uptake assay, uptake buffer containing 2 mM glucose and 1.5 μCi of [14C]glucose ml−1 in 50 mM sodium phosphate buffer (pH 5) was prepared. For a concentration-dependent uptake assay, uptake buffers with 11iter glucose concentrations (0.2, 1, 2, 5, 10, 20, 40, 100, 200, and 500 mM glucose in 50 mM sodium phosphate buffer, pH 5) with same molar ratio of μCi of [14C]glucose were prepared. For competition assays, uptake buffers (2 mM glucose with 2 μCi of [14C]glucose ml−1) without other sugars (control) or supplemented with 10-fold concentrations of various cold sugars (20 mM glucose, galactose, fructose, mannose, sucrose, and maltose, respectively) were prepared. For analysis under various pH values, uptake buffer medium with various pH values were prepared using 50 mM sodium phosphate containing 2 mM glucose with 2 μCi of [14C]glucose ml−1. Before the uptake assay, cells were washed once with corresponding cold pH buffers. For treatment with the protonophore NH4Cl, yeast cells were pre-treated with 10 mM NH4Cl for 10 min at 30 °C, then uptake buffer of pH 5 (2 mM glucose with 2 μCi of [14C]glucose ml−1) was added. To start the uptake assay, 110 μl of corresponding uptake buffers were always added into an equal volume of cells that was incubated at 30 °C. At the indicated time points in the time-dependent assay or at 10 min in all other assays, cells were collected on filter paper (MCE Membrane Filter, 0.45 μm) and washed three times with ice-cold 50 mM sodium phosphate buffer by vacuum filtration. Filter paper (with cells) was transferred into scintillation vials containing 4 ml of Rotiszint® eco plus (Carl Roth, Germany) and incubated at room temperature for 16–18 h. Radioactivity of lysed cells was quantified in a Tri-Carb 4810TR scintillation counter (Perkin Elmer, USA) and a kinetic curve determined using single rectangular regression (Sigmaplot Version 13).
Induction of VIGS (virus-induced gene silencing) in tomato leaves
VIGS was carried out as described (Lu et al., 2003). In general, the 300 bp fragment of SlSWEET1a for gene silencing was selected via the VIGS tool on the Sol Genomics Network (https://solgenomics.net/) and amplified from the pDONR221-f1-SlSWEET1a plasmid using Phusion polymerase with specific primers (forward, 5'-CACCATGGGTGTTGTTCATACT-3'; reverse, 5'-CTTCTTCTCTTTAGTTGG-3'). The resulting fragment was first cloned into pENTR-D-TOPO (Invitrogen, USA) via TOPO cloning technology, then transferred into pTRV2 via an LR reaction (Lu et al., 2003). Then, Agrobacterium tumefaciens GV3101 was transformed with the resulting pTRV2-SlSWEET1a construct via the freeze–thaw method (Höfgen and Willmitzer, 1988). Positive transformants were selected on LB medium (5 g of yeast extract, 10 g of tryptone, 10 g of NaCl in 1 liter) supplemented with kanamycin (50 mg l−1) and gentamycin (50 mg l−1). A portion of overnight culture (2.5 ml) derived from an Agrobacterium transformant was added to 25 ml of LB fresh medium containing antibiotic, 10 mM MES, and 200 μM acetosyringone (Sigma-Aldrich, USA) and grown overnight at 30 °C. Cells were collected by centrifugation at 4000 g for 5 min and re-suspended to an OD600 of 1.5 with infiltration medium (10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone). An aliquot (1 ml of culture) was injected into cotyledons of 2-week-old tomato plants with a 1 ml syringe. To estimate silencing efficiency, Agrobacterium cells carrying the silencing construct, pTRV2-SlPDS-VIGS that would result in reduced expression of the phytoene desaturase gene so as to inhibit chlorophyll biosynthesis and cause etiolation (Liu et al., 2002), was also injected into other plants.
Analysis of sugar contents
Young leaves (the terminal leaflet <2 cm, <50% of the final size) and mature leaves (the terminal leaflet ~4 cm), including all three leaflets, from VIGS-silenced tomato plants were harvested 2 weeks after injection and stored at −70 °C before extraction. Frozen leaves (0.2–0.5 g) were ground into fine powder and 480 μl of extraction buffer medium [containing 460 μl of methanol (HPLC grade) and 20 μl of 0.2 mg ml−1 ribitol] was added. Mixtures were incubated at 70 °C for 10 min and centrifuged at 14 000 g for 10 min at room temperature. The supernatant was transferred into a glass vial, with 250 μl of cold chloroform (HPLC grade) and 500 μl of cold MQ water added in the given order. Mixtures were vortexed for 15 s and then centrifuged at 4000 g for 15 min at 20 °C. An aliquot of 200 µl of supernatant was transferred into a new 1.5 ml Eppendorf tube and freeze-dried in an LyoLab 5 LT freeze dryer (LSI, USA). Dry samples were derivatized with 20 μl of methoxyamination solution (0.02 g of methoxyamine hydrochloride in 1 ml of pyridine) at 30 °C for 90 min and then the mixture was added to 100 μl of 99:1 BSTFA+TMCS solution (Macherey-Nagel, Germany) and incubated at 70 °C for another 120 min. All extracts were then transferred into 250 μl pulled conical glass insert vials (Agilent, Germany) and analyzed with an Agilent 7890A/5975C GC-MS System with a 60 m×0.250 mm column (Agilent, Germany), as described (Ruiz-Matute et al., 2011; Uri et al., 2014). Sugars were quantified relative to the internal ribitol standard, using the following retention times: glucose, 23.286 min and 23.521 min; fructose, 23.001 min and 23.093 min; galactose, 23.235 min and 23.487 min; mannose, 23.168 min and 23.378 min; and sucrose, 30.869 min.
Results
SlSWEET1a was highly expressed in aerial sink organs
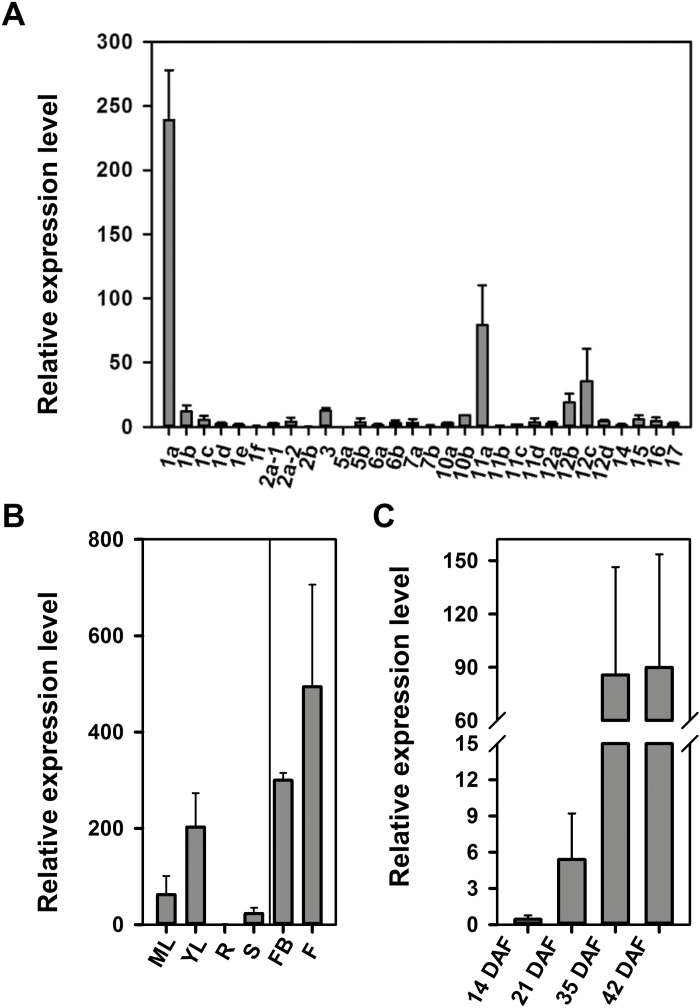
The genome of tomato cultivar ‘Heinz 1706’ was sequenced in 2012 (Tomato Genome Consortium, 2012) and 31 tomato SlSWEET genes have been identified (Feng et al., 2015; Shammai et al., 2018). However, these genes were not well annotated to the SWEET family, which makes it difficult to compare orthologous genes. Therefore, the phylogenic tree of all 31 SlSWEET genes was generated and classified into four clades (Supplementary Fig. S2). To identify the key SlSWEET member responsible for sugar unloading into sink tomato leaves, qRT-PCR was performed for all 31 SlSWEET genes using cDNA isolated from whole new developing leaves (the terminal leaflet was smaller than 2 cm) of 4-week-old hydroponic-grown tomato plants. Based on relative expression levels in young tomato leaves, transcript levels of SlSWEET1a (Solyc04g064610) were markedly higher compared with other genes (Fig. 1A). Interestingly, SlSWEET1a was also highly expressed in reproductive sink organs, such as flower buds and flowers (Fig. 1B), but only weakly expressed during the maturation stage of tomato fruits (Fig. 1C). Nevertheless, expression of SlSWEET1a was increased >10-fold during the maturation stage (35–42 d after flowering) of fruit development.
Fig. 1.
Expression of SlSWEET genes in tomato organs. (A) Stable mRNA transcripts of 31 SlSWEET genes in young leaves. (B) Developmental expression of SlSWEET1a in young leaves (YL), mature leaves (ML), roots (R), stems (S), flowers (F), and flower buds (FB) from 3- (vegetative organs) to 5-week- (reproductive organs) old hydroponically grown tomato plants. (C) Gene expression of SlSWEET1a in several stages of tomato fruits from soil-grown plants. Total RNA was isolated from various organs, and cDNA was used for qRT-PCR with gene-specific primers. Relative expression level by normalizing to an internal control, SlActin7, is shown. Results are means ±SE from three independent biological repeats. DAF=days after flowering.
SlSWEET1a proteins are highly abundant in unloading tissues
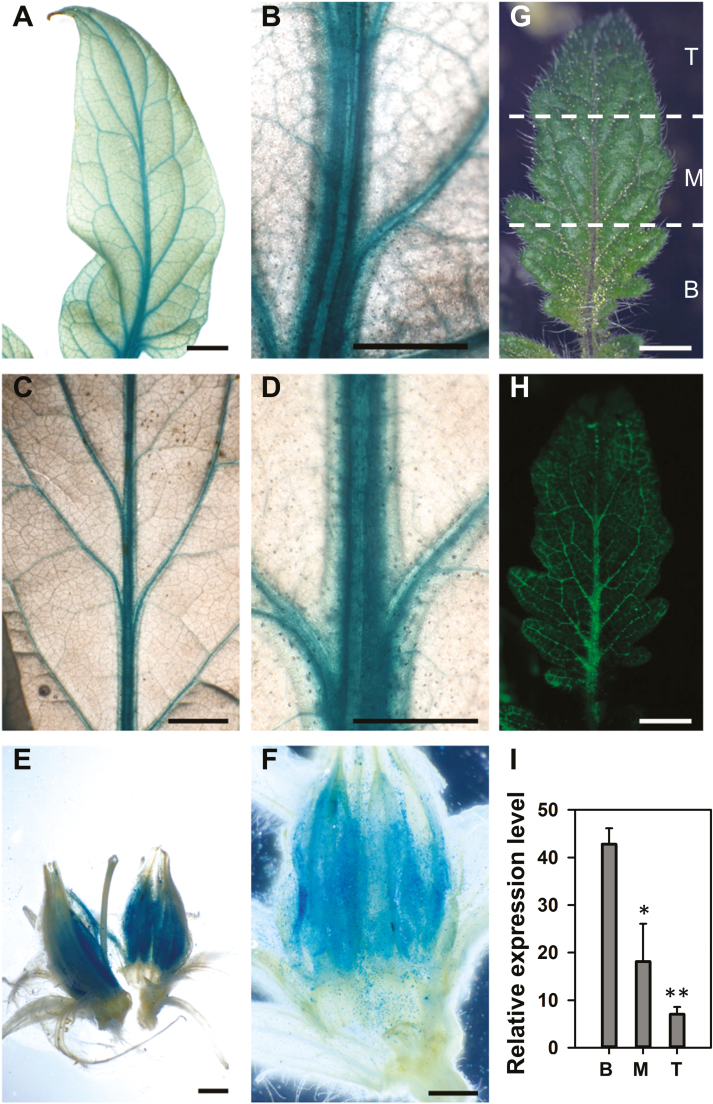
To explore tissue-specific localization of SlSWEET1a proteins, we generated transgenic tomato plants expressing a C-terminal translational GUS gene fusion with the complete genomic SlSWEET1a DNA sequence, containing all introns driven by the native SlSWEET1a promoter (SlSWEET1a–GUS). After shoot regeneration and transfer to potting mix for 1.5 months, mature and new leaves from T1 tomato plants were stained to detect GUS activity. Predominant blue staining was observed in the first- to third-order veins in both young (Fig. 2A, B) and mature leaves (Fig. 2C, D). Dominant expression of the SlSWEET1a–GUS fusion in leaf veins was also observed in the other four independent T1 transgenic tomato plants (Supplementary Fig. S3A). The same localization to leaf veins was also observed in the three T2 transgenic plants, although with less staining (data not shown). Low expression of SlSWEET1a–GUS in T2 plants may be the result of an unknown silencing effect, occurring in the tomato Micro-tom cultivar (Khuong et al., 2013). In flowers derived from T2 plants, distinct staining was obvious in stamens (Fig. 2E, F). To determine whether the expression of SlSWEET1a–GUS fusion coincides with sugar unloading, we exposed a nearby mature terminal leaflet to CFDA that, on accumulation in the phloem, is de-esterfied into the membrane-impermeant and fluorescent carboxyfluorescein (CF), to trace carbohydrate unloading in developing leaves (Roberts et al., 1997). After 3 h of CF loading in the nearby mature leaves, fluorescence was clearly observed in the first- (midrib), second-, and third-order veins in a young leaflet (<2 cm) (Fig. 2G, H), but not in mature leaves (Supplementary Fig. S3B). The overall fluorescence intensities exhibited a high to low gradient from leaflet base to tip, indicating that the highest sugar unloading activity was located in the base of the young leaflet. Interestingly, relatively higher mRNA levels of SlSWEET1a were measured in the leaflet base compared with the middle and tip that had reached 80% of their full size (Fig. 2I) and probably undergone sink–source transition (Roberts et al., 1997).
Fig. 2.
Tissue-specific expression of SlSWEET1a proteins. (A–F) Histochemical staining of GUS activity in transgenic tomato plants expressing SlSWEET1a–GUS fusion proteins driven by the SlSWEET1a native promoter. Independent transgenic plants were grown in soil for >4 weeks, and selected organs were harvested for staining for 16 h in (A, B) young leaves and (C, D) mature leaves, and 9 h for (E, F) flowers. Leaves and flowers were harvested from T1 and T2 plants, respectively. Representative terminal leaflets are shown in (A–D). Images in (B), (D), and (F) are magnified images of (A), (C), (E), respectively. (G and H) Fluorescence images of (H) CF dye unloaded into the terminal leaflet of a young leaf (G). The same pattern had been observed in three independent plants. (I)Transcript levels (by qRT-PCR) of SlSWEET1a in terminal leaflets of tomato young leaves. Relative expression levels by normalizing to an internal control, SlActin7, is shown. B, M, and T indicated samples harvested from the leaflet base, middle, and tip, as shown in (G). Results are means ±SE from four independent biological repeats. Significant differences from expression in the leaflet base (B) were determined by Student’s t-test and indicated by one (P<0.05) or two (P<0.01) asterisks. Scale bars are 1 mm in (B), (D), and (F); 2 mm in (A), (C), (E), (G), and (H).
Plasma membrane-specific localization of SlSWEET1a proteins
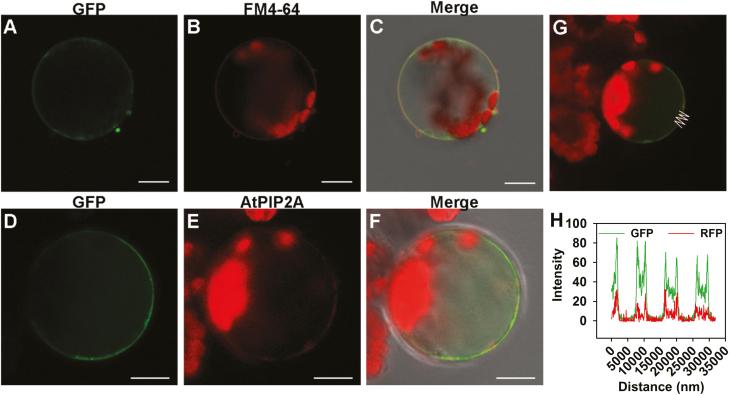
To observe the subcellular localization of SlSWEET1a proteins at a single-cell resolution, the C-terminal SlSWEET1a–GFP was transiently expressed in isolated Arabidopsis mesophyll protoplasts. Green fluorescence derived from SlSWEET1a–GFP enclosed cytosolic chloroplasts indicated by red autofluorescence, and largely overlapped with red fluorescence derived from the plasma membrane dye, FM4-64 (Fig. 3A–C; Wayne, 2009). Furthermore, when Arabidopsis protoplasts were co-transformed with SlSWEET1a–GFP and a plasma membrane marker, AtPIP2–RFP, based on path-analysis of fluorescence intensities, there was co-localization of green and red fluorescence (Fig. 3D–H). Similarly, plasma membrane-specific localization of SlSWEET1–GFP proteins was present in isolated tobacco BY2 cells (Supplementary Fig. S4A). However, N-terminal fusion of GFP to SlSWEET1a resulted in mislocalization and aggregation of the fusion proteins in the cytosol (Supplementary Fig. S4B), suggesting the localization signal was at the N-terminus of SlSWEET1a.
Fig. 3.
SlSWEET1a proteins co-localized with plasma membrane proteins. (A–C) The SlSWEET1a–GFP fusion proteins were transiently expressed in Arabidopsis mesophyll protoplasts driven by the 35S promoter. Fluorescence derived from (A) SlSWEET1a–GFP fusion proteins, (B) FM4-64 staining, and (C) merged signals from the same focal plane are shown. (D–F) Fluorescence images of mesophyll protoplast expressing both SlSWEET1a–GFP fusion proteins and the Arabidopsis membrane marker, AtPIP2A–RFP fusion proteins, are shown. (G and H) Intensity analysis of the indicated line path in the protoplast shown in (F). Scale bar=10 µm.
SlSWEET1a activity complements glucose uptake deficiency
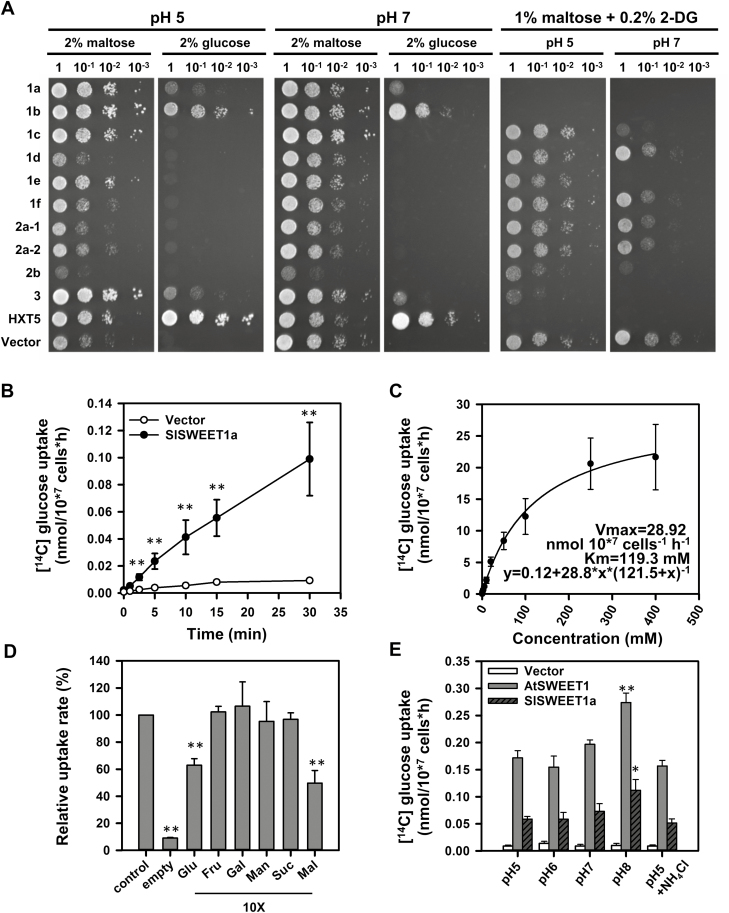
Based on genome structure analysis, the four clade I genes, SlSWEET1a, 1b, 1c, and 1d were clustered in a tandem repeat, implying that these clade I homologs could have been derived from gene duplication (Shammai et al., 2018). Furthermore, due to high identity (31–81%) between amino acid sequences of clade I SlSWEET genes (Supplementary Fig. S5), perhaps all clade I members have similar transport properties. Therefore, cDNAs of clade I SlSWEET genes, 10 genes in total, were isolated and expressed in the baker’s yeast (Saccharomyces cerevisiae) mutant YSL2-1, which lacks all endogenous hexose transporters but expresses a cytosolic invertase and can grow only on medium containing maltose, but not monosaccharides or sucrose (Guo et al., 2014). Expressing the yeast hexose transporter HXT5 allows YSL2-1 to grow on media containing glucose (Fig. 4A). Under acidic growth conditions (pH 5), expression of only SlSWEET1a, 1b, and 3 complemented growth deficiency of YSL2-1 on glucose- and galactose-containing media. However, there was no growth on media with fructose, mannose, or sucrose (Fig. 4A; Supplementary Fig. S6). There were similar trends on medium buffered at neutral pH (pH 7), suggesting that the transport activity of clade I SlSWEETs was not dependent on a pH gradient.
Fig. 4.
Transport activity of SlSWEET1a in yeast. (A) Complementary growth assay in the YSL2-1 yeast mutant line. Yeast transformants expressing clade I SlSWEET genes, yeast hexose transporter (Hxt5), or the empty vector (Vector) were cultured on solid media supplemented with 2% maltose, 2% glucose, or 1% maltose together with 0.2% 2-deoxyglucose (2-DG) under pH 5 and 7 conditions. Images were taken after incubation at 30 °C for 4–6 d. (B) Time-dependent [14C]glucose uptake activity was measured in YSL2-1 yeast cells expressing SlSWEET1a or the empty vector. Result are means ±SE (n=3). Significant differences from cells expressing the empty vector were determined. (C) Kinetics of [14C]glucose uptake activity of SlSWEET1a are shown. Results are means ±SE (n=4–5). (D) Competition binding ability of other sugars was measured by incubating cells with both [14C]glucose and 10-fold concentrations of the indicated sugars (Glu, glucose; Gal, galactose; Fru, fructose; Man, mannose; Suc, sucrose; Mal, maltose). Relative uptake rate was calculated compared with cells incubated with only 1 mM hot glucose (control). The background uptake of non-transformed cells (empty) was also shown. Results are means ±SE (n=5). Significant differences from the control were determined. (E) Effect of various pH conditions and treatment of NH4Cl on SlSWEET1a uptake activity in EBY4000 cells. Yeast cells expressing the empty vector, AtSWEET1, or SlSWEET1a were analyzed. Results are means ±SE (n=3). Significant differences from the pH 5 condition were determined. All statistics were performed by Student’s t-test and indicated by one (P<0.05) or two (P<0.01) asterisks.
YSL2-1 cells transformed with an empty vector grew on 2-deoxyglucose (2-DG) medium. In contrast, cells expressing HXT5 had no detectable growth due to uptake of 2-DG that is a poorly metabolized glucose analog and causes cell death in low amounts (Fig. 4A; Chen et al., 2015). Similarly, only yeast cells expressing SlSWEET1a, 1b, 2b, and 3 had a significant 2-DG-dependent growth inhibition, especially on medium buffered at neutral pH (pH 7). Furthermore low uptake activity of SlSWEET1c and 1e was only observed at neutral pH. These results indicated that SlSWEET1a, as well as some homologs in clade I, probably functions as a glucose-specific passive transporter.
SWEET1 functions as a glucoses-specific low-affinity carrier
To confirm the glucose-specific transport activity of SlSWEET1a, indicated by the growth assay in YSL2-1 yeast cells (Fig. 4A), a time-dependent glucose uptake assay was done using [14C]glucose in YSL2-1 yeast cells expressing SlSWEET1a. Radioactivity had accumulated in transformed yeast cells after 5 min, with a linear increase until 30 min, whereas there was limited glucose uptake in cells expressing the empty vector (Fig. 4B). The kinetics of the glucose transport activity of SlSWEET1a were determined by exposure to glucose concentrations of 0.1–400 mM for 10 min. Significant uptake of [14C]glucose occurred with exposure to 0.1 mM, with saturation of glucose uptake at a concentration close to 200 mM (Fig. 4C). Therefore, the estimated Km and Vmax of SlSWEET1a activities for glucose were 119.3 mM and 28.9 nmol 107 cells−1 h−1, respectively, indicating a low-affinity transport mode.
To address further the substrate specificity of SlSWEET1a, a 10-fold excess of unlabeled sugars was used to compete with [14C]glucose. Only a 10-fold excess of unlabeled glucose, but no other hexoses or sucrose, competed the binding and significantly reduced the amount of [14C]glucose imported (Fig 4D). The lower uptake rate to [14C]glucose under 10-fold higher maltose was attributed to background uptake activity in the YSL2-1 yeast strain (Chen et al., 2010). The specific competition ability of unlabeled glucose was also measured by treatment with a 30-fold excess of sugar substrates (Supplementary Fig. S7). To verify whether SlSWEET1a proteins functioned as other known SWEET transporters, the EBY4000 yeast strain was used. Similar to AtSWEET1 (Chen et al., 2010), glucose uptake activity of SlSWEET1a was not repressed by alkaline pH (Fig. 4E) and only slightly decreased by NH4Cl (10 mM) (Fig. 4E), perhaps due to indirect toxicity.
SlSWEET1a contributes to sugar distribution to sink leaves
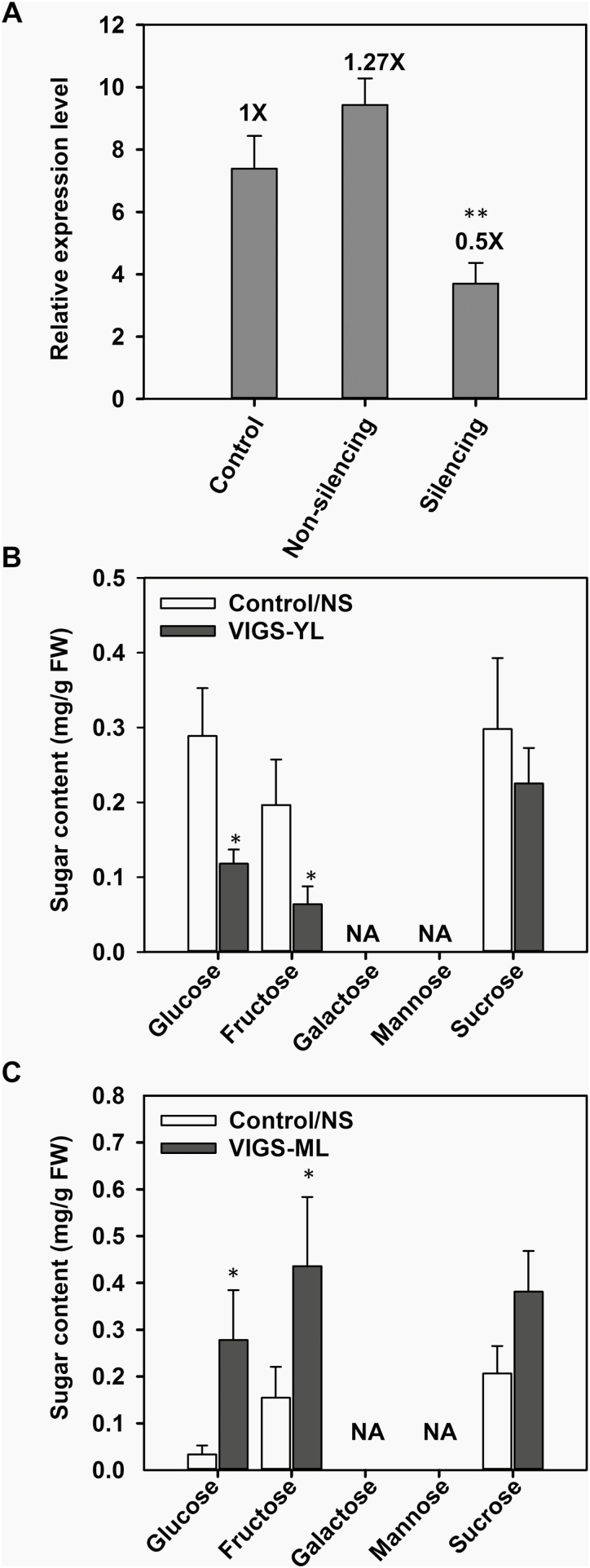
To explore the physiological role of SlSWEET1a in leaves, VIGS was used to transiently silence expression of SlSWEET1a in leaves. The efficiency of VIGS was confirmed by expressing VIGS-SlPDS in parallel plants with leaf etiolation due to silenced expression of the phytoene desaturase gene (Supplementary Fig. S8). Samples collected from non-silenced and control plants (injected with infiltration buffers only, Control/NS) were compared with silenced plants in which SlSWEET1a expression in mature leaves was reduced by 50% (Fig. 5A). In silenced young leaves (VIGS-YL), glucose contents were significantly reduced (>50%) compared with control leaves (Fig. 5B). The same reduction was also observed for fructose, but not for sucrose. In contrast, glucose and fructose contents in silenced mature leaves were increased >2-fold compared with controls (Fig. 5C).
Fig. 5.
Effect of SlSWEET1a deficiency on sugar allocation ability in tomato leaves. (A) Gene expression of SlSWEET1a in matures leaves of tomato plants injected with buffer (Control) or Agrobacterium cells containing the VIGS–SlSWEET1a construct (non-silencing and silencing). Total RNA were isolated and cDNA was used for qRT-PCR with gene-specific primers. Relative expression by normalizing to an internal control, SlActin7. Numbers indicate fold changes of SlSWEET1a expression compared with control leaves. Results are means ±SE (n=5–11). (B and C) Sugar contents in young (B) and mature (C) leaves of tomato plants subjected to the VIGS-SlSWEET1a construct. Two samples from control plants and non-silenced plants in (A) were used as the control/NS for comparison. Results are means ±SE (n=3–4). Significant differences from the control (A) or control/NS leaves (B and C) were determined by Student’s t-test and indicated by one (P<0.05) or two (P<0.01) asterisks.
Discussion
For decades, possible membrane carriers involved in apoplasmic sugar unloading in developing leaves have been the subject of debate. In our study, distinct high expression of SlSWEET1a was detected (qRT-PCR) in young leaves of vegetative tomato plants (Fig. 1), suggesting that the SlSWEET1a proteins may mainly function in young leaves compared with source (mature) leaves. In addition, expression of a whole-gene SlSWEET1a–GUS translational fusion further demonstrated that SlSWEET1a proteins were co-localized in leaf vein tissues (Fig. 2; Supplementary Fig. S3). The GUS staining was most evident in the first- to the third-order veins, representing major sites for unloading of photoassimilates from the transport phloem in both tomato sink leaves (Fig. 2G, H) and developing tobacco leaves (Turgeon, 1987; Roberts et al., 1997). Moreover, the carbohydrate unloading activity in a sink leaflet co-occurred with relatively high levels of SlSWEET1a transcripts in the leaflet base, compared with the tip (Fig. 2G–I), which is the first region to switch to sugar export during sink/source transition (Turgeon, 1987; Roberts et al., 1997). Based on these results, we suspect that SlSWEET1a probably functions in mediating sugar flows along the phloem unloading pathway in sink leaves. The phloem unit is composed of SE–CC complexes and phloem parenchyma, with central control of sugar loading, transport, and unloading in plants (Oparka and Turgeon, 1999; Lalonde et al., 2003). Before post-phloem unloading to neighboring sink cells, sucrose needs to be exported out of SE–CC complexes and into phloem parenchyma (Patrick, 1997). Sucrose movement between SEs and CCs occurs mostly symplasmically (Turgeon, 1987; Kempers and van Bel, 1997; Patrick, 1997; Lalonde et al., 2003). In contrast, the sucrose efflux between CCs and phloem parenchyma can be mediated in parallel via sym- and apoplasmic routes (with the latter requiring sugar-transporting proteins, such as SlSWEET1a).
To confer apoplasmic sugar unloading to sink leaves, the plasma membrane would be the primary site to control sugar fluxes. Co-localization of SlSWEET1a–GFP fusion protein with a plasma membrane protein, namely AtPIP2–RFP (Nelson et al., 2007), at single-cell resolution was evidence that SlSWEET1a was located at the plasma membrane (Fig. 3; Supplementary Fig. S4). The specific subcellular localization of SlSWEET1a in single cells was consistent with transient expression of SlSWEET1a–GFP in leaves of tobacco and Arabidopsis (Shammai et al., 2018). All major sugar loaders, SUCs/SUTs and STPs, are located in the plasma membrane (Hackel et al., 2006; Ayre, 2011; Cheng et al., 2015; Baker et al., 2016; Pommerrenig et al., 2018). In contrast, SWEETs expressed in sink organs were present in plasma or vacuolar membranes (Chen et al., 2010; Chardon et al., 2013; Chen, 2014; Chen et al., 2015). Similar to key SWEET exporters to supply sugars in reproductive organs, SlSWEET1a locates at the correct cellular site, the plasma membrane, to be involved in sugar unloading. Taken together, dominant accumulation of SlSWEET1a proteins in the plasma membrane of vein tissues in young leaves supported its putative role to mediate sugar fluxes across the apoplasm between CCs and phloem parenchyma during sugar unloading to sink leaves.
Although strong SlSWEET1a RNA accumulation also occurred in flowers during the reproductive stage, most SlSWEET1a protein accumulated in stamens (Fig. 2E, F), suggesting that SlSWEET1a is probably not involved in phloem sugar unloading during the reproductive stage. Furthermore, AtSWEET8 (also named AtRPG1) has been demonstrated to be strongly expressed in Arabidopsis pollen, possibly to mediate import of apoplasmic hexoses required for microspore development (Guan et al., 2008). Therefore, whether SlSWEET1a participates in post-phloem unloading required for microsporogenesis in flowers requires further investigation. Interestingly, induced expression of SlSWEET1a during fruit maturation implies that SlSWEET1a is involved in sugar homeostasis during fruit maturation. This conclusion correlates with the previous observation that variation of SlSWEET1a expression correlates with hexose composition of ripening tomato fruits (Shammai et al., 2018). Taken together, these results suggest that SlSWEET1a probably has multiple physiological functions, depending on tissue localization and developmental stages.
Transport analyses on yeast cells using complementary growth assays or a radiotracer confirmed that SlSWEET1a was a glucose-specific transporter, consistent with its closest Arabidopsis homolog, AtSWEET1 (Fig. 4; Supplementary Fig. S6, S7; Chen et al., 2010) and previous speculations (Shammai et al., 2018). In contrast, many hexose transporters, such as TMT/TST (tonoplast monosaccharide transporter/tonoplast sugar transporter) or some STPs, have a broad substrate range (Slewinski, 2011). For example, Arabidopsis AtTST1 transports glucose, fructose, and sucrose (Wingenter et al., 2010; Schulz et al., 2011). Sugar beet BvTST1 also transports both glucose and sucrose (Jung et al., 2015), whereas CsHT1 (hexose transporter from Cucumis sativus) transports glucose, galactose, mannose, and xylose (Cheng et al., 2015). Functional redundancy between MSTs would be consistent with MST proteins not having a confirmed physiological role, despite their dominant expression in heterotrophic sink tissues (Sherson et al., 2000; Slewinski, 2011; Yamada et al., 2011). Thus, high specificity of SlSWEET1a to glucose reflects its putative importance in hexose transport during sugar unloading in young leaves.
In contrast to most plasma membrane-localized MSTs exhibiting a low Km (2–10 mM) (Büttner and Sauer, 2000; Büttner et al., 2000; Klepek et al., 2005; Poschet et al., 2010), SlSWEET1a import activity exhibited a relatively high Km (119 mM) for glucose (Fig. 4C). Although this low-affinity feature may be due to a high turnover rate of SlSWEET1a proteins (Chen et al., 2015a), it also highlights the possibility that SlSWEET1a actively operates under high physiological concentrations of apoplasmic glucose. Due to significantly higher expression and activities of cell wall invertases observed in young leaves compared with mature leaves (Sherson et al., 2003; Jin et al., 2009), sucrose unloaded to the leaf apoplasm could be hydrolyzed and glucose taken up via the hexose carrier. These observations further support the need for a low-affinity glucose importer, such as SlSWEET1a, operating along the initial sugar unloading path from CCs to parenchyma cells in developing leaves.
Moreover, SlSWEET1a was a proton-independent bidirectional uniporter (Fig. 4A, E), in agreement with all other AtSWEET members (Chen et al., 2012; Guo et al., 2014). In that case, a passive glucose carrier, such as SlSWEET1a, would possess a more efficient transport system in response to instant sugar fluxes to assist continuous sugar unloading without the delay for energy biosynthesis. This was nicely in line with passive sugar fluxes observed in young leaves, where the sucrose unloading is insensitive to anoxia and PCMBS, a metabolite inhibitor (Schmalstig and Geiger, 1985; Turgeon, 1987; Fisher and Wang, 1995). Furthermore, proton ATPase is not expressed in the protophloem of growing sinks, such as root tips and expanding region of leaves (Stadler et al., 1995; Truernit and Sauer, 1995), indicating a lack of energized transport. Taken together, all transport features of SlSWEET1a supported its putative role in participating in initial sugar unloading from SE–CC complexes to phloem parenchyma in sink leaves.
Our physiological evidence supported the assertion that SlSWEET1a import activity regulated hexose accumulation in young leaves. During the vegetative growth stage of tomato plants, suppressed expression of SlSWEET1a by VIGS significantly reduced accumulation of glucose and fructose in young leaves, with concurrent increased concentrations of these sugars in source leaves (Fig. 5). Since sucrose is the dominant type of phloem-transported sugar (Swanson and El-Shishiny, 1958; Liu et al., 2012), it follows that apoplasmic glucose in sink leaves is mainly derived from hydrolysis of sucrose unloaded into the sink leaf apoplasm. Based on its low affinity, vein localization, and glucose-specific uniport feature, our results were best explained by the model that some SlSWEET1a on the plasma membrane of phloem parenchyma significantly contributes to hexose import in sink leaves. The high glucose concentrations derived from sucrose hydrolysis during phloem unloading drive the import activity of SlSWEET1a to enable glucose uptake. Once in the cytosol, glucose can be quickly distributed to sink cells via symplasmic connections (Schmalstig and Geiger, 1985), or it can be compartmentalized into corresponding vacuoles (Pommerrenig et al., 2018). The finding that knock out of SlSWEET1a resulted in hexose concentrations being reduced in sink leaves is consistent with SlSWEET1a playing a key role in the phloem unloading of hexoses. Similar conclusions have been reached for knock down of the hexose symporter that drives hexose uptake from the apoplasm into storage parenchyma cells of developing tomato fruit (McCurdy et al., 2010) and of ZmSWEET4, located on the membrane of basal endosperm transfer cells, mediating hexose uptake into the endosperm of developing maize seeds (Sosso et al., 2015).
These results also concur nicely with the report that reduced sink strength by removing active sinks, such as developing leaves and fruits, stimulated sucrose accumulation in mature leaves of a citrus tree (Iglesias et al., 2002), and with the observation that removal of young axillary leaves increased the size of mature leaves and the number of nodes in tomato plants (Decoteau, 1990). Furthermore, as observed in wheat grains, once sugar import into sink efflux was diminished by detaching the sink, sucrose efflux out of the SE–CC complex declined immediately (Fisher and Wang, 1995). As a consequence, the resulting high sucrose contents in the phloem will suppress expression of the phloem loader, SUC/SUT2 symporter (Chiou and Bush, 1998; Vaughn et al., 2002), and thus reduce whole-plant sugar loading and lead to increased carbohydrates in sucrose mesophyll cells (Vaughn et al., 2002). We inferred that the import activity of SlSWEET1a may be an indicator for sink strength and is thus a novel element in whole-plant carbon partitioning from source leaves to sink leaves.
Summary
Herein, we uncovered important functions of the plasma membrane-located uniporter SlSWEET1a in glucose uptake during apoplasmic phloem unloading to young leaves (Supplementary Fig. S9). We propose that in vegetative tomato plants, once sucrose is allocated to sink leaves via phloem pressure flow, it will passively exit out of the SE–CC complexes (symplasmically or apoplasmically). In the apoplasmic route, some sucrose may be exported by an unknown sucrose exporter and be directly taken up through an unidentified sucrose carrier, possibly a SUT member. Yet, a significant portion of sucrose is hydrolyzed to glucose and fructose by extracellular invertase in the apoplasm around phloem parenchyma, partially contributing to formation of a sucrose gradient from SE–CC complexes to facilitate efflux (Osorio et al., 2014). Then, SlSWEET1a, partially located in the plasma membrane of unloading cells, such as phloem parenchyma cells, promotes glucose import to increase apoplasmic solute potential so as to maintain lower turgor pressure toward the SE–CC symplast for continuous sucrose unloading. The fructose-specific carrier remains to be identified. Further studies will be needed to dissect how SlSWEET1a import activity is coordinated with sink strengths during transition of leaf development and transition from the vegetative stage to the reproductive stage. We also speculated that efficient unloading to sink leaves was important for whole-plant growth. Thus, engineering-efficient unloading of hexoses in phloem parenchyma, for example by increasing SlSWEET1a import activity, coupling symport with higher source import capacity (Ainsworth and Bush, 2011), may be key for future crop enhancement.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used for qRT-PCR.
Table S2. Primers used to generate yeast-expressing constructs for clade I SlSWEET genes.
Fig. S1. Images of the tomato organs analyzed.
Fig. S2. Phylogenetic tree of 31 tomato SlSWEET genes.
Fig. S3. Tissue-specific expression of SlSWEET1a proteins in five T1 plants.
Fig. S4. Subcellular localization of SlSWEET1a in tobacco and Arabidopsis protoplasts.
Fig. S5. Amino acid identity between clade I SlSWEET genes.
Fig. S6. Transport activities of clade I SlSWEETs for various sugars in yeast.
Fig. S7. Competition binding ability of sugars to SlSWEET1a transport.
Fig. S8. Images of tomato plants subject to VIGS.
Fig. S9. Proposed model of SlSWEET1a function.
Acknowledgements
We thank Dr Yi-Min Chen at the National Cheng Kung University for his assistance in the GC-MS analysis. We acknowledge the confocal core facility at the National Cheng Kung University for fluorescence observations, and the transgenic plant core lab in Academia Sinica for generating transgenic tomato plants. This work was financially supported by grants from the Ministry of Science and Technology, Taiwan (MOST 105-2628-B-006-001-MY3) and from the DAAD/NSC PPP program (106-2911-I-006-506; 107-2911-I-006-506) to WJG.
References
- Ainsworth EA, Bush DR. 2011. Carbohydrate export from the leaf: a highly regulated process and target to enhance photosynthesis and productivity. Plant Physiology 155, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayre BG. 2011. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Molecular Plant 4, 377–394. [DOI] [PubMed] [Google Scholar]
- Baker RF, Leach KA, Boyer NR, Swyers MJ, Benitez-Alfonso Y, Skopelitis T, Luo A, Sylvester A, Jackson D, Braun DM. 2016. Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiology 172, 1876–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB. 2000. SUT2, a putative sucrose sensor in sieve elements. The Plant Cell 12, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezrutczyk M, Hartwig T, Horschman M, Char SN, Yang J, Yang B, Frommer WB, Sosso D. 2018. Impaired phloem loading in zmsweet13a,b,c sucrose transporter triple knock-out mutants in Zea mays. New Phytologist 218, 594–603. [DOI] [PubMed] [Google Scholar]
- Braun DM, Slewinski TL. 2009. Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiology 149, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Wang L, Ruan YL. 2014. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. Journal of Experimental Botany 65, 1713–1735. [DOI] [PubMed] [Google Scholar]
- Büttner M, Sauer N. 2000. Monosaccharide transporters in plants: structure, function and physiology. Biochimica et Biophysica Acta 1465, 263–274. [DOI] [PubMed] [Google Scholar]
- Büttner M, Truernit E, Baier K, Scholz-Starke J, Sontheim M, Lauterbach C, Huss VAR, Sauer N. 2000. AtSTP3, a green leaf-specific, low affinity monosaccharide–H+ symporter of Arabidopsis thaliana. Plant, Cell & Environment 23, 175–184. [Google Scholar]
- Chardon F, Bedu M, Calenge F, et al. 2013. Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Current Biology 23, 697–702. [DOI] [PubMed] [Google Scholar]
- Chen H-Y, Huh J-H, Yu Y-C, Ho L-H, Chen L-Q, Tholl D, Frommer WB, Guo W-J. 2015. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. The Plant Journal 83, 1046–1058. [DOI] [PubMed] [Google Scholar]
- Chen LQ. 2014. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytologist 201, 1150–1155. [DOI] [PubMed] [Google Scholar]
- Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. 2015a Transport of sugars. Annual Review of Biochemistry 84, 865–894. [DOI] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, et al. 2010. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Lin IW, Qu XQ, Sosso D, McFarlane HE, Londoño A, Samuels AL, Frommer WB. 2015b A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. The Plant Cell 27, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-Q, Qu X-Q, Hou B-H, Sosso D, Osorio S, Fernie AR, Frommer WB. 2012. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. [DOI] [PubMed] [Google Scholar]
- Cheng J, Wang Z, Yao F, Gao L, Ma S, Sui X, Zhang Z. 2015. Down-regulating CsHT1, a cucumber pollen-specific hexose transporter, inhibits pollen germination, tube growth, and seed development. Plant Physiology 168, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Bush DR. 1998. Sucrose is a signal molecule in assimilate partitioning. Proceedings of the National Academy of Sciences, USA 95, 4784–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoteau DR. 1990. Tomato leaf development and distribution as influenced by leaf removal and decapitation. HortScience 25, 681–684. [Google Scholar]
- Eom JS, Chen LQ, Sosso D, Julius BT, Lin IW, Qu XQ, Braun DM, Frommer WB. 2015. SWEETs, transporters for intracellular and intercellular sugar translocation. Current Opinion in Plant Biology 25, 53–62. [DOI] [PubMed] [Google Scholar]
- Feng CY, Han JX, Han XX, Jiang J. 2015. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 573, 261–272. [DOI] [PubMed] [Google Scholar]
- Fisher DB, Wang N. 1995. Sucrose concentration gradients along the post-phloem transport pathway in the maternal tissues of developing wheat grains. Plant Physiology 109, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel Q, Diretto G, Monforte AJ, Fortes AM, Granell A. 2017. Use of natural diversity and biotechnology to increase the quality and nutritional content of tomato and grape. Frontiers in Plant Science 8, 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. 2007. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 35–37. [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. 2000. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proceedings of the National Academy of Sciences, USA 97, 13979–13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. 2010. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. The Plant Journal 64, 355–365. [DOI] [PubMed] [Google Scholar]
- Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN. 2008. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiology 147, 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WJ, Nagy R, Chen HY, Pfrunder S, Yu YC, Santelia D, Frommer WB, Martinoia E. 2014. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiology 164, 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel A, Schauer N, Carrari F, Fernie AR, Grimm B, Kühn C. 2006. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. The Plant Journal 45, 180–192. [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. 1988. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Research 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias DJ, Lliso I, Tadeo FR, Talon M. 2002. Regulation of photosynthesis through source:sink imbalance in citrus is mediated by carbohydrate content in leaves. Physiologia Plantarum 116, 563–572. [Google Scholar]
- Jiang K, Liberatore KL, Park SJ, Alvarez JP, Lippman ZB. 2013. Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine-tunes shoot architecture. PLoS Genetics 9, e1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ni DA, Ruan YL. 2009. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. The Plant Cell 21, 2072–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius BT, Leach KA, Tran TM, Mertz RA, Braun DM. 2017. Sugar transporters in plants: new insights and discoveries. Plant & Cell Physiology 58, 1442–1460. [DOI] [PubMed] [Google Scholar]
- Jung B, Ludewig F, Schulz A, et al. 2015. Identification of the transporter responsible for sucrose accumulation in sugar beet taproots. Nature Plants 1, 14001. [DOI] [PubMed] [Google Scholar]
- Kempers R, van Bel AJE. 1997. Symplasmic connections between sieve element and companion cell in the stem phloem of Vicia faba L. have a molecular exclusion limit of at least 10 kDa. Planta 201, 195–201. [Google Scholar]
- Khuong TT, Crété P, Robaglia C, Caffarri S. 2013. Optimisation of tomato Micro-tom regeneration and selection on glufosinate/Basta and dependency of gene silencing on transgene copy number. Plant Cell Reports 32, 1441–1454. [DOI] [PubMed] [Google Scholar]
- Klepek YS, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N. 2005. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribose. The Plant Cell 17, 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Tegeder M, Throne-Holst M, Frommer WB, Patrick JW. 2003. Phloem loading and unloading of sugars and amino acids. Plant, Cell & Environment 26, 37–56. [Google Scholar]
- Le Hir R, Spinner L, Klemens PA, et al. 2015. Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Molecular Plant 8, 1687–1690. [DOI] [PubMed] [Google Scholar]
- Lin IW, Sosso D, Chen LQ, et al. 2014. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508, 546–549. [DOI] [PubMed] [Google Scholar]
- Liu DD, Chao WM, Turgeon R. 2012. Transport of sucrose, not hexose, in the phloem. Journal of Experimental Botany 63, 4315–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. 2002. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. The Plant Journal 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. 2003. Virus-induced gene silencing in plants. Methods 30, 296–303. [DOI] [PubMed] [Google Scholar]
- McCurdy DW, Dibley S, Cahyanegara R, Martin A, Patrick JW. 2010. Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit. Molecular Plant 3, 1049–1063. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Turgeon R. 1999. Sieve elements and companion cells—traffic control centers of the phloem. The Plant Cell 11, 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Ruan YL, Fernie AR. 2014. An update on source-to-sink carbon partitioning in tomato. Frontiers in Plant Science 5, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang K, Li J, Huang H, Que Q, Li P, Chen X. 2014. A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnology, Biotechnological Equipment 28, 1008–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JW. 1997. Phloem unloading: sieve element unloading and post-sieve element transport. Annual Review of Plant Physiology and Plant Molecular Biology 48, 191–222. [DOI] [PubMed] [Google Scholar]
- Pommerrenig B, Ludewig F, Cvetkovic J, Trentmann O, Klemens PAW, Neuhaus HE. 2018. In concert: orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance. Plant & Cell Physiology 59, 1290–1299. [DOI] [PubMed] [Google Scholar]
- Poschet G, Hannich B, Büttner M. 2010. Identification and characterization of AtSTP14, a novel galactose transporter from Arabidopsis. Plant & Cell Physiology 51, 1571–1580. [DOI] [PubMed] [Google Scholar]
- Rennie EA, Turgeon R. 2009. A comprehensive picture of phloem loading strategies. Proceedings of the National Academy of Sciences, USA 106, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AG, Cruz SS, Roberts IM, Prior D, Turgeon R, Oparka KJ. 1997. Phloem unloading in sink leaves of Nicotiana benthamiana: comparison of a fluorescent solute with a fluorescent virus. The Plant Cell 9, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Bermudez L, Carrari F. 2015. Crop yield: challenges from a metabolic perspective. Current Opinion in Plant Biology 25, 79–89. [DOI] [PubMed] [Google Scholar]
- Ruan YL. 2014. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annual Review of Plant Biology 65, 33–67. [DOI] [PubMed] [Google Scholar]
- Ruan YL, Patrick JW, Bouzayen M, Osorio S, Fernie AR. 2012. Molecular regulation of seed and fruit set. Trends in Plant Science 17, 656–66. [DOI] [PubMed] [Google Scholar]
- Ruiz-Matute AI, Hernández-Hernández O, Rodríguez-Sánchez S, Sanz ML, Martínez-Castro I. 2011. Derivatization of carbohydrates for GC and GC–MS analyses. Journal of Chromatography B 879, 1226–1240. [DOI] [PubMed] [Google Scholar]
- Sauer N. 2007. Molecular physiology of higher plant sucrose transporters. FEBS Letters 581, 2309–2317. [DOI] [PubMed] [Google Scholar]
- Schmalstig JG, Geiger DR. 1985. Phloem unloading in developing leaves of sugar beet: I. Evidence for pathway through the symplast. Plant Physiology 79, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A, Beyhl D, Marten I, Wormit A, Neuhaus E, Poschet G, Büttner M, Schneider S, Sauer N, Hedrich R. 2011. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. The Plant Journal 68, 129–136. [DOI] [PubMed] [Google Scholar]
- Shammai A, Petreikov M, Yeselson Y, et al. 2018. Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. The Plant Journal 96, 343–357. [DOI] [PubMed] [Google Scholar]
- Sherson SM, Alford HL, Forbes SM, Wallace G, Smith SM. 2003. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. Journal of Experimental Botany 54, 525–531. [DOI] [PubMed] [Google Scholar]
- Sherson SM, Hemmann G, Wallace G, Forbes S, Germain V, Stadler R, Bechtold N, Sauer N, Smith SM. 2000. Monosaccharide/proton symporter AtSTP1 plays a major role in uptake and response of Arabidopsis seeds and seedlings to sugars. The Plant Journal 24, 849–857. [DOI] [PubMed] [Google Scholar]
- Shwartz I, Levy M, Ori N, Bar M. 2016. Hormones in tomato leaf development. Developmental Biology 419, 132–142. [DOI] [PubMed] [Google Scholar]
- Slewinski TL. 2011. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Molecular Plant 4, 641–662. [DOI] [PubMed] [Google Scholar]
- Slewinski TL, Meeley R, Braun DM. 2009. Sucrose transporter1 functions in phloem loading in maize leaves. Journal of Experimental Botany 60, 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D, Luo D, Li QB, et al. 2015. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nature Genetics 47, 1489–1493. [DOI] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. 2008. Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiology 148, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Brandner J, Schulz A, Gahrtz M, Sauer N. 1995. Phloem loading by the PmSUC2 sucrose carrier from Plantago major occurs into companion cells. The Plant Cell 7, 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CA, El-Shishiny ED. 1958. Translocation of sugars in the concord grape. Plant Physiology 33, 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N. 1995. The promoter of the Arabidopsis thaliana SUC2 sucrose–H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196, 564–570. [DOI] [PubMed] [Google Scholar]
- Turgeon R. 1987. Phloem unloading in tobacco sink leaves: insensitivity to anoxia indicates a symplastic pathway. Planta 171, 73–81. [DOI] [PubMed] [Google Scholar]
- Uri C, Juhász Z, Polgár Z, Bánfalvi Z. 2014. A GC-MS-based metabolomics study on the tubers of commercial potato cultivars upon storage. Food Chemistry 159, 287–292. [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR. 2002. Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proceedings of the National Academy of Sciences, USA 99, 10876–10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente MH, Zsögön A, de Sá AFL, Ribeiro RV, Peres LEP. 2015. Semi-determinate growth habit adjusts the vegetative-to-reproductive balance and increases productivity and water-use efficiency in tomato (Solanum lycopersicum). Journal of Plant Physiology 177, 11–19. [DOI] [PubMed] [Google Scholar]
- Voitsekhovskaja OV, Koroleva OA, Batashev DR, Knop C, Tomos AD, Gamalei YV, Heldt HW, Lohaus G. 2006. Phloem loading in two Scrophulariaceae species. What can drive symplastic flow via plasmodesmata? Plant Physiology 140, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw IF. 1990. Tansley Review No. 27 The control of carbon partitioning in plants. New Phytologist 116, 341–381. [DOI] [PubMed] [Google Scholar]
- Wayne RO. 2009. The vacuole. In: Plant cell biology: from astronomy to zoology. New York: Academic Press, 101–118. [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM. 2000. A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. The Plant Cell 12, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingenter K, Schulz A, Wormit A, Wic S, Trentmann O, Hoermiller II, Heyer AG, Marten I, Hedrich R, Neuhaus HE. 2010. Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiology 154, 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. 2009. Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Kanai M, Osakabe Y, Ohiraki H, Shinozaki K, Yamaguchi-Shinozaki K. 2011. Monosaccharide absorption activity of Arabidopsis roots depends on expression profiles of transporter genes under high salinity conditions. Journal of Biological Chemistry 286, 43577–43586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Luo D, Yang B, Frommer WB, Eom JS. 2018. SWEET11 and 15 as key players in seed filling in rice. New Phytologist 218, 604–615. [DOI] [PubMed] [Google Scholar]
- Zhang C, Turgeon R. 2018. Mechanisms of phloem loading. Current Opinion in Plant Biology 43, 71–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.