Flower color alterations often occur in ornamental plants. Changes in the expression of a dose-dependent anthocyanin repressor, CmMYB#7, accounts for this process in chrysanthemum.
Keywords: Activator, anthocyanin, bHLH, chrysanthemum, flower color, MYB, repressor
Abstract
‘Jimba’, a well-known white flowered chrysanthemum cultivar, occasionally and spontaneously produces red colored petals under natural cultivation, but there is little information about the molecular regulatory mechanism underlying this process. We analysed the expression patterns of 91 MYB transcription factors in ‘Jimba’ and ‘Turning red Jimba’ and identified an R3 MYB, CmMYB#7, whose expression was significantly decreased in ‘Turning red Jimba’ compared with ‘Jimba’, and confirmed it is a passive repressor of anthocyanin biosynthesis. CmMYB#7 competed with CmMYB6, which together with CmbHLH2 is an essential component of the anthocyanin activation complex, for interaction with CmbHLH2 through the bHLH binding site in the R3 MYB domain. This reduced binding of the CmMYB6–CmbHLH2 complex and inhibited its ability to activate CmDFR and CmUFGT promoters. Moreover, using transient expression assays we demonstrated that changes in the expression of CmMYB#7 accounted for alterations in anthocyanin content. Taken together, our findings illustrate that CmMYB#7 is a negative regulator of anthocyanin biosynthesis in chrysanthemum.
Introduction
Conspicuous flower colors attract pollinators, and particular color traits have been perceived and selected by multiple species during evolution (Renoult et al., 2017). McEwen and Vamosi (2010) showed that plant species flowering simultaneously tend to have more divergent floral colors than expected by chance. Furthermore, color variation can often occur even in the same cultivars of ornamental plants under natural cultivation (Ohmiya et al., 2006; Ohno et al., 2011; Han et al., 2012). It is thought that alterations in these traits may result in the attraction of different pollinators, genetic isolation, and ultimately speciation (Quattrocchio et al., 1999).
Chrysanthemum (Chrysanthemum morifolium Ramat.) is one of the most popular ornamentals, with rich variations in floral color deriving from spontaneous or induced changes (Zhang et al., 2013). According to Zhang et al. (2018) flower color variation occurs in 202 out of 317 chrysanthemum cultivars and is most common in white cultivars. Ohmiya et al. (2006) showed that white-flowered cultivars could mutate to yellow due to the low expression level of a carotenoid cleavage dioxygenase (CmCCD4a), and suppression of CmCCD4a by RNAi constructs in the white cultivar ‘Jimba’ successfully produced a yellow ‘Jimba’. White-flowered cultivars are the most popular in Japan and have a specific use for funerals (Ohmiya et al., 2009). ‘Jimba’ produces white flowers due to the accumulation of flavones instead of anthocyanins (Chen et al., 2012). Flowers on some ‘Jimba’ plants also turn red spontaneously under natural cultivation conditions, which may influence their commodity value.
Flower colors from orange to blue are generated by qualitative and quantitative differences in anthocyanin pigments, and several genetic and molecular investigations indicate that alterations in floral color result mostly from differences in expression of pigment-related structural or regulatory genes (Tanaka et al., 2008; Schiestl and Johnson, 2013). In all plant species studied to date, the temporal and spatial distribution of anthocyanins has been shown to be regulated by an MYB–bHLH–WD40 (MBW) transcriptional complex, and MYB levels have been thought to be the most specific and conspicuous regulators determining anthocyanin patterns (Zimmermann et al., 2004; Quattrocchio et al., 2006). In petunia, several R2R3 MYBs (including both activators and repressors) coordinately regulate anthocyanin pigmentation patterning. AN2, which encodes an anthocyanin-related MYB protein expressed only in flower petals, and two other MYBs, PHZ and DPL, are required for light-induced anthocyanin biosynthesis in vegetative tissues (Albert et al., 2011). In Antirrhinum, the venation flower pigmentation patterning is closely associated with the spatial expression of Venosa (encoding an R2R3 MYB) (Shang et al., 2011). The MYB and bHLH transcription factors regulating anthocyanin biosynthesis in chrysanthemum flowers have also been identified (Liu et al., 2015; Xiang et al., 2015). There is, however, no information available about the mechanism involved in the change of petal color in ‘Turning red Jimba’.
To investigate the mechanism that switches on anthocyanin biosynthesis in ‘Turning red Jimba’ (TRJ), the expression patterns of 91 MYBs expressed in ‘Jimba’ flower tissue were analysed in white ‘Jimba’ (WJ) and TRJ. One R3 MYB gene, named CmMYB#7, was identified as an anthocyanin repressor and CmMYB6 was identified as an activator. Spontaneous changes in expression of these activator and repressor MYBs were found to play an essential role in the formation of the red flowers in TRJ.
Materials and methods
Plant materials
Chrysanthemum flowers of ‘Jimba’ used in this study produced either white (WJ) or red (TRJ) colored flowers when cultivated outdoors from March 2016 to January 2017 in Hangzhou, Zhejiang Province, China, located at 120°2′E and 30°3′N. Both WJ and TRJ were collected at full-bloom stage. TRJ petals in each inflorescence were divided into three groups: inner, middle, and outer layers within the inflorescence, based on the extent of their color. Three replicates were collected for each sample and frozen with liquid nitrogen, then stored in −80 °C for future use.
RNA extraction and cDNA synthesis
The cetyl trimethylammonium bromide (CTAB) method was used for RNA extraction and the intactness of the RNA was analysed by agarose gel electrophoresis. DNA contamination was removed by DNase I (Invitrogen, Lithuania), and then first-strand cDNA was produced by reverse transcriptase (Bio-Rad, USA). For real-time quantitative PCR, 1 μg RNA samples were used for first-strand cDNA synthesis, and then cDNAs were diluted 10-fold. The small differences between cDNA concentrations were monitored with a reference gene, CmACT (GenBank AB770471).
Gene cloning and amino acid sequence analysis
All of the MYB sequences were obtained from the chrysanthemum RNA-Seq database (accession number: SRP174546), which was constructed using data from a mixed RNA pool from different developmental stages of flower, young stems, leaves, and roots. The library sequencing and data analysis were conducted by Novogene in Beijing, China. Full-length mRNA sequences were obtained by rapid amplification of cDNA ends (RACE) PCR using the SMARTTM RACE cDNA Amplification Kit (Clontech, USA). The full-length proteins were predicted with a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed 3 April 2019 and the amino acids sequences were deduced by https://web.expasy.org/translate/, accessed 3 April 2019) and the amino acids sequences were deduced by the translate algorithm (https://web.expasy.org/translate, accessed 3 April 2019). FastStart High Fidelity kits (Roche, Switzerland) were used for gene cloning.
A neighbor-joining tree was constructed using MEGA7 (Kumar et al., 2016) to analyse the phylogenetic relationship of CmMYBs with reported MYBs of the phenylpropanoid pathway in other plant species.
ClustalX (Thompson et al., 1997) was used for CmMYB#7 protein sequence alignment with AtCPC (Arabidopsis, GenBank accession number NP_182164.1), AtMYBL2 (Arabidopsis, NP_177259.1), FaMYB1 (Fragaria × ananassa, AAK84064.1), FcMYB1 (F. chiloensis, ADK56163.1), MtMYB2 (Medicago truncatula, XP_003616388.1), PhMYB27 (Petunia hybrida, AHX24372.1), PhMYBx (P. hybrida, AHX24371.1), PtMYB182 (Populus tremula × Populus tremuloides, AJI76863.1), SlMYBATV-X1 (Solanum lycopersicum, AUG72362.1), VvMYC2-L1 (Vitis vinifera, XP_002273328.1) and GENEDOC (Nicholas et al., 1997) was used for comparison of conserved amino acids, indicated by shading in the figures.
qPCR analysis
Expression of specific genes was quantified by qPCR using the fast EvaGreen supermix (Bio-Rad, USA). Firstly, the full-bloom stage of WJ and TRJ was used to detect the expression pattern changes of anthocyanin biosynthetic genes from CmCHS to CmUFGT and 91 MYBs expressed in flower tissues. Subsequently, the three different types of petal of TRJ were collected to study the transcript levels of the genes including CmDFR, CmUFGT, CmbHLH2, and seven MYBs that showed quite significant changes between WJ and TRJ. qPCR primers are listed in Supplementary Table S1 at JXB online and were verified by melting curve and PCR product sequencing. The reference gene, CmACT, was used to evaluate the expression of target genes. No-template reactions were used as negative controls.
Anthocyanin component analysis
Flower petals were ground into powder under liquid nitrogen. Samples of 1 g were added to 5 ml methanol–0.05% HCl and placed in the dark for 12 h; the supernatant was collected and the extraction process was repeated (Wrolstad et al., 1982). The combined supernatants were filtered through Millipore membranes for further analysis by HPLC.
The HPLC (Agilent 1269) analytical column was an SB-C18 (Agilent Technologies, 4.6×250 mm, 5 µm). The temperature was 30 °C. The analytical column was eluted with mobile phase A (formic acid: water, 5:95, v/v) and mobile phase B (methanol) at a rate of 0.8 ml min−1. The gradient of mobile phase B was: 0–2 min, 5%; 2–7 min, 5–15%; 7–20 min, 15–20%; 20–25 min, 20–27%; 25–32 min, 27%; 32–41 min, 27–35%; and 41.01–43 min, 5% (Cheng et al., 2014). The detection wavelength was set at 520 nm.
For LC, a Waters UPLC (Waters Corp., Milford, MA, USA) and Agilent SB-C18 column (3.5 μm, 2.1×100 mm; agilent Corp.) were used in all the chromatographic experiments. The mobile phases were 0.1% formic acid–water (A) and 0.1% formic acid–acetonitrile (B). The linear gradient program was 0/5, 35/95, 36/5 (min/B%); sample injection volume, 3 μl; column oven temperature, 30 °C; flow rate, 0.5 ml min−1; and the UV detector was set at 254 nm.
Mass spectrometry was carried out using an AB TripleTOF 5600plus System (AB SCIEX, Framingham, MA, USA) with a scan range m/z of 100–2000 in negative ion mode; source voltage was −4.5 kV and the source temperature was 550 °C. The pressure of gas 1 (air) and gas 2 (air) was set to 50 psi; the curtain gas (N2) pressure was set to 30 psi. Injection volume was 10 μl; flow rate, 0.2 ml min−1; maximum allowed error was set to ±5 ppm; declustering potential, 100 V; collision energy, 10 V. For MS/MS acquisition mode, the IDA-based auto-MS2 was performed on the eight most intense metabolite ions; the parameters were almost the same except that the collision energy was set at −40±20 V, ion release delay at 67, ion release width at 25 min for a full scan cycle (1 s). The scan range of m/z of precursor ion and product ion were set as 100–2000 Da and 50–1500 Da, respectively.
The exact mass calibration was performed automatically before each analysis employing the automated calibration delivery system.
Anthocyanin content
The total petal anthocyanin content was determined by the pH difference method (Wrolstad et al., 1982; Huang et al., 2013). Briefly, a 1 g sample was added into 5 ml methanol–0.05% HCl, placed in the dark for 12 h, and the supernatant transferred to a 50 ml tube. This step was repeated twice to ensure anthocyanins were fully extracted. Then absorbance of the solution was measured in a UV-2550 spectrophotometer at 510 and 700 nm.
For tobacco leaves, the content of anthocyanin was measured using the method described previously (Carrie and Gregory, 2009; Liu et al., 2015). Briefly, frozen leaves were extracted using methanol–1% HCl overnight in the dark at 4 °C. Chloroform was used to remove chlorophyll. Absorbance at 530 and 657 nm was measured using a UV-2550 spectrophotometer (SHI-MADZU). Anthocyanin content on a per gram fresh weight basis was calculated with the formula ((A530−A657)/mg FW tissue)×1000.
Dual-luciferase assays
Dual-luciferase assays were used to analyse trans-activation of target gene promoters by transcription factors according to Xiang et al. (2015). Full-length regulatory genes and target promoters were cloned into pGreenII0029 62-SK and pGreenII 0800-LUC vectors, respectively, with the primers listed in Supplementary Table S2. The recombinant plasmids were transformed into Agrobacterium strain GV3101 (PM90). Transcription factors and promoters were mixed (10:1 v/v) and then infiltrated into tobacco leaves (Nicotiana benthamiana). Enzyme activities of firefly luciferase and Renilla luciferase were tested using the Dual-Luciferase Reporter Assay System (Promega, USA).
Site-directed mutagenesis
A mutant protein, CmMYB#7-m, was created by replacement of the nucleotides AGG by GAC at the bHLH binding site of CmMYB#7 by overlap extension PCR, which introducing a targeted mutation, GAC, in the primer. Two overlapping PCR products were mixed (1:1 v/v) and used as a template for the third PCR. The PCR product was confirmed by sequencing (Dai et al., 2010).
Yeast two-hybrid assay
Protein–protein interactions were tested by yeast two-hybrid assays following the manufacturer’s instructions (Matchmaker Gold Yeast Two-Hybrid System, Clontech, USA) as described in our previous publication (Xiang et al., 2015). CmMYB#7, CmMYB#7-m, and CmbHLH2 were cloned into GAL4-AD and GAL4-BD vectors, respectively. The appropriate concentrations of aureobasidin A (AbA) to inhibit self-transactivation were tested on SD/−Ura+AbAx medium. GAL4-AD and GAL4-BD vectors containing target genes were co-transformed into the yeast two-hybrid strain and the interactions were detected on quadruple medium (SD/−Ade/−His/−Leu/−Trp+ X-α-Gal+ AbA).
Transient overexpression assays
Transient overexpression of genes in tobacco (Nicotiana tabacum) leaves was a fast and effective method for verification of gene function. CmbHLH2, CmMYB6, and CmMYB#7 were cloned into pGreenII0029 62-SK vectors and then transformed separately into Agrobacterium strain GV3101 (PM90). Cultures were grown to OD600=0.75 and 1 ml infiltrated into tobacco leaves. Anthocyanin contents were tested 7 d after infiltration. Empty vector infiltrations (SK) were used as negative controls.
Firefly luciferase complementation imaging assays
CmMYB6 was added into pCAMBIA1300-NLuc and CmbHLH2 into pCAMBIA1300-CLuc vectors and then transformed into Agrobacterium GV3101 and cultured using a selection medium containing kanamycin. They were grown to OD600=0.5 and then CmMYB6-NLuc, CLuc-CmbHLH2, and CmMYB#7-SK/SK were mixed 1:1:1 by volume. The mixed agrobacteria were cultured for 3 h and then infiltrated into the tobacco leaves (N. benthamiana). Forty-eight hours after infiltration, 0.2 mM luciferase substrates were infiltrated into the same part of the leaves and allowed to stand for 5 min in the dark before detecting the luciferase activity using a low-light cooled CCD imaging apparatus.
Results
Anthocyanin component analysis of white ‘Jimba’ and ‘Turning red Jimba’
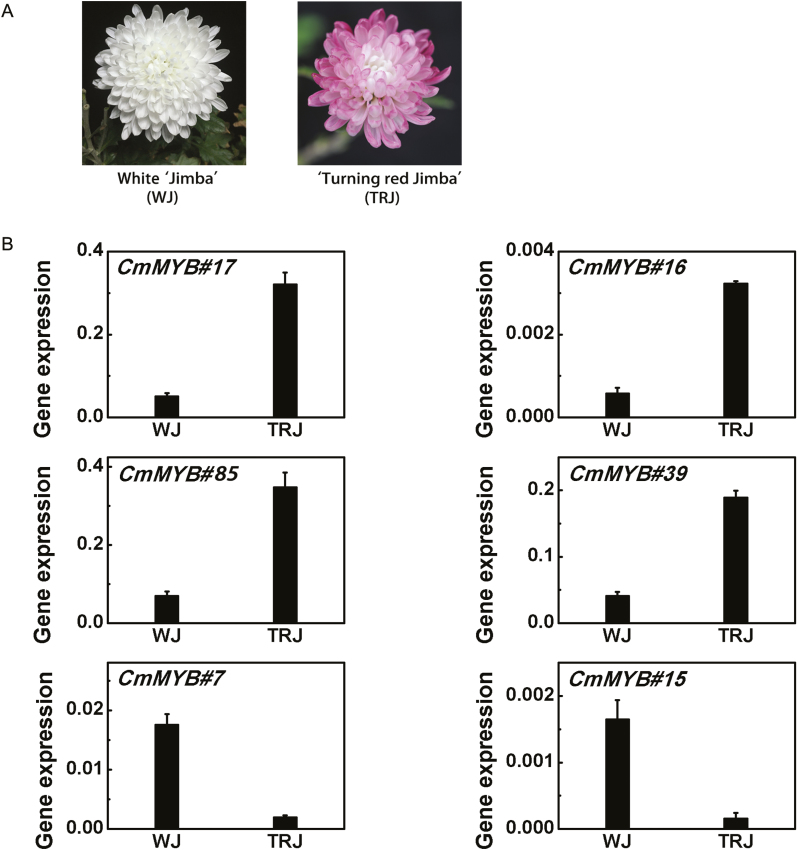
Flower colors of chrysanthemum cultivar ‘Jimba’ are usually white, but in natural cultivation both white and red flowers are produced by the same cultivar, sometimes even by the same plant (Fig. 1A). The petal pigments were detected by HPLC analysis. Results showed that the red petal color in TRJ was due to accumulation of cyanidin pigments included cyanidin-3-glucoside, cyanidin-3-O-(6-O-acetyl-β-D-glucoside) and cyanidin 3-O-(6″-O-succinyl-β- glucopyranoside) according to LC-MS, while no anthocyanin component could be found in the WJ petals (Supplementary Fig. S1).
Fig. 1.
Expression levels of six MYBs differentially expressed in white ‘Jimba’ (WJ) and ‘Turning red Jimba’ (TRJ) flower petals. (A) Photos of the WJ and TRJ at full-bloom stage. (B) Expression levels of six MYBs in flower petals of WJ and TRJ. Error bars are the SE of three independent experiments. (This figure is available in color at JXB online.)
Expression patterns of anthocyanin-related genes in WJ and TRJ
As expected, the transcript levels of anthocyanin biosynthetic genes including CmCHS (chalcone synthase), CmCHI (chalcone isomerase), CmF3H (flavanone 3-hydroxylase), CmF3′H (flavanone 3′-hydroxylase), CmDFR (dihydroflavonol 4-reductase), CmANS (anthocyanidin synthase), and CmUFGT (UDP-flavonoid glucosyl transferase) were dramatically up-regulated in TRJ flowers compared with WJ (Supplementary Fig. S2), implying that the process of ‘Jimba’ flowers turning from white to red was regulated at the transcriptional level. The reported CmMYB6 (Liu et al., 2015) was expressed weakly in WJ and increased in TRJ, but the difference was not significant (Supplementary Fig. S2).
Analysis of MYBs associated with ‘Jimba’ flower color alteration
To identify key factors that could account for this process, differentially expressed MYBs were screened using qPCR, since MYBs are often found to be the most important regulators of the anthocyanin biosynthetic pathway (Zimmermann et al., 2004; Quattrocchio et al., 2006).
Among the RNA-seq database of chrysanthemum, there are 163 unigenes annotated as MYBs according to their structural or functional characteristics. A total of 91 of these MYBs have been verified as being expressed in flower tissues, and their expression pattern changes were determined. Measurements of the transcript levels of these MYBs in WJ and TRJ indicated that 6 out of 91 MYBs were changed over 4-fold (Supplementary Fig. S3), with CmMYB#17, CmMYB#16, CmMYB#39, and CmMYB#85 being up-regulated and CmMYB#7 and CmMYB#15 down-regulated in TRJ compared with WJ (Fig. 1B).
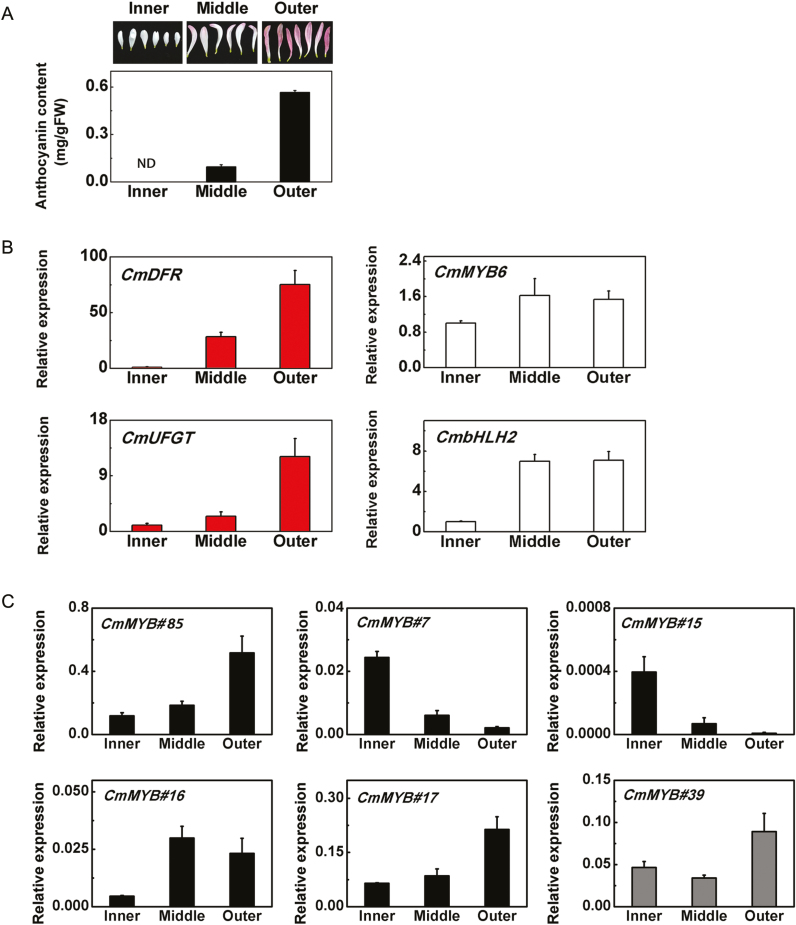
Petals of TRJ were further divided into three different groups, outer, middle, and inner (Fig. 2A), based on their color. The outer petals were purple with 0.6 mg/g FW anthocyanin, the middle ones were colored at the tips, with 0.1 mg/g FW anthocyanin, while the inner petals appeared white without any detectable anthocyanin (Fig. 2A). The expression patterns of CmDFR and CmUFGT were strongly correlated with anthocyanin content (Fig. 2B). There was no strong correlation between CmMYB6 expression patterns in inner, middle and outer petals of TRJ and anthocyanin pathway genes. CmMYB6 transcripts only increased slightly in the middle petals, where substantial anthocyanin accumulation occurred (Fig. 2B). However, expression of CmbHLH2 increased considerably in the middle petals compared with the inner petals and remained at a high level in the outer petals, around 7-fold higher than in the inner petals (Fig. 2B).
Fig. 2.
Anthocyanin content and gene expression in different colored petals of ‘Turning red Jimba’ (TRJ) flowers. Error bars are the SE of three independent experiments. (A) Anthocyanin content in different petal types of TRJ flowers. (B) Transcripts of CmDFR, CmUFGT, CmMYB6, and CmBHLH2 in different petal types of TRJ. (C) Expression patterns of six candidate MYBs in different petals types of TRJ. (This figure is available in color at JXB online.)
Further analysis of the expression patterns of the six candidate MYBs, which showed differentially expressed patterning in WJ and TRJ, in the three different colored petal types indicated three out of four up-regulated MYBs (CmMYB#85, CmMYB#16, and CmMYB#17) were positively correlated and two down-regulated MYBs (CmMYB#7 and CmMYB#15) were negatively correlated with anthocyanin accumulation and were selected for further studies, excluding CmMYB#39 whose expression pattern was poorly correlated with anthocyanin biosynthetic gene expression (Fig. 2C).
Transcriptional activation of anthocyanin biosynthetic gene promoters by MYBs
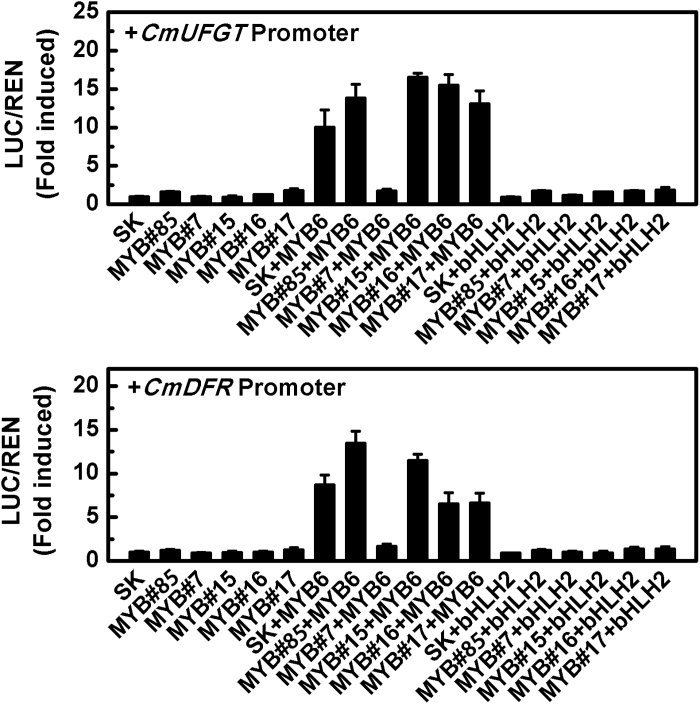
Dual-luciferase assays were used to analyse the activity of the five MYBs (CmMYB#16, CmMYB#17, CmMYB#85, CmMYB#7, and CmMYB#15) that were positively or negatively correlated with anthocyanin content on transcription from the CmDFR and CmUFGT promoters. The results indicated none of them could activate or inhibit CmDFR and CmUFGT promoters directly (Fig. 3). Since MYB proteins generally form a transcriptional complex that regulates anthocyanin biosynthetic genes, a combination of two reported anthocyanin activators (CmMYB6 and CmbHLH2) (Liu et al., 2015; Xiang et al., 2015) was tested. Unexpectedly, there were no regulatory effects on target promoters when co-expressed with CmbHLH2, but when co-expressed with CmMYB6, CmMYB#7 significantly inhibited the activation of both CmDFR and CmUFGT. Other MYBs tested had little or no effect (Fig. 3).
Fig. 3.
Analysis of the regulatory effects of five selected MYBs alone or in combination with CmMYB6 or CmbHLH2 on anthocyanin biosynthetic gene promoters. SK refers to ‘empty vector’ and serves as control. Error bars are the SE of three independent experiments.
The CmMYB#7 protein sequence has a conserved bHLH binding domain
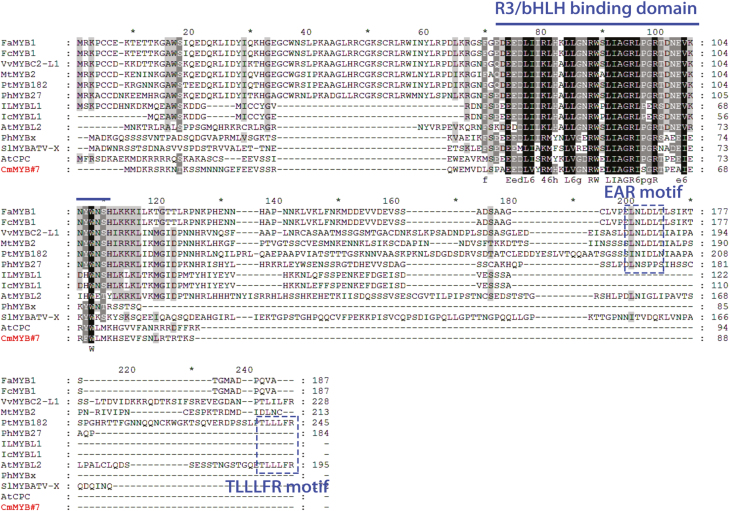
The phylogenetic analysis suggested that CmMYB#7 was grouped with R3-type anthocyanin repressors, while other CmMYBs were divided into other groups (Supplementary Fig. S4). The CmMYB#7 protein sequence was then compared with those of other reported anthocyanin repressors from other species. The predicted protein sequence of CmMYB#7 was 88 amino acids, which including an R3 repeat and a conserved bHLH binding domain ([D/E]LX2[RK]X3LX6LX3R) required for interaction with IIIf bHLHs (Zimmermann et al., 2004). No common repressive domain, such as an EAR or TLLLFR motif, was found in the CmMYB#7 protein sequence, as was also the case for other reported R3 type anthocyanin repressors AtCPC and PhMYBx, with which CmMYB#7 showed 50% and 30% similarity, respectively (Fig. 4).
Fig. 4.
Protein sequence alignment of CmMYB#7 with other reported anthocyanin-related MYBs. The R3/bHLH binding domain is marked with a line. Repressive motifs EAR and TLLLFR are enclosed in dashed boxes. Protein sequences were aligned using ClustalX and conserved amino acids are shaded using GeneDoc. (This figure is available in color at JXB online.)
Protein sequence alignment showed the anthocyanin repressor MYBs can be divided into two types (Fig. 4). The R2R3 MYB type, including FaMYB1, FcMYB1, VvMYBC2-L1, MtMYB2, PtMYB182, and PhMYB27, contains a conserved R2R3 domain and an EAR domain at the C terminus, while the other type, R3 MYB, which includes IlMYBL1, IcMYBL1, PhMYBx, SlMYBATV-X, AtCPC, and AtMYBL2, possesses only an R3 domain. All of these R3 MYBs have a conserved bHLH binding domain but no repressive domain, with the exception of AtMYBL2, which contains a reported TLLLFR repressive domain (Matsui et al., 2008).
CmMYB#7 interacts with CmbHLH2 to inhibit the activation effect of CmMYB6–CmbHLH2 on their target genes
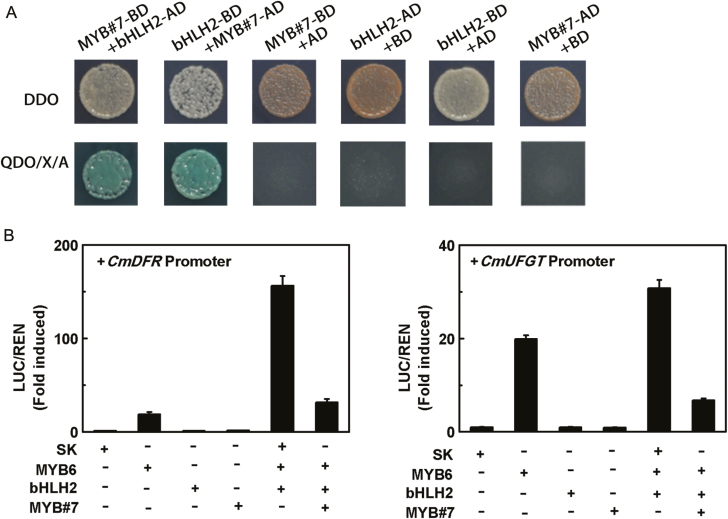
The interaction between CmMYB#7 and CmbHLH2 was investigated using yeast two-hybrid assays. Control experiments suggested that the minimal concentration of aureobasidin A (AbA) required to inhibit the self-activities of BD-CmbHLH2 and BD-CmMYB#7 was 400 and 200 ng/ml, respectively. When transformed with both AD-CmMYB#7 and BD-CmbHLH2, or AD-CmbHLH2 and BD-CmMYB#7, the yeast two-hybrid yeast could grow on QDO (SD/−Ade/−His/−Leu/−Trp) media with appropriate AbA concentrations and X-α-Gal, indicating protein interactions between these two transcription factors (Fig. 5A).
Fig. 5.
CmMYB#7 inhibited transcription of anthocyanin biosynthetic genes by interacting with CmbHLH2. (A) CmMYB#7 formed a transcriptional complex with CmbHLH2 detected by yeast two-hybrid assays. The image shown represents growth on selective media. QDO, quadruple medium, SD/−Ade/−His/−Leu/−Trp+ X-α-Gal+ AbA. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. (B) The interaction between CmMYB#7 and CmbHLH2 and the effects on target gene promoters were studied by dual-luciferase assays. SK refers to ‘empty vector’ and serves as control. Error bars are the SE of three independent experiments. (This figure is available in color at JXB online.)
Enhanced transcription from the promoters of the anthocyanin biosynthetic genes CmDFR and CmUFGT could be detected in the presence of both CmMYB6 and CmbHLH2 transcription factors (Fig. 5B). Although CmMYB#7 alone had no effect on the activation of either CmDFR or CmUFGT promoters, when co-transformed with CmMYB6 and CmbHLH2, it significantly reduced the activity of the CmMYB6–CmbHLH2 complex by 73% and 82%, respectively (Fig. 5B), whereas CmMYB#15, CmMYB#16, CmMYB#17, and CmMYB#85 co-transformed with CmMYB6 and CmbHLH2 had no significant activation effect on the promoters of CmDFR and CmUFGT (Supplementary Fig. S5).
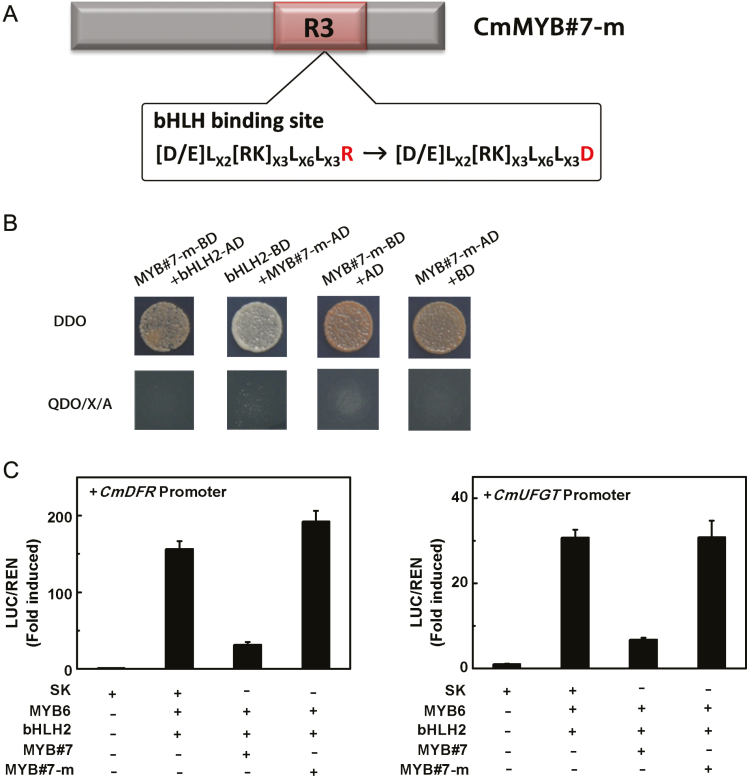
To verify whether the bHLH binding site located in CmMYB#7 is essential for repression, a mutated protein, designated CmMYB#7-m, was created by changing the crucial amino acid R to D (residue 58) in the bHLH binding site according to Zimmermann et al. (2004) (Fig. 6A). No interaction could be detected between CmMYB#7-m and CmbHLH2 in yeast (Fig. 6B). Dual luciferase assays showed that this mutation leads to loss of the ability to inhibit the activation of target promoters by the CmMYB6–CmbHLH2 complex (Fig. 6C). This suggests that the interaction between CmMYB#7 and CmbHLH2 interferes with the transcriptional activation of the anthocyanin biosynthetic genes by the CmMYB6–CmbHLH2 complex.
Fig. 6.
Role of CmMYB#7 R3 domain in inhibiting the transcription of target gene promoters. (A) The structure of CmMYB#7-m. CmMYB#7-m was created by replacing an arginine by aspartic acid in the bHLH binding site. (B) CmMYB#7-m was unable to interact with CmbHLH2 in a yeast two-hybrid assay. The image shown represents growth on selective media. QDO, quadruple medium (SD/−Ade/−His/−Leu/−Trp+ X-α-Gal+ AbA); AD, GAL4 activation domain; BD, GAL4 DNA binding domain). (C) CmMYB#7-m was unable to repress transcription from target gene promoters induced by CmMYB6–CmbHLH2 complex measured by dual-luciferase assays. SK refers to ‘empty vector’ and serves as control. Error bars are the SE of three independent experiments. (This figure is available in color at JXB online.)
CmMYB#7 inhibits the transcriptional activation of anthocyanin biosynthetic genes by the CmMYB6–CmbHLH2 complex in a dose-dependent manner
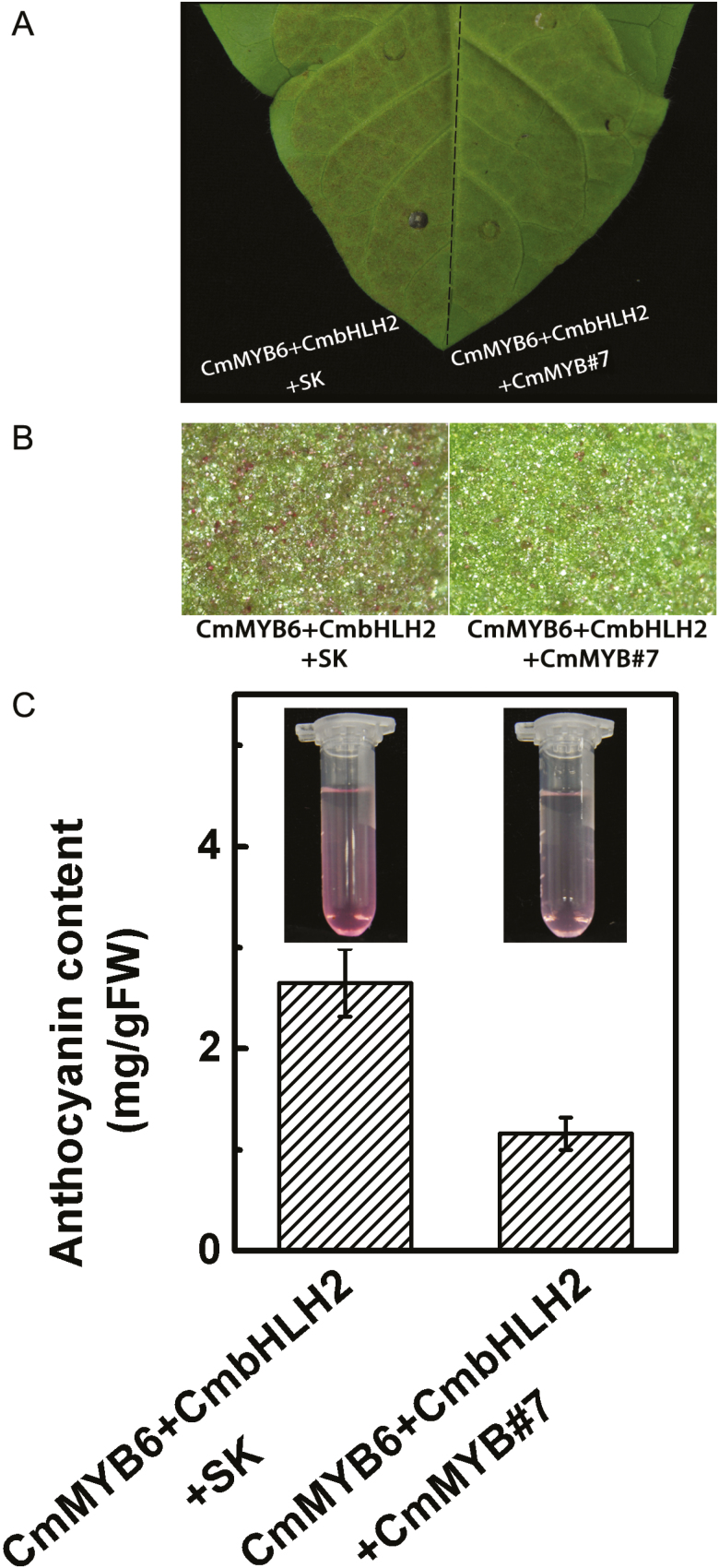
Transient co-expression of CmMYB6–CmbHLH2 together with SK (empty vector) in tobacco leaves caused the accumulation of 2.8 mg/g FW anthocyanin and addition of CmMYB#7 reduced anthocyanin content by half (Fig. 7A–C).
Fig. 7.
Production of anthocyanin by the transient overexpression of CmMYB6, CmbHLH2 with CmMYB#7 or SK (empty vector) in tobacco leaves. Leaves were photographed by camera (A) and stereomicroscope (B) 7 d after injection for transient expression of CmMYB6, CmbHLH2 with SK (left) or CmMYB6, CmbHLH2 with CmMYB#7 (right). (C) Quantitative measurement of anthocyanin content of tobacco leaves (seeing Materials and methods). Error bars are the SE of three independent experiments. (This figure is available in color at JXB online.)
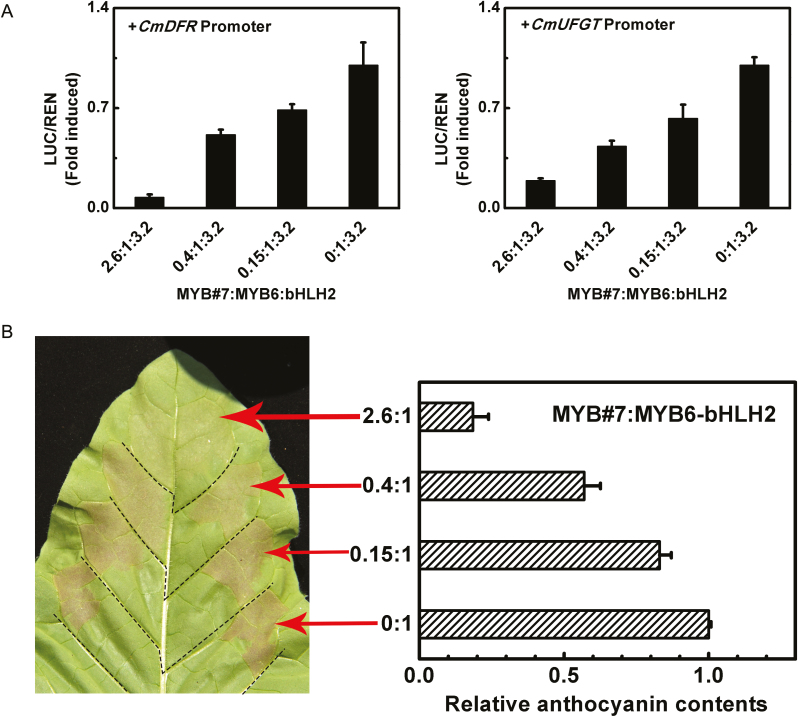
CmMYB#7 transcript levels were significantly different in petals of different colors whereas CmMYB6 changed very little (Fig. 2B). To verify the dose effect of CmMYB#7, dual-luciferase assays were carried out by adding increasing amounts of CmMYB#7 to a fixed volume of CmMYB6 and CmbHLH2. The ratio of CmMYB#7:CmMYB6 set as 2.6:1, 0.4:1, and 0.15:1 was based on their expression in inner, middle, and outer petals, respectively (Fig. 8A). When the ratio of CmMYB#7:CmMYB6 ranged from 0:1 to 2.6:1, the induction of CmDFR and CmUFGT promoters decreased from 1 to 0.1 and 0.18 respectively (Fig. 8A). This indicated that the change in repressor concentration accounts for the overall activities of the transcriptional MYB activators on their target promoters. Transient overexpression assays were carried out in tobacco leaves with the same CmMYB#7 and CmMYB6 ratios set in dual luciferase assays. The results suggested that anthocyanin accumulation in infiltrated patches was negatively correlated with the dose of CmMYB#7. When the ratio of CmMYB#7:CmMYB6 decreased from 2.6:1 to 0.4:1, anthocyanin content was increased 3-fold and increased further as the ratio of CmMYB#7:CmMYB6 was reduced from 0.4:1 to 0:1 (Fig. 8B).
Fig. 8.
The dose-dependent relationship between anthocyanin transcriptional activator and repressor. (A) Extent of transcription from anthocyanin biosynthetic gene promoters induced by different proportions of CmMYB6 and CmMYB#7, ranging from 2.6:1 to 0:1. SK refers to ‘empty vector’ and serves as control. Error bars are the SE of three independent experiments. (B) Anthocyanin content in infiltrated regions was measured 7 d after infiltration. Error bars are the SE of three independent experiments. (This figure is available in color at JXB online.)
CmMYB#7 weakens the binding force of CmMYB6–CmbHLH2
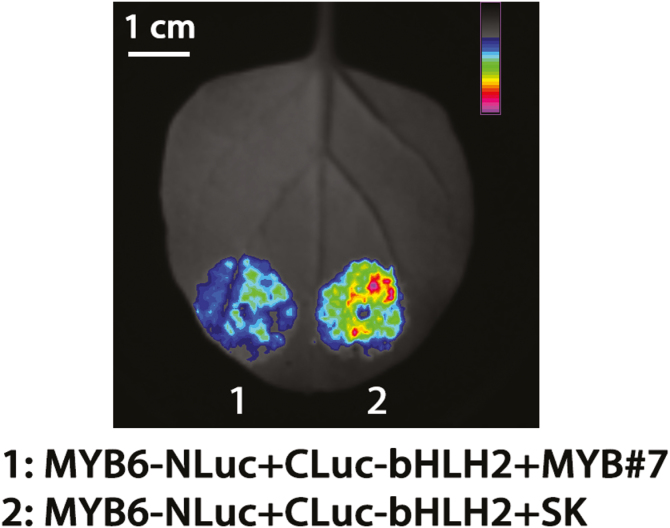
Since CmMYB6 and CmMYB#7 could interact with CmbHLH2 accorded to the yeast two-hybrid assay (Xiang et al. (2015) and Fig. 5A), the protein–protein interaction affinities were analysed using firefly luciferase complementation imaging assays. Transiently overexpressed CmMYB6-NLuc and CLuc-CmbHLH2 in tobacco leaves could produce a functional firefly luciferase protein which could be observed qualitatively and quantitatively using a low-light cooled CCD imaging apparatus (Fig. 9). When CmMYB#7-SK was added to the mixture of CmMYB6-NLuc and CLuc-CmbHLH2 in a proportion 1:1:1, fluorescence intensity was reduced visibly (Fig. 9). Moreover, this effect was not observed when adding empty vector SK into the mixture of CmMYB6-NLuc and CLuc-CmbHLH2 in the same proportions (data not shown). This suggested that CmMYB#7 could weaken the binding force between CmMYB6 and bHLH2 and prevent CmMYB6-NLuc and CLuc-CmbHLH2 from combining to produce a functional protein.
Fig. 9.
Protein–protein interaction affinities were analysed using firefly luciferase complementation imaging assays. Agrobacterium GV3101 strain harboring different constructs was infiltrated into tobacco leaves. Luminescence signals in the infiltrated region were measured 48 h after infiltration. Color bar ranges from blue to purple indicating the strength of the luciferase activities from weak to strong. The experiment was carried out with at least three replicates.
Discussion
MYBs are the important factors in anthocyanin biosynthetic regulation
In many ornamental plants, flower color transitions to white are accounted for by the loss-of-function of R2R3 MYB transcription factors (Quattrocchio et al., 1999; Schwinn et al., 2006). For example, a loss-of-function mutation of ANTHOCYANIN2 (AN2), a well-defined MYB-type transcription factor, is the major determinant of flower color variation between Petunia axillaris with white flowers and P. integrifolia with purple ones (Quattrocchio et al., 1999; Hoballah et al., 2007). Besides anthocyanin activators, several repressors have also been isolated. For example, the transition of flower color from blue to white was shown to be associated with an R3 MYB repressor in Iochroma loxense (Gates et al., 2018).
‘Jimba’ produces white flowers due to the accumulation of flavones instead of anthocyanin (Chen et al., 2012). The expression patterns of anthocyanin structural genes in WJ and TRJ (Supplementary Fig. S2) suggested that the limiting step was from CmF3H and this needs further verification. These results suggest transcription factors may play an important role in anthocyanin biosynthesis in TRJ. Based on the specificity and importance of MYB transcription factors in anthocyanin regulation, we speculated that the formation of TRJ was likely to have been caused by MYBs. In previous studies, CmMYB6 was verified to be an anthocyanin activator (Liu et al., 2015; Xiang et al., 2015). The transcript levels of CmMYB6 were tightly correlated with the expression of anthocyanin pathway genes during the developmental stages of red-flowered ‘Amadea’. We also found, however, that CmMYB6 could be expressed normally in many non-anthocyanin-accumulating cultivars sometimes at a relative low level, which suggested there may be other factors that inhibit the effect of CmMYB6 (Xiang et al. 2015). Furthermore, CmMYB6 transcripts only showed a weak correlation with anthocyanin accumulation in different colored petals of TRJ (Fig. 2B), again supporting the possibility that another factor was involved. Thus, we searched for additional MYBs that co-regulated anthocyanin accumulation in TRJ flower petals. Based on these results, we identified an R3 MYB, CmMYB#7(Fig. 4), whose transcripts were significantly decreased in red flower petals compared with white flowers, and showed a negative correlation with expression of anthocyanin pathway genes in different colored petals of TRJ (Fig. 2). We performed additional experiments to analysis the expression of CmMYB#7 in different types of petals in WJ (Supplementary Fig. S6). The results indicated that the expression patterns of CmMYB#7 were opposite in red and white petal types (Supplementary Fig. S6).
Furthermore, besides the WJ and TRJ, expression patterns of CmMYB6 and CmMYB#7 were also studied in three cultivars named ‘Monalisa white’ (MW), ‘Monalisa yellow’ (MY) and ‘Monalisa pink’ (MP), with white, yellow, and pink floral colors, respectively. The transcript levels of CmMYB6 showed no significant difference among these three cultivars (Supplementary Fig. S7), while the levels of CmMYB#7 decreased dramatically in MP, which could accumulate anthocyanin, compared with the others. This suggested that the suppression activity of CmMYB#7 might be conserved for other non-red-flowered chrysanthemum cultivars, but such a conclusion still requires further confirmation in more cultivars.
Interactions between activator and repressor MYBs is an essential aspect of metabolic pathway regulation
MYBs are one of the largest families of transcription factors in plants and regulate multiple plant-specific processes such as epidermal cell fate, responses to biotic or abiotic stresses, and primary or secondary metabolism (Albert et al., 2011). Beside activators, MYBs also act as transcriptional repressors, including the anthocyanin biosynthetic pathway. Activator MYBs usually interact with bHLH and WD repeat proteins to form a regulatory complex (Albert et al., 2014). Anthocyanin repressor MYBs can be divided into two different groups based on the repressive patterns, namely those with active repression and those with passive steric hindrance (Aharoni et al., 2001). The active repressors usually contain a repressive domain located in the C-terminus, which is essential for the repressive effects. For example, PhMYB27 (P. hybrida) and MtMYB2 (M. truncatula) have been characterized as anthocyanin repressors though their C-terminal EAR motif and deletion or mutation of the repressive domain diminishes their inhibitory effects (Albert et al., 2014; Jun et al., 2015).
The passive repressors are mainly R3 MYBs, which only have an R3 domain and lack either a repressive domain or a DNA-binding domain. The first identified R3 anthocyanin repressor was PhMYBx in petunia (Koes et al., 2005; Albert et al., 2014). Subsequently, AtMYBL2 and AtCPC from Arabidopsis were shown to be anthocyanin repressors and an Atcpc mutant caused ectopic anthocyanin accumulation in seedlings (Dubos et al., 2008; Matsui et al., 2008; Zhu et al., 2009). Here CmMYB#7 was identified as the R3 anthocyanin repressor in chrysanthemum. Such repressor MYBs usually compete with the activators during regulation. For example, AtCPC competed with AtPAP1/2 for binding GL3/EGL3 to regulate anthocyanin biosynthesis in Arabidopsis (Zhu et al., 2009), and CmMYB#7 formed a complex with CmbHLH2 (Fig. 5) and further weakened the binding forces of the CmMYB6–CmbHLH2 complex (Figs 3, 6–9). Such interactions between activators and repressors are a frequent aspect of metabolic pathway regulation.
Genetic regulation of flower color change in ornamental plants
Flower color variations often occur spontaneously in ornamental plants and produce various color hues and pigmentation patterning (venation, stripes, spots, and blotches) (Shang et al., 2011; Morita and Hoshino, 2018). Such floral color alterations are associated mainly with expression changes in pigment-related genes. Many factors could cause the changes of gene expression patterns such as DNA methylation in gene promoter regions or transposon insertions and deletions. For example, DNA methylation of the MdMYB1 promoter region affects the transcript levels of MdMYB1 and determines anthocyanin content in skin of two apple cultivars (Ma et al., 2018). Also, a histone H3K9 demethylase, JMJ25, directly affects the transcript levels of MYB182, an anthocyanin repressor, to modulate anthocyanin biosynthesis in poplar (Fan et al., 2018). In morning glory, flower color variation can be caused by transposon insertions or excisions into anthocyanin biosynthetic or regulatory genes (Inagaki et al., 1994; Hoshino et al., 2001; Morita et al., 2014, 2015). In dahlia, insertion of a 5.4-kb transposable element of the CACTA superfamily in the fourth intron of DvIVS resulted in a yellow cultivar, MJY, which was unable to accumulate anthocyanin (Ohno et al., 2011). In ‘Turning red Jimba’, anthocyanin biosynthetic genes CmDFR and CmUFGT were significantly induced when the repressive effect of CmMYB#7 weakened (Fig. 2). However, the reason for the changes of CmMYB#7 transcript levels among three different types of petals remains unknown. This change in expression of CmMYB#7 may be caused by some underlying epigenetic change or transposon in MYB promoter regions, and these will be the focus of future experiments.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. HPLC analysis of extracts of ‘Turning red Jimba’ and white ‘Jimba’ petals.
Fig. S2. The expression patterns of anthocyanin biosynthetic genes and two reported transcription factors were studied in white ‘Jimba’ and ‘Turning red Jimba’.
Fig. S3. Log fold change expression of 91 MYBs in flower petals of white ‘Jimba’ and ‘Turning red Jimba’.
Fig. S4. Phylogenetic analysis of seven CmMYBs and reported MYBs related to phenylpropanoid pathway in other plant species.
Fig. S5. Regulatory effects of transiently overexpressing CmMYB#85, CmMYB#15, CmMYB#16, and CmMYB#17 with CmMYB6 or CmbHLH2 on transcription from CmDFR and CmUFGT promoters.
Fig. S6. The expression patterns of CmMYB#7 in different petal types of white ‘Jimba’ and ‘Turning red Jimba’ measured by real-time PCR.
Fig. S7. The expression patterns of CmMYB6 and CmMYB#7 in three cultivars with different floral colors.
Table S1. Primers used in qPCR.
Table S2. Primers used in cloning genes into SK, NLuc, CLuc, and LUC vectors.
Acknowledgements
This research was supported by the National Key Research and Development Program (2018YFD1000405) and the 111 Project (B17039).
References
- Aharoni A, De Vos C, Wein M, Sun Z, Greco R, Kroon A, Mol JM, O’Connell A. 2001. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal 28, 319–332. [DOI] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. 2014. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 26, 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. 2011. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal 65, 771–784. [DOI] [PubMed] [Google Scholar]
- Carrie CS, Gregory NH. 2009. The impact of supplemental carbon sources on Arabidopsis thaliana growth, chlorophyll content and anthocyanin accumulation. Plant Growth Regulation 59, 255–271. [Google Scholar]
- Chen SM, Li CH, Zhu XR, Deng YM, Sun W, Wang LS, Chen FD, Zhang Z. 2012. The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biologia Plantarum 56, 458–464. [Google Scholar]
- Cheng J, Wei G, Zhou H, Gu C, Vimolmangkang S, Liao L, Han Y. 2014. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiology 166, 1044–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Miao CX, Lu GX. 2010. Site-directed mutagenesis based on overlap extension PCR. Progress in Modern Biomedicine 10, 1673–6273. [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. 2008. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. The Plant Journal 55, 940–953. [DOI] [PubMed] [Google Scholar]
- Fan D, Wang XQ, Tang XF, Ye X, Ren S, Wang DH, Luo KM. 2018. Histone H3K9 demethylase JMJ25 epigenetically modulates anthocyanin biosynthesis in poplar. The Plant Journal 96, 1121–1136. [DOI] [PubMed] [Google Scholar]
- Gates DJ, Olson BJSC, Clemente TE, Smith SD. 2018. A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytologist 217, 1346–1356. [DOI] [PubMed] [Google Scholar]
- Han KT, Zhao L, Tang XJ, Hu K, Dai SL. 2012. The relationship between the expression of key genes in anthocyanin biosynthesis and the color of Chrysanthemum. Acta Horticulturae Sinica 39, 516–524. [Google Scholar]
- Hoballah ME, Gübitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell’Olivo A, Arnold M, Kuhlemeier C. 2007. Single gene-mediated shift in pollinator attraction in Petunia. The Plant Cell 19, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Johzuka-Hisatomi Y, Iida S. 2001. Gene duplication and mobile genetic elements in the morning glories. Gene 265, 1–10. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Song S, Allan AC, Liu XF, Yin XR, Xu CJ, Chen KS. 2013. Differential activation of anthocyanin biosynthesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry. Plant Cell Tissue and Organ Culture 113, 491–499. [Google Scholar]
- Inagaki Y, Hisatomi Y, Suzuki T, Kasahara K, Iida S. 1994. Isolation of a Suppressor-mutator/Enhancer-like transposable element, Tpn1, from Japanese morning glory bearing variegated flowers. The Plant Cell 6, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Liu C, Xiao X, Dixon RA. 2015. The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. The Plant Cell 27, 2860–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 236–242. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Xiang LL, Yin XR, Grierson D, Li F, Chen KS. 2015. The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chrysanthemum flowers. Scientia Horticulturae 194, 278–285. [Google Scholar]
- Ma CQ, Jing CJ, Chang B, Ya JY, Liang BW, Liu L, Yang YZ, Zhao ZY. 2018. The effect of promoter methylation on MdMYBl expression determines the level of anthocyanin accumulation in skins of two non-red apple cultivars. BMC Plant Biology 18, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. 2008. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. The Plant Journal 55, 954–967. [DOI] [PubMed] [Google Scholar]
- McEwen JR, Vamosi JC. 2010. Floral colour versus phylogeny in structuring subalpine flowering communities. Proceedings of the Royal Society. B, Biological Sciences 277, 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Hoshino A. 2018. Recent advances in flower color variation and patterning of Japanese morning glory and petunia. Breeding Science 68, 128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Ishiguro K, Tanaka Y, Iida S, Hoshino A. 2015. Spontaneous mutations of the UDP-glucose:flavonoid 3-O-glucosyltransferase gene confers pale- and dull-colored flowers in the Japanese and common morning glories. Planta 242, 575–587. [DOI] [PubMed] [Google Scholar]
- Morita Y, Takagi K, Fukuchi-Mizutani M, Ishiguro K, Tanaka Y, Nitasaka E, Nakayama M, Saito N, Kagami T, Hoshino A, Iida S. 2014. A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. The Plant Journal 78, 294–304. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HBJ, Deerfield DWI. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS 4, 14. [Google Scholar]
- Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. 2006. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiology 142, 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A, Sumitomo K, Aida R. 2009. “Yellow Jimba”: suppression of carotenoid cleavage dioxygenase (CmCCD4a) expression turns white chrysanthemum petals yellow. Journal of the Japanese Society for Horticultural Science 78, 450–455. [Google Scholar]
- Ohno S, Hosokawa M, Hoshino A, Kitamura Y, Morita Y, Park KI, Nakashima A, Deguchi A, Tatsuzawa F, Doi M. 2011. A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis). Journal of Experimental Botany 62, 5105–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. 2006. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. The Plant Cell 18, 1274–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. 1999. Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. The Plant Cell 11, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoult JP, Kelber A, Schaefer HM. 2017. Colour spaces in ecology and evolutionary biology. Biological Reviews 92, 292–315. [DOI] [PubMed] [Google Scholar]
- Schiestl FP, Johnson SD. 2013. Pollinator-mediated evolution of floral signals. Trends in Ecology & Evolution 28, 307–315. [DOI] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. 2006. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. The Plant Cell 18, 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE, Martin CR, Davies KM. 2011. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytologist 189, 602–615. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. 2008. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. The Plant Journal 54, 733–749. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrolstad RE, Culbertson JD, Cornwell CJ, Mattick LR. 1982. Detection of adulteration in blackberry juice concentrates and wines. Journal of the Association of Official Analytical Chemists 65, 1417–1423. [PubMed] [Google Scholar]
- Xiang LL, Liu XF, Li X, Yin XR, Grierson D, Li F, Chen KS. 2015. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE 10, e0143892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FJ, Sheng YL, Zhang Y. 2013. Standardization production techniques of cut Chrysanthemum morifolium Ram. Modern Agricultural Science and Technology 21, 181–182. [Google Scholar]
- Zhang YQH, Wang YG, Fang WM, Guan ZY, Zhang F, Chen FD. 2018. Changes of colors and pigment compositions during the senecence process of Chrysanthemum morifolium. Acta Horticulturae Sinica 45, 519–529. [Google Scholar]
- Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG. 2009. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Molecular Plant 2, 790–802. [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. The Plant Journal 40, 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.