NO regulates cold acclimation through C-repeat binding factor-dependent and -independent pathways by regulating anthocyanin biosynthesis and ABA sensitivity.
Keywords: ABA, anthocyanins, Arabidopsis, CBFs, cold acclimation, freezing tolerance, nitric oxide
Abstract
Plant tolerance to freezing temperatures is governed by endogenous components and environmental factors. Exposure to low non-freezing temperatures is a key factor in the induction of freezing tolerance in the process called cold acclimation. The role of nitric oxide (NO) in cold acclimation was explored in Arabidopsis using triple nia1nia2noa1-2 mutants that are impaired in the nitrate-dependent and nitrate-independent pathways of NO production, and are thus NO deficient. Here, we demonstrate that cold-induced NO accumulation is required to promote the full cold acclimation response through C-repeat Binding Factor (CBF)-dependent gene expression, as well as the CBF-independent expression of other cold-responsive genes such as Oxidation-Related Zinc Finger 2 (ZF/OZF2). NO deficiency also altered abscisic acid perception and signaling and the cold-induced production of anthocyanins, which are additional factors involved in cold acclimation.
Introduction
When facing low non-freezing temperatures, plants respond by expressing a wide array of cold-responsive genes and by reprogramming metabolism and diverse non-metabolic processes that include the production of cryoprotective proteins and metabolites, changes in membrane structure and function, and growth arrest (Pollock and Eagles, 1988; Fowler and Thomashow, 2002; Cook et al., 2004; Renaut et al., 2004; Hannah et al., 2005; Lee et al., 2005; Chinnusamy et al., 2007; Kaplan et al., 2007; Guy et al., 2008). Tolerance to freezing is the result of endogenous components of the plant and inducible environmental factors. Among the environmental factors that enhance freezing tolerance, the previous exposure of plants to low non-freezing temperatures, or cold acclimation, is likely the most efficient way to induce further freezing tolerance (Thomashow, 1999; Knight and Knight, 2012). Cold acclimation is an adaptive process that allows plants to survive freezing winters. It is mainly controlled by changes in gene expression that are mostly regulated at the transcriptional and post-transcriptional levels (Barrero-Gil and Salinas, 2013; Miura and Furumoto, 2013). Among the cold-induced signaling pathways, the one mediated by the C-repeat binding factors (CBFs)/dehydration-responsive element binding factors (DREBs) is probably the most important and the best characterized (Gilmour et al., 2004; Medina et al., 2011). This pathway is controlled by inducer of CBF expression 1 (ICE1), a MYC-type transcriptional activator that confers increased freezing tolerance upon binding to the promoters of CBF genes (Chinnusamy et al., 2003). The CBF regulon is regulated by the ICE1–CBF pathway as well as other transcription factors that are rapidly induced by cold and work as a complex regulatory network only partially dependent on CBFs (Shi et al., 2018). Both CBF-dependent and -independent pathways converge onto the activation of cold-responsive (COR) genes (Gilmour et al., 1998) that are critical for cold acclimation and the enhancement of freezing tolerance. COR genes code for dehydrins, heat shock proteins, glucanases, and chitinases that are important to prevent membrane alterations and protein aggregation commonly by altering water properties (Ferreira et al., 2018). Moreover, some of the antifreeze properties of these intrinsically disordered proteins seem to be due to the effect on slowing the growth of ice crystals and avoiding recrystallization (Griffith et al., 2005).
As mentioned above, in addition to being determined genetically, cold acclimation is also controlled by environmental factors such as temperature and photoperiod (Kohn and Levitt, 1966; Weiser, 1970) that, in turn, regulate plant hormone biosynthesis and signaling (Adams and Carré, 2011; Atamian and Harmer, 2016). Besides abscisic acid (ABA), whose quantitative and qualitative contribution is decisive (Nakashima et al., 2014), other phytohormones, including jasmonates, salicylates, auxins, cytokinins, gibberellins, and brassinosteroids, have important functions in this adaptive process (Jeon et al., 2010; Miura and Ohta, 2010; Rahman, 2013; Richter et al., 2013; Sharma and Laxmi, 2015; Eremina et al., 2016). Sugars have been also reported to enhance freezing tolerance of plants by stabilizing protein structure (Arakawa and Timasheff, 1982), and by avoidance of freezing through supercooling mechanisms in Arabidopsis (Reyes-Díaz et al., 2006). Other non-hormone regulatory molecules such as polyamines, lipids, anthocyanins/flavonoids, reactive oxygen species and nitric oxide (NO) have been described as being involved in freezing tolerance and cold acclimation (Cuevas et al., 2008; Zhao et al., 2009; Puyaubert and Baudouin, 2014; Chen and Thelen, 2016; Takahashi et al., 2016; van Buer et al., 2016). In addition, anthocyanins/flavonoids have been characterized as essential factors to achieve full development of cold acclimation in Arabidoopsis (Catalá et al., 2011; Schulz et al., 2016; Perea-Resa et al., 2017) because of their antioxidative functions protecting from photoinhibition (Krol et al., 1995; Harvaux and Kloppstech, 2001; Korn et al., 2008; Schulz et al., 2016).
We previously reported that NO-deficient nia1nia2noa1-2 mutant Arabidopsis plants displayed an enhanced constitutive freezing tolerance due to the accumulation of osmolytes, antioxidants, and hormones such as ABA and jasmonates (Costa-Broseta et al., 2018), thus pointing to NO as a negative regulator of constitutive freezing tolerance. In turn, genetic and pharmacological approaches allowed the proposal of a positive regulatory role for NO in cold acclimation in Arabidopsis that was achieved through NO-regulated expression of CBF genes (Zhao et al., 2009, 2011; Cantrel et al., 2011; Puyaubert and Baudouin, 2014; Fan et al., 2015). Since the contribution of NO produced independently of nitrate to cold acclimation has not been sufficiently addressed, in this work we have studied the capacity to cold acclimate of triple nia1nia2noa1-2 mutant plants that are impaired in nitrate-dependent and nitrate-independent nitric oxide associated 1 (NOA1)-associated pathways (Lozano-Juste and León, 2010). We found that, besides the already reported regulation of CBF gene expression, NO can also promote cold acclimation trough CBF-independent pathways involving altered ABA perception and anthocyanin/flavonoid accumulation and likely through the CBF-independent expression of other cold-responsive genes such as the Oxidation-Related Zinc Finger 2 (ZF/OZF2).
Materials and methods
Plant materials and growth conditions
The Arabidopsis Col-0 ecotype was the wild-type genetic background used in this work. Seeds from nia1nia2 and noa1-2 alleles were obtained from the NASC seed bank (N2356 and T-DNA insertion SAIL_507_E11, respectively). The triple nia1nia2noa1-2 mutant seeds were obtained by crossing as previously reported (Lozano-Juste and León, 2010). To ensure that triple homozygous mutant plants were used, every seed stock obtained was genotyped by PCR and cleaved amplified polymorphic sequences (CAPS) as described previously (Lozano-Juste and León, 2010) by using specific primers (Supplementary Table S1 at JXB online). Seeds were grown in peat substrate:vermiculite (3:1)-containing pots and allowed to develop for 7 d under a long-day photoperiod (16 h light at 21 °C: 8 h darkness at 20 °C, cool-white fluorescent light, photon flux of 100 μmol m−2 s−1) light regime as previously described (Castillo and León, 2008). Experiments were performed with 2-week-old plants.
Freezing tolerance assays
Seeds from the different genotypes were sown and grown as described in the previous section. Plants were removed from each pot to leave a similar number (25–30) of plants, homogeneously distributed in all pots. Before being subjected to freezing temperatures, plants acclimated at 4 °C for 7 d were exposed for an additional 1 h to 4 °C in the freezing chamber. Then, temperature was progressively decreased (−1 °C/30 min) until reaching the indicated freezing temperatures. After exposing plants to the appropriate freezing temperature for 6 h, temperature was gradually increased to 4 °C (+1 °C/30 min). One hour later, plants were transferred to 21 °C under a long-day photoperiod as described above for recovery and subsequent survival evaluation 7 d later. Freezing tolerance was determined as the percentage of surviving plants after exposure to different freezing temperatures for 6 h. LT50, defined as the freezing temperature causing 50% lethality in a plant population, was calculated after plotting survival rates versus temperature.
RNA isolation and quantitative transcript analysis
Total RNA was isolated from 10- to 12-day-old seedlings, separated, and analysed by RT-qPCR techniques as previously described (Castillo and León, 2008) with specific primers. Supplementary Table S1 shows the oligonucleotide sequence, the identity and full name annotation of every gene analysed in this work. For microarray-based transcriptomic analyses, Col-0 and nia1nia2noa1-2 seedlings were grown for 14 d as described above. Then, plants were either kept at the above described growing conditions (control non-acclimated) or incubated at 4 °C (acclimated) in a freezer chamber at similar light conditions to control plants for 1 h. After that, non-acclimated and cold-acclimated plant samples were collected, frozen in liquid nitrogen and RNA was extracted with the Nucleo Spin RNA Plus kit from Macherey-Nagel (Düren, Germany). RNA integrity was assessed with a 2100 Bioanalyzer (Agilent). Three independent biological replicates for each genotype and condition were used for the trancriptomic analyses. Total RNA samples of 0.5 μg were amplified and labelled with the Agilent Low Input Quick Amp Labelling Kit in a two color design. Hybridization was performed on an Agilent Arabidopsis (V4) Gene Expression 4×44K Microarray. After washing and drying, slides were scanned in an Agilent G2565AA microarray scanner, at 5 μm resolution using the double scanning, as recommended. Image files were analysed with Feature Extraction software 9.5.1. with GeneSpring 11.5 software.
NO detection by fluorescence and confocal microscopy
The endogenous levels of NO in shoots were determined by staining with 10 µM 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA) as described (Guo et al., 2003) with some modifications. Plants were grown in soil under standard long day phoroperiod light conditions and 21 °C for 6 d. Then, plants were either maintained at standard non-acclimated growing conditions or cold acclimated at 4 °C for 7 d. At day 13, plants were sprayed with DAF-FM DA to run-off, kept under darkness for 1 h, and later, after extensive washing, transferred to growing conditions at either 4 or 21 °C for an additional 23 h. Fluorescence was detected by confocal microscopy with a Leica TCS SP5 confocal laser scanning microscope, using unchanged parameters for every measurement. The specificity of NO-related fluorescence detection was assessed by treatment with 0.5 mM of the NO scavenger 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO, Sigma-Aldrich, USA). The DAF-FM DA fluorescence intensities were analysed using Adobe Photoshop by quantifying green pixels in three to six replicate images taken from independent plants in at least three different pots for every genotype and condition.
Quantification of anthocyanins
Anthocyanins were spectrophotometrically determined in methanolic extracts by reading their absorbance at 530 nm as described (Solfanelli et al., 2006).
Phytohormone quantification
Four independent biological replicate samples of around 150–200 mg fresh weight of either non-acclimated or cold-acclimated Col-0 and nia1nia2noa1-2 whole seedlings were suspended in 80% methanol–1% acetic acid containing internal standards and mixed by shaking for 1 h at 4 °C. The extract was kept a −20 °C overnight, centrifuged, the supernatant dried in a vacuum evaporator, and the dry residue was dissolved in 1% acetic acid and passed through an Oasis HLB (reverse phase) column as described in Seo et al. (2011). The dried eluate was dissolved in 5% acetonitrile–1% acetic acid, and the hormones were separated using an autosampler and reversed phase UHPLC chromatography (2.6 µm Accucore RP-MS column, 50 mm length×2.1 mm i.d., Thermo Fisher Scientific) with a 5–50% acetonitrile gradient containing 0.05% acetic acid, at 400 µl min−1 over 14 min. The phytohormones were analysed with a Q-Exactive mass spectrometer (Orbitrap detector, Thermo Fisher Scientific) by targeted selected ion monitoring. The concentrations of hormones in the extracts were determined using embedded calibration curves and the Xcalibur 2.2 SP1 build 48 and TraceFinder programs. The internal standards for quantification of each of the different plant hormones were the deuterium-labelled hormones.
In silico analyses of gene ontology and transcriptome profiles
Gene Ontology (GO) enrichment of functional categories in gene lists was performed by the Gene Ontology Consortium tools (http://www.geneontology.org/) and the GO Analysis Toolkit and Database for Agricultural Community (AgriGO, http://bioinfo.cau.edu.cn/agriGO/analysis.php). Comparison of transcriptome profiles with publicly available datasets was performed with the AtCAST3.1 tool (http://atpbsmd.yokohama-cu.ac.jp/cgi/atcast/search_input.cgi).
Results
Impaired accumulation of NO in nia1nia2noa1-2 mutant plants correlated with defective cold acclimation
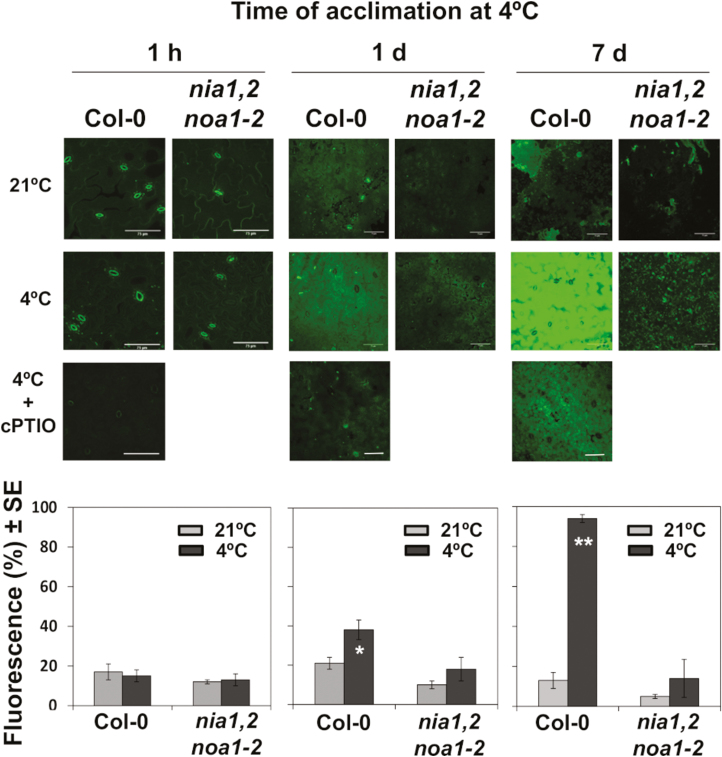
Wild-type but not NO-deficient nia1nia2noa1-2 mutant plants responded to the incubation at 4 °C by increasing their endogenous NO content (Fig. 1). Significant changes in endogenous NO content could not be detected by an incubation time as short as 1 h (Fig. 1). However, by 1 d at 4 °C, Col-0 leaves accumulated slightly less than 2-fold NO over the levels detected in plants growing at 21 °C, and, as expected, no significant increase was observed in nia1nia2noa1-2 mutant leaves (Fig. 1). After exposure for 7 d at 4 °C, wild-type leaves accumulated 16-fold more NO compared with leaves grown under non-acclimated conditions. In turn, cold-acclimated nia1nia2noa1-2 leaves did not accumulate significantly different NO from leaves growing at 21 °C (Fig. 1).
Fig. 1.
NO levels in cotyledons of Col-0 and nia1nia2noa1-2 plants in response to low temperature. Plants were either maintained at standard 21 °C growing conditions or exposed for different times to 4 °C. The fluorescence of DAF-FM DA-treated plants was detected by confocal microscopy. As controls for checking that fluorescence was due to NO, plants exposed to 4 °C were treated with 0.5 mM of the NO scavenger cPTIO. Images shown are representative of four to six different analysed plants per genotype and condition, and the quantification values are the mean ±standard error. Asterisks indicate significant differences between nia1nia2noa1-2 and wild-type plants in Student’s t-test (*P<0.05, **P<0.01). Scale bars: 75 μm.
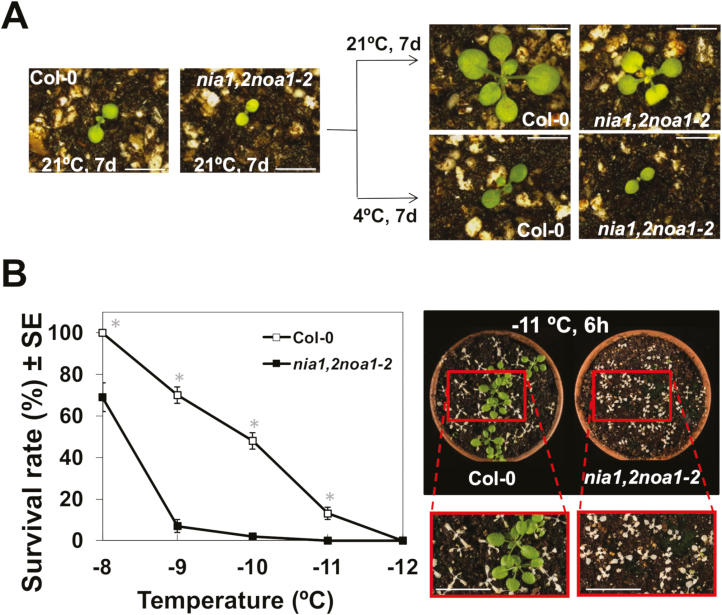
The analysis of the freezing tolerance of cold acclimated (7 d at 4 °C) Col-0 and NO-deficient nia1nia2noa1-2 plants revealed significant differences. As previously reported, nia1nia2noa1-2 plants display reduced size with a smaller rosette and shorter roots than Col-0 plants when growing under control conditions (Lozano-Juste and León, 2010), but both had retarded growth on exposure at 4 °C (Fig. 2A). Cold-acclimated mutant plants exhibited a significantly lower capacity to tolerate freezing temperatures than cold-acclimated wild-type plants, the LT50 being −8.3 °C and −9.8 °C, respectively (Fig. 2B). Moreover, since changes in NO content were already significant by 1 d upon cold treatment (Fig. 1), the earliest processes involved in NO-mediated low temperature-induced freezing tolerance should occur from minutes to a few hours after treatment.
Fig. 2.
Cold acclimation capacity of Col-0 and nia1nia2noa1-2 plants. (A) Representative 7-day-old wild-type and mutant plants grown under standard conditions before being shifted to 4 °C for an additional 7 d (cold-acclimated) or kept for 7 d more at 21 °C (non-acclimated as control). (B) Freezing tolerance of 2-week-old plants exposed for 6 h to the indicated freezing temperatures after being acclimated for 7 d at 4 °C. In all cases, freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 d of recovery under control conditions. Data are expressed as means of three independent experiments with around 50 plants each ±standard deviation. *Significant differences between nia1nia2noa1-2 and wild-type plants in Student’s t-test (P<0.05). The right panel in (B) shows a representative image of cold-acclimated plants from both genotypes after exposure to −11 °C for 6 h and recovery under control conditions for an additional 7 d. Close-up images corresponding to the framed areas are included. Scale bars: 5 mm (A) and 1 cm (B). (This figure is available in color at JXB online.)
Cold-induced expression of CBF genes in wild-type and NO-deficient Arabidopsis
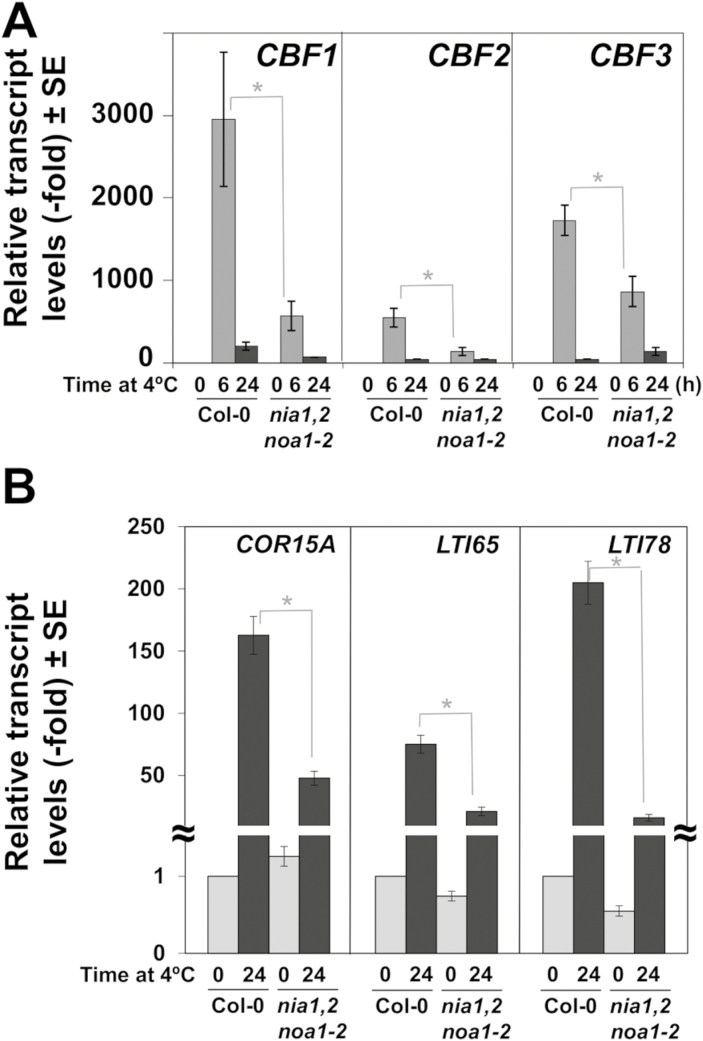
We analysed whether the impaired ability to cold acclimate of nia1nia2noa1-2 plants might be due to a reduced cold induction of the CBF genes as reported for the nia1,2 mutant (Zhao et al., 2009). Figure 3A shows that, after 6 h of exposure to 4 °C, the induction of CBF1, 2, and 3 in nia1nia2noa1-2 was significantly lower than in wild-type plants. This reduced induction was specially accentuated in the case of the CBF1 gene. Accordingly, COR15A, LTI65, and LTI78 genes, which are well characterized targets of CBFs (Thomashow, 1999), were significantly less induced upon cold treatment in nia1nia2noa1-2 than in Col-0 plants (Fig. 3B). These data confirm that NO functions as a positive regulator of cold acclimation likely by promoting the cold-induced expression of CBF genes and their corresponding gene targets. However, it is noteworthy that NO-deficient nia1nia2noa1-2 plants were not fully blocked in cold-induced CBF expression (Fig. 3A), thus suggesting CBFs might be also up-regulated by cold independently of NO, and also that NO-regulated cold acclimation might proceed through CBF-independent pathways.
Fig. 3.
Expression levels of the CBFs and their target genes in Col-0 and nia1nia2noa1-2 plants exposed to 4 °C for different times. (A,B) CBF1, CBF2, CBF3 (A) and COR15A, LTI65, and LTI78 (B) transcript levels were quantified by RT-qPCR from total RNAs isolated from the indicated plants at the indicated times of exposition at 4 °C. Values are the mean of three independent biological replicate samples for each genotype and condition ±standard error. *Significantly different with P≤0.05 in Student’s t-test.
Early events in gene expression during cold acclimation
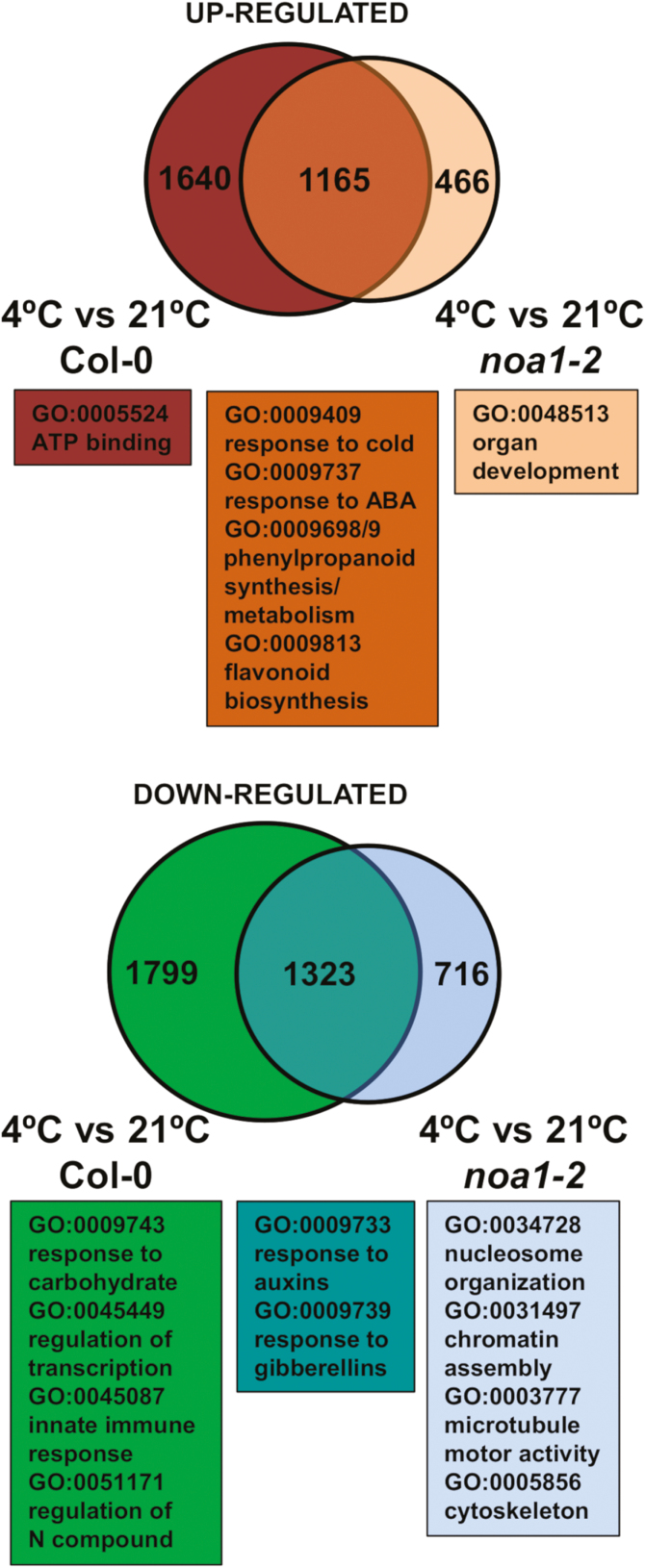
To unveil early NO-related factors and processes involved in low temperature triggering cold acclimation, we performed a transcriptomic analysis of 2-week-old Col-0 and nia1nia2noa1-2 plants after 1 h exposure to 4 °C in comparison with the corresponding plants grown under control conditions. Supplementary Table S2 shows that extensive changes in their transcriptomes (P-value corrected for false discovery rate (FDR) lower than 0.05 and fold change>|1.5|) were detected by 1 h after the 4 °C treatment, with a significant attenuation of the low temperature response in the NO-deficient genotype. Of the 5927 genes that respond differentially to the cold treatment compared with plants maintained at 21 °C, 2805 were up-regulated and 3122 were down-regulated (Fig. 4). In turn, 3670 differentially expressed genes (1631 and 2039 up- and down-regulated, respectively) were detected when comparing cold-treated with control nia1nia2noa1-2 plants (Fig. 4). The transcript levels of randomly selected differentially expressed genes were quantified by qRT-PCR from independent RNAs extracted from samples under the same conditions as those used to perform the transcriptome analysis. We observed 22 out of 28 genes tested by qRT-PCR correlated with data obtained from the microarray analysis (Supplementary Table S3), thus suggesting microarray data were truly representative of the transcriptomes of each genotype and condition. Around 40% and 70% of the differentially expressed genes in cold-treated versus untreated wild-type and NO-deficient plants were common for Col-0 and nia1nia2noa1-2 (Fig. 4), thus suggesting that both genotypes shared most targets affected by cold. However, wild-type plants showed 1640 up-regulated genes and 1799 down-regulated genes that were not differentially expressed in cold-treated NO-deficient plants (Fig. 4). Moreover, nia1nia2noa1-2 plants displayed 466 up-regulated genes and 716 down-regulated genes that were not differentially expressed in Col-0 plants (Fig. 4). These subsets of genes likely include specific regulatory targets that might explain the different capacity of wild-type and NO-deficient plants to cold acclimate.
Fig. 4.
Up- and down-regulated genes in cold-treated and non-treated Col-0 and nia1nia2noa1-2 plants. Venn diagrams show the intersection between genes differentially expressed in plants of the indicated genotypes exposed 1 h to 4 °C. Gene Ontology analysis of the genes remaining out of the intersection was performed at the Gene Ontology Consortium and AgriGO Platform Analysis. The most significant functional categories are shown.
To identify potential early factors or processes impairing cold acclimation in nia1nia2noa1-2 plants, we first performed GO analyses of the differential transcriptomes and the above mentioned gene subsets. Cold exposed Col-0 and nia1nia2noa1-2 plants overrepresented functional categories related to cold responses and other ABA-related abiotic stress processes among up-regulated genes (Supplementary Table S4). Among the down-regulated genes, the GO analysis showed a significant enrichment of light and phytohormone related stimuli as well as photosynthesis in Col-0 plants, whereas sulfur and glucosinolate metabolism was over-represented in nia1nia2noa1-2 plants (Supplementary Table S4). The GO analysis for genes differentially expressed only in wild-type plants yielded similar functional categories of enrichment to the above mentioned for the global analysis with the exemption of the category of ATP binding among up-regulated genes and RNA-related processes and regulation of transcription among down-regulated genes (Supplementary Table S4). For those genes differentially expressed exclusively in NO-deficient triple mutant plants, only the categories related to nucleosome and chromatin organization and to cytoskeleton components, were differentially overrepresented among down-regulated genes (Supplementary Table S4). Despite the differences described above in the GO categories represented in Col-0 and nia1nia2noa1-2 plants upon cold treatment, we found that the categories more over-represented among the up-regulated genes in both genotypes were those related to ABA-regulated abiotic stress responses and those involved in phenylpropanoid-derived synthesis of flavonoids and anthocyanins (Fig. 4; Supplementary Table S4). Interestingly, some of the genes in those categories were only up-regulated in wild-type but not in NO-deficient plants, thus suggesting that ABA signaling and flavonoid/anthocyanin production might be key differential factors in NO-modulated cold-triggered responses.
NO regulates the sensitivity of Arabidopsis to ABA in response to low temperature
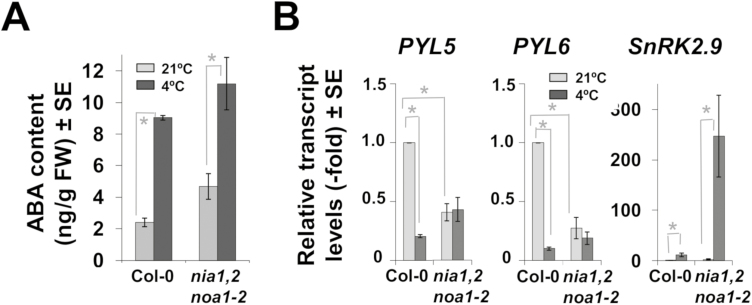
As mentioned above, the early responses to cold in wild-type and NO-deficient plants included different ABA-related genes. Table 1 shows the differentially expressed ABA-related genes upon cold treatment in both genotypes. Among the up-regulated genes, only wild-type plants showed enhanced expression of genes encoding negative ABA regulatory components such as the phosphatases PP2C5 and HAI1 and the transcription factor ANAC002/ATAF1, which activates the expression of the ABA biosynthetic gene NCED3 (Jensen et al., 2013) but negatively regulates the expression of ABA-related stress responsive genes (Lu et al., 2007) (Table 1). In turn, only NO-deficient plants displayed enhanced expression of the Ninja family AFP3 and AFP4 transcription factors, which act as repressors of ABA response (Lynch et al., 2017). It is noteworthy that only Col-0 plants showed down-regulated expression of several ABA receptor encoding genes such as PYL5, PYL6, PYL7, and PYL9 (Table 1). As many of the genes differentially expressed by cold only in one of the genotypes coded for either receptors or regulatory components of ABA, our findings suggest that perception and/or signaling of ABA might be altered in NO-deficient plants. To check this hypothesis, we first analysed whether the altered ABA-related response might be due to changes in ABA homeostasis in NO-deficient plants. Figure 5A shows that, although nia1nia2noa1-2 plants have enhanced basal ABA content relative to Col-0 plants, both wild-type and NO-deficient plants increased their ABA content after exposure to 4 °C reaching values that were not significantly different, thus suggesting the ABA homeostasis was not significantly altered in NO-deficient plants during the early events of the cold acclimation process.
Table 1.
ABA-related genes differentially expressed upon cold treatment in Col-0 and nia1nia2noa1-2
| Probe name | Fold change 4 °C versus 21 °C | AGI | Gene name | |
|---|---|---|---|---|
| Col-0 | nia1nia2noa1-2 | |||
| A_84_P12528 | 9.844977 | n.s.d. | AT2G40180 | ATHPP2C5, PP2C5 |
| A_84_P203128 | n.s.d. | 11.643681 | AT3G29575 | AFP3 |
| A_84_P760385 | 6.7814 817 | n.s.d. | AT3G55610 | P5CS2 |
| A_84_P21026 | 6.094576 | n.s.d. | AT2G33380 | AtCLO3, RD20 |
| A_84_P12575 | n.s.d. | 8.8540325 | AT2G23030 | SNRK2.9, SNRK2-9 |
| A_84_P11248 | 4.8641524 | n.s.d. | AT5G59220 | HAI1, SAG113 |
| A_84_P275730 | n.s.d. | 4.1209784 | AT3G02140 | AFP4, TMAC2 |
| A_84_P12143 | 4.412484 | n.s.d. | AT5G45340 | CYP707A3 |
| A_84_P18843 | 3.659796 | n.s.d. | AT5G66400 | ATDI8, RAB18 |
| A_84_P13815 | 3.4428418 | n.s.d. | AT4G19030 | ATNLM1 |
| A_84_P23021 | 3.3664775 | n.s.d. | AT2G29090 | CYP707A2 |
| A_84_P861274 | 3.212613 | n.s.d. | AT5G66400 | ATDI8, RAB18 |
| A_84_P172093 | n.s.d. | 3.030949 | AT1G12480 | SLAC1, OZS1, RCD3 |
| A_84_P19986 | 3.0502687 | n.s.d. | AT1G01720 | ANAC002, ATAF1 |
| A_84_P10329 | 2.8910208 | n.s.d. | AT5G65280 | GCL1 |
| A_84_P807853 | 2.580678 | n.s.d. | AT2G36530 | ENO2, LOS2 |
| A_84_P23380 | 2.5109425 | n.s.d. | AT5G01540 | LECRKA4.1 |
| A_84_P11990 | n.s.d. | 1.9472115 | AT4G34240 | ALDH3, ALDH3I1 |
| A_84_P809296 | n.s.d. | 1.8705412 | AT2G05520 | ATGRP3 |
| A_84_P531660 | n.s.d. | 1.4853065 | AT4G21670 | ATCPL1, CPL1, FRY2 |
| A_84_P20725 | 1.7062953 | n.s.d. | AT5G63980 | ALX8, ATSAL1, FRY1 |
| A_84_P23165 | 1.6769094 | n.s.d. | AT3G50500 | SNRK2.2 |
| A_84_P593088 | 1.4889517 | n.s.d. | AT5G13680 | ABO1, AtELP1, ELO2 |
| A_84_P23632 | −1.4289963 | n.s.d. | AT1G35670 | ATCDPK2, CPK11 |
| A_84_P14981 | −1.543499 | n.s.d. | AT5G46240 | KAT1 |
| A_84_P14532 | −1.6045026 | n.s.d. | AT1G01360 | PYL9, RCAR1 |
| A_84_P808612 | n.s.d. | −2.3673778 | AT3G26520 | GAMMA-TIP2 |
| A_84_P816097 | −1.7132156 | n.s.d. | AT1G01360 | PYL9, RCAR1 |
| A_84_P10468 | −1.9757272 | n.s.d. | AT1G05630 | 5PTASE13, AT5PTASE13 |
| A_84_P20389 | −2.0552506 | n.s.d. | AT4G01026 | PYL7, RCAR2 |
| A_84_P23439 | −2.1560888 | n.s.d. | AT5G23350 | ABA-responsive protein |
| A_84_P795995 | −3.5029442 | n.s.d. | AT3G26520 | GAMMA-TIP2 |
| A_84_P23593 | −5.856309 | n.s.d. | AT5G08350 | ABA-responsive protein |
| A_84_P813773 | −5.9837146 | n.s.d. | AT1G75380 | ATBBD1, BBD1 |
| A_84_P10533 | −6.5018597 | n.s.d. | AT1G75380 | ATBBD1, BBD1 |
| A_84_P174151 | −6.7487288 | n.s.d. | AT2G40330 | PYL6, RCAR9 |
| A_84_P852047 | −12.006997 | n.s.d. | AT5G05440 | PYL5, RCAR8 |
| A_84_P817892 | −13.558136 | n.s.d. | AT5G05440 | PYL5, RCAR8 |
| A_84_P146199 | −14.759594 | n.s.d. | AT5G05440 | PYL5, RCAR8 |
n.s.d, not significantly different.
Fig. 5.
ABA homeostasis and signaling in cold-treated wild-type and NO-deficient plants. (A) Quantification of ABA in control (light gray) and cold-treated for 24 h at 4 °C (dark gray) Col-0 and nia1nia2noa1-2 plants. Values represent the mean values of four independent biological replicate samples for each genotype and condition ±standard error. (B) The transcript levels of the ABA signaling genes PYL5, PYL6, and SnRK2.9 were quantified by RT-qPCR from three independent total RNAs isolated from control plants and plants exposed to 4 °C for 24 h. Values are the mean of three independent biological replicate samples for each genotype and condition ±standard error. *Significantly different with P≤0.05 in Student’s t-test.
We examined the capacity of nia1nia2noa1-2 mutants to correctly sense and signal ABA by analysing the levels of different ABA-related signaling transcripts. As shown in Fig. 5B, the transcripts of two closely related genes coding for the ABA receptors PYL5 and PYL6 were strongly down-regulated in response to 4 °C exposure in wild-type plants. In turn, no such repression by cold was observed in the nia1nia2noa1-2 plants, which indeed showed levels in control plants comparable to those detected under cold conditions in Col-0 plants (Fig. 5B), thus confirming the above mentioned transcriptome data (Table 1). The levels of SnRK2.9 transcripts in nia1nia2noa1-2 plants were more than 20-fold higher than in Col-0 plants after cold treatment (Fig. 5B), thus also confirming the microarray data (Table 1).
NO activates anthocyanin accumulation in Arabidopsis during cold acclimation
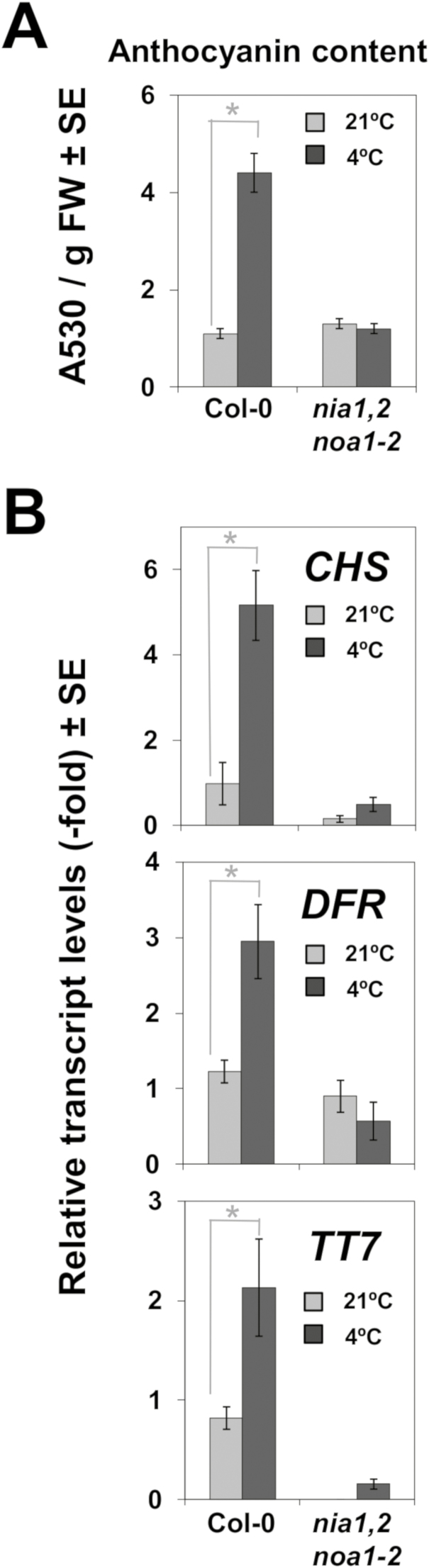
The cold-regulated transcriptomes of Col-0 and nia1nia2noa1-2 plants also contained an over-representation of flavonoid and anthocyanin biosynthetic and metabolic genes (Supplementary Table S4). However, as shown in Supplementary Table S2, only wild-type plants up-regulated the MYB75/PAP1 gene coding for the master regulator of flavonoid and anthocyanin biosynthesis (Borevitz et al., 2000), as well as several other early and late flavonoid/anthocyanin biosynthetic genes (Petroni and Tonelli, 2011) coding for the UDP-glycosyltransferases UGT75C1, UGT78D3, UF3GT and UGT89C1; the dihydroflavonol 4-reductase DFR/TT3; the caffeic acid/5-hydroxyferulic acid O-methyltransferase OMT1; the flavonoid 3′-monooxygenase CYP75B1/TT7; and the ANAC078/NAC2 transcription factor that regulates flavonoid biosynthesis under high light (Morishita et al., 2009). Hence, we analysed the content of anthocyanins in nia1nia2noa1-2 and Col-0 plants after they were exposed to 4 °C for 24 h. Wild-type plants increased their anthocyanin content more than 4-fold in response to low temperature (Fig. 6A). In contrast, nia1nia2noa1-2 plants did not accumulate anthocyanins when exposed to cold (Fig. 6A). As expected from these findings, the cold induction of several genes involved in anthocyanin biosynthesis, such as CHS, DFR, and TT7, was blocked in the NO-deficient plants (Fig. 6B).
Fig. 6.
Effects of low temperature on anthocyanin biosynthesis in Col-0 and nia1nia2noa1-2 plants. (A) Anthocyanin content was spectrophotometrically quantified. Data represent the means of three independent experiments with 25 and 10 plants each. (B) Transcript levels of anthocyanin biosynthetic genes chalcone synthase (CHS), dihydroflavonol reductase (DFR), and flavonol 3-hydroxylase (TT7/F3H) were quantified by RT-qPCR from total RNAs isolated from plants of the indicated genotype grown under control conditions (light gray) or exposed to 4 °C for 24 h (dark gray). Values are the mean of three independent biological replicate samples for each genotype and condition ±standard error. *Significantly different with P≤0.05 in Student’s t-test.
NO activates cold acclimation also through CBF-independent signaling
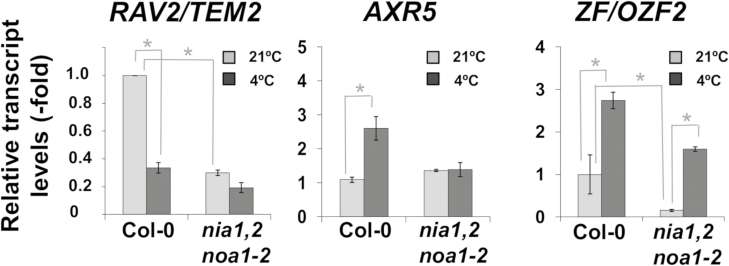
The results reported above indicated that NO regulates the process of cold acclimation in Arabidopsis by modulating CBF expression, ABA sensitivity and anthocyanin accumulation, indicating that it mediates this adaptive process through several independent pathways. It has been recently reported that the cold-induced expression of the CBF regulon requires an extensive co-regulation by other so-called first wave transcription factors (Park et al., 2015). We analysed the transcript levels of some of those genes in wild-type and nia1nia2noa1-2 plants exposed to 21 and 4 °C. Three of them, RAV2/TEM2, AXR5, and ZF/OZF2, were differentially expressed upon cold treatment in both genotypes. Figure 7 shows that the RAV2/TEM2 gene was down-regulated (0.3-fold) upon cold treatment in wild-type plants but the down-regulation was abolished in the nia1nia2noa1-2 plants. Conversely, the AXR5 gene was up-regulated (2.5-fold) upon cold treatment in wild-type plants but not in nia1nia2noa1-2 plants (Fig. 7). Finally, reduced levels of ZF/OZF2 transcript were detected in nia1nia2noa1-2 compared with wild-type plants both at 21 °C and at 4 °C, and the up-regulation by cold treatment was significantly higher in nia1nia2noa1-2 (10.3-fold) than in Col-0 (2.8-fold) plants (Fig. 7). Differential expression was specific for those genes, as other members of the first wave transcription factors described by Park et al. (2015) such as HSFC1, RAV1, CRF2, DEAR1, ZAT10, DOF1.10, MYB44, ATHB2, and ANAC62 were similarly regulated by cold in wild-type and NO-deficient plants (Supplementary Fig. S1).
Fig. 7.
Effect of cold treatment on the transcript levels of cold-related RAV2/TEM2, AXR5, and ZF/OZF2 transcription factors in Col-0 and nia1nia2noa1-2 plants. Transcript levels were quantified by RT-qPCR from three independent RNAs for each genotype grown under control conditions (light gray) or exposed to 4 °C for 1 h (dark gray). Values are the mean of three independent biological replicate samples for each genotype and condition ±standard error. *Significantly different with P≤0.05 in paired Student’s t-test.
Although we found a differential expression pattern in nia1nia2noa1-2 and Col-0 plants for RAV2/TEM2, AXR5, and ZF/OZF2 genes under both control and cold conditions, an in silico analysis with data from the AtGenExpress Visualization Tool (http://jsp.weigelworld.org/expviz/expviz.jsp) showed that only ZF/OZF2 was largely induced by cold treatment in both shoots and roots (Supplementary Fig. S2A), and specifically induced by ABA (Supplementary Fig. S2B).
Discussion
NO promotes cold acclimation through CBF-dependent and CBF-independent processes and ABA perception and signaling
Genetic and pharmacological approaches suggest NO is required for plants to cold acclimate (Zhao et al., 2009, 2011; Cantrel et al., 2011; Puyaubert and Baudouin, 2014; Fan et al., 2015). Nevertheless, our knowledge of the early events occurring in plants during the exposure to low temperatures and, more specifically, on the NO involvement in that process is rather limited. Here, we have used nia1nia2noa1-2 triple mutant plants, which are impaired not only in nitrate reductase-mediated but also in nitrate-independent NOA1-associated production of NO (Lozano-Juste and León, 2010), to explore differential fast responses upon cold exposure of wild-type and NO-deficient plants (Fig. 1). We first confirmed that nia1nia2noa1-2 plants are also impaired in their ability to cold acclimate (Fig. 2). Then, transcriptome changes in wild-type and NO-deficient plants were analysed as soon as 1 h after cold exposure of plants to identify potential targets involved in NO-regulated activation of cold acclimation. Besides genes related to cold-triggered responses, a significant enrichment of the functional categories related to the control of transcription and transcription factors occurred only in wild-type plants among the cold up-regulated genes (Fig. 4; Supplementary Table S4). The process of cold acclimation involves many physiological and biochemical changes, most of them being controlled through changes in gene expression (Knight and Knight, 2012). In Arabidopsis, the best characterized and, likely, the most relevant is the pathway mediated by the CBFs. Regarding this, the reduced capacity of nia1,2 plants to cold acclimate has been proposed to be due to an impaired cold-induced expression of genes coding for the CBF transcription factors and their downstream targets (Cantrel et al., 2011). Our data with nia1nia2noa1-2 plants further support an important function of NO in promoting the induction of CBFs in response to low temperature (Fig. 3), but also suggest that NO controls the process of cold acclimation through CBF-independent regulatory pathways. A significant contribution to the transcriptional control of cold acclimation in nia1nia2noa1-2 plants could come from the altered expression pattern of some of the so-called ‘first wave’ genes coding for transcription factors that participate in the co-regulation of CBF-independent cold triggered processes (Park et al., 2015). In particular, RAV2/TEM2, AXR5, and ZF/OZF2 were differentially regulated by cold acclimation in Col-0 and nia1nia2noa1-2 plants (Fig. 7). The in silico analysis of the transcript levels of those genes in the cold treatment experiments in shoots and roots (Kilian et al., 2007) showed a strong up-regulation of the ZF/OZF2 gene in both shoots and roots and to a lesser extent of RAV2/TEM2 and AXR5 only in roots or shoots, respectively (Supplementary Fig. S2A). Our previous transcriptome analysis of the non-acclimated nia1nia2noa1-2 vs Col-0 plants (GEO database identification number GSE41958; Gibbs et al., 2014) showed that neither RAV2/TEM2 nor AXR5 was differentially expressed in nia1nia2noa1-2 plants but, in turn, the ZF/OZF2 transcript levels were significantly down-regulated (−2.86-fold and P-value corrected for FDR of 0.00007) in nia1nia2noa1-2 plants. This is consistent with the reduced ZF/OZF2 transcript levels we detected in nia1nia2noa1-2 plants both at 21 °C and at 4 °C (Fig. 7).
It has been reported that OZF2 is involved in ABA-triggered responses through ABI2-mediated signaling (Huang et al., 2012), thus suggesting the potential involvement of OZF2 connecting the deficient cold acclimation phenotype of nia1nia2noa1-2 plants with ABA signaling. By using AtCAST (Arabidopsis thaliana: DNA Microarray Correlation Analysis Tool) 3.0 (Kakei and Shimada, 2015), we found that the ZF/OZF2 gene was also up-regulated by hypoxia, different ABA-related abiotic stresses, and darkness. Both ABA- and hypoxia-related processes are targets of the regulation exerted by changes in the endogenous NO content (Gibbs et al., 2014). Regarding this, the expression of the OZF2 gene was strongly up-regulated by ABA treatment (Supplementary Fig. S2B; Huang et al., 2012). Thus our findings suggest ZF/OZF2 might be an important node in the NO- and ABA-related regulation of cold acclimation responses through a complex network of CBF-dependent and -independent processes. It is noteworthy that our transcriptome analysis pointed to a significant over-representation of functional categories related to ABA-modulated processes and flavonoid/anthocyanin biosynthesis and metabolism in both wild-type and NO-deficient plants (Fig. 4). However, a number of genes in those subsets were specifically regulated by cold only in wild-type plants, thus suggesting that somehow NO-deficient plants were partially impaired in proper signaling of those regulators. We previously reported that nia1nia2noa1-2 plants display an ABA hypersensitive phenotype in seed germination, root elongation, and stomatal closure (Lozano-Juste and León, 2010). This work confirmed that ABA perception and signaling, but not homeostasis, were altered in nia1nia2noa1-2 plants under low temperature conditions (Fig. 5). We found a differential expression pattern of several genes encoding positive regulators of the ABA core signaling pathway, such as the PYL5 and 6 receptors and the SnRK2.9 kinase, in nia1nia2noa1-2 compared with wild-type plants, suggesting that the NO control on ABA perception and signaling is also a relevant factor for cold acclimation.
Anthocyanins/flavonoids are key antioxidant factors for NO-triggered cold acclimation
Besides the regulation of CBF gene expression and the modulation of ABA perception and signaling, NO also positively regulates the induction of genes involved in anthocyanin/flavonoid biosynthesis and, consequently, the biosynthesis of these pigments in a differential way in nia1nia2noa1-2 plants (Fig. 6). It is well documented that, in response to low temperature, anthocyanins accumulate to protect photosystems from photoinhibition, avoiding the concentration of high levels of reactive oxygen species (Krol et al., 1995; Harvaux and Kloppstech, 2001; Korn et al., 2008; Schulz et al., 2016). Consistent with these results, it has been shown that the expression of genes coding for critical enzymes in the anthocyanin and flavonoid biosynthetic pathway are induced by low temperature, and that the accumulation of these pigments is required to ensure full development of cold acclimation in Arabidopsis (Catalá et al., 2011; Schulz et al., 2016; Perea-Resa et al., 2017). Under low temperature conditions, the reduced ability to cold acclimate of nia1nia2noa1-2 plants could be due to the reduced content of anthocyanins and flavonoids, which as efficient antioxidants, usually prevent the accumulation of reactive oxygen species.
It has been previously reported that anthocyanins accumulate under conditions of deficient nitrogen assimilation (Diaz et al., 2006) by up-regulating transcription factor-encoding genes for positive (MYBs) and negative (LBDs) regulators of the flavonoid pathway (Soubeyrand et al., 2014). A search for MYB- and LBD-encoding genes among cold-regulated plants, specifically in wild-type but not in NO-deficient plants, indicated MYB102, MYB75, MYB31, MYB4, MYB44, and MYB38 that were up-regulated and MYBC1 that was down-regulated by cold treatment only in Col-0 plants, and LBD41, LBD37, and LBD38 that were down-regulated by cold also only in wild-type plants (Supplementary Table S5). Impaired cold acclimation of NO-deficient plants correlated with the lack of up-regulation of genes encoding MYB75/PAP1 (Supplementary Table S5), which has been characterized as a major determinant of flavonoid production and freezing tolerance (Borevitz et al., 2000; Schulz et al., 2016). Besides the major role of MYB75/PAP1, it has been reported that MYB96 integrates cold and abscisic acid signaling to activate the CBF–COR pathway in Arabidopsis (Lee and Seo, 2015). However, it is not likely to be a target of NO in regulating cold acclimation as we found up-regulation by cold-treatment in both wild-type and NO-deficient plants (Supplementary Table S5). MYBC1 negatively regulates freezing tolerance in Arabidopsis (Zhai et al., 2010), and we found that only wild-type plants down-regulated the MYBC1 gene upon cold treatment (Supplementary Table S5), thus suggesting this transcription factor may be relevant for NO-triggered cold acclimation. MYB44 has been reported to interact with the PYL9 ABA receptor modulating the interaction of the receptor with the phosphatase ABI1 and thus modulating ABA signaling (Li et al., 2014), which represents an interesting functional link between NO, cold acclimation responses and ABA signaling. Moreover, MYB44 has been characterized as an enhancer of tolerance to abiotic stresses presumably through an improvement in the antioxidative capacity of the stressed plants (Persak and Pitzschke, 2014), thus representing also a potential link of NO-induced cold acclimation with the oxidative status of the plant. Regarding LBDs, LBD37 and LBD38 have been already characterized as negative regulators of anthocyanin biosynthesis in Arabidopsis (Rubin et al., 2009). All these data suggest that the full disability of NO-deficient plants in up-regulating anthocyanin biosynthetic genes and, consequently, in producing anthocyanins in response to low temperatures (Fig. 6) support the essential requirement of NO for the cold-induced anthocyanin accumulation and further enhanced plant freezing tolerance.
Deficient nitrate assimilation may account also for impaired cold acclimation of NO-deficient plants
Although our findings suggest a decisive role for NO in regulating different pathways involved in promoting cold-induced freezing tolerance, we cannot rule out the possibility that some of the effects we observed in nia1nia2noa1-2 plants exposed to 4 °C are actually due to defective nitrogen assimilation and/or increased C:N ratio. Under low temperature conditions, protein turnover is potentiated (Wang et al., 2012, Guiboileau et al., 2013). The degradation of key positive regulators of cold acclimation in NO-deficient mutant plants may be a determinant of the impaired capacity to cold acclimate of nia1nia2noa1-2 plants. We have observed that wild-type shoots produced more NO when growing in nitrate-containing media than when growing on nitrite or ammonium as unique N sources, or subjected to N starvation (Supplementary Fig. S3). This enhanced NO production in nitrate-grown plants was dependent on nitrate reductase activity as it was attenuated in nia1,2 mutant plants (Supplementary Fig. S3). Since nitrite also induces NO accumulation, although to a lesser extent than nitrate (Supplementary Fig. S3), an alternative nitrate-independent biosynthesis of NO cannot be ruled out. Mitochondrial electron transport chain-derived NO production from nitrite has been reported under low oxygen conditions (Igamberdiev et al., 2014). Therefore, the N status of the plants is likely relevant for the NO-modulated cold acclimation process.
Regulation of cold acclimation through NO-triggered post-translational modifications
Data presented above suggest that NO seems to regulate cold acclimation through different signaling pathways. However, a question remains unanswered: how is NO exerting those regulatory roles? NO regulates plant physiology and metabolism mainly through post-translational modification of proteins such as S-nitrosylation of cysteines and nitration of tyrosines (Astier and Lindermayr, 2012). We have previously reported the identification of more than 120 in vivo nitrated proteins in Arabidopsis (Lozano-Juste et al., 2011). Among them, more than 60% of the identified nitrated proteins were involved in primary and secondary metabolism. Nitration of those enzymes potentially alters their activities, suggesting that NO-triggered nitration of enzymes represent a new level of metabolic regulation. Interestingly, in our proteomic analysis (Lozano-Juste et al., 2011) we found nitrated the anthocyanin/flavonoid-related enzyme quercetin-3-O-methyltransferase 1, supporting the regulatory action of NO on altered anthocyanin/flavonoid levels reported in this work. Moreover, we have previously reported that NO performs a negative regulation on ABA signaling through the post-translational nitration of key Y residues of the PYR/PYL/RCAR ABA receptors, which makes them inactive in the inhibition of type 2C phosphatases upon ABA binding (Castillo et al., 2015). ABA receptors such as PYR1 and PYL4 were degraded by the proteasome after being nitrated through a NO-dependent mechanism (Castillo et al., 2015), which may be relevant for ABA-regulated processes such as cold responses. Therefore, our data suggest that NO would regulate metabolism and hormone signaling and homeostasis, presumably through post-translational modifications of key regulatory proteins in the different signaling pathways. However, more work will be necessary to document whether the NO-dependent nitration of CBFs or other transcription factors, such as ZF/OZF2, occurs also during cold acclimation, and if these modifications are relevant to activate cold acclimation through the different proposed pathways.
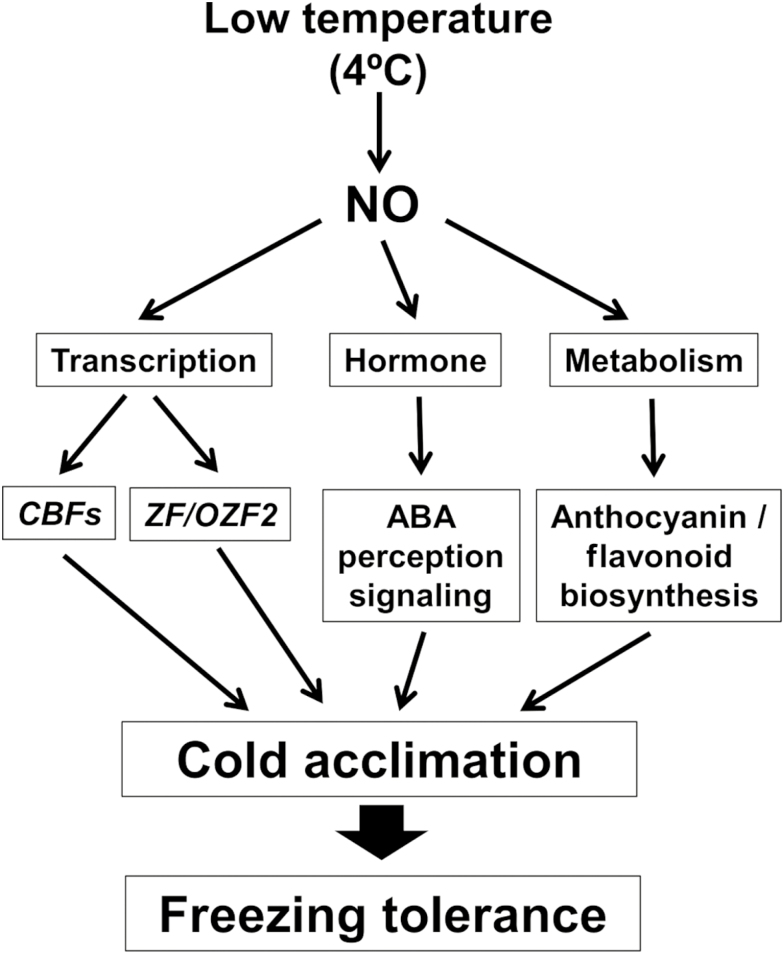
Together, our findings suggest that NO regulates cold acclimation at the transcriptional level, through CBF-dependent and CBF-independent likely ZF/OZF2-mediated pathways, at the metabolic level, by regulating the production of anthocyanins and flavonoids, and at the hormonal level, by modulating the sensitivity to hormones such as ABA (Fig. 8). The functional interactions between NO and ABA do not apply only to cold-triggered responses but are relevant for many other stress-triggered responses and developmental processes in plants (León et al., 2014; Prakash et al., 2019). We have previously reported that NO-deficient nia1nia2noa1-2 plants are constitutively more freezing tolerant than wild-type plants (Costa-Broseta et al., 2018). The opposite roles of NO as a negative and positive regulator of constitutive and cold acclimation-induced freezing tolerance allow the suggestion that the endogenous NO levels might function as a sensor determining the levels of freezing tolerance in plants.
Fig. 8.
Proposed model for the function of NO in cold acclimation.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Effect of cold treatment on the transcript levels of non-CBF cold-related transcription factor encoding genes in Col-0 and nia1nia2noa1-2 plants.
Fig. S2. Normalized transcript levels of RAV2/TEM2, AXR5, and ZF/OZF2 genes in response to cold, ABA, and JA.
Fig. S3. Effect of different nitrogen sources on NO production.
Table S1. Oligonucleotides used in this work.
Table S2. Differential up- or down-regulated transcriptomes in the comparisons between cold acclimated versus non-acclimated plants for both Col-0 and nia1nia2noa1-2 genotypes.
Table S3. Ratios of gene transcript levels comparing cold acclimated (Acc) versus non-acclimated (NoAcc) in wild-type Col-0 and NO-deficient nia1nia2noa1-2 plants based on qRT-PCR and microarray analysis.
Table S4. Gene Ontology analyses of the functional categories overrepresented among up- and down-regulated genes in cold acclimated versus non-acclimated plants of wild-type Col-0 and NO-deficient nia1nia2noa1-2 genotypes.
Table S5. MYB and LBD encoding gene transcript levels in cold-treated versus control untreated Col-0 and nia1nia2noa1-2 NO-deficient plants.
Author contributions
JL, AC-B, CP-R and JS conceived and designed the experiments; AC-B, CP-R, MCC, MFR and JL performed the experiments; JL analysed the transcriptome data; JL wrote the article with the contribution of JS.
Acknowledgements
We thank Isabel Lopez-Diaz and Esther Carrera for the hormone quantification carried out at the Plant Hormone Quantification Service, IBMCP, Valencia, Spain. This work was supported by grants from MINECO of Spain Government and FEDER EU funds [BIO2014-56067-P, BIO2017-82945-P to JL and BIO2016-79187-R to JS].
References
- Adams S, Carré IA. 2011. Downstream of the plant circadian clock: output pathways for the control of physiology and development. Essays in Biochemistry 49, 53–69. [DOI] [PubMed] [Google Scholar]
- Arakawa T, Timasheff SN. 1982. Stabilization of protein structure by sugars. Biochemistry 21, 6536–6544. [DOI] [PubMed] [Google Scholar]
- Astier J, Lindermayr C. 2012. Nitric oxide-dependent posttranslational modification in plants: an update. International Journal of Molecular Sciences 13, 15193–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian HS, Harmer SL. 2016. Circadian regulation of hormone signaling and plant physiology. Plant Molecular Biology 91, 691–702. [DOI] [PubMed] [Google Scholar]
- Barrero-Gil J, Salinas J. 2013. Post-translational regulation of cold acclimation response. Plant Science 205–206, 48–54. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell 12, 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrel C, Vazquez T, Puyaubert J, Rezé N, Lesch M, Kaiser WM, Dutilleul C, Guillas I, Zachowski A, Baudouin E. 2011. Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytologist 189, 415–427. [DOI] [PubMed] [Google Scholar]
- Castillo MC, León J. 2008. Expression of the β-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. Journal of Experimental Botany 59, 2171–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo MC, Lozano-Juste J, González-Guzmán M, Rodriguez L, Rodriguez PL, León J. 2015. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Science Signaling 8, ra89. [DOI] [PubMed] [Google Scholar]
- Catalá R, Medina J, Salinas J. 2011. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 16475–16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Thelen JJ. 2016. Acyl-lipid desaturase 1 primes cold acclimation response in Arabidopsis. Physiologia Plantarum 158, 11–22. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. 2003. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes & Development 17, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. 2007. Cold stress regulation of gene expression in plants. Trends in Plant Science 12, 444–451. [DOI] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. 2004. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 15243–15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Broseta Á, Perea-Resa C, Castillo MC, Ruíz MF, Salinas J, León J. 2018. Nitric oxide controls constitutive freezing tolerance in Arabidopsis by attenuating the levels of osmoprotectants, stress-related hormones and anthocyanins. Scientific Reports 8, 9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas JC, López-Cobollo R, Alcázar R, Zarza X, Koncz C, Altabella T, Salinas J, Tiburcio AF, Ferrando A. 2008. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiology 148, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C. 2006. Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant & Cell Physiology 47, 74–83. [DOI] [PubMed] [Google Scholar]
- Eremina M, Unterholzner SJ, Rathnayake AI, et al. 2016. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proceedings of the National Academy of Sciences, USA 113, E5982–E5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Chen K, Amombo E, Hu Z, Chen L, Fu J. 2015. Physiological and molecular mechanism of nitric oxide (NO) involved in bermudagrass response to cold stress. PLoS One 10, e0132991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LA, Walczyk Mooradally A, Zaslavsky B, Uversky VN, Graether SP. 2018. Effect of an intrinsically disordered plant stress protein on the properties of water. Biophysical Journal 115, 1696–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. 2002. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, et al. 2014. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Molecular Cell 53, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. 2004. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Molecular Biology 54, 767–781. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. 1998. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. The Plant Journal 16, 433–442. [DOI] [PubMed] [Google Scholar]
- Griffith M, Lumb C, Wiseman SB, Wisniewski M, Johnson RW, Marangoni AG. 2005. Antifreeze proteins modify the freezing process in planta. Plant Physiology 138, 330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiboileau A, Avila-Ospina L, Yoshimoto K, Soulay F, Azzopardi M, Marmagne A, Lothier J, Masclaux-Daubresse C. 2013. Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytologist 199, 683–694. [DOI] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM. 2003. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103. [DOI] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. 2008. Metabolomics of temperature stress. Physiologia Plantarum 132, 220–235. [DOI] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK. 2005. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genetics 1, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvaux M, Kloppstech K. 2001. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 213, 953–966. [DOI] [PubMed] [Google Scholar]
- Huang P, Ju HW, Min JH, Zhang X, Chung JS, Cheong HS, Kim CS. 2012. Molecular and physiological characterization of the Arabidopsis thaliana Oxidation-related Zinc Finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2-mediated signaling pathway. Plant and Cell Physiology 53, 193–203. [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Ratcliffe RG, Gupta KJ. 2014. Plant mitochondria: source and target for nitric oxide. Mitochondrion 19 Pt B, 329–333. [DOI] [PubMed] [Google Scholar]
- Jensen MK, Lindemose S, de Masi F, et al. 2013. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Biology 3, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Kim NY, Kim S, et al. 2010. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. The Journal of Biological Chemistry 285, 23371–23386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakei Y, Shimada Y. 2015. AtCAST3.0 update: a web-based tool for analysis of transcriptome data by searching similarities in gene expression profiles. Plant and Cell Physiology 56, e7. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL. 2007. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. The Plant Journal 50, 967–981. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Knight MR, Knight H. 2012. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytologist 195, 737–751. [DOI] [PubMed] [Google Scholar]
- Kohn H, Levitt J. 1966. Interrelations between photoperiod, frost hardiness and sulfhydryl groups in cabbage. Plant Physiology 41, 792–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn M, Peterek S, Mock HP, Heyer AG, Hincha DK. 2008. Heterosis in the freezing tolerance, and sugar and flavonoid contents of crosses between Arabidopsis thaliana accessions of widely varying freezing tolerance. Plant, Cell & Environment 31, 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol M, Gray GR, Huner NPA, Hurry VM, Öquist G, Malek L. 1995. Low-temperature stress and photoperiod affect an increased tolerance to photoinhibition in Pinus banksiana seedlings. Canadian Journal of Botany 73, 1119–1127. [Google Scholar]
- Lee BH, Henderson DA, Zhu JK. 2005. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. The Plant Cell 17, 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Seo PJ. 2015. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. The Plant Journal 82, 962–977. [DOI] [PubMed] [Google Scholar]
- León J, Castillo MC, Coego A, Lozano-Juste J, Mir R. 2014. Diverse functional interactions between nitric oxide and abscisic acid in plant development and responses to stress. Journal of Experimental Botany 65, 907–921. [DOI] [PubMed] [Google Scholar]
- Li D, Li Y, Zhang L, et al. 2014. Arabidopsis ABA receptor RCAR1/PYL9 interacts with an R2R3-type MYB transcription factor, AtMYB44. International Journal of Molecular Sciences 15, 8473–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, Colom-Moreno R, León J. 2011. In vivo protein tyrosine nitration in Arabidopsis thaliana. Journal of Experimental Botany 62, 3501–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, León J. 2010. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiology 152, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC. 2007. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Molecular Biology 63, 289–305. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Erickson BJ, Miller DR, Finkelstein RR. 2017. ABI5-binding proteins (AFPs) alter transcription of ABA-induced genes via a variety of interactions with chromatin modifiers. Plant Molecular Biology 93, 403–418. [DOI] [PubMed] [Google Scholar]
- Medina J, Catalá R, Salinas J. 2011. The CBFs: three arabidopsis transcription factors to cold acclimate. Plant Science 180, 3–11. [DOI] [PubMed] [Google Scholar]
- Miura K, Furumoto T. 2013. Cold signaling and cold response in plants. International Journal of Molecular Sciences 14, 5312–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ohta M. 2010. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. Journal of Plant Physiology 167, 555–560. [DOI] [PubMed] [Google Scholar]
- Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S. 2009. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant and Cell Physiology 50, 2210–2222. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. 2014. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Frontiers in Plant Science 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF. 2015. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. The Plant Journal 82, 193–207. [DOI] [PubMed] [Google Scholar]
- Perea-Resa C, Rodríguez-Milla MA, Iniesto E, Rubio V, Salinas J. 2017. Prefoldins negatively regulate cold acclimation in Arabidopsis thaliana by promoting nuclear proteasome-mediated HY5 degradation. Molecular Plant 10, 791–804. [DOI] [PubMed] [Google Scholar]
- Persak H, Pitzschke A. 2014. Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and hypersensitivity to abiotic stress. International Journal of Molecular Sciences 15, 2517–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K, Tonelli C. 2011. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Science 181, 219–229. [DOI] [PubMed] [Google Scholar]
- Pollock CJ, Eagles CF. 1988. Low temperature and the growth of plants. Symposia of the Society for Experimental Biology 42, 157–180. [PubMed] [Google Scholar]
- Prakash V, Singh VP, Tripathi DK, Sharma S, Corpas FJ. 2019. Crosstalk between nitric oxide (NO) and abscisic acid (ABA) signaling molecules in higher plants. Environmental and Experimental Botany, doi: 10.106/j.envexpbot.2018.10.033. [DOI] [Google Scholar]
- Puyaubert J, Baudouin E. 2014. New clues for a cold case: nitric oxide response to low temperature. Plant, Cell & Environment 37, 2623–2630. [DOI] [PubMed] [Google Scholar]
- Rahman A. 2013. Auxin: a regulator of cold stress response. Physiologia Plantarum 147, 28–35. [DOI] [PubMed] [Google Scholar]
- Renaut J, Lutts S, Hoffmann L, Hausman JF. 2004. Responses of poplar to chilling temperatures: proteomic and physiological aspects. Plant Biology 6, 81–90. [DOI] [PubMed] [Google Scholar]
- Reyes-Díaz M, Ulloa N, Zúñiga-Feest A, Gutiérrez A, Gidekel M, Alberdi M, Corcuera LJ, Bravo LA. 2006. Arabidopsis thaliana avoids freezing by supercooling. Journal of Experimental Botany 57, 3687–3696. [DOI] [PubMed] [Google Scholar]
- Richter R, Bastakis E, Schwechheimer C. 2013. Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiology 162, 1992–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. 2009. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. The Plant Cell 21, 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E, Tohge T, Zuther E, Fernie AR, Hincha DK. 2016. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Scientific Reports 6, 34027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Jikumaru Y, Kamiya Y. 2011. Profiling of hormones and related metabolites in seed dormancy and germination studies. Methods in Molecular Biology 773, 99–111. [DOI] [PubMed] [Google Scholar]
- Sharma M, Laxmi A. 2015. Jasmonates: emerging players in controlling temperature stress tolerance. Frontiers in Plant Science 6, 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S. 2018. Molecular regulation of CBF signaling in cold acclimation. Trends in Plant Science 23, 623–637. [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. 2006. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiology 140, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyrand E, Basteau C, Hilbert G, van Leeuwen C, Delrot S, Gomès E. 2014. Nitrogen supply affects anthocyanin biosynthetic and regulatory genes in grapevine cv. Cabernet-Sauvignon berries. Phytochemistry 103, 38–49. [DOI] [PubMed] [Google Scholar]
- Takahashi D, Kawamura Y, Uemura M. 2016. Cold acclimation is accompanied by complex responses of glycosylphosphatidylinositol (GPI)-anchored proteins in Arabidopsis. Journal of Experimental Botany 67, 5203–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50, 571–599. [DOI] [PubMed] [Google Scholar]
- van Buer J, Cvetkovic J, Baier M. 2016. Cold regulation of plastid ascorbate peroxidases serves as a priming hub controlling ROS signaling in Arabidopsis thaliana. BMC Plant Biology 16, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Zou H, Sun SS, He JX. 2012. A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. Journal of Proteome Research 11, 2301–2315. [DOI] [PubMed] [Google Scholar]
- Weiser CJ. 1970. Cold resistance and injury in woody plants: knowledge of hardy plant adaptations to freezing stress may help us to reduce winter damage. Science 169, 1269–1278. [DOI] [PubMed] [Google Scholar]
- Zhai H, Bai X, Zhu Y, Li Y, Cai H, Ji W, Ji Z, Liu X, Liu X, Li J. 2010. A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochemical and Biophysical Research Communications 394, 1018–1023. [DOI] [PubMed] [Google Scholar]
- Zhao MG, Chen L, Zhang LL, Zhang WH. 2009. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiology 151, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Sheng J, Lv S, et al. 2011. Nitric oxide participates in the regulation of LeCBF1 gene expression and improves cold tolerance in harvested tomato fruits. Postharvest Biology and Technology 62, 121–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.