Arabidopsis var2 mutants with defective PSII repair trigger chloroplast-to-nucleus retrograde signaling that results in a response resembling the unfolded protein response, leading to accumulation of proteins involved in proteostasis and detoxification.
Keywords: Photosystem II, PSII repair, proteostasis, retrograde signaling, singlet oxygen, unfolded protein response, damaged protein response
Abstract
Cellular protein homeostasis (proteostasis) is maintained through the balance between de novo synthesis and proteolysis. The unfolded/misfolded protein response (UPR) that is triggered by stressed endoplasmic reticulum (ER) also plays an important role in proteostasis in both plants and animals. Although ER-triggered UPR has been extensively studied in plants, the molecular mechanisms underlying mitochondrial and chloroplastic UPRs are largely uncharacterized despite the fact that these organelles are sites of production of harmful reactive oxygen species (ROS), which damage proteins. In this study, we demonstrate that chloroplasts of the Arabidopsis yellow leaf variegation 2 (var2) mutant, which lacks the metalloprotease FtsH2, accumulate damaged chloroplast proteins and trigger a UPR-like response, namely the accumulation of a suite of chloroplast proteins involved in protein quality control (PQC). These PQC proteins include heat-shock proteins, chaperones, proteases, and ROS detoxifiers. Given that FtsH2 functions primarily in photosystem II proteostasis, the accumulation of PQC-related proteins may balance the FtsH2 deficiency. Moreover, the apparent up-regulation of the cognate transcripts indicates that the accumulation of PQC-related proteins in var2 is probably mediated by retrograde signaling, indicating the occurrence of a UPR-like response in var2.
Introduction
The unfolded/misfolded protein response (UPR) was first characterized in the endoplasmic reticulum (ER) (Kozutsumi et al., 1988). This ER-mediated UPR (erUPR) is activated when protein folding is impaired at the lumenal side under ER stress conditions. This response is ubiquitously conserved in eukaryotic cells and is crucial to eliminate misfolded/unfolded proteins, thereby maintaining protein homeostasis (proteostasis) (Walter and Ron, 2011). Similarly, mitochondria also activate a mitochondrial UPR (mtUPR) under oxidative stress conditions (Aldridge et al., 2007; Pellegrino et al., 2013). Both erUPR and mtUPR lead to the accumulation of proteins involved in proteostasis (Aldridge et al., 2007; Iwata et al., 2008; Pellegrino et al., 2013). These proteins include various chaperones and proteases, which are induced via a process called organelle-to-nucleus retrograde signaling (RS). A chloroplast-mediated UPR (cpUPR) has been found in the green unicellular alga Chlamydomonas reinhardtii through use of a repressible chloroplast gene expression system (Pérez-Martin et al., 2014; Ramundo et al., 2014; Ramundo and Rochaix, 2014). The repression of ClpP, a plastid-encoded catalytic subunit of the ATP-dependent caseinolytic protease (Clp), results in the accumulation of proteins involved in proteostasis in chloroplasts, which resembles the typical signature of UPR. A similar response was recently identified in plants treated with a pharmacological inhibitor of plastid gene expression (PGE) (Llamas et al., 2017). Treatment of Arabidopsis wild-type (WT) plants with the chloroplast translation inhibitor lincomycin (LIN) results in the up-regulation of a subset of nuclear-encoded genes that encode proteins involved in chloroplast proteostasis (Llamas et al., 2017). It has been shown that the heat-shock transcription factor HSFA2, which specifically binds to heat-shock promoter elements (Nishizawa et al., 2006; Schramm et al., 2006), mediates the cpUPR in LIN-treated plants (Llamas et al., 2017). Given that the LIN treatment leads to protein aggregation in the chloroplasts and increases the levels of proteins involved in protein quality control (PQC) via transcriptional regulation (Llamas et al., 2017), it is likely that chloroplasts in higher plants are able to trigger cpUPR.
In addition to Clp, the processive protease FtsH, an AAA-type ATP-dependent metalloprotease localized in the thylakoid membranes, plays a pivotal role in chloroplast PQC (Patel and Latterich, 1998; Ogura and Wilkinson, 2001; Yu et al., 2004; Nishimura et al., 2016). In plants, this membrane-bound FtsH protease is present as a hexameric heterocomplex composed of four subunits of two major isoforms, namely Type A, which includes FtsH2 (also called VAR2) and FtsH8, and Type B, which includes FtsH1 and FtsH5 (also called VAR1) (Sakamoto et al., 2003; Zaltsman et al., 2005). FtsH2 and FtsH5 are the major subunits, and functional loss of either of them results in impaired acclimation to light stress (Sakamoto et al., 2003; Zaltsman et al., 2005). Indeed, var1 and var2 mutant plants exhibit a higher susceptibility to mild photooxidative stress, whereas ftsh1 and ftsh8 mutant plants acclimate like the WT. The FtsH protease functions primarily in the degradation of photodamaged photosystem II (PSII) reaction center (RC) proteins such as D1 and D2, followed by their de novo synthesis and subsequent PSII reassembly (Zaltsman et al., 2005; Kato et al., 2009, 2015; Malnoë et al., 2014). Interestingly, despite the disruption in PSII repair, which is a default process regardless of light intensity, var2 mutant plants are sustainable under moderate light conditions. This suggests the existence of some adaptive system that compensates for chloroplast dysfunction in var2.
In the present study, we investigated the molecular basis of this putative adaptive mechanism in the var2 mutant. We found that the impaired proteostasis in the chloroplasts of var2 mutant plants induces a UPR-like response conceptually similar to the erUPR, which leads to the accumulation of chaperones, proteases, and proteins associated with detoxification.
Materials and methods
Plant material and growth conditions
All the Arabidopsis thaliana seeds were derived from the Columbia (Col-0) ecotype and were harvested on the same day from plants grown together under of conditions continuous light (CL; 80 µmol m–2 s–1 at 20±2 °C). Seeds for the var2 knock-out allele (SAIL_253_A03) were obtained from the Nottingham Arabidopsis Stock Centre (NASC). The WT and var2 seeds were surface-sterilized and plated on Murashige and Skoog medium (Duchefa Biochemie) with 0.8% (w/v) agar,
supplemented with 0.5% (w/v) sucrose. Seeds were stratified for 3 d at 4 °C in darkness and then placed under CL. At 5 d old, seedlings were transferred to soil and grown under CL until sampling.
Chloroplast isolation and tandem mass spectrometry
Chloroplasts were isolated from 3-week-old plants of the WT and var2 grown under CL as described previously (Kauss et al., 2012). Briefly, rosette leaves of mature plants (90 plants for WT and 180 plants for var2) were homogenized in a Waring blender in chloroplast isolation buffer [50 mM Hepes-KOH, pH 8, 5 mM MgCl2, 5 mM EDTA pH8, 5 mM EGTA pH 8, 10 mM NaHCO3, and 0.33 M D-sorbitol, supplemented with SIGMAFASTTM Protease Inhibitor (1 tablet per 100 ml)]. The homogenate was filtered through four layers of Miracloth and centrifuged at 400 g for 8 min at 4 °C. The pellets were suspended in isolation buffer and loaded onto a two-step Percoll gradient (40:80%) to separate intact and broken chloroplasts. Intact chloroplasts enriched between the two Percoll steps were carefully collected and washed twice with HS buffer (50 mM Hepes-KOH, pH 8, and 0.33 M D-sorbitol). The integrity of the chloroplasts was checked under a microscope (Supplementary Fig. S1 at JXB online). Intact chloroplasts corresponding to equal amounts of chlorophyll were lysed, and the proteins extracted using 6 M guanidine hydrochloride buffer (guanidine hydrochloride dissolved in 100 mM Tris, pH 8.5). The lysed samples were sonicated in an ice bath for 1 min with a pulse of 3 s on and 5 s off, heated at 95 °C for 5 min, and then centrifuged at 21 000 g for 30 min at 4 °C. Total protein content was estimated using a PierceTM BCA protein assay kit (ThermoFisher Scientific). For MS analysis, equal amounts of total protein (2 µg µl–1) from three independent biological samples were denatured using 10 mM DTT at 56 °C for 30 min followed by alkylation in 50 mM iodoacetamide at room temperature for 40 min in the dark. The proteins were then desalted using a Nanosep membrane (Pall Corporation, MWCO 10K) in 200 µl of 100 mM NH4HCO3 buffer. Desalted proteins were incubated in digestion buffer (40 ng µl–1 trypsin in 100 mM NH4HCO3, corresponding to an enzyme-to-protein ratio of 1:50) for 20 h at 37 °C. Finally, the digested peptides were dried in a refrigerated CentriVap concentrator (Labconco, Kansas, MO). The dried peptides were resuspended in 0.1% (v/v) formic acid (FA) solution, and separated using a nanoAcquity Ultra Performance LC (Waters, Milford, MA) equipped with a 20-mm trap column (C18 5 μm resin, 180 μm I.D., Waters) and a 250-mm analytical column (C18 1.7 μm resin,75 μm I.D., Waters). The peptide mixture reconstituted in 0.1% FA was loaded onto the trap column with a flow rate of 3 μl min–1 for 10 min, followed by elution to the analytical column for further separation under the following conditions with a flow rate of 250 nl min–1: (i) 140 min gradient from 8–25% of solvent B (Acetonitrile, ACN), (ii) 15 min gradient from 25–40% of solvent B, (iii) 5 min gradient from 40–90% of solvent B, (iv) 5 min washing at 90% of solvent B, and finally (v) equilibrating with 97% of solvent A for 15 min (solvent A: 0.1% FA; solvent B: 99.9% ACN/0.1% FA). The separated peptides were analysed using a Q Exactive Mass Spectrometer (ThermoFisher Scientific). A full MS survey scan was carried out at a resolution of 70 000 at 400 m/z over an m/z range of 300–1800, with an automatic gain controls (AGC) target of 3×106 and a maximum ion injection time (IT) of 30 ms. The top 20 multiply charged parent ions were selected using data-dependent MS/MS mode, and fragmented by higher-energy collision dissociation (HCD) with a normalized collision energy of 27% in the m/z scan range of 200–2000. MS/MS detection was carried out at a resolution of 17 500 with an AGC target value of 5×106 and a maximum IT of 120 ms. Dynamic exclusion was enabled for 30 s.
Label-free quantitation
Raw MS data files were processed and analysed using the MaxQuant software (v. 1.5.8.3) with label-free quantitation (LFQ) and the intensity-based absolute quantification (iBAQ) algorithm enabled as described previously (Luber et al., 2010; Schwanhäusser et al., 2011). Parent ion and MS/MS spectra were searched against the FASTA format database at TAIR (http://www.arabidopsis.org/). The precursor ion tolerance was set at 7 ppm with an allowed fragment mass deviation of 20 ppm. Carbamidomethylation of cysteine was set as a fixed modification while N-terminal acetylation and oxidation of methionine and tryptophan were defined as variable modifications. Peptides of a minimum of six amino acids and a maximum of two missed cleavages were allowed. False discovery rate (FDR) was set to 0.01 for both peptide and protein identification. The LFQ and iBAQ intensity values were used to calculate the protein expression and abundance. Proteins were considered as being expressed if the intensity values were detected in at least two of the three replicates in at least one of the independent biological samples. Expression matrices of the proteins were represented as heat maps prepared using Multi-Experiment viewer (MeV4.9.0). After log2-transformation of the intensity values and data imputation (replacing missing values by normal distribution), proteins exhibiting at least a 2-fold accumulation with P<0.05 (Student’s t-test) were considered as differentially accumulated in var2 in comparison with the WT. The oxidation (Oxi-PTM) in the proteins was calculated using the intensities of the individual oxidized peptides of the respective proteins. Gene Ontology (GO) enrichment analysis of differentially expressed proteins was carried out using the Generic GO Term Finder tool (http://go.princeton.edu/cgi-bin/GOTermFinder) to determine the significantly enriched GO terms in the category of biological processes (Katari et al., 2010) with a significance of P<0.05.
Western blot analyses
The total chloroplast proteins were separated by 10% SDS-PAGE gels and blotted onto Immun-Blot PVDF membrane (Bio-Rad). HSP70, CPN60A, CPN60B, and RbcL proteins were immunochemically detected using rabbit anti-HSP70 (1:10 000 dilution), rabbit anti-CPN60A (1:10 000), rabbit anti-CPN60B (1:10 000), and rabbit anti-RbcL (1:10 000) antibodies, respectively (all obtained from Agrisera).
RNA extraction and quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from three independent biological replicates of 3-week-old plants of the WT and var2 grown under CL using a Spectrum Plant Total RNA Kit (Sigma-Aldrich). Samples of 1 µg RNA were treated with RQ1 RNase-free DNase I (Promega). First-strand cDNA was synthesized with oligo(dT)15 primers (Promega) and Improm II reverse transcriptase (Promega) according to the manufacturer’s protocol. qRT-PCR was carried out using a QuantStudioTM 6 Flex Real-Time PCR System (Applied Biosystems) and iTaq Universal SYBR Green PCR master mix (Bio-Rad). The relative transcript level of each gene was determined with the comparative delta-CT method and normalized to the transcript level of PP2A (At1g13320). The primer sequences used in this study are listed in Supplementary Table S1.
Results
Changes in the chloroplast proteome in var2
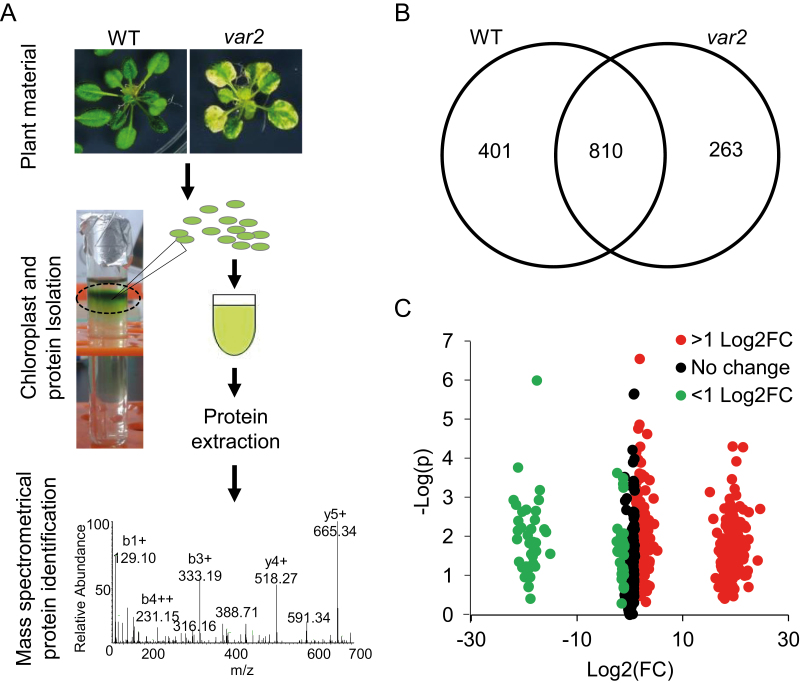
Inactivation of the FtsH2 protease disturbs PSII proteostasis (Patel and Latterich, 1998; Ogura and Wilkinson, 2001; Yu et al., 2004; Nishimura et al., 2016). In spite of this, var2 mutant plants lacking FtsH2 are viable, indicating that chloroplasts may activate certain signaling pathway(s) to compensate for the defective proteostasis. We were therefore interested in learning more about this adaptive mechanism. To this end, we compared the total chloroplast proteome of var2 with that of the WT by using a label-free quantitation assay (Fig. 1A). We normalized each protein sample based on total protein content, since there was an apparent correlation between the WT and var2 samples with regards to chloroplast number (or chlorophyll content) and protein amount (Supplementary Fig. S1). Equal amounts of total protein extracted from intact chloroplasts were subjected to MS followed by protein identification using the MaxQuant quantitative proteomics software package (Fig. 1A) (Luber et al., 2010; Schwanhäusser et al., 2011). A total of 1474 protein were detected in at least two replicates in either var2 or the WT. Among these, 263 proteins were specifically detected in var2, 401 proteins were WT-specific, and 810 proteins were present in both the WT and var2 (Fig. 1B, Supplementary Table S2). This quantitative analysis identified 603 differentially accumulated proteins (Log2 FC>1, P<0.05), of which 317 proteins had higher and 286 proteins had lower accumulation in var2 as compared to WT (Fig. 1C; Supplementary Tables S3, S5). To gain insights into these differentially accumulated proteins in var2, the 603 proteins were subjected to Gene Ontology (GO) term enrichment analyses (P<0.05) for Biological Processes. The results obtained indicated that proteins involved in stress responses and chloroplast PQC were significantly overrepresented among the up-regulated proteins in var2 (Fig. 2A; Supplementary Fig. S2A). Conversely, the representation of proteins involved in photosynthesis, chlorophyll metabolism, and protein import was reduced (Supplementary Tables S4, S6). Many other biological processes, including carbohydrate and amino acid metabolism, ATP synthesis, and transcription/translation-related components, were also affected. Among the up-regulated proteins, increased accumulation of chloroplast transcription/translation-related proteins such as TypA translation elongation GTPase (SVR3), pentatricopeptide repeat-containing protein (SVR7), and elongation factor-G (EF-G) was consistent with previous reports where knockdown or complete loss of either of these proteins has been found to suppress the leaf variegation in var2 (Miura et al., 2007; Liu et al., 2010; Zoschke et al., 2013). Proteins involved in photosynthetic electron transport and in the degradation of PSII core proteins (such as FtsH proteases) were considerably down-regulated (Supplementary Table S5). The observation that all subunits of the FtsH protease were reduced in var2 confirms that FtsH2 is indispensable for the assembly of the FtsH protease. Strikingly, a suite of light-harvesting antenna proteins in PSII were markedly under-represented in var2, implying that proteins involved in genome-coupled expression may function in var2 to repress the expression of photosynthesis-associated nuclear genes (PhANGs). Although a substantial number of proteins involved in various biological processes were differentially accumulated, we focused our analyses on those that were directly involved in maintaining or reinstating proteostasis.
Fig. 1.
Impaired PSII proteostasis alters the chloroplast proteome in the Arabidopsis var2 mutant. (A) Schematic representation of MS-based analysis of the total chloroplast proteins isolated from 3-week-old plants of var2 and the WT grown under continuous light (80 µmol m–2 s–1) at 20±2 °C. (B) Distribution of 1474 proteins detected in either var2 or the wild-type (WT) (see Supplementary Table S2). (C) Label-free quantitation identified a total of 603 differentially accumulated proteins (Log2 FC ≥ 1; P<0.05, Student’s t-test), among which, 317 were increased and 286 were reduced in var2 as compared to the WT (Supplementary Tables S3, S5).
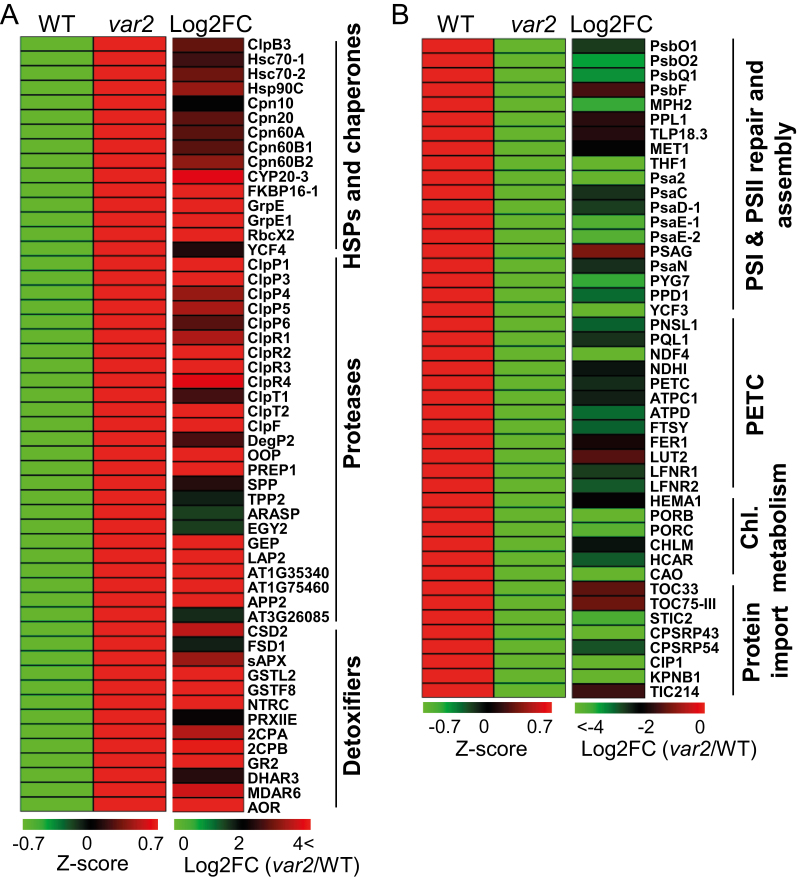
Fig. 2.
Proteins related to protein quality control (PQC) are highly accumulated whereas photosynthesis-related proteins are significantly reduced in the Arabidopsis var2 mutant. (A) Heat maps showing the expression of PQC-related proteins in var2 as compared with the wild-type (WT). GO analysis of the proteins highly accumulated in var2 compared with the WT revealed a significant enrichment in protein folding, proteolysis, detoxification, and chloroplast organization processes (Supplementary Fig. S2A). A complete GO analysis of proteins accumulated in var2 is shown in Supplementary Table S4. (B) Heat maps showing the expression of photosynthesis-related proteins in var2 as compared with the WT. GO analysis of the proteins reduced in var2 revealed a significant enrichment of proteins involved in photosynthetic protein import, PSI and PSII assembly, PSII repair, photosynthetic electron transport chain (PETC), and chlorophyll biosynthesis (Supplementary Fig. S2B). A complete GO analysis of down-regulated proteins in var2 is shown in Supplementary Table S6.
Proteins involved in chloroplast PQC accumulate in the chloroplasts of var2
The GO enrichment analysis of proteins highly accumulated in var2 revealed a suite of proteins associated with PQC (Fig. 2A, Supplementary Fig. S2A, Supplementary Tables S3, S4), the functions of which were similar to those of proteins implicated in the erUPR (Iwata et al., 2008; Walter and Ron, 2011; Duwi Fanata et al., 2013). For example, the heat-shock protein Hsp60 family is linked to folding and assembly of their target proteins (Koumoto et al., 2001; Bonshtien et al., 2007; Salvucci, 2008). Indeed, HSPs such as Cpn10, Cpn20, Cpn60A, Cpn60B1, Cpn60B2, and Cpn60B4 were highly accumulated in var2, and there was also notable accumulation of the heat-shock proteins ClpB3 (an Hsp100 family protein), Hsc70-1 (Hsp70-6), Hsc70-2 (Hsp70-7), and Hsp90C, which prevent the misfolding of functional proteins or refolding of damaged proteins (Pulido et al., 2016) (Fig. 2A; Supplementary Fig. S2A, B; Fig. 3; Supplementary Tables S3, S4). In addition, two peptidyl-prolyl cis-trans isomerases, cyclophilin 20–3 (CYP20-3) and FKBP16-1, which facilitate protein folding by catalysing the cis-trans isomerization of proline imidic peptide bonds in oligopeptides, were accumulated (Fig. 2A). CYP20-3 also regulates cellular redox homeostasis under certain stress conditions by activating sulfur assimilation, leading to increased cellular thiol content and reduction potential (Park et al., 2013).
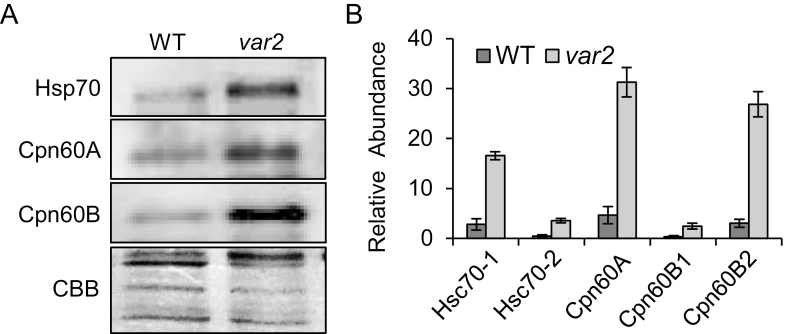
Fig. 3.
Validation of the label-free protein quantitation. Proteins including Hsp70, Cpn60A, and Cpn60B exhibited higher steady-state levels in the Arabidopsis var2 mutant compared with the wild-type (WT) in both western blots (A) and label-free quantitation data (B). (A) Total chloroplast proteins were extracted from intact chloroplasts isolated from 3-week-old plants grown under continuous light and were assessed by western blotting using protein-specific antibodies. (B) Label-free quantitation-based steady-state levels (expressed as relative abundance) of Hsp70 (Hsc70-1, Hsc70-2), Cpn60A, and Cpn60B (Cpn60B1 and Cpn60B2) in var2 as compared to the WT. Relative abundance was calculated using mean intensity values. Data shown are means (±SD) of n=3 replicates.
In addition to these chaperones and HSPs, several proteases involved in protein degradation and processing also exhibited higher levels in var2 (Fig. 2A, Supplementary Fig. S2A, Supplementary Tables S3, S4). These included Clp, Deg-protease, presequence protease 1 (PreP1), stromal processing peptidase (SPP), and organellar oligopeptidase (OOP). Interestingly, the protein levels of all subunits comprising the Clp protease were elevated (Fig. 2A, Supplementary Fig. S2A, Supplementary Tables S2–S4), including the five catalytic subunits (ClpP1, ClpP3, ClpP4, ClpP5, ClpP6), four non-catalytic subunits (ClpR1, ClpR2, ClpR3, ClpR4), two accessory subunits (ClpT1, ClpT2), and one adaptor subunit (ClpF) (Adam et al., 2006). By contrast, the auxiliary components ClpC1, ClpC2, and ClpD, which are involved in unfolding the substrates for subsequent degradation by the Clp protease (Rosano et al., 2011), remained unchanged in var2 compared to the WT (Supplementary Table S2). Since chloroplasts also require ClpC1/2 and ClpD for protein import (Rosano et al., 2011), unchanged levels of these chaperones probably ensures the regulation of protein import into the chloroplasts. The accumulated Clp protease may degrade the damaged/misfolded proteins in var2, counterbalancing the deficiency of FtsH2. The proteome analysis consistently revealed a reduced accumulation of some of the known targets of the Clp protease involved in chlorophyll biosynthesis, including chlorophyllide a oxygenase (CAO) (Sakuraba et al., 2009), HEMA1/GluTR (Apitz et al., 2016), and phytoene synthase (PSY) (Welsch et al., 2018) (Fig. 2B; Supplementary Table S5). This augmented Clp activity might produce a large quantity of short peptides, which then undergo further degradation in the var2 mutant as a result of the concomitant accumulation of OOP (Kmiec et al., 2013). The increased levels of SPP and PreP1, involved in the processing of signal peptides (Stahl et al., 2002; Zhong et al., 2003), may promote maturation of newly imported proteins involved in PQC in var2.
The Deg proteases, which are ATP-independent serine endopeptidases, function in the degradation of photodamaged PSII proteins under excess light conditions (Haussühl et al., 2001; Chi et al., 2012). Among the five chloroplast Deg proteases, DegP1, DegP5, and DegP8 are present at the luminal side, whereas DegP2 and DegP7 are located in the stroma (Chi et al., 2012). We anticipated an accumulation of DegP1, DegP2, DegP5, and DegP8 because of their involvement in D1 degradation (Haussühl et al., 2001), which would balance the FtsH2 deficiency in var2. However, we found that the luminal Deg proteases were either down-regulated (DegP1 and DegP5) or remained unchanged (DegP8). Only the stromal DegP2 showed a higher accumulation in var2 (Fig. 2A, Supplementary Tables S2, S3, S5). Despite controversary as to whether DegP2 cleaves damaged D1 or not (Haussühl et al., 2001), it might be interesting to examine the role of DegP2 in var2 in this regard in future studies. Collectively, our results indicated that the lack of FtsH2 lead to remarkable changes in the accumulation of proteins associated with chloroplast PQC.
Proteins related to detoxification and redox homeostasis are accumulated in var2
Impaired proteostasis results in the alteration of the redox state in chloroplasts, which may thus impair ROS homeostasis (Kato et al., 2009; Rochaix and Ramundo, 2018). Indeed, chloroplasts of var2 exhibit increased levels of both superoxide radicals (O2–) and hydrogen peroxide (H2O2) relative to the WT (Kato et al., 2009). These increased levels of ROS may cause the accumulation of damaged proteins in the chloroplasts. Thus, not surprisingly, a group of proteins primarily involved in ROS detoxification were accumulated in var2, including CuZn-superoxide dismutase 2 (CSD2), Fe-superoxide dismutase 1 (FSD1), stromal ascorbate peroxidase (sAPX), monodehydroascorbate reductase 6 (MDAR6), dehydroascorbate reductase 3 (DHAR3), and glutathione reductase 2 (GR2) (Fig. 2A, Supplementary Fig. S2A, Supplementary Tables S3, S4). Similarly, var2 exhibited an elevated level of the glutathione S-transferase F8 (GSTF8), which scavenges secondary and tertiary oxidized derivatives of lipids and flavonoids, thereby mitigating the deleterious effects of reactive carbonyl species derived from lipid peroxide (Dixon and Edwards, 2010; Yamauchi et al., 2011). Proteins acting in redox maintenance, such as 2-Cys peroxiredoxin A and B (2CPA and 2CPB), peroxiredoxin-II-E (PRXIIE), NADPH-dependent thioredoxin reductase C (NTRC), and thioredoxin-like protein CDSP32 (CDSP32), were also up-regulated in var2. Under oxidative stress conditions, peroxiredoxins containing a redox-active cysteine directly reduce H2O2 and organic hydroperoxides into water and alcohol. During this scavenging process, the cysteine is oxidized into sulfenic acid, which is then reduced back into its active form by thioredoxins, including CDSP32 (Broin et al., 2002) and NTRC (Moon et al., 2006). CDSP32 induced under oxidative stress conditions protects the photosynthetic apparatus against oxidative damage (Broin et al., 2002). NTRC is also involved in ROS-scavenging during chlorophyll biosynthesis in chloroplasts, thus possibly protecting the function of Mg-protoporphyrin monomethyl ester cyclase (CHL27) (Stenbaek et al., 2008). Increasing the amount of proteins involved in detoxification and redox maintenance might be crucial for mitigating photodamage in var2 chloroplasts.
Proteins associated with photosynthesis are reduced in var2
Whilst many proteins involved in protection, maintenance, and detoxification were up-regulated in var2, it was apparent that proteins involved in PSI and PSII assembly, the photosynthetic electron transport chain, chlorophyll synthesis, and chloroplast protein import were down-regulated in var2 compared to the WT (Fig. 2B, Supplementary Fig. S2B, Supplementary Tables S5, S6). Even though many of the proteins associated with photosynthesis were reduced in var2 compared to the WT, it was noteworthy that the PSII RC proteins PsbA/D1, PsbB/CP47, PsbC/CP43, and PsbD/D2, as well as some of the PSI RC proteins, including PsaA, PsaB, and PsaL, were accumulated in var2. Like PSII RC proteins, PSI RC proteins are prone to oxidative damage (Zivcak et al., 2015; Takagi et al., 2016). Therefore, the accumulation of PSI RC proteins together with PSII RC proteins in var2 suggests that the FtsH protease might also be involved in PSI repair or assembly. Interestingly, proteins involved in the dark reaction or the Calvin cycle, including Rubisco subunits, phopshoglycerate kinase 1 (PGK1), and glyceraldehyde 3-phosphate dehydrogenase subunits, also accumulated in var2 (Supplementary Fig. S2A, Supplementary Table S3, S5). This could be an adaptive strategy of energy-deprived var2 plants to improve carbon fixation, thus producing more sugars to fulfill the energy demand, at least to some extent.
The outer envelope translocon protein TOC33, which is involved in the import of photosynthesis-associated proteins (PhAPs) (Jarvis and López-Juez, 2013), was down-regulated in var2 (Fig. 2B, Supplementary Fig. S2B, Supplementary Table S5). TOC33, TOC34, and TOC159 act as receptors for preproteins, whereas TOC75-III forms a β-barrel cation-selective channel through which the preproteins cross the outer membrane (Jarvis and López-Juez, 2013). The TOC33/159/75 complex is responsible for the import of PhAPs, while the TOC34/120/75 complex imports housekeeping proteins (Jarvis and López-Juez, 2013). The down-regulation of the TOC complex, especially of those subunits involved in the import of photosynthetic preproteins, may be one of the photoprotection mechanisms in var2 in addition to the increased levels of proteins related to PQC. Indeed, it has previously been demonstrated that under oxidative stress conditions the chloroplast import machinery undergoes a rapid E3 ligase-dependent turnover, which reduces the supply of photosynthetic proteins and subsequently minimizes ROS production by the photosystems (Ling et al., 2012; Ling and Jarvis, 2015).
Accumulation of PQC-related proteins in var2 is transcriptionally regulated
Since the majority of chloroplast proteins are encoded in the nucleus, any perturbations in the chloroplast may activate the process of retrograde signaling (RS) to regulate gene expression and to sustain chloroplastic homeostasis (Apel and Hirt, 2004; Fischer et al., 2007; Tripathy and Oelmüller, 2012; Ashraf and Harris, 2013; Chan et al., 2016). It is possible that the up-regulation of nuclear-encoded plastid proteins involved in PQC and detoxification and the down-regulation of photosynthesis-related proteins in the dysfunctional chloroplasts of var2 results from RS. To test whether this was the case, the transcript levels of ClpR2, ClpR4, CPN10, CPN60A, FSD1, and CSD2 were assessed in the WT and var2 seedlings using qRT-PCR. All the examined transcripts were significantly up-regulated in var2 compared to the WT (Fig. 4A, B), indicating that the accumulation of these PQC- and detoxification-related proteins seemed to be transcriptionally controlled.
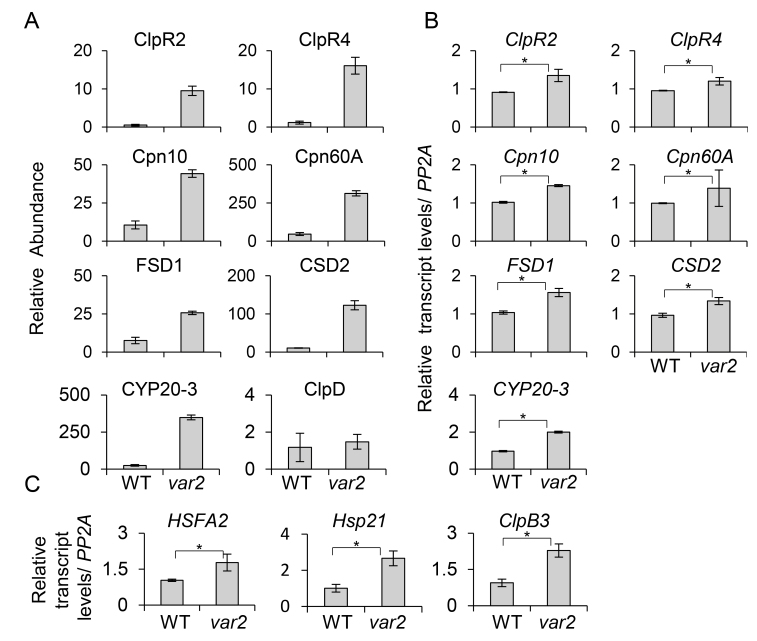
Fig. 4.
The accumulation of proteins related to proteostasis in the Arabidopsis var2 mutant is transcriptionally regulated. (A) Relative abundance of proteins exhibiting a higher accumulation in var2 compared with the wild-type (WT). The ClpD protein is shown as the control in which the steady-state levels remained unchanged. (B) Relative transcript levels of the proteins accumulated in var2 were examined using qRT-PCR. (C) The expression of -shock transcription factor A2 (HSFA2) and its downstream target genes Hsp21 and ClpB3 are up-regulated in var2. The relative abundance of the proteins was calculated using mean protein intensities. The relative transcript levels were calculated by qRT-PCR using PPA2 as a control. The data are means (±SD) of n=3 replicates. Significant differences between mean values were determined using Student’s t-test (*P<0.05).
The heat-stress transcription factor A-2 (HSFA2) drives the expression of HSPs by binding to the palindromic HSF-binding motif present in their promoter regions (Nishizawa et al., 2006; Schramm et al., 2006). Interestingly, HSFA2 was up-regulated in var2 (Fig. 4C), coinciding with a substantial accumulation of HSPs. Previous reports of the existence of a chloroplast UPR were based on the analysis of both transcript and protein abundance in cells in which protein-folding stress was elicited by knockdown of the stromal Clp protease and/or treatment with LIN (Ramundo et al. 2014; Llamas et al. 2017). In both studies, the genes encoding ClpB3 and Hsp21 (direct targets of HSFA2) were the two most highly up-regulated. While ClpB3 was up-regulated in var2 (Fig. 2A), the Hsp21 protein could not be detected in our chloroplast proteome data, probably due to the limitations of MS in detecting either small-sized or low-abundant (or rapidly turned-over) proteins. Nevertheless, the confirmation of higher transcript levels of both ClpB3 and Hsp21 in var2 compared to the WT was indicative of a possible function of RS in priming a cpUPR-like response.
Chloroplast proteome changes induced by clp and var2 are largely comparable
The accumulation Clp in the var2 mutant may compensate for the deficiency in var2, which includes impaired PSII repair. This would suggest that a deficiency of either of these proteases may induce a cpUPR-like response. To test this hypothesis, we compared the chloroplast proteome of var2 with the available chloroplast proteomes of different Arabidopsis clp mutants, such as clpr2, clpr4, and clpp3 (Kim et al., 2009, 2013a; Zybailov et al., 2009). Loss of any of these subunits results in the loss of Clp activity. A total of 82 proteins were significantly changed in these mutants, among which the abundances of 57 proteins were increased whilst those of 25 proteins were reduced by at least 2-fold (Supplementary Table S7). Among the 57 up-regulated proteins in the clp mutants, 54 were also up-regulated in var2 and of the 25 significantly down-regulated proteins, 16 were also down-regulated in var2. The majority of the 54 proteins up-regulated in both clp and var2 function in protein folding and ROS detoxification, whereas the down-regulated proteins are mostly implicated in photosynthesis.
Loss of FtsH2 also affected the abundance of plastid-encoded proteins. For instance, ClpP, and PSI and PSII core proteins were up-regulated, whereas a significant portion of plastid-encoded proteins involved in photosynthesis as well as in housekeeping were down-regulated (Supplementary Table S2). This down-regulation of plastid-encoded proteins could be due to the decreased accumulation of the plastid-encoded DNA polymerases (PEP, RPOA, RPOC1, and RPOC2) (Supplementary Table S5). However, in the clp mutants, except for the photosynthetic apparatus, other plastid-encoded proteins remained almost unchanged (Kim et al., 2013a). Interestingly, two plastid-encoded proteins, YCF3 and YCF4, which are involved in the PSI assembly (Krech et al., 2012), showed opposing expression: while YCF3 was reduced, YCF4 was strongly accumulated in var2 (Fig. 2A, B). This difference could be attributable to the relative relevance of these proteins. For instance, YCF4 acts as an auxiliary and a non-essential assembly factor, whereas YCF3 is indispensable for PSI assembly in higher plants (Krech et al., 2012). This also suggests that, besides PSI assembly, YCF4 may have additional functions in chloroplasts deficient in PSII repair.
The var2 cpUPR-like response results in the accumulation of enzymes involved in the MEP pathway
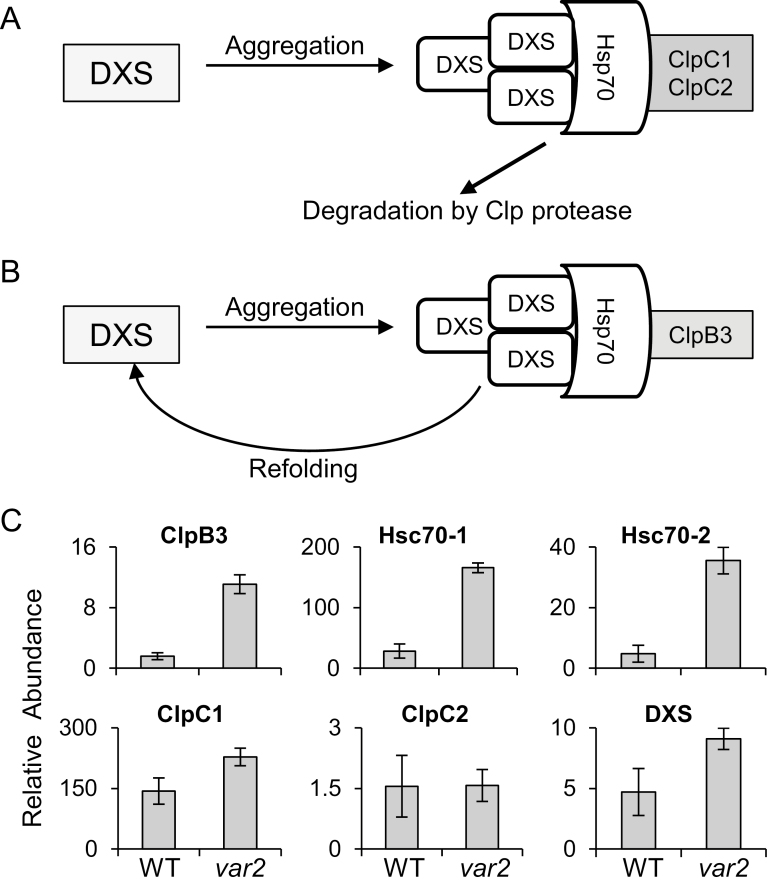
1O2 mainly generated by chlorophyll molecules in PSII is the prime cause of PSII damage (Aro et al., 1993; Santabarbara et al., 2007; Triantaphylidès et al., 2008; Telfer, 2014; Dogra et al., 2018). Therefore, chlorophyll biosynthesis needs to be tightly regulated (Meskauskiene et al., 2001; Krieger-Liszkay, 2005; Triantaphylidès and Havaux, 2009). Chlorophyll is composed of a chlorin ring and an isoprenoid phytol tail that are synthesized via the tetrapyrrole and methylerythritol 4-phosphate (MEP) pathways, respectively (Kim et al., 2013b). At the final step of chlorophyll biosynthesis, the chlorophyll synthase catalyses the esterification of chlorophyllide with the geranylgeranyl diphosphate (GGPP) synthesized via the MEP pathway (Oster et al., 1997; Wu et al., 2007). Inhibition of the MEP pathway results in the loss of the stoichiometric ratio between GGPP and chlorophyllide, causing the accumulation of free chlorophyllide (Arakane et al., 1996; Meskauskiene et al., 2001). Because free tetrapyrrole molecules generate 1O2 in the presence of light, the coordinated regulation of the MEP and tetrapyrrole pathways is essential to avoid its generation (Kim et al., 2013b). In addition, several enzymes in the MEP pathway are tightly regulated (Sauret-Güeto et al., 2006; Kim et al., 2013b; Pulido et al., 2016). For example, deoxyxylulose 5-phosphate synthase (DXS), the first enzyme in the MEP pathway, readily aggregates under oxidative stress conditions, resulting in its inactivation. The J-protein J20 interacts with the inactive DXS and enables association with Hsp70 for either refolding (reactivation) or degradation (Pulido et al., 2013). This regulatory process largely relies on additional Hsp100 chaperones such as ClpB3 and ClpC1. When Hsp70-DXS interacts with ClpC1, DXS is unfolded and subsequently degraded via the Clp protease (Fig. 5A). In contrast, when Hsp70-DXS interacts with CLPB3 it leads to the reactivation of DXS (Fig. 5B) (Pulido et al., 2016). DXS and other MEP pathway enzymes, including DXR, ISPD, ISPE, ISPG, and ISPH, are potential substrates of the Clp protease, as manifested by their accumulation in the clp mutants (Kim et al., 2013a). Given the increased accumulation of the catalytic core subunits of the Clp protease in var2, we anticipated that there would be concurrently reduced levels of the MEP enzymes. However, they either remained stable or instead accumulated (Supplementary Tables S2, S3). The DXS level was almost comparable between var2 and the WT, which was consistent with a previous report (Pulido et al., 2016). This unforeseen phenotype may be partly explained by the higher accumulation of ClpB3 and HSP70 (HSC70-1 and HSC70-2), which may protect these enzymes against proteolysis (Fig. 5C).
Fig. 5.
The cpUPR-like response contributes to the refolding of enzymes involved in the MEP pathway in the Arabidopsis var2 mutant. (A, B) DXS is the first enzyme in the MEP pathway and is a prone-to-aggregate protein under oxidative stress conditions. Interaction of Hsp70 with ClpC1/ClpC2 results in the unfolding and degradation of DXS (A). In contrast, Hsp70–ClpB3 interaction assists the refolding and reactivation of DXS (B). (C) Label-free quantitation indicating the steady-state levels of ClpB3, Hsp70 (Hsc70-1 and Hsc70-2), ClpC1, ClpC2, and DXS in var2 as compared to the wild-type (WT). The data are means (±SD) of n=3 replicates.
Accumulation of damaged chloroplast proteins in var2
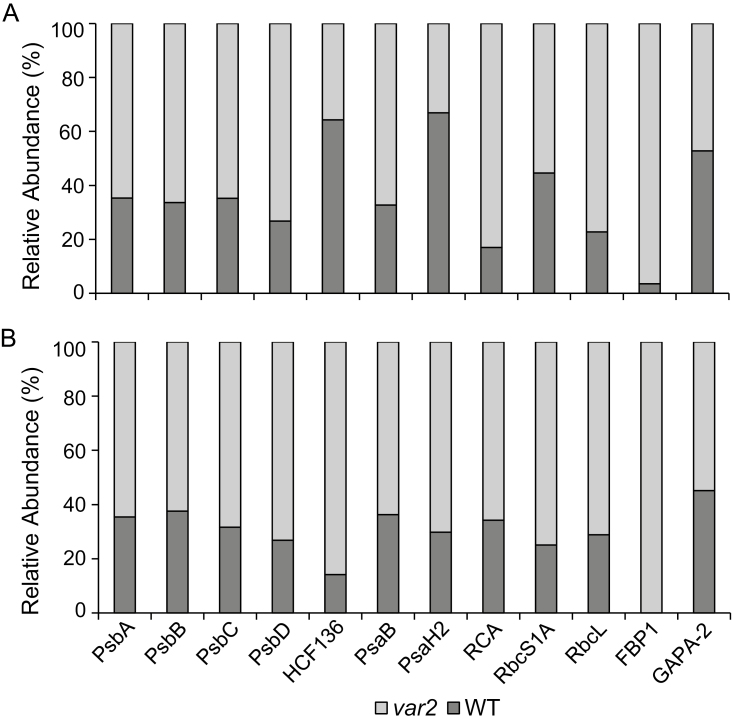
Transcriptional induction and subsequent accumulation of proteins related to proteostasis suggested a probable action of chloroplast-to-nucleus RS in var2. We further assumed that this RS pathway, if triggered, may have been partly caused by an accumulation of the substrates of the FtsH protease, including the photodamaged PSII RC proteins. In response to excess light, PSII core proteins as well as light-harvesting proteins of the PSII RC are prone to oxidation at certain tryptophan (Trp) residues (Dreaden Kasson et al., 2012; Kale et al., 2017). Various oxidized forms of Trp, namely oxindolylalanine, N-formylkynurenine, and kynurenine with their corresponding mass shifts of +16, +32, and +4 Da, respectively, were detected for those proteins (Supplementary Table S8). Given that PSII repair is a default process and that ROS are by-products of photosynthesis, photodamaged PSII proteins may accumulate in the chloroplasts of var2 mutant plants grown under normal light conditions. In agreement with this, we found at least 1.8-fold higher accumulation of PSII proteins in var2 compared to the WT (Fig. 6A; Supplementary Table S2). The relative proportion of oxidized to non-oxidized PSII proteins was higher in var2 compared to the WT (Fig. 6B; Supplementary Table S8). In addition to PSII proteins, the HCF136 protein involved in the assembly of PSII (Meurer et al., 1998) exhibited a similar Trp oxidation, which was more intensified in var2. Two PSI proteins, PsaH2 and PsaB, as well as four proteins involved in the Calvin-Benson cycle, namely RCA, RbcL, RbcS1A, and FBP1, also exhibited higher levels of Trp oxidation.
Fig. 6.
Trp-oxidized photosynthetic proteins are significantly accumulated in the Arabidopsis var2 mutant. (A) Steady-state levels photosynthetic proteins and GAPA-2 in var2 and the wild-type (WT). All proteins except HCF136 and PsaH exhibited at least 1.8-fold higher levels in var2. The data represent mean intensity values of the related peptides (n=3 replicates) (Supplementary Table S2). (B) Oxidation levels of the proteins. These proteins exhibited at least 1.6-fold higher oxidation levels in var2 as compared with the WT. The levels of oxidation were calculated using total intensities of peptides carrying oxidized Trp residues (Supplementary Table S8). The data represent the means of n=3 replicates. GAPA-2, a non-photosynthetic protein, was also found to undergo Trp-oxidation but showed no change in expression or in oxidation levels in var2 as compared with the WT.
Discussion
Under unfavorable environmental conditions, plant cells often produce ROS because of disturbed oxygenic metabolism. This leads to changes in the cellular redox status and the accumulation of unfolded/misfolded/damaged proteins in various subcellular compartments (Martínez and Chrispeels, 2003; Duwi Fanata et al., 2013). These stressed compartments, such as the ER and mitochondria, then activate the UPR to eradicate the inactivated proteins (Martínez and Chrispeels, 2003; Aldridge et al., 2007; Iwata et al., 2008; Walter and Ron, 2011; Duwi Fanata et al., 2013; Pellegrino et al., 2013). Recent studies have shown that chloroplasts lacking Clp protease activity also exhibit a similar response (Schmollinger et al., 2013; Ramundo et al., 2014; Llamas et al., 2017). Since the Clp protease is one of the major components of PQC, its failure presumably results in the accumulation of its misfolded/unfolded or aggregated substrates (Kim et al., 2009, 2013a; Zybailov et al., 2009). In return, chloroplasts trigger a UPR-like response to reinstate proteostasis. Like the Clp protease, the membrane-bound FtsH metalloprotease also contributes to chloroplast proteostasis, especially for PSII RC proteins (Zaltsman et al., 2005; Kato et al., 2009; Nishimura et al., 2016). Hence, inactivation of the FtsH protease impairs PSII proteostasis, potentiating ROS accumulation in the chloroplasts (Kato et al., 2007; Miura et al., 2010), which then results in an altered chloroplast proteostasis in terms of both quality and quantity (Fig. 1B, C). One of the changes (which takes place in this organelle) is the accumulation of damaged proteins. In turn, enhanced expression of proteins related to PQC and detoxification in the chloroplasts of var2 might be required in order to eliminate both ROS and damaged proteins (Figs 2A, 3) and to maintain redox homeostasis.
The substantial accumulation of catalytic core subunits of the Clp protease (Fig. 2A, Supplementary Tables S2, S3) suggests its complementary role in var2. Contrary to this notion, however, some MEP pathway enzymes, including DXS, that have previously been demonstrated to be substrates of Clp remained unchanged or even accumulated in var2 (Supplementary Table S2, S3). One explanation might be that the up-regulation of ClpB3 and other chaperones may protect DXS and other MEP enzymes from the Clp-mediated degradation (Figs 4, 5). However, some other known targets of Clp, including CAO, GluTR, and PSY, did not seem to be protected. The ORANGE (OR) protein, a holdase chaperone, protects PSY from Clp-mediated degradation (Welsch et al. 2018). Even though the OR protein was highly accumulated in var2 (Supplementary Table S3), it did not seem to protect PSY from degradation. One possible explanation for the down-regulation of PSY is that the increased Clp activity prevails over the OR activity. Zybailov et al. (2009) suggested that reduced or impaired photosynthetic electron transport (PET) may facilitate the accumulation of enzymes involved in the MEP pathway. Since PET is reduced in var2 due to impaired PSII repair, this may consequently cause the accumulation of MEP enzymes.
The chloroplast proteome in var2 was largely similar to that observed in different clp mutants (Supplementary Table S7). This suggests that any deficiency of PQC may generally affect chloroplast proteostasis, which consequently triggers cpUPR to reinstate proteostasis. The onset of cpUPR in the clp and var2 mutants, caused by the reduced PQC, might explain the previously demonstrated epistatic complementation of the leaf variegation in var2 by the loss of ClpC2 activity (Park and Rodermel, 2004). The deficiency of ClpC2 may potentiate cpUPR in var2, leading to the suppression of the leaf variegation. Despite the great similarity between the var2- and the clp-induced chloroplast proteomes, we noticed that whilst FtsH proteases remained unchanged in the clp mutants (Kim et al., 2013a), almost all subunits of the Clp protease were accumulated in var2 compared to the WT (Fig. 2; Supplementary Table S3). This indicates that the var2 cpUPR-like response is slightly different from the cpUPR established in the clp mutants, which coincides with their phenotypic differences (e.g. var2 leaf variegation). Given that the inactivation of FtsH2 resulted in the accumulation of damaged proteins (Fig. 6B) and ROS (Kato et al., 2007) in chloroplasts, the var2-conferred cpUPR-like response may be more precisely referred to as a damaged protein response (DPR).
Despite the non-stressful light conditions, the accumulation of Trp-oxidized PSII proteins was apparent in both the WT and var2 (Fig. 6B), supporting the notion that generation of 1O2 and photodamage of PSII are inevitable during photosynthesis (Krieger-Liszkay, 2005). The 1O2-induced oxidation of PSII RC proteins has been previously reported as a signature of photodamage under conditions of photoinhibition (Dreaden Kasson et al., 2012; Kale et al., 2017). In our study, we observed this Trp-oxidation for the first time also in other proteins, such as those constituting PSI and enzymes involved in the Calvin–Benson cycle. Given that 1O2 is mainly generated at the PSII RC in the appressed region of the grana (the grana core) and that it is unable to travel long distances owing to its extremely short life span (Gorman and Rodgers, 1992), this result was quite puzzling. Furthermore, the vast majority of the PSI complex is located at the non-appressed regions of the grana (the grana margin) and the stroma lamellae (Andersson and Anderson, 1980; Wang et al., 2016). Although it is possible that the Trp-oxidation of these proteins resulted from in vitro oxidation, the non-appressed region of grana may serve as an alternative source of 1O2 generation, as has been proposed recently (Wang et al., 2016; Dogra et al., 2018). In this regard, perhaps enzymes involved in the Calvin–Benson cycle may reside close to the non-appressed region of the grana in order to use the chemical energy generated via the light-dependent photosynthetic reaction. This proximity may cause Trp-oxidation. Highly enriched chlorophyll-synthesis enzymes along with PSII proteins and the de novo protein synthesis machinery in the grana margin also suggest that chlorophyll or its precursors synthesized during PSII reassembly may act as photosensitizers (Wang et al., 2016; Dogra et al., 2018).
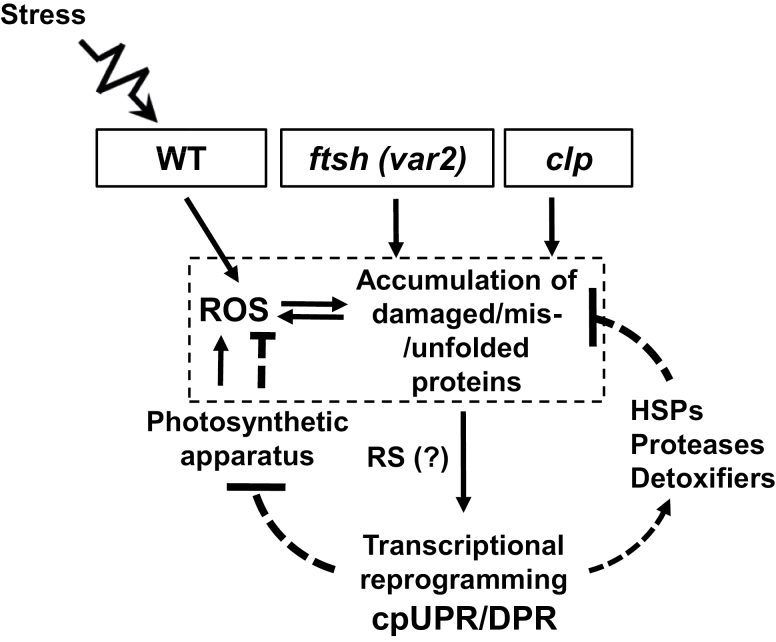
Since photooxidative stress conditions directly affect the chloroplast PQC, cpUPR/DPR-like responses must play important roles in WT plants (Fig. 7). Hence, determining the underlying mechanisms of cpUPR/DPR-like responses, especially in the context of RS, would shed light on adaptive responses of plants to photooxidative stress. Even though we are unable to provide any direct evidence as to whether chloroplast-to-nucleus RS triggers a cpUPR-like response in var2 (Fig. 7), given the repression of light-harvesting antenna proteins of PSII (Supplementary Table S2) it is rational to assume that Genomes Uncoupled 1 (GUN1), a central regulator of the expression of PhANGs, may be involved in the cpUPR-like response in var2. However, given that GUN1 has no obvious function in either the clp- or LIN-conferred cpUPR (Llamas et al., 2017), it is possible that, if GUN1 acts in var2, its function may be restricted to the repression of photosynthesis-associated nuclear genes. In addition, the enhanced levels of ROS in var2 (Kato et al., 2007) may lead to an accumulation of oxidized products of lipids and carotenoids, some of which are known to trigger RS. Reactive electrophile species such as OPDA and phytoprostanes can also induce detoxification-related genes (Mueller et al., 2008). In addition, H2O2 may diffuse out of the chloroplast to activate HSFAs (Yu et al., 2012). Given that Trp-oxidized proteins accumulated in var2 and that Trp-oxidation is mainly dependent on 1O2, RS mediated by EXECUTER1 (EX1, a putative 1O2 sensor)- (Wang et al., 2016) may contribute to the var2-conferred cpUPR-like response. However, since EX1-mediated 1O2 signaling requires an active FtsH2 protease, this signaling is likely to be dormant in var2. Forward genetic studies such as second-site mutagenesis of the var2 mutant may provide a pathway towards revealing the molecular components involved in RS priming of a cpUPR/DPR-like response. In addition, inactivation and/or overexpression of the PQC-related proteins that differentially accumulate in var2 as compared to the WT would provide further insights into how chloroplasts adapt to photooxidative stress conditions.
Fig. 7.
A model depicting the cpUPR-like damaged protein response (DPR) triggered by impaired proteostasis in the Arabidopsis var2 mutant. Inactivation of FtsH2 results in the accumulation of damaged proteins in the chloroplasts of var2. Simultaneously, it also leads to accumulation of reactive oxygen species (ROS), as demonstrated previously (Kato et al., 2009), which may subsequently alter the redox status in the chloroplasts, exaggerating the impairment of proteostasis. This then triggers chloroplast-to-nucleus retrograde signaling (RS) to activate the DPR, which is comprised of increased expression of heat-shock proteins (HSPs), proteases, and detoxifiers in order to restore the protein and redox homeostasis. In addition, this retrograde response also includes the repression of the photosynthetic apparatus to reduce the generation of ROS. Impaired Clp protease also exhibits a similar phenotype and activates cpUPR. In wild-type (WT) plants, stress conditions cause a burst of ROS in chloroplasts, which may also lead to the accumulation of damaged/misfolded/unfolded chloroplast proteins. As an adaptive mechanism, chloroplasts of WT plants may also initiate a similar retrograde cpUPR/DPR to reinstate chloroplastic homeostasis.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Amounts of chlorophyll and proteins are shown to be comparable with respect to chloroplast number in var2 and the wild-type.
Fig. S2. GO analysis of up- and down-regulated proteins in var2 relative to the wild-type.
Table S1. List of the primers used in the qRT-PCR analysis.
Table S2. List of the chloroplast proteins detected in var2 and/or the wild-type.
Table S3. List of the proteins with at least 2-fold higher accumulation in var2 relative to the wild-type.
Table S4. GO analysis of proteins with at least 2-fold higher accumulation in var2 relative to the wild-type.
Table S5. List of proteins with at least 2-fold down-regulation in var2 relative to the wild-type.
Table S6. GO analysis of proteins with at least twofold down-regulation in var2 relative to the wild-type.
Table S7. Comparison of clp- and var2-conferred proteomes.
Table S8. List of proteins showing increased levels of Trp-oxidation in var2 as compared with the wild-type.
Acknowledgements
We thank the Core Facility of Proteomics at the Shanghai Center for Plant Stress Biology (PSC) for carrying out the proteome profiling. We thank Rosa Lozano-Durán for critical comments on the manuscript. This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, Grant No. XDB27040102, the 100 Talent Program of the Chinese Academy of Sciences and the National Natural Science Foundation of China (NSFC) Grant No. 31570264 to CK. The authors have no conflicts of interest to declare.
References
- Adam Z, Rudella A, van Wijk KJ. 2006. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Current Opinion in Plant Biology 9, 234–240. [DOI] [PubMed] [Google Scholar]
- Aldridge JE, Horibe T, Hoogenraad NJ. 2007. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE 2, e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B, Anderson JM. 1980. Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochimica et Biophysica Acta 593, 427–440. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Apitz J, Nishimura K, Schmied J, Wolf A, Hedtke B, van Wijk KJ, Grimm B. 2016. Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiology 170, 2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakane K, Ryu A, Hayashi C, Masunaga T, Shinmoto K, Mashiko S, Nagano T, Hirobe M. 1996. Singlet oxygen (1Δg) generation from coproporphyrin in Propionibacterium acnes on irradiation. Biochemical and Biophysical Research Communications 223, 578–582. [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. 1993. Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochimica et Biophysica Acta 1143, 113–134. [DOI] [PubMed] [Google Scholar]
- Ashraf M, Harris PJC. 2013. Photosynthesis under stressful environments: an overview. Photosynthetica 51, 163–190. [Google Scholar]
- Bonshtien AL, Weiss C, Vitlin A, Niv A, Lorimer GH, Azem A. 2007. Significance of the N-terminal domain for the function of chloroplast cpn20 chaperonin. The Journal of Biological Chemistry 282, 4463–4469. [DOI] [PubMed] [Google Scholar]
- Broin M, Cuiné S, Eymery F, Rey P. 2002. The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. The Plant Cell 14, 1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Mabbitt PD, Phua SY, et al. 2016. Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proceedings of the National Academy of Sciences, USA 113, E4567–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L. 2012. The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochimica et Biophysica Acta 1817, 239–246. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. 2010. Roles for stress-inducible lambda glutathione transferases in flavonoid metabolism in plants as identified by ligand fishing. The Journal of Biological Chemistry 285, 36322–36329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V, Rochaix JD, Kim C. 2018. Singlet oxygen-triggered chloroplast-to-nucleus retrograde signalling pathways: an emerging perspective. Plant, Cell & Environment 41, 1727–1738. [DOI] [PubMed] [Google Scholar]
- Dreaden Kasson TM, Rexroth S, Barry BA. 2012. Light-induced oxidative stress, N-formylkynurenine, and oxygenic photosynthesis. PLoS ONE 7, e42220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwi Fanata WI, Lee SY, Lee KO. 2013. The unfolded protein response in plants: a fundamental adaptive cellular response to internal and external stresses. Journal of Proteomics 93, 356–368. [DOI] [PubMed] [Google Scholar]
- Fischer BB, Krieger-Liszkay A, Hideg E, Snyrychová I, Wiesendanger M, Eggen RI. 2007. Role of singlet oxygen in chloroplast to nucleus retrograde signaling in Chlamydomonas reinhardtii. FEBS Letters 581, 5555–5560. [DOI] [PubMed] [Google Scholar]
- Gorman AA, Rodgers MA. 1992. Current perspectives of singlet oxygen detection in biological environments. Journal of Photochemistry and Photobiology. B, Biology 14, 159–176. [DOI] [PubMed] [Google Scholar]
- Haussühl K, Andersson B, Adamska I. 2001. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. The EMBO Journal 20, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. 2008. Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. The Plant Cell 20, 3107–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews Molecular Cell Biology 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Kale R, Hebert AE, Frankel LK, Sallans L, Bricker TM, Pospisil P. 2017. Amino acid oxidation of the D1 and D2 proteins by oxygen radicals during photoinhibition of Photosystem II. Proceedings of the National Academy of Sciences, USA 114, 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, et al. 2010. VirtualPlant: a software platform to support systems biology research. Plant Physiology 152, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W. 2009. The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiology 151, 1790–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Matsushima R, Sakamoto W. 2007. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiology 144, 952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Ozawa S, Takahashi Y, Sakamoto W. 2015. D1 fragmentation in photosystem II repair caused by photo-damage of a two-step model. Photosynthesis Research 126, 409–416. [DOI] [PubMed] [Google Scholar]
- Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R. 2012. FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg++-branch of this pathway. FEBS Letters 586, 211–216. [DOI] [PubMed] [Google Scholar]
- Kim J, Olinares PD, Oh SH, Ghisaura S, Poliakov A, Ponnala L, van Wijk KJ. 2013a. Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis. Plant Physiology 162, 157–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PD, van Wijk KJ. 2009. Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. The Plant Cell 21, 1669–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Schlicke H, Van Ree K, Karvonen K, Subramaniam A, Richter A, Grimm B, Braam J. 2013b. Arabidopsis chlorophyll biosynthesis: an essential balance between the methylerythritol phosphate and tetrapyrrole pathways. The Plant Cell 25, 4984–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Berntsson RP, et al. 2013. Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proceedings of the National Academy of Sciences, USA 110, E3761–E3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumoto Y, Shimada T, Kondo M, Hara-Nishimura I, Nishimura M. 2001. Chloroplasts have a novel Cpn10 in addition to Cpn20 as co-chaperonins in Arabidopsis thaliana. The Journal of Biological Chemistry 276, 29688–29694. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332, 462–464. [DOI] [PubMed] [Google Scholar]
- Krech K, Ruf S, Masduki FF, Thiele W, Bednarczyk D, Albus CA, Tiller N, Hasse C, Schöttler MA, Bock R. 2012. The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiology 159, 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A. 2005. Singlet oxygen production in photosynthesis. Journal of Experimental Botany 56, 337–346. [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P. 2012. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659. [DOI] [PubMed] [Google Scholar]
- Ling Q, Jarvis P. 2015. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Current Biology 25, 2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Rodermel SR, Yu F. 2010. A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biology 10, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas E, Pulido P, Rodriguez-Concepcion M. 2017. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genetics 13, e1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, et al. 2010. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289. [DOI] [PubMed] [Google Scholar]
- Malnoë A, Wang F, Girard-Bascou J, Wollman FA, de Vitry C. 2014. Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. The Plant Cell 26, 373–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ. 2003. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. The Plant Cell 15, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. 2001. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 98, 12826–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P. 1998. A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. The EMBO Journal 17, 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W. 2007. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. The Plant Cell 19, 1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E, Kato Y, Sakamoto W. 2010. Comparative transcriptome analysis of green/white variegated sectors in Arabidopsis yellow variegated2: responses to oxidative and other stresses in white sectors. Journal of Experimental Botany 61, 2433–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JC, Jang HH, Chae HB, et al. 2006. The C-type Arabidopsis thioredoxin reductase ANTR-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochemical and Biophysical Research Communications 348, 478–484. [DOI] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S. 2008. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. The Plant Cell 20, 768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Kato Y, Sakamoto W. 2016. Chloroplast proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiology 171, 2280–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. 2006. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. The Plant Journal 48, 535–547. [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. 2001. AAA+ superfamily ATPases: common structure–diverse function. Genes to Cells 6, 575–597. [DOI] [PubMed] [Google Scholar]
- Oster U, Bauer CE, Rüdiger W. 1997. Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. The Journal of Biological Chemistry 272, 9671–9676. [DOI] [PubMed] [Google Scholar]
- Park S, Rodermel SR. 2004 Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proceedings of the National Academy of Sciences, USA 101, 12765–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Li W, Viehhauser A, et al. 2013. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proceedings of the National Academy of Sciences, USA 110, 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Latterich M. 1998. The AAA team: related ATPases with diverse functions. Trends in Cell Biology 8, 65–71. [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Haynes CM. 2013. Signaling the mitochondrial unfolded protein response. Biochimica et Biophysica Acta 1833, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín M, Pérez-Pérez ME, Lemaire SD, Crespo JL. 2014. Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiology 166, 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P, Llamas E, Llorente B, Ventura S, Wright LP, Rodríguez-Concepción M. 2016. Specific Hsp100 chaperones determine the fate of the first enzyme of the plastidial isoprenoid pathway for either refolding or degradation by the stromal Clp protease in Arabidopsis. PLoS Genetics 12, e1005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P, Toledo-Ortiz G, Phillips MA, Wright LP, Rodríguez-Concepción M. 2013. Arabidopsis J-protein J20 delivers the first enzyme of the plastidial isoprenoid pathway to protein quality control. The Plant Cell 25, 4183–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Casero D, Mühlhaus T, et al. 2014. Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. The Plant Cell 26, 2201–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Rochaix JD. 2014. Chloroplast unfolded protein response, a new plastid stress signaling pathway? Plant Signaling & Behavior 9, e972874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD, Ramundo S. 2018. Chloroplast signaling and quality control. Essays in Biochemistry 62, 13–20. [DOI] [PubMed] [Google Scholar]
- Rosano GL, Bruch EM, Ceccarelli EA. 2011. Insights into the Clp/HSP100 chaperone system from chloroplasts of Arabidopsis thaliana. The Journal of Biological Chemistry 286, 29671–29680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. 2003. Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. The Plant Cell 15, 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Tanaka R, Yamasato A, Tanaka A. 2009. Determination of a chloroplast degron in the regulatory domain of chlorophyllide a oxygenase. The Journal of Biological Chemistry 284, 36689–36699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME. 2008. Association of Rubisco activase with chaperonin-60β: a possible mechanism for protecting photosynthesis during heat stress. Journal of Experimental Botany 59, 1923–1933. [DOI] [PubMed] [Google Scholar]
- Santabarbara S, Agostini G, Casazza AP, Syme CD, Heathcote P, Böhles F, Evans MC, Jennings RC, Carbonera D. 2007. Chlorophyll triplet states associated with Photosystem I and Photosystem II in thylakoids of the green alga Chlamydomonas reinhardtii. Biochimica et Biophysica Acta 1767, 88–105. [DOI] [PubMed] [Google Scholar]
- Sauret-Güeto S, Botella-Pavía P, Flores-Pérez U, Martínez-García JF, San Román C, León P, Boronat A, Rodríguez-Concepción M. 2006. Plastid cues posttranscriptionally regulate the accumulation of key enzymes of the methylerythritol phosphate pathway in Arabidopsis. Plant Physiology 141, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S, Schulz-Raffelt M, Strenkert D, Veyel D, Vallon O, Schroda M. 2013. Dissecting the heat stress response in Chlamydomonas by pharmaceutical and RNAi approaches reveals conserved and novel aspects. Molecular Plant 6, 1795–1813. [DOI] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P. 2006. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Molecular Biology 60, 759–772. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Stahl A, Moberg P, Ytterberg J, Panfilov O, Brockenhuus Von Lowenhielm H, Nilsson F, Glaser E. 2002. Isolation and identification of a novel mitochondrial metalloprotease (PreP) that degrades targeting presequences in plants. The Journal of Biological Chemistry 277, 41931–41939. [DOI] [PubMed] [Google Scholar]
- Stenbaek A, Hansson A, Wulff RP, Hansson M, Dietz KJ, Jensen PE. 2008. NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Letters 582, 2773–2778. [DOI] [PubMed] [Google Scholar]
- Takagi D, Takumi S, Hashiguchi M, Sejima T, Miyake C. 2016. Superoxide and singlet oxygen produced within the thylakoid membranes both cause Photosystem I photoinhibition. Plant Physiology 171, 1626–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A. 2014. Singlet oxygen production by PSII under light stress: mechanism, detection and the protective role of β-carotene. Plant & Cell Physiology 55, 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C, Havaux M. 2009. Singlet oxygen in plants: production, detoxification and signaling. Trends in Plant Science 14, 219–228. [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, Van Breusegem F, Mueller MJ. 2008. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiology 148, 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy BC, Oelmüller R. 2012. Reactive oxygen species generation and signaling in plants. Plant Signaling & Behavior 7, 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang L, Kim C, Xu X, Piskurewicz U, Dogra V, Singh S, Mahler H, Apel K. 2016. Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proceedings of National Academy of Sciences, USA 113, E3792–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Zhou X, Yuan H, et al. 2018. Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Molecular Plant 11, 149–162. [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhang X, He B, et al. 2007. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiology 145, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Hasegawa A, Taninaka A, Mizutani M, Sugimoto Y. 2011. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. The Journal of Biological Chemistry 286, 6999–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. 2004. The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. The Plant Journal 37, 864–876. [DOI] [PubMed] [Google Scholar]
- Yu HD, Yang XF, Chen ST, Wang YT, Li JK, Shen Q, Liu XL, Guo FQ. 2012. Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genetics 8, e1002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Ori N, Adam Z. 2005. Two types of FtsH protease subunits are required for chloroplast biogenesis and Photosystem II repair in Arabidopsis. The Plant Cell 17, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Wan J, Jin R, Lamppa G. 2003. A pea antisense gene for the chloroplast stromal processing peptidase yields seedling lethals in Arabidopsis: survivors show defective GFP import in vivo. The Plant Journal 34, 802–812. [DOI] [PubMed] [Google Scholar]
- Zivcak M, Brestic M, Kunderlikova K, Olsovska K, Allakhverdiev SI. 2015. Effect of photosystem I inactivation on chlorophyll a fluorescence induction in wheat leaves: does activity of photosystem I play any role in OJIP rise? Journal of Photochemistry and Photobiology. B, Biology 152, 318–324. [DOI] [PubMed] [Google Scholar]
- Zoschke R, Qu Y, Zubo YO, Börner T, Schmitz-Linneweber C. 2013. Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. Journal of Plant Research 126, 403–414. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Friso G, Kim J, Rudella A, Rodríguez VR, Asakura Y, Sun Q, van Wijk KJ. 2009. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Molecular & Cellular Proteomics 8, 1789–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.