In contrast to the light requirement for Arabidopsis seed germination, the germination of several Aethionema arabicum accessions is inhibited by light, due to antipodal transcriptional regulation of hormone balance.
Keywords: Aethionema arabicum, light inhibition, model plant, natural variation, seed germination, transcriptional regulation
Abstract
The timing of seed germination is crucial for seed plants and is coordinated by internal and external cues, reflecting adaptations to different habitats. Physiological and molecular studies with lettuce and Arabidopsis thaliana have documented a strict requirement for light to initiate germination and identified many receptors, signaling cascades, and hormonal control elements. In contrast, seed germination in several other plants is inhibited by light, but the molecular basis of this alternative response is unknown. We describe Aethionema arabicum (Brassicaceae) as a suitable model plant to investigate the mechanism of germination inhibition by light, as this species has accessions with natural variation between light-sensitive and light-neutral responses. Inhibition of germination occurs in red, blue, or far-red light and increases with light intensity and duration. Gibberellins and abscisic acid are involved in the control of germination, as in Arabidopsis, but transcriptome comparisons of light- and dark-exposed A. arabicum seeds revealed that, upon light exposure, the expression of genes for key regulators undergo converse changes, resulting in antipodal hormone regulation. These findings illustrate that similar modular components of a pathway in light-inhibited, light-neutral, and light-requiring germination among the Brassicaceae have been assembled in the course of evolution to produce divergent pathways, likely as adaptive traits.
Introduction
Proper timing of germination is a critical step for the survival and propagation of seed plants. Light is a major environmental factor regulating seed germination, which provides information about the position in the soil, the presence of competitors, day length, and the season. Plants living in various habitats have different optima for light conditions at the time of germination. Seeds can be categorized based on their response to white light during germination (Takaki, 2001). Light-requiring (positive photoblastic) seeds germinate only after a minimal exposure to light, while light-inhibited (negative photoblastic) seeds germinate only in the dark. A third category, light-neutral seeds, germinate in both light and darkness. The categories are not mutually exclusive: germination of the ricegrass species Oryzopsis miliacea and the salt cress (Thellungiella halophila) is promoted by a short period of illumination but inhibited by continuous light (Koller and Negbi, 1959; Negbi and Koller, 1964; Li et al., 2015). The photoblastic classification considers only responses to the whole spectrum of white light, regardless of wavelength-specific effects. For example, germination of Brachypodium seeds and those of other monocotyledonous species is inhibited by blue light via cryptochrome receptors (Barrero et al., 2014) but induced by red light. In white light, the seeds germinate and therefore belong to the light-requiring seed category (Barrero et al., 2012). Seeds of many accessions of Arabidopsis thaliana and lettuce (Lactuca sativa), the model plants for research in this field, require a minimal light exposure for complete germination. Therefore, most insight into the role of light for seed germination originates from these light-requiring seed types (Shropshire et al., 1961; Shinomura et al., 1994; Casal and Sanchez, 1998). Only a limited number of plant species with light-inhibited seeds have been described, for example, Phacelia tanacetifolia (Chen, 1968; Chen, 1970) or Citrullus lanatus, for which seed germination is inhibited by the whole spectrum, including white, blue, red, and far-red light (Botha and Small, 1988; Thanos et al., 1991). The different photoblastic responses are likely an adaptive trait to harsh or quickly changing habitats: species with light-inhibited seeds often grow on sea coasts or in deserts where germination on the surface might be risky or deleterious. Light-inhibited germination might be advantageous to avoid direct sunlight, so that germination occurs when the seeds are buried at various depths under shifting sand dunes (Koller, 1956; Thanos et al., 1991; Lai et al., 2016), although this germination strategy is not strictly correlated with specific habitats (Vandelook et al., 2018).
In seeds of all photoblastic categories, light-regulated molecular changes during germination are associated with the perception of light through phytochromes, which regulates hormonal levels (Casal et al., 1998; Takaki, 2001). Gibberellic acid (GA) and abscisic acid (ABA) play a central role: GA induces germination and helps to break seed dormancy, while ABA is involved in the establishment and maintenance of dormancy. The balance of these two hormones determines seed fate (Finch-Savage and Leubner-Metzger, 2006). After light perception by phytochrome B (phyB), a cascade including several transcription factors and repressors leads to GA synthesis and ABA degradation in light-requiring seeds (Seo et al., 2009). A dual role for light has been shown in salt cress, where weak light promotes GA accumulation, but strong light inhibits it (Li et al., 2015). In Arabidopsis, in red light, the expression of the GA biosynthesis genes GA3 OXIDASE 1 (AtGA3ox1) and GA3 OXIDASE 2 (AtGA3ox2) as well as the ABA-degrading CYTOCHROME P450 gene family member AtCYP707A2 are enhanced, whereas the ABA biosynthesis gene NCED6 and the GA-degrading GA2 OXIDASE 2 (AtGA2ox2) are repressed (reviewed in Seo et al., 2009; Shu et al., 2016). In contrast, knowledge about the molecular mechanisms of light-inhibited and light-insensitive germination is lacking, as no species with this seed trait has so far been established as a model for molecular and genetic approaches.
Aethionema arabicum (L.) Andrz. ex DC. (Brassicaceae) is an annual spring plant with a relatively small (203–240 Mbp), diploid genome that was recently sequenced (Franzke et al., 2011; Haudry et al., 2013). The Aethionemeae tribe, with approximately 57 species, is the earliest-diverged tribe within the Brassicaceae and shares 70–80% genetic information with Arabidopsis. Aethionema arabicum (hereafter Aethionema) is distributed in the Middle East and eastern Mediterranean regions.
In this study, we show that the seeds of one Aethionema accession from Turkey (TUR) germinate well in light, while the seeds of another accession from Cyprus (CYP) are strongly inhibited by the entire spectrum of visible light, in a quantitative manner. We characterize the physiological and molecular properties of seed germination in these two accessions and demonstrate that inhibition of germination in light is associated with a decreased GA:ABA ratio, in contrast to the situation in light-requiring Arabidopsis. Transcriptome analysis revealed the involvement of similar regulatory components as in Arabidopsis but with opposite responses to light. In addition, we identified large natural variation of the photoblastic phenotype within the Aethionemeae tribe. This makes Aethionema a very suitable model to investigate the variation in seed germination responses to light that exist in closely related species.
Materials and methods
Plant material
Experiments were conducted with Aethionema arabicum (L.) Andrz. ex DC. accessions TUR ES1020 and CYP (obtained from Eric Schranz, Wageningen), Iran8456-1, Iran8456-2, and Iran8458 (obtained from Setareh Mohammadin, Wageningen), Aethionema carneum (Banks & Sol.) B.Fedtsch. accession KM2496, and Aethionema heterocarpum Trev. accessions KM2491 and KM2614 (obtained from Klaus Mummenhoff, Osnabrück). All seed material was produced by plants grown under 16 h light/19 °C and 8 h dark/16 °C diurnal cycles, under ~50 μmol m−2 s−1 light intensity. Indehiscent and dehiscent fruits encompassing non-mucilaginous and mucilaginous seeds, respectively, were manually separated and sieved. After seed harvesting, seed stocks were kept dry at 24 °C for a minimum of 2 months.
Germination test
All germination tests were conducted at the optimal temperature of 14 °C in Petri dishes on four-layer filter paper wetted with distilled H2O and supplemented with 0.1% plant preservative PPM (Plant Cell Technology). Germination assays shown in Fig. 6 were carried out with the addition of 10 µM GA4 + 7 (Duchefa), 10 µM fluridone, 10 µM norflurazon (Sigma Aldrich and Duchefa), or ABA (Cayman Chemical) as indicated. All assays were done in triplicate with a minimum of 15–20 seeds each. Except for the experiments reported in Fig. 1C and D and Fig. 2, white, red, and blue light exposure was uniformly set to 100 μmol m−2 s−1 light intensity, and far-red exposure was set to 15 μmol m−2 s−1 for all experiments. For dark treatments, seeds were placed on wet filter paper in complete darkness. Diurnal and high-light tests were carried out under an LED NS1 lamp with a wide sun-like spectrum (Valoya). Light spectra and intensity were measured by using an LED Meter MK350S (UPRtek). Except for the seeds shown in Supplementary Fig. S1 at JXB online, only mucilaginous seeds were used for all germination assays.
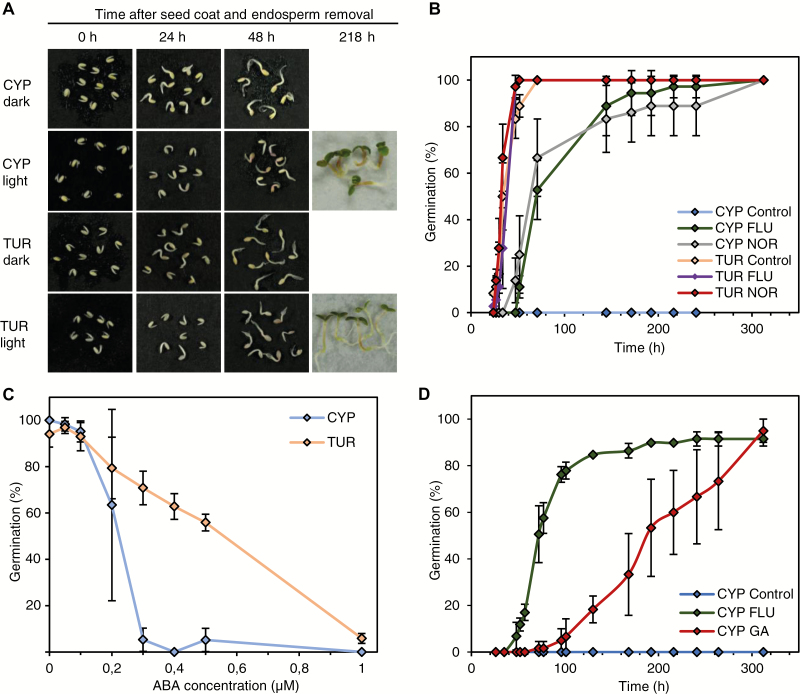
Fig. 6.
Rescue of CYP seed germination in light by interference with ABA and GA signaling. (A) Development of CYP and TUR seedlings in darkness or 100 µmol m−2 s−1 light after removal of the ABA-producing seed coat and endosperm. 0 h represents the status directly after seedling isolation. (B) Percentage of germination over time when seeds were kept on plates supplemented with the ABA inhibitors 10 µM fluridone (FLU) or 10 µM norflurazon (NOR), or 0.01% DMSO as a control, under continuous 100 µmol m−2 s−1 light. (C) Percentage of germination scored after 6 days for seeds kept in the dark on plates with external application of ABA at concentrations of 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, or 1 μM, or 0.01% DMSO as a control. (D) Percentage of germination over time for seeds kept under continuous 100 µmol m−2 s−1 light on plates with external application of 10 µM GA4 + 7 (GA) or 0.01% DMSO as a control. Error bars represent SD (three independent replicates).
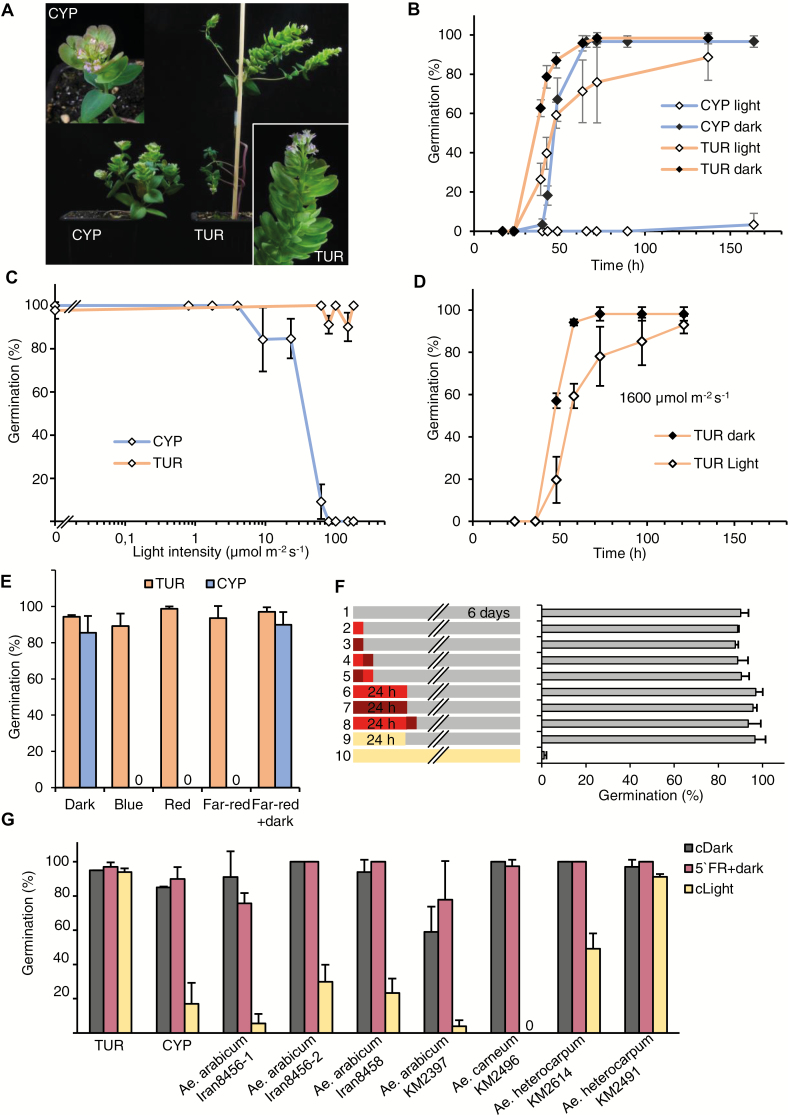
Fig. 1.
Light inhibition of seed germination in Aethionema arabicum accession CYP. (A) Eight-week-old A. arabicum TUR (Turkey) and CYP (Cyprus) plants grown in a growth chamber. (B) Percentage of germinating seeds kept in darkness or under 100 μmol m−2 s−1 white light, scored over time. (C) Percentage of germinating seeds kept at various light intensities: (0.8, 1.74, 4, 9.2,23, 62, 80, 100, 150 and 180 μmol m−2 s−1 white light) or in darkness (indicated as 0 μmol m−2 s−1), scored after 6 d. (D) Percentage of germinating TUR seeds kept in darkness or under 1600 μmol m−2 s−1 white light, scored over time. (E) Percentage of germinating seeds in continuous white, red, blue, or far-red light, dark, or exposed to 5 min far-red light followed by darkness, scored after 6 d. (F) Percentage of germinating CYP seeds after various light treatments. The left panel indicates the treatment; the right panel indicates the percentage of germinating seeds, scored after 6 d. Red, dark red, and cream rectangles indicate red, far-red, and white light exposure, respectively. Grey bars indicate dark periods. Short and long red/far-red rectangles indicate 5 min and 24 h exposure, respectively. (G) Percentage of germinating seeds from different A. arabicum accessions and closely related species, kept in the dark (black columns), light (yellow columns), or in the dark after a 5 min far-red pulse at imbibition (red columns), scored after 6 d. Error bars represent SD (three independent replicates).
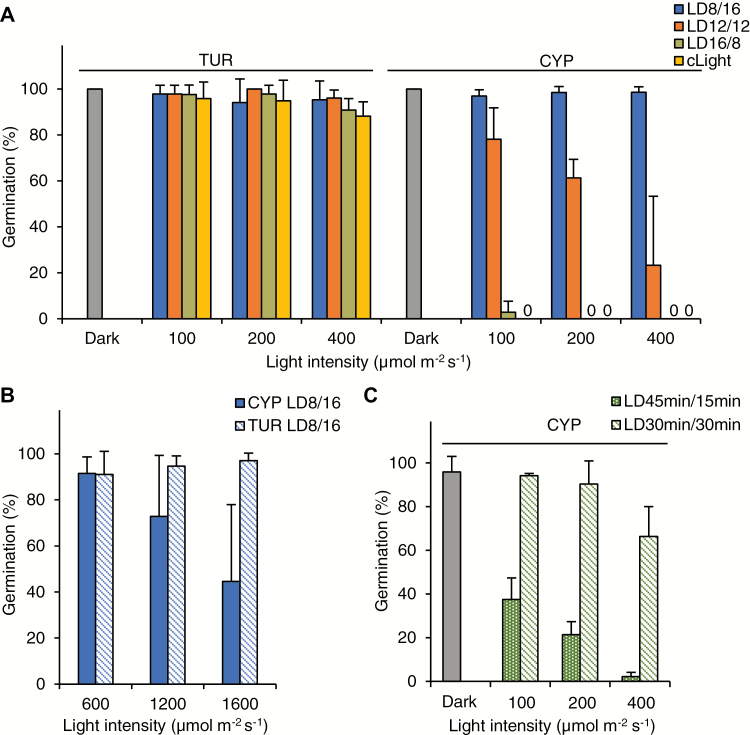
Fig. 2.
Diurnal regulation of CYP seed germination. (A) Percentage of germinating CYP and TUR seeds kept under different diurnal regimes, scored after 6 d. LD8/16, 8 h light/16 h dark; LD12/12, 12 h light/12 h dark, LD16/8, 16 h light/8 h dark. In addition, seeds were kept under continuous dark (Dark) or light (cLight). (B) Percentage of germinating CYP and TUR seeds kept under different light intensities with a LD8/16 regime. 0 indicates no germination. (C) Percentage of germinating CYP seeds under continuously repeated 30 min light/30 min dark cycles (LD30min/30min) or 45 min light and 15 min dark cycles (LD45min/45min), scored after 6 d. Error bars represent SD (three independent replicates).
Quantitative RT–PCR
Before imbibition, seeds were sterilized with chlorine gas for 10 minutes. After 23 h incubation in darkness or light, seeds with intact seed coats were collected for RNA extraction (three biological replicates for each sample). RNA was extracted as described by Oñate-Sánchez and Vicente-Carbajosa (2008). Total RNA (3 µg) was treated with DNase I (Thermo Scientific) and precipitated with 1/10 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol. DNase I-treated RNA (1 µg) was used for cDNA synthesis with random hexamer primers using the RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Scientific). Quantitative reverse transcription–PCR (qRT–PCR) reactions were performed in a Lightcycler® 96 System (Roche) in duplicate, using FastStart Essential DNA Green Master mix (Roche) and primer pairs listed in Supplementary Table S2, with the following parameters: 95 °C for 10 min, 45 cycles of 95 °C for 10 s and 60 °C for 30 s, and one cycle of 95 °C for 10 s, 60 °C for 30 s, and 97 °C for 1 s to obtain the melting curve for each reaction. Cycle threshold values were calculated using Lightcycler® 96 Software (Roche). The geometric mean of Aethionema orthologs of ACTIN2 (AearACT2, AA26G00546), POLYUBIQUITIN10 (AearUBQ10, AA6G00219), and ANAPHASE-PROMOTING COMPLEX2 (AearAPC2, AA61G00327) was used for normalization. For each gene, the expression level under white light is presented as fold change relative to the level of the dark-treated samples where the mean expression was set to a value of 1.
Aethionema arabicum genome and annotations
Genome scaffolds and the accompanying gene feature format file of the A. arabicum genome version 2.5 was obtained from Haudry et al. (2013). The gene models were searched against the non-redundant database of NCBI (release 13-05-2015) using BLAST (Altschul et al., 1990). Gene ontology (GO) terms were retrieved using BLAST2GO version 2.5 (Conesa et al., 2005) along with the best hit and its description. The coding DNA sequence of each gene model was translated into amino acid sequence using the R package biostrings version 2.32.0 (https://www.bioconductor.org/packages/release/bioc/html/Biostrings.html) and then searched against the uniprot database (release 2015_10) and The Arabidopsis Information Resource (TAIR, TAIR10_pep_20110103_representative_gene_model_updated) using BLAST for extracting the best hit and its description. Results were filtered for having at least 80% query coverage, according to Rost (1999), to unambiguously detect homologous sequences. Sequence data from A. arabicum can be found in the CoGe database (https://genomevolution.org/coge/) under the following genome ID: v2.5, id33968. Accession numbers used in this study are listed in Supplementary Table S3. RNA-Seq information and files are deposited on GEO with accession number GSE125854 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125854).
RNA extraction for RNA-Seq
Light incubation of seed material was done as for the qRT–PCR. Total RNA was isolated as described by Chang et al. (1993). Genomic DNA was removed by DNase-I (Qiagen) digestion in solution, followed by additional purification using columns (Qiagen RNeasy Kit). RNA quantity and purity were determined using a NanoDrop™ spectrophotometer (ND-1000, Thermo Scientific™, Wilmington, DE, USA) and an Agilent 2100 Bioanalyzer with the RNA 6000 Nano Kit (Agilent Technologies, CA, USA) using 2100 Expert Software to calculate RNA Integrity Number values. At least three biological replicate RNA samples were used for downstream applications. Sequencing was performed at the Vienna BioCenter Core Facilities GmbH Next Generation Sequencing Unit, Vienna, Austria (www.vbcf.ac.at). Libraries were sequenced in 50 bp single-end mode on Illumina HiSeq 2000 Analyzers using the manufacturer’s standard cluster generation and sequencing protocols.
RNA-Seq data trimming and filtering
The cDNA sequence libraries were processed using Trimmomatic (Bolger et al., 2014) with the options ‘ILLUMINACLIP:adaptors:2:20:8, SLIDINGWINDOW:4:15, TRAILING:15, HEADCROP:12, MINLENGTH:20’ to remove poor quality, adapters, and other technical sequences. Chloroplast, mitochondrial, and ribosomal RNA sequences were filtered out using Bowtie 2 version 2.2.3 (Langmead and Salzberg, 2012) and by mapping the reads against the chloroplast (GenBank: AP000423.1), mitochondrial (GenBank: Y08501.2), and rRNA (GenBank: X52320.1) sequences of A. thaliana.
Read mapping and feature counting
Trimmed and filtered reads were mapped against the Aethionema genome version 2.5 using Bowtie2. Uniquely mapped reads were retained. Mapped reads per feature were counted using HTSeq-count version 0.6.1 (Anders et al., 2015).
Differentially expressed genes
R (http://www.R-project.org) and the Bioconductor packages Deseq2 version 1.14.1 (Love et al., 2014), edgeR version 2.18 (Robinson et al., 2010), and NOISeq version 3.16.5 (Tarazona et al., 2011) were used in combination with the feature counts to identify differentially expressed genes. For Deseq2, edgeR (classic approach, ‘exactTest’) and NOISeq default parameters were used. Adjusted p-value (q-value) cut-offs were set to 0.001 for Deseq2 and edgeR. For NOISeq, which uses probabilities of differential expression, a cut off value of >0.9 was used. The overlap (strict consensus) of the output from the three packages was used for further analysis.
Heatmap and GO term enrichment
Heatmaps were created using Morpheus visualization software (https://software.broadinstitute.org/morpheus). GO term enrichment was analyzed using AGI gene identifiers of the Arabidopsis orthologs of Aethionema genes in ThaleMine (https://apps.araport.org/thalemine/begin.do). Pvalues indicate Benjamini–Hochberg test correction.
Phylogenetic analysis
To verify the ortholog status of phytochromes and gibberelin-2-oxidases in Aethionema, Arabidopsis query protein sequences were searched with BLASTP (Altschul et al., 1990) against a plant-specific, custom-made protein database that included genomes of the species listed in Supplementary Dataset S7. Results were filtered for having at least 80% query coverage according to Rost (1999). Resulting sequences were aligned using MAFFT version 7.037b (Katoh and Standley, 2013) in automatic mode, and resulting alignments were inspected manually and trimmed using Jalview version 2.8 (Clamp et al., 2004). Based on these alignments, neighbor-joining guide trees were built using quicktree_sd (http://hdl.handle.net/10013/epic.33164.d001) with 1000 bootstrap samples. Sequences with very long branches, potentially representing flawed gene models, were removed upon inspection of initial trees. Afterwards, the appropriate models were selected based on AIC/BIC using ProtTest 3.4 (Guindon and Gascuel, 2003; Darriba et al., 2011). Final phylogenies were constructed by Bayesian inference using Mr. Bayes 3.2.5 (Ronquist et al., 2012). Bayesian inference analysis was run with two hot and two cold chains, discarding 25% of trees as burn-in, for 1 688 500 generations (standard deviation of split frequencies 0.009992) and 2 000 000 generations (standard deviation of split frequencies 0.063371) for phytochrome and gibberellin-2-oxidase family trees, respectively. Trees were displayed, colored, and midpoint-rooted with FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Measurement of hormone levels
For ABA and GA analysis, seed samples were collected as described for RNA extraction, except that five biological replicates were prepared per sample. For ABA analysis, 13 mg of seed material was homogenized and extracted for 1 h in 1 ml ice-cold methanol/water/acetic acid (10/89/1, v/v). Deuterium-labelled standard (20 pmol of (+)-3′,5′,5′,7′,7′,7′-2H6-ABA) was added to each of the samples. The homogenates were centrifuged at 30 000 × g for 5 min at 4 °C, and the pellets were then re-extracted in 0.5 ml extraction solvent for 30 min. The combined extracts were purified by solid-phase extraction on Oasis® HLB cartridges (60 mg, 3 ml, Waters, Milford, MA, USA) and then evaporated to dryness in a Speed-Vac (UniEquip). Subsequently, the evaporated samples were methylated, purified by ABA-specific immunoaffinity extraction (Hradecka et al., 2007), and analyzed by UPLC-ESI(+)-MS/MS (Tureckova et al., 2009).
The sample preparation and analysis of GAs was performed as described by Urbanová et al. (2013), with some modifications. Briefly, 13 mg of seed material per sample was homogenized in 1 ml ice-cold 80% acetonitrile containing 5% formic acid. After adding 17 internal GA standards ([2H2]GA1, [2H2]GA3, [2H2]GA4, [2H2]GA5, [2H2]GA6, [2H2]GA7, [2H2]GA8, [2H2]GA9, [2H2]GA15, [2H2]GA19, [2H2]GA20, [2H2]GA24, [2H2]GA29, [2H2]GA34, [2H2]GA44, [2H2]GA51 and [2H2]GA53; purchased from Lewis Mander, Australia), the samples were extracted overnight at 4 °C. The homogenates were centrifuged at 36 670 × g for 10 min at 4 °C. Supernatants were further purified using reversed-phase and mixed-mode solid phase extraction cartridges (Waters, Milford, MA, USA) and analyzed by ultra-high-performance chromatography–tandem mass spectrometry (UHPLC-MS/MS; Micromass, Manchester, UK). GAs were detected using multiple-reaction monitoring mode of the transition of the ion [M–H]- to the appropriate product ion. MassLynx 4.1 software (Waters, Milford, MA, USA) was used to analyze the data, and the standard isotope dilution method (Rittenberg and Foster, 1940) was used to quantify the levels of GAs.
Results
Light inhibits seed germination in Aethionema arabicum
The Aethionema accession TUR originates from Konya, Turkey (accession ES1020) and was used to generate the reference genome (Haudry et al., 2013) and to characterize its interesting seed and fruit dimorphism (Lenser et al., 2016). The Aethionema accession CYP comes from the Kato-Moni region of Cyprus (Mohammadin et al., 2018). Both accessions were propagated for several generations under the same conditions in a growth chamber (Fig. 1A). Seeds of both accessions germinate optimally at 14 °C, and all experiments were performed at that temperature. Testing the light dependence, we found that TUR seeds germinated under white light or in darkness. CYP seeds germinated well in darkness but were strongly inhibited under white light (Fig. 1B). Comparing the dimorphic seed types, we did not find any difference between mucilaginous and non-mucilaginous seeds in response to light, therefore we used mucilaginous seeds for further experiments (see Supplementary Fig. S1). Species with light-requiring seeds need various periods of illumination for germination induction, ranging from seconds to days (Bewley et al., 2013). Conversely, in light-inhibited seeds, light inhibition of germination can be exerted with a wide range of photon irradiance, from a relatively weak ~17 µmol m−2 s−1 light intensity in Citrullus lanatus (Botha and Small, 1988) to strong irradiance with 1000 µmol m−2 s−1 in some desert plants (Botha and Small, 1988; Lai et al., 2016). Therefore, we tested the germination of TUR and CYP seeds with different light intensities ranging from 0.8 to 180 µmol m−2 s−1 (Fig. 1C). Increased light intensity gradually decreased the proportion of CYP seeds that germinated: at 62 µmol m−2 s−1 only ~10% of the seeds germinated, and at 80 µmol m−2 s−1 the inhibition was complete. Germination of TUR seeds was light-neutral in this range (Fig. 1C). As the irradiance can be much stronger at the geographic origin of the species, we further tested the germination of TUR seeds under 1600 µmol m−2 s−1 white light with a wide, sun-like spectrum. Although the germination of TUR seeds was slower, almost all seeds germinated under this strong light, therefore we categorized TUR seeds as neutral to light (Fig. 1D).
Light-dependent germination often requires exposure to a specific part of the light spectrum. Therefore, we tested whether the inhibition of CYP seed germination was wavelength-dependent. Continuous monochromatic blue, red, and far-red light were equally effective at inhibiting the germination of CYP seeds (Fig. 1E), while TUR seeds could germinate under any light condition, including continuous far-red light, which inhibits phytochrome-mediated light-induced germination in Arabidopsis and lettuce (Borthwick et al., 1952; Shropshire et al., 1961). A short (5 min) far-red pulse also effectively inhibits germination in Arabidopsis and lettuce, as it converts phyB to the inactive form, while a following red pulse allows germination again by converting phyB to the active form (Borthwick et al., 1952; Shropshire et al., 1961). Importantly, a short (5 min) or longer (24 h) exposure to either far-red or red light at the time of imbibition, followed by 6 days in darkness, allowed CYP seeds to germinate equally well (Fig. 1F). This indicates that (i) the induction of germination is independent of the active form of phyB and (ii) the light inhibition is not established in this time scale (Fig. 1E, F).
TUR and CYP accessions cluster closely together in a network analysis of several A. arabicum accessions (Mohammadin et al., 2018). To determine whether the inhibition of CYP seed germination by light is a unique trait in the genus Aethionema, we investigated the germination phenotype of other available accessions, including the closest relatives, Aethionema heterocarpum and Aethionema carneum (Lenser et al., 2016; Mohammadin et al., 2018; Supplementary Table S1), after propagating the seeds under the same controlled conditions as for TUR and CYP. None of the tested accessions required light, as all of them germinated similarly well in constant darkness or after a 5 min far-red pulse followed by darkness (Fig. 1G). We observed variations in the response to white light: germination of A. carneum and two A. arabicum (Iran 8456-1 and KM2397) accessions was clearly light-inhibited, one A. heterocarpum accession (KM2491) had light-neutral seeds, while the germination of three accessions (A. arabicum Iran 8458 and Iran 8456-2, and A. heterocarpum KM2614) was partially inhibited by light (Fig. 1G). These findings reveal the presence of natural variation for the negative or neutral photoblastic phenotype, suggesting that light-inhibited or light-neutral seed germination may be an adaptive trait in the Aethionemeae tribe, and provides interesting material for genetic analysis of the phenomenon.
Diurnal regulation of CYP seed germination
To better understand the ecological relevance of light-inhibited germination of CYP seeds, we also tested germination under different diurnal conditions. Again, TUR seeds were unaffected and germinated well under any day length. Interestingly, at a lower range of light intensity, the CYP seeds germinated well under short-day conditions (cycles of 8 h light/16 h darkness, LD8/16), compared to 12 h light/12 h dark cycles (LD12/12), which produced partial inhibition, or long-day conditions (16 h light/8 h dark cycles, LD16/8), which resulted in complete inhibition (Fig. 2A). Remarkably, at higher light intensity, similar to conditions in the plant’s natural habitat (1600 µmol m−2 s−1), ~40% of the CYP seeds still germinated under short-day conditions (Fig. 2B). Gradual inhibition was also observed with hourly alternating light exposure (Fig. 2C). However, longer uninterrupted light periods resulted in stronger inhibition than more frequent alternations between light and dark, despite equal daily fluence in LD12/12 compared with 30 min light/30 min dark cycles (Fig. 2A and Fig. 2C). These data indicate that CYP seeds integrate the regime, duration, and intensity of illumination into their germination regulation.
Light-neutral and light-inhibited seeds differ in their transcriptomes
To better understand the light-inhibited seed germination phenotype in Aethionema, we performed transcriptome analysis of the TUR and CYP accessions. Seeds were imbibed and kept in darkness (D) or under 100 µmol m−2 s−1 white light (WL) for 23 h, which was determined as the start point for the completion of germination (~1% seeds in the responsive populations with emerged radicles). It is important to note that CYP seeds are fully able to germinate if transferred to darkness after 23 h of light exposure (Fig. 1F). Only seeds with intact seed coats were sampled and used for the preparation of RNA libraries. Comparison of dark- and light-exposed samples revealed 168 differentially expressed genes in the TUR accession, comprising 51 up-regulated and 117 down-regulated genes in seeds kept in darkness compared with those kept in light (Fig. 3A; Supplementary Dataset S1). In the CYP accession, we found 214 differentially expressed genes, comprising 105 up-regulated and 109 down-regulated genes in seeds kept in darkness (Fig. 3A; Supplementary Dataset S2). Considering the close relation between the accessions, the overlap of 93 genes commonly differentially regulated in TUR and CYP was surprisingly small, while 75 genes were light-dependent only in TUR, and 121 only in CYP (Fig. 3A). This comparison, however, did not consider the genetic differences between the two accessions. We therefore compared the transcriptome of each condition between the TUR and CYP samples. In seeds kept in darkness, 564 genes were differentially expressed between the two accessions (Fig. 3B; Supplementary Dataset S3). The number of differentially expressed genes between light-exposed TUR and CYP samples was much higher (969), matching the expectation of a larger difference upon light exposure and a different physiological state regarding the capability for germination. Among the 969 genes, 613 were expressed at higher levels in TUR and 356 were expressed at higher levels in CYP (Fig. 3B; Supplementary Dataset S4). Nearly half of the genes (469) were found in the overlap between the two genotypes, indicating transcriptional differences independent of the light conditions. However, these genes might undergo further light regulation that could contribute to the phenotypes.
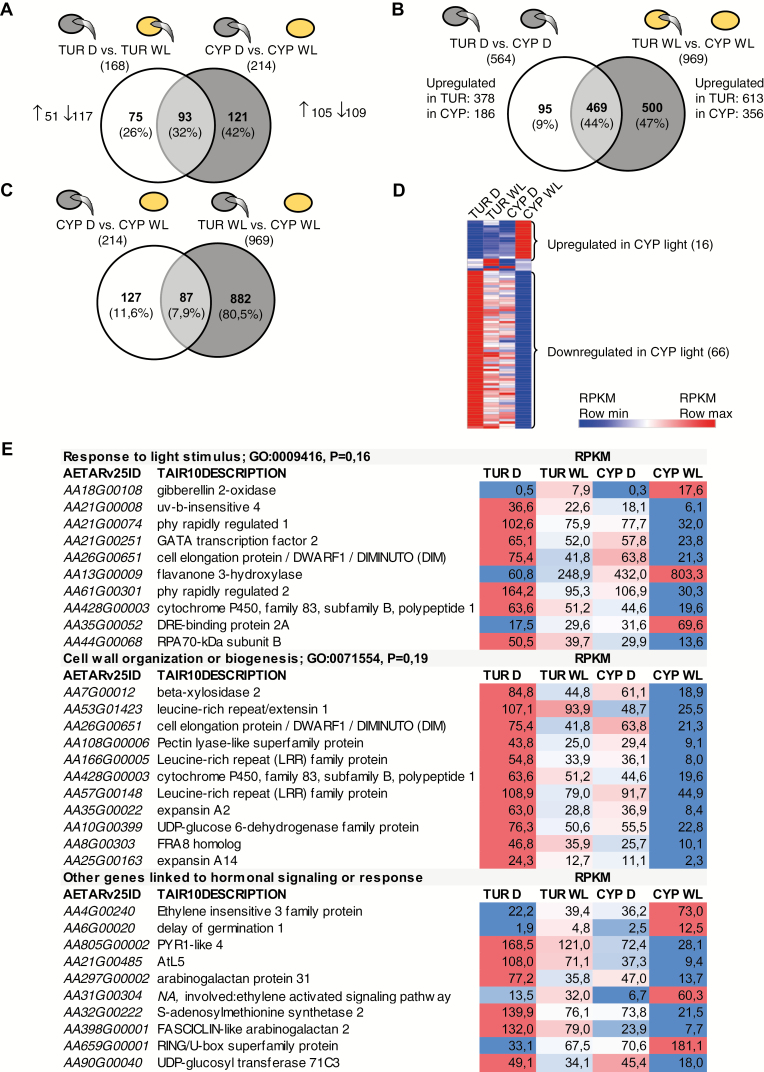
Fig. 3.
Transcriptome analysis of Aethionema TUR and CYP seeds by RNA-Seq. (A–C) Number of differentially expressed genes and Venn diagrams showing the proportion of overlapping genes in pairwise comparisons between TUR and CYP, and between exposure to darkness (D, grey seed icon), or white light (WL, yellow icon). Icons showing germinating seeds indicate the capability of samples for germination; note that the seeds had not yet germinated at the point of sampling. (D) Heatmap indicating the relative expression of the overlapping 87 genes in the four treatments. Coloring is based on reads per kilobase of transcript per million mapped reads (RPKM) values. A detailed list is provided in Supplementary Fig. S1. (E) Gene list of three selected GO terms with average RPKM values and relative expression differences, indicated by shades of blue (lowest RPKM value) or red (highest RPKM value).
To distinguish these possibilities, we hypothesized that the light-inhibited germination in CYP should be associated with genes that are (i) light-regulated in CYP seeds and (ii) differentially expressed in light-exposed TUR and CYP seeds. This selection would include genes that are possibly also light-regulated in TUR seeds, to a lesser extent than in CYP seeds, or genes whose induction/repression would be similar in both accessions but their absolute level results in different expression upon light induction. Both criteria were fulfilled for 87 genes (Fig. 3C, D; Supplementary Dataset S5, Supplementary Fig. S2). Of the 87 genes, 16 were up-regulated and 66 were down-regulated in light-exposed CYP seeds (Fig. 3D). Interestingly, 15 of the 87 genes were also significantly up- or down-regulated in light-exposed TUR seeds, and the direction of the changes in transcript levels was the same in both accessions (Fig. 3D; Supplementary Dataset S5, Supplementary Fig. S2). For 85 of the 87 genes we could identify the Arabidopsis orthologs (Supplementary Dataset S5). Based on the TAIR10 database description, 10 of these genes are linked to light stimuli and 11 to cell wall organization or biogenesis (Fig. 3E). Additionally, 10 genes are involved in hormonal signaling or response (Fig. 3E). Genes associated with ABA biosynthesis or degradation were not present among these genes (Fig. 3E; Supplementary Dataset S5). However, the most strongly up-regulated transcript [>50-fold induction in CYP seeds exposed to white light (CYP WL in Fig. 3)] was AA18G00108, encoding a gibberellin-2-oxidase. Phylogenetic analysis and synteny (Supplementary Fig. S3) of the genomic position confirmed that Aethionema AA18G00108 is the ortholog of Arabidopsis AtGA2ox3 (AT2G34555), which encodes a protein with C-19 gibberellin 2-beta-dioxygenase activity that is involved in the degradation of GA, and therefore expected to negatively influence germination.
Transcriptome analysis reveals differences in light-mediated hormonal responses
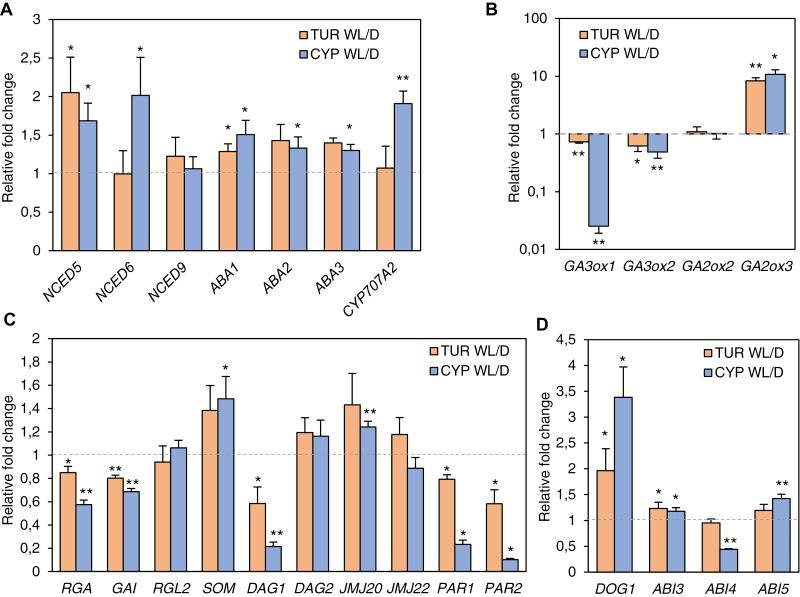
In most species, the balance of the hormones GA and ABA is an important component of light-regulated germination. We therefore tested individual changes in the transcript levels of genes involved in GA and ABA biosynthesis in dark- and white light-germinated TUR and CYP seeds. Overall, upon light exposure of seeds, we found slightly increased transcript levels of the genes encoding ABA biosynthetic enzymes and a decrease of the main GA biosynthetic enzymes (Fig. 4A, B). Expression of the Aethionema orthologs of ABA1, ABA2, and ABA3 was slightly but significantly up-regulated upon light exposure, in both the light-neutral TUR and the light-inhibited CYP seeds (Fig. 4A). NCED6 and NCED9, which encode the 9-cis epoxycarotenoid dioxygenase gating the catalysis of 9′-cis neoxanthin, the rate-limiting step of ABA synthesis, are known to be transcriptionally down-regulated in Arabidopsis upon red light induction (Seo et al., 2006, 2009; Oh et al., 2007). AearNCED5 was significantly up-regulated in both TUR and CYP accessions, and AearNCED6 only in CYP seeds kept in light. The level of AearNCED9 was similar in dark and light (Fig. 4A). In contrast, expression of the gene for the ABA-deactivating enzyme AearCYP7072A was also elevated, in this case matching the observations in Arabidopsis seeds (Fig. 4A) (Seo et al., 2006; Oh et al., 2007).
Fig. 4.
Analysis of the Aethionema light-regulated transcriptome network in the TUR and CYP accessions. (A–D) Quantitative RT–PCR for selected genes. The expression level under white light (WL) is presented as fold change relative to the average expression level in darkness (D), which is set to a value of 1 (indicated by the grey dashed line). Asterisks indicate significant differences between the WL and D expression level of a gene, based on the Welch test: *P<0.05, **P<0.01. Error bars represent SD (three independent biological replicates).
Light-induced GA accumulation in Arabidopsis seeds is mediated by the enhanced expression of GA biosynthetic enzymes, encoded by AtGA3ox1 and AtGA3ox2, and the decrease of the GA-deactivating gibberellin-2-oxidase encoded by AtGA2ox2 (Yamaguchi et al., 1998; Seo et al., 2006). We found a reciprocal situation in Aethionema seeds: the expression of both AearGA3ox1 and AearGA3ox2 was decreased on exposure to white light (Fig. 4B). In good agreement with the RNA-Seq results, the expression of AearGA2ox3 was increased upon illumination with white light in both accessions (Fig. 4B). Remarkably, the repression of AearGA3ox1 was considerably more pronounced in the light-inhibited CYP seeds than in TUR seeds (Fig. 4B). This also suggests that the TUR and CYP accessions might differ in the regulatory network upstream of GA-related genes. Therefore, we further investigated the transcriptome for regulation by factors known to be involved in light-mediated hormonal responses in seeds.
In Arabidopsis, upon light reception, the active Pfr form of phyB interacts with PIL5, a basic helix-loop-helix (bHLH) protein, facilitating its degradation by the 26S proteasome (Oh et al., 2004, 2007; Shen et al., 2008). Under conditions of darkness or far-red light, PIL5 mediates the stable expression of GAI and RGA, two genes encoding DELLA proteins, which are negative components of GA signaling (Oh et al., 2007; Piskurewicz et al., 2009). In parallel, PIL5 directly stimulates the expression of SOM (SOMNUS), a negative regulator of light-dependent germination that controls the expression of ABA and GA metabolic genes (Kim et al., 2008). Both pil5 and som mutants of Arabidopsis germinate in a light-insensitive manner (Oh et al., 2004; Kim et al., 2008). The link between SOM and the GA3ox1/2 genes is formed by the two jumonji-domain proteins JMJ20 and JMJ22 that are directly repressed by SOM and support germination by removing the repressing histone H4 arginine 3 methylation from GA3ox1/2, allowing their expression (Cho et al., 2012). Another negative regulator of seed germination is DAG1 (DOF AFFECTING GERMINATION1), which is under indirect positive control downstream of PIL5 and directly represses the transcription of GA3ox1 and DAG2, which was recently identified as a positive regulator of light-induced germination (Gabriele et al., 2010; Boccaccini et al., 2014; Santopolo et al., 2015). We found significant changes in the transcript levels of the Aethionema orthologs of RGA, GAI, SOM, DAG1, and JMJ20 in light-exposed seeds compared with those in the dark, either in both TUR and CYP accessions or only in CYP (Fig. 4C). The expression of DAG1 was decreased in light-exposed seeds of both accessions, similar to its response in Arabidopsis (Fig. 4C). PHYTOCHROME RAPIDLY REGULATED 1 and 2 (PAR1 and PAR2) were both down-regulated in seeds exposed to white light (Fig. 3E; Supplementary Dataset S5). In Arabidopsis, PAR1 and PAR2 are negative factors in shade avoidance (Roig-Villanova et al., 2007) and promote seedling de-etiolation under different light conditions, likely through interaction with PIF proteins (Zhou et al., 2014). Their role during seed germination has not been elucidated, although available transcriptome data suggest that PAR2 is repressed by PIL5 and up-regulated in seeds exposed to red light (Shi et al., 2013). Importantly, we also confirmed the RNA-Seq results that AearPAR1 and AearPAR2 are down-regulated in seeds exposed to white light (Figs 3E and 4C; AA21G00074 and AA61G00301).
Germination under unfavorable conditions can be avoided by the establishment of seed dormancy, which is strongly correlated with the key dormancy protein DOG1 (DELAY OF GERMINATION 1) (Bentsink et al., 2006, 2010; Footitt et al., 2011; Graeber et al., 2014; Kerdaffrec et al., 2016). The expression of the DOG1 gene is regulated by environmental signals and highly variable among Arabidopsis accessions (Chiang et al., 2011; Kendall et al., 2011; Finch-Savage and Footitt, 2017). Therefore, we tested whether the exposure of TUR or CYP seeds to light enhances DOG1 expression. AearDOG1 was indeed among the light-responsive genes up-regulated in CYP (Fig. 3E, AA6G00020). qRT–PCR data confirmed that DOG1 expression was significantly enhanced in both accessions in light-exposed seeds, but the increase was more pronounced in the light-inhibited CYP seeds (Fig. 4D). These results indicate that light can indeed influence the level of dormancy of light-responsive seeds. The expression of Aethionema orthologs of ABA-responsive transcription factors linked to dormancy, including ABI3, ABI4, and ABI5 (ABA INSENSITIVE 3, 4, 5) (Shu et al., 2013; Dekkers et al., 2016) showed significant, but only moderate, differences in light-exposed CYP seeds (Fig. 3D). Taking these findings together, (i) Aethionema uses similar key regulatory components to control germination as Arabidopsis, but (ii) the direction of change of transcript levels for several genes in light-exposed seeds are opposite in seeds of both Aethionema accessions compared with Arabidopsis, and (iii) at least two genes related to GA biosynthesis or degradation (GA3ox1 and GA2ox3) show a much more pronounced response in the light-inhibited CYP seeds compared with the light-neutral TUR seeds and might be responsible for the observed contrasting effect of light on germination.
The GA:ABA ratio decreases during light inhibition of Aethionema seed germination
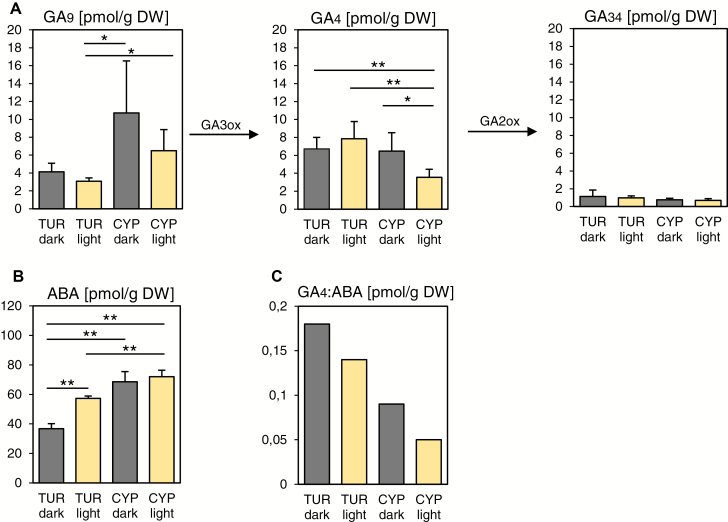
To test whether the differential expression of the GA- or ABA-related genes would indeed affect the levels of the respective bioactive hormones, we compared GA and ABA levels in TUR and CYP seeds during the different germination regimes. For induction of germination of Arabidopsis seeds, the most active form of GA is GA4, among other GAs studied (Derkx et al., 1994). The absolute GA4 hormone level was similar in both Aethionema accessions in seeds kept in darkness and in light-exposed TUR seeds (Fig. 5A). In contrast, and in good correlation with the light-induced repression of GA3ox1 expression (Fig. 4B), we observed the lowest level of the bioactive GA4 in CYP seeds exposed to light for 23 h (Fig. 5A; Supplementary Fig. S4). Moreover, the level of GA9, the biosynthetic precursor of GA4, was also significantly higher in light-exposed CYP seeds compared with TUR seeds, indicating that the GA9 → GA4 conversion might be less efficient in CYP seeds exposed to light (Fig. 5A). Given the enhanced expression of the catabolic GA2ox3 in light-exposed CYP seeds, we expected an increased level of the catabolic GA34 in CYP seeds. We did not observe this tendency during the 23 h period, but this might have been due to the slower turnover of GA (Fig. 5A). Absolute ABA levels were higher in CYP than TUR seeds under both regimes, in agreement with the later onset of CYP germination in the dark compared with TUR and significantly higher ABA levels in light-exposed CYP seeds compared with light-exposed TUR (Fig. 5B).
Fig. 5.
Decreased GA:ABA hormone ratio in light-exposed Aethionema CYP seeds. Hormone concentrations [in pmol g–1 dry weight (DW)] of GA9 precursor, bioactive GA4, and GA34 catabolite (A) and ABA (B) are shown for dark-exposed (grey columns) and light-exposed (cream columns) TUR and CYP seeds. (C) Ratio of the averages from GA4 and ABA measurements. Significance was tested using the Welch test in pairwise comparisons; asterisks indicate significant differences: *P<0.05, **<0.01. Error bars represent SD (five independent replicates). Values for other GA metabolites are presented in Supplementary Fig. S4.
As seed germination is determined by the balance of the two antagonistically acting hormones, we calculated the ratio of average GA and ABA levels. Interestingly, the GA:ABA ratio decreased in both accessions under light (Fig. 5C), but to a much larger extent in light-exposed CYP seeds, which had the lowest GA:ABA ratio of all. This indicates a threshold for the hormonal control below which germination of CYP seeds under continuous light exposure is not possible.
GA and ABA are involved in light inhibition of CYP seed germination
In Arabidopsis, germination of far-red-exposed seeds can be rescued by removal of the seed coat and the endosperm layer, as these extraembryonal tissues release ABA in response to far-red light, thereby inhibiting germination (Lee et al., 2012; Yan et al., 2014). Therefore, we tested whether the light inhibition of germination in Aethionema CYP seeds was also mediated by the seed coat and endosperm. Indeed, after mechanical removal of the extraembryonal tissue 24 h after imbibition under light exposure, development of CYP and TUR seedlings was similar, and 100% of the seedlings grew normally, even under continuous light (Fig. 6A). These data indicate that although Arabidopsis and Aethionema CYP seeds respond differently to light, the role of the seed coat and endosperm, and likely the involvement of ABA, appear similar.
Germination of light-requiring lettuce seeds in the dark or under far-red light could also be rescued by the addition of norflurazon [4-chloro-5-methylamino-2-(3-trifluoromethylphenyl)pyridazin-3-one] (Widell et al., 1981). Fluridone [1-methyl-3-phenyl-5-(3-trifluoromethyl-phenyl)-4-(1H)-pyridinone] restored the germination of lettuce and other seeds at suboptimal temperatures (Yoshioka et al., 1998; Debeaujon and Koornneef, 2000; Argyris et al., 2008). Both chemicals are inhibitors of carotenoid biosynthesis, which is required for de novo ABA synthesis (Bartels and Watson, 1978). We applied fluridone and norflurazon to both Aethionema accessions under continuous light exposure. CYP seed germination was completely rescued (Fig. 6B), indicating that de novo ABA synthesis induced by light is an important component of the negative control of germination in light-exposed CYP seeds.
The difference in the GA:ABA ratio between TUR and CYP seeds exposed to the dark (Fig. 5C) suggested that exogenously applied ABA might inhibit the germination of CYP seeds in darkness, which is otherwise optimal for the germination of both accessions. When this was tested by applying increasing concentrations of ABA, germination of CYP seeds was inhibited by 0.3 µM ABA while for TUR seeds 1 µM ABA was needed to produce the same level of inhibition (Fig. 6C).
Finally, we tested whether externally applied GA could overcome the inhibitory effect of light on germination. The addition of 10 µM GA4 + 7 allowed CYP seeds to germinate under continuous light, although the germination was slower than that of seeds treated with fluridone (Fig. 6D). These data suggest that GA and ABA are indeed involved in the control of germination, as in other plants. However, the signaling pathways downstream of light reception to the transcriptional control of these two key hormones must be antipodal to those in Arabidopsis.
Discussion
Ecological significance of light-regulated germination
The first observations on photoblastic differences were reported more than a century ago (Kinzel, 1913), and since then, several species have been described to have a light-inhibited or light-neutral germination phenotype (Grime et al., 1981). Despite this, research in the past decades about the molecular control of seed germination focused nearly exclusively on the light-requiring germination of Arabidopsis. Here, we present an initial physiological and molecular characterization of light-neutral and light-inhibited germination in two accessions of Aethionema, another Brassicaceae species.
The light requirement for seed germination is often considered to be a depth-sensing strategy associated with small seed size (Grime et al., 1981; Fenner and Thompson, 2005). As light penetrates only a few millimeters into the soil, light dependence of small seeds ensures that the elongating hypocotyl will reach the surface before its resources are exhausted (Woolley and Stoller, 1978; Fenner and Thompson, 2005). Additionally, the light quality, sensed by the phytochrome photoreceptors as the ratio of red:far-red wavelengths, provides information about the leaf canopy, as leaves absorb more red than far-red light. Therefore, the minimum red:far-red ratio required for seed germination of a certain species may determine the optimal season when competition is reduced, as was shown for Cirsium palustre (Pons, 1984).
In contrast, light-inhibited germination is plausible in species originating from open, arid, or semi-arid habitats where high light intensity is likely coupled with drought conditions, which could be unfavorable for young seedlings (Thanos et al., 1991; Lai et al., 2016). These conditions occur at many original habitats of A. arabicum. Our data indicate that the light-inhibitable germination of the CYP accession might be a photoperiod-sensing mechanism, as the seeds germinate well under short-day but not long-day conditions. Short-day conditions correspond to the day length of early spring days, when Aethionema germinates in its natural habitat. As the average lifespan of Aethionema is around 4 months, germination in early spring is necessary in order to complete the life cycle and seed production before the dry and warm season. Similar to the CYP accession of Aethionema, seeds of the light-inhibited garden variant of Nemophila insignis germinate preferentially in short-day conditions (Black and Wareing, 1960; Chen, 1968). Seasonal adaptation of germination via opposite photoperiod sensitivity was described for arctic tundra species, which are inhibited by short days and prefer to germinate under long days, corresponding to the short summer season in Alaska (Densmore, 1997). Although only these few examples are known, photoperiod dependence of seed germination is likely more common for plants in habitats where optimal timing of germination is crucial.
Germination in most of the examined Aethionema species was at least partially inhibited by light, whereas the Turkish accession of Aethionema (TUR) and one A. heterocarpum accession germinated independently of light. The occurrence of both phenotypes among close relatives in the Aethionemeae indicates that it is likely an adaptive trait that appeared more than once during evolution, although the exact environmental cues that favor light-neutral germination is unknown. Given the limited number of Aethionema accessions available that have been propagated under controlled conditions, it is too early to conclude which of the phenotypes is ancestral. Based on the habitats of most Aethionema species and the identified transcriptome changes of key regulatory genes, in the same direction but to different degrees, in TUR and CYP, it is tempting to speculate that light-inhibited germination is the ancestral mechanism that has been desensitized in some instances. Collection and amplification of seed material and analysis of further Aethionema species and accessions for which detailed phylogenetic data are available (Mohammadin et al., 2017) are expected to provide an answer to this question.
Light-dependent versus light-inhibited germination control
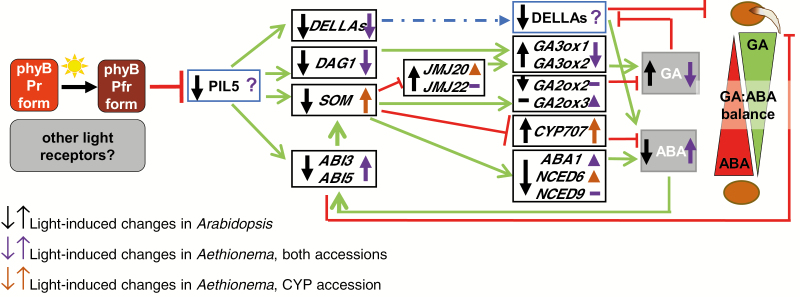
The species for which light inhibition of seed germination have been previously described (Chen, 1968, 1970; Botha and Small, 1988; Thanos et al., 1991) are phylogenetically distant from the model plants lettuce and Arabidopsis, which show light-dependent germination. Some molecular aspects of light-inhibited germination have been investigated for monocotyledonous plants (Barrero et al., 2012; Hoang et al., 2014), and in both cases were clearly restricted to blue light. The comparisons within the triangle of closely related species with different germination phenotypes—light-requiring Arabidopsis, light-neutral Aethionema TUR, and Aethionema CYP that is inhibited by the full spectrum of visible light—may allow us to understand the mechanistic and evolutionary divergence of the light-controlled signaling network that induces germination. Previous studies in numerous species (Finch-Savage and Leubner-Metzger, 2006) together with the data presented here leave no doubt about the central role of ABA and GA in the inhibition and stimulation, respectively, of germination. The positive, essential stimulus of light in Arabidopsis and its negative, blocking role in Aethionema CYP are expected to reflect a fundamental and qualitative difference between light reception and hormonal control. Although many other factors, including other hormones, are known to modulate germination (Argyris et al., 2008; Linkies and Leubner-Metzger, 2012; Meng et al., 2016), the antipodal changes in transcript levels in Aethionema upon light exposure for some of the same ABA/GA key regulatory components as in Arabidopsis indicate a major source of the difference in this signaling pathway (Fig. 7). Among these components are AearSOM, AearABI3, AearABI5, AearABA1, AearNCED6, AearGA3ox1, AearGA3ox2, and AearGA2ox3. However, the expression of many other genes responds to light in a similar fashion in Aethionema and Arabidopsis (e.g. RGA, GAI, DAG1, CYP707A2, and JMJ20), indicating that the light response is partially conserved (Fig. 7). In Arabidopsis, PIL5/PIF1, a key regulator in the light-induced transcriptional cascade, undergoes rapid protein degradation (Shen et al., 2008). As the antibody to Arabidopsis PIL5/PIF1 did not recognize the Aethionema protein from the orthologous gene (AA33G00286) (our unpublished results), we were unable to test its light-responsive protein degradation in Aethionema seeds. However, it is remarkable that the direct downstream target genes (DAG1, SOM, DELLAs, and ABI5) are either up- or down-regulated in Aethionema, whereas their transcriptional repression by light is rather uniform in Arabidopsis (Fig. 7). A study based on chromatin immunoprecipitation in Arabidopsis with the PIL5/PIF1 antibody and microarray data revealed 166 genes that are under the direct control of PIL5/PIF1 (Oh et al., 2009). The Aethionema orthologs of the direct PIL5 target genes that could be identified (132 out of the 166) were found to have relatively stable and light-independent expression in Aethionema seeds; only eight genes in TUR and seven genes in CYP showed more than 2-fold changes in either direction (Supplementary Dataset S6). Therefore, one possible divergence between Arabidopsis and Aethionema might be in the regulation of PIL5/PIF1 protein activity or stabilization. The germination of Arabidopsis pil5 mutant seeds in darkness and far-red light further indicates that PIL5/PIF1 is the most upstream element in the network that is possibly associated with germination in the dark.
Fig. 7.
Antipodal transcriptional changes in Arabidopsis and Aethionema of key hormone regulatory genes. Summary of light-regulated changes in transcript levels in seeds, based on published data for Arabidopsis (after Stawska and Oracz 2015) and with the data for Aethionema presented in this study. Blue boxes indicate proteins whose stability is regulated by light. Black boxes indicate genes whose expression is light-regulated. Changes in expression upon light exposure are indicated by black (Arabidopsis), purple (Aethionema, both accessions), and orange (Aethionema, CYP accession only) arrows. The role of other photoreceptors in Aethionema seed germination is not yet known. As indicated by question marks, the stability of PIL5 and DELLA proteins is unknown in Aethionema.
Light-independent versus light-inhibited germination control
Remarkably, among the 87 genes that were light-responsive in CYP and differentially regulated between the TUR and CYP accessions, 15 nevertheless showed the same direction of change in transcript levels in both accessions. One possible explanation for this observation might be a reduced sensitivity of germination of TUR seeds to light. In Arabidopsis, screens for light-hyposensitive mutants have often identified PHYB or PHYA mutations, indicating that the primary reason for the hyposensitive response may be variations within the phytochrome protein. As in Arabidopsis, there are five phytochromes in Aethionema, which are highly conserved (Supplementary Fig. S5A). The protein sequences of PHYB, PHYC, and PHYD are identical in the CYP and TUR accessions, while the PHYA and PHYE proteins harbor a few missense single nucleotide polymorphisms (SNPs) (Supplementary Fig. S5B–F). However, the second, light-inhibited accession from Turkey (KM2397) shares most of the same SNPs with the light-neutral TUR accession. It is therefore unlikely that allelic variations of phytochromes are responsible for the different responses (Supplementary Fig. S5B–F). Similarly, there are no SNPs in the PIL5/PIF1 coding sequences that would cause non-synonymous amino acid changes and that diverge between the light-neutral TUR and the two light-inhibited accessions (Supplementary Fig. S6). A detailed analysis of phytochrome actions may help to understand and identify the photoreceptors involved in light-inhibited germination.
As the qRT–PCR data indicated, many genes of the light-regulated network are differentially expressed either slightly or strongly in TUR and CYP seeds under dark and light conditions, resulting in substantial differences in AearGA3ox1 and AearGA2ox2 expression and lower levels of GA4 in CYP seeds under light. The differential GA:ABA ratio is likely determined by more than one upstream event early in the transcriptional cascade. Our data show that the PAR1 and PAR2 genes are significantly differentially expressed in TUR and CYP seeds under dark and light conditions. PAR1 and PAR2 both encode bHLH proteins, but do not possess DNA binding activity and are involved in the shade avoidance response (Wray et al., 2003; Roig-Villanova et al., 2007). Although their precise mechanism of action and role in seed germination is still unknown, it has been speculated that they form heterodimers with other bHLH proteins, such as PIFs, and modulate their activity as transcriptional cofactors (Roig-Villanova et al., 2007). Therefore, the differential expression of AearPAR1 and AearPAR2 in TUR and CYP seeds might play a role in the regulation of PIL5/PIF1 activity, influencing the network downstream of PIL5/PIF1.
While our data did not identify a unique point of divergence in the molecular control of light over seed germination between the investigated Aethionema accessions, the identified natural variation within the genus, and the phylogenetic relationship with the conversely responding Arabidopsis, provide great opportunities to elucidate the mechanism of an ecologically important but underinvestigated trait. Based on the current evidence, the basic components seem to be conserved but connected in a different cascade of events. Growth conditions, genome size, and generation time of Aethionema are similar to those of Arabidopsis, allowing for future forward mutant screens and genetic association studies.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Germination of dimorphic seed types in response to light.
Fig. S2. Heatmap of all 87 genes light-regulated in Aethionema arabicum CYP seeds and differentially expressed in light-exposed TUR and CYP seeds based on RPKM values.
Fig. S3. Identification of the Arabidopsis orthologue of Aethionema AA18G00108 as GA2ox3.
Fig. S4. Accumulation of GA forms in Aethionema arabicum TUR and CYP seeds under dark and light conditions.
Fig. S5. Identification and alignments of phytochromes in Aethionema arabicum.
Fig. S6. Alignment of the PIL5/PIF1 protein sequence of three Aethionema arabicum accessions.
Table S1. Information about the geographic origin of Aethionema arabicum accessions.
Table S2. List of primers used for quantitative RT–PCR analysis.
Table S3. List of Aethionema accession numbers used for this study.
Dataset S1. List of differentially expressed Aethionema arabicum genes in TUR Dark versus TUR Light.
Dataset S2. List of differentially expressed Aethionema arabicum genes in CYP Dark versus CYP Light.
Dataset S3. List of differentially expressed Aethionema arabicum genes in CYP Dark versus TUR Dark.
Dataset S4. List of differentially expressed Aethionema arabicum genes in CYP Light versus TUR Light.
Dataset S5. List of common differentially expressed Aethionema arabicum genes in CYP Light versus TUR Light and TUR Dark versus TUR Light.
Dataset S6. List of target genes of Arabidopsis PIL5/PIF1 and transcriptional changes of orthologues in the Aethionema experiments.
Dataset S7: List of plant species for which protein sequences were considered for phylogenetic tree constructions.
Acknowledgements
This work is part of the European Research Area Network for Coordinating Action in Plant Sciences (ERA-CAPS) ‘SeedAdapt’ consortium project (https://www.seedadapt.eu/). We thank all members for fruitful cooperation and discussion. We are especially grateful to Eric M. Schranz, Klaus Mummenhof, and Setareh Mohammadin for providing various Aethionema seed stocks. We also thank the staff of the Vienna BioCenter Core Facilities GmbH (VBCF), a member of Vienna BioCenter (VBC), Austria, especially the Plant Sciences Facility for growth of the plants and the wavelength-specific light experiments, and the Next Generation Sequencing Facility for generating the RNA-Seq data. We thank Nicole Lettner and Sarhan Khalil for technical support. We further acknowledge critical reading of the manuscript by J. Matthew Watson, Frederic Berger, Peter Hedden, and the anonymous reviewers. The work was funded by the Austrian Science Fund (FWF) to OMS (FWF I1477) and to ZM (FWF I3979), the Deutsche Forschungsgemeinschaft (DFG) to SAR (RE1697/8-1), the Biotechnology and Biological Sciences Research Council (BBSRC) to GLM (BB/M00192X/1), a Natural Environment Research Council (NERC) Doctoral Training Grant to WA (NE/L002485/1), the Czech Grant Agency to DT (18-10349S), and the European Regional Development Fund Project ‘Centre for Experimental Plant Biology’ (CZ.02.1.01/0.0/0.0/16_019/0000738) to DT and MS.
Author contributions
ZM, KG, MS, SAR, GLM, and OMS planned and designed the research; ZM, KG, WA, DT, and VT performed the experiments; ZM, KG, PW, KKU, WA, CG, DT, VT, MS, SAR, GLM, and OMS analyzed and interpreted the data; ZM, GLM, and OMS wrote the paper. All authors approved the submitted version.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris J, Dahal P, Hayashi E, Still DW, Bradford KJ. 2008. Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiology 148, 926–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Downie AB, Xu Q, Gubler F. 2014. A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. The Plant Cell 26, 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Jacobsen JV, Talbot MJ, White RG, Swain SM, Garvin DF, Gubler F. 2012. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytologist 193, 376–386. [DOI] [PubMed] [Google Scholar]
- Bartels PG, Watson CW. 1978. Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Science 26, 198–203. [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, et al. 2010. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proceedings of the National Academy of Sciences, USA 107, 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. 2006. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proceedings of the National Academy of Sciences, USA 103, 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Bradford K, Hilhorst H, Nonogaki H. 2013. Seeds. Physiology of development, germination and dormancy, 3rd edn. New York: Springer. [Google Scholar]
- Black M, Wareing PF. 1960. Photoperiodism in the light-inhibited seed of Nemophila insignis. Journal of Experimental Botany 11, 28–39. [Google Scholar]
- Boccaccini A, Santopolo S, Capauto D, Lorrai R, Minutello E, Belcram K, Palauqui JC, Costantino P, Vittorioso P. 2014. Independent and interactive effects of DOF affecting germination 1 (DAG1) and the Della proteins GA insensitive (GAI) and Repressor of ga1-3 (RGA) in embryo development and seed germination. BMC Plant Biology 14, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK. 1952. A reversible photoreaction controlling seed germination. Proceedings of the National Academy of Sciences, USA 38, 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha FC, Small JGC. 1988. The germination response of the negatively photoblastic seeds of Citrullus lanatus to light of different spectral compositions. Journal of Plant Physiology 132, 750–753. [Google Scholar]
- Casal JJ, Sanchez RA. 1998. Phytochromes and seed germination. Seed Science Research 8, 3. [Google Scholar]
- Casal JJ, Sanchez RA, Botto JF. 1998. Modes of action of phytochromes. Journal of Experimental Botany 49, 127–138. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116. [Google Scholar]
- Chen SSC. 1968. Germination of light-inhibited seed of Nemophila insignis. American Journal of Botany 55, 1177–1183. [Google Scholar]
- Chen SS. 1970. Influence of factors affecting germination on respiration of Phacelia tanacetifolia seeds. Planta 95, 330–335. [DOI] [PubMed] [Google Scholar]
- Chiang GC, Bartsch M, Barua D, Nakabayashi K, Debieu M, Kronholm I, Koornneef M, Soppe WJ, Donohue K, De Meaux J. 2011. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Molecular Ecology 20, 3336–3349. [DOI] [PubMed] [Google Scholar]
- Cho JN, Ryu JY, Jeong YM, Park J, Song JJ, Amasino RM, Noh B, Noh YS. 2012. Control of seed germination by light-induced histone arginine demethylation activity. Developmental Cell 22, 736–748. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. 2004. The Jalview Java alignment editor. Bioinformatics 20, 426–427. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M. 2000. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiology 122, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, He H, Hanson J, Willems LA, Jamar DC, Cueff G, Rajjou L, Hilhorst HW, Bentsink L. 2016. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. The Plant Journal 85, 451–465. [DOI] [PubMed] [Google Scholar]
- Densmore R. 1997. Effect of day length on germination of seeds collected in Alaska. American Journal of Botany 84, 274. [PubMed] [Google Scholar]
- Derkx MPM, Verneer E, Karssen CM. 1994. Gibberellins in seeds of Arabidopsis thaliana: biological activities, identification and effects of light and chilling on endogenous levels. Plant Growth Regulation 15, 223–234. [Google Scholar]
- Fenner M, Thompson K. 2005. The ecology of seeds. Annals of Botany 97, 151–152. [Google Scholar]
- Finch-Savage WE, Footitt S. 2017. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. Journal of Experimental Botany 68, 843–856. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. 2006. Seed dormancy and the control of germination. New Phytologist 171, 501–523. [DOI] [PubMed] [Google Scholar]
- Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE. 2011. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proceedings of the National Academy of Sciences, USA 108, 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. 2011. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Science 16, 108–116. [DOI] [PubMed] [Google Scholar]
- Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. 2010. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. The Plant Journal 61, 312–323. [DOI] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Steinbrecher T, et al. 2014. DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proceedings of the National Academy of Sciences, USA 111, E3571–E3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP, Mason G, Curtis AV, Rodman J, Band SR. 1981. A comparative study of germination characteristics in a local flora. Journal of Ecology 69, 1017–1059. [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, et al. 2013. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nature Genetics 45, 891–898. [DOI] [PubMed] [Google Scholar]
- Hoang HH, Sechet J, Bailly C, Leymarie J, Corbineau F. 2014. Inhibition of germination of dormant barley (Hordeum vulgare L.) grains by blue light as related to oxygen and hormonal regulation. Plant, Cell & Environment 37, 1393–1403. [DOI] [PubMed] [Google Scholar]
- Hradecká V, Novák O, Havlícek L, Strnad M. 2007. Immunoaffinity chromatography of abscisic acid combined with electrospray liquid chromatography-mass spectrometry. Journal of Chromatography 847, 162–173. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall SL, Hellwege A, Marriot P, Whalley C, Graham IA, Penfield S. 2011. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. The Plant Cell 23, 2568–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdaffrec E, Filiault DL, Korte A, Sasaki E, Nizhynska V, Seren U, Nordborg M. 2016. Multiple alleles at a single locus control seed dormancy in Swedish Arabidopsis. eLife 5, e22502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G. 2008. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. The Plant Cell 20, 1260–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzel W. 1913. Frost und Licht als beeinflussende Kräfte bei der Samenkeimung. Stuttgart: Ulmer. [Google Scholar]
- Koller D. 1956. Germination-regulating mechanisms in some desert seeds. Ecology 37, 430–433. [Google Scholar]
- Koller D, Negbi M. 1959. The regulation of germination in Oryzopsis miliacea. Ecology 40, 20–36. [Google Scholar]
- Lai LM, Chen LJ, Jiang LH, Zhou JH, Zheng YR, Shimizu H. 2016. Seed germination of seven desert plants and implications for vegetation restoration. AoB Plants 8, plw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Tureckova V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L. 2012. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes and Development 26, 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenser T, Graeber K, Cevik ÖS, et al. 2016. Developmental control and plasticity of fruit and seed dimorphism in Aethionema arabicum. Plant Physiology 172, 1691–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WQ, Khan MA, Yamaguchi S, Liu XJ. 2015. Hormonal and environmental regulation of seed germination in salt cress (Thellungiella halophila). Plant Growth Regulation 76, 41–49. [Google Scholar]
- Linkies A, Leubner-Metzger G. 2012. Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Reports 31, 253–270. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YJ, Chen F, Shuai HW, et al. 2016. Karrikins delay soybean seed germination by mediating abscisic acid and gibberellin biogenesis under shaded conditions. Scientific Reports 6, 22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadin S, Peterse K, van de Kerke SJ, Chatrou LW, Dönmez AA, Mummenhoff K, Pires JC, Edger PP, Al-Shehbaz IA, Schranz ME. 2017. Anatolian origins and diversification of Aethionema, the sister lineage of the core Brassicaceae. American Journal of Botany 104, 1042–1054. [DOI] [PubMed] [Google Scholar]
- Mohammadin S, Wang W, Liu T, et al. 2018. Genome-wide nucleotide diversity and associations with geography, ploidy level and glucosinolate profiles in Aethionema arabicum (Brassicaceae). Plant Systematics and Evolution 304, 619–630. [Google Scholar]
- Negbi M, Koller D. 1964. Dual action of white light in the photocontrol of germination of Oryzopsis miliacea. Plant Physiology 39, 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. 2009. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. The Plant Cell 21, 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. 2004. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. The Plant Cell 16, 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G. 2007. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. The Plant Cell 19, 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Turecková V, Lacombe E, Lopez-Molina L. 2009. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. The EMBO Journal 28, 2259–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons TL. 1984. Possible significance of the light requirement of Cirsium palustre seeds after dispersal in ash coppice. Plant, Cell & Environment 7, 263–268. [Google Scholar]
- Rittenberg D, Foster GL. 1940. A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. Journal of Biological Chemistry 133, 737–744. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF. 2007. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. The EMBO Journal 26, 4756–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. 1999. Twilight zone of protein sequence alignments. Protein Engineering 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Santopolo S, Boccaccini A, Lorrai R, Ruta V, Capauto D, Minutello E, Serino G, Costantino P, Vittorioso P. 2015. DOF AFFECTING GERMINATION 2 is a positive regulator of light-mediated seed germination and is repressed by DOF AFFECTING GERMINATION 1. BMC Plant Biology 15, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, et al. 2006. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. The Plant Journal 48, 354–366. [DOI] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S. 2009. Interaction of light and hormone signals in germinating seeds. Plant Molecular Biology 69, 463–472. [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. 2008. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. The Plant Cell 20, 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW. 2013. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. The Plant Cell 25, 3770–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M. 1994. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiology 104, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire W, Klein WH, Elstad VB. 1961. Action spectra of photomorphogenic induction and photoinactivation of germination in Arabidopsis thaliana. Plant & Cell Physiology 2, 63–69. [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH. 2016. Two faces of one seed: hormonal regulation of dormancy and germination. Molecular Plant 9, 34–45. [DOI] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q. 2013. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genetics 9, e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawska M, Oracz K. 2015. Network of signal transduction pathways mediated by phytochromes, cryptochromes and regulators of growth and development in seed biology. Postepy Biologii Komorki 42, 687–706. [Google Scholar]
- Takaki M. 2001. New proposal of classification of seeds based on forms of phytochrome instead of photoblastism. Revista Brasileira de Fisiologia Vegetal 13, 104–108. [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. 2011. Differential expression in RNA-seq: a matter of depth. Genome Research 21, 2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos CA, Georghiou K, Douma DJ, Marangaki CJ. 1991. Photoinhibition of seed germination in Mediterranean maritime plants. Annals of Botany 68, 469–475. [Google Scholar]
- Turecková V, Novák O, Strnad M. 2009. Profiling ABA metabolites in Nicotiana tabacum L. leaves by ultra-performance liquid chromatography–electrospray tandem mass spectrometry. Talanta 80, 390–399. [DOI] [PubMed] [Google Scholar]
- Urbanová T, Tarkowská D, Novák O, Hedden P, Strnad M. 2013. Analysis of gibberellins as free acids by ultra performance liquid chromatography–tandem mass spectrometry. Talanta 112, 85–94. [DOI] [PubMed] [Google Scholar]
- Vandelook F, Newton RJ, Carta A. 2018. Photophobia in Lilioid monocots: photoinhibition of seed germination explained by seed traits, habitat adaptation and phylogenetic inertia. Annals of Botany 121, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley JT, Stoller EW. 1978. Light penetration and light-induced seed germination in soil. Plant Physiology 61, 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. 2003. The evolution of transcriptional regulation in eukaryotes. Molecular Biology and Evolution 20, 1377–1419. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T. 1998. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. The Plant Cell 10, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan DW, Duermeyer L, Leoveanu C, Nambara E. 2014. The functions of the endosperm during seed germination. Plant and Cell Physiology 55, 1521–1533. [DOI] [PubMed] [Google Scholar]
- Yoshioka T, Endo T, Satoh S. 1998. Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant and Cell Physiology 39, 307–312. [Google Scholar]
- Zhou P, Song M, Yang Q, et al. 2014. Both PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiology 164, 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.