Competitive binding and complex formation of MBW proteins has a functional relevance for anthocyanidin production and trichome development across a range of different species, which can be explained by changes in one amino acid.
Keywords: Arabidopsis thaliana, Arabis alpina, competitive complex formation, evolution, Gossypium hirsutum, MBW complex, Petunia hybrida, Zea mays
Abstract
A protein complex consisting of a MYB, basic Helix-Loop-Helix, and a WDR protein, the MBW complex, regulates five traits, namely the production of anthocyanidin, proanthocyanidin, and seed-coat mucilage, and the development of trichomes and root hairs. For complexes involved in trichome and root hair development it has been shown that the interaction of two MBW proteins can be counteracted by the respective third protein (called competitive complex formation). We examined competitive complex formation for selected MBW proteins from Arabidopsis thaliana, Arabis alpina, Gossypium hirsutum, Petunia hybrida, and Zea mays. Quantitative analyses of the competitive binding of MYBs and WDRs to bHLHs were done by pull-down assays using ProtA- and luciferase-tagged proteins expressed in human HEC cells. We found that some bHLHs show competitive complex formation whilst others do not. Competitive complex formation strongly correlated with a phylogenetic tree constructed with the bHLH proteins under investigation, suggesting a functional relevance. We demonstrate that this different behavior can be explained by changes in one amino acid and that this position is functionally relevant in trichome development but not in anthocyanidin regulation.
Introduction
Flavonoid biosynthesis is controlled by a gene regulatory network involving MYB factors, basic Helix-Loop-Helix factors (bHLH), and a WDR protein, referred to as MBW (Ramsay and Glover, 2005; Xu et al., 2015; Zhang and Schrader, 2017). This function of the network is evolutionarily conserved in plants and is found in angiosperms (Ramsay and Glover, 2005; Xu et al., 2015; Zhang and Schrader, 2017) and also recently in gymnosperms (Nemesio-Gorriz et al., 2017). Most plant MYB proteins belong to the R2R3 class of MYBs that contain two DNA-binding R motives (Braun and Grotewold, 1999; Kranz et al., 2000). bHLH proteins have been identified in all eukaryotes and are characterized by a DNA-binding basic domain that is 13–17 amino acids long and two amphipathic alpha helices separated by a loop, which are considered to mediate protein interactions (Feller et al., 2011). WDR proteins have several WD40 domain repeats that are important for protein–protein interactions (van Nocker and Ludwig, 2003). The three protein classes form a ternary complex in which both the R2R3 MYB and the WDR proteins bind to the bHLH protein (Payne et al., 2000; Zhang et al., 2003; Zimmermann et al., 2004; Feller et al., 2006). Analysis of protein fragments suggests that the WDR and MYB proteins bind to different regions of the bHLH protein (Payne et al., 2000). The regulation of flavonoid production by MBW proteins is likely to be the most ancient function of these complexes and it is thought that additional functions have evolved from gene duplication and subsequent diversification (Serna and Martin, 2006). In the rosid clade, in addition to the anthocyanidin and proanthocyanidin pathways, distinct MBW protein combinations also regulate three other traits, namely the development of trichomes and root-hairs, and the production of seed-coat mucilage. This is evident from mutants in the WDR gene TTG1 in various species including Arabidopsis thaliana, Arabis alpina, and Matthiola incana (Koornneef, 1981; Dressel and Hemleben, 2009; Chopra et al., 2014).
The function of the MBW proteins is best studied in Arabidopsis. Here, TTG1 regulates the production of anthocyanidin, proanthocyanidin (seed color), and seed-coat mucilage, and the development of trichomes and root hairs (Koornneef, 1981). The regulation of each of these traits is controlled in combination with specific bHLH and MYB genes. The bHLH genes include GL3 (Hülskamp et al., 1994; Payne et al., 2000; Bernhardt et al., 2003; Zhang et al., 2003; Gonzalez et al., 2008; Feyissa et al., 2009), EGL3 (Bernhardt et al., 2003; Zhang et al., 2003; Gonzalez et al., 2008), MYC1 (Symonds et al., 2011; Bruex et al., 2012), and TT8 (Nesi et al., 2000; Zhang et al., 2003; Baudry et al., 2006). Each bHLH gene is involved in the regulation of two or more traits in a partially redundant manner. The R2R3 MYB genes specifically control one trait each, with GL1 and MYB23 controlling trichome development (Herman and Marks, 1989; Marks and Feldmann, 1989; Oppenheimer et al., 1991; Kirik et al., 2005), WER controlling root hair development (Lee and Schiefelbein, 1999), MYB61 controlling seed-coat mucilage (Penfield et al., 2001; Zhang et al., 2003), TT2 proanthocyanidin controlling biosynthesis (Baudry et al., 2004; Gonzalez et al., 2009), and PAP1 and PAP2 controlling anthocyanidin production (Borevitz et al., 2000; Teng et al., 2005; Gonzalez et al., 2008).
It has been shown for TTG1 and most of the MYB proteins that they interact with bHLH proteins in yeast two-hybrid assays and/or LUMIER (luminescence-based mammalian interactome) pull-down experiments (Payne et al., 2000; Zhang et al., 2003; Baudry et al., 2004, 2006; Zhao et al., 2008; Symonds et al., 2011; Pesch et al., 2013), which suggests that various combinations of ternary complexes can be formed. However, it is likely that this is still a simplified view as the bHLH proteins can dimerize, leading to higher-order complexes (Payne et al., 2000; Spelt et al., 2002; Bernhardt et al., 2003; Zhang et al., 2003; Feller et al., 2006; Quattrocchio et al., 2006; Kong et al., 2012; Albert et al., 2014; Shangguan et al., 2016). The concept of ternary complex formation has been challenged by the demonstration that GL1 or WER can reduce the interaction of GL3 with TTG1 and that TTG1 can counteract the interaction of GL3 with GL1 (Pesch et al., 2015). Moreover, this study also showed that GL1 can counteract the GL3- and TTG1-induced transcriptional regulation of the TRY promoter, and that TTG1 can repress the activation of the CPC promoter by GL3 and GL1. Together, these data suggest both the presence of alternative dimers and their functional differentiation.
These findings raise the question as to whether alternative complex formation is a general property of all bHLH and MYB proteins in the pathways under consideration or whether this protein interaction behavior is specific to some combinations—and, if so, whether this is a property of the bHLH or MYB proteins. To address these questions, we systematically tested all Arabidopsis bHLH MYB combinations for competitive complex formation. In addition, we analysed the competition behavior for selected homologs from Arabis alpina, cotton (Gossypium hirsutum), petunia (Petunia × hydrida), and maize (Zea mays). These homologs were chosen either by sequence similarity (A. alpina) or because they have already been reported to be related to the traits studied here.
Materials and methods
Constructs
All CDS entry clones were generated by amplifying the CDSs from the start to stop codon (for confirmed sequences see Supplementary Table S9) from cDNA of Arabidopsis thaliana (At) Col-0, Arabis alpina (Aa) Pajares, Gossypium hirsutum (Gh), Petunia × hybrida (Ph), and Zea mays (Zm) or from available plasmids (Supplementary Table S10) followed by BP recombination in donor pDONR201/207.
Point-mutations of AtGL3 were introduced to pDONR201 AtGL3 using the primers listed in Supplementary Table S11 in a PCR protocol that amplified the entire plasmid template (Zheng et al., 2004). The parent template was removed using a methylation-dependent nuclease DpnI, and bacteria were transformed with the nuclease-resistant nicked plasmid. Plasmids were isolated from the resulting colonies and screened for the desired modification. Finally, the positive clones were sequenced to confirm the desired modification.
CDSs of the AtGL3 wild-type and mutant AtGL3 variants were cloned into donor pDONR201 vectors by BP Reactions (Invitrogen) and introduced into pAMPAT-35S-GW (GenBank accession no. AY436765) (Pesch et al., 2015). Rescue constructs were introduced in the gl3 egl3 tt8 triple-mutant using the floral dip method (Clough and Bent, 1998).
Bioinformatic analysis
Alignments were done using the Optimal Global Alignment with PAM40 Similarity Matrix in BioEdit Sequence Alignment Editor (Hall, 1999).
Phylogenetic relationships were inferred using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and selected for the best likelihood values. The tree with the highest log-likelihood (–6100.8152) is shown. The analysis involved 15 proteins. All positions containing gaps and missing data were eliminated, leaving a total of 342 positions for comparison. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013).
Yeast vectors
Fusions of the CDSs to the GAL4 binding and activation domains were produced through an LR Reaction in pAS-attR and pC-ACT2-attR, respectively. As a negative control, the vectors pAS-attRans pC-ACT2-attR were recombined with pENTR1A-w/o-ccdB.
LUMIER vectors
Three different destination vectors were used for subsequent LR Reactions. pcDNA3-Rluc-GW and pTREX-dest30 (Invitrogen) that enable the N-terminal fusion of Renilla reniformis and Staphylococcus aureus protein, respectively, have been described previously (Pesch et al., 2013).
Defined genes were N-terminal fused to the S. aureus protein A sequence in pTREX-dest30-ntProtA by LR Reactions. As a negative control, the vector pTREX-dest30-ntProtA was recombined with pENTR1A-w/occdB (Pesch et al., 2015).
The full-length Renilla reniformis luciferase-gene was fused to the N-terminus of selected genes by LR reaction in pcDNA3-Rluc-GW. pENTR1A-w/o-ccdB was also recombined to this vector as a negative control.
Yellow fluorescent protein (YFP)-tagged proteins and the control without any CDSs were created by LR recombination of pTREX-dest30-YFP with the respective entry clones.
Yeast two-hybrid assays
Yeast two-hybrid assays, using the strain AH109, were carried out as described previously (Gietz et al., 1995). The transformed yeast cells were selected by plating them onto synthetic defined (SD) selection medium lacking Leu and Trp (SD-LW). Interactions were examined by analysing co-transformed yeast cells on SD interaction medium lacking Leu, Trp, and His, and supplemented with 5, 10, and 15 mM 3-amino-1, 2, 3-triazole (SD-LWH).
LUMIER assays
Six-well plate for pairwise assay
For LUMIER (luminescence-based mammalian interactome) assays, S. aureus protein A with or without the R. reniformis luciferase was fused to the N-terminus of each protein and transiently expressed in HEK293TM cells (BioCat/SBI: LV900A-1). Transfection and cell harvesting were carried out as described previously (Pesch et al., 2013, 2015). After 48 h, cells were washed three times with PBS, lysed in 150–250 μl lysis buffer, and extracts for different combinations were mixed after 1 h of lysis. Each combination was prepared in duplicate. Protein-immunoprecipitation and luminescence measurements were done as described previously (Pesch et al., 2013) using non-transfected cells or cells expressing the Renilla luciferase protein (Rluc) as controls. The percentage of Rluc on the beads compared with the lysate was calculated by dividing the Rluc activity on the beads by the Rluc activity in the same amount of lysate used in the pull-down assay (Input).
Nine-cm Petri dish for triple-components assay
To test whether the addition of another protein affected the binding behavior in the LUMIER assays described above, we expressed a third protein fused to YFP at the N-terminus using the backbone of pTREXdest30. Lysis of cells was done with 750–1000 μl lysis buffer for each plate. Extracts were normalized with respect to the YFP signal (TECAN) and combined after 1 h lysis. The total volume was kept constant by adding non-transfected cell lysate. Each combination was prepared in duplicate. Probes without additional YFP-fused protein were used for normalization. Cells expressing YFP-protein were included in the analysis to exclude non-specific interference of signals.
Results
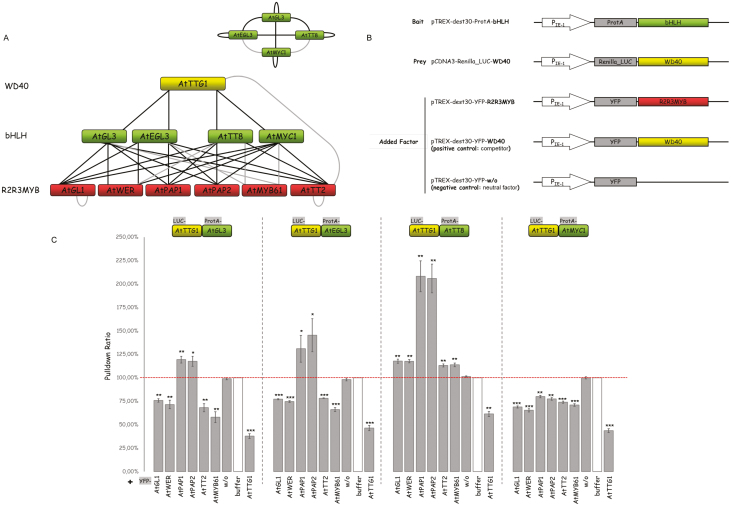
Competitive complex formation between MBW proteins in Arabidopsis
Our previous finding that the AtGL1 and AtWER proteins can interfere with the interaction of AtTTG1 and GL3 (Pesch et al., 2015) raises the question as to whether competitive complex formation is a general property of all MBW proteins or if it is specific to only some combinations. To examine this, we studied TTG1 combinations with the bHLH proteins GL3, EGL3, MYC1, and TT8, and the R2R3 MYB proteins GL1, MYB23, MYB61, WER, TT2, PAP1, and PAP2. As a reference we performed pull-down experiments using LUMIER assays (Blasche and Koegl, 2013; Supplementary Table S1) to confirm previously published interaction data (Payne et al., 2000; Zhang et al., 2003; Baudry et al., 2004; Zimmermann et al., 2004; Zhao et al., 2008; Bruex et al., 2012; Pesch et al., 2015). ,The interaction network including the dimerization behavior of the bHLH proteins is summarized in Fig. 1A. We used ProtA-tagged bHLH proteins (AtGL3, AtEGL3, AtTT8, and AtMYC1) to co-immunoprecipitate R. luciferase-fused AtTTG1 (LUC-AtTTG1) with and without YFP-tagged R2R3MYB proteins. TheAtTTG1–AtGL3 interaction substantially dropped in the presence of AtGL1, AtWER, AtTT2, or AtMYB61 (Fig. 1C), indicating that AtTT2 and AtMYB61 can also counteract AtTTG1 binding to AtGL3. On the other hand, addition of AtPAP1 and AtPAP2 resulted in significantly increased immunoprecipitation. We found similar competition behavior when using AtEGL3 instead of AtGL3 in these experiments. Strikingly, the interaction patterns of R2R3 MYB proteins were different in combination with the two bHLH proteins AtTT8 and AtMYC1. In the case of AtTT8, the addition of all R2R3 MYB proteins caused significantly increased co-immunoprecipitation of AtTTG1, which we henceforth term synergistic behavior. When testing the interaction between AtMYC1 and AtTTG1, the addition of all MYB proteins led to significantly reduced precipitation of AtTTG1. These results indicated that competitive complex formation of MBW proteins was a property of bHLH and R2R3 MYB proteins, and that it was only found in specific combinations. In addition, the data showed that the AtTTG1–bHLH interactions could be modulated positively and negatively by the addition of R2R3 MYB proteins.
Fig. 1.
Interactions and competition between MBW proteins in Arabidopsis. (A) Interaction network of MBW proteins based on yeast two-hybrid and pairwise LUMIER pull-down assays. Grey lines indicate weak interactions among the proteins. The homo- and hetero-dimerization of bHLHs is summarized at the top-right. (B) Schematic representation of the constructs driven by human promoter cytomegalovirus immediate early 1 (IE-1) that were used in the triple LUMIER assays. (C) Competition analysis of R2R3 MYB and bHLH proteins. Triple LUMIER pull-down assays were used to determine the pull-down efficiency of Renilla-tagged AtTTG1 by ProtA-tagged AtbHLH proteins with and without AtR2R3 MYB proteins. The results obtained in the presence of AtR2R3 MYB proteins are presented in relation to the pull-down values without addition of AtR2R3 MYB proteins (the white bar and horizontal dashed line indicates 100%). Yellow fluorescent protein (YFP) was included as a negative control (w/o). YFP-tagged AtTTG1 served as a positive control for competition. Data are means (±SE), n=3 experiments. Significant differences compared to the negative control (w/o) were determined using Student’s t-test: ***P<0.001; **P<0.01; *P<0.05. (This figure is available in color at JXB online.)
Competitive complex formation between MBW proteins in Arabis alpina and Gossypium hirsutum
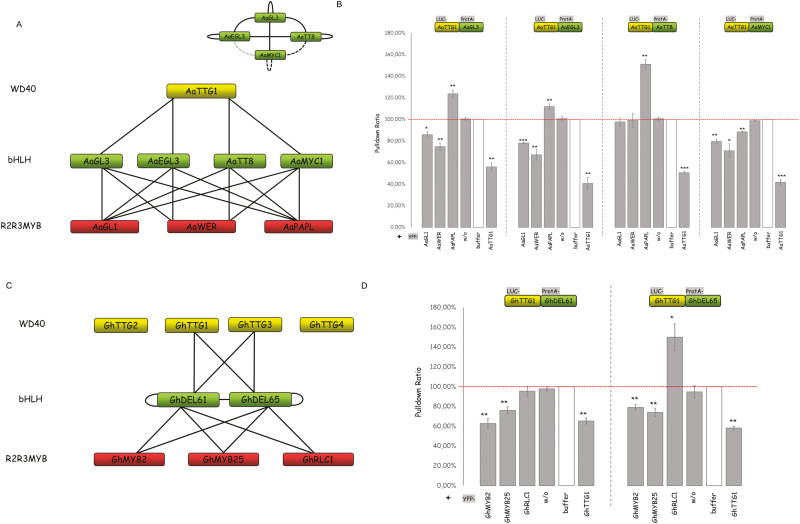
A comparison of the functions of MBW proteins in different species has suggested that they were initially relevant in the context of anthocyanidin and proanthocyanidin production, and that additional functions evolved in the rosid clade (Serna and Martin, 2006). We therefore studied competitive complex formation of MBW proteins in two well-examined rosid species, A. alpina and G. hirsutum.
Arabis alpina is a member of the crucifer family with an evolutionary distance of ~26–40 million years from Arabidopsis (Koch et al., 2006; Beilstein et al., 2010). This enabled the identification of most corresponding orthologs by sequence similarity and synteny (Table 1, Supplementary Tables S2–S4, Supplementary Fig. S1). Arabis alpina has one AaTTG1 gene and orthologs of the four bHLH genes considered here. Orthologs of most R2R3 MYB genes were also found. We found no PAP1 or PAP2 orthologs; instead, we identified a PAP-like (PAPL) gene that shares high sequence similarity to PAP1 and PAP2. Similar to Arabidopsis, we found interactions of all bHLH proteins with AaTTG1 and all MYB proteins (Fig. 2A, Supplementary Table S5, Supplementary Fig. S2A). In addition, homo- and hetero-dimerization was found for all combinations (Fig. 2A, Supplementary Table S5). The competition analysis revealed a similar behavior such that AaGL1 and AaWER counteracted the interaction of AaTTG1 and AaGL3/EGL3 whereas the addition of PAPL caused increased co-immunoprecipitation (Fig. 2B). AaTT8 differed from AaGL3 and AaEGL3 in that AaGL1 and AaWER did not interfere with the interaction of AaTT8 and AaTTG1. AaMYC1 showed a similar behavior to that found in Arabidopsis in that the addition of all MYB proteins caused a reduced interaction.
Table 1.
Selected CDSs for MBW proteins in five plant species*
| Gene name used in this study | Gene ID | Reference sequence | |
|---|---|---|---|
| Arabidopsis thaliana, Col-0 | |||
| R2R3 MYB | AtGL1 | AT3G27920 | NM_113708.2 |
| AtWER | AT5G14750 | NM_121479.3 | |
| AtPAP1 | AT1G56650 | NM_104541.4 | |
| AtPAP2 | AT1G66390 | NM_105310.4 | |
| AtTT2 | AT5G35550 | NM_122946.3 | |
| AtMYB61 | AT1G09540 | NM_100825.5 | |
| bHLH | AtGL3 | AT5G41315 | NM_148067.4 |
| AtEGL3 | AT1G63650 | NM_202351.2 | |
| AtMYC1 | AT4G00480 | NM_001160722.2 | |
| AtTT8 | AT4G09820 | NM_117050.3 | |
| WD40 | AtTTG1 | AT5G24520 | NM_122360.2 |
| Arabis alpina, Pajares | |||
| R2R3 MYB | AaGL1 | AALP_AA5G050100 | A_alpina_V4.cds** |
| AaWER | AALP_AA8G149800 | A_alpina_V4.cds** | |
| AaPAPL | AALP_AAs71396U000200*** | A_alpina_V4.cds** | |
| bHLH | AaGL3 | AALP_AA6G320200 | A_alpina_V4.cds** |
| AaEGL3 | AALP_AA6G002800 | A_alpina_V4.cds** | |
| AaMYC1 | AALP_AA6G006500.1 | A_alpina_V4.cds** | |
| AaTT8 | AALP_AA6G192900 | A_alpina_V4.cds** | |
| WD40 | AaTTG1 | AALP_AA8G421800 | A_alpina_V4.cds** |
| Gossypium hirsutum, TM-1 | |||
| R2R3 MYB | GhMYB2 | AF034130.1 | |
| GhMYB25 | AY464054.1 | ||
| GhRLC1 | NM_001327615.1 | ||
| bHLH | GhDEL65 | AF336280.1 | |
| GhDEL61 | AF336279.1 | ||
| WD40 | GhTTG1 | AF530907.1 | |
| GhTTG2 | AF530909.1 | ||
| GhTTG3 | AF530911.1 | ||
| GhTTG4 | AF530912.1 | ||
| Petunia × hybrida, cultivar R27 | |||
| R2R3 MYB | PhAN2 | AF146704 | |
| PhAN4 | HQ428105.1 (V30) or EB175066 (R27) | ||
| PhPH4 | AY973324.1 | ||
| bHLH | PhAN1 | AF260919.1 | |
| PhJAF13 | AF260918 | ||
| WD40 | PhAN11 | U94748.1 | |
| Zea mays, cultivar B73 | |||
| R2R3 MYB | ZmC1 | GRMZM2G005066 | AY237128 |
| ZmPL | GRMZM2G701063 | KJ727236.1 | |
| ZmP1 | GRMZM2G084799 | KJ728395.1 | |
| bHLH | ZmR(Lc) | GRMZM5G822829 | KJ726800.1 |
| ZmR(S) | Zm00001d026147 | XM_008663066.3 (6s) | |
| ZmB | GRMZM2G172795 | KJ727396.1 | |
| WD40 | ZmPAC1 | GRMZM2G058292 | KJ728394.1 |
| ZmMP1 | GRMZM2G099334 | AY339884.1 (Carey et al., 2004) |
* Data presented in this table have been extracted from the following sources. For A. thaliana: https://www.ncbi.nlm.nih.gov/nuccore;www.arabidopsis.org. For A. alpina: http://www.arabis-alpina.org/refseq.html (Willing et al., 2015). For cotton: Gossypium hirsutum cultivar TM-1 seed was kindly provided by Dr Jing Qu (Institute of Microbiology, Chinese Academy of Sciences, China). Examples for reference sequences of plasmids were selected from NCBI BLASTn (Cedroni et al., 2003; Wang et al., 2004; Gao et al., 2013; Guan et al., 2014; Wan et al., 2014). For petunia: plasmids were kindly provided by R. Koes. Examples for reference sequences of plasmids were selected from NCBI BLASTn (Altschul and Gish, 1996; Zhang et al., 2000; Morgulis et al., 2008). For maize (both plasmids and own cloning reference): https://www.maizegdb.org/;http://plants.ensembl.org/Zea_mays/Info/Index?db=core;https://www.ncbi.nlm.nih.gov/nuccore; genome assembly AGPv4, B73 RefGen_v4 (information on the assembly at http://plants.ensembl.org/Zea_mays/Info/Annotation/#genebuild); plasmids from the maize TFome (transcription factor ORF collection) (Burdo et al., 2014) were selected from https://grassius.org/index.php (Yilmaz et al., 2009).
** A_alpina_V4.cds.fasta from http://www.arabis-alpina.org/refseq.html (Willing et al., 2015).
*** AaPAPL was first identified at the protein level using A. alpina protein sequences, http://www.arabis-alpina.org/refseq.html (Willing et al., 2015).
Fig. 2.
Interactions and competition between MBW proteins in Arabis alpina and Gossypium hirsutum. (A) Interaction network of MBWs in A. alpina based on yeast two-hybrid (Y2H) and pairwise LUMIER pull-down assays. The homo- and hetero-dimerization of AabHLHs are shown at the top-right: solid lines indicate interactions that were supported by both methods, and dashed lines indicate interactions that were supported by either Y2H or LUMIER assays. Grey indicates a weak interaction. (B) Competition analysis of AaR2R3MYBs and AabHLH proteins by triple LUMIER pull-down assays, as detailed in Fig. 1. (C) Interaction network of MBWs in G. hirsutum based on Y2H and pairwise LUMIER pull-down assays. (D) Competition analysis of GhR2R3MYBs and GhbHLH proteins by triple LUMIER pull-down assays, as detailed in Fig. 1. Data in (B, D) are means (±SE), n=3 experiments, and significant differences compared to the negative control (w/o) were determined using Student’s t-test: ***P<0.001; **P<0.01; *P<0.05. (This figure is available in color at JXB online.)
In Gossypium hirsutum we considered four WD40 proteins (GhTTG1, GhTTG2, GhTTG3, GhTTG4) (Table 1, Supplementary Tables S2–S4). Among these, GhTTG1 and GhTTG3 have been shown to be involved in fiber formation (Humphries et al., 2005). We selected two bHLH proteins, GhDEL65 and GhDEL61, both of which are important in fiber development (Wang et al., 2013; Shangguan et al., 2016). In addition, we analysed three R2R3 MYB proteins: GhMYB2 and GhMYB25 are relevant for seed trichome formation, and GhRLC1 plays a role in the regulation of anthocyanidin production (Cedroni et al., 2003; Wang et al., 2004; Machado et al., 2009; Gao et al., 2013; Guan et al., 2014; Wan et al., 2014). We found that only the WD40 proteins GhTTG1 and GhTTG3 showed interactions with the two bHLH proteins (Fig. 2C) (Humphries et al., 2005). Both the bHLH proteins interacted with the three R2R3 MYB proteins ( Supplementary Table S6, Supplementary Fig. S2B). As GhTTG1 and GhTTG3 can functionally replace each other, we focused our analysis of the competition properties on GhTTG1. We found that the addition of GhMYB2 and GhMYB25 both interfered with GhTTG1 binding to both bHLH proteins (Fig. 2D). GhRLC1 showed a differential behavior in that it had no effect on the GhTTG1–GhDEL61 interaction but strengthened the GhTTG1–GhDEL65 interaction.
Competitive complex formation between MBW proteins in Petunia hybrida and Zea mays
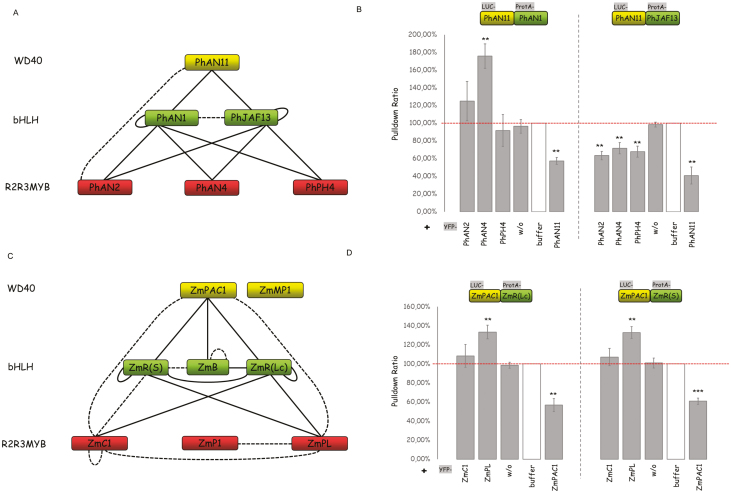
We next analysed the MBW complexes in Petunia hybrida and Zea mays. The MBW genes in petunia are functionally well-characterized and have been shown to act as controllers of anthocyanidin biosynthesis pathways (Quattrocchio et al., 1993, 1998, 2006; de Vetten et al., 1997; Spelt et al., 2000; Albert et al., 2011). Trichome development does not appear to depend on MBW genes but is instead controlled by a different pathway (Ramsay and Glover, 2005; Serna and Martin, 2006; Zhang and Schrader, 2017). Similarly, the MBW genes in maize are controllers of anthocyanidin production.
Our analysis of the petunia MBW proteins examined the WD40 protein PhAN11, the bHLH proteins PhAN1 and PhJAF13, and the R2R3 MYB proteins AN2, PhAN4, and PhPH4 (Table 1, Supplementary Tables S2–S4). Yeast two-hybrid and LUMIER interactions were found for both the bHLH proteins with the WD40 protein, with each other, and with all the R2R3 MYB proteins (Fig. 3A, Supplementary Table S7, Supplementary Fig. S2C) (Spelt et al., 2002; Albert et al., 2014). The analysis of competitive complex formation revealed that the interaction of PhAN1 with PhAN11 could not be repressed by the R2R3 MYB proteins. Addition of PhAN4 resulted in better binding (Fig. 3B). The interaction of PhJAF13 with PhAN11 was significantly reduced in the presence of each of the three R2R3 MYB proteins.
Fig. 3.
Interactions and competition between MBW proteins in Petunia hybrid and Zea mays. (A) Interaction network of MBWs in P. hybrid based on yeast two-hybrid (Y2H) and pairwise LUMIER pull-down assays. Solid lines indicate interactions that were supported by both methods, and dashed lines indicate interactions that were supported by either the Y2H or LUMIER assays. (B) Competition analysis of PhR2R3MYBs and PhbHLH proteins by triple LUMIER pulldown assays, as detailed in Fig. 1. (C) Interaction network of MBWs in Z. mays based on Y2H and pairwise LUMIER pull-down assays. Solid lines indicate interactions that were supported by both methods, and dashed lines indicate interactions that were supported by either Y2H or LUMIER assays. (D) Competition analysis of ZmR2R3MYBs and ZmbHLH proteins by triple LUMIER pull-down assays, as detailed in Fig. 1. Data in (B, D) are means (±SE), n=3 experiments, and significant differences compared to the negative control (w/o) were determined using Student’s t-test: ***P<0.001; **P<0.01; *P<0.05. (This figure is available in color at JXB online.)
For the analysis of the Zea mays MBW proteins, we examined the WD40 proteins ZmPAC1 and ZmMP1, the bHLH proteins ZmR(S), ZmB, and ZmR(Lc), and the R2R3 MYB proteins ZmC1, ZmP1, and ZmPL (Table 1, Supplementary Tables S2–S4). Yeast-two hybrid and LUMIER interactions were found for all the bHLH proteins with ZmPAC1 and between each other (Fig. 3C, Supplementary Table S8). We found no interactions of the bHLH proteins with ZmMP1. ZmR(S) and ZmR(Lc) showed interactions with the R2R3 MYB proteins ZmC1 and ZmPL. The bHLH protein ZmB did not interact with any of the R2R3 MYBs used in this study (Goff et al., 1992; Hernandez et al., 2004). We found no competitive behavior for ZmR(S) and ZmR(Lc) in combination with any of the R2R3 MYB proteins (Fig. 3D). Addition of the ZmPL protein caused improved binding of the ZmR proteins to ZmPAC1.
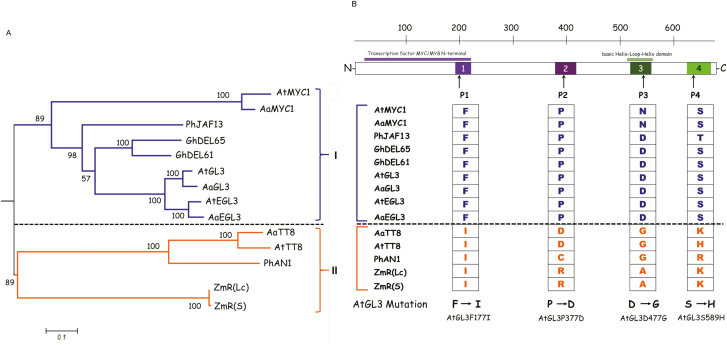
The phylogenetic relationship of bHLH proteins correlates with their ability to form competitive complexes
Our findings that competitive complex formation was found only in a subset of the bHLH proteins prompted us to examine the correlation of this property with the evolutionary relationship as assessed by a maximum-likelihood analysis. In constructing the phylogenetic tree, we used only those proteins for which we had data on their ability to form competitive complexes. Using the JTT matrix-based mode (Jones et al., 1992), we created a phylogenetic tree for the bHLH proteins considered here using MEGA6 (Tamura et al., 2013), and found that they fell into two clades (Fig. 4A). The first clade consisted of AtMYC1, AaMYC1, PhJAF13, GhDEL65, GhDEL61, AtGL3, AaGL3, AtEGL3, and AaEGL3, all of which showed competitive complex formation in combination with one or several R2R3 MYB proteins. With the exceptions of AtMYC1, AaMYC1, and PhJAF13, all the bHLH proteins in this clade also exhibited synergistic interactions with at least one MYB protein. The second clade consisted of AaTT8, AtTT8, PhAN1, ZmR(Lc), and ZmR(S), all of which exhibited no competitive complex formation with any of the tested MYB proteins but showed synergistic interactions in combination with one or more MYBs.
Fig. 4.
Phylogenetic analysis of bHLH proteins used in this study. (A) The phylogenetic tree was constructed using the alignment of full-length bHLH proteins. The tree is drawn to scale with branch lengths measured as the number of substitutions per site, and it was created by mid-point rooting. Bootstrap values are given at the branch nodes, determined from 500 bootstrap repetitions. The interaction behaviors are indicated on the right: clade I, competitive complex formation; clade II, non-competitive complex formation. (B) Analysis of bHLH protein motifs. Four motifs were identified by MEME (http://alternate.meme-suite.org;(Bailey and Gribskov, 1998), shown as numbered boxes in the construct diagram of the bHLH protein. The sequence information for each motif is provided in the Supplementary Fig. S4). The amino acids shared in clade I in the four motifs are shown for the bHLH proteins analysed in this study and compared to those in clade II. The changes in amino acid that were introduced into wild-type AtGL3 are shown at the bottom. (This figure is available in color at JXB online.)
Identification of sites required for competitive complex formation
We extended our analysis by performing a detailed sequence comparison to identify possible regions or sites correlating with the property to mediate competitive complex formation. As a first step, we selected four motifs with high sequence similarity that are present in similar positions in the bHLH proteins using MEME Suite 4.12.0 (Bailey and Gribskov, 1998) (Fig. 4B, Supplementary Figs S3, S4). Next, we searched for differences in conserved amino acids that were specific to one clade. We found four amino acids at different positions of the protein. In motif 1, all the bHLH proteins of the first clade had a phenylalanine in common; in motif 2, the first clade was characterized by a proline; in motif 3, we found the basic amino acid aspartate or an asparagine in the first clade; and in motif 4, the first clade had a serine or a threonine in common (Fig. 4B).
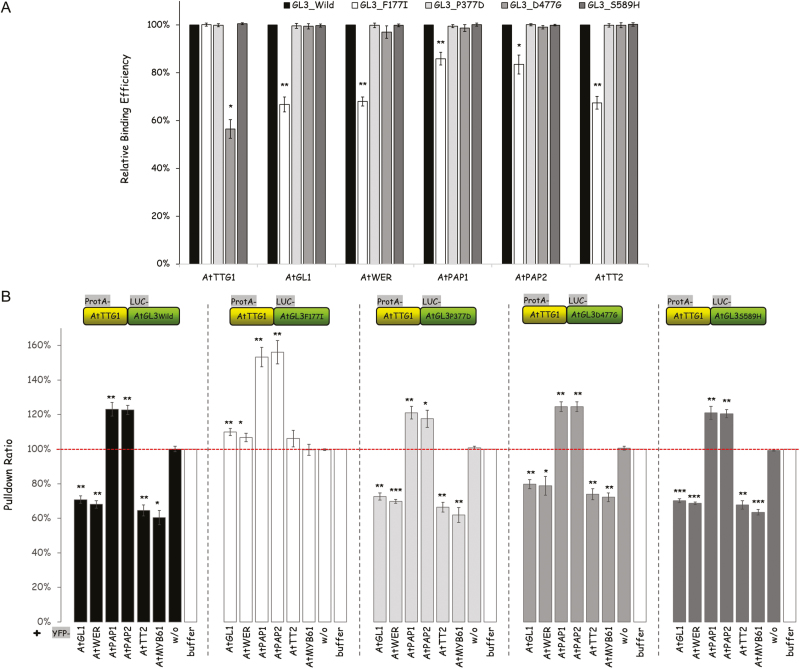
To test whether one of these amino acids was indeed relevant for competitive complex formation, we decided to introduce mutations in the AtGL3 protein to render it non-competitive. To do this, we substituted the four amino acids with an amino acid found at the same position in the non-competitive bHLH proteins (Fig. 4B): the phenylalanine in motif 1 was changed to an isoleucine [AtGL3 (F177I)]; the proline in motif 2 to an aspartate [AtGL3 (P377D)]; the aspartate in motive 3 to a glycine [AtGL3 (D477G)]; and the serine in motif 4 to a histidine [AtGL3 (S589H)]. We then tested whether the mutations lead to different binding behaviors of AtGL3 to AtTTG1 or to AtR2R3 MYB proteins (Fig. 5A). We found significantly reduced binding of AtGL3 (D477G) to AtTTG1 and significantly reduced binding of AtGL3 (F177I) to all the AtR2R3 MYB proteins tested.
Fig. 5.
Analysis of mutant AtGL3 protein variants. (A) Binding analysis of the wild-type and mutant AtGL3 alleles with AtTTG1 or AtR2R3 MYB proteins by pairwise LUMIER assays. Data are shown as relative values compared to the wild-type AtGL3, which was defined as 100% (the inputs of the AtGL3 wild-type and mutant alleles were normalized). (B) Competition behavior of the wild-type and AtGL3 mutants. The interaction strength of the ProtA_ATTG1 and 4 LUC_AtGL3 alleles (as indicated at the top) was analysed in the presence of different AtR2R3 MYB proteins, as detailed in Fig. 1. The data are shown as relative values with reference to the probes without any AtR2R3 MYB proteins (the white bar and horizontal dashed line indicates 100%). Data are means (±SE), n=3 experiments, and significant differences compared to the negative control (w/o) were determined using Student’s t-test: ***P<0.001; **P<0.01; *P<0.05. (This figure is available in color at JXB online.)
We next examined the four mutant proteins for competitive binding behavior. We compared ProtA-AtTTG1 and LUC-AtGL3 binding with and without different AtR2R3 MYB proteins (Fig. 5B). While AtGL3 (P377D), AtGL3 (D477G), and AtGL3 (S589H) showed the same binding behavior as the wild-type AtGL3, AtGL3 (F177I) was not able to mediate competition. This was true for all four MYBs namely AtGL1, AtWER, AtTT2, and AtMYB61. Interestingly, the synergistic interaction with AtPAP1 and AtPAP2 was not altered, indicating that competitive complex formation and synergistic interactions can be separate and therefore mechanistically distinct.
AtGL3 (F177I) rescues anthocyanidin production but not trichome development
To examine the relevance of the induced point-mutations, we performed rescue experiments. Wild-type AtGL3, AtGL3(F177I), and AtGL3(D477G) were expressed under the 35S promoter in the gl3 egl3 tt8 triple-mutant. This mutant shows a phenotype reminiscent of ttg1, including defects in trichome and root hair development, and in anthocyanidin, proanthocyanidin, and seed-coat mucilage production (Zhang et al., 2003). T1 seeds were initially grown on agar containing 4% sugar to assess rescue of the anthocyanidin phenotype. This phenotype was effectively rescued by overexpression of all three proteins (Fig. 6). Rescued plants were transferred to soil and rosette leaves were inspected for rescue of the trichome phenotype. All T1 plants expressing wild-type AtGL3 or AtGL3 (D477G) exhibited a rescue of the phenotype; however, none of the plants expressing AtGL3 (F177I) showed rescue. This indicated that this amino acid is essential for the regulation of trichome production but not for anthocyanidin production.
Fig. 6.
Phenotypic rescue of the Arabidopsis gl3 egl3 tt8 triple-mutant by mutant AtGL3 protein variants. The triple-mutant was transformed with 35S::AtGL3, 35S::AtGL3 (F177I), and 35S::AtGL3 (D477G) and analysed in the T1 generation. The top row shows seedlings grown on half-strength Murashige and Skoog medium with 4% sugar under Basta selection at 4 d after germination. Scale bars are 2 mm. Representative images are shown and the proportions of rescued plants/Basta-resistant plants are indicated below. The bottom row shows rosette leaves of 2-week-old T1 plants that had previously been analysed for the anthocyanidin phenotype at 9 d after germination. The proportion of plants showing trichome rescue in the plants that exhibited anthocyanidin rescue are indicated below. Scale bars are 5 mm.
Discussion
All three components of the MBW complexes are ancient and evolutionarily conserved proteins that are present in all eukaryotes (Ramsay and Glover, 2005), and the regulation of flavonoid biosynthesis in plants is considered to be the most ancient function of the MBW complexes (Ramsay and Glover, 2005; Zhang and Schrader, 2017). Additional functions in the regulation of seed-coat mucilage production and in trichome and root hair development appear to have evolved in dicotyledon plants after the asteroid–rosid split (Serna and Martin, 2006). Competitive complex formation of the MBW proteins was discovered for proteins involved in an evolutionarily more recent trait, namely trichome formation (Pesch et al., 2015). This raises the question as to whether competitive complex formation is also a new biochemical property or whether it is a general behavior of MBW proteins. In an attempt to answer this, we used a biochemical approach in the form of the LUMIER assay (Blasche and Koegl, 2013). This technique has the advantage that the interactions are determined in the absence of other plant proteins that may stabilize or destabilize the interactions. By the same token, it is possible that the interactions are different in plants because of other proteins. However, we are convinced that this technique provides a solid basis for future in planta studies.
Modulation of MBW complex formation in Arabidopsis depends on the individual combinations of bHLH and R2R3 MYB proteins
When comparing the ability of the four Arabidopsis bHLH proteins, namely AtGL3, AtEGL3, AtMYC1, and AtTT8, to mediate competitive complex formation, three types of interaction behaviors were seen (Fig. 1C). While AtGL3 and AtEGL3 showed competitive complex formation with some R2R3 MYBs and enhanced complex formation with others, AtMYC1 showed competitive complex formation with all R2R3 MYBs. By contrast, AtTT8 exhibited a better interaction with AtTTG1 in the presence of all the tested R2R3 MYBs. These results also indicated that the R2R3 MYB proteins differed in their ability to modulate the binding of the bHLH proteins with AtTTG1 (Fig. 1C). For example, AtGL1, AtTT2, AtMYB61, and AtWER could counteract the binding of AtTTG1 to AtGL3 and AtEGL3, whereas AtPAP1 and AtPAP2 improved AtTTG1 binding. In combination with AtTT8, however, AtGL1, AtTT2, AtMYB61, and AtWER could enhance AtTTG1 binding. Thus, the modulation of TTG1 binding to the bHLH protein depends on the individual bHLH–R2R3 MYB combinations.
How can this modulation of AtTTG1 binding to bHLH proteins by R2R3 MYBs be explained at the molecular level? As AtTTG1 and R2R3 MYBs are considered to bind to different regions of the bHLH proteins (Payne et al., 2000), it is likely that the binding of a R2R3 MYB protein leads to an allosteric change. One possibility is that a conformational change of the bHLH protein leads to modulation of the binding affinity of AtTTG1. It is conceivable that this might work in both directions, such that the binding affinity is either reduced or enhanced.
Evolutionary analysis of the modulation of MBW complex formation
The finding that the modulation of MBW complex formation is a property of specific combinations of bHLH and R2R3 MYB proteins in Arabidopsis raises the question as to how this behavior has evolved. Interestingly, we found the same interaction behavior for the orthologous proteins in A. alpina (Fig. 2B). The finding that ttg1 mutants in A. alpina share the same spectrum of phenotypes with Arabidopsis suggests that the overall function of TTG1-dependent traits is conserved (Chopra et al., 2014). Arabis alpina is in the same family as Arabidopsis, with an evolutionary distance of ~26–40 million years (Koch et al., 2006; Beilstein et al., 2010). While this evolutionary distance enables the identification of orthologous genes by sequence similarity and synteny, it would be expected that a conservation of the protein behavior would point to a functional relevance. In this respect, it is plausible that the same complex formation behavior of orthologous MBW proteins suggests some functional relevance.
In cotton, we found that the two bHLH proteins examined, GhDEL6 and GhDEL65, mediated competitive complex formation (Fig. 2D). In petunia, PhJAF13 and PhAN1 behaved differently, such that PhJAF13 showed competitive complex formation with all the R2R3 MYBs tested whereas PhAN1 did not (Fig. 3B). In maize, however, two bHLH proteins, ZmR(Lc) and ZmR(S), did not show competitive complex formation but did show better ZmPAC1 binding in the presence of the ZmPL protein (Fig. 3D). This suggests that competitive complex formation might have evolved in eudicots; however, more data from more species are needed to confirm this.
One possible way to assess the evolution of competitive complex formation is to correlate this trait with a phylogenetic tree of bHLH proteins. In our analysis we found two main clades (Fig. 4A). One clade consisted of the two R genes from maize, the AN1 gene from petunia, and TT8 from A. alpina and Arabidopsis, all of which did not show competitive complex formation. The second clade consisted of GL3, EGL3, and MYC1 from A. alpina and Arabidopsis, JAF13 from petunia, and DEL65 and DEL61 from cotton, all of which exhibited competitive complex formation. Together, these data suggest that competitive complex formation can be considered as a trait that correlates well with the evolutionary distance of the respective proteins.
Functional relevance of competitive complex formation
Our mutational analyses (Fig. 5) revealed that an exchange of the phenylalanine in position 177 of AtGL3 for an amino acid found in non-competitive bHLH reduced binding to R2R3 MYBs and rendered AtGL3 non-competitive. Whether the loss of competitive complex formation was caused by the 30% reduced binding of the R2R3 MYBs is not known. However, as reduced binding of GL3 to AtPAP1/AtPAP2 did not affect the binding of AtTTG1 to AtGL3 (F177I) in the presence of AtPAP1 or AtPAP2 (Fig. 5B) it is conceivable that the loss of competitive complex formation was not the immediate consequence of reduced R2R3 MYB binding.
The phenylalanine 177 in AtGL3 also appears to be functionally relevant. Strikingly, mutations in this position did not affect rescue of the anthocyanidin phenotype but only the rescue of the trichome phenotype (Fig. 6). It is therefore tempting to speculate that competitive complex formation is an evolutionary innovation in the context of patterning processes. Competitive complex formation might lead to a much more complex regulation network in which different complexes may adopt different functions. This is consistent with earlier findings that GL1 can counteract the activation of the TRY-promoter by GL3 and TTG1, and that TTG1 can counteract the activation of the CPC promoter by GL3 and GL1 (Pesch et al., 2015). Thus, this protein behavior could, in principle, translate different ratios of the proteins in the MBW complex into differential gene regulation. This turn could be favorable for the formation of new regulatory networks for creating a de novo pattern.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Phylogenetic tree of the WD40 and R2R3 MYB proteins used in this study.
Fig. S2. Protein–protein interactions among members of the MBW regulatory complex in A. alpina, G. hirsutum, P. hybrida, and Z. mays as determined by yeast two-hybrids assays.
Fig. S3. Sequence comparison of bHLH proteins.
Fig. S4: Common motifs in bHLH proteins.
Table S1. Pairwise interaction of MBW components in Arabidopsis.
Table S2. Shared sequence identity of WD40 proteins.
Table S3. Shared sequence identity of bHLH proteins.
Table S4. Shared sequence identity of R2R3 MYB proteins.
Table S5. Pairwise interaction of MBW components in A. alpina.
Table S6. Pairwise interaction of MBW components in G. hirsutum.
Table S7. Pairwise interaction of MBW components in P. hybrida.
Table S8. Pairwise interaction of MBW components in Z. mays.
Table S9. CDS sequence information for MBW.
Table S10. Details for plasmids used in this study.
Table S11. List of primers used in this study.
Acknowledgements
We thank Mona Mapar and Hanna Bechtel for critical reading of the manuscript. We thank Prof. Dr Ronald Koes for providing the petunia cDNA constructs of AN11, AN1, JAF13, AN2, AN4, and PH4. We are grateful for Dr Jing Qu (Chinese Academy of Sciences, China) for providing us with the seed for the cotton cultivar TM-1. We thank Sabine Lohmer for technical help, and Heike Wolff and Johannes Span for providing the Arabis alpina clones. This work was funded by the DFG Priority Program SFB 680.
Author contributions
BZ performed the experiments and analysed the data; MH supervized the experiments; DC and AS identified and cloned the A. alpina orthologous genes; MH and BZ wrote the manuscript.
References
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. 2014. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 26, 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. 2011. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal 65, 771–784. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W. 1996. Local alignment statistics. Methods in Enzymology 266, 460–480. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14, 48–54. [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L. 2006. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant Journal 46, 768–779. [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. 2004. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant Journal 39, 366–380. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. 2010. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 18724–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. 2003. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. [DOI] [PubMed] [Google Scholar]
- Blasche S, Koegl M. 2013. Analysis of protein–protein interactions using LUMIER assays. Methods Molecular Biology 1064, 17–27. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell 12, 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EL, Grotewold E. 1999. Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiology 121, 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, et al. . 2012. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genetics 8, e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo B, Gray J, Goetting-Minesky MP, et al. . 2014. The Maize TFome – development of a transcription factor open reading frame collection for functional genomics. The Plant Journal 80, 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey CC, Strahle JT, Selinger DA, Chandler VL. 2004. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. The Plant Cell 16, 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedroni ML, Cronn RC, Adams KL, Wilkins TA, Wendel JF. 2003. Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Molecular Biology 51, 313–325. [DOI] [PubMed] [Google Scholar]
- Chopra D, Wolff H, Span J, Schellmann S, Coupland G, Albani MC, Schrader A, Hülskamp M. 2014. Analysis of TTG1 function in Arabis alpina. BMC Plant Biology 14, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- de Vetten N, Quattrocchio F, Mol J, Koes R. 1997. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes & Development 11, 1422–1434. [DOI] [PubMed] [Google Scholar]
- Dressel A, Hemleben V. 2009. Transparent Testa Glabra 1 (TTG1) and TTG1-like genes in Matthiola incana R. Br. and related Brassicaceae and mutation in the WD-40 motif. Plant Biology 11, 204–212. [DOI] [PubMed] [Google Scholar]
- Feller A, Hernandez JM, Grotewold E. 2006. An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. The Journal of Biological Chemistry 281, 28964–28974. [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal 66, 94–116. [DOI] [PubMed] [Google Scholar]
- Feyissa DN, Løvdal T, Olsen KM, Slimestad R, Lillo C. 2009. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta 230, 747–754. [DOI] [PubMed] [Google Scholar]
- Gao Z, Liu C, Zhang Y, Li Y, Yi K, Zhao X, Cui ML. 2013. The promoter structure differentiation of a MYB transcription factor RLC1 causes red leaf coloration in Empire Red Leaf Cotton under light. PLoS ONE 8, e77891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. 1992. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes & Development 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. 2009. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Developmental Biology 325, 412–421. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ. 2014. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nature Communications 5, 3050. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98. [Google Scholar]
- Herman PL, Marks MD. 1989. Trichome development in Arabidopsis thaliana. II. Isolation and complementation of the GLABROUS1 gene. The Plant Cell 1, 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JM, Heine GF, Irani NG, Feller A, Kim MG, Matulnik T, Chandler VL, Grotewold E. 2004. Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. The Journal of Biological Chemistry 279, 48205–48213. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Misŕa S, Jürgens G. 1994. Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Humphries JA, Walker AR, Timmis JN, Orford SJ. 2005. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Molecular Biology 57, 67–81. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer D, Schiefelbein J, Hulskamp M. 2005. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132, 1477–1485. [DOI] [PubMed] [Google Scholar]
- Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K. 2006. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Molecular Ecology 15, 825–839. [DOI] [PubMed] [Google Scholar]
- Kong Q, Pattanaik S, Feller A, Werkman JR, Chai C, Wang Y, Grotewold E, Yuan L. 2012. Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proceedings of the National Academy of Sciences, USA 109, E2091–E2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M. 1981. The complex syndrome of ttg mutants. Arabidopsis Information Service 18, 45–51. [Google Scholar]
- Kranz H, Scholz K, Weisshaar B. 2000. c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. The Plant Journal 21, 231–235. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. 1999. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES. 2009. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. The Plant Journal 59, 52–62. [DOI] [PubMed] [Google Scholar]
- Marks MD, Feldmann KA. 1989. Trichome development in Arabidopsis thaliana. I. T-DNA tagging of the GLABROUS1 gene. The Plant Cell 1, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schaffer AA. 2008. Database indexing for production MegaBLAST searches. Bioinformatics 24, 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemesio-Gorriz M, Blair PB, Dalman K, Hammerbacher A, Arnerup J, Stenlid J, Mukhtar SM, Elfstrand M. 2017. Identification of Norway Spruce MYB-bHLH-WDR transcription factor complex members linked to regulation of the flavonoid pathway. Frontiers in Plant Science 8, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. 2000. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. The Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. 1991. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. 2000. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. 2001. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. The Plant Cell 13, 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Schultheiß I, Digiuni S, Uhrig JF, Hülskamp M. 2013. Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140, 3456–3467. [DOI] [PubMed] [Google Scholar]
- Pesch M, Schultheiß I, Klopffleisch K, Uhrig JF, Koegl M, Clemen CS, Simon R, Weidtkamp-Peters S, Hülskamp M. 2015. TRANSPARENT TESTA GLABRA1 and GLABRA1 compete for binding to GLABRA3 in Arabidopsis. Plant Physiology 168, 584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. 2006. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. The Plant Cell 18, 1274–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen H, Mol J, Koes RE. 1993. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. The Plant Cell 5, 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R. 1998. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. The Plant Journal 13, 475–488. [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. 2005. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science 10, 63–70. [DOI] [PubMed] [Google Scholar]
- Serna L, Martin C. 2006. Trichomes: different regulatory networks lead to convergent structures. Trends in Plant Science 11, 274–280. [DOI] [PubMed] [Google Scholar]
- Shangguan XX, Yang CQ, Zhang XF, Wang LJ. 2016. Functional characterization of a basic helix-loop-helix (bHLH) transcription factor GhDEL65 from cotton (Gossypium hirsutum). Physiologia Plantarum 158, 200–212. [DOI] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol J, Koes R. 2002. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. The Plant Cell 14, 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JNM, Koes R. 2000. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. The Plant Cell 12, 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds VV, Hatlestad G, Lloyd AM. 2011. Natural allelic variation defines a role for ATMYC1: trichome cell fate determination. PLoS Genetics 7, e1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. 2005. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology 139, 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Ludwig P. 2003. The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Zhang H, Ye W, Wu H, Zhang T. 2014. Genome-wide transcriptome profiling revealed cotton fuzz fiber development having a similar molecular model as Arabidopsis trichome. PLoS ONE 9, e97313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhao GH, Jia YH, Du XM. 2013. Identification and characterization of cotton genes involved in fuzz-fiber development. Journal of Integrative Plant Biology 55, 619–630. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY. 2004. Control of plant trichome development by a cotton fiber MYB gene. The Plant Cell 16, 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing EM, Rawat V, Mandáková T, et al. . 2015. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nature Plants 1, 14023. [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends in Plant Science 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Nishiyama MY Jr, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E. 2009. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiology 149, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Schrader A. 2017. TRANSPARENT TESTA GLABRA 1-dependent regulation of flavonoid biosynthesis. Plants 6, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology 7, 203–214. [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. 2008. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135, 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Research 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant Journal 40, 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.