Plasma membrane aquaporin PIP1;3 of rice interacts with type III translocator Hpa1 of bacterial blight pathogen to facilitate translocation of the TAL effector PthXo1 from the bacteria into rice cells.
Keywords: Aquaporin, bacterial pathogen, PIP1, rice, translocation, type III effector
Abstract
Varieties of Gram-negative bacterial pathogens infect their eukaryotic hosts by deploying the type III translocon to deliver effector proteins into the cytosol of eukaryotic cells in which effectors execute their pathological functions. The translocon is hypothetically assembled by bacterial translocators in association with the assumed receptors situated on eukaryotic plasma membranes. This hypothesis is partially verified in the present study with genetic, biochemical, and pathological evidence for the role of a rice aquaporin, plasma membrane intrinsic protein PIP1;3, in the cytosolic import of the transcription activator-like effector PthXo1 from the bacterial blight pathogen. PIP1;3 interacts with the bacterial translocator Hpa1 at rice plasma membranes to control PthXo1 translocation from cells of a well-characterized strain of the bacterial blight pathogen into the cytosol of cells of a susceptible rice variety. An extracellular loop sequence of PIP1;3 and the α-helix motif of Hpa1 determine both the molecular interaction and its consequences with respect to the effector translocation and the bacterial virulence on the susceptible rice variety. Overall, these results provide multiple experimental avenues to support the hypothesis that interactions between bacterial translocators and their interactors at the target membrane are essential for bacterial effector translocation.
Introduction
Many effector proteins of Gram-negative bacterial pathogens of plants (Alfano and Collmer, 2004) and animals (Büttner, 2012) are secreted by the type III (T3) secretion system (T3SS). Subsequently, effectors must be translocated from bacteria into the cytosol of eukaryotic cells so that they can play virulent or avirulent roles depending on host–pathogen interactions (Bogdanove and Voytas, 2011; Chen et al., 2017). Effector translocation is generally assumed to take place through the T3 translocon, which is hypothetically made up of hydrophilic and hydrophobic proteinaceous translocators from bacteria (Ji and Dong, 2015a). The distinct translocators make up the sophisticated translocon theoretically by monogenous and heterogeneous molecular interactions and by associations with recognition compounds at the plasma membranes (PMs) of eukaryotic cells (Chatterjee et al., 2013). In a current model (Ji and Dong, 2015a), hydrophilic translocators form complexes at the tip (Espina, et al., 2006; Mueller, et al., 2008) of proteinaceous pili in plant pathogens or proteinaceous needles in animal pathogens. The complexes of hydrophilic translocators further associate with hydrophobic counterparts, which then oligomerize into the target PM (Büttner, 2012). The resulting translocon consists of an inner trafficking conduit that is connected to the bacterial pilus or needle channel with an opening inward to the eukaryotic cytoplasm, thereby allowing the so-called ‘direct injection’ of bacterial effectors into the cytosol of eukaryotic cells immediately following secretion by the pilus or needle pathway (Kvitko, et al., 2007; Bocsanczy et al., 2008; Chakravarthy et al., 2017).
All hydrophilic components of T3 translocators so far identified in plant-pathogenic bacteria belong to a group of T3SS accessory proteins called harpins, which are required for pilus growth or translocon formation (Choi et al., 2013; Ji and Dong, 2015a). These proteins fall into two categories: one-domain harpins that contain a unitary hydrophilic domain, and two-domain harpins that carry an additional lytic transglycosylase or pectate lyase enzymatic domain directly ligated to the upstream hydrophilic sequence (Charkowski et al., 1998; Kvitko et al., 2007). This structural difference makes one-domain and two-domain harpins distinct from each other in terms of subcellular localization and molecular recognition in plants under bacterial infection. While two-domain harpins may target the bacterial periplasm or plant cell walls to facilitate pilus growth (Mushegian et al., 1996; Charkowski et al., 1998; Kim and Beer, 1998; Koraimann, 2003; Zahrl et al., 2005; Dik et al., 2017), one-domain harpins localize to plant PMs, with a potential role in translocon assembly (Oh and Beer, 2007; Bocsanczy et al., 2008). Specific recognition compounds of either lipids (Haapalainen et al., 2011) or proteins (Oh and Beer, 2007; Li et al., 2015) located at eukaryotic PMs are thought to be indispensable for translocator recognition and translocon formation (Ji and Dong, 2015a).
Membrane lipids are believed to be common interactors of bacterial T3 translocators (Matteï et al., 2011; Cortes et al., 2014). In animals, cholesterol is involved in animal cellular signaling and is required for the PM binding of IpB, a hydrophobic translocator from Shigella (Cortes et al., 2014). Cholesterol is also needed for the bilayer binding of PopB and PopD, hydrophobic translocators from Pseudomonas aeruginosa (Cortes et al., 2014). In plants, phosphatidic acid is implicated in sensing HrpZ1, a one-domain harpin (He et al., 1993) and an effector translocator (Kvitko et al., 2007) of Pseudomonas syringae, a bacterial pathogen that infects a variety of plant species. HrpZ1 interacts with synthetic phosphatidic acid and induces it to create pores in artificial lipid vesicles and in designer vesicles made from Arabidopsis thaliana PMs (Haapalainen et al., 2011). In all cases, the function of lipids in bacterial effector translocation remains elusive.
In addition to membrane lipids, PM-integral proteins are also implicated in plant recognition of bacterial T3 translocators, especially one-domain harpins (Ji and Dong, 2015a, b; Li et al., 2015). Recently characterized harpin interactors belong to the PM intrinsic proteins (PIPs) (Li et al., 2015), which constitute a unique aquaporin (AQP) family in plants (Gomes et al., 2009; Verkman, 2013). It has been shown that PIPs from A. thaliana and Oryza sativa L. (rice) interact with Hpa1 (Ji and Dong, 2015a; Li et al., 2015), a one-domain harpin from Xanthomonas oryzae pv. oryzae (Xoo). Xoo causes bacterial blight in rice through effector proteins, either transcription activator-like effectors (TALEs) or non-TALEs such as Xanthomonas outer proteins (Xops) as designated with the bacterial genus landmark (White et al., 2009; Büttner, 2016). TALEs and non-TALEs are secreted along with harpins by the T3SS in a chronological pattern (Roden et al., 2004; Büttner, 2012) and then translocated into plant cells to play a virulent or avirulent role depending on plant varieties (Bogdanove and Voytas, 2011; Büttner, 2016).
Like all bacterial effectors, TALE and non-TALE translocation is a transient passage through the T3 translocon hypothetically made up of translocators (Büttner et al., 2002; Büttner, 2012), which include one-domain harpins such as Hpa1 from bacteria (Kvitko et al., 2007; Wang et al., 2018). Hpa1 can interact with AtPIP1;4 at Arabidopsis PMs (Li et al., 2015) and with OsPIP1;3 in a split-ubiquitin yeast two-hybrid (SUB-Y2H) system (Ji and Dong, 2015a), but whether both proteins also interact in plants is unclear. Indeed, there is as yet no study to validate the hypothetic role of plant PMs in translocator recognition and effector translocation. This study demonstrates that OsPIP1;3 is a Hpa1 interactor required for the translocation of PthXo1, a TALE that determines virulence of the famous Xoo strain PXO99A on the susceptible rice variety Nipponbare (Yang et al., 2006).
Materials and methods
Plant growth
Rice seeds were germinated in flat plastic trays filled with a substrate containing peat, sand, and vermiculite (1:1:1, v/v/v). Three days later, the germinated seedlings were moved into 12 liter pots (3‒5 plants per pot) filled with soil from a local rice grower field. Seeds were incubated and the plants were grown in environment-controlled chambers under 28 °C, 12 h light at 250±50 μmol quanta m–2 s–1 and a relative humidity of 85%.
Microbes and vectors
Bacterial and yeast (Saccharomyces cerevisiae) strains and plasmid vectors used and created in the present study and information on antibiotic resistance are listed in Supplementary Table S1 at JXB online. Escherichia coli was grown at 37 °C in Luria–Bertani (LB) broth or on LB agar (LA) plates with the appropriate antibiotics. Xoo strains were cultured at 28 °C on nutrient broth (NB) or NA agar (NA) medium (Li et al., 2011). Bacteria were cultured on medium supplemented with 100 μg ml–1 ampicillin, 100 μg ml–1 spectinomycin, or 50 μg ml–1 kanamycin. Agrobacterium tumefaciens strain GV3101 was used in plant transformations. The yeast strain NMY51 was used in SUB-Y2H (Dualsystems) assays and grown on synthetic dropout (SD) amino acid nutrient media.
Protein interaction assays
The SUB-Y2H (Li et al., 2015), in vitro glutathione S-transferase (GST) pulldown (Yamaguchi et al., 2013), co-immunoprecipitation (Co-IP), and bimolecular fluorescence complementation (BiFC) (Li et al., 2015) assays were performed as previously described.
Xoo gene modifications
The hpa1 and pthXo1 genes were deleted from the PXO99A genome by using the unmarked deletion method (Li et al., 2011). Upstream and downstream flanking partial sequence fragments of hpa1 or pthXo1 were amplified from the PXO99A genomic DNA and connected together by overlapped fusion-PCR using specific primers (see Supplementary Table S2). Every PCR product was confirmed by sequencing and then cloned into the vector pK18sacB by digestion with BamHI and XbaI, and ligation with T4 ligase (Thermo Scientific). Every recombinant vector was introduced into PXO99A cells by electroporation, followed by single-colony selection on kanamycin-containing and sugar-lacking NA plates. Colonies from single crossovers were transferred to NB broth, grown at 28 °C for 12 h, and then transferred onto plates containing NA and 10% sucrose. Sucrose-resistant colonies were replica streaked onto NA plates with and without kanamycin supplementation. Colonies resulting from double crossover events were selected based on kanamycin-negative and sucrose-positive traits, and unmarked mutants were confirmed by PCR amplification of hpa1 and pthXo1, respectively. To create double mutants, pK18sacB:∆hpa1 was transformed into the ∆pthXo1 mutant.
Different tags were attached to the 3ʹ terminus of the pthXo1 sequence in the pZWpthXo1 plasmid vectors. To create a cya-fused gene, a 1218 bp cya fragment encoding amino acids 2–406 of the calmodulin-dependent adenylate cyclase (Cya) protein was amplified from plasmid pMS107 and pre-fixed with the last 51 bp region of pthXo1 that contained a SacI recognition site (Supplementary Table S2). The recombinant sequence was inserted into pZWpthXo1 at the SacI site. To construct pthXo1–blaM, blaM was amplified from plasmid pBR322 using specific primers that contained a SalI site. The confirmed PCR product was inserted into the pthXo1 sequence at the SalI site in pZWpthXo1 (Makino et al., 2006). Every recombinant vector was linearized with HindIII and cloned into the pHM1 vector for genetic complementation. The hpa1pthXo1 double complementary vectors were constructed using two steps. First, the hpa1 sequence that was linked to its own promoter was cloned into pHM1 between the PstI and KpnI sites. Secondly, pZWpthXo1 was linearized using HindIII and inserted into the HindIII site of pHM1hpa1. Complementation or transformation was performed by electroporation.
OsPIP1;3 overexpression
The 867 nucleotide full-length OsPIP1;3 sequence was cloned by PCR using specific primers (Supplementary Table S2). The confirmed OsPIP1;3 cDNA sequence was fused to the yellow fluorescent protein (YFP) gene, yielding the OsPIP1;3:YFP fusion gene. OsPIP1;3:YFP was cloned into the plant binary vector pCAMBIA1300 that carried the Cauliflower mosaic virus 35S promoter (Supplementary Fig. S4). The recombinant vector was transferred into A. tumefaciens and introduced into the Nipponbare genome by a previously described protocol (Hiei et al., 1994). A total of 21 OsPIP1;3-YFP lines were characterized in terms of the transgene integration and expression. PM and cytoplasmic proteins were isolated from leaves or protoplasts and analyzed by immunoblotting as previously described (Yamaguchi et al., 2013). T3 homozygous progenies of transgenic OsPIP1;3-YFP plants were used in the present study.
OsPIP1;3 knockout
Target sites of OsPIP1;3 TALENs were predicted by using the online software, TALE Nucleotide Targeter 2.0 (https://tale-nt.cac.cornell.edu/node/add/talen) and by referencing BioEdit alignments of OsPIP1 nucleotide sequences. TALENs were constructed using a previously described method (Li et al., 2012). The synthesized TALE domain was ligated into the SphI and AatII sites of the psk/TALEN vector, while the TALEN gene was ligated into the pCAMBIA1300 and pEH3 vectors at the KpnI and SacI sites. Both vectors were ligated together at the HindIII site to form an intact TALEN construct, which was used in A. tumefaciens-mediated rice transformation. Rice transformation was conducted, and transgenic plants were generated by a regular protocol. To characterize transgenic plants, DNA was sequenced to identify regions in the OsPIP1;3 sequence targeted by TALENs. Genomic DNA from individual plants was analyzed by PCR using TALEN-F/R primers to amplify a region of 554 bp, with the pre-selected target sites located in the middle. PCR products were sequenced using an internal primer. Each sequencing chromatogram was manually analyzed for polymorphisms (Li et al., 2012). T3 homozygous progenies of TALEN (OsPIP1;3‒) lines were used in the present study.
PthXo1 translocation assays
The Cya reporter (Wang et al., 2018) and the β-lactamase (BlaM) reporter (Mills et al., 2008; Mills et al., 2013) assays were performed on 2-week-old rice seedlings following inoculation with Cya-related Xoo strains.
Additional materials and methods
Bioinformatics analysis, protein interaction assay procedures, bacterial virulence test, gene expression analysis, Cya and BlaM reporter assay procedures, protein preparation and statistical analysis, and virus-mediated gene overexpression are described in the Supplementary data.
Results
Hpa1 directly interacts with OsPIP1;3 at plant PMs
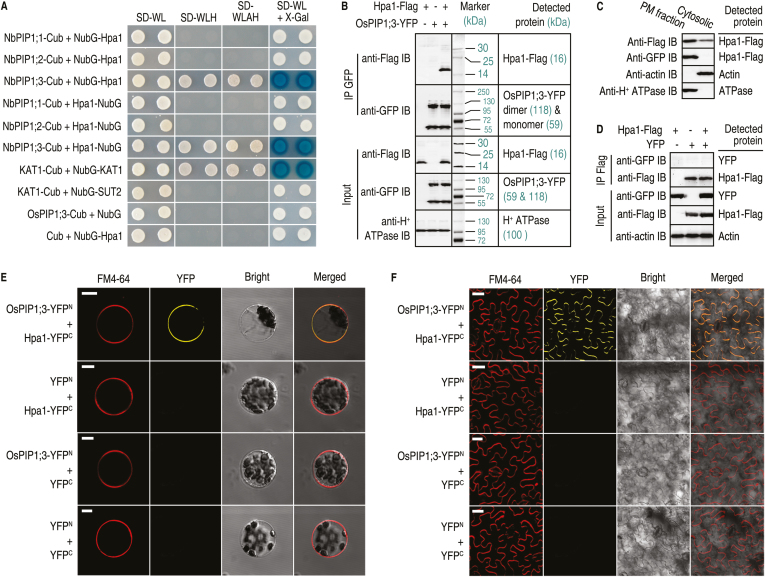
Previously, Y2H assays indicated that Hpa1 interacted with Arabidopsis PIP1;4 and not other PIP family members (Li et al., 2015). As rice lacks orthologs of Arabidopsis PIP1;4 and PIP1;5 (Gomes et al., 2009), SUB-Y2H was used to assay Hpa1 interactions with PIP1;1, PIP1;2, and PIP1;3 from rice variety Nipponbare. In this study, SUB-Y2H assays confirmed a direct interaction between Hpa1 and OsPIP1;3, but not between Hpa1 and OsPIP1;1 or OsPIP1;2 (Fig. 1A). In OsPIPs, OsPIP1;3 is closely related to AtPIP1;4 (Supplementary Fig. S1), explaining the evolutionary basis of Hpa1 interactions with AtPIP1;4 (Ji and Dong, 2015a) and OsPIP1;3 (Li et al., 2015). This interaction was confirmed with Co-IP of PM proteins from rice protoplasts after transformation with both hpa1-flag and OsPIP1;3-YFP. Apparent Hpa1-flag‒OsPIP1;3–YFP complexes were precipitated using an anti-green fluorescent protein (GFP) antibody that cross-reacts with YFP. Hpa1-flag and OsPIP1;3–YFP in the complex were detected by immunoblotting with antibodies against flag and GFP (Fig. 1B). Interestingly, OsPIP1;3–YFP appeared as both a monomer and dimer, whereas only monomeric Hpa1-flag was observed (Fig. 1B). Hpa1 was present alone in the cytosolic fraction, as detected by immunoblotting with anti-Flag antibody, or appeared in the complex with OsPIP1;3 in the PM fraction, as detected by immunoblotting with anti-Flag and anti-GFP antibodies, respectively (Fig. 1C). This indicated that Hpa1 and OsPIP1;3 interacted specifically at rice PMs. The specificity of OsPIP1;3–Hpa1 interaction was confirmed by the absence of interaction between Hpa1 and YFP (Fig. 1D). Furthermore, the OsPIP1;3–Hpa1 interaction was monitored by YFP BiFC. In rice protoplasts, the OsPIP1;3–Hpa1 interaction was localized to PMs, where the BiFC signal was co-localized with the PM marker, FM4-64 (Fig. 1E). In contrast, no BiFC signal was found in combinations between Hpa1 and OsPIP1;3, or for Hpa1 and OsPIP1;2 (Supplementary Fig. S2). In tobacco Nicotiana benthamiana leaves, Hpa1 was localized to PMs and interacted with the introduced OsPIP1;3 (Fig. 1F). These findings indicate that Hpa1 and OsPIP1;3 directly interact with each other at plant PMs.
Fig. 1.
Hpa1 and OsPIP1;3 interact in vitro and in vivo. (A) SUB-Y2H tests of Hpa1 and OsPIP1s. KAT1-Cub+NubG-KAT1 and KAT1-Cub+NubG-SUT2 were used as positive and negative controls, respectively. (B) Co-IP of PM proteins from rice protoplasts co-transformed with OsPIP1;3-YFP and hpa1-flag. GFP antibody IP products and input proteins were detected by immunoblotting (IB) with the indicated antibodies. H+-ATPase was detected as a PM-localized protein reference. Weak signals of non-specific hybridization appear, presumably due to protein degradation and/or the presence of homologous proteins. (C) Immunoblotting of PM and cytosolic proteins isolated from rice protoplasts treated with the Hpa1-Flag fusion protein. (D) Co-IP of PM proteins from rice protoplasts co-transformed with YFP and hpa1-flag. (E) YFP BiFC of OsPIP1;3 and Hpa1 expressed in rice protoplasts. Scale bars=10 μm. (F) YFP BiFC of OsPIP1;3 and Hpa1 expressed in N. benthamiana leaves. Scale bars=20 μm. In (E, F) the red fluorescent PM marker FM4-64 was used to show cell outlines.
OsPIP1;3 is a host disease-susceptible component
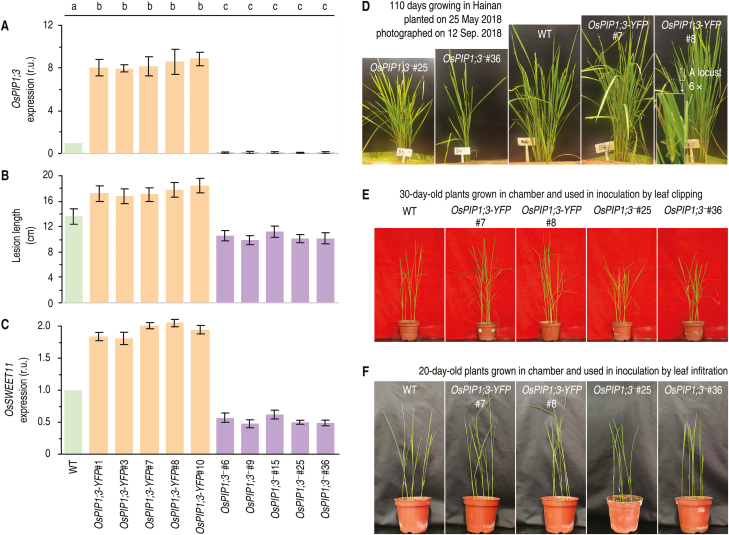
To determine if OsPIP1;3 plays a role in the regulation of bacterial virulence associated with translocation of PthXo1, OsPIP1;3 expression levels were altered by two methods. First, TALEN DNA sequence editing (Li et al., 2012) was used to disrupt OsPIP1;3 in Nipponbare (Supplementary Fig.S2 A–C) as demonstrated in subsequent genetic analyses of progeny (Supplementary Fig. S2D, E). Secondly, Nipponbare was transformed with the chimeric gene OsPIP1;3:YFP and transgenic plants were created, which expressed the OsPIP1;3–YFP fusion gene under a constitutive promoter (Supplementary Fig. S3). These plants were referred to as OsPIP1;3-YFP or OsPIP1;3-overexpressing lines (Fig. 2A–C) for the sake of convenient description. Five lines of OsPIP1;3 knockout (OsPIP1;3‒) plants and five lines of OsPIP1;3-overexpressing (OsPIP1;3-YFP) plants were characterized in terms of genetic characteristics of gene overexpression and transcriptional nullification, respectively, in comparison with the steady-state expression level in the wild-type (WT) plants (Fig. 2A). Then, OsPIP1;3‒ and OsPIP1;3-YFP plants were further compared with the WT in terms of susceptibility to the Xoo virulent strain PXO99A, which was prepared as a bacterial suspension and used as an inoculum applied by the leaf-top clipping method. As observed at the 12th day post-inoculation (dpi), the different genotypes of the plant displayed a high degree of variation in severity of the bacterial blight symptoms. The disease severity quantified as blight lesion length was significantly aggravated (P<0.05) in OsPIP1;3-YFP plants but this was significantly (P<0.05) alleviated in OsPIP1;3‒ lines compared with the WT (Fig. 2B). Moreover, the aggravation and alleviation of bacterial blight severity coincided with enhanced and weakened expression of OsSWEET11 (Fig. 2C), the host susceptible gene used as the regulatory target of PthXo1 in Nipponbare (Yang et al., 2006). These data suggest that OsPIP1;3 is a disease-facilitating factor supporting the virulent role of PthXo1 in the susceptible rice variety.
Fig. 2.
Altered OsPIP1;3 expression levels affect the virulent function of PthXo1 and growth extents of plants. (A) Relative units (r.u.) of OsPIP1;3 expression in rice leaves based on RT–qPCR analyses in which the average expression level was set as 1 in the WT plant to estimate relative levels of gene expression in other plants. (B) Lesion length of bacterial blight on leaves at 12 dpi by leaf-top clipping and the areas of hypersensitive cell death on leaves at 5 dpi by leaf infiltration with each of the bacterial suspensions. (C) Relative levels of gene expression in leaves 12 hpi. (A–C) All data are shown as mean values ±SD error bars; different letters on bar graphs indicate significant differences by Duncan’s multiple range tests; P<0.01; n=30 plants from six independent experiments each including five repetitions in (A); n=30 leaves from six independent experiments in (B); n=9 repetitions from three independent experiments in (C). (D) Plants grown in an environment-protected breeding base located at Hainan University, Haikou, Hainan Province, China. (E, F) Appearance of plants at two ages used in inoculation experiments.
OsPIP1;3 and hpa1 cooperate to promote the virulent role of PthXo1
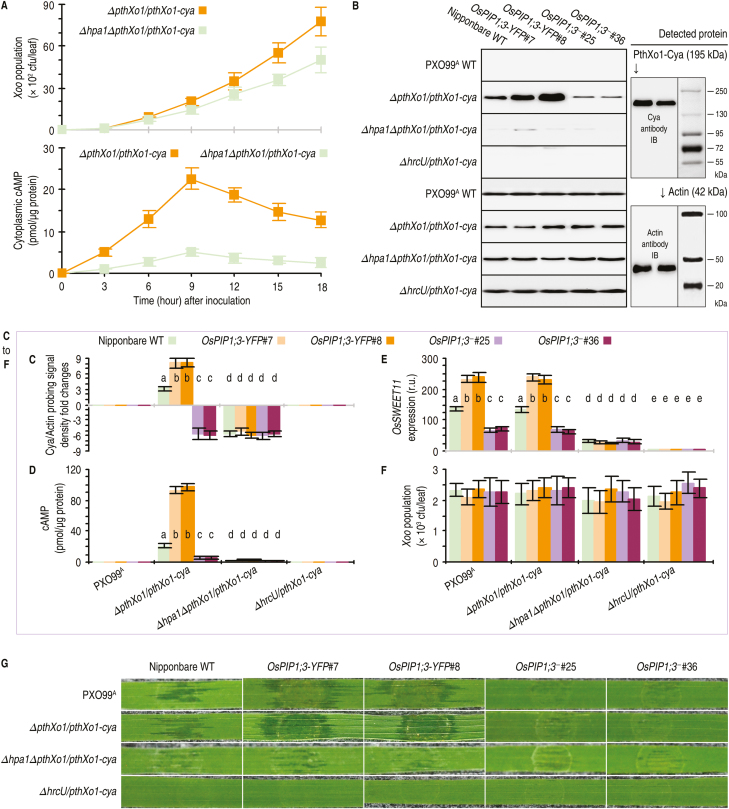
Two randomly selected OsPIP1;3-YFP lines (#7 and #8) and two OsPIP1;3‒ lines (#25 and #36) were further studied. Compared with the WT plant, OsPIP1;3-YFP and OsPIP1;3‒ lines displayed growth enhancement and impairment, respectively (Fig. 2D), suggesting that OsPIP1;3 is needed for plant growth in addition to disease susceptibility. The designed variations in OsPIP1;3 expression levels were confirmed by real-time reverse transcription–PCR (RT–PCR) analysis (Fig. 3A). The OsPIP1;3–YFP fusion protein was detected only in OsPIP1;3-YFP lines (Fig. 3B). The susceptibility of OsPIP1;3 to the bacterial blight disease was verified by leaf-top clipping inoculation experiments performed on 30-day-old plants (Fig. 2E). As measured at 5 dpi, the pathogen had higher and lower populations accordingly in OsPIP1;3-YFP and OsPIP1;3‒ lines than in the WT plant (Fig. 3C). In the subsequent period up to 12 dpi when the bacterial blight disease was assessed, the OsPIP1;3-YFP plants were found to incur more severe blight symptoms while the disease severity was strikingly alleviated in OsPIP1;3‒ lines (Fig. 3D). Furthermore, the in planta bacterial populations (Fig. 3C) and leaf blight symptom severities (Fig. 3D), both of which reflect levels of bacterial virulence, were weakened similarly by hpa1 knockout and almost nullified by knockout of pthXo1 or both pthXo1 and hpa1 (Fig. 3C, D). In contrast, levels of bacterial virulence were increased to the highest extents in OsPIP1;3-YFP plants inoculated with the recombinant PXO99A strain containing both hpa1 and pthXo1 genes compared with the other combinations between plant genotypes and Xoo strains (Fig. 3C, D). Therefore, the bacterial hpa1 gene and the plant OsPIP1;3 gene are concomitantly involved in the virulent function of PthXo1.
Fig. 3.
Hpa1 and OsPIP1;3 influence the role of PthXo1 in Xoo virulence on the susceptible rice variety Nipponbare. The experiments were performed on the wild-type (WT) plant and two types of transgenic plants, OsPIP1;3-overexpressing (OsPIP1;3-YFP) and TALEN-based OsPIP1;3 knockout (OsPIP1;3‒). Transgenic plants were characterized in terms of OsPIP1;3 expression levels and the presence and absence of OsPIP1;3 protein production. Transgenic plants were further compared with the WT to evaluate the virulence of hpa1- and pthXo1-related bacterial strains. (A) OsPIP1;3 expression in leaves of the different plants. Gene expression was analyzed by RT–qPCR using the constitutively expressed EF1α gene as a reference. (B) Immunoblotting of rice PM fractions. H+-ATPase was detected as a PM-localized protein reference. (C) Bacterial populations in leaf tissues at 5 dpi by the leaf-top clipping method. (D) Graduated presentation of bacterial blight symptoms on rice leaves photographed at 12 dpi by the leaf-top clipping method. In (A, C), quantitative data are given as the means ±SEM. Different letters on bar graphs indicate significant differences among data from the different plants inoculated with the indicated bacterial strain (P<0.01). Repeat number (n)=30 plants from six independent experiments each including five plants in (A); n=24 repetitions from eight independent experiments each involving three repetitions in (C).
OsPIP1;3 and Hpa1 cooperate to mediate PthXo1 translocation
Translocation of PthXo1 was analyzed by fusion to Cya, a widely used eukaryotic cytoplasmic import marker with no detectable effect on pathological roles of the tested effectors in the fused form (Chakravarthy et al., 2017). The Cya reporter system allows for accurate quantification of an effector moved into eukaryotic cells based on the cytosolic concentration of cAMP as an exclusive product of the effector–Cya activity. The effects of OsPIP1;3 and Hpa1 on PthXo1 translocation were assessed on 20-day-old WT Nipponbare, OsPIP1;3-YFP, and OsPIP1;3‒ plants. These plants were inoculated in advance by leaf infiltration separately with bacterial suspensions of the PXO99A WT strain and its recombinant strains ∆pthXo1/pthXo1-cya, ∆hpa1∆pthXo1/pthXo1-cya, and ∆hpa1∆pthXo1/hpa1/pthXo1-cya generated previously (Ji and Dong, 2015a; Wang et al., 2018). In addition, HrcU is an inner membrane protein essential for substrate docking into the T3SS in Xanthomonas bacteria (Hartmann and Büttner, 2013). Thus, the previously generated recombinant PXO99A strain ∆hrcU/pthXo1-cya (Wang et al., 2018) was used as a negative control for effector translocation in the inoculation experiments. Inoculated leaves were sampled at 9 hours post-infection (hpi to analyze the assumed translocation of the PthXo1–Cya fusion protein from the bacterial cells into the cytosol of rice cells. This time point was chosen with the aim of accurately quantifying the hypothesized regulatory role of OsPIP1;3 or Hpa1 i PthXo1 translocation and to exclude the possible effect of bacterial multiplication in leaf tissues on the TALE translocation. Within 12 hpi, recombinant PXO99A strains ∆pthXo1/pthXo1-cya and ∆hpa1∆pthXo1/pthXo1-cya had an equivalent population in leaf tissues. However, high levels of PthXo1 translocation from ∆pthXo1/pthXo1-cya bacteria were detected at 9 hpi (Fig. 4A). Therefore, the time point of 9 hpi was used consistently in order to correlate the virulence role of PthXo1 with associated molecular responses in rice, such as PthXo1-activated expression of the rice OsSWEET11 gene (Yang et al., 2006).
Fig. 4.
OsPIP1;3 and hpa1 cooperate for TALE translocation and function in rice leaves. The WT and OsPIP1;3-related plants were inoculated by leaf infiltration with a bacterial suspension of hpa1- and pthXo1-related Xoo strains. Inoculated leaves were sampled at the indicated time points in (A) or at 9 hpi in (B, F) for use in the indicated measurements. (A) Changes of the bacterial population in leaf tissues and concentrations of cAMP resulting from the PthXo1–Cya activity in leaf cells over the course of time after plant inoculation with the indicated bacterial strains. Data shown are mean values ±SEMs (n=3 independent experiments each involving five leaves). (B) Immunoblotting (IB) of cytoplasmic proteins from rice leaves. As shown on the right, protein blots were hybridized either with the specific anti-Cya antibody, or with the specific antibody against monomeric actin, which was used as a cytoplasmic protein marker. (C) Relative levels of the PthXo1–Cya protein in leaf samples based on densities of hybridization signals in blots from (B, D). Concentrations of cAMP from PthXo1–Cya activity in the cytosol of leaf cells. (E) Relative levels of OsSWEET11 expression in leaves. Quantities of the gene transcript in the corresponding plants inoculated with ΔhrcU/TALE-cya were defined as 1 to evaluate relative levels of the gene expression in other plants. (F) Bacterial populations in leaf tissues. (G) Segments of leaves photographed at 3 dpi. In (C, F), all data are provided as the means ±SEMs. On bar graphs, different letters indicate significant differences in multiple comparisons of data from the different combinations of plants and bacterial strains; P<0.01; n=15 repetitions from five independent experiments each involving three repetitions.
Immunoblotting with an anti-Cya antibody revealed that PthXo1–Cya was present in cytosolic proteins isolated from inoculated rice leaves of the different plant genotypes (Fig. 4B). Cytosolic PthXo1–Cya signal coincided with actin, as shown by hybridization with the antibody against monomeric actin (Fig. 4B), suggesting that the PthXo1–Cya fusion protein had moved into the cytosol of rice cells within 9 hpi. The Cya to actin ratio of immunoblotting signal densities was used to assess relative levels of the protein translocation. Based on this estimate, a substantial quantity of PthXo1–Cya translocation was provided by the presence of the hpa1 gene, whereas the protein translocation amount was significantly (P<0.01) decreased due to the absence of hpa1 in the recombinant PXO99A strains (Fig. 4C). In contrast, the largest amount of PthXo1–Cya translocation was associated with the concomitant presence of the canonical hpa1 gene and overexpressed OsPIP1;3 (Fig. 4C).
The cooperative role of hpa1 and OsPIP1;3 in PthXo1 translocation was confirmed by measuring cAMP concentrations in rice leaf cells (Fig. 4C). High concentrations of cAMP were detected in leaves of OsPIP1;3-YFP plants following inoculation with the recombinant PXO99A strain ΔpthXo1/pthXo1-cya or Δhpa1ΔpthXo1/hap1/pthXo1-cya. In contrast, Cya activity was significantly (P<0.01) decreased due to deletion of the hpa1 gene, as evidenced by a marked reduction in the cAMP content in leaves inoculated with the bacterial Δhpa1ΔpthXo1/pthXo1-cya strain (Fig. 4D). In agreement with the role of PthXo1 in OsSWEET11 activation (Roche and Törnroth-Horsefield, 2017), OsSWEET11 expression was highly increased in plants inoculated with bacterial strains that contained both hpa1 and pthXo1. Conversely, OsSWEET11 expression was almost eliminated with hpa1 or pthXo1 deletion (Fig. 4E). Furthermore, both PthXo1 translocation and OsSWEET11 expression were enhanced to the greatest extent in OsPIP1;3-overexpressing plants inoculated with the bacterial strain that carries the canonical hpa1 gene in addition to pthXo1 (Fig. 4C–E).
In all experiments, neither PthXo1 translocation nor OsSWEET11 expression was found with ∆hrcU/pthXo1-cya (Fig. 4B-E), confirming the reliability of our experimental system in assessing the effector translocation. In all experiments, moreover, bacterial populations in leaves had little effect on changes in PthXo1 translocation within 9 hpi. At this time point, cAMP concentrations were measured to quantify translocated PthXo1–Cya protein, while in planta bacterial populations of the different strains were almost similar regardless of hpa1 deletion or OsPIP1;3 expression levels (Fig. 4F). Indeed, the biggest difference in the in planta bacterial populations between different strains was <20%. However, cAMP concentrations were increased by ~3.5 times in OsPIP1;3-overexpressing lines and decreased by nearly 70% in OsPIP1;3-knockout lines compared with the WT plant (Fig. 4D). Deletion of the hpa1 gene from the bacteria caused an ~90% reduction of the cAMP content (Fig. 4D). Thus, the quantitative changes in PthXo1 translocation were attributable to hpa1 deletion and OsPIP1;3 overexpression or knockout, rather than differences in bacterial populations. In addition, the leaf infiltration method reflected well the bacterial virulence determined by PthXo1 and modulated by both Hpa1 and OsPIP1;3 (Fig. 4G). These results support the notions that Hpa1 and OsPIP1;3 work together to facilitate PthXo1 translocation from Xoo cells into the cytosol of rice cells, and that the functional interaction contributes to the bacterial virulence on the susceptible rice variety.
The α-helix repeat of Hpa1 is an OsPIP1;3-interacting motif essential for PthXo1 translocation
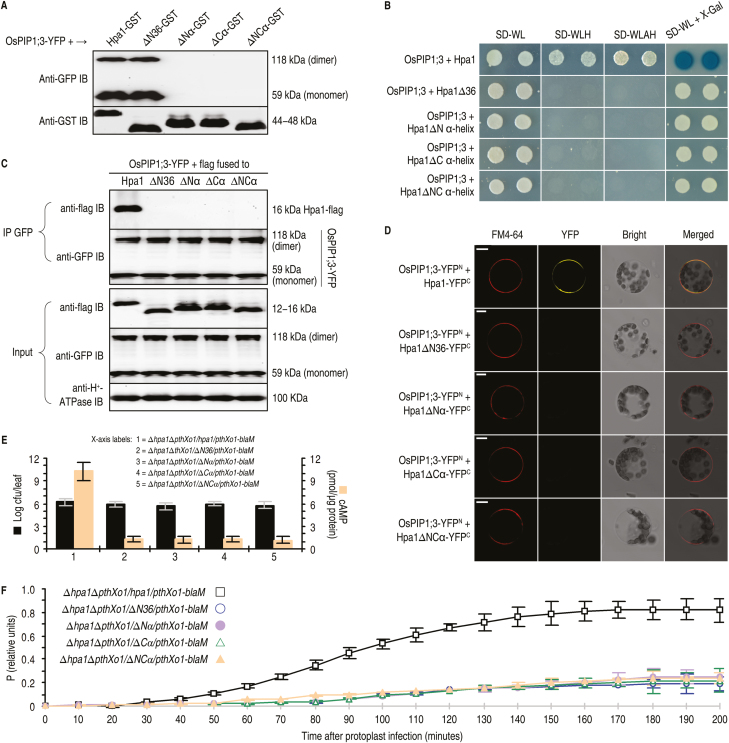
T3 translocators, including one-domain harpins, rely on α-helical coiled-coil motifs to interact with themselves or with functional partners (Zhu et al., 2000; Espina et al., 2006; Ji and Dong, 2015a, b). To identify OsPIP1;3-interacting motifs in the Hpa1 sequence, hpa1 mutant versions ∆N36, ∆Nα, ∆Cα, and ∆NCα were generated by deleting the first 36 residues, the N-terminal α-helix, the C-terminal α-helix, and both α-helices, respectively. All mutant versions attenuated the virulent role of PthXo1 (Supplementary Fig. S4).
In the in vitro pulldown assay, an interaction was detected in the combination of OsPIP1;3 either with Hpa1 or with Hpa1∆N36, but no interaction was found between OsPIP1;3 and the other mutant versions of Hpa1 (Fig. 5A). In the in vivo assays using SUB-Y2H (Fig. 5B), Co-IP (Fig. 5C), and BiFC (Fig. 5D) techniques, an interaction was found between OsPIP1;3 and Hpa1, but all mutant versions of Hpa1 lost the ability to interact with OsPIP1;3. The different behaviors of Hpa1∆N36 in the in vitro and in vivo interacting assays are consistent with the predicted role of the N-terminal region in directing Hpa1 secretion or targeting to plant PMs (SignalP4.1 Server at http://www.cbs.dtu.dk/services/SignalP/). In agreement with the demonstrated importance of the interaction Hpa1 and OsPIP1;3 for PthXo1 translocation from the bacteria into rice cells, the cytosolic import of PthXo1–Cya was severely compromised by Hpa1 mutations compared with that observed with the canonical protein (Fig. 5E).
Fig. 5.
The pair of α-helices in the Hpa1 sequence is an OsPIP1;3-interacting motif playing a critical role in PthXo1 translocation. (A–D) Hpa1 mutant versions ∆N36, ∆Nα, ∆Cα, and αNC were generated by deleting the N-terminal region made up of 36 residues, the N-terminal α-helix, the C-terminal α-helix, and both α-helices, respectively. The canonical and mutant versions of Hpa1 were fused to the GST tag, followed by molecular interaction assays in single combinations with OsPIP1s, which were used either in the canonical form or in a fusion to YFP, depending on the experimental methods. (A) Pulldown assays. Every GST-linking protein was purified and then immobilized on glutathione affinity resins. The eluates were analyzed by immunoblotting with GST or GFP antibody. (B) SUB-Y2H tests of Hpa1 mutants and OsPIP1;3. (C) Co-IP of PM proteins from OsPIP1;3-YFP#8 rice protoplasts transformed with the hpa1 mutants. (D) YFP BiFC of OsPIP1;3 and Hpa1 mutant versions expressed in rice protoplasts. Scale bars=10 μm. (E, F) The hpa1 gene variants ∆N36, ∆Nα, ∆Cα, and αNC were introduced into the ∆hpa1 or ∆hpa1∆pthXo1/pthXo1-cya mutant of Xoo strain PXO99A. Recombinant bacteria were used in Nipponbare inoculation by the leaf-top clipping method. Inoculated plants were subjected to the following analyses. (E) Blight lesion length on leaves 9 dpi and the content of cAMP from PthXo1–Cya activity in cytoplasm of leaf cells at 12 hpi. (F) Concentrations of P from PthXo1–BlaM activity in rice protoplasts infected by co-incubation with bacteria of the indicated strains. In (E, F) data shown are the mean values ±SEM bars; n=9 repetitions from three independent experiments each involving three repetitions.
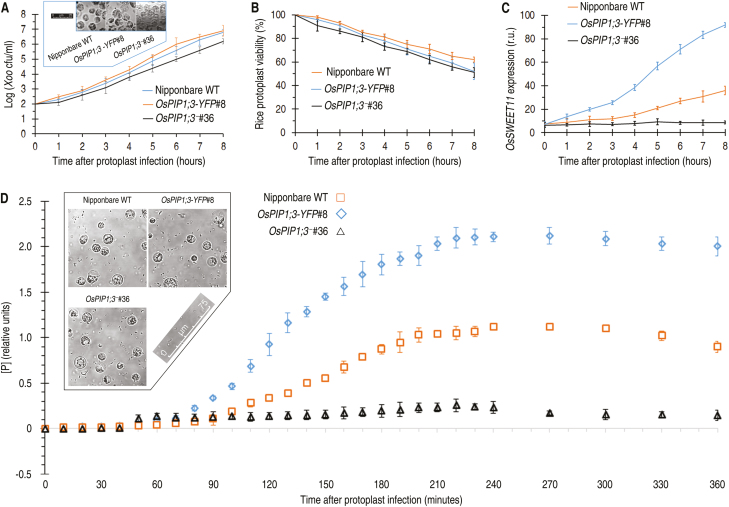
Similar results were obtained by analysis with the BlaM reporter system, which is a real-time, high-throughput translocation assay previously developed for animal-pathogenic bacteria (Mills et al., 2008, 2013). Plant protoplasts have not been used in studying effector translocations during bacterial infections. However, PXO99A bacteria displayed virulence on rice protoplasts after pre-activation in a medium which induces the expression of hrp genes and assembly of the T3SS (Tsuge et al., 2002). Pre-activated bacteria were able to induce OsSWEET11 expression in rice protoplasts, while non-induced bacteria were not (Supplementary Fig. S5. This result suggests that PthXo1 had been translocated into host cells and that rice protoplasts are feasible for use in the study of effector translocation.
The BlaM reporter system was used in monitoring of PthXo1 translocation into rice protoplasts. The blaM reporter gene fused to pthXo1 did not affect the virulent role of pthXo1 in Nipponbare (Supplementary Fig. S6A). At 200 min post-infection (mpi), the viability rate of protoplasts declined, but ensured assessments of the effector translocation (Supplementary Fig. S6B). At 150 mpi, a perfect correlation was observed between the effector translocation level of [P] and the bacterial inoculum dosage, with a saturation point of [P] at a dosage of OD600=0.5 (Supplementary Fig. S6C). However, the high bacterial dosage caused a quick decline in protoplast viability, and an adequate time course of assays was impossible. The short time might cover only a few cycles of bacterial replication and could not ensure sufficient secretion and translocation of the effector. Thus, the protoplast viability was increased by reducing the bacterial amount in the incubation. In this case, bacteria multiplied well (Fig. 6A) and rice protoplasts gained high levels of viability (Fig. 6B) at least with 8 h of incubation. OsSWEET11 was more highly expressed in protoplasts of OsPIP1;3-YFP#8 than in the WT, while gene expression was maintained at low levels in OsPIP1;3−#36 (Fig. 6C). The amount of PthXo1–BlaM translocation (Fig. 6D) was increased in protoplasts of the OsPIP1;3–YFP plant from 80 mpi and abolished in those of the OsPIP1;3− line. In these assays, >70% of rice protoplasts retained their integrity (Fig. 6A, D insets). These analyses confirm the technical feasibility of analyzing the effector translocation and the role of OsPIP1;3 and Hpa1 cooperation. In addition, the cytosolic import of PthXo1–BlaM in rice protoplasts was highly reduced by Hpa1 mutations as compared with that observed with the canonical protein, full-length Hpa1 (Fig. 5F). Overall, these findings indicate that the pair of α-helical coiled-coil motifs present in the Hpa1 sequence comprise an OsPIP1;3-interacting determinant, and also determines the role of Hpa1 in PthXo1 translocation.
Fig. 6.
The Blam reporter confirms the regulatory role of OsPIP1;3 in PthXo1 translocation. (A–C) Changes of Xoo population, rice protoplast viability, and OsSWEET11 expression levels after 8 h of incubation with bacteria of the recombinant PXO99A strain containing the pthXo1-blaM fusion gene. Insert in (A) shows protoplasts photographed after an 8 h incubation. In (C), gene expression was analyzed by RT–qPCR using the EF1α gene as a reference to quantify OsPIP1;3 expression levels in protoplasts of the different plants. (D) Concentrations of Pfrom PthXo1–BlaM activity in rice protoplasts infected by co-incubation with bacteria of the recombinant PXO99A strain. The inset shows rice protoplasts photographed after 6 h of incubation. (A–D) All quantitative data are given as means ±SEMs; n=9 repetitions from three independent experiments each involving three repetitions.
LE of OsPIP1;3 is a Hpa1-interacting determinant and also determines PthXo1 translocation and virulence performance
The primary roles of PIPs in substrate transport are determined by their topological structures. In the current model, PIPs consist of six α-helical transmembrane (TM) domains (TM1–TM6) that are tilted along the plane of the PM and are linked to each other by five connecting loops (LA–LE) (Gomes et al., 2009). LB, LD, and both termini are located in the cytoplasm and potentially bind to cytosolic substrates (Lindsey et al., 2006; Hu et al., 2012). In contrast, LA, LC, and LE face the apoplasm and come into contact with apoplastic substrates and signals (Gomes et al., 2009; Jin et al., 2012), and protein-interacting motifs might be located in these extramembrane regions (Ji and Dong, 2015a, b). Hpa1 interacts with OsPIP1;3, but not with OsPIP1;1 and OsPIP1;2 (Fig. 1A; see Supplementary Fig. S1B). Therefore, either OsPIP1;1 or OsPIP1;2 is pertinent for site and fragment substitutions with OsPIP1;3 to look for Hpa1-interacting motifs in the OsPIP1;3 sequence.
A series of site-directed mutations were performed within LA, LC, and LE at amino acid residues that are different between OsPIP1s (see Supplementary Fig. S7, showing residue differences in LE, for example). Unfortunately, none of the site substitutions affected OsPIP1;3 interaction with Hpa1. Then, mutagenicity was used to create different proteins by swapping each of the six extramembrane-related regions between OsPIP1;3 and OsPIP1;1 (Fig. 7A). This approach was conceived since TMs, connecting loops, and particular residues depend on three-dimensional intramolecular interactions to establish a functional architecture (Maurel et al., 2008; Gomes et al., 2009; Qin and Boron, 2013).
Fig. 7.
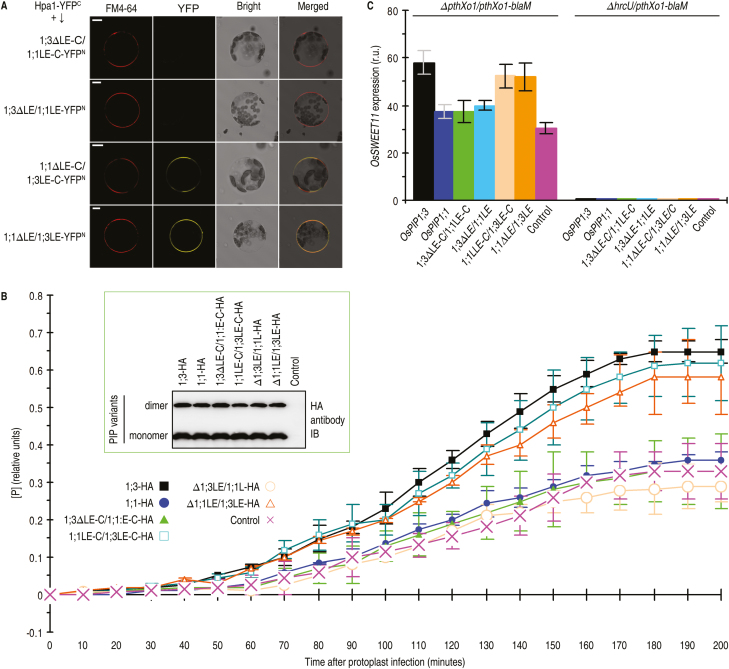
OsPIP1;3 LE is a Hpa1-interacting motif critical for PthXo1 translocation. (A) Diagrams of predicted OsPIP1 structures and sequence region swapping between OsPIP1;3 and OsPIP1;1 to yield substituted proteins. TMs are shown as rectangles and indicated by numbers according to the predicted location in PIP sequences. The nomenclature for substituted proteins is, as for example in 1;3∆LE-C/1;1LE-C, created by switching the LE-linking C-end region from OsPIP1;1 to OsPIP1;3. (B) SUB-Y2H tests of Hpa1 and OsPIP1 substituted proteins. (C) Co-IP of PM proteins from Nipponbare protoplasts transformed with each pair of genes encoding the indicated proteins.
In SUB-Y2H, the OsPIP1;3-Hpa1 interaction was not affected until replacements were extended to the LE-containing C-terminal sequence (Fig. 7B). LE has a different amino acid composition between OsPIP1;3 and its isoforms (Supplementary Fig. S7), and is likely to be critical for OsPIP1;3 interacting with Hpa1. In line with this idea, interactions were not observed between Hpa1 and the protein 1;3∆LE-C/1;1LE-C (Fig. 7B), which was created by switching the LE-linking C-end region from OsPIP1;1 to OsPIP1;3 (Fig. 7A). More importantly, no interaction occurred between Hpa1 and the protein 1;3∆LE/1;1LE (Fig. 7B), which was generated by switching LE from OsPIP1;1 to OsPIP1;3 (Fig. 7A). Furthermore, OsPIP1;1 acquired the ability to interact with Hpa1 when the LE-linking C-end region or LE only was switched from OsPIP1;3 (Fig. 7B). BiFC of Nipponbare protoplasts indicated a direct interaction between Hpa1 and OsPIP1;3 or between Hpa1 and other proteins that contained OsPIP1;3 LE (Fig. 8A). In contrast, no interaction occurred between Hpa1 and the new proteins with LE switching from OsPIP1;1 to OsPIP1;3 (Fig. 8A). These results were confirmed by Co-IP assays, which were based on the co-transformation of Nipponbare protoplasts with the hpa1-flag fusion gene and the canonical or mutant OsPIP genes that were separately fused to the influenza HA epitope. Hpa1 interactions with OsPIP1;3 or with new proteins containing OsPIP1;3 LE were detected by HA or flag immunoblotting of flag antibody IP products, but no interactions took place with OsPIP1;1 or with new proteins containing OsPIP1;1 LE (Fig. 7C). Therefore, LE of OsPIP1;3 determines its interaction with Hpa1.
Fig. 8.
The Hpa1-interacting motif in OsPIP1;3 is critical for TALE translocation. (A) BiFC in Nipponbare protoplasts. Scale bars=10 µm. (B) PthXo1 translocation assessments based on [P] from PthXo1–BlaM activity. Protoplasts of Nipponbare TALEN line OsPIP1;3−#25 were transformed with the HA-fused canonical or mutant form of OsPIP genes and infected with ∆pthXo1/pthXo1-blaM. Control refers to the vector used in gene recombination. Data are the means ±SEMs; asterisks indicate significant differences between proteins carrying OsPIP1;3 LE and lacking the loop, with respect to [P] at the time points from 50 min to 200 min; P<0.01; n=9 repetitions from three independent experiments. The inset is an immunoblotting analysis. (C) OsSWEET11 expression in protoplasts. Protoplasts of the Nipponbare TALEN line OsPIP1;3−#25 were transformed with the HA-fused canonical or mutant form of OsPIP genes and infected with ∆PthXo1/PthXo1-blaM or ∆hrcU/PthXo1-blaM. Data shown are mean values ±SEM bars; different letters indicate significant differences by Duncan’s multiple range tests; P<0.01; n=9 repetitions from three independent experiments.
To assess the effect of OsPIP1;3 LE on PthXo1 translocation, transformed protoplasts of a Nipponbare TALEN OsPIP1;3‒ line were transformed with canonical or mutant OsPIP genes, protoplasts were inoculated by co-incubation with ∆pthXo1/pthXo1-blaM bacteria, and real-time [P] was determined at 200 mpi. At 50 mpi, [P] significantly increased with time in the presence of OsPIP1;3 or substituted mutants carrying OsPIP1;3 LE as compared with that in the control (Fig. 8B). However, [P] showed minimal changes with OsPIP1;1 or mutants carrying OsPIP1;1 LE (Fig. 8B). As measured at 150 mpi, the different proteins of OsPIP1;1, OsPIP1;3, and their variants were produced in similar quantities in infected protoplasts (Fig. 8B inset). Thus, it is the presence of OsPIP1;3 LE, instead of protein production levels, that determines PthXo1 translocation. In accordance, OsSWEE11 was up-regulated with OsPIP1;3 or substituted mutants carrying OsPIP1;3 LE, whereas OsSWEE11 expression was equivalent to that of OsPIP1;1 or OsPIP1;1 LE relative to that of the control (Fig. 8C).
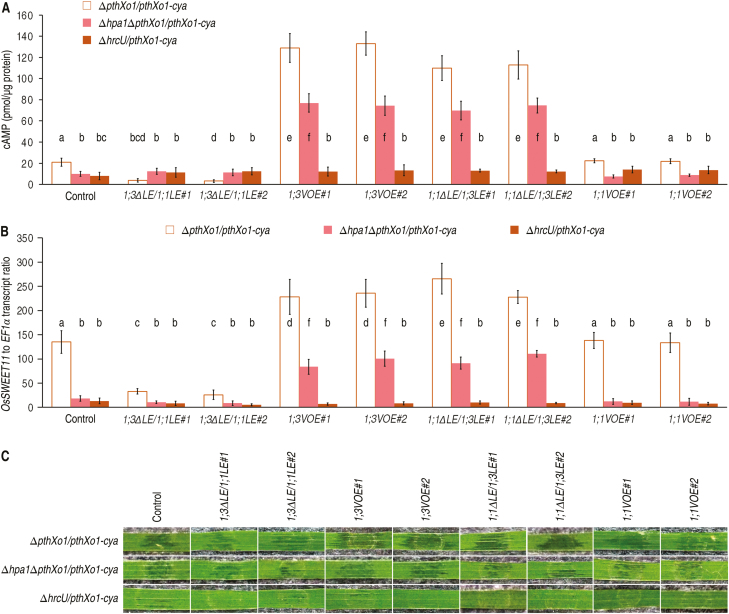
The Rice tungro bacilliform virus-mediated gene overexpression (VMGOE) system (Purkayastha et al., 2010; Kant and Dasgupta, 2017) was then used to analyze the pathological role of the LE of OsPIP1;3 in rice plants. The VMGOE protocol (see Supplementary Fig. S8A) was applied to OsPIP1;3 (Supplementary Fig. S8B) and OsPIP1;1 (Supplementary Fig. S8C) in the presence and absence of the LE of OsPIP1;3s, respectively. Compared with the level in control (transformation with the empty vector), significant (P<0.01) increases of PthXo1–Cya translocation, shown as cytoplasmic cAMP, were provided by overexpression of OsPIP1;3 or by de novo expression of the LE of OsPIP1;3 in OsPIP1;1 via exchange of the region (Fig. 9A). Coincidently, overexpression of OsPIP1;3 or de novo expression of LE of OsPIP1;3 in OsPIP1;1 caused enhancements of OsSWEET11 expression (Fig. 9B). In contrast, both PthXo1 translocation and OsSWEET11 expression incurred significant (P<0.01) reductions by LE switching from OsPIPl;1 to OsPIP1;3. However, both PthXo1 translocation and OsSWEET11 expression did have an evident change with OsPIP1;1 overexpression compared with the control, suggesting that OsPIP1;1 does not relate to the pathological process. The pathological process led to disease, shown as water-soaked symptoms as observed at 5 dpi by the leaf infiltration method (Fig. 9C). Disease severity was manifestly aggrevated by OsPIP1;3 overexpression or de novo expression of the LE of OsPIP1;3 in OsPIP1;1 but was noticeably alleviated by LE switch from OsPIPl;1 to OsPIP1;3.
Fig. 9.
The function of the LE of OsPIP1;3 to support virulence in plants. (A) PthXo1–Cya translocation measured as cytoplasmic cAMP concentrations in leaves of 30-day-old Nipponbare seedlings. Fifteen days previously, plants were transformed with the empty VMGOE vector (control) or each of the indicated genes inserted into the vector. Plants were inoculated by leaf infiltration with every suspension of the recombinant PXO99A strains shown on top, and inoculated leaves were excised at 9 hpi for use in cAMP measurements. (B) OsSWEET11 expression in leaves equivalent to those in (A). (A, B) Data show are means ±SEMs; different letters indicate significant differences by Duncan’s multiple range tests; P<0.01; n=15 plants tested in three independent experiments. (C) Segments of leaves photographed at 5 dpi.
Overall, these data provide evidence supporting LE as a determinant of OsPIP1;3 interaction with Hpa1 and plays a key role in PthXo1 translocation from Xoo cells into the cytosol of rice cells. This functional relationship determines the virulence performance of PthXo1 to cause bacterial blight disease in susceptible rice varieties.
Discussion
This article presents a large amount of data to delineate the interaction between rice and Xoo at the interface, as well as virulence-relevant consequences in the plant. Molecular interaction assays reveal that the Xoo T3 translocator Hpa1 (Wang et al., 2018) directly interacts with the rice membrane AQP OsPIP1;3, but not with OsPIP1 orthologs (Fig. 1), suggesting the specificity in PM sensing of the translocator. Genetic analyses confirm that OsPIP1;3 is a disease susceptibility factor, partly supporting Xoo virulence on the susceptible rice variety. Evidence has been found in virulence aggravation with OsPIP1;3 overexpression and in virulence reduction by TALEN-based knockout of the OsPIP1;3 gene (Figs 2, 3). This agrees with a recent demonstration of the virulence-deterrent effect of OsPIP1;3 silencing (Zhang et al., 2018). Biochemical data suggest the indirect effects of OsPIP1;3 and its interaction with Hpa1 on, but not causality for, PthXo1 translocation and OsSWEET11 expression (Fig. 4). Analyses of Hpa1 (Figs 5, 6) and OsPIP1;3 (Fig 7, 8) variants corroborate the role of OsPIP1;3–Hpa1 cooperation in PthXo1 translocation. The α-helix repeat of Hpa1 (Ji et al., 2011) is an OsPIP1;3-interacting motif (Fig. 5), LE of OsPIP1;3 is a Hpa1-interacting determinant (Fig. 7), and the presence of both motifs is essential for PthXo1 translocation from bacteria into rice protoplasts (Figs 6, 8). The protoplast BlaM reporter allows for high-throughput translocation assays and convenient detection of OsSWEET11 expression. Timely use of VMGOE proves that the LE of OsPIP1;3 is crucial for the disease susceptibility. The native LE in OsPIP1;3 determines PthXo1 translocation, which induces OsSWEET11 expression, and contributes to virulence, leading to the disease phenotype (Fig. 9).
These demonstrations update the current understandings of T3 effector translocation at least in two aspects: the involvement of eukaryotic PM proteins and the regulation schemes. In the first, it was not until recently that eukaryotic PM proteins were appreciated as being as important as lipids for the T3 translocon assembly or pore formation in target PMs (Ji and Dong, 2015a, b; Guignot and Tran Van Nhieu, 2016). With respect to translocator recognition by eukaryotic PMs, OsPIP1;3 was characterized as a Hpa1 interactor, and both proteins were found to interact at plant PMs. This interaction was further correlated with PthXo1 translocation, as well as Xoo virulence on the susceptible rice variety. These studies offer a novel angle for the exploration of PM sensing of T3 translocators in regard to previous studies that have focused on membrane lipids (Büttner et al., 2002; Haapalainen et al., 2011; Chatterjee et al., 2013). Potential roles of membrane lipids have been extensively tested with synthetic lipid bilayer systems (Lee et al., 2001a, b; Büttner et al., 2002; Haapalainen et al., 2011; Cortes et al., 2014). HrpF is able to bind to artificial lipids in silicon beads and to induce pore formation shown as electrical current fluxes in a synthetic planar lipid bilayer (Büttner et al., 2002). Synthetic phosphatidic acid binds HrpZ1 in a lipid stripe and is required for oligomerization of HrpZ1 and induction of plant immune responses (Haapalainen et al., 2011). Possibly, the PM-integral phosphatidic acid serves as an interactor of HrpZ1 and contributes to the effector translocation, but this hypothesis has not been validated to date.
In the second aspect, regulation mechanisms of T3 effector translocation have been explored in depth in animal-pathogenic bacteria (Domingues et al., 2016; Guignot and Tran Van Nhieu, 2016; McQuate et al., 2017; Tejeda-Dominguez et al., 2017), but remain almost stagnant in plant-pathogenic bacteria during the past 20 years (Büttner et al., 2002; Scheibner et al., 2017). Recent literature tackles translocon-independent pore formation by Salmonella SteA (Domingues et al., 2016), endocytosis of S. enterica SopB by membrane trafficking (Dong et al., 2016), and the canonical translocon-dependent delivery by several animal pathogens (Ji and Dong, 2015a; Büttner, 2016). The composition of the T3 translocon and the number of translocators present in a species or a strain of plant-pathogenic bacteria are still not known (Ji and Dong, 2015a; Scheibner et al., 2017; Wang et al., 2018). Regarding Hpa1, it may not be required only for PthXo1 or AvrXa10 (Wang et al., 2018) but also contributes to the translocation of all effectors of a given bacterial strain. For example, a Xoo strain produces near 30 Xops (Xanthomonas outer proteins; White et al., 2009), but none of them has been studied with respect to translocation. In addition to Hpa1, HrpF also functions in TALE translocation (Büttner et al., 2002; Li et al., 2011).
In an early report, the C-terminal region of HrpF was found to be essential for AvrBs3 entry into plant cells, whereas the N-terminal part containing a secretion signal did not affect the effector translocation (Büttner et al., 2002). This finding suggests the translocon-dependent manner of T3 effector translocation. A recent report proposed a translocon-independent route (Scheibner et al., 2017). The N-terminal 10 and 50 amino acids are required for T3 secretion and AvrBs3 translocation. Additional signals in the N-terminal 30 amino acids and the region between amino acids 64 and 152 promote translocation of AvrBs3. AvrBs3 translocation occurs not only in the absence of the T3 secretion chaperone HpaB, but also in the absence of HrpF. The authors’ explanation is that presumably, the delivery of AvrBs3 starts at early stages of the infection process, before the activation of HpaB or the translocon integration into the plant PM (Scheibner et al., 2017). It is more likely that a different translocator, which is in the reserve stock and whose function is suspended when HrpF is present, is employed in the case of loss of HrpF. Hpa1 may also have substitutes, which may completely or partly execute the translocator function originally performed by Hpa1. This hypothesis explains why loss of Hpa1 does not eliminate PthXo1 translocation and virulence.
Plant receptors for T3 translocators of the one-domain harpin group have attracted long-term attention. An early study identified plant proteins that interacted with HrpN, a one-domain harpin secreted by the fire blight pathogen Erwinia amylovora of rosaceous plants (Wei et al., 1992). HrpN is a translocator for the T3 effector DspA/E (Bocsanczy et al., 2008). HrpN-interacting protein in apple Malus (HIPM) contains a single TM domain and associates with plant PMs (Oh and Beer, 2007). Whether HIPM affects DspA/E translocation remains elusive. HIPM and its orthologs in Arabidopsis and rice share this single TM domain (Oh and Beer, 2007), which is distinct from the six TMs present in PIPs (Gomes et al., 2009; Hu et al., 2012). By testing OsPIP1s and screening the Arabidopsis genome (Li et al., 2015), the present study has identified OsPIP1;3 and AtPIP1;4, instead of HIPM, as Hpa1-interacting proteins. Therefore, it is pertinent to propose that both PIP1s are specific Hpa1 interactors in plant PMs. Indeed, OsPIP1;3 makes a substantial contribution to PthXo1 translocation. However, silencing (Zhang et al., 2018) and knockout of the OsPIP1;3 gene only impair but do not eliminate the effector translocation. This suggests the role of additional PM compounds in translocator recognition. Presumably, a different compound in PMs, whose function is suspended when OsPIP1;3 is workable, is employed in the case of loss of OsPIP1;3.
The pair of α-helix coiled-coil motifs present in the Hpa1 sequence (Ji et al., 2011) was characterized as an OsPIP1;3-interacting domain. This is in agreement with the critical effects of the motifs for Hpa1 bioactivity in plants (Wang et al., 2008; Li et al., 2014; Li et al., 2015) and its interaction with PIP1s (Ji and Dong, 2015a; Li et al., 2015). This protein-interacting domain is essential for the effector translocation. In support of the hypothesis that protein-interacting motifs exist in extramembrane regions of PIPs (Ji and Dong, 2015a), LE was identified as an Hpa1-interacting motif, which was not only a determinant for Hpa1–OsPIP1;3 interaction but also indispensable for PthXo1 translocation. These findings confirm that Hpa1–OsPIP1;3 interaction in rice PMs governs the effector translocation from bacterial cells into the cytosol of rice cells. The VMGOE data further suggest that native LE is essential for OsPIP1;3 to function as a disease susceptibility factor in the plant.
How the OsPIP1;3–Hpa1 interaction mediates bacterial effector translocation is presently unresolved. Proteins with nanometer size are never permeable to any AQP channel, either the intramolecular gate (2.8–3.4 Å) of a PIP or the wider intermolecular space in a PIP tetramer situated at PMs (Wang et al., 2007; Wudick et al., 2009; Otto et al., 2010; Kaldenhoff et al., 2014). However, very recent studies find that specific channels can be created owing to conformational changes triggered by biochemical or mechanical signals in contact with AQPs, enabling them to perform more functions than expected (Sutka et al., 2017). Of relevance to this, the OsPIP1;3–Hpa1 interaction which occurred at the plant PM may lead to the PM opening resembling the translocon pore (Ji and Dong, 2015b).
In summary, data detailed in this study demonstrate that the Xoo T3 translocator Hpa1 interacts with the rice AQP OsPIP1;3 at the plant PM to regulate TALE translocation from the bacterial cells into the cytosol of plant cells. Questions remain concerning the possibility that OsPIP1;3 experiences a topological change upon interacting with Hpa1 to facilitate translocon assembly, which also involves PM phospholipids beyond the scope of this study and perhaps is not limited to a single effector such as PthXo1. With a broad horizon, questions are also related to the hypothesis that the translocon model may be superseded by the translocon-independent models in different pathosystems.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Phylogenetic tree of Arabidopsis and rice PIP1s established by the MEGA4 program and BiFC tests of OsPIP1s and Hpa1.
Fig. S2. Analyses of OsPIP1;3 knockout by TALENs.
Fig. S3. Schematic diagram of OsPIP1;3 overexpression constructs.
Fig. S4. Deletions of hpa1 sequence regions affect the virulent function of PthXo1.
Fig. S5. Xoo pre-activation is required for infection of rice protoplasts.
Fig. S6. Optimization of BlaM reporter assays.
Fig. S7. Compositions in the LE of OsPIP1s.
Fig. S8. Efficiency of the rice VMGOE.
Table S1. Strains and plasmids used and created in this study.
Table S2. Information on genes tested and primers used in this study.
Table S3. ORF sequences of two designer TALENs.
Acknowledgments
The authors thank Professors Bing Yang (Iowa State University) for the gift of pthXo1 and TALEN vectors, Indranil Dasgupta (University of Delhi South Campus) for the gift of VMGOE vector, Ling Jiang (Nanjing Agricultural University) for assistance in rice transformation, and Adam J. Bogdanove (Cornell University), Jian Hua (Cornell University), Bing Yang, and Huanbin Zhou (Chinese Academy of Agricultural Sciences) for comments on the experimentation and manuscript. This study was supported by the China National Key Research and Development Plan (grant no. 2017YFD0200901), the Natural Science Foundation of China 31772247) and Talent Recruitment Funding of Shandong Agricultural University (0171226) to HD, and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_0663) to PL.
References
- Alfano JR, Collmer A. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annual Review of Phytopathology 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Baumgart F, Rossi A, Verkman AS. 2012. Light inactivation of water transport and protein–protein interactions of aquaporin–Killer Red chimeras. Journal of General Physiology 139, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocsanczy AM, Nissinen RM, Oh CS, Beer SV. 2008. HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Molecular Plant Pathology 9, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. 2011. TAL effectors: customizable proteins for DNA targeting. Science 333, 1843–1846. [DOI] [PubMed] [Google Scholar]
- Büttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiology and Molecular Biology Reviews 76, 262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D. 2016. Behind the lines—actions of bacterial type III effector proteins in plant cells. FEMS Microbiology Reviews 40, 894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D, Nennstiel D, Klüsener B, Bonas U. 2002. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. Journal of Bacteriology 184, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Huot B, Kvitko BH. 2017. Effector translocation: Cya reporter assay. Methods in Molecular Biology 1615, 473–487. [DOI] [PubMed] [Google Scholar]
- Charkowski AO, Alfano JR, Preston G, Yuan J, He SY, Collmer A. 1998. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. Journal of Bacteriology 180, 5211–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Chaudhury S, McShan AC, Kaur K, De Guzman RN. 2013. Structure and biophysics of type III secretion in bacteria. Biochemistry 52, 2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen J, Li M, et al. . 2017. A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host & Microbe 22, 777–788.e7. [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim W, Lee C, Oh CS. 2013. Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Molecular Plant-Microbe Interactions 26, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Cortes VA, Busso D, Maiz A, Arteaga A, Nervi F, Rigotti A. 2014. Physiological and pathological implications of cholesterol. Frontiers in Bioscience 19, 416–428. [DOI] [PubMed] [Google Scholar]
- Dik DA, Marous DR, Fisher JF, Mobashery S. 2017. Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Critical Reviews in Biochemistry and Molecular Biology 52, 503–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues L, Ismail A, Charro N, Rodríguez-Escudero I, Holden DW, Molina M, Cid VJ, Mota LJ. 2016. The Salmonella effector SteA binds phosphatidylinositol 4-phosphate for subcellular targeting within host cells. Cellular Microbiology 18, 949–969. [DOI] [PubMed] [Google Scholar]
- Dong N, Niu M, Hu L, Yao Q, Zhou R, Shao F. 2016. Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4-kinase activity of the Legionella LepB effector. Nature Microbiology 2, 16236. [DOI] [PubMed] [Google Scholar]
- Espina M, Ausar SF, Middaugh CR, Picking WD, Picking WL. 2006. Spectroscopic and calorimetric analyses of invasion plasmid antigen D (IpaD) from Shigella flexneri reveal the presence of two structural domains. Biochemistry 45, 9219–9227. [DOI] [PubMed] [Google Scholar]
- Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F. 2009. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochimica et Biophysica Acta 1788, 1213–1228. [DOI] [PubMed] [Google Scholar]
- Guignot J, Tran Van Nhieu G. 2016. Bacterial control of pores induced by the type III secretion system: mind the gap. Frontiers in Immunology 7, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapalainen M, Engelhardt S, Küfner I, Li CM, Nürnberger T, Lee J, Romantschuk M, Taira S. 2011. Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Molecular Plant Pathology 12, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann N, Büttner D. 2013. The inner membrane protein HrcV from Xanthomonas spp. is involved in substrate docking during type III secretion. Molecular Plant-Microbe Interactions 26, 1176–1189. [DOI] [PubMed] [Google Scholar]
- He SY, Huang HC, Collmer A. 1993. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hu S, Wang B, Qi Y, Lin H. 2012. The Arg233Lys AQP0 mutation disturbs aquaporin0–calmodulin interaction causing polymorphic congenital cataract. PLoS One 7, e37637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Dong H. 2015a. Key steps in type III secretion system (T3SS) towards translocon assembly with potential sensor at plant plasma membrane. Molecular Plant Pathology 16, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Dong H. 2015b. Biological significance and topological basis of aquaporin-partnering protein–protein interactions. Plant Signaling & Behavior 10, e1011947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song C, Lu X, Wang J. 2011. Two coiled-coil regions of Xanthomonas oryzae pv. oryzae harpin differ in oligomerization and hypersensitive response induction. Amino Acids 40, 381–392. [DOI] [PubMed] [Google Scholar]
- Jin M, Berrout J, Chen L, O’Neil RG. 2012. Hypotonicity-induced TRPV4 function in renal collecting duct cells: modulation by progressive cross-talk with Ca2+-activated K+ channels. Cell Calcium 51, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldenhoff R, Kai L, Uehlein N. 2014. Aquaporins and membrane diffusion of CO2 in living organisms. Biochimica et Biophysica Acta 1840, 1592–1595. [DOI] [PubMed] [Google Scholar]
- Kant R, Dasgupta I. 2017. Phenotyping of VIGS-mediated gene silencing in rice using a vector derived from a DNA virus. Plant Cell Reports 36, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Kim JF, Beer SV. 1998. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. Journal of Bacteriology 180, 5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cellular and Molecular Life Sciences 60, 2371–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko BH, Ramos AR, Morello JE, Oh HS, Collmer A. 2007. Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. Journal of Bacteriology 189, 8059–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Klessig DF, Nürnberger T. 2001a A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. The Plant Cell 13, 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Klusener B, Tsiamis G, et al. . 2001. b HrpZPsph from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proceedings of the National Academy of Sciences, USA 98, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang H, Gago J, et al. . 2015. Harpin Hpa1 interacts with aquaporin PIP1;4 to promote the substrate transport and photosynthesis in Arabidopsis. Scientific Reports 5, 17207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. 2012. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nature Biotechnology 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Li X, Han L, Zhao Y, You Z, Dong H, Zhang C. 2014. Hpa1 harpin needs nitroxyl terminus to promote vegetative growth and leaf photosynthesis in Arabidopsis. Journal of Biosciences 39, 127–137. [DOI] [PubMed] [Google Scholar]
- Li YR, Che YZ, Zou HS, Cui YP, Guo W, Zou LF, Biddle EM, Yang CH, Chen GY. 2011. Hpa2 required by HrpF to translocate Xanthomonas oryzae transcriptional activator-like effectors into rice for pathogenicity. Applied and Environmental Microbiology 77, 3809–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey Rose KM, Gourdie RG, Prescott AR, Quinlan RA, Crouch RK, Schey KL. 2006. The C terminus of lens aquaporin 0 interacts with the cytoskeletal proteins filensin and CP49. Investigative Ophthalmology & Visual Science 47, 1562–1570. [DOI] [PubMed] [Google Scholar]
- Makino S, Sugio A, White F, Bogdanove AJ. 2006. Inhibition of resistance gene-mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Molecular Plant-Microbe Interactions 19, 240–249. [DOI] [PubMed] [Google Scholar]
- Matteï PJ, Faudry E, Job V, Izoré T, Attree I, Dessen A. 2011. Membrane targeting and pore formation by the type III secretion system translocon. The FEBS Journal 278, 414–426. [DOI] [PubMed] [Google Scholar]
- Maurel C, Verdoucq L, Luu DT, Santoni V. 2008. Plant aquaporins: membrane channels with multiple integrated functions. Annual Review of Plant Biology 59, 595–624. [DOI] [PubMed] [Google Scholar]
- McQuate SE, Young AM, Silva-Herzog E, Bunker E, Hernandez M, de Chaumont F, Liu X, Detweiler CS, Palmer AE. 2017. Long-term live-cell imaging reveals new roles for Salmonella effector proteins SseG and SteA. Cellular Microbiology 19, e10.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E, Baruch K, Aviv G, Nitzan M, Rosenshine I. 2013. Dynamics of the type III secretion system activity of enteropathogenic Escherichia coli. Mbio 4, e00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E, Baruch K, Charpentier X, Kobi S, Rosenshine I. 2008. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host & Microbe 3, 104–113. [DOI] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Cornelis GR. 2008. The type III secretion system tip complex and translocon. Molecular Microbiology 68, 1085–1095. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Fullner KJ, Koonin EV, Nester EW. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proceedings of the National Academy of Sciences, USA 93, 7321–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Beer SV. 2007. AtHIPM, an ortholog of the apple HrpN-interacting protein, is a negative regulator of plant growth and mediates the growth-enhancing effect of HrpN in Arabidopsis. Plant Physiology 145, 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto B, Uehlein N, Sdorra S, et al. . 2010. Aquaporin tetramer composition modifies the function of tobacco aquaporins. Journal of Biological Chemistry 285, 31253–31260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Mathur S, Verma V, Sharma S, Dasgupta I. 2010. Virus-induced gene silencing in rice using a vector derived from a DNA virus. Planta 6, 232–1531. [DOI] [PubMed] [Google Scholar]
- Qin X, Boron WF. 2013. Mutation of a single amino acid converts the human water channel aquaporin 5 into an anion channel. American Journal of Physiology. Cell Physiology 305, C663–C672. [DOI] [PubMed] [Google Scholar]
- Roche JV, Törnroth-Horsefield S. 2017. Aquaporin protein–protein interactions. International Journal of Molecular Sciences 18, e2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden JA, Belt B, Ross JB, Tachibana T, Vargas J, Mudgett MB. 2004. A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proceedings of the National Academy of Sciences, USA 101, 16624–16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibner F, Marillonnet S, Büttner D. 2017. The TAL effector AvrBs3 from Xanthomonas campestris pv. vesicatoria contains multiple export signals and can enter plant cells in the absence of the type III secretion translocon. Frontiers in Microbiology 8, 2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutka M, Amodeo G, Ozu M. 2017. Plant and animal aquaporins crosstalk: what can be revealed from distinct perspectives. Biophysical Reviews 9, 545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejeda-Dominguez F, Huerta-Cantillo J, Chavez-Dueñas L, Navarro-Garcia F. 2017. A novel mechanism for protein delivery by the type 3 secretion system for extracellularly secreted proteins. MBio 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge S, Furutani A, Fukunaka R, Oku T, Tsuno K, Ochiai H, Inoue K, Kaku H, Kubo Y. 2002. Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. Journal of General Plant Pathology 68, 363–371. [Google Scholar]
- Verkman AS. 2013. Aquaporins. Current Biology 23, R52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang L, Ji H, Mo X, Li P, Wang J, Dong H. 2018. Hpa1 is type III translocator in Xanthomonas oryzae pv. oryzae. BMC Microbiology 18, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XY, Song CF, Miao WG, Ji ZL, Wang X, Zhang Y, Zhang JH, Hu JS, Borth W, Wang JS. 2008. Mutations in the N-terminal coding region of the harpin protein Hpa1 from Xanthomonas oryzae cause loss of hypersensitive reaction induction in tobacco. Applied Microbiology and Biotechnology 81, 359–369. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cohen J, Boron WF, Schulten K, Tajkhorshid E. 2007. Exploring gas permeability of cellular membranes and membrane channels with molecular dynamics. Journal of Structural Biology 157, 534–544. [DOI] [PubMed] [Google Scholar]
- Wei ZM, Lacy RJ, Zumoff CH, Bauer DW, He SY, Collmer A, Beer SV. 1992. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257, 85–88. [DOI] [PubMed] [Google Scholar]
- White FF, Potnis N, Jones JB, Koebnik R. 2009. The type III effectors of Xanthomonas. Molecular Plant Pathology 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick MM, Luu DT, Maurel C. 2009. A look inside: localization patterns and functions of intracellular plant aquaporins. New Phytologist 184, 289–302. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, et al. . 2013. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host & Microbe 13, 347–357. [DOI] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF. 2006. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proceedings of the National Academy of Sciences, USA 103, 10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrl D, Wagner M, Bischof K, Bayer M, Zavecz B, Beranek A, Ruckenstuhl C, Zarfel GE, Koraimann G. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Frontiers in Microbiology 151, 3455–3467. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hu Y, Li P, Wang X, Dong H. 2018. Silencing of an aquaporin gene diminishes bacterial blight disease in rice. Australasian Plant Pathology 10, 1448–6032. [Google Scholar]
- Zhu WG, Magbanua MM, White FF. 2000. Identification of two novel hpaG-associated genes in the hpaG gene cluster of Xanthomonas oryzae pv. oryzae. Journal of Bacteriology 182, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.