Abstract
Thirty-two actinomycetes strains were isolated from sediment samples from 12 different sites at Lagos Lagoon and identified using standard physiological and biochemical procedures as well as 16S rDNA gene sequence analysis. Secondary metabolites were extracted from the strains and their anticancer activity on the K562 (Human acute myelocytic leukemia), HeLa (cervical carcinoma), AGS (Human gastric), MCF-7 (breast adenocarcinoma) and HL-60 (Human acute promyelocytic leukemia) cell lines was determined. The metabolic extracts exhibited cytotoxicity with IC50 values ranging from 0.030 mg/mL to 4.4 mg/mL. The Streptomyces bingchenggensis ULS14 extract was cytotoxic against all the cell lines tested. The bioactivity-guided extraction and purification of the metabolic extracts from this strain yielded two purified anticancer compounds: ULDF4 and ULDF5. The structures of the extracted compounds were determined using spectroscopic analyses, including electrospray ionization mass spectrophotometer and nuclear magnetic resonance (1 Dimensional and 2 Dimensional), and were shown to be structurally similar to staurosporine and kigamicin. The IC50 of ULDF4 and ULDF5 against the HeLa cell line was 0.034 μg/mL and 0.075 μg/mL, respectively. This study is the first to reveal the anticancer potential of actinomycetes from Lagos Lagoon, which could be exploited for therapeutic purposes.
Keywords: Streptomyces bingchenggensis ULS14, Metabolic extracts, Cytotoxicity, Spectroscopy, Kigamicin, Staurosporine
1. Introduction
There was an estimated 12 million new cases of cancer and 7.6 million cancer-related deaths in 2008, and its incidence is expected to rise to 26.4 million annual cases worldwide, with 17 million deaths by 2030. Most of these new cancer cases are expected to occur in low-income African countries [1]. Parkin et al. [2] reported that an estimated 650,000 of 965 million indigenous Africans are diagnosed with cancer annually, and the lifetime risk of dying from cancer for African women is greater than the risk for women in developed countries. In Nigeria, there are approximately 100,000 new cancer cases every year, with a high case fatality ratio. The six most common cancers in Nigeria are breast, cervical, prostate, colorectal, and liver cancers, and non-Hodgkin Lymphoma [3].
Intense efforts have been made to develop cancer therapeutics, and natural products have been proven to be promising sources of novel anti-cancer drugs. In the past few decades, there have been many reports of anticancer activity in actinobacteria isolated from marine environments.
It has been reported that over 10,000 bioactive secondary metabolites are produced by actinomycetes, accounting for 45% of all discovered bioactive microbial metabolites, with approximately 7600 of the actinomycetes compounds produced by Streptomyces spp [4]. Many of these secondary metabolites are clinically useful antitumor drugs, such as anthracyclines (aclarubicin, daunomycin, and doxorubicin), peptides (bleomycin and actinomycin D), aureolic acids (mithramycin), enediynes (neocarzinostatin), antimetabolites (pentostatin), carzinophilin, and mitomycins [[5], [6], [7]]. It is therefore imperative that the search for novel natural microbial products be continued in underexplored habitats. It has been hypothesized that since these microorganisms can thrive in the marine environment, some species may produce novel bioactive compounds for their survival, which could also serve as new pharmaceutical compounds. The search for novel anticancer drugs is a priority, as the high toxicity and undesirable side effects associated with chemotherapy drugs have increased the demand for drugs with fewer side effects and/or with greater therapeutic efficiency against recalcitrant tumors [8]. The Lagos Lagoon is a complex system of waterways linked to the Atlantic Ocean along the West African Gulf of Guinea coastline. Several streams and rivers, such as the Ogun river, empty into the Lagoon. We have previously reported the antimicrobial potential of actinomycetes strains isolated from Lagos Lagoon sediment [9]. The actinomycetes showed bioactivity against CoN Staphylococcus warneri, methicillin-resistant S. aureus, and Candida albicans due to the synthesis of bioactive compounds such as erythromycin, nystatin, oxytetracycline, tylosin, marinomycins A-D, chloramphenicol, glaciapyrroles, and cycloheximide by the actinomycetes. We hypothesized that the marine actinomycetes strains present in Lagos Lagoon sediment produce metabolic compounds with potent anticancer potential. This study therefore aimed to isolate, purify, and identify anticancer compounds from the metabolic extracts of marine actinomycetes strains from Lagos Lagoon sediment. The anticancer potential of two compounds extracted from the metabolites of these actinomycetes from Lagos marine sediments is also reported.
2. Materials and methods
2.1. Isolation and identification of actinomycetes from sediment samples
Sediment samples were collected from 12 different locations in Lagos Lagoon, at Okobaba, Offin, Folawiyo, Iddo, Ejirin, Imoru, Imope, Ikosi, Egbin, Ijede, Palaver Island, and Bayeku, which are located between 3°22′4″E and 3°90.384′E, and 6°26′4″N and 6°60.886′N. Samples obtained using a Van Veen grab were immediately taken to the laboratory for analysis and dried at 25 to 29 °C ambient temperature for two weeks. The isolation of actinomycetes was carried out using the spread plate method by serially diluting 1 g of each sample in 9 mL of distilled water, then spreading 100 μL each of this diluted sample on the surface of five different agar media (Kuster's (glycerol 10 mL, casein 0.3 g, KNO3 2 g, NaCl 2 g, K2HPO4 2 g, MgSO4·7H2O 0.05 g, CaCO3 0.02 g, FeSO4·7H2O 0.01 g (Sigma-Aldrich, Germany), agar (BD, Difco, U.S.A) 18 g)), starch-casein (starch (Fisher Scientific Education, U.S.A) 10 g, casein 0.3 g, KNO3 2 g, NaCl 2 g, K2HPO4 2 g, MgSO4·7H2O 0.05 g, CaCO3 0.02 g, FeSO4·7H2O 0.01 g (Sigma-Aldrich, Germany), agar 18 g), Gauze 1 (starch 20.0 g, FeSO4·7H2O 0.01 g, KNO3 1.0 g, K2HPO4 0.5 g, MgSO4·7H2O 0.5 g, NaCl 2 g, agar 15 g), Gauze 2 (glucose 10.0 g, NaCl 5.0 g, peptone 5.0 g, tryptone 3.0 g (Sigma-Aldrich, Germany), agar 15.0 g), marine (starch 10.0 g, yeast extract 4.0 g, peptone 2.0 g, agar 18.0 g) and actinomycetes isolation agar (Biomark, India). Plates were incubated at 29 °C for 1–5 weeks. Pure cultures of representative actinobacterial colonies were obtained by repeated streaking on sterile plates containing different media [10]. Cultural, morphological, and biochemical characterizations were carried out as described previously by Davies et al. [9]. Genomic DNA extraction, PCR amplification, and 16S rDNA gene sequencing were carried out according to the methods of Stach et al. [11]. Phylogenetic analysis was carried out using the neighbour-joining method with MEGA 5 software [12]. Sequences were submitted to GenBank (www.ncbi.nlm.nih.gov/Genbank) and accession numbers were generated.
2.2. Production and bioactivity screening of metabolites
2.2.1. Small scale fermentation
Pure cultures of actinomycetes were transferred to test tubes containing 10 mL sterile culture broth (Kuster's, starch-casein, marine, or Guaze 1 media) and cultured for 3 days. They were then transferred to flasks containing 200 mL sterile culture broth and shaken at 180 rpm, pH 7, and 28 °C for 3 days. The fermentation broth was scaled up by transferring to flasks containing 1000 mL sterile culture broth and was shaken at 180 rpm for 10 days [13].
2.2.2. Extraction of secondary metabolites
Cells were separated from the broth by centrifugation at 6000 rpm and 10 °C for 30 min. The metabolites were extracted from the culture supernatant using the liquid-liquid extraction method [13] with an equal amount (1:1) of ethyl acetate and concentrated by rotary evaporation to yield crude extracts. The mycelial cake was washed with methanol and cells were separated. The methanolic fraction was obtained and concentrated to yield crude extract.
2.3. Cytotoxicity assay
The anticancer properties of the crude extracts were determined using K562 (human acute myelocytic leukemia), HeLa (cervical carcinoma), AGS (human gastric), MCF-7 (breast adenocarcinoma), and HL-60 (human acute promyelocytic leukemia) cell lines according to the methods of Ravikumar et al. [14], with slight modifications. Different concentrations (0.01–5 mg/mL) of the crude extracts were prepared and screened for cytotoxicity on the cancer cell lines in vitro using the CCK8 assay. CCK8 assays were carried out using 96-well microtiter plates, with cells grown in each well until 70% confluence was reached before adding the extract (0.01–5 mg/mL), except for the positive (cell culture medium with standard drug SAHA) and negative (cell culture medium) controls. The cells were grown for 2 days in an incubator at 5% CO2 and 37 °C. CCK8 (100 μL) was added to each well, and the plates were incubated for 2 h and then read at 450 nm using a FLUOstar microplate reader. All experiments were performed in triplicate. Microsoft Excel 2013 and GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA) were used for statistical analysis. The data obtained are expressed as the mean ± standard error. P-values ≤ 0.05 were considered statistically significant. Log IC50 calculations were carried out using algorithms for a dose-response curve with a variable slope.
2.4. Isolation and identification of bioactive metabolites
2.4.1. Large-scale fermentation
The actinomycetes strain Streptomyces bingchenggensis ULS14 had the best overall cytotoxic activity and was therefore selected for large-scale fermentation. The strain was cultured for 72 h in five 250mL flasks containing sterile starch-casein broth. The cells were transferred to five 2-L flasks containing sterile starch-casein broth and fermented for 10 days at 29 °C and 180 rpm. The cells were next separated by centrifugation at 6000 rpm and 10 °C, then the adsorbent resins Amberlite XAD7 and Amberlite XAD16 were added to the 10 L cell-free broth. The bound bioactive metabolites were eluted from the resins with acetone. The cells were washed with methanol and both extracts (methanol and acetone extracts) were dried and combined to yield 8 g crude extract [15].
2.4.2. Extraction and purification of bioactive compounds
To separate the crude extract by partitioning, the method of Kwon et al. [15] was used with some modifications, with the solvents, dichloromethane, water, and 10% dichloromethane in 2-propanol. Each of the three fractions was screened for bioactivity against the HeLa cell line (by CCK8 assay). Further fractionation was performed by flash column chromatography (Biotage, U.S.A) using a SNAP KP sil-10 g column. This was run using n-hexane/ethyl acetate (0–100%), which was changed to ethyl acetate/methanol (60%: 40%, v/v), and then monitored at UV 254 nm to obtain sub-fractions which were collected in 15mL tubes at UV 280 nm. Each fraction was dried, re-dissolved in DMSO, and subjected to a bioactivity assay (cytotoxic assay). The purity of the fraction was checked using a C18 reversed-phase HPLC column (Shimadzu) with an acetonitrile/water gradient solvent system (50% acetonitrile/water) for 15 min, a 20 mm × 250 mm Waters C18 column, and a 1 mL/min flow rate [15].
2.4.3. Structural elucidation
Electrospray ionization mass spectrometry (ESI-MS) experiments were carried out using a Waters Micromass Q-TOF micro (ESI-Q-TOF, Milford, MA, USA) instrument in positive ion mode. The mobile phase was solvent A (0.1% formic acid (FA) in water) and solvent B (acetonitrile (CH3CN) with 95% A) for 5 min and had a linear gradient between 5% and 40% CH3CN (from A to B for 30 min). The capillary voltage was −3317 V and the sample cone voltage was −50 V. The samples were directly injected into the ion source at a 1 mL/min flow rate. Full scan MS spectra were recorded in the 100–1200 mass/charge region [16].
Purified compounds were dissolved in chloroform, a drop of which was placed on the plate, and infrared spectra (IR) were obtained using a Nicolet Magna-IR Fourier-Transform 560 Spectrometer. Absorption was recorded and reported in wavenumber (cm−1) [13].
NMR analysis was carried out by dissolving the pure compounds in chloroform, and then pipetting 600 μL of the solution into the NMR tubes. NMR spectra (1H, 13C, COSY, and HMBC) were recorded on a Brucker Avance 500 spectrometer. Chemical shifts for 1H NMR were referenced relative to tetramethylsilane (0.00 ppm) and chloroform (7.24 ppm). Chemical shifts for 13C NMR were referenced relative to chloroform (77.23 ppm) [16].
3. Results
3.1. Strain identification
A total of 32 suspected actinomycetes strains were isolated from sediment from the 12 sampling points in Lagos Lagoon, using five different culture media. Twenty-three isolates had tough, velvety, or leathery, white, grey, green, purple, or cream colored aerial mycelia on the surface of the culture media. Three isolates had dry, tough, yellow, wrinkled colonies while four isolates had tough and waxy, orange or black colonies. Two isolates appeared as orange and purple mucoid colonies on the agar medium (Table 1). All the suspected actinomycetes strains were gram-positive, had a filamentous cellular morphology, and exhibited varying physiological characteristics (Table S1). All isolates fermented glucose and lactose, and hydrolyzed starch except for ULM32, ULM33, and ULM36. Approximately 70% of the isolates fermented lactose, saccharose, and maltose, with ULS14, UL19b, UL31a, ULM32, ULM36, ULMa40, ULG1.08, ULG2.17, ULM33, and ULK3 also able to hydrolyze gelatin.
Table 1.
Culture and morphological characteristics of the actinomycete isolates.
| Isolates/Labcodes | Medium | Colour and colonial characteristics | Pigment in medium | Cellular morphology |
|---|---|---|---|---|
| ULS12 | SCA | White powdery surface colonies turns faint greenish with age | Present | Filamentous fragments into cocci |
| ULS13 | SCA | White leathery surface which turns greyish with age | None | Filamentous |
| ULK3 | KA | Faint grayish powdery surface colonies | None | Filamentous fragments into cocci |
| ULK2 | KA | White powdery surface which turns cream with age | Present | Filamentous |
| ULS14 | SCA | White powdery surface turns pinkish purple with age | Present | Filamentous |
| ULK7 | KA | White powdery surface colonies turn grey with age | Present | Filamentous |
| ULMa26 | MA | Yellow dry tough and wrinkled | None | Filamentous |
| ULS7 | SCA | White convex powdery surface turns grey with age with red colour behind colonies | None | Filamentous |
| ULK11 | KA | Grayish green, leathery | None | Filamentous, fragments into rods |
| UL19b | SCA | White powdery colonies | None | Filamentous |
| UL31a | SCA | White powdery colonies | None | Filamentous |
| UL31b | SCA | White powdery colonies | None | Filamentous |
| ULM32 | MA | Translucent mucoid colonies which turn faint purple with age | None | Filamentous fragments into cocci |
| ULM33 | MA | Orange waxy colonies | None | Filamentous |
| ULM36 | MA | Yellow dry tough and wrinkled | None | Filamentous |
| ULMa40 | MA | Orange waxy colonies, surface darken with age | None | Filamentous |
| ULM27 | MA | Orange waxy colonies | None | Filamentous |
| ULG1.08 | G1A | Orange waxy colonies, surface darken with age | None | Filamentous |
| ULG2.23 | G2A | Orange mucoid colonies | None | Filamentous |
| ULG2.17 | G2A | White powdery colonies | None | Filamentous |
| UL28a | SCA | White powdery colonies | None | Filamentous |
| UL28f | AIA | White powdery colonies | None | Filamentous |
| UL6a | SCA | White powdery colonies | None | Filamentous |
| UL030 | SCA | Yellow dry tough and wrinkled | None | Filamentous |
| UL7b | SCA | White leathery colonies | None | Filamentous |
| ULT1 | AIA | White powdery colonies | None | Filamentous |
| ULA9 | SCA | White leathery colonies | None | Filamentous |
| UL23a | SCA | White powdery colonies | None | Filamentous |
| ULK10 | KA | White leathery convex powdery colonies which turn grey with age and dark back colony observed | None | Filamentous |
| UL28d | SCA | White leathery powdery colonies which turn grey | None | Filamentous |
| ULAct2 | SCA | White powdery colonies | None | Filamentous |
| ULMa30 | MA | Orange waxy colonies | None | Filamentous |
SCA Starch-casein agar, KA- Kuster's agar, MA- Marine agar, G1A- Guaze 1 agar, G2A- Guaze 2 agar, AIA- Actinomycetes isolation agar.
The isolates were identified based on the molecular identification of their 16S rDNA gene sequences, and were found to be Micromonospora spp. and Streptomyces spp. The closest known identity, percentage similarity, and frequency of occurrence of each isolate is shown in Table 2. Streptomyces spp. were isolated from all the samples, while Micromonospora spp. were only isolated from eight samples. Streptomyces albus, S. avermitilis, and S. coelicolor were isolated from all the samples, while S. bingchenggensis was isolated from 16.7% of the samples. The other species isolated include S. fulvissimus, S. pratensis, Micromonospora aurantica, M. sediminicola, and M. humi.
Table 2.
Identification of actinomycetes isolated from Lagos Lagoon based on 16Sr DNA gene sequences.
| Strains | Sampling location | 16 Sr RNA gene of closest known relative | % Similarity | Accession numbers |
|---|---|---|---|---|
| ULMa27 | Imope, Ikosi | Micromonospora aurantica | 97 | KX352083 |
| ULG2.23 | Imope, Ikosi, | Micromonospora sp. | 100 | KX352058 |
| ULMa40 | Imope, Ikosi, Egbin | Micromonospora sediminicola | 99 | KX352076 |
| ULMa33 | Okobaba, Offin, Iddo, Ikosi, Egbin | Micromonospora humi | 99 | KX352075 |
| ULMa30 | Imope, Ikosi, Egbin | Micromonospora sp. | 100 | KX352073 |
| ULG1.08 | Ejirin, Imoru | Micromonospora sp. | 100 | KX352080 |
| ULMa32 | Ejirin, Imoru, Itokin | Agromyces sp. | 94 | KX352074 |
| ULK2 | Folawiyo, Ejirin, Imoru | Streptomyces albus | 100 | KX352059 |
| ULK3 | Okobaba, Offin, Folawiyo, Iddo, Ejirin, Imoru, Imope, Ikosi, Egbin, Ijede, Bayeku | Streptomyces avermitilis | 99 | KX352077 |
| ULS7 | Okobaba, Offin, Folawiyo, Iddo, Ejirin, Imoru, Imope, Ikosi, Egbin, Ijede | Streptomyces coelicolor | 100 | KX352086 |
| ULS14 | Folawiyo, Iddo | Streptomyces bingchenggensis | 98 | KX352065 |
| ULS13 | Offin | Streptomyces fulvissimus | 100 | KX352087 |
| ULK11 | Offin | Streptomyces albus | 99 | KX352062 |
| ULK7 | Ikosi, Egbin | Streptomyces pratensis | 98 | KX352060 |
| ULG2.17 | Imope, Ikosi, Bayeku | Streptomyces albus | 100 | KX352081 |
| UL28f | Imope, Ikosi | Streptomyces albus | 99 | KX352082 |
| UL28a | Ikosi, Egbin | Streptomyces albus | 99 | KX352079 |
| ULT1 | Okobaba, Egbin | Streptomyces albus | 100 | KX352088 |
| ULMa 36 | Imope, Ikosi, Egbin | Micromonospora sp. | 100 | KX352085 |
| UL7B | Okobaba, Offin, Folawiyo, Iddo, Ejirin, Imoru, Imope, Ikosi, Egbin, Ijede, Bayeku | Streptomyces albus | 99 | KX352066 |
| UL28d | Ejirin, Imoru, Itokin | Streptomyces pratensis | 98 | KX352068 |
| UL31b | Folawiyo, Ejirin, Imoru | Streptomyces albus | 99 | KX352070 |
| ULA9 | Ikosi, Egbin | Streptomyces avermitilis | 99 | KX352071 |
| ULK10 | Folawiyo | Streptomyces sp. | 98 | KX352061 |
| UL23a | Imope, Ikosi | Streptomyces albus | 99 | KX352057 |
| UL19b | Offin | Streptomyces fulvissimus | 100 | KX352067 |
| UL030 | Offin | Micromonospora sp. | 99 | KX352063 |
| ULAct2 | Ikosi, Egbin | Streptomyces pratensiss | 98 | KX352072 |
| UL6a | Imope, Ikosi | Streptomyces albus | 100 | KX352078 |
| ULS12 | Imope, Ikosi | Streptomyces sp. | 100 | KX352064 |
| UL31a | Ikosi, Egbin | Streptomyces albus | 99 | KX352069 |
| ULMa26 | Okobaba, Egbin | Streptomyces albus | 99 | KX352084 |
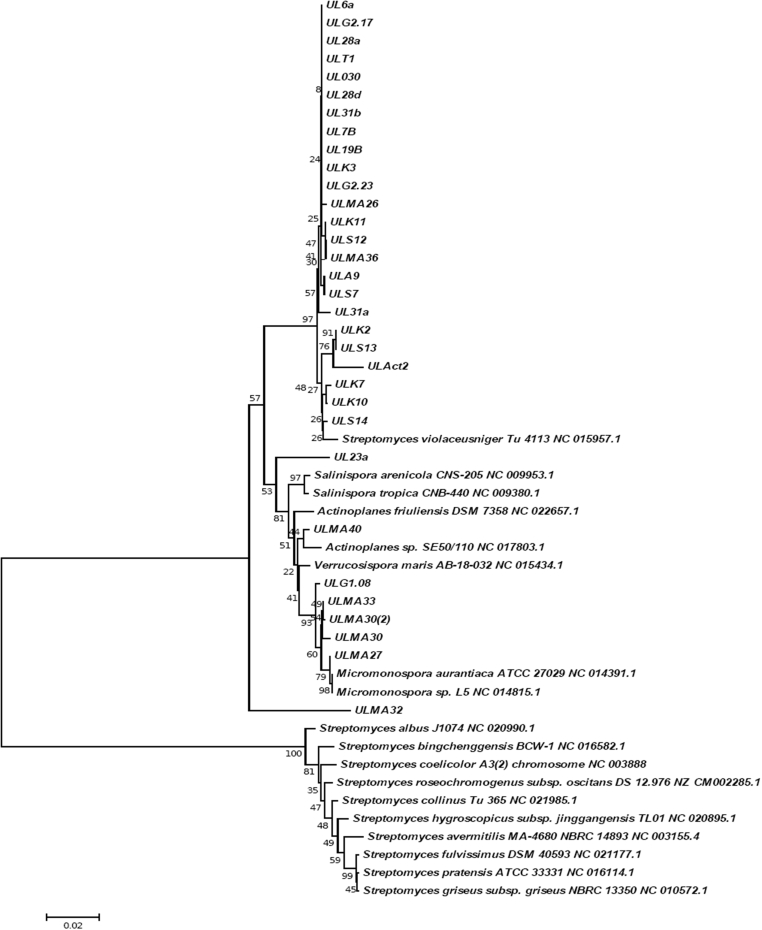
Based on the dendogram generated, partial nucleotide sequences of the 16S rDNA gene of representative actinomycetes strains were compared to those of species from the Genbank database to elucidate the evolutionary relationships between the species isolated in this study. The 16S rDNA gene sequences were also used to construct a phylogenetic tree of the actinobacteria strains obtained from the Lagos Lagoon sediments, which demonstrated the diversity of the strains. The phylogenetic tree also revealed the closest relative of the divergent Agromyces sp. ULMa32 isolate was Agromyces sp. However, the phylogenetic distance between the 16S rDNA sequences of Agromyces sp. ULMa32 and the most related Agromyces species does not rule out this strain being a novel species. Careful analysis revealed that the strains belonged mainly to Streptomyces sp. and Micromonospora sp.; only one was found to be a close relative of Agromyces sp. (Fig. 1). Cluster analysis showed that the S. violaceusniger Tu 4113 strains were closely related to over 70% of the isolates while ULG1.08, ULMa33, ULMa30, and ULMa27 were more closely related to M. aurantica ATCC 27029 and Micromonospora sp. L5 NC 014816.1.
Fig. 1.
Phylogenetic tree obtained by distance matrix analysis of 16S rDNA sequences and constructed using the neighbour-joining method, showing the phylogenetic position of the actinomycetes strains among related species. Numbers on branch nodes are bootstrap values (1000 resamplings). Bar and 0.02 Knuc unit are shown at the branch points.
3.2. Cytotoxic effects of crude actinomycetes extracts against cancer cell lines
The crude extracts of 9 isolates showed cytotoxic activity against at least one of the cell lines screened, with the IC50 ranging from 0.030 mg/mL to 4.4 mg/mL. Four of the extracts (S. albus UL7B, S. fulvissimus ULS13, S. bingcheggensis ULS14, and S. albus ULK2) were active against all the cell lines (Table 3). Extracts from S. fulvissimus ULS13 had the highest cytotoxicity at 0.030 mg/mL against the AGS cell line. The S. fulvissimus ULS13 and S. bingchenggensis ULS14 extracts were active against all five cell lines at concentrations below 1 mg/mL. Crude extracts from S. pratensis ULK7, Streptomyces sp. ULS12, and Streptomyces sp. ULK10 were observed to have cytotoxic activity against all cell lines except HL-60, while S. coelicolor ULS7 extracts displayed cytotoxicity against AGS, MCF-7, and HeLa cells. Crude extracts from M. aurantica ULMa27 were only cytotoxic against HeLa cells.
Table 3.
IC50 value of actinomycete crude extracts on cell lines (mg/mL).
| Extracts | HL-60 | AGS | K562 | MCF-7 | HeLa |
|---|---|---|---|---|---|
| Streptomyces albus UL7B | 0.083 | 0.095 | 0.045 | 1.315 | 2.277 |
| Streptomyces coelicolor ULS7 | – | 2.320 | – | 1.181 | 0.312 |
| Streptomyces pratensis ULK7 | – | 1.231 | 1.250 | 2.251 | 1.251 |
| Streptomyces sp. ULS12 | – | 1.981 | 2.371 | 2.082 | 2.031 |
| Streptomyces fulvissimus ULS13 | 0.096 | 0.030 | 0.101 | 0.125 | 0.070 |
| Streptomyces sp. ULK10 | – | 2.14 | 3.124 | 2.163 | 0.240 |
| Micromonospora aurantica ULMa27 | – | – | – | – | 4.401 |
| Streptomyces bingcheggensis ULS14 | 0.640 | 0.075 | 0.203 | 0.139 | 0.040 |
| Streptomyces albus ULK2 | 0.083 | 1.543 | 0.078 | 2.176 | 0.034 |
| SAHA (μg/mL) | 0.053 | 0.031 | 0.03 | 0.04 | 0.04 |
3.3. Extraction, purification, and cytotoxic assay of compounds
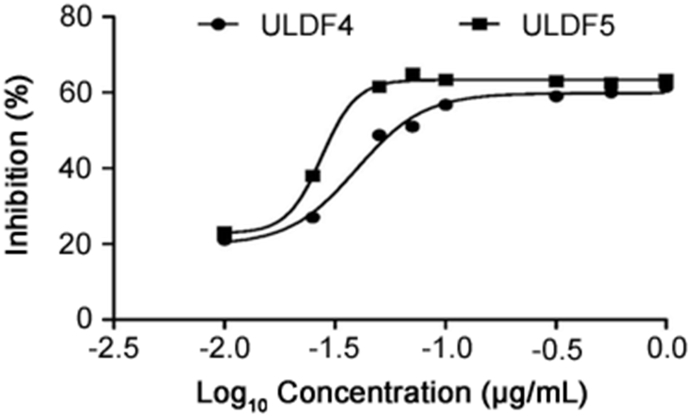
The crude extract of the most bioactive strain, S. bingchengensis ULS14, was partitioned using dichloromethane, water, and 10% dichloromethane in 2-propanol, as well as bioactivity-guided fractionation using HeLa cells. Consequently, two purified bioactive compounds, ULDF4 (5 mg) and ULDF5 (6 mg), were isolated. The antitumor activity of ULDF5 against the HeLa cell line was observed to be higher (0.034 μg/mL) than that of ULDF4 (0.075 μg/mL; Fig. 2).
Fig. 2.
Inhibition activity of ULDF4 and ULDF5 against HeLa cell line.
3.4. Structural elucidation of the cytotoxic compounds purified from Streptomyces bingchenggensis ULS14
The structures of ULDF4 and ULDF5 were determined using ESI-MS, IR, and 1D (1H and 13C) and 2D (COSY and HMBC) NMR (Figs. S1-S12) as well as comparison with spectral data from previous reports [17]. ULDF4 was obtained as yellow oil and its IR absorption bands indicated the presence of a hydroxyl group (2925.73 cm−1 due to hydrogen bonding), a conjugated carbonyl (1553.89 cm−1), an aromatic C C stretch (1456.67–1600 cm−1), and a C—H bond (1080.28 cm−1) (Fig. S3). Its molecular formula was found to be C34H35NO13 by ESI-MS (m/z 665.23 [M]) (Fig. S1). The 1H NMR spectrum of ULDF4 indicated the presence of four deuterium exchangeable protons (δH1.678, 4.159, 5.133, and 7.283) and sp3 methylene proton (δH4.159) (Fig. S5). The 13C NMR spectrum (Fig. S6) showed a carbonyl carbon (δC171.1). The 1H and 13C NMR spectra, 2D NMR (1H—1H COSY and HMBC) (Figs. S7 and S8), and comparison with the MS Library and literature, also showed ULDF4 to be structurally similar to the polycyclic xanthone, kigamicin (Fig. 3).
Fig. 3.
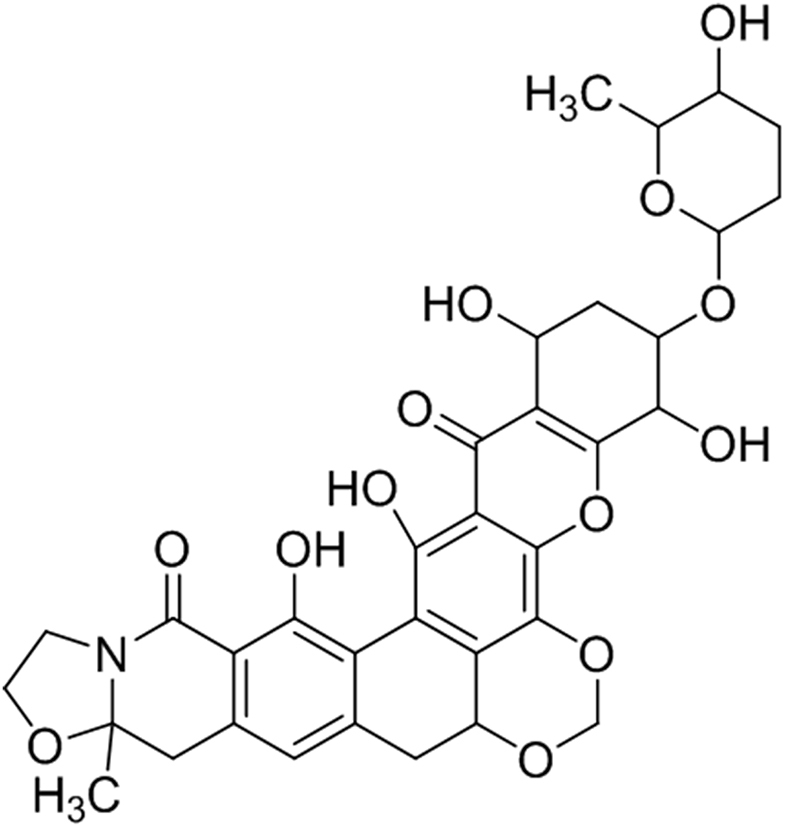
Structure of compound ULDF4.
ULDF5 was also obtained as yellow oil and its molecular formula was found to be C28H26N4O3 by ESI-MS (m/z 467.21 [M]) (Fig. S2). The 1H NMR spectrum of ULDF5 (Fig. S9) showed the presence of aromatic protons (δH7.28, 7.30, 7.33, and 7.32), while the 13C NMR spectrum (Fig. S10) showed the presence of a carbonyl carbon (δC170.13) which was more visible in the HMBC spectrum. The combined data from these spectral analyses including 2D NMR (1H—1H COSY and HMBC; Figs. S11 & S12) and comparison with the MS library and literature indicated that the compound was structurally similar to the indolocarbazole, staurosporine (Fig. 4).
Fig. 4.
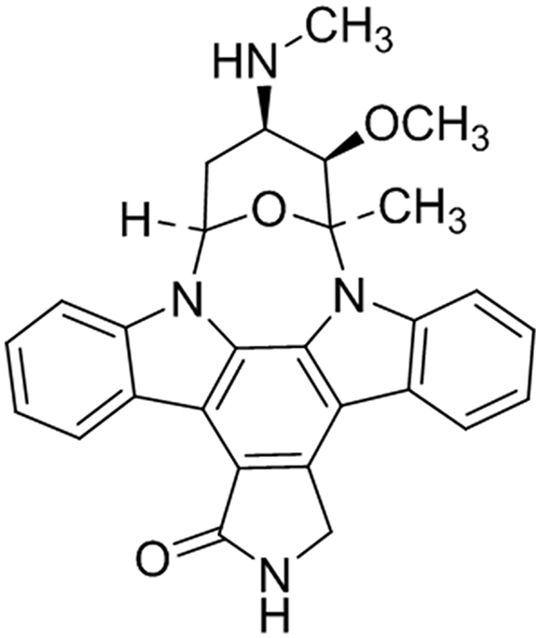
Structure of compound ULDF5.
4. Discussion
Actinomycetes have gained unprecedented relevance in recent years due to their various biological activities and ability to produce novel, pharmaceutically useful compounds including antimicrobials, antitumor agents, and immunosuppressant chemotherapeutics [18]. Streptomyces was the major bacterial group isolated, which has exceptional bioactive product synthesis capabilities. This report gives further credence to reports suggesting that sporulating, non-motile actinomycetes, which have long been considered terrestrial, are also present in marine environments in their mycelial, physiologically active phase [19,20]. Prior to this study, there were no reports on the anticancer potentials of metabolic extracts from S. fulvissimus, S. bingcheggensis, or S. albus; however, the cytotoxic activity of other actinomycetes strains has been reported. In this study, the percentage of bioactive strains with anticancer potential was 28.1%, higher than that reported previously in the literature. Becerril-Espinosa et al. [21] found that 19.2% of actinobacteria isolates possessed anticancer activity, whilst El-Shatoury et al. [22] showed that 17.5% of actinomycetes isolated from the marine environment in Egypt showed anticancer activity. Hong et al. [23] found that 20% of the bacteria isolated from different mangrove sites in China were active against colonic cancer cells (HCT-116), and that the Streptomyces genus showed the highest activity when in vitro assays were carried out. However, the increased percentage of actinomycetes with cytotoxicity isolated in this study is not surprising due to the unusual environmental conditions of the Lagos Lagoon, such as intense UV radiation from sunlight and high tropical temperatures. Marine actinomycetes present in these habitats could therefore have developed unique biosynthetic pathways which enable the synthesis of unique metabolic compounds for their survival in such an extreme environment [24]. The crude extracts of eight of the actinomycetes evaluated were found to have metabolites with cytotoxic activities against all five cancer cell lines tested. The lowest crude extract concentration with cytotoxic activity in this study was 0.03 μg/mL, against the AGS cellline. Ravikumar et al. [14] reported that the lowest IC50 value of crude extracts from the marine actinomycete strain Act 01 was 10.13 μg/mL against the MCF-7 cell line, a lower cytotoxicity than that reported here. Suthindhiran and Kannabiran [25] used an MTT assay to show that the crude extract from a Streptomyces strain isolated from the Marakkanam coast was cytotoxic, with an IC50 value of 26.2 mg/mL against HeLa cells, which is also lower than that observed in this study. The cytotoxic activities of the crude extract could be attributed to partial cellular differentiation, the induction of apoptosis and degradation of fusion transcripts, antiproliferation effects, or the inhibition of angiogenesis [14]. The bioactive metabolites in this study were toxic to the cell lines at concentrations below 1 mg/mL. Therefore, they could potentially be developed as anticancer therapeutics for human use, because cytotoxicity analysis is a crucial step in the development of new therapeutic drugs for clinical application.
Since kigamicin was first isolated from Amycolatopsis sp. ML630-mF1 by Kumimoto et al. [26,27], it has also been isolated from different Amycolatopsis species such as A. regifaucium [28]. The IR spectra of ULDF5 showed the presence of functional groups such as amines (—NH2), which have been previously associated with antifungal activity [29]. Kigamicins are known mainly for their antitumor bioactivities against tumor cell lines and antibacterial activities. Since it was first isolated from Streptomycesstaurosporeus by Omura et al. [30], staurosporine has also been isolated from other Streptomycesspecies such as S. roseoflavus LS-A24 [31]. Indolocarbazole alkaloids are known mainly for their antitumor bioactivities against cancer cell lines. However, in this study compounds structurally similar to kigamicin and staurosporine were isolated from S. bingchenggensis ULS14 for the first time; hence, this strain could be another source of these highly potent bioactive compounds. The cytotoxicity of ULDF5 against HeLa cells (34 ng/mL) was higher than the value of 10 μM against HeLa cells reported by Lovborg et al. [32] who evaluated the cytotoxicity of staurosporine against HeLa cells. However, this suggests that while ULDF5 is structurally similar to staurosporine, it has a higher cytotoxicity against HeLa cells at a very low dose, hence could be a better alternative as a highly potent cervical cancer drug.
5. Conclusion
The findings of this study confirm our hypothesis that marine actinomycetes isolated from Lagos Lagoon sediments are highly diverse, cultivatable actinobacteria with anticancer potential. We purified two novel compounds, ULDF4 and ULDF5, from the metabolic extracts of these marine actinomycetes which exhibited cytotoxicity at a lower concentration than SAHA. The results of this study, which is the first to be conducted in this sub-region, justify the anticancer therapeutic potential of actinomycetes from the West African marine environment, which could immensely benefit the development of useful chemotherapeutics. Further work will focus on the mechanisms of action of these compounds and the culture-independent diversity of these marine actinomycetes using metabolomics. These studies could improve our understanding of the distribution of actinobacteria in underexplored tropical marine habitats and help develop high-throughput screening programs for anticancer drugs and chemotherapeutics.
Acknowledgments
This work was supported by the University of Lagos Central Research Committee Grant (Grant No: ULCRC 2012/08).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2019.03.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Akarolo-Anthony S.N., Ogundiran T.O., Adebanwo C.A. Emerging breast cancer epidemic: evidence from Africa. Breast Cancer Res. 2010;12:8–11. doi: 10.1186/bcr2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin D.M., Ferlay J., Hamdi-Cherif M., et al. IARC Scientific Publications; Lyon: 2003. Cancer in Africa—Epidemiology and Prevention. pp. 153. [PubMed] [Google Scholar]
- 3.Ferlay J., Shin H.R., Bray F., et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Berdy J. Bioactive microbial metabolites. J. Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 5.Olano C., Méndez C., Salas J.A. Antitumor compounds from actinomycetes: from gene clusters to new derivatives by combinatorial biosynthesis. Nat. Prod. Rep. 2009;26:628–660. doi: 10.1039/b822528a. [DOI] [PubMed] [Google Scholar]
- 6.Olano C., Méndez C., Salas J.A. Antitumor compounds from marine actinomycetes. Mar. Drugs. 2009;7:210–248. doi: 10.3390/md7020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 8.Demain A.L., Sánchez S. Microbial drug discovery: 80 years of progress. J. Antibiot (Tokyo) 2009;62:5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies O.F., Adeleye I.A., Wang P.G. Hyoscamine-producing marine actinomycetes from Lagos Lagoon sediment. Asian Pac. J. Trop. Biomed. 2015;3:1–10. [Google Scholar]
- 10.Elliah P., Ramana T., Bapi Raju K.V.V.S., et al. Investigation on marine actinomycetes from Bay of Bengal near Karnataka coast of Andhra Pradesh. Asian J. Microbiol. Biotechnol. Environ. Sci. 2004;6:53–56. [Google Scholar]
- 11.Stach J.E.M., Maldonado L.A., Ward A.C., et al. New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol. 2003;5:828–841. doi: 10.1046/j.1462-2920.2003.00483.x. [DOI] [PubMed] [Google Scholar]
- 12.Tamura K., Peterson D., Peterson N., et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolution distance and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janardhan A., Kumar A.P., Viswanath B., et al. Production of bioactive compounds by actinomycetes and their antioxidant properties. Biotechnol. Res. Int. 2014;20:1–8. doi: 10.1155/2014/217030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravikumar S., Gnanadesigan M., Thajuddin N., et al. Anticancer property of sponge associated actinomycetes along Palk strait. J. Pharm. Res. 2010;3:2415–2417. [Google Scholar]
- 15.Kwon H.C., Kauffman C.A., Jensen P.R., et al. Marinisporolides, polyene-polyol macrolides from a marine actinomycete of the new genus “Marinispora”. J. Org. Chem. 2009;74:675–684. doi: 10.1021/jo801944d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis E.A., Munde M., Wang S., et al. Complexity in the binding of minor groove agents: netropsin has two thermodynamically different DNA binding modes at a single site. Nucleic Acids Res. 2011;39:9649–9658. doi: 10.1093/nar/gkr699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R., Zhu T., Li D., et al. Two indolocarbazole alkaloids with apoptosis activity from a marine-derived actinomycete Z2039-2. Arch Pharm. Res. 2007;30:270–274. doi: 10.1007/BF02977605. [DOI] [PubMed] [Google Scholar]
- 18.Saurav K., Kannabiran K. Cytotoxicity and antioxidant activity of 5-(2,4-dimethylbenzyl)pyrrolidin-2-one extracted from marine Streptomyces VITSVK5 spp. Saudi J. Biol. Sci. 2012;19:81–86. doi: 10.1016/j.sjbs.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran M.A., Rutherford L.T., Hodson R.E. Evidence for indigenous Streptomyces populations in a marine environment determined with a 16S rRNA probe. Appl. Environ. Microbiol. 1995;61:3695–3700. doi: 10.1128/aem.61.10.3695-3700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malet-Cascon L., Romero F., Espliego-Vazquez F., et al. IB-00208, a new cytotoxic polycyclic xanthone produced by a marine-derived actinomadura, I. Isolation of the strain, taxonomy and biological activites. J. Antibiot (Tokyo) 2003;56:219–225. doi: 10.7164/antibiotics.56.219. [DOI] [PubMed] [Google Scholar]
- 21.Becerril-Espinosa A., Guerra-Rivas G., Ayala-Sánchez N., et al. Antitumor activity of actinobacteria isolated in marine sediment from Todos Santos Bay, Baja California, Mexico. Rev. Biol. Mar. Oceanogr. 2012;47:317–325. [Google Scholar]
- 22.El-Shatoury S.A., El-Shenawy N.S., El-Salam I.M.A. Antimicrobial, antitumor and in vivo cytotoxicity of actinomycetes inhabiting marine shellfish. World J. Microbiol. Biotechnol. 2009;25:1547–1555. [Google Scholar]
- 23.Hong K., Gao A., Xie Q., et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs. 2009;7:24–44. doi: 10.3390/md7010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao X., Lu Y., Xing Y., et al. A novel anticancer and antifungus phenazine derivative from a marine actinomycete BM-17. Microbiol. Res. 2012;167:616–622. doi: 10.1016/j.micres.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Suthindhiran K., Kannabiran K. Cytotoxic and antimicrobial potential of actinomycete species Saccharopolyspora salina VITSDK4 isolated from the Bay of Bengal coast of India. Am. J. Infect. Dis. 2009;5:90–98. [Google Scholar]
- 26.Kunimoto S., Lu J., Esumi H., et al. Novel antitumor antibiotics I. Taxonomy, isolation, physico-chemical properties and biological activities. J. Antibiot (Tokyo) 2003;56:1004–1011. doi: 10.7164/antibiotics.56.1004. [DOI] [PubMed] [Google Scholar]
- 27.Kunimoto S., Someno T., Yamazaki Y., et al. Novel antitumor antibiotics. II. Structure determination. J. Antibiot (Tokyo) 2003;56:1012–1017. doi: 10.7164/antibiotics.56.1012. [DOI] [PubMed] [Google Scholar]
- 28.Tan A.Y.G., Robinson S., Lacey E., et al. Amycolatopsis regifaucium sp. nov., a novel actinomycete that produces kigamicins. Int. J. Syst. Evol. Microbiol. 2007;57:2562–2567. doi: 10.1099/ijs.0.64974-0. [DOI] [PubMed] [Google Scholar]
- 29.Dhanasekaran D., Thajuddi N., Panneerselvam A. Antifungal compound: 4' phenyl -1-napthyl –phenyl acetamide from Streptomyces sp. DPTB16. Med. Biol. 2008;15:7–12. doi: 10.1016/j.compbiomed.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Omura S., Iwai Y., Hirano A., et al. A new alkaloid AM-2282 of Streptomyces origin taxonomy, fermentation, isolation and preliminary characterization. J. Antibiot (Tokyo) 1977;30:275–282. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 31.Park H.J., Lee J.Y., Hwang I.S., et al. Isolation and antifungal and antioomycete activities of staurosporine from Streptomyces roseoflavus strain LS-A24. J. Agric. Food Chem. 2006;54:3041–3046. doi: 10.1021/jf0532617. [DOI] [PubMed] [Google Scholar]
- 32.Lovborg H., Nygren P., Larsson R. Multiparametric evaluation of apoptosis: effects of standard cytotoxic agents and the cyanoguanidine CHS 828. Mol. Cancer Ther. 2004;3:521–526. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.