Abstract
Introduction
Bovine parainfluenza virus-3 (BPIV3) and bovine respiratory syncytial virus (BRSV) are the cause of respiratory disease in cattle worldwide. With other pathogens, they cause bovine respiratory disease complex (BRDC) in ruminants. The aim of the study was the detection and molecular characterisation of BPIV3 and BRSV from nasal swabs and lung samples of cows in and around the Erzurum region of eastern Turkey.
Material and Methods
In total, 155 samples were collected. Of animals used in the study 92 were males and 63 females. The age of the animals was between 9 months and 5 years, mean 1.4 years. Most males were in the fattening period and being raised in open sheds; females were in the lactating period and kept in free stall barns. All samples were tested for the presence of viral genes using RT-PCR. Gene-specific primers in a molecular method (RT-PCR) identified BRSV (fusion gene) and BPIV3 (matrix gene) strains at the genus level.
Results
RNA from BRSV and BPIV3 was detected in two (1.29%) and three (1.93%) samples, respectively, one of each of which was sequenced and the sequences were aligned with reference virus strains. Phylogenetic analyses clustered the strains in genotype C/BPIV3 and subgroup III/BRSV.
Conclusion
The results indicate that BRSV and BPIV3 contribute to bovine respiratory disease cases in Turkey. This is the first report on their detection and molecular characterisation in ruminants in Turkey.
Keywords: cattle, bovine parainfluenza virus-3, bovine respiratory syncytial virus, molecular characterisation, Turkey
Introduction
Bovine respiratory disease complex (BRDC) is a major problem for cattle breeders worldwide, causing serious economic losses. BRDC is associated with infection by certain viruses, bacteria, and parasites (33). In addition to these infectious agents, stress factors such as transport, gestation, and poor management conditions play an important role in the onset of the disease (30). Bovine herpes virus 1 (BHV-1), bovine respiratory syncytial virus (BRSV), and bovine parainfluenza virus-3 (BPIV3) are the most common viral agents of the respiratory system. Some opportunistic agents (Mannheimia haemolytica, Pasteurella multocida, Haemophilus somnus, and Mycoplasma spp.) contribute to the appearance of clinical signs and thus increase mortality and cause losses in the herds (18). Suppressed immunity also has an important role in the prognosis. Diseases such as bovine leucosis and bovine viral diarrhoea suppress immunity and lead to more animal loss by worsening clinical symptoms.
BPIV3 (the new name of which is bovine respirovirus 3) is an RNA virus assigned to the Paramyxoviridae family under the Respirovirus genus. BRSV (the new name of which is bovine orthopneumovirus) is in the Pneumoviridae family under the Orthopneumovirus genus. To date, three genotypes of BPIV3 have been described. These genotypes, termed A, B, and C, were differentiated based on phylogenetic analysis. Genotype A strains have been isolated in North America, China, and Japan. Genotype B was originally found in Australia. Isolations of genotype C were in China, South Korea, and Japan. In addition, all three genotypes have been reported in Argentina (23).
Initially BRSV subgroups were identified (A, B, and AB or intermediary) based on monoclonal antibody and polyclonal sera analyses against F and G proteins (31). Additionally, Valarcher et al. (36) proposed that six genetic subgroups may be found in BRSV strains, when F, G, and nucleoprotein sequences are phylogenetically analysed by maximum-likelihood algorithms. Therefore, six subgroups were detected in BRSV. These subgroups termed I (the subgroup B prior to the recommendation of Valarcher et al. (36)), III (subgroup A), and II, IV, V, and VI (subgroup AB) were differentiated based on phylogenetic analysis. Subgroup I consists of European strains (UK and Switzerland). Subgroup III includes viruses exclusively from the USA. Subgroup II aggregates strains from the Netherlands, Belgium, France, Denmark, Sweden, and Japan. Subgroup IV is of European and USA strains while subgroups V and VI are found only in French and Belgian isolates (29, 36). Subgroup VII was detected in later years (9) and some strains are known which are still not classified (these are regarded as untyped) (10).
BPIV3 and BRSV can cause mild symptoms or subclinical disease when present alone. However, when there is a co-infection, they may cause bronchopneumonia, severe cough, high fever, and nasal discharge and contribute to a more serious clinical course of infection (33). Regardless of the infecting agent in BRDC, clinical symptoms may be similar and the process of detecting the underlying primal agent may be hindered due to mixed bacterial infections. This situation makes viral diagnosis difficult and decreases the specificity and sensitivity of the molecular methods (when compared to immunofluorescence antibody tests) (15).
Data on virological detection of these agents in Turkey is limited (2, 6), but there are more studies on seroprevalence of these viruses among cattle herds. The studies reported lowest and highest seropositivity of 11% (1) and 92.8% (13) for BPIV3 and 28% (1) and 94% (13) for BRSV. Serological studies on BRSV and BPIV3 were previously conducted in different geographic regions of our country. In these studies the following percentage values for BRSV and BPIV3 prevalence were determined respectively: Alpay et al. (5) 26.6% and 44.6%, Alkan et al. (3) 62.0% and 44.6%, Avci et al. (7) 78.2% and 85.6%, Çabalar and Can Sahna (11) 67.3% and 18%, Yavru et al. (40) 46% and 53.9%, and Yesilbag and Gungor (41) 73.0% and 43%. These studies were conducted either countrywide (3) or in selected regions (40, 41).
The aim of this study was the detection and molecular characterisation of BPIV3 and BRSV strains retrieved from nasal swabs and lung samples of cows in the eastern region of Turkey. The determination of BRSV and BPIV3 types and associated co-infections for respiratory system infections was conducted.
Material and Methods
Sampling. Nasal swabs and lung tissue samples were gathered from cattle from the Erzurum district and its neighbouring provinces. Table 1 shows the origin and quantity of the collected samples. A total of 155 animals were used in the study (92 males and 63 females). The herd consisted of cross-breeds of brown Swiss, Holstein, and Simmental. The age of the animals ranged between 9 months and 5 years and their mean age was 1.4 years. The males were in the fattening period and kept in open sheds; the lactating females were housed in free stall barns. Samples were acquired from animals with nasal discharge and severe coughing. In total 155 samples were collected including 28 lung tissue samples and 127 nasal swabs. Although sampling was performed throughout the year, the winter and autumn months were generally preferred as the sampling period (Table 1).
Table 1.
Sampling locations, number of samples, and type of material
| No | Sampling region | Number of samples | Material |
|---|---|---|---|
| 1 | Erzurum slaughterhouse (1st period, Dec 2013) | 8 | Lung |
| 2 | Erzurum slaughterhouse (2nd period, Apr 2014) | 10 | Lung |
| 3 | Erzurum slaughterhouse (3rd period, Sept 2014) | 7 | Lung |
| 4 | Erzurum slaughterhouse (4th period, Jan 2015) | 3 | Lung |
| 5 | Erzurum Centre (1st period, Mar 2014) | 7 | Nasal swab |
| 6 | Erzurum Centre (2nd period, Dec 2014) | 5 | Nasal swab |
| 7 | Erzurum–Aziziye Province | 23 | Nasal swab |
| 8 | Erzurum–Aşkale Province | 22 | Nasal swab |
| 9 | Erzurum–Hasankale Province | 12 | Nasal swab |
| 10 | Erzurum–Horasan Province | 15 | Nasal swab |
| 11 | Erzurum–Oltu Province | 5 | Nasal swab |
| 12 | Erzurum–Tekman Province | 2 | Nasal swab |
| 13 | Erzurum–Şenkaya Province | 1 | Nasal swab |
| 14 | Bayburt Centre | 10 | Nasal swab |
| 15 | Tokat Centre | 10 | Nasal swab |
| 16 | Gümüşhane Centre | 15 | Nasal swab |
| Total | 155 |
Molecular analysis
Extraction of the viral nucleic acid. Nasal swabs were vortexed in 1 mL of PBS and centrifuged at 3,000 rpm for 5 min. Supernatant was collected and 200 μL was used for extraction. A total of 25 mg of each tissue sample was homogenised in 500 μL PBS using a Tissue Lyser II homogenizer (Qiagen, Germany) and centrifuged at 3,000 rpm at room temperature, and then 200 μL of the supernatant was used for extraction. A GF-1 viral nucleic acid kit (Vivantis, Malaysia) was used for nucleic acid extraction according to the manufacturer’s recommendations. All extracts were kept at −20°C before testing.
Reverse transcription (RT). Viral RNA was subjected to reverse transcription using a First Strand cDNA synthesis kit (Thermo Fisher Scientific, Germany) according to the manufacturer’s instructions.
PCR. Samples were tested by PCR using gene-specific primer pairs. Primers specific for the gene-encoding fusion region for BRSV and matrix gene for BPIV3 were used. Primer sequences, their related gene regions, and the size of estimated amplicons are shown in Table 2. Taq DNA polymerase (Thermo Fisher Scientific, Germany) was used for the PCR which was run under previously described conditions (22, 37).
Table 2.
Primers used in the study
| Virus/Gene | Primers | Primer sequences (5’-3’) | Product size (bp) | References |
|---|---|---|---|---|
| BPIV3/Matrix | M1 | AGTGATCTAGATGATGATCCA | 328 | (22) |
| M2 | GTT ATT GAT CCA ATT GCT GT | |||
| BRSV/Fusion | B1 | AATCAACATGCAGTGCAGTTAG | 710 (1st round) | |
| B2A | TTTGGTCATTCGTTATAGGCAT | (37) | ||
| B3 | GTGCAGTTAGTAGAGGTTATCTTAGT | |||
| B4A | TAGTTCTTTAGATCAAGTACTTTGCT | 481 (2nd round) |
Sequencing. After PCR, the samples with the clearest virus-specific PCR bands were subjected to sequencing. Samples were sequenced (with the Sanger sequencing method, bi-directionally with forward and reverse primers) by the Pendik Veterinary Control Institute, Istanbul, Turkey. Obtained sequences were submitted to GenBank as bovine respirovirus 3 isolate BPIV3/TR/Erz/2014 under accession number KY511410 and bovine orthopneumovirus isolate BRSV/TR/Erz/2014 under accession number KY499619, and then were analysed phylogenetically.
Phylogenetic analysis. The raw sequence data and reference virus sequences obtained from GenBank were aligned using Bioedit version 7.0.5.3 software (17). The aligned data were subjected to phylogenetic analysis using MEGA version 6 (32) to give bootstrap analyses (1,000 replicates) by the neighbour-joining method. Tamura’s two-parameter model was used and bootstrap values are shown in the phylogenetic tree.
Results
Polymerase chain reaction (PCR). Among 155 samples, two were BRSV- and three BPIV3-positive. Four of the positive samples were swabs, and one was a lung sample. The BPIV3-positive samples originated from Erzurum-Aşkale Province (swab), Erzurum-Aziziye Province (swab), and Erzurum slaughterhouse (2nd period, Apr. 2014) (lung sample). BRSV-positive samples originated from Erzurum-Aziziye Province (swab) and Bayburt-Centre (swab). All animals identified as positive were young cattle.
Phylogenetic analysis. Nucleotide sequences of the viruses identified in the study were closely related to Asian and European strains (Figs 1 and 2). The phylogenetic analysis of the fusion (F) protein of BRSV showed that the identified BRSV strain belonged to subgroup III (subgroup A) and was in the same cluster as reference viruses (KU159366 strain USII/S1/USA and FJ543092 strain 375/Brazil). The matrix gene sequence of the BPIV3 strain sequenced from a BPIV3-positive sample was classified into a cluster of genotype C.
Fig. 1.
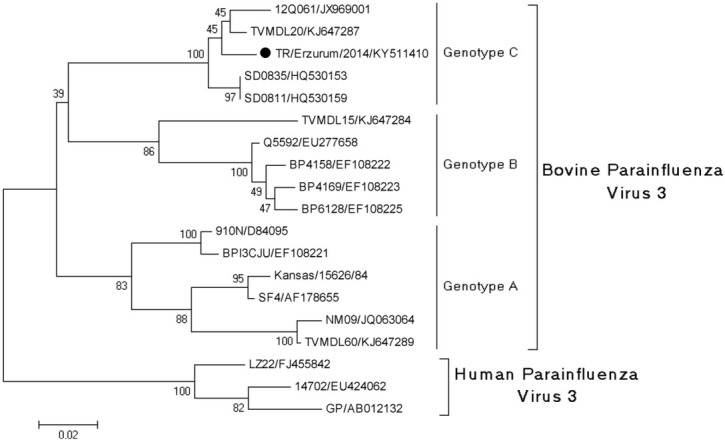
Phylogenetic tree generated based on the nucleotide sequence alignment of the BPIV3 matrix (M) gene region of the virus obtained in the study and viral strains from GenBank
Fig. 2.
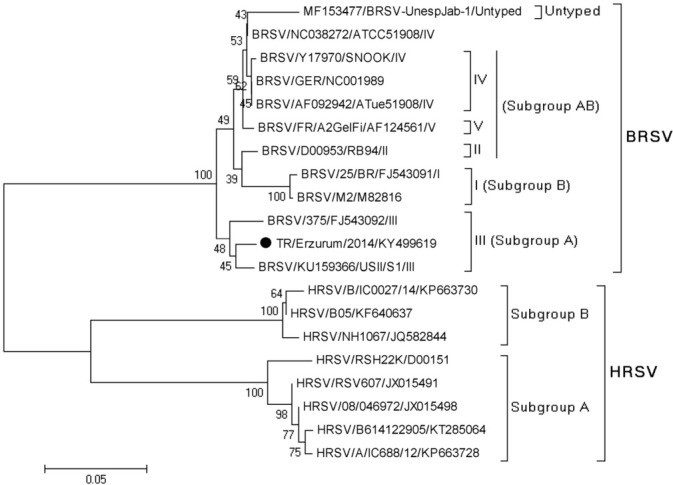
Phylogenetic tree generated on the basis of the nucleotide sequence alignment of the BRSV fusion (F) gene region of virus strain obtained in the study and viral strains from GenBank including human respiratory syncytial virus (HRSV)
Discussion
There are various diseases affecting young and old cattle worldwide. These diseases can impair the respiratory, digestive, and genital systems (4, 25, 34, 35). However, it is viral diseases associated with the respiratory system of cattle which are regarded as one of the main problems in cattle breeding, affecting both adults and young animals. This holds especially true when mortality rates increase in case of mixed infections. In this study, BRSV and BPIV3 virus strains were detected in pool of 155 lung tissue and nasal swab samples collected from cattle from Erzurum and neighbouring provinces.
BPIV3 and BRSV were detected by other researchers with serological and virological methods in cases of lower respiratory tract infections in cattle in Turkey (2, 6, 41). However, these studies were not performed by molecular methods and did not include genetic characterisation as we have done. Although it was proved that in cases of lower respiratory tract infections lungs could contain a high number of virus particles (8), we could detect only BPIV3, even though samples were collected in late winter. This may be because the animals were in a convalescent phase of infection or a majority of tissue samples suspected of being from pneumonic animals did not contain virus particles.
Enveloped viruses, paramyxoviruses in particular, are generally susceptible when exposed to the outside environment. Pirtle and Beran (27) reported that when diluted to 105.5 CCID50/mL and used to contaminate different kinds of media, the viability time was 1.5 h on rubber gloves, 20 min on skin, and a maximum of 6 h on an indoor surface. This is why samples must be admitted to the laboratory in a short time and tested in the shortest time possible. When low positivity rates are considered, both in this study and worldwide, there is always the possibility of false negativity because of virus sensitivity. PCRs specific for the gene-encoding fusion region for BRSV and the matrix gene for BPIV3 were used. The fusion gene of BRSV is a region that determines antigenic variations (26), and the matrix gene for BPIV3 is responsible for viral assembly, pathogenesis, and antigenicity of strains (20, 21). Phylogenetic analysis of BRSV based on the F protein sequence classified the isolates into seven different subgroups. The topology of the phylogeny was retained when an analysis of the N and G protein gene sequences was conducted (36). In our study, the determined strain was found to belong to subgroup III. Knowing the genotypes helps to determine the type-specific vaccine selection in the geographic region. Valarcher et al. (36) identified vaccine failure among animals infected with BRSV groups V and VI, indicating that commercial vaccines act poorly against infections caused by such viral groups. Therefore, selection of vaccines is important and should be specific to the type of strains present in the region. For example in the 2000s in Europe, vaccines including Rispoval RS (strain RB-94, subgroup II), Bayovac (strain Lehmkuhl 375, subgroup III), and Vacores (strain 220/69, subgroup II) were used. If these vaccines were currently in use, Bayovac could be suitable for our country. However, in vivo trials would be required for other vaccines. Therefore, genotyping studies of viruses are necessary for molecular epidemiology and vaccine studies.
It is well established that respiratory tract infections are mostly mixed. These types of infections may contain viruses, bacteria, and other agents or may be caused by virus-virus or bacteria-bacteria interaction (18, 33). We investigated the samples with single or mixed (BRSV + BPIV3) infections and no other mixed infections were detected. Therefore, when viruses are the sole cause of infection, it is highly likely that they will cause subclinical infections; this may explain the low rate of detection in this study.
Aetiology in respiratory infection is quite important (14, 38). A fast and reliable method must be selected to identify the underlying agent. Genotyping studies may provide information about strains that cause serious clinical onsets related to vaccine strains and preventive status. Phylogenetic analysis of the BRSV strain was performed (Fig. 2). Analysis of the partial sequence of BRSV shows that our strains were more closely related to American and Brazilian (USA: KU159366; and Brazil: FJ543092) strains instead of European ones. Although the strains identified in the study are expected to be close to the European strains (because of the geographical location of Turkey), the increase in meat and live animal imports into our country over the last 5 years may be a factor in this situation, because our country imports meat from USA, Canada, Brazil, and Uruguay. Many infectious diseases may be spread among cities, countries, and continents as a result of animal or animal product transportation (12, 16). In our study, BRSV sequences showed high genetic similarity with the American and Brazilian strains. This genetic similarity among BRSV strains suggests that it could have come from Brazil or America, as a result of live animal exports.
Infection with BPIV3 is common in cattle worldwide. The virus is divided into three genotypes: A, B, and C (19, 24, 39, 42). Genotype A is most common in America, genotype B in Australia and genotype C is seen mostly in Asia (24, 28, 42) (Fig. 1). Our samples clustered in genotype C in the phylogenetic tree. This is the first study to report BPIV3 of this type (genotype C) in Turkey. This also may be regarded as the first report in Europe since no published data is available as far as we are aware.
In conclusion, this paper is the first report of BRSV and BPIV3 infections in cattle in the east Anatolian region of Turkey and specifically Erzurum and its neighbouring provinces by molecular methods. There is a need for additional epidemiological studies to better understand the prevalence and control of the infections. Regional and local infections should be investigated and properly addressed through random sampling. For implementing control and eradication studies at a national level, full genome sequence data should be provided in further studies.
Footnotes
Conflict of Interest
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: This research was supported by the Atatürk University Scientific Research Project Foundation (BAP) project no: 2013/035.
Animal Rights Statement: None required.
References
- 1.Akca Y., Burgu I., Gur S., Bilge-Dagalp S.. A study on investigations of occurrence of some virus infection in buffaloes in Turkey. Rev Med Vet. 2004;156:268–271. [Google Scholar]
- 2.Alkan F., Ozkul A., Bilge-Dagalp S., Yesilbag K., Oguzoglu C., Akca Y., Burgu I.. Virological and serological studies on the role of PI-3 virus, BRSV, BVDV, and BHV- 1 on respiratory infections of cattle. I. The detection of etiological agents by direct immunofluorescence technique. Dtsch Tierarztl Wochenschr. 2000;107:193–195. [PubMed] [Google Scholar]
- 3.Alkan F., Özkul A., Karaoglu M.T., Bilge S., Akça Y., Burgu I., Yesilbag K., Oğuzoğlu T.C.. Sığırlarda viral nedenli solunum sistemi enfeksiyonlarinin seroepidemiyolojisi. Ankara Üniv Vet Fak Derg. 1997;44:73–80. [Google Scholar]
- 4.Alkan F., Ozkul A., Oguzoglu T.C., Timurkan M.Ö., Caliskan E., Martella V., Burgu I.. Distribution of G (VP7) and P (VP4) genotypes of group A bovine rotaviruses from Turkish calves with diarrhea, 1997–2008. Vet Microbiol. 2010;141:231–237. doi: 10.1016/j.vetmic.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Alpay G., Tuncer P., Yeşilbağ K.. Bir ada ekosistemindeki sığır, koyun ve keçilerde bazı viral enfeksiyonların serolojik olarak araştırılması. Ankara Üniv Vet Fak Derg. 2014;61:43–48. [Google Scholar]
- 6.Avci O., Yavru S., Ekik M.. Detection of respiratory viral antigens in cattle lung tissues by direct ELISA. Anim Vet Sci. 2014;2:146–149. [Google Scholar]
- 7.Avci O., Yavru S., Sevik M.. Antibody prevalence against respiratory viruses in naturally infected cattle in Central Anatolia. Eurasian J Vet Sci. 2014;30:80–84. [Google Scholar]
- 8.Baker J.C., Werdin R.E., Ames T.R., Markham R.J.F., Larson V.L.. Study on the etiologic role of bovine respiratory syncytial virus in pneumonia of dairy calves. J Am Vet Med Assoc. 1986;189:66–86. [PubMed] [Google Scholar]
- 9.Bertolotti L., Giammarioli M., Rosati S.. Genetic characterization of bovine respiratory syncytial virus strains isolated in Italy: evidence for the circulation of new divergent clades. J Vet Diagn Invest. 2018;30:300–304. doi: 10.1177/1040638717746202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bortolin Affonso I., de Souza A., Cavalheiro Martini M., Bianchi dos Santos M., Rosado Spilki F., Weis Arns C., Issa Samara S.. Detection of an untyped strain of bovine respiratory syncytial virus in a dairy herd. Semina: Ciências Agrárias. 2014;35:2539–2549. [Google Scholar]
- 11.Çabalar M., Can Şahna K.. Doğu ve güneydoğu anadolu bölgesinde süt sığırlarında parainfluenza virus-3, bovine herpes virus-1 ve respiratory syncytial virus enfeksiyonlarının seroepidemiyolojisi. YYÜ Vet Fak Derg. 2000;11:101–105. [Google Scholar]
- 12.Dean A.S., Fournié G., Kulo A.E., Boukaya G.A., Schelling E., Bonfoh B.. Potential risk of regional disease spread in West Africa through cross-border cattle trade. Plos One. 2013;8:e75570. doi: 10.1371/journal.pone.0075570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duman R., Yavru S., Kale M., Avcı O.. Seroprevalence of viral upper respiratory infections in dairy cattle. Kafkas Üniv Vet Fak Derg. 2009;15:539–542. [Google Scholar]
- 14.Fulton R.W.. Bovine respiratory disease research (1983–2009) Anim Health Res Rev. 2009;10:131–139. doi: 10.1017/S146625230999017X. [DOI] [PubMed] [Google Scholar]
- 15.Fulton R.W., Confer A.W.. Laboratory test descriptions for bovine respiratory disease diagnosis and their strengths and weaknesses: gold standards for diagnosis, do they exist? Can Vet J. 2012;53:754–761. [PMC free article] [PubMed] [Google Scholar]
- 16.Graham J.P., Leibler J.H., Price L.B., Otte J.M., Pfeiffer D.U., Tiensin T., Silbergeld E.K.. The animal-human interface and infectious disease in industrial food animal production: rethinking biosecurity and biocontainment. Publ Health Rep. 2008;123:282–299. doi: 10.1177/003335490812300309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall T.A.. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 18.Hodgson P.D., Aich A., Manuja A., Hokamp H., Roche F.M., Brinkman F.S.I., Potter A., Babiuk L.A., Griebel P.J.. Effect of stress on viral-bacterial synergy in bovine respiratory disease; novel mechanisms to regulate inflammation. Comp Func Genom. 2005;6:244–250. doi: 10.1002/cfg.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwood P.F., Gravel J.L., Mahony T.J.. Identification of two distinct bovine parainfluenza virus type 3 genotypes. J Gen Virol. 2008;89:1643–1648. doi: 10.1099/vir.0.2008/000026-0. [DOI] [PubMed] [Google Scholar]
- 20.Kawano M., Bando H., Ohgimoto S., Okamoto K., Kondo K., Tsurudome M., Nishio M., Ito Y.. Complete nucleotide sequence of the matrix gene of human parainfluenza type 2 virus and expression of the M protein in bacteria. Virology. 1990;179:857–861. doi: 10.1016/0042-6822(90)90155-k. [DOI] [PubMed] [Google Scholar]
- 21.Kondo K., Fujii M., Nakamura T., Bando H., Kawano M., Tsurudome M., Komada H., Kusakawa S., Nishio M., Ito Y.. Sequence characterization of the matrix protein genes of parainfluenza virus types 4A and 4B. J Gen Virol. 1991;72:2283–2287. doi: 10.1099/0022-1317-72-9-2283. [DOI] [PubMed] [Google Scholar]
- 22.Maidana S.S., Lomonaco P.M., Combessies G., Craig M.I., Diodati J., Rodriguez D., Parreño V., Zabal O., Konrad J.L., Crudelli G., Mauroy A., Thiry E., Romera S.A.. Isolation and characterization of bovine parainfluenza virus type 3 from water buffaloes Bubalus bubalis in Argentina. BMC Vet Res. 2012;20:83. doi: 10.1186/1746-6148-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neill J.D., Ridpath J.F., Valayudhan B.T.. Identification and genome characterization of genotype B and genotype C of bovine parainfluenza type 3 viruses isolated in the United States. BMC Vet Res. 2015;11:112. doi: 10.1186/s12917-015-0431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oem J.K., Lee E.Y., Lee K.K., Kim S.H., Lee M.H., Hyun B.H.. Molecular characterization of a Korean bovine parainfluenza virus type 3 isolate. Vet Microbiol. 2013;162:224–227. doi: 10.1016/j.vetmic.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Oğuzoğlu T.C., Muz D., Yılmaz V., Timurkan M.Ö., Alkan F., Akça Y., Burgu İ.. Molecular characteristics of bovine virus diarrhoea virus 1 isolates from Turkey: approaches for an eradication programme. Transbound Emerg Dis. 2012;59:303–310. doi: 10.1111/j.1865-1682.2011.01272.x. [DOI] [PubMed] [Google Scholar]
- 26.Pastey M.K., Samal S.K.. Analysis of the bovine respiratory syncytial virus fusion protein (F) using monoclonal antibodies. Vet Microbiol. 1997;58:175–185. doi: 10.1016/s0378-1135(97)00171-5. [DOI] [PubMed] [Google Scholar]
- 27.Pirtle E.C., Beran G.W.. Virus survival in the environment. Rev Sci Tech. 1991;10:733–748. doi: 10.20506/rst.10.3.570. [DOI] [PubMed] [Google Scholar]
- 28.Ren J.L., Zhu Y.M., Zhou Y.H., Lv C., Yan H., Ma L., Shi H.F., Xue F.. Identification of three antigen epitopes on the nucleocapsid protein of the genotype C of bovine parainfluenza virus type 3. Vet Microbiol. 2015;178:61–69. doi: 10.1016/j.vetmic.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Sarmiento-Silva R.E., Nakamura-Lopez Y., Vaughan G.. Epidemiology, molecular epidemiology, and evolution of bovine respiratory syncytial virus. Viruses. 2012;4:3452–3467. doi: 10.3390/v4123452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snowder G.D., Van Vleck L.D., Cundiff L.V., Bennett G.. Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J Anim Sci. 2006;84:1999–2008. doi: 10.2527/jas.2006-046. [DOI] [PubMed] [Google Scholar]
- 31.Spilki F.R., Almeida R.S., Domingues H.G., D’Arce R.C.F., Ferreira H.L., Campalans J., Costa S.C., Arns C.W.. Phylogenetic relationships of Brazilian bovine respiratory syncytial virus isolates and molecular homology modeling of attachment glycoprotein. Virus Res. 2006;116:30–37. doi: 10.1016/j.virusres.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thonur L., Maley M., Gilray J., Crook T., Laming E., Turnbull D., Nath M., Willoughby K.. One-step multiplex real time RT-PCR for the detection of bovine respiratory syncytial virus, bovine herpesvirus 1, and bovine parainfluenza virus 3. BMC Vet Res. 2012;28:37. doi: 10.1186/1746-6148-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timurkan M.O., Alcigir M.E.. Phylogenetic analysis of a partial L1 gene from bovine papillomavirus type 1 isolated from naturally occurring papilloma cases in the northwestern region of Turkey. Onderstepoort J Vet Res. 2017;84:1–6. doi: 10.4102/ojvr.v84i1.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timurkan M.O., Aydın H., Belen S.. The detection and molecular characterization of bovine respiratory coronavirus infection by RT-PCR in Erzurum. Atatürk Üniv Vet Bil Derg. 2015;10:186–192. [Google Scholar]
- 36.Valarcher J.F., Schelcher F., Bourhy H.. Evolution of bovine respiratory syncytial virus. J Virol. 2000;74:10714–10728. doi: 10.1128/jvi.74.22.10714-10728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilcek S., Elvander M., Ballagi-Pordány A., Belák S.. Development of nested PCR assays for detection of bovine respiratory syncytial virus in clinical samples. J Clin Microbiol. 1994;32:2225–2231. doi: 10.1128/jcm.32.9.2225-2231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willoughby K., Thomson K., Maley M., Gilray J., Scholes S., Howie F., Caldow G., Nettleton P.F.. Development of a real time reverse transcriptase polymerase chain reaction for the detection of bovine respiratory syncytial virus in clinical samples and its comparison with immunohistochemistry and immunofluorescence antibody testing. Vet Microbiol. 2008;126:264–270. doi: 10.1016/j.vetmic.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y., Wang F.X., Sun N., Cao L., Zhang S.Q., Zhu H.W., Guo L., Cheng S.P., Wen Y.J.. Development and evaluation of two truncated recombinant NP antigen-based indirect ELISAs for detection of bovine parainfluenza virus type 3 antibodies in cattle. J Virol Methods. 2015;222:47–54. doi: 10.1016/j.jviromet.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Yavru S., Şimşek A., Yapkıç O., Kale M.. Serological evaluation of viral infections in bovine respiratory tract. Acta Vet-Beograd. 2005;55:219–226. [Google Scholar]
- 41.Yesilbag K., Gungor B.. Seroprevalence of bovine respiratory viruses in North-Western Turkey. Trop Anim Health Prod. 2008;40:55–60. doi: 10.1007/s11250-007-9053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y.M., Shi H.F., Gao Y.R., Xin J.Q., Liu N.H., Xiang W.H., Ren X.G., Feng J.K., Zhao L.P., Xue F.. Isolation and genetic characterization of bovine parainfluenza virus type 3 from cattle in China. Vet Microbiol. 2011;149:446–451. doi: 10.1016/j.vetmic.2010.11.011. [DOI] [PubMed] [Google Scholar]


