Abstract
The present study demonstrates the miquelianin or quercetin 3-O-glucuronide (compound 1) isolated from aerial parts of Euphorbia schimperi exhibited significant results for antioxidant and antidiabetic potential. The compound 1 along with kaempferol 3-O-glucuronide (compound 2) and quercetin 3-O-rhamnoside (compound 3) isolated from the same source were quantified by validated HPTLC method. Antioxidant activity was determined by chemical means in terms of ABTS radical cation and DPPH radical scavenging activity. Compound 1 showed significant scavenging activity in both ABTS and DPPH assays as compared to standard BHA. In ABTS method IC50 values of compound 1 and standard BHA is found to be 58.90 ± 3.40 µg/mL and 28.70 ± 5.20 µg/mL respectively while in DPPH assay IC50 values of Compound 1 and standard BHA is 47.20 ± 4.90 µg/mL and 34.50 ± 6.20 µg/mL respectively. Antidiabetic effect was studied through α-amylase and α-glucosidase inhibitory activity. The mechanistic approach through molecular modelling also support the strong binding sites of compound 1 which showed significant α-amylase and α-glucosidase inhibitory activities with IC50 values 128.34 ± 12.30 and 89.20 ± 9.20 µg/mL respectively as compared to acarbose 64.20 ± 5.60 and 52.40 ± 4.60 µg/mL respectively. The results of validated RP-HPTLC analyses revealed the concentration of compound 1 found to be 16.39 µg/mg and for compound 2 and compound 3 as 3.92 and 14.98 µg/mg of dried extract, respectively.
Keywords: Miquelianin, Antidiabetic, Antioxidant, HPTLC, Quantification
1. Introduction
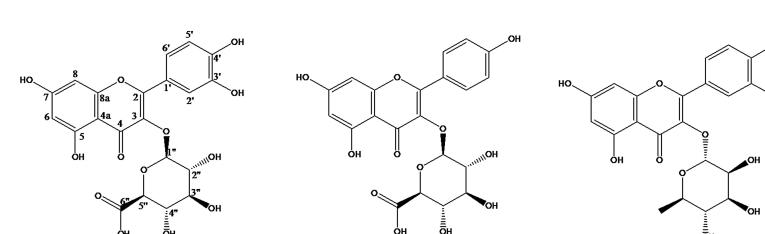
The Euphorbiaceae is one of the largest plant family (comprising 283 genera and 7300 species) having cosmopolitan distribution in both temperate and tropical regions, of which Euphorbia (2000 species) is the biggest genus (Jassbi, 2006). It is well known for chemical diversity of their phytoconstituents mainly Isoprenoid type (Shi et al., 2008). The plants of Euphorbia produce latex containing several phytoconstituents which has good therapeutic as well as commercial value. The latices of Euphorbia plant are unpleasant in taste and having poisonous property and protects the plant from grazing animals (AL-Sultan and Hussein, 2006). The latex also contains a number of chemical entities along with triterpene alcohol which is used as chemotaxonomic marker. Moreover, steroid, flavonoid and sesquiterpenes are also reported (Giner, 2015). Euphorpia schimperi C. Presl. grown in Wadi-e-Gama, Saudi Arabia (Chaudhary, 2001) apparently possess bioactivity against cancer cells, free radicals and different microorganisms (Abdel-Monem et al., 2008, Mothana et al., 2009) and also reported to contain several biologically active phytoconstituents like luteolin, α-amyrin, scopoletin, kaempferol, etc. (Abdel-Monem et al., 2008). Earlier, authors have reported that methanol extract of E. schimperi exhibited excellent wound healing activity (Ahmed et al., 2016) and further explored the wound healing property of its different fractions. The active fraction leads to the isolation of a major compound quercetin-3-O-glucuronide (Compound 1) along with two minor compounds kaempferol-3-O-glucuronide (compound 2) and quercetin-3-O-rhamnoside (compound 3) (Fig. 1) using different chromatographic techniques (Ahmed et al., 2017). Kaempferol 3-O-glucuronide and quercetin-3-O-rhamnoside have also been reported from E. guyoniana and E. hirta (Smara et al., 2014, Mallavadhani et al., 2002).
Fig. 1.
Chemical structures of compounds 1–3.
Since the compounds were previously isolated from the extract having potential wound healing activity. Keeping in view present paper demonstrates the antioxidant and antidiabetic potential of natural isolate (compound 1) from E. schimperi. A further molecular docking study is also performed to check the reliability of antidiabetic potential. The experiments were performed because in diabetic conditions the wound healing is found to be very challenging but the natural antioxidants play a vital role in protective mechanisms by their radical scavenging and lipid peroxidation ability (Singh et al., 2014). The role of free radical reactions is well established in pathological findings particularly in various acute and chronic human ailments such as atherosclerosis diabetes, immunosuppression, neurodegeneration and aging etc. (Harman, 1998). An antioxidant can be broadly explained as any substance that inhibits or delays oxidative injury to a target molecule (Yamagishi and Matsui, 2011). The compound such as phenolic acids, polyphenols and flavonoids scavenges the free radicals like hydroperoxide and peroxide and curbing the oxidative damage caused by free radical that may lead to degenerative diseases (Wu et al.2011).
Diabetes mellitus is a metabolic disorder in which rapid rising of blood sugar level is observed. This rise in blood sugar level is either due to deficiency in making of insulin by β cells (Type 1) or somehow the produced insulin becomes inert (Type 2) (WHO, 1999). Although a lot of research has been performed to manage the diabetes and many oral hypoglycemic agents have been developed but due to certain limitations of these agents the scientific community is still trying to explore some new oral hypoglycemic agents (Kavishanker et al., 2011). Since the diabetes has been treated with medicinal plants from the ancient time, therefore the area of exploration of new antidiabetic drugs via herbs is still the field of interest and scope (Gupta et al., 2005). Various chromatographic techniques are in trend to quantify the active constituents in herbal extracts. High performance thin layer chromatography (HPTLC) is one of the reliable analytical techniques used for detection and quantification of biomarkers in plant extracts as well as marketed formulations (Siddiqui et. al., 2015). Herein authors also develop a validated HPTLC method for quantification of all three compounds (1, 2 and 3) simultaneously in ESME (methanol extract of E. schimperi) in order to standardize the extract.
2. Experimental
2.1. Apparatus and reagents
Compounds 1, 2 and 3 (standards) were isolated from E. schimperi (Ahmed et al., 2017). Analytical grade reagents such as ABTS, DPPH, p-anisaldehyde, acarbose, α‐Amylase, α‐glucosidae and BHA were purchased from Sigma Aldrich. Other chemicals like dinitrosalicylic acid, sodium carbonate, p-nitrophenyl glucopyranoside and potassium persulphate are procured from different sources. Glass-backed silica gel 60F254 HPTLC plates (10 × 20 cm) were purchased from Merck (Darmstadt, Germany). CAMAG automatic TLC sampler-4 (CAMAG, Muttenz, Switzerland) was used to apply the standards and samples band wise. Development was done in ADC2 (automatic development chamber), scanning in CATS 4 (CAMAG) and documentation by CAMAG TLC Reprostar 3.
2.2. Methods
2.2.1. Antioxidant activity
2.2.1.1. ABTS radical cation scavenging activity
The ABTS [2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid)] scavenging property of the compound 1 was evaluated according to the method of Re et al., 1999. Aqueous solutions of ABTS (7 mmol/L) and potassium persulfate (2.45 mmol/L) were prepared, mixed (in the ratio of 1:1) and stored in dark (for 6hrs at room temperature) to produce cation radicals. The ABTS stock solution was further diluted with ethanol and equilibrated at 30 °C (λmax = 734 nm). Aliquots of compound 1 (0.1 mL) of different concentrations (20, 40, 60, 80 and 100 µg/mL) were mixed with diluted ABTS radical cation solution (2.9 mL), incubated (at 30 °C for 20 min) and, absorbance was measured (at λ = 734 nm) using butylated hydroxyanisole (BHA) as reference compound. The ability of compound 1 to reduce the ABTS cation radicals was estimated using formula:
where Ac = absorbance of control; Aa = absorbance of sample.
2.2.1.2. DPPH free radical scavenging activity
The free radical scavenging property of compound 1 was assessed as per Braca et al. (2001). Aliquots of 30 µL of various concentrations (20, 40, 60, 80 and 100 µg/mL) of compound 1 were added in 3 mL DPPH (2,2-diphenyl-1-picrylhydrazyl) solution (0.004%) and absorbance was taken at λ max = 520 nm after 30 min using Butylated hydroxyanisole (BHA) as reference and methanol as control. The free radical scavenging ability of compound 1 was estimated as % inhibition of free radicals by following formula:
where At = absorbance of compound; Ao = absorbance of control.
2.2.2. Antidiabetic effect
2.2.2.1. Inhibition assay for amylase activity
The amylase inhibitory activity of compound 1 was evaluated as per Kwon et al. (2008). A total of 500 µL compound 1 of concentration 20, 40, 60, 80 and 100 µg/mL and 500 µL of 0.02 M sodium phosphate buffer with 0.006 M sodium chloride, containing 20 µL of amylase (0.5 mg/mL) were incubated (at 25 °C for 10 min). Further, 1% starch solution (500 μL) in buffer (0.02 M sodium phosphate, pH 6.9 with 0.006 M sodium chloride) was added to all test tubes, incubated (for 15 min) and reaction was stopped by adding dinitrosalicyclic acid (1 mL). After that all test tubes were incubated (for 5 min) in boiling water bath, cooled, diluted with distilled water (10 mL) and absorbance was measured at λmax = 540 nm).
2.2.2.2. α-Glucosidase inhibitory assay
The α-glucosidase inhibitory activity of compound 1 was evaluated as per Kwon et al. (2008). A 20 mM phosphate buffer (pH 6.9) was used to prepare the p-nitrophenyl glucopyranoside (pNPG) substrate solution. Various concentrations (20, 40, 60, 80 and 100 µg/mL) of compound 1 (50 μL) was preincubated (for10 min.) with α-glucosidase (100 μL) and the reaction started by adding up 3.0 mM pNPG (50 μL) solution while stopped by adding 0.1 M Na2CO3 (2 mL) solution. The reaction mixtures were incubated (at 37 °C for 20 min) and assessed for p-nitrophenol release from pNPG at λ max = 405 nm. The percentage inhibition was estimated as:
2.2.3. Molecular docking
Autodock 4.2 was employed to perform molecular docking of compound 1 with α-amylase and α-glucosidase as described earlier (Morris et al., 2009, Rehman et al., 2016). The X-ray crystal structures of α-amylase (PDB ID: 3BAW; Resolution 2.0 Å) (Maurus et al., 2008) and α-glucosidase (PDB ID: 4NN4; Resolution 1.6 Å) (Roig-Zamboni et al., 2017) were retrieved from RCSB database (https://www.rcsb.org/). The protein molecules were prepared by removing heterogeneous and water molecules for molecular docking. Polar hydrogen atoms and assigning Kollman charges were added by using AutoDock Tools (ADT). Affinity grid maps of 50 × 70 × 60 Å dimensions centered at 35 × 30 × 15 Å were generated for α-amylase. Similarly, for α-glucosidase, affinity grid maps of 60 × 75 × 70 Å dimensions centered at 20 × -1 × 18 Å were generated. The SDF file of compound 1 (CID: 5274585) was downloaded from NCBI database (https://pubchem.ncbi.nlm.nih.gov/) and converted to pdbqt using Open Babel after minimizing its energy with MMFF94 force field. The non-polar hydrogen atoms were added to the compound 1 and rotatable bonds were defined. Further, Gasteiger partial chargers were added using ADT. Molecular docking was executed using Lamarck Genetic Algorithm (LGA) and Solis-Wets search procedure with default parameters. For each run of the docking, a maximum of 2.5 × 106 energy calculations was performed. The binding affinity (Kd) of compound 1 towards proteins was estimated by means of the subsequent relation (Rehman et al., 2014):
where ΔG is Gibb’s free energy of interaction between protein and ligand, R is universal gas constant (=1.987 cal/mol/K), and T is the experiment temperature (298°K).
2.2.4. Quantitative analysis of compounds 1, 2 and 3 by HPTLC
2.2.4.1. HPTLC instrumentation and conditions
The HPTLC analysis of compounds 1, 2 and 3 in ESME was performed on RP-HPTLC plates (20 × 10 cm). All the compounds along with ESME were applied on the plate by ATS-4 (Automatic TLC sampler-4; 160 nL/s) and the plate development was accomplished in Automatic Development Chamber at controlled temperature (25 ± 2 °C) and humidity (60 ± 5%). The plate was dried and analyzed quantitatively (at λmax = 350 nm) in absorbance mode (Siddiqui et al.; 2018).
2.2.4.2. Preparation of standard stock solutions
The stock solutions of compounds 1, 2 and 3 (1 mg/mL) were prepared in methanol and further diluted with methanol to get seven different concentrations (20, 40, 60, 80, 100, 120 and140 μg/mL). 10 μL of each concentration was applied on plate to provide linearity range of 200–1400 ng/spot.
2.2.4.3. Validation of method
The latest ICH Guideline for validation of analytical procedure (ICH, 2005) was adopted to determine LOD (limit of detection), LOQ (limit of quantification), linearity range, precision, robustness and recovery.
2.2.4.4. Concurrent analysis of compounds 1, 2 and 3 in ESME
The compounds 1, 2 and 3 were spotted on RP-HPTLC plate along with ESME and the peak areas were measured to find out its concentration (µg/mg of dried extract) in ESME.
2.2.5. Statistical analysis
It was carried out by ANOVA (one-way analysis) and Dunnet’s test for the evaluation of overall variation in a set of data. Results expressed in mean ± SD. P < 0.05 considered as significant.
3. Results and discussion
3.1. Antioxidant activity
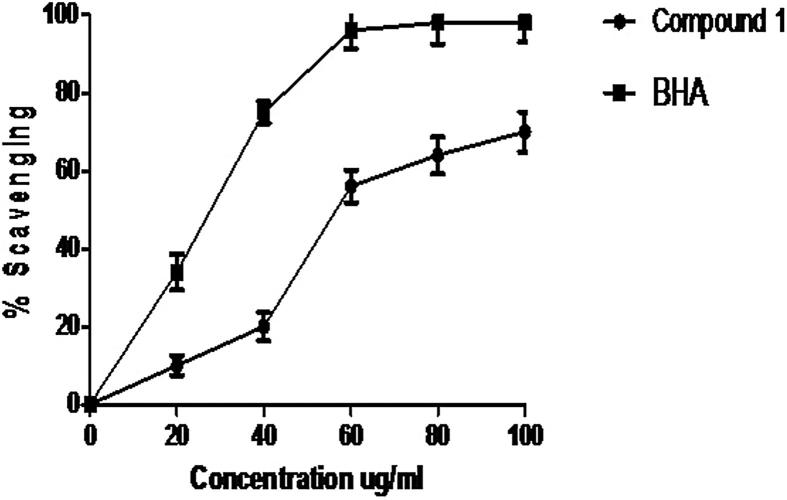
ABTS assay is frequently employed to measure the antioxidant capacities of natural products. Fig. 2 shows ABTS radical scavenging activity of compound 1 and standard BHA. The compound 1 exhibited 78.00 ± 2.80% scavenging property comparable with BHA (98.20 ± 4.60%) at 100 µg/mL concentration. The IC50 value (µg/mL) for compound 1 and BHA were found as 58.90 ± 3.40 and 28.70 ± 5.20, respectively.
Fig. 2.
ABTS scavenging activity of compound 1 and BHA. Values are means of three replicates.
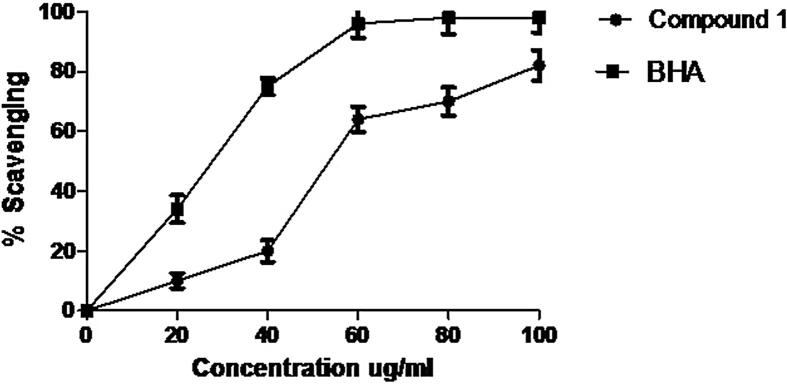
Compound 1 exhibited 82.90 ± 4.30% (IC50: 47.20 ± 4.90 µg/mL) DPPH free radical scavenging property at 100 µg/mL compared to BHA 93.4 ± 1.6% (IC50: 34.50 ± 6.20 µg/mL) (Fig. 3). The antioxidant property of compound 1 found to increase with its increased concentration. The hydrogen or electron donated to DPPH leads to its reduction and change in its color from violet to yellow. The ability of a substance to carry out such reaction will be considered as antioxidants (Bondet et al., 1997). It is observed that several compound such as ascorbic acid, tocopherol, glutathione, pyrogallol, gallic acid and aromatic amines etc. act as hydrogen donors and decolorize 1,1 diphenyl 2 picrylhydrazyl (Blois, 1958). Several scientific reports proved the correlation between the antioxidant potential of vegetables and fruits and their total phenol contents (Deighton et.al., 2000).
Fig. 3.
DPPH scavenging activity of compound 1 and BHA. Values are means of three replicates.
3.2. Antidiabetic effect
The compound 1 inhibitory property against α-amylase and α- glucosidase was evaluated and compared with acarbose. Compound 1 showed modest inhibitory effect against α-amylase (IC50: 128.34 ± 12.30 μg/mL; Fig. 4) and α-glucosidase (IC50: 89.20 ± 9.20 μg/mL; Fig. 5) compared to acarbose (IC50: 64.20 ± 5.60 and 52.40 ± 4.60 µg/mL, respectively) given in Table 1, which is in accordance with data recently published on the inhibitory property of flavonoids against α-amylase and α-glucosidase (Kim et al., 2000). The α-Glucosidase is a renowned catalyzing enzyme used in carbohydrate digestion and glucose release. Inhibiting the action of α- glucosidase will delay the absorption of glucose and lowers the postprandial blood glucose level, which leads to the suppression and progression of Diabetes mellitus. Although, compound 1 in this experiment demonstrated average inhibitory property on both the enzymes but it required further research to clarify their potential selective α-amylase and α-glucosidase activities.
Fig. 4.
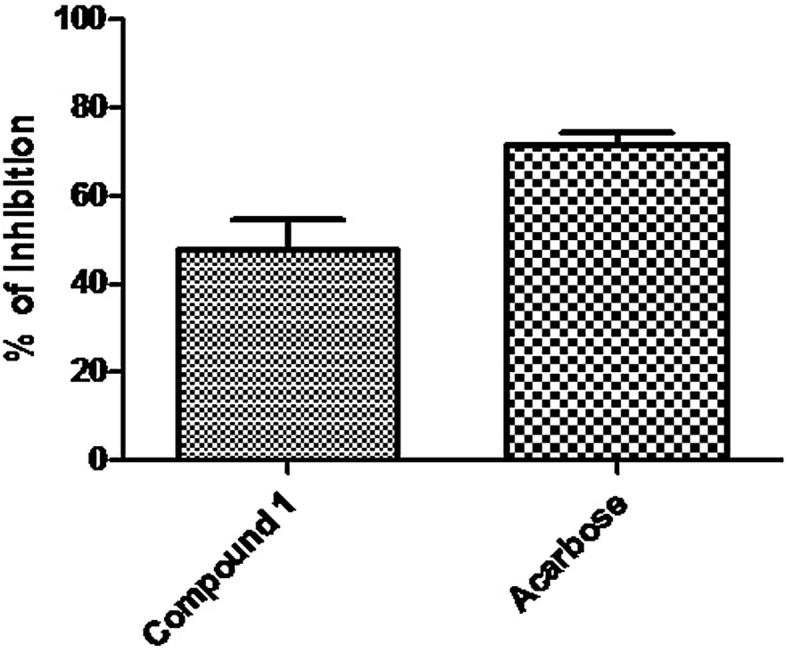
IC50 value of compound 1 and acarbose against α amylase. Data are presented as mean ± standard deviation values of triplicate determinations.
Fig. 5.
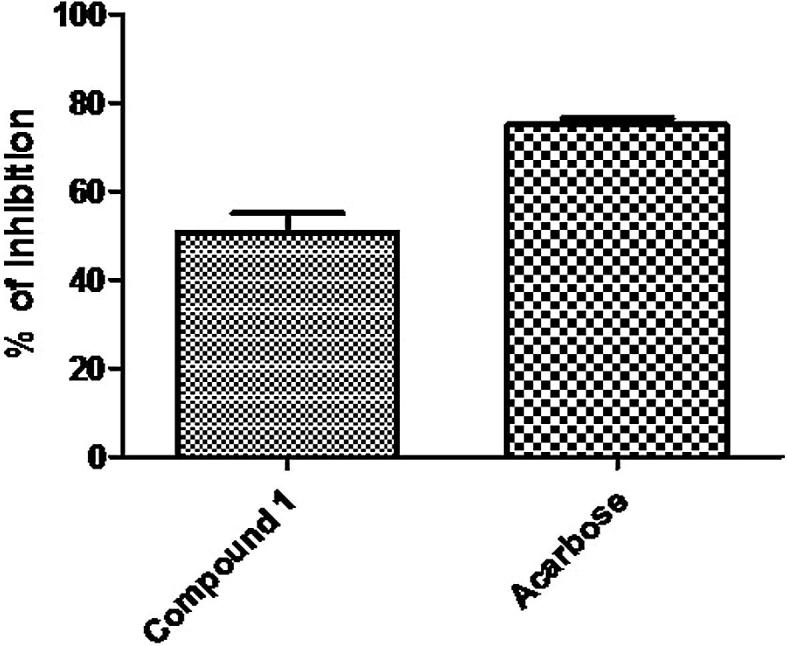
IC50 value of compound 1 and acarbose gainst α glucosidase. Data are presented as mean ± standard deviation values of triplicate determinations.
Table 1.
Comparison of IC50 value of compound 1 and Acarbose. Values are means of three replicates ± SD.
| Sample | α-Amylase | α-Glucosidase |
|---|---|---|
| Compound 1 | 128.34 ± 12.30 | 89.20 ± 9.20 |
| Acarbose | 64.20 ± 5.60 | 52.40 ± 4.60 |
3.3. Molecular docking studies
AutoDock is an extensively used platform for analyzing the interaction between a protein and its ligand/inhibitor. In order to get close to the mechanism of α-amylase and α-glucosidase inhibition by compound 1, we performed molecular docking using AutoDock. We also performed molecular docking of Acarbose (an inhibitor) with α-amylase and α-glucosidase for the comparative analysis.
3.3.1. Molecular docking analysis of compound 1 with α-amylase
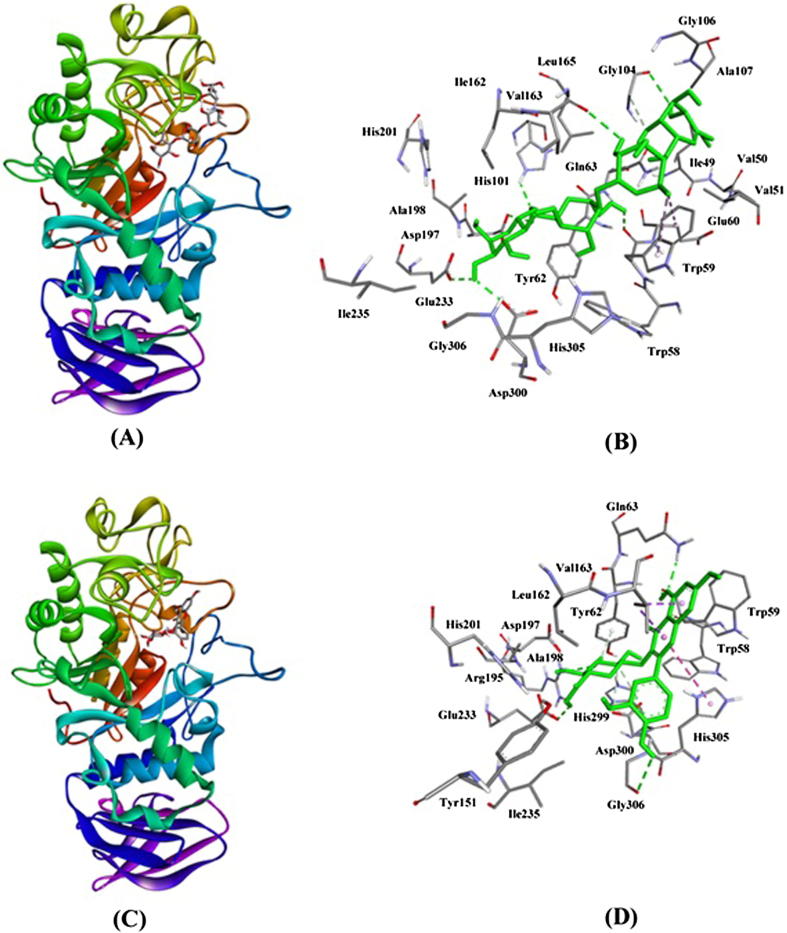
The results reported in Fig. 7 and Table 4 clearly revealed that compound 1 fits perfectly at the active site of α-amylase. The compound 1–α-amylase complex was stabilized by eight hydrogen bonds and three hydrophobic interactions. The amino acid residues involved in hydrogen bond formation were Trp59, Tyr62, Gln63, Arg195, Glu233, Asp300, and Gly306 (Table 2). It is very exciting to note that Asp300 formed two hydrogen bonds. Moreover, Val163 formed two hydrophobic interactions (Pi-Sigma) with compound 1 while His305 interacted with compound 1 through Pi-Pi (T-shaped) hydrophobic interaction (Fig. 6; Table 2). Other amino acid residues involved in stabilizing compound 1–α-amylase complex were Trp59, Tyr151, Leu162, Asp197, Ala198, His201, Ile235, His299, etc. The Gibb’s free energy for the interaction between compound 1 and α-amylase was estimated to be −8.2 kcal/mol, which corresponded to a binding affinity of 1.03 × 106 M−1. Similarly, Acarbose formed eight hydrogen bonds and two hydrophobic interactions with α-amylase. The amino acid residues Trp59, His101, Val163, Asp197, Glu233, and Asp300 formed only one hydrogen bond with α-amylase, while Gly104 formed two hydrogen bonds with α-amylase. On the other hand, Trp59 was involved in the formation of two hydrophobic (Pi-Alkyl) interactions with α-amylase (Fig. 7; Table 4). Other amino acid residues stabilizing the Acarbose-α-amylase were Ile49, Val50, Val51, Trp58, Glu60, Tyr62, Gln63, Gly106, Ala107, Ile162, Leu165, Ala198, His201, Ile235, His305, and Gly306. The Gibbs’s free energy and binding affinity of Acarbose-α-amylase complex formation were −8.0 kcal/mol and 7.37 × 105 M−1 respectively. It is evident that the binding energy and affinity of compound 1 were comparable to a known inhibitor (Acarbose).
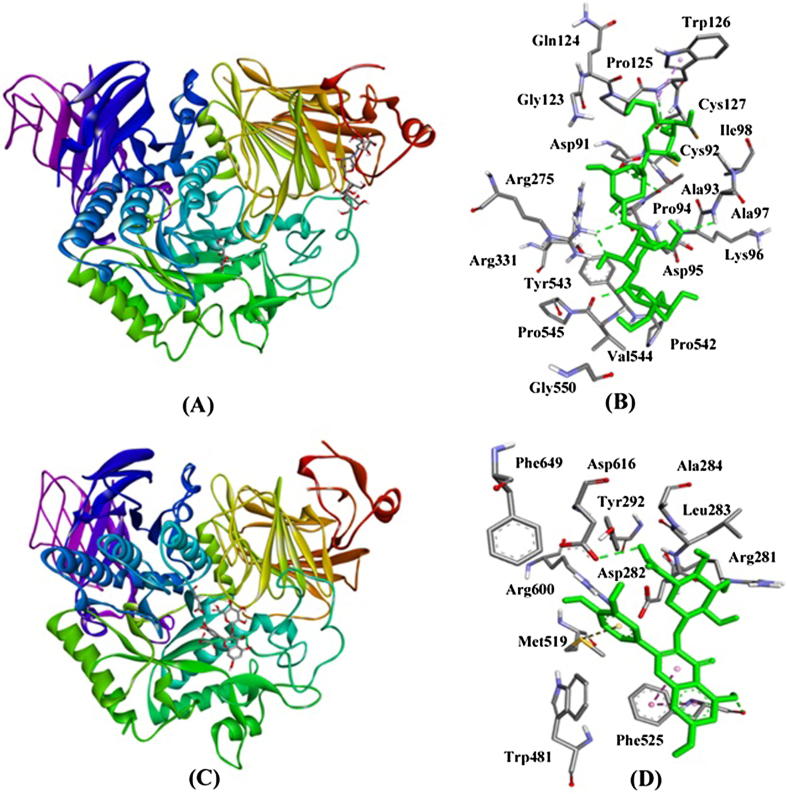
Fig. 7.
Inhibition of α-glucosidase activity by Acarbose and compound 1. (A) binding of Acarbose at Trefoil type P domain of α-glucosidase, (B) Interaction of Acarbose with the residues of α-glucosidase, (C) binding of compound 1 at the active site of α-glucosidase, and (D) Interaction of compound 1 with the active site residues of α-glucosidase.
Table 4.
Rf, Linear regression data for the calibration curve of compound 1, compound 2 and compound 3 (n = 6).
| Parameters | Compound 1 | Compound 2 | Compound 3 |
|---|---|---|---|
| Linearity range (ng/spot) | 200–1400 | 200–1400 | 200–1400 |
| Regression equation | Y = 7.517X + 518.11 | Y = 7.593X + 2193.06 | Y = 4.822X + 221.71 |
| Correlation (r2) coefficient | 0.9964 ± 0.001 | 0.9942 ± 0.002 | 0.9972 ± 0.001 |
| Slope ± SD | 7.517 ± 0.016 | 7.593 ± 0.035 | 4.822 ± 0.048 |
| Intercept ± SD | 518.11 ± 9.18 | 2193.06 ± 45.91 | 221.71 ± 16.15 |
| Standard error of slope | 0.006 | 0.014 | 0.019 |
| Standard error of intercept | 3.74 | 18.73 | 6.592 |
| Rf | 0.66 ± 0.002 | 0.60 ± 0.003 | 0.46 ± 0.003 |
| LOD (ng) | 7.42 | 15.51 | 33.21 |
| LOQ (ng) | 22.43 | 47.01 | 100.66 |
Table 2.
Inhibition of human pancreatic α-amylase by compound 1 (Miquelianin) and Acarbose.
| Interacting residues | Type of interactions | Bond length (Å) | Binding free energy (ΔG), kcal/mol | Binding affinity, Kd (M−1) |
|---|---|---|---|---|
| Acarbose | ||||
| HIS101:HE2 – UNK:O | Hydrogen Bond | 2.31 | −8.0 | 7.37 × 105 |
| UNK:H – GLY104:O | Hydrogen Bond | 2.85 | ||
| UNK:H – VAL163:O | Hydrogen Bond | 2.84 | ||
| UNK:H – TRP59:O | Hydrogen Bond | 2.41 | ||
| UNK:H – GLU233:OE2 | Hydrogen Bond | 2.41 | ||
| UNK:H – ASP300:OD2 | Hydrogen Bond | 2.28 | ||
| UNK:H – ASP197:OD2 | Hydrogen Bond | 2.70 | ||
| GLY104:CA – UNK:O | Hydrogen Bond | 3.49 | ||
| TRP59 – UNK:C | Hydrophobic (Pi-Alkyl) | 5.16 | ||
| TRP59 – UNK:C | Hydrophobic (Pi-Alkyl) | 4.45 | ||
| Miquelianin | ||||
| GLN63:HE22 – UNK:O | Hydrogen Bond | 2.25 | −8.2 | 1.03 × 106 |
| ARG195:HH21 – UNK:O | Hydrogen Bond | 2.80 | ||
| UNK:H – GLU233:OE2 | Hydrogen Bond | 2.29 | ||
| UNK:H – GLY306:O | Hydrogen Bond | 3.09 | ||
| UNK:H – TRP59:O | Hydrogen Bond | 2.62 | ||
| UNKC – ASP300:OD1 | Hydrogen Bond | 3.57 | ||
| UNK:C – ASP300:OD2 | Hydrogen Bond | 3.55 | ||
| UNK:H – TYR62 | Hydrogen Bond | 2.89 | ||
| VAL163:CG2 – UNK | Hydrophobic (Pi-Sigma) | 3.85 | ||
| VAL163:CG2 – UNK | Hydrophobic (Pi-Sigma) | 3.82 | ||
| HIS305 – UNK | Hydrophobic (Pi-Pi T-shaped) | 5.82 | ||
Fig. 6.
Inhibition of α-amylase activity by Acarbose and compound 1. (A) binding of Acarbose at the active site of α-amylase, (B) Interaction of Acarbose with the active site residues of α-amylase, (C) binding of compound 1 at the active site of α-amylase, and (D) Interaction of compound 1 with the active site residues of α-amylase.
The structural and mechanistic analysis of human pancreatic α-amylase revealed that it comprises three domains (A–C) (Rydberg et al., 2002). Domain A is the largest and main domain encompassing residues 1–99, and 169–404. It forms a central eight-stranded parallel β-barrel structure. At the end domain A, active site residues namely Asp197, Glu233, and Asp300 are located. A chloride ion is also bound in the vicinity of the active site and interacts with Agr195, Asn298, and Arg337. Domain B is the smallest domain of α-amylase comprising residues 100–168, and serves as the binding site of calcium ions. Domain C (residues 405–496) is mainly composed of anti-parallel β-structure, which is loosely associated with Domains A and B (Li, 2005). Our molecular docking analysis revealed that compound 1 form hydrogen bonds with two out of the three active site residues (namely Glu233 and Asp300). Moreover, it also interacted through hydrogen bonding with Arg195, one of the residue which binds with the chloride ion (Fig. 6). These results together with the findings of in-vitro α-amylase activity suggest that compound 1 is a potent inhibitor of α-amylase activity.
3.3.2. Molecular docking analysis of compound 1 with α-glucosidase
The X-ray crystal structure of α-glucosidase revealed that its substrate-binding active site pocket is formed by the C-terminal ends of the catalytic GH31 or (βα)8 domain, followed by a loop from N-terminal β-sheet domain both the inserts I and II (Roig-Zamboni et al., 2017). It harbors two active site residues namely Asp518 and Asp616 which act as a catalytic nucleophile and acid/base respectively in the classical Koshland double displacement reaction mechanism. We performed molecular docking of compound 1 to evaluate its potential as an inhinitor of α-glucosidase (Fig. 7; Table 3). The results evidently suggest that compound 1 formed a hydrogen bond (2.84 Å) with OD2 of Asp616, which is an active site residue. In addition, it also formed two more hydrogen bonds with the oxygen atoms of Asp282 (2.29 Å) and Phe525 (2.33 Å). The standard 1-α-glucosidase complex was further stabilized by two hydrophobic interactions (Pi-Pi Stacked) with Phe525 and one Pi-Sulphur interaction with the terminal Sulphur atom of Met519 (Fig. 7; Table 3). Other amino acid residues involved in the interaction were Arg281, Leu283, Ala284, Tyr292, Trp481, Arg600, and Phe649. The Gibb’s free energy (ΔG) of interaction between compound 1 and α-glucosidase was estimated to be −7.3 kcal/mol, corresponding to a binding affinity (Kd) of 2.26 × 105 M−1. Recently, Roig-Zamboni et al., 2017) also confirmed that 1-deoxynojirimycin (a well-establish inhibitor of α-glucosidase) binds at the active site of α-glucosidase and interacts with Trp376, Asp404, Ile441, Tp516, Asp518, Met519, Arg600, Trp613 Asp616, Phe649, and His674 through multiple hydrogen bonds and hydrophobic interactions. Conversely, Acarbose binds at the Trefoil type P domain of α-glucosidase and the complex was stabilized by eleven hydrogen bonds (with Asp91, Ala93, Pro94, Asp95, Ala97, Trp126, Cys127, Arg275, Pro542, and Val544) and one hydrophobic interaction with Trp126 (Fig. 7). It should be noted that Arg275 formed two hydrogen bonds with the Oxygen atoms of α-glucosidase. The Gibb’s free energy (ΔG) and binding affinity (Kd) for the standard 1-α-glucosidase complex formation were estimated to be −7.6 kcal/mol, and 3.75 × 105 M−1 respectively. From these results, we infer that the binding sites of Acarbose and compound 1 on α-glucosidase were different but they both interact with the key residues of α-glucosidase and act as an inhibitor.
Table 3.
Inhibition of lysosomal α-glucosidase by compound 1 (Miquelianin) and Acarbose.
| Interacting residues | Type of interactions | Bond length (Å) | Binding free energy (ΔG), kcal/mol | Binding affinity, Kd (M−1) |
|---|---|---|---|---|
| Acarbose | ||||
| ALA97:HN – UNK:O | Hydrogen Bond | 2.80 | −7.6 | 3.75 × 105 |
| TRP126:HN – UNK:O | Hydrogen Bond | 2.64 | ||
| ARG275:HH12 – UNK:O | Hydrogen Bond | 2.28 | ||
| ARG275:HH12 – UNK:O | Hydrogen Bond | 2.13 | ||
| UNK:H – ASP91:OD2 | Hydrogen Bond | 2.43 | ||
| UNK:H – ALA93:O | Hydrogen Bond | 2.81 | ||
| UNK:H – CYS127:O | Hydrogen Bond | 2.73 | ||
| UNK:H – PRO94:O | Hydrogen Bond | 2.59 | ||
| UNK:H – VAL544:O | Hydrogen Bond | 1.92 | ||
| UNK:H – PRO542:O | Hydrogen Bond | 2.14 | ||
| UNK:C – ASP95:O | Hydrogen Bond | 3.41 | ||
| UNK:C – TRP126 | Hydrophobic (Pi-Sigma) | 3.87 | ||
| Miquelianin | ||||
| UNK:H – ASP616:OD2 | Hydrogen Bond | 2.84 | −7.3 | 2.26 × 105 |
| UNK:H – ASP282:O | Hydrogen Bond | 2.29 | ||
| UNK:H – PHE525:O | Hydrogen Bond | 2.33 | ||
| MET519:SD – UNK | Other (Pi-Sulphur) | 5.10 | ||
| PHE525 – UNK | Hydrophobic (Pi-Pi Stacked) | 3.77 | ||
| PHE525 – UNK | Hydrophobic (Pi-Pi Stacked) | 3.84 | ||
3.4. HPTLC method development and validation
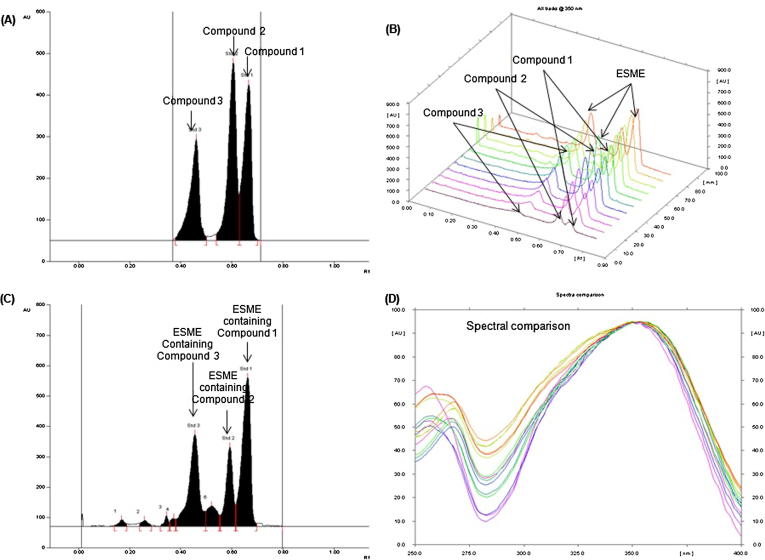
The mobile phase for the HPTLC investigation of compounds 1, 2 and 3 in ESME was selected by trying combinations of different solvents. Of these a mixture of methanol and water (60:40, v/v) was found suitable to furnish sharp, compact and intense peak of compounds 1, 2 and 3 (at Rf = 0.66 ± 0.002, 0.60 ± 0.003 and 0.46 ± 0.003, respectively; Fig. 8A). This method clearly separated the compounds 1, 2 and 3 in addition to the constituents of ESME (Fig. 8B). The above method was fairly selective with high baseline resolution. The peaks of standard and the extracts were compared by aligning each other at wavelength 350 nm (Fig. 8D). The regression equation/correlation co-efficient (r2) for compounds 1, 2 and 3 were obtained as Y = 7.517X + 518.11/0.9964, Y = 7.593X + 2193.06/0.9942 and Y = 4.822X + 221.71/0.9972, respectively in 200–1400 ng/spot linearity range. The LOD/ LOQ (ng/band) for compounds 1, 2 and 3 were calculated as 7.42/22.43, 15.51/47.01 and 33.21/100.66 ng/band, respectively (Table 4). The recoveries as accuracy study of above method were calculated and reported in Table S1 (Table attached as suppl. material). The (%) recovery/RSD for compounds 1, 2 and 3 were found as 98.21–99.41/1.05–1.19, 98.71–99.54/ 1.37–1.44 and 98.72–99.39/1.27–1.33, respectively. The intra-day/ inter-day precision (% RSD; n = 6) of the above method for the compounds 1, 2 and 3 were calculated and recorded as 1.17–1.24/ 1.09–1.22, 1.32–1.37/1.31–1.35 and 1.04–1.12/1.03–1.10, respectively and recorded in Table S2 (Table attached as suppl. material) which demonstrated good precision of proposed method. The robustness for the above method was tested by creating a small intentional change in composition of mobile phase, saturation time and volume of mobile phase used and the obtained data were reported in Table S3 (attached as suppl. material). The low SD and % RSD indicated that above method was robust.
Fig. 8.
Quantification of compound 1, compound 2 and compound 3 in ESME by RP-HPTLC at λ = 350 nm [mobile phase: methanol: water (60:40)]. (A) Chromatogram of standard compound 3 (Rf = 0.46), compound 2 (Rf = 0.60) and compound 1 (Rf = 0.66) (B) 3-D display of all tracks (C) Chromatogram of ESME [compound 3, spot 5, Rf = 0.46; compound 2, spot 7, Rf = 0.60; compound 1, spot 8, Rf = 0.66)]. (D) Spectral comparison of all the tracks at 350 nm.
3.5. Concurrent HPTLC analysis of compounds 1, 2 and 3 in ESME
The selected HPTLC method was used for concurrent analysis of compounds 1, 2 and 3 in the ESME (Fig. 8C). Using the above method the quantity of compounds 1, 2 and 3 was found as 16.39, 3.92 and 14.98 µg/mg of dried extract, respectively (Table 5).
Table 5.
HPTLC analysis of compound 1, compound 2 and compound 3 in ESME.
| S. No. | Sample | Compound 1 content (µg/mg of dried weight of extract) | Compound 2 content (µg/mg of dried weight of extract) | Compound 3 content (µg/mg of dried weight of extract) |
|---|---|---|---|---|
| 1 | ESME | 16.39 ± 0.89 | 3.92 ± 0.019 | 14.98 ± 0.79 |
The results of HPTLC analysis of Compound 1 (miquelianin; 16.39 µg/mg) strongly support the therapeutic uses of E. schimperi. Quantification of compound 2 (kaempferol 3-O-glucuronide) and compound 3 (quercetin 3-O-rhamnoside) in methanol extract of E. schimperi provides additional information about their quantity which will facilitate the formulation designing.
4. Conclusion
The study revealed significant antidiabetic and antioxidant property of miquelianin which is strongly aligned with its molecular docking results, and approves our previous work on wound healing. This was a maiden study on the concurrent HPTLC analysis of miquelianin, kaempferol-3-O-glucuronide and quercetin-3-O-rhamnoside in ESME. The chromatographic quantification proves the ample amount of miquelianin present in ESME which offers the medicinal use of extract as such or as a component in a poly herbal formulation. The proposed HPTLC method can be applied in the investigation of miquelianin, kaempferol 3-O-glucuronide and quercetin 3-O-rhamnoside in other plant species extracts and marketed herbal preparations.
Acknowledgement
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University, Riyadh, Saudi Arabia for funding the work through the research group project number (RGP - 073).
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2019.03.008 .
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abdel-Monem A.R., Abdel-Sattar E., Harraz F.M., Petereit F. Chemical investigation of Euphorbia schimperi C. Presl. Rec. Nat. Prod. 2008;2:39–45. [Google Scholar]
- Ahmed S., Nur-e-alam M., Mothana R.A., Yousaf M., Al-Rehaily A.J. Activity guided isolation of chemical constituents from the biologically active methanol extract of Euphorbia schimperi c. presl. Bull. Chem. Soc. Ethiop. 2017;31:471–479. [Google Scholar]
- Ahmed S., Yousaf M., Mothana R.A., Al-Rehaily A.J. Studies on wound healing activity of some Euphorbia species on experimental rats. Afr. J. Tradit. Complement. Altern. Med. 2016;13:45–52. doi: 10.21010/ajtcam.v13i5.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AL-Sultan S.I., Hussein Y.A. Acute toxicity of Euphorbia heliscopia in Rats. Pak. J. Nut. 2006;5:135–140. [Google Scholar]
- Blois M.S. Antioxidant determinations by the use of a stable free radical. Nature. 1958;29:1199–1200. [Google Scholar]
- Bondet V., Brand-Williams W., Berset C. Kinetics and mechanisms of antioxidant activity using the DPPH free radical method. Lebensm.-Wiss. u.-Technol. 1997;30:609–615. [Google Scholar]
- Braca A., De Tommasi N., Di Bari L., Pizza C., Politi M., Morelli I. Antioxidant principles from Bauhinia tarapotensis. J. Nat. Prod. 2001;64:892–895. doi: 10.1021/np0100845. [DOI] [PubMed] [Google Scholar]
- Chaudhary S.A. Ministry of Agriculture and Water, National Herbarium, National Agriculture and Water Research Center; Riyadh: 2001. Flora of Kingdom of Saudi Arabia I Llustrated. [Google Scholar]
- Deighton N., Brennan R., Finn C., Davies H.V. Antioxidant properties of domesticated and wild Rubus species. J. Sci. Food Agric. 2000;80:1307–1313. [Google Scholar]
- Giner J.L., Schroeder T.N. Polygonifoliol, a new tirucallane triterpene from the latex of the seaside sandmat Euphorbia polygonifolia. Chem. Biodivers. 2015;12:1126–1129. doi: 10.1002/cbdv.201400426. [DOI] [PubMed] [Google Scholar]
- Gupta R.K., Kesari A.N., Murthy P.S., Chandra R., Tandon V., Watal G. Hypoglycemicand anti-diabetic effect of ethanolic extract of leaves of Annona squamosa L. in experimental animals. J Ethnopharmacol. 2005;99:75–81. doi: 10.1016/j.jep.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Harman D. Elsevier; Amster-dam: 1998. Free Radical Theory of Aging. Current Status; pp. 3–7. [Google Scholar]
- International Conference on Harmonization (ICH) of Technical Requirements for Registration of Pharmaceuticals for Human use, Harmonised Triplicate Guideline on Validation of Analytical Procedures: Text and Methodology Q2 (R1), Complementary Guideline on Methodology incorporated in November 2005 by the ICH Steering Committee, IFPMA, Geneva.
- Jassbi A.R. Chemistry and biological activity of secondary metabolites in Euphorbia from Iran. Phytochemistry. 2006;67:1977–1984. doi: 10.1016/j.phytochem.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Kavishankar G.B., Lakshmidevi N., Murthy S.M., Prakash H.S., Niranjana S.R. Diabetes and medicinal plants: a review. Int. J. Pharm. Biomed. Sci. 2011;2:65–80. [Google Scholar]
- Kim J.S., Kwon C.S., Son K.H. Inhibition of alpha-glucosidase and amylase by luteolin, a flavonoid. Biosci. Biotechnol. Biochem. 2000;64:2458–2461. doi: 10.1271/bbb.64.2458. [DOI] [PubMed] [Google Scholar]
- Kwon Young-In, Apostolidis E., Shetty K. Inhibitory potential of wine and tea against α-amylase and α-glucosidase for management of hyperglycemia linked to type 2 diabetes. J. Food Biochem. 2008;32:15–31. [Google Scholar]
- Mallavadhani U.V., Sahu G., Narasimhan K., Muralidhar J. Quantitative estimation of an antidiarrhoeic marker in euphorbia hirta samples. Pharmaceut. Bio. 2002;40:103–106. [Google Scholar]
- Maurus R., Begum A., Williams L.K., Fredriksen J.R., Zhang R., Withers S.G., Brayer G.D. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity. Biochemistry. 2008;47:3332–3344. doi: 10.1021/bi701652t. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput. Chem. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothana R.A., Gruenert R., Bednarski P.J., Lindequist U. Evaluation of the in vitro anticancer, antimicrobial and antioxidant activities of some Yemeni plants used in folk medicine. Die Pharmazie. 2009;64:260–268. [PubMed] [Google Scholar]
- Re R., Pellegrini N., Proteggente A., Pannala A., Yang M., Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999;26:1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- Rehman M.T., Ahmed S., Khan A.U. Interaction of meropenem with 'N' and 'B' isoforms of human serum albumin: a spectroscopic and molecular docking study. J. Biomol. Struct. Dyn. 2016;34:1849–1864. doi: 10.1080/07391102.2015.1094411. [DOI] [PubMed] [Google Scholar]
- Rehman M.T., Shamsi H., Khan A.U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharmaceut. 2014;11:1785–1797. doi: 10.1021/mp500116c. [DOI] [PubMed] [Google Scholar]
- Roig-Zamboni V.R., Cobucci-Ponzano B., Iacono R., Ferrara M.C., Germany S., Bourne Y., Parenti G., Moracci M., Sulzenbacher G. Structure of human lysosomal acid α-glucosidase – a guide for the treatment of Pompe disease. Nat. Commun. 2017;8:1111. doi: 10.1038/s41467-017-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg E.H., Li C., Maurus R., Overall C.M., Brayer G.D., Withers S.G. Mechanistic analyses of catalysis in human pancreatic alpha-amylase: detailed kinetic and structural studies of mutants of three conserved carboxylic acids. Biochemistry. 2002;41:4492–4502. doi: 10.1021/bi011821z. [DOI] [PubMed] [Google Scholar]
- Shi Q.W., Su X.H., Kiyota H. Chemical and pharmacological research of the plants in genus Euphorbia. Chem. Rev. 2008;108:4295–4327. doi: 10.1021/cr078350s. [DOI] [PubMed] [Google Scholar]
- Singh A., Singh P.K., Singh R.K. Antidiabetic and wound healing activity of catharanthus roseus l. in streptozotocininduced diabetic mice. Am. J. Phytomed. Clin. Therapeut. 2014;2:686–692. [Google Scholar]
- Siddiqui N.A., Alam P., Al-Rehaily A.J., Al-Oqail M.M., Parvez M.K. Simultaneous quantification of biomarkers bergenin and menisdaurin in the methanol extract of aerial parts of Flueggea virosa by validated HPTLC densitometric method. J. Chromatogr. Sci. 2015;53(5):824–829. doi: 10.1093/chromsci/bmu231. [DOI] [PubMed] [Google Scholar]
- Siddiqui N.A., Al-Yousef H.M., Alhowiriny T.A., Alam P., Hassan W.H.B., Musarat A., Hussain A., Abdelaziz S., Abdallah R.H. Concurrent analysis of bioactive triterpenes oleanolic acid and b-amyrin in antioxidant active fractions of Hibiscus calyphyllus, Hibiscus deflersii and Hibiscus micranthus grown in Saudi Arabia by applying validated HPTLC method. Saudi Pharmaceut. J. 2018;26:266–273. doi: 10.1016/j.jsps.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smara O., Julia A., Moral-Salmi C., Vigor C., Joseph V., Legseir B. Flavonoïds from Euphorbia guyoniana Boissier & Reuter. J. Life Sci. 2014;8:544–551. [Google Scholar]
- WHO . World Health Organization; Geneva: 1999. Expert Committee on Diabetes Mellitus: Diagnosis and Classification of Diabetes Mellitus. Technical Report Series. Part 1. [Google Scholar]
- Wu Y.Y., Li W., Xu Y., Jin E.H., Tu Y.Y. Evaluation of the antioxidant effects of four main theaflavin derivatives through chemiluminescence and DNA damage analyses. J. Zhejiang Univ. Sci. B. 2011;12:744–751. doi: 10.1631/jzus.B1100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi S., Matsui T. Nitric oxide, a Janus-faced therapeutic target for diabetic microangiopathy-friend or foe? Pharmacol. Res. 2011;64:187–194. doi: 10.1016/j.phrs.2011.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.