Abstract
Background
Heavy metal toxicity has become a major threat to sustainable crop production worldwide. Thus, considerable interest has been placed on deciphering the mechanisms that allow plants to combat heavy metal stress. Strategies to deal with heavy metals are largely focused on detoxification, transport and/or sequestration. The P1B subfamily of the Heavy Metal-transporting P-type ATPases (HMAs) was shown to play a crucial role in the uptake and translocation of heavy metals in plants. Here, we report the locus-specific expression changes in the rice HMA genes together with several low-copy cellular genes and transposable elements upon the heavy metal treatment and monitored the transgenerational inheritance of the altered expression states. We reveal that plants cope with heavy metal stress by making heritable changes in gene expression and further determined gene-specific responses to heavy metal stress.
Results
We found most HMA genes were upregulated in response to heavy metal stress, and furthermore found evidence of transgenerational memory via changes in gene regulation even after the removal of heavy metals. To explore whether DNA methylation was also altered in response to the heavy metal stress, we selected a Tos17 retrotransposon for bisulfite sequencing and studied its methylation state across three generations. We found the DNA methylation state of Tos17 was altered in response to the heavy metal stress and showed transgenerational inheritance.
Conclusions
Collectively, the present study elucidates heritable changes in gene expression and DNA methylation in rice upon exposure to heavy metal stress and discusses implications of this knowledge in breeding for heavy metal tolerant crops.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1887-7) contains supplementary material, which is available to authorized users.
Keywords: Heavy metal stress, Transgenerational memory, Gene expression, DNA methylation
Background
Plants are sessile organisms and are often confronted with a variety of stress factors simultaneously, which can dramatically decrease their yield and quality. In the recent years, heavy metal pollution, i.e., contamination of the natural environment with cadmium (Cd), chromium (Cr), copper (Cu), mercury (Hg), and zinc (Zn) has become a global problem, affecting about 235 million hectares of the arable land worldwide [1]. Heavy metals compromise crop productivity and pose a threat to human health via heavy metal accumulation in the food chain [2]. In plants, heavy metals interfere with several metabolic processes including photosynthesis, water relations, and nutrient uptake, resulting in reduced plant growth, stunting, and in some instances, death [3, 4]. Cu is an essential micronutrient; however, if present in excess it also causes toxicity to plants [5]. Cr is also a common metal contaminant in the Earth’s crust. While naturally occurring, Cr does not cause toxicity to plants, but excessive amounts can cause injury [6]. Cd and Hg are both non-essential and toxic elements for plant growth and human health. These elements are almost ubiquitously present at low levels in the environment but have now started to accumulate due to anthropogenic activities. In its 25-year plan for the comprehensive prevention and control of heavy metals the Ministry of Environmental Protection of the People’s Republic of China listed Cd, Pb, Hg, and Cr as the major environmental pollutants, and pledged efforts to control their release to the environment (www.cleanairchina.org/file/loadFile/9.html). Parallelly, in view of the public health concern, in the report on the National Food Safety Standard Limits on contaminants in food (GB 2762–2017) the National Standards of the People’s Republic of China, made recommendations on the maximum tolerable amount of Cu (10 mg kg− 1), Cr (1.0 mg kg− 1), Cd (0.2 mg kg− 1), and Hg (0.02 mg kg− 1) in rice grains.
Since heavy metal toxicity has become one of the major challenges in increasing crop productivity, investigating heavy metal tolerance genes and stacking them in a single genetic background, have become a major theme of plant breeding research. Over the course of evolution, plants have developed different strategies to overcome heavy metal toxicity. For example, relatively low levels of metals are present in shoots by either restricting translocation of toxic metals, sequestration to vacuoles, or detoxification [7–12]. Conversely, some plants have developed exceptional abilities to translocate and accumulate heavy metals in their aboveground organs [13].
Recent research has revealed that the P1B subfamily of Heavy Metal-transporting P-type ATPases (HMAs) play a crucial role in the uptake and translocation of heavy metals in plants [14, 15]. There are eight and nine members of P1B-ATPases in Arabidopsis thaliana and rice (Oryza sativa L.), respectively [16, 17]. Based on the metal-substrate specificity these ATPases can be divided into two subgroups: a zinc (Zn)/cobalt (Co)/cadmium (Cd)/lead (Pb) group and a copper (Cu)/silver (Ag) group [18]. In A. thaliana and rice, AtHMA1-AtHMA4 and OsHMA1-OsHMA3 belong to the former group whereas AtHMA5-AtHMA8 and OsHMA4-OsHMA9 belong to the latter group [18]. All members of the HMA family in A. thaliana have been functionally well characterized. The HMA family members exhibit differences in expression sub-cellular localization, and metal specificity and regulation, which all indicate unique functions within the gene family. For instance, AtHMA1, AtHMA5-AtHMA8 were reported to play a role in Cu homeostasis [19–22]. AtHMA2-AtHMA4 were involved in Cd translocation and sequestration [23–25]. In contrast, the rice HMA transporter family is not as well characterized. For instance, OsHMA1 and OsHMA9 were postulated to play a role in Zn transport [26, 27]. OsHMA2 and OsHMA3 were reported to be involved in the transportation of Cd [28–30], OsHMA4 and OsHMA5 have a function in Cu transport, loading, and detoxification [31, 32]. However, little research has been performed on OsHMA6, OsHMA7, and OsHMA8.
Modulation of gene expression is one rapid strategy to respond to environmental stresses. It has been repeatedly shown that heavy metal stress induces changes in gene expression. For instance, transcript profiling of the Cd-tolerant cultivar of Chinese flowing cabbage revealed numerous changes in gene expression in response to Cd treatment including upregulation of HMA3 and HMA4 [33]. Research in Sedum plumbizincicola showed elevated expression of the SpHMA3 gene in response to Cd stress suggesting a role in Cd detoxification and normal growth of young leaves under Cd stress [34]. Similarly, in Lycopersicum esculentum, heavy metal transporters COPT1 and COPT2 could be induced to express under Cu stress [35]. Functional genomics tools have been extensively used to examine mechanisms conferring tolerance to various heavy metal stresses. In a recent report, genome-wide transcriptome analysis in rice showed dose-dependent changes in expression of metal ion transporter genes in response to Cd stress [36].
One way to maintain changes in gene expression is via epigenetic modification. Indeed, epigenetic variation contributes to phenotypic plasticity in response to the environmental changes [37]. In particular, DNA methylation is an important epigenetic marker, which regulates gene expression as an adaptive mechanism for survival under stress. In a recent study, genome-wide single-base resolution maps of methylated cytosines and transcript profile of Cd-treated rice was reported [38]. The study showed that most of the epigenetically regulated genes were transcriptionally activated under Cd stress, and many of these genes represent formerly characterized stress responders, metal transporters and transcription factors [38]. Despite initial progress, implementation of these epigenetic markers in plant breeding has stalled because the heritability of these makers has not yet been tested [37].
Since rice (O. sativa L.) is one of the major staple grains worldwide, increasing its productivity and nutritional quality is one of the foremost priorities. In the interest of ensuring food security and better nutritional quality, it is important to reduce the accumulation of toxic elements in rice grains [39, 40]. A deep understanding of the genes responsible for the sequestration of toxic elements can enable the development of crop varieties with reduced content of these elements in the edible plant parts. Our previously, work has shown that heavy metal stress (Cd, Cr, Cu, and Hg) could inhibit further shoot and root development of the ten-day-old rice seedlings and induce transgenerational changes in their DNA methylation pattern at specific loci [41]. Rice plants were treated with two different concentrations of Cd, Cr, Cu, or Hg to determine dose-dependent responses to these heavy metals. As expected, more hypomethylations were observed at specific-loci on the higher doses of Cd, Cr, and Cu but no change in DNA methylation pattern was witnessed upon Hg treatment. Remarkably, the progeny of the stressed plants exhibited enhanced tolerance to the same stress their progenitors experienced and showed the transgenerational inheritance of changes in the DNA methylation patterns [41]. The aim of this study was to address whether locus-specific changes in gene expression also take place in response to the heavy metal stress and whether different classes of genes have common or specific responses to heavy metal stress.
Results
Heavy metal stress induced locus-specific gene expression changes in the S0 plants
We previously showed that heavy metals elicit epigenetic changes in DNA methylation patterns of specific loci and in a transgenerational manner [41]. In the present study, we addressed whether locus-specific changes in gene expression also take place in response to the heavy metal stress and whether different classes of genes have common or specific responses to the heavy metal stress. To test this possibility, we assessed the expression of 18 randomly-distributed and functionally diverse genes by reverse transcription (RT)-PCR in the heavy-metal stressed rice seedlings (Fig. 1). Out of these 18 genes, two (Tos17 and Osr42) were formerly tested by us to respond epigenetically to the heavy metal stress, seven (Homeobox gene, DNA-binding protein, Elongation factor, HSP70, SNF-FZ14, S3, and YF25) were randomly distributed cellular genes, and nine genes (OsHMA1-OsHMA9) were known to be heavy metal transporters. This panel of genes allows testing if global or specific transcriptional changes are involved in heavy-metal stress avoidance or mitigation in rice. In the S0 generation, plants for expression analysis were selected on the basis of the gel-blot analysis. Specifically, S0 plants that showed the most conspicuous modifications in DNA methylation patterns under Cu2+ (1000 μM), Cd2+ (1000 μM), Cr3+ (1000 μM) and Hg2+ (50 μM) treatments were selected for the expression analysis [41].
Fig. 1.
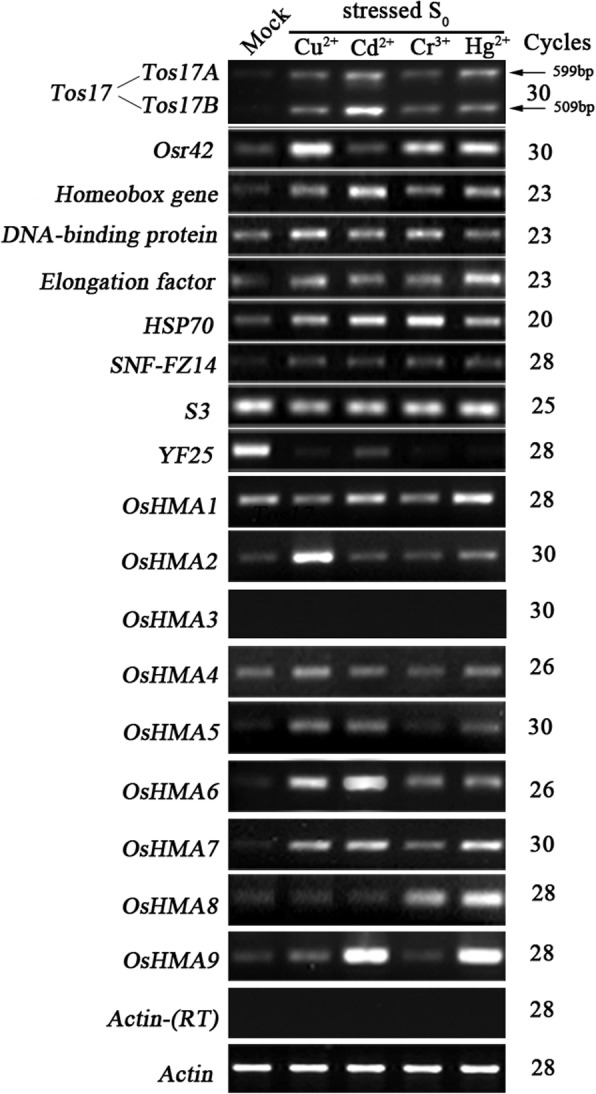
Alteration in the steady-state transcript abundance determined by semi-quantitative RT-PCR analysis in a set of 18 randomly selected genes, which include two transposable element genes (Tos17 and Osr42), seven cellular genes (homeobox gene, DNA-binding protein, Elongation factor, HSP70, SNF-FZ14, S3, and YF25), and nine rice Heavy Metal-transporting P-type ATPases (OsHMA1-OsHMA9). The results were highly reproducible among the three independent RNA batches, and hence, only one was presented. Gene names are listed to the left and amplification cycles are labeled to the right of the gel. The rice Actin gene (Genbank accession # X79378) was used as a control for normalization of RNA input. Lack of genomic DNA was validated by the Actin gene on the template without RT
Interestingly, we found two rice TE (transposable element) genes, the Tos17 and Osr42 that showed significantly up-regulated expression under all or three of the four heavy metal treatments (Fig. 1 and Table 1). Specifically, for Tos17, there are two copies in wild-type rice cv. Nipponbare, one located on chromosome 10 dubbed Tos17A, and the other located on chromosome 7 called Tos17B. The two Tos17 copies are identical except for a 90 bp insertion in Tos17A [42]. We designed gene-specific primers to study expression changes in the two copies under heavy metal stress. The results showed that the two copies of Tos17 seldom exhibit activation of gene expression under all four (100%) heavy-metal treatments (S0 plants), particularly under Cd stress. Similarly, Osr42 showed a significantly up-regulated expression under three (Cu, Cr, and Hg) of the four (75%) heavy metal treatments. The two TE genes exhibited contrasting expression patterns in Cd-treated plants, while Tos17 showed the most conspicuous activation of gene expression, Osr42 exhibited no change in expression.
Table 1.
Gene expression changes observed for the 18 functionally diverse random genes in heavy metal treated seedlings of rice cv. Matsumae (S0 generation)
| Gene name | Genbank acc.a | Chr.a | Gene expression changes observed in heavy metal treated plants of S0 generationb | ||||
|---|---|---|---|---|---|---|---|
| Cu2+(1000 μm . L−1) | Cd2+ (1000 μm . L−1) | Cr3+ (1000 μm . L− 1) | Hg2+ (50 μm . L− 1) | Freq. (%)c | |||
| Transposable elements (TEs) | |||||||
| Tos17 | AC087545(Tos17A) | 10 | U | U | U | U | 100 |
| AP008213(Tos17B) | 7 | U | U | U | U | 100 | |
| Osr42 | AF458768 | 4 | U | N | U | U | 75 |
| Low copy protein-coding genes | |||||||
| Homeobox gene | AB007627 | 2 | U | U | U | U | 100 |
| DNA-binding protein | X88798 | 5 | U | U | U | U | 100 |
| Elongation factor | D12821 | 7 | U | U | U | U | 100 |
| HSP70 | X67711 | 11 | U | U | U | U | 100 |
| SNF-FZ14 | DQ239432 | 7 | U | U | U | U | 100 |
| S3 | AY328087 | 12 | N | N | N | N | 0 |
| YF25 | DQ239435 | 11 | D | D | D | D | 100 |
| Rice P1B subfamily of Heavy Metal-transporting P-type ATPases (HMAs) | |||||||
| OsHMA1 | AP003935 | 6 | N | U | N | U | 50 |
| OsHMA2 | AP004278 | 6 | U | N | N | N | 25 |
| OsHMA3 | AP005246 | 7 | – | – | – | – | – |
| OsHMA4 | AP004184 | 2 | N | N | N | N | 0 |
| OsHMA5 | AL606647 | 4 | U | U | N | U | 75 |
| OsHMA6 | AP004836 | 2 | U | U | U | U | 100 |
| OsHMA7 | AP004376 | 8 | U | U | U | U | 100 |
| OsHMA8 | AC125472 | 3 | N | N | U | U | 50 |
| OsHMA9 | AP008212 | 6 | N | U | N | U | 50 |
| Totald (%) | 72.2 | 72.2 | 66.7 | 83.3 | |||
Note: aDetermined by BlastN searches performed at NCBI
bChanges in gene expression pattern were defined as: N = No change in gene expression; U = Significantly up-regulated expression; D = Significant down-regulated expression; and - = No expression
CNumber of times a gene responded similarly to different heavy metal stresses; represented as percentage in the table
d Number of times a heavy metal stress affected different genes in a similar fashion; represented as percentage in the table. For calculations the two copies of the Tos17 were treated separately
In addition, among seven low-copy cellular genes (Homeobox gene, DNA-binding protein, Elongation factor, HSP70, SNF-FZ14, S3, and YF25), five of the genes (Homeobox gene, DNA-binding protein, Elongation factor, HSP70, and SNF-FZ14) showed transcriptional upregulation in all (100%) heavy metal treated plants (Fig. 1 and Table 1). Whereas, YF25 showed significant down-regulation under Cd treatment to complete suppression under other heavy metal treatments (Cu, Cr, and Hg), and S3 exhibited no change in expression under any of the tested heavy metal treatments.
We also tested the nine rice HMAs (OsHMA1- OsHMA9) and found that 7 HMAs showed significant up-regulation under at least one of the four heavy metal treatments (Fig. 1 and Table 1). Specifically, OsHMA1 showed up-regulated expression in Cd and Hg-treated plants (two of the four heavy metal treatments; 50%). Similarly, OsHMA2 showed significantly up-regulated expression in Cu-treated plants (one of the four heavy metal treatments; 25%). OsHMA5 showed significant transcriptional activation under Cu, Cd, and Hg treatments (three of the four heavy metal treatments; 75%). OsHMA6 and OsHMA7 showed transcriptional activation under all four (100%) heavy metal treatments. OsHMA8 showed significant transcriptional activation in Hg and Cr treated plants (two of the four heavy metal treatments; 50%), whereas OsHMA9 showed significant transcriptional activation in Cd and Hg treated plants (two of the four heavy metal treatments; 50%). OsHMA4 did not show significant transcriptional changes under any of the four heavy metal treatments, and OsHMA3 showed no expression either in plants treated with any of the heavy metals or mock plants.
Taking the results of all four heavy metal treatments together, (i) different genes responded from none (0%) to all (100%) studied heavy metal treatments by exhibiting alterations in their respective expression patterns. Specifically, 10 of the 18 genes responded to all four heavy metal treatments by transcriptional upregulation. Interestingly, TEs and the low-copy number protein-coding genes showed more transcriptional plasticity than HMAs under heavy metal stress. (ii) With respect to the number of genes that showed transcriptional changes in response to heavy metal stress, Hg treatment induced changes in expression patterns of the maximum (83.3%) number of genes followed by Cu/Cd (72.2%), and Cr (66.7%) treatments. (iii) With respect to type (up- or down-regulation) of the gene expression changes occurring in response to the heavy metal treatment, all genes responded by up-regulation of expression, except YF25 that showed transcriptional downregulation and S3, which exhibited no change in expression pattern (Table 1).
The altered gene expression patterns were transgenerationally inherited, coupled with additional alterations in the S1 generation
To test if the altered gene expression state of the S0 plants would be maintained in the next generation, we selfed a single Hg2+ (50 μM) treated plant, as this treatment induced gene expression changes in the majority of the studied genes (83.3%) (Table 1). Later, the leaf-tissue collected from the S1 seedlings growing under optimal conditions was subjected to transcript profiling of 14 genes including two transposable element genes, four cellular genes, and eight OsHMAs. All fourteen genes tested here showed transcriptional changes in Hg-treated S0 plants. We divided the expression state of S1 progeny into three patterns of expression: inheritance of Hg-treated S0 pattern, reversion to the mock pattern, and a differential expression pattern. The last category was further divided into two sub-categories: transgenerational memory (further up-regulated expression pattern) and other (cf. Fig. 2 and Table 2).
Fig. 2.
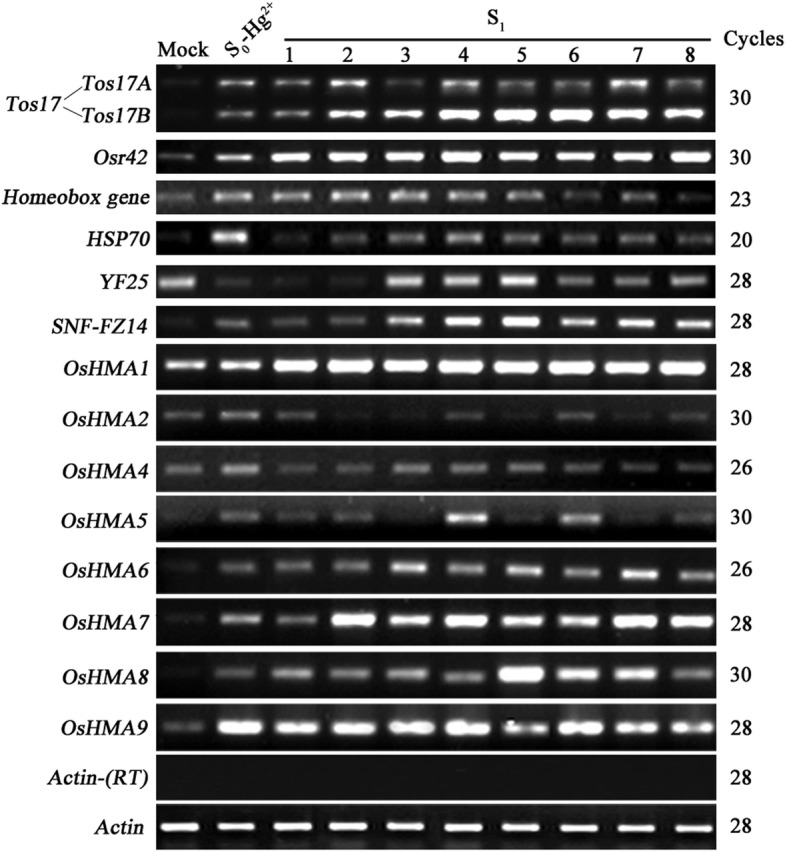
Transgenerational inheritance of altered expression states of 14 genes in a single Hg-treated S0 rice plant. The mock-treated plant serves as a control, and the S0 parental line is the reference for changes in the gene expression in response to Hg-treatment. RNA was isolated from eight S1 individual progeny derived from the S0 parent. The results were highly reproducible among the three independent RNA batches, and hence, only one was presented. Gene names are listed to the left and amplification cycles are labeled to the right of the gel. Relative band intensities were used to calculate the percent progeny falling in either of the three gene expression categories: i) inheritance of Hg-treated S0 pattern, ii) reversion to the mock pattern, and iii) a differential expression pattern (predominately up-regulated expression compared to the S0 progenitor). The rice Actin gene (Genbank accession # X79378) was used as a control for normalization of RNA input. Lack of genomic DNA was validated by the Actin gene on the template without RT
Table 2.
Transgenerational alteration and inheritance of gene expression patterns in 8 randomly chosen S1 plants derived from a Hg2+(50 μm . L−1)-treated S0 individual
| Gene name | Alteration of gene expression pattern in the S0 plant and its S1 progenies | Type and Freq. (%) of patterna | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S0-Hg2+ | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Inherit. of S0 pat. | Rev. to mock Pat. | New pat. (Trans. memory/others) | |
| Tos17A | U | i | +U | i | +U | i | i | +U | i | 62.5 | 0.0 | 37.5 (37.5/0.0) |
| Tos17B | U | i | +U | +U | +U | +U | +U | +U | +U | 12.5 | 0.0 | 87.5 (87.5/0.0) |
| Osr42 | U | +U | +U | +U | +U | +U | +U | +U | +U | 0.0 | 0.0 | 100.0 (100.0/0.0) |
| Homeobox gene | U | i | i | i | i | i | r | i | r | 75.0 | 25.0 | 0.0 (0.0/0.0) |
| HSP70 | U | r | i | i | i | i | i | i | i | 87.5 | 12.5 | 0.0 (0.0/0.0) |
| YF25 | D | i | i | r | r | r | -r | -r | -r | 25.0 | 37.5 | 37.5 (0.0/37.5) |
| SNF-FZ14 | U | i | i | +U | +U | +U | +U | +U | +U | 25.0 | 0.0 | 75.0 (75.0/0.0) |
| OsHMA1 | U | +U | +U | +U | +U | +U | +U | +U | +U | 0.0 | 0.0 | 100.0 (100.0/0.0) |
| OsHMA2 | U | i | D | D | i | D | i | D | i | 50.0 | 0.0 | 50.0 (0.0/50.0) |
| OsHMA4 | U | r | r | i | i | i | r | r | r | 37.5 | 0.0 | 62.5 (0.0/62.5) |
| OsHMA5 | U | i | i | r | +U | i | +U | r | i | 50.0 | 25.0 | 25.0 (25.0/0.0) |
| OsHMA6 | U | i | i | +U | +U | +U | +U | +U | +U | 25.0 | 0.0 | 75.0 (75.0/0.0) |
| OsHMA7 | U | i | +U | +U | +U | +U | +U | +U | +U | 12.5 | 0.0 | 87.5 (87.5/0.0) |
| OsHMA8 | U | i | i | i | i | +U | +U | +U | i | 62.5 | 0.0 | 37.5 (37.5/0.0) |
| OsHMA9 | U | i | i | i | i | i | i | i | i | 100.0 | 0.0 | 0.0 (0.0/0.0) |
| Average Freq. (%)a | 41.7 | 6.7 | 41.7/10.0 | |||||||||
Explanation of symbols: U = up-regulated gene expression in the S0 plant;
+U denotes further up-regulated gene expression in the S1 progeny plant;
D denotes down-regulated gene expression in the S1 progeny plants compared to the mock control
i denotes inheritance of S0 expression pattern in the S1 progeny;
r denotes reversal to Mock control expression pattern in the S1 progeny;
Note: a The average frequency of the specified pattern in S1 progeny plants. For calculations the two copies of Tos17 were treated separately,
aRelative band intensities were used to calculate the percent progeny following in either of the three gene expression categories: i) inheritance of Hg- treated S0 pattern, ii) reversion to the mock pattern, and iii) a differential expression pattern (cf. Fig. 2)
Specifically, for the two copies of Tos17 (Tos17A and Tos17B), the S1 progeny either exhibited inheritance of the S0 expression pattern (62.5% for Tos17A and 12.5% for Tos17B) or further up-regulation of it (37.5% for Tos17A and 87.5% for Tos17B) (Fig. 2 and Table 2). Similarly, for Osr42, 100% S1 progeny showed further up-regulation of the S0 expression pattern.
Out of four low-copy number protein-coding genes (Fig. 2 and Table 2), for Homeobox gene and HSP70, the majority of S1 progeny (75% for Homeobox gene and 87.5% for HSP70) exhibited stable inheritance of the S0 expression pattern, and the remainder (25% for Homeobox gene and 12.5% for HSP70) showed reversal to the mock expression pattern. On the other hand, YF25 which showed significant down-regulation in the S0 generation, exhibited inheritance of the altered expression state, reversal and novel gene expression pattern in the S1 progeny at frequencies of 25, 37.5, and 37.5%, respectively. For SNF-FZ14, which showed transcriptional activation in S0 generation exhibited further up-regulated expression pattern in the majority (75%) of the S1 plants and exhibited inheritance of the altered expression state in the remaining 25% of the progeny.
For the eight OsHMAs tested (Fig. 2 and Table 2), all showed up-regulated expression in S0 plants compared to the mock-treated plants, but differences were found in the S1 generation: OsHMA1 showed further up-regulated expression in 100% progeny. OsHMA2 showed 50% inheritance of up-regulated expression and reversal to the basal expression state in 50% of the progeny. OsHMA4 showed the inheritance of the S0 expression state in 37.5% of the progeny and reversal to the basal expression state in 62.5% of the progeny. OsHMA5 showed inheritance, reversal and further up-regulated expression patterns in 50, 25, and 25% of the S1 plants, respectively; OsHMA6, OsHMA7, and OsHMA8 showed inheritance of the altered expression state in 25, 12.5, and 62.5% of the S1 progeny, and further up-regulated expression in 75, 87.5, and 37.5% of the progeny. OsHMA9 showed significantly up-regulated expression in the S0 plants, and all S1 progeny (100%) inherited the expression pattern.
In summary, we found that for those genes that showed changes in expression in the S0, two major gene expression patterns were manifest in the S1 progeny: either inheritance of the S0 expression pattern (41.7%) or adaptation to a new expression pattern (51.7%). However, the maintenance of change in gene expression varied among the genes tested. For instance, some genes (Tos17A, Homeobox gene, HSP70, OsHMA2, OsHMA5, OsHMA8 and OsHMA9) exhibited inheritance of the expressed state from S0 to S1 generations in ≥50% progeny plants, whereas other genes (Tos17B, Osr42, SNF-FZ14, OsHMA1, OsHMA2, OsHMA6, and OsHMA7) showed a further up-regulated expression in ≥50% progeny plants suggesting genetic memory of the altered expression pattern gained in response to the heavy metal treatment that is transmitted to the next generation.
The altered gene expression states were transgenerationally persistent, coupled with the genetic memory in the S2 generation
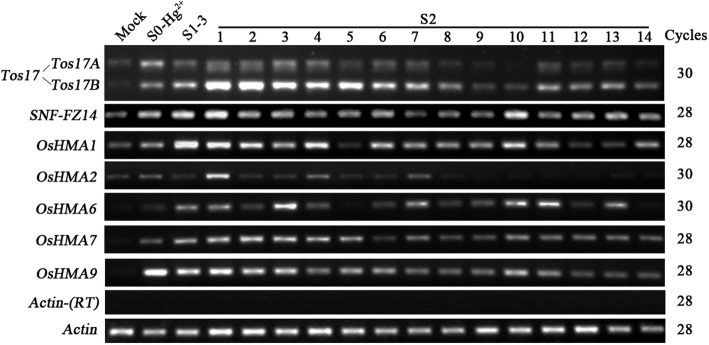
To further test if the altered expression states are transgenerationally persistent, we selected one S1 plant (plant #3) that exhibited all three expression patterns for several of the tested genes, i.e., inheritance of the S0 expression pattern, reversal to the basal expression pattern and adaption of a new expression pattern, to obtain S2 progeny. To study the expression pattern, we performed the RT-PCR analysis of seven genes (Tos17, SNF-FZ14, OsHMA1, OsHMA2, OsHMA6, OsHMA7, and OsHMA9) in the leaf-tissue of 14 randomly selected S2 individuals grown under optimal conditions. The seven genes selected for RT-PCR analysis showed increased expression in the S0 generation and exhibited different expression patterns in the S1 generation. Of the seven genes tested, we identified four gene expression patterns in the S2 progeny, i.e., the inheritance of the S1 expression state, reversion to the S0 expression state, reversion to the mock expression state, and a novel expression pattern (Fig. 3 and Table 3). We observed the majority of S2 progeny inherited the expression state of the S1 progenitor, 36.6% progeny showed inheritance of the S1 expression state, 22.3% progeny reverted to the S0 expression state, 22.3% progeny showed reversal to the basal expression state (similar to mock), and the remaining 18.8% progeny adopted a new expression pattern.
Fig. 3.
Transgenerational inheritance of altered expression states of seven genes in a single S1 rice plant. As evidence of inheritance of the expression states, the S0 and S1 plants are used as a reference as well as the mock control (no metal treatment). A total of 14 S2 individuals were examined to determine the expression of the Tos17A, Tos17B, SNF-FZ14 and five OsHMA transporters in the second generation. Gene names are listed to the left and amplification cycles are labeled to the right of the gel. Relative band intensities were used to calculate the percent progeny falling in either of the four gene expression categories: i) inheritance of the S1 expression state, ii) reversion to the S0 expression state, iii) reversion to the mock expression state, and iv) a novel expression pattern. The rice Actin gene (Genbank accession # X79378) was used as a control for normalization of RNA input. Lack of genomic DNA was validated by the Actin gene on the template without RT
Table 3.
Transgenerational alteration and inheritance of gene expression patterns in the 14 randomly chosen S2 plants derived from one S1 individual #3 (S1–3), which was derived from a single Hg2+ (50 μm)-treated S0 individual
| Gene name | Gene expression patterns in the S2 progeny | Frequency (%)b | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S0-Hg2+ | S1–3 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | Inherit. of S1 pat. | Rev. to S0 pat. | Rev. to mock pat. | New pat. (Trans. Memory/others) | |
| Tos17A | U | U | U | U | U | U | r | U | U | r | r | r | U | r | U | r | 57.1 | 0.0 | 42.9 | 0.0 (0.0/0.0/) |
| Tos17B | U | +U | ++U | ++U | ++U | ++U | ++U | ++U | ++U | +U | U | U | +U | +U | +U | +U | 35.7 | 0.0 | 14.3 | 50.0 (50.0/0.0) |
| SNF-FZ14 | U | +U | ++U | U | U | U | U | U | r | r | r | ++U | U | U | U | U | 0.0 | 21.4 | 64.3 | 14.3 (14.3/0.0) |
| OsHMA1 | U | +U | +U | +U | U | +U | r | U | U | U | U | +U | U | r | r | U | 28.6 | 50.0 | 21.4 | 0.0 (0.0/0.0) |
| OsHMA2 | U | D | +U | r | r | U | r | r | U | -D | -D | -D | -D | -D | -D | -D | 0.0 | 14.3 | 28.6 | 57.1 (0.0/57.1) |
| OsHMA6 | U | +U | +U | U | ++U | U | ++U | U | +U | U | U | ++U | ++U | U | +U | r | 21.4 | 42.9 | 7.1 | 28.6 (28.6/0.0) |
| OsHMA7 | U | +U | +U | +U | +U | +U | +U | U | +U | U | U | +U | U | U | U | U | 50.0 | 50.0 | 0.0 | 0.0 (0.0/0.0) |
| OsHMA9 | U | U | U | U | U | U | U | U | U | U | U | U | U | U | U | U | 100.0 | 0.0 | 0.0 | 0.0 (0.0/0.0) |
| Average Freq. (%)a | 36.6 | 22.3 | 22.3 | 18.8 (11.6/7.2) | ||||||||||||||||
Explanation of symbols: U denotes up-regulated gene expression in the S0 plant, and kept the trend in S1–3 and S2 individuals;
+U denotes further up-regulated gene expression in the S1–3 and kept the trend in S2 individuals;
++U denotes further up-regulated gene expression in S2 individuals compared to their S1 progenitor;
D =denotes down-regulated gene expression in the S1–3 compared to the Mock control;
-D denotes further down-regulated gene expression in S2 individuals compared to their S1 progenitor;
r denotes reversal to Mock control expression pattern in the S2 progeny;
Note: aThe average frequency of the specified pattern in S2 progeny plants. For calculations the two copies of Tos17 were treated separately,
bRelative band intensities were used to calculate the percent progeny following in either of the four gene expression categories (cf. Fig. 3)
On gene by gene basis, the proportions of S2 progeny following one of the four expression patterns (see above) also varied, for instance, in case of Tos17A, OsHMA7, and OsHMA9, ≥50% S2 progeny exhibited inheritance of the S1 expressed state. For OsHMA1 and OsHMA7, ≥50% S2 progeny showed reversal to the expression state of the S0 progenitor. Similarly, for SNF-FZ14 64.3% S2 progeny showed a reversal to the basal expression state. Whereas, in the case of Tos17B and OsHMA6 respectively 50 and 28.6% S2 progeny showed a further up-regulation of the S1 expression pattern.
Collectively, these results suggested that the altered gene expression states induced by heavy metal stress are heritable (11.6%; Table 3), and hence indicates transgenerational memory is involved. Additionally, the progeny also appears to maintain the upward trend of induced expression in response to heavy metal stress.
DNA methylation changes of Tos17 and its transgenerational effect
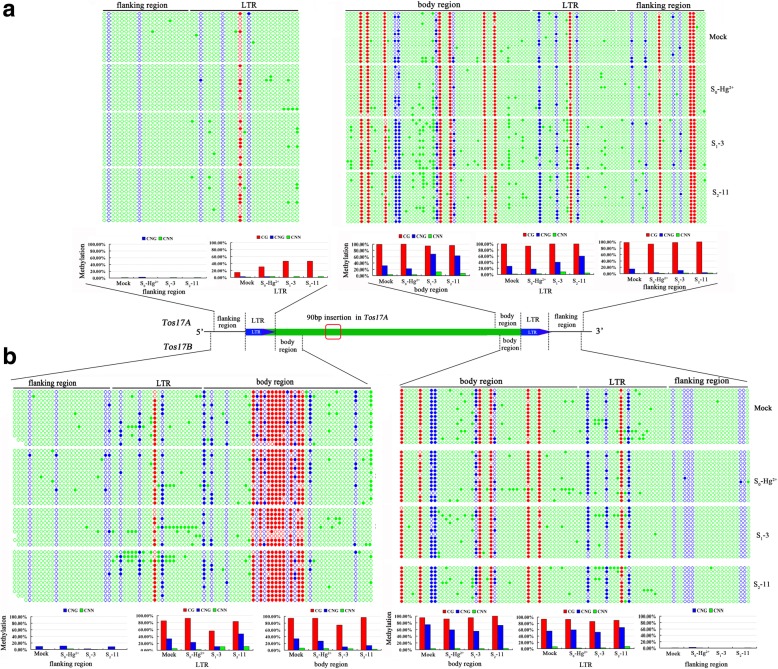
To further explore whether DNA methylation was also altered due to heavy metal stress and to explain its inheritance across generations, we chose Hg-treated S0 plants, one S1 individual (#3) and one S2 individual (#11) to investigate the methylation state and its transmission. We chose Tos17 as a representative gene to test because both copies of Tos17 showed induced expression in the S0 and the progeny kept the trend through two successive generations. We analyzed cytosine methylation patterns of Tos17A and Tos17B by bisulfite sequencing (Fig. 4). Specifically, we inspected the 5′- LTR and its immediate upstream and downstream regions as well as the 3′-LTR and its immediate upstream and downstream regions for Tos17A and Tos17B located on chromosomes 7 and 10, respectively. The results of bisulfite sequencing are presented in Fig. 4, and some salient observations are described: (i) The region immediately upstream of 5′-LTR in Tos17A showed no change in DNA methylation in the S0 plants and the S1/S2 progeny; the LTR region was slightly methylated at CG and CNG regions in the mock-treated plants and showed CG hypermethylation in S0 plants, further hypermethylation in S1 progeny and inheritance of methylation state in S2 plants. (ii) The 3′-LTR and its flanking regions in Tos17A showed CG hypermethylation and partial methylation for CNG and CNN sequences in the mock plants. However, the CG methylation pattern remained unchanged in the S0, S1, and S2 plants. A slight loss of CNG methylation was observed in the body and LTR regions in S0 plants, but increased methylation levels were observed in the S1 progeny. In the S2 progeny, a slight decrease in methylation pattern in the body region and hypermethylation in the LTR region was observed (Fig. 4a). (iii) The flanking region upstream of the 5′-LTR of Tos17B was unmethylated in the mock plants and showed slight de novo methylation in CNG sequences in the S0 plants, a pattern which disappeared in the S1 progeny. In contrast, the 5′-LTR and the downstream body regions of Tos17B showed heavy methylation in CG sequences, and slight to moderate increases in CNN and CNG methylation compared to the mock control. A decrease of CG methylation was observed in the S1, as well as a decrease in CNG methylation in both S0 and S1, but an increase in CNG methylation was found in the S2 progeny (Fig. 4b). Taken together, the results of bisulfite sequencing at Tos17A and Tos17B confirmed that DNA methylation changes occur in response to the heavy metal treatment and also showed transgenerational inheritance. Furthermore, the major pattern of DNA methylation changes is CNG hypomethylation in the S0, which showed different transgenerational effects in either the 3′-region of Tos17A or 5′-region of Tos17B.
Fig. 4.
DNA methylation status of the Tos17A (a) and Tos17B (b) determined by bisulfite sequencing, respectively, in mock and the Hg-treated S0 plant, and its two successive offspring: S1–3 (S1 generation plant # 3) and S2–11 (S2 generation plant # 11). Specific primers were used on the bisulfite-treated rice genomic DNA to amplify six sites from the two Tos17 (Transposon of Oryza sativa 17) copies in the rice genome (cf. Additional file 2: Table S2). Each copy of Tos17 was amplified from six genomic sites: 3 from the 5′-LTR region (i.e., flanking region, LTR, and body region, expect the body region of Tos17A) and 3 from the 3′-LTR region (i.e., flanking region, LTR, and body region). Subsequently, 10 to 15 clones for each PCR product were sequence analyzed, and the methylation levels per site for each of the three cytosine contexts (CG, CHG, and CHH) were calculated and expressed as a percentage (%). Methylation level was calculated by dividing the number of non-converted (methylated) cytosines with the total number of cytosines underlying a sequenced region. In the picture, each DNA sequences was represented by a string of dots, where filled dots represent methylated cytosines and the open dots represent unmethylated cytosines
The gene expression and DNA methylation of two copies of Tos17 changed under heavy metal stress and showed transgenerational memory of the stress. In addition, under certain circumstances, some of the epigenetically silenced TEs are known to become activated and then transpose. TE activity is often causally linked to the compromised repressive epigenetic state in which cytosine DNA methylation is a critical component. We, therefore, analyzed Tos17 mobility in the S0, S1, and S2 generations by Southern blotting. The results showed that Tos17 stayed inactive, which is evident from the consistent copy number maintained in individuals from the S0, S1, and S2 generations (Fig. 5).
Fig. 5.
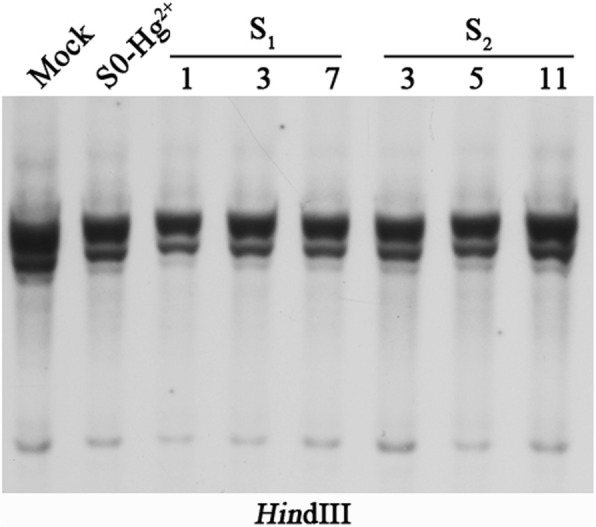
Determination of the Tos17 copy number using gel-blot analysis in a Hg-treated S0 plant and its two successive offspring S1 (1, 3, and 7) and S2 (3, 5, and 11). The results showed that Tos17 stayed inactive, which is evident from the consistent copy number maintained in individuals of the S0, S1, and S2 generations observed using a single LTR retrotransposon Tos17 specific probe (cf. Additional file 1: Table S1)
Discussion and conclusions
In this study, locus-specific gene expression changes and the transgenerational effect of heavy metal stress in rice were analyzed. For this purpose, we chose two retrotransposons, seven protein-coding genes, and nine rice OsHMAs, most of them except seven OsHMAs were analyzed in a previous study of the transgenerational inheritance of modified DNA methylation patterns in response to heavy metal stress [41]. In the present study, we addressed whether the altered expression state of the target genes in response to heavy metal stress is transgenerationally inherited and whether different kinds of genes have common or specific responses to the same heavy metal stress. Based on our previous findings, we chose a single dose of each heavy metal that induced maximum DNA methylation changes [41], and also included a lower dose of heavy metal to study its effect on the transcription and methylation states. The results showed that 16 of 18 genes exhibited up-regulated expression upon treatment with at least one heavy metal (Fig. 1), indicating that a common response might exist for most genes upon heavy metal stress. Our previous data showed that Tos17 and Osr42 exhibit up-regulated expression in response to nitric oxide (NO) treatment in rice [43]. It indicates that these two retrotransposons exhibit transcriptional plasticity to cope with stress. For Tos17, we examined the transcriptional response of the two genomic copies, and both of them showed activation in response to the heavy metal treatment (Fig. 1). It is the first time that the transcriptional activation of both copies of Tos17 was demonstrated in response to abiotic stress.
All OsHMAs except OsHMA3 showed significantly up-regulated expression in response to at least one kind of heavy metal treatment, which suggested that these might all be involved in the heavy metal transport. Previous reports suggested OsHMA1 to be exclusively involve in Zn transport [26], however, in the present study, it showed significantly up-regulated expression in Hg treated rice plants, implicating that it might be also involved in transporting Hg. Similarly, OsHMA2 was formerly reported to be expressed in the root maturation zone and to function in the root-shoot translocation of Zn and Cadmium (Cd) [28, 44]. In the present study, OsHMA2 showed transcriptional activation in Cu treated rice plants, suggesting its potential role in copper (Cu) transport. OsHMA3 was localized to tonoplast in the root cells and was found to be responsible for Cd sequestration in vacuoles [29, 30, 45]. In the present study, OsHMA3 showed no expression in rice shoots or induction after Cu, Cd, Cr or Hg treatment, which is consistent with a recent report that it was not induced in roots and shoots of Cr-treated rice plants [46]. However, overexpression of OsHMA3 was shown to enhance Cd tolerance in rice [47], and a loss-of-function allele was shown to accumulate Cd in grains and shoots [48]. Interestingly, it was recently shown that OsHMA3 driven under the control of the OsHMA2 promoter was successful at reducing Cd accumulation in rice grains [28]. OsHMA4 is localized to the vacuolar membrane, and its expression was shown to be induced by long-term Cu treatment and suppressed by Cu deficiency [31] suggesting its role in Cu sequestration in vacuoles and consequently Cu tolerance. In the present study, OsHMA4 was only slightly induced by Cu treatment, which is in conformity with the previous reports where OsHMA4 was only shown to be induced by long-term Cu treatment [27, 31]. OsHMA5 was mainly expressed in the roots at the vegetative stage, and its expression was shown to be up-regulated by the excess of Cu and other metals such as Zn, Fe, and Mn [32]. Here, we report that OsHMA5 is not expressed in the shoots of mock-treated plants, but is induced in the presence of Cu, which is consistent with a previous study [32]. Additionally, we noticed that OsHMA5 exhibits induced expression in the presence of Cd and Hg as well. There are few reports on the function of OsHMA6, OsHMA7, and OsHMA8. These genes are largely silent in the shoots and only exhibited transcriptional activation under heavy metal stress. Although detailed functions are not known for these genes, our data suggest they may also play a role in heavy metal detoxification. Previous reports showed that OsHMA9 is mainly expressed in vascular tissues and its expression could be induced by high concentrations of Cu, Zn or Cd [27]. In the present study, OsHMA9 showed significant transcriptional activation in Cd and Hg treated plants, and a slight up-regulation in Cu treated plants. Our data support an additional role for OsHMA9 in Hg efflux.
To confirm and extend our findings, we tested whether the altered gene expression state of S0 plants was transgenerationally inherited by the S1 and S2 progeny. We reported an average inheritance rate of 41.7% in the S1 and 36.6% in the S2 (Figs. 2, 3 and Tables 2, 3). However, the rate of inheritance varied depending on gene in question. A majority of the genes tested showed up-regulated expression in the S1 (41.7%) and about 11.6% maintained the trend of up-regulated expression and exhibited further up-regulation in the S2. It indicates that the progeny maintained a memory of the altered expression state of the progenitors even after removal of the heavy metal. Recently, some studies showed a clear connection between the ethylene signaling and response to heavy metal stress in diverse plant species [49–51]. We have not evaluated this aspect in the present study, but believe it is worthy of checking the transcriptional pattern of ethylene biosynthesis and signaling genes in heavy metal treated plants and study the transgenerational inheritance of the expression pattern.
The traditional concept of epigenetics refers to heritable changes in gene expression without an accompanying change in the DNA sequence. Recent research advocates inclusion of the ‘memory concept’ in the formal definition of epigenetics, as even after the disappearance of the initial stress signal, the DNA and/or chromatin modifications are transmitted to maintain the altered transcriptional state from one generation to another [52, 53]. Several studies showed that epigenome is remodeled in plants upon exposure to diverse stresses and DNA methylation pattern is most likely to respond [54–59]. It has been proposed that the DNA methylation state is only partially transmitted to the immediate offspring, as part of it resets during sexual reproduction, which in turn limits the transmission of the acquired epigenetic alterations from parents to offspring [60, 61]. However, our previous research demonstrated that the heavy metal-induced DNA methylation changes in rice are inheritable for at least two successive generations [41]. Here, we monitored the DNA methylation changes under heavy-metal stress in two copies of Tos17 and studied the transgenerational inheritance of epigenetic changes by bisulfite sequencing (Fig. 4). We observed that the major DNA methylation change in Tos17 is CNG hypomethylation, which showed variable inheritance patterns in the 3′- and 5′-regions of the two genomic copies of Tos17 (Tos17A and Tos17B). These observations conform with our previous findings where CNG hypomethylation was most prevalent in response to heavy metal stress and showed at least partial inheritance of the epigenetic changes [41, 43]. DNA methylation changes are associated with changes in gene expression. For instance, A. thaliana mutants defective in DNA methylation showed that regulation of phosphate-starvation-responsive genes requires changes in the DNA methylation pattern [59]. Thus, we set out to find the relationship between DNA methylation and gene expression. Our data suggest that there is no direct correlation between the methylation status and gene expression for Tos17. Moreover, Tos17 stayed silent over three generations, which indicates that the methylation changes in Tos17 are not sufficient for its activation followed by transposition. However, it is unclear whether the heritable change in gene expression is related to methylation changes as there can be locus-specific changes in methylation. Moreover, our study was limited to Tos17A and Tos17B.
Interestingly, recent research has proposed a key role for dynamic changes in chromatin substructure in transgenerational memory of gene expression changes in response to various stresses [62–64]. In line with this research, maize-researchers showed that stress-induced changes in chromatin structure activate transposable elements, and new transposition events contribute to altered phenotypes observed in the progeny [65]. Several studies indicated that DNA methylation and small interfering (si) RNAs might play a role in transgenerational epigenetic memory, i.e., modification in gene expression patterns that are transmittable across generations via the germline [37, 66–69]. Therefore, we expect a role for siRNA in the observed transgenerational memory of heavy-metal induced transcriptional and epigenetic changes in the rice genome. However, as noted by Probst and Mittelsten [63], while the concept of transgenerational memory is attractive, it is difficult to determine the actual mechanism contributing to it and the number of generations in which it persists.
Methods
Plant material
O. sativa L. ssp. japonica, cv. Matsumae, a cultivated rice, used in the present study was initially obtained from Japan and has since been propagated for more than twenty generations in our laboratory. For the experiments elaborated here, seeds were thoroughly washed with distilled water and germinated in the dark at 28 °C in Petri dishes containing distilled water. After two days incubation, seedlings were transferred to a greenhouse maintained at 26 °C under a 12 h photoperiod.
Heavy metal treatment
The ten-day-old, seedlings were subjected to different heavy metal treatments: Cu2+ (50 μM or 1000 μM CuSO4), Cd2+ (50 μM or 1000 μM CdCl2), Cr3+ (50 μM or 1000 μM CrCl3) or Hg2+ (50 μM or 1000 μM HgCl2) in Hoagland nutrient solution for a week. As several microelements in Hoagland nutrient solution are either used as sulfates or chlorides, and the pH of the solution is also adjusted using sulfuric acid, so we made no attempts to balance the sulfate and chloride ions in the Hoagland solution. Additionally, the treatments are similar to the one reported in our previous work [41]. Mock controls were grown in parallel in the Hoagland nutrient solution. After treatment, seedlings were transplanted to the field. Leaf samples were harvested at different time points in liquid nitrogen and stored at − 80 °C until used. The plants were marked “stressed S0”. Panicles of several selected stressed and mock plants were bagged for self-pollination and seeds were collected to produce the next generation of plants, which were labeled as S1. In a similar way, S2 generation plants were produced, and the seeds were harvested.
Reverse-transcription PCR (RT-PCR) analysis
RT-PCR was performed essentially as reported in Liu et al. [70]. In brief, total RNA was isolated from expanded young leaves using Trizol reagent (Invitrogen) following the manufacturer’s instructions. RNA was converted to cDNA using Super ScriptTM RNase H reverse transcriptase kit (Invitrogen), and subjected to RT-PCR analysis using gene-specific primers (Additional file 1: Table S1). The rice Actin gene (Genbank accession # X79378) was used as the control for normalization of RNA input. Gene-specific primers were designed using Primer 3 (http://bioinfo.ut.ee/primer3/) and are listed in Additional file 1: Table S1. Different cycle numbers were used for different genes to ensure amplifications stay within the linear range for each gene. For S0 samples, we pooled seedings and used three technical replications to check the gene expression changes. Whereas, for the S1 and S2 individuals, three batches of independently prepared total RNAs were used as technical replications. The amplified products were visualized via agarose gel electrophoresis and ethidium bromide staining.
Bisulfite sequencing of the Tos17 loci
Genomic DNA was extracted from fully expended rice leaves and was given a bisulfite treatment [71]. Briefly, an EZ DNA Methylation-Gold Kit from Zymo Research was used to treat 5 μg of genomic DNA. The PCR primers, which were used to amplify bisulfite-converted genomic DNA for the two copies of the Tos17 (Transposon of Oryza sativa 17), are listed in Additional file 2: Table S2. From 10 to 15 clones for each sample were sequence analyzed. The methylation level was expressed as the percentage (%) per site for each of the three cytosine contexts (CG, CHG, and CHH). Methylation level was calculated by dividing the number of non-converted (methylated) cytosines with the total number of cytosines underlying a sequenced region. The sequences were analyzed by the Kismeth program (http://katahdin.mssm.edu/kismeth/revpage.pl), and the results were presented as histograms.
Southern blotting
Genomic DNA was isolated from fully expanded leaves of heavy metal-stressed and mock control rice plants by a modified CTAB method [72] and purified by phenol extraction. For the transposon activity analysis, 5 μg of genomic DNA was digested with Hind III (NEB) and resolved on 1% agarose gel. Subsequently, DNA was transferred to Hybond N+ nylon membranes (Amersham Pharmacia Biotech, Piscataway, New Jersey) via alkaline transfer, as recommended by the manufacturer. Only one Tos17 copy was used as a probe in the present study (see Additional file 1: Table S1). For probe preparation, the Tos17 fragments were amplified via PCR at annealing temperature 59 °C. The authenticity of the PCR products was confirmed by DNA sequencing. The fragments were gel-purified and labeled with fluorescein-11-dUTP using the Gene Images random prime-labeling module from Amersham Pharmacia Biotech. Hybridization signal was detected by the Gene Images CD2+P-Star detection module (Amersham Pharmacia Biotech) after two stringent washes with 0.2 × SSC and 0.1% SDS for 50 min each. Subsequently, the membrane was exposed to X-ray film.
Additional files
Table S1. List of gene-specific primers used for RT-PCR analysis and amplification of probe used for Southern blotting. (DOC 50 kb)
Table S2. List of primers used for bisulfite sequencing of Tos17. (DOC 39 kb)
Acknowledgments
We greatly thank for the support of “National college student life science competition committee”, as four college students (YCL, WJM, LCH and ZR) contributing to this study.
Abbreviations
- Cd
Cadmium
- Co
Cobalt
- Cr
Chromium
- CTAB
Cetyltrimethylammonium bromide
- Cu
Copper
- Hg
Mercury
- HMA
Heavy Metal-transporting P-type ATPases
- NO
Nitric oxide
- Pb
Lead
- RT-PCR
Reverse transcription-polymerase chain reaction
- SDS
Sodium dodecyl sulfate
- SSC
Saline sodium citrate
- TE
Transposable element
- Zn
Zinc
Authors’ contributions
CWX, XL, MYL performed RT-PCR analysis and they contributed equally to this work; ZYH performed bisulfite sequencing; ZYH, YCL, WJM, LCH, ZR are responsible for the heavy metal treatment of rice plants; ZTT, LXY, JLL, WNN, MJ are responsible for rice filed propagation; LB participated in the design, SR and OXF conceived the study, participated in the design and wrote the manuscript with help from KAS. All authors have read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 31200198 and No. 31400256), National Students’ platform for innovation and entrepreneurship training program (No.201810200055), National college student life science competition committee, and NIFA Hatch/Multi-state grant (S009) to SR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Author Bao Liu is a Section Editor of BMC Plant Biology and no other authors have competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Weixuan Cong, Yiling Miao and Lei Xu contributed equally to this work.
Contributor Information
Weixuan Cong, Email: congwx377@nenu.edu.cn.
Yiling Miao, Email: 974748096@qq.com.
Lei Xu, Email: 1051783110@qq.com.
Yunhong Zhang, Email: 247933082@qq.com.
Chunlei Yuan, Email: 858727590@qq.com.
Junmeng Wang, Email: 815198354@qq.com.
Tingting Zhuang, Email: dooby110@126.com.
Xiuyun Lin, Email: 1499769757@qq.com.
Lili Jiang, Email: jiangll269@nenu.edu.cn.
Ningning Wang, Email: wangnn826@163.com.
Jian Ma, Email: winter0106@163.com.
Karen A. Sanguinet, Email: karen.sanguinet@wsu.edu
Bao Liu, Email: baoliu@nenu.edu.cn.
Sachin Rustgi, Email: srustgi@clemson.edu.
Xiufang Ou, Email: ouxf074@nenu.edu.cn.
References
- 1.Bermudez GMA, Jasan R, Pla R, Pignata ML. Heavy metals and trace elements in atmospheric fallout: their relationship with topsoil and wheat element composition. J Hazard Mater. 2012;213:447–456. doi: 10.1016/j.jhazmat.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan O, Ince M, Yaman M. Sequential extraction of cadmium in different soil phases and plant parts from a former industrialized area. Environ Chem Lett. 2011;9:397–404. doi: 10.1007/s10311-010-0292-0. [DOI] [Google Scholar]
- 3.Shanker AK, Djanaguiraman M, Venkateswarlu B. Chromium interactions in plants: current status and future strategies. Metallomics. 2009;1:375–383. doi: 10.1039/b904571f. [DOI] [PubMed] [Google Scholar]
- 4.Mayor DJ, Gray NB, Elver-Evans J, Midwood AJ, Thornton B. Metalmacrofauna interactions determine microbial community structure and function in copper contaminated sediments. PLoS One. 2013;8:64–94. doi: 10.1371/journal.pone.0064940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guzel S, Terzi R. Exogenous hydrogen peroxide increases dry matter production, mineral content and level of osmotic solutes in young maize leaves and alleviates deleterious effects of copper stress. Bot Stud. 2013;54:26. doi: 10.1186/1999-3110-54-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabata-Pendias Alina. Trace Elements in Soils and Plants. 2010. [Google Scholar]
- 7.Kazakou E, Dimitrakopoulos PG, Baker AJM, Reeves RD, Troumbis AY. Hypotheses, mechanisms and trade-offs of tolerance and adaptation to serpentine soils: from species to ecosystem level. Biol Rev. 2008;83:495–508. doi: 10.1111/j.1469-185X.2008.00051.x. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Wu YR, Ling HQ, Clu CC. OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol. 2009;70:219–229. doi: 10.1007/s11103-009-9466-1. [DOI] [PubMed] [Google Scholar]
- 9.Clemens S. Molecular mechanisms of plant metal tolerance and homeostasis. Planta. 2001;212:475–486. doi: 10.1007/s004250000458. [DOI] [PubMed] [Google Scholar]
- 10.Mganga N, Manoko MLK, Rulangaranga ZK. Classification of plants according to their heavy metal content around North Mara gold mine, Tanzania: implication for phytoremediation. Tanzania J Sci. 2011;37:109–119. [Google Scholar]
- 11.Sytar O, Kumar A, Latowski D, Kuzynska P, Strzalka K, Prasad MNK. Heavy metal induced oxidative damage, defense reactions, and detoxification mechanisms in plants. Acta Physiol Plant. 2013;35:985–999. doi: 10.1007/s11738-012-1169-6. [DOI] [Google Scholar]
- 12.Song WY, Mendoza-Cozatl DG, Lee Y. Phytochelatin-metal (loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant Cell Enviro. 2014;37:1192–1201. doi: 10.1111/pce.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rascio N, Navari-Izzo F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011;180:169–181. doi: 10.1016/j.plantsci.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Colangelo EP, Guerinot ML. Put the metal to the petal: metal uptake and transport throughout plants. Curr Opin Plant Biol. 2006;9:322–330. doi: 10.1016/j.pbi.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Kraemer U, Talke IN, Hanikenne M. Transition metal transport. FEBS Lett. 2007;581:2263–2272. doi: 10.1016/j.febslet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Axelsen KB, Palmgren MG. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001;126:696–706. doi: 10.1104/pp.126.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams LE, Mills RF. P1B-ATPases-an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa NK, et al. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and cd in rice. Plant Cell Environ. 2012;35:1948–1957. doi: 10.1111/j.1365-3040.2012.02527.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim YY, Choi H, Segami S, Cho HT, Martinoia E, Maeshima M, Lee Y. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 2009;58:737–753. doi: 10.1111/j.1365-313X.2009.03818.x. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi Yuriko, Kuroda Keishi, Kimura Keisuke, Southron-Francis Jennafer L., Furuzawa Aya, Kimura Kazuhiko, Iuchi Satoshi, Kobayashi Masatomo, Taylor Gregory J., Koyama Hiroyuki. Amino Acid Polymorphisms in Strictly Conserved Domains of a P-Type ATPase HMA5 Are Involved in the Mechanism of Copper Tolerance Variation in Arabidopsis. Plant Physiology. 2008;148(2):969–980. doi: 10.1104/pp.108.119933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell. 2005;17:1233–1251. doi: 10.1105/tpc.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binder BM, Rodríguez FI, Bleecker AB. The copper transporter RAN1 is essential for biogenesis of ethylene receptors in Arabidopsis. J Biol Chem. 2010;285:37263–37270. doi: 10.1074/jbc.M110.170027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong Chong Kum Edwin, Cobbett Christopher S. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytologist. 2008;181(1):71–78. doi: 10.1111/j.1469-8137.2008.02638.x. [DOI] [PubMed] [Google Scholar]
- 24.Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, et al. AtHMA3, a P1B-ATPase allowing cd/Zn/co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong Chong Kum Edwin, Jarvis Renée S., Sherson Sarah M., Cobbett Christopher S. Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytologist. 2008;181(1):79–88. doi: 10.1111/j.1469-8137.2008.02637.x. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki M, Bashir K, Inoue H, Takahashi M, Nakanishi H, Nishizawa NK. Accumulation of starch in Zn-deficient rice. Rice. 2012;5:9. doi: 10.1186/1939-8433-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Kim Y, Lee Y, An G. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007;145:831–842. doi: 10.1104/pp.107.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao Ji Feng, Xia Jixing, Yamaji Naoki, Shen Ren Fang, Ma Jian Feng. Effective reduction of cadmium accumulation in rice grain by expressing OsHMA3 under the control of the OsHMA2 promoter. Journal of Experimental Botany. 2018;69(10):2743–2752. doi: 10.1093/jxb/ery107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tezuka K, Miyaadte H, Katou K, Kodama I, Matsumoto S, Kawamoto T, et al. A single recessive gene controls cadmium translocation in the cadmium hyperaccumulating rice cultivar Cho-Ko-Koku. Theor Appli Genet. 2010;120:1175–1182. doi: 10.1007/s00122-009-1244-6. [DOI] [PubMed] [Google Scholar]
- 30.Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, et al. Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci U S A. 2010;107:16500–16505. doi: 10.1073/pnas.1005396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang XY, Deng F, Yamaji N, Pinson ARM, Fujii-Kashino M, Danku J, et al. A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat Commun. 2016;7:12138. doi: 10.1038/ncomms12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng F, Yamaji N, Xia J, Ma JF. A member of the heavy metal P-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 2013;163:1353–1362. doi: 10.1104/pp.113.226225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Li H, Zou D, Zhao J, Fan L, Wu T. Transcriptome profile analysis of cadmium tolerance in Chinese flowering cabbage. Hortic Environ Biotechnol. 2017;58:56–65. doi: 10.1007/s13580-017-0075-7. [DOI] [Google Scholar]
- 34.Liu Huan, Zhao Haixia, Wu Longhua, Liu Anna, Zhao Fang-Jie, Xu Wenzhong. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytologist. 2017;215(2):687–698. doi: 10.1111/nph.14622. [DOI] [PubMed] [Google Scholar]
- 35.Zhang JH, Zeng L, Sun HL, Wu H, Chen SY. Adversity stress-related responses at physiological attributes, transcriptional and enzymatic levels after exposure to cu in Lycopersicum esculentm seedlings. Scientia Hortic. 2017;222:213–220. doi: 10.1016/j.scienta.2017.05.027. [DOI] [Google Scholar]
- 36.Oono Y, Yazawa T, Kanamori H, Sasaki H, Mori S, Handa H, et al. Genome-wide transcriptome analysis of cadmium stress in rice. BioMed Res Intern. 2016:9739505–5. [DOI] [PMC free article] [PubMed]
- 37.Gallusci Philippe, Dai Zhanwu, Génard Michel, Gauffretau Arnaud, Leblanc-Fournier Nathalie, Richard-Molard Céline, Vile Denis, Brunel-Muguet Sophie. Epigenetics for Plant Improvement: Current Knowledge and Modeling Avenues. Trends in Plant Science. 2017;22(7):610–623. doi: 10.1016/j.tplants.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Feng Sheng Jun, Liu Xue Song, Tao Hua, Tan Shang Kun, Chu Shan Shan, Oono Youko, Zhang Xian Duo, Chen Jian, Yang Zhi Min. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant, Cell & Environment. 2016;39(12):2629–2649. doi: 10.1111/pce.12793. [DOI] [PubMed] [Google Scholar]
- 39.Sebastian A, Prasad MNV. Cadmium minimization in rice. A review. Agron Sustain Dev. 2014;34:155–173. doi: 10.1007/s13593-013-0152-y. [DOI] [Google Scholar]
- 40.Mitra A, Chatterjee S, Moogouei R, Gupta D. Arsenic accumulation in Rice and probable mitigation approaches: a review. Agronomy. 2017;7:67. doi: 10.3390/agronomy7040067. [DOI] [Google Scholar]
- 41.Ou X, Zhang Y, Xu C, Lin X, Zang Q, Zhuang T, et al. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in Rice (Oryza sativa L.) PLoS One. 2012;7:e41143. doi: 10.1371/journal.pone.0041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Y, Wang X, Su LB, Zhai J, Cao S, Zhang D, et al. SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell. 2007;19:9–22. doi: 10.1105/tpc.106.048124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ou X, Zhuang T, Yin W, Miao Y, Wang B, Zhang Y, et al. DNA methylation changes induced in rice by exposure to high concentrations of the nitric oxide modulator, sodium Nitroprusside. Plant Mol Biol Rep. 2015;33:1428–1440. doi: 10.1007/s11105-014-0843-9. [DOI] [Google Scholar]
- 44.Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, Sakurai K, et al. Mutations in rice (Oryza sativa) heavy metal ATPase2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012;53:213–224. doi: 10.1093/pcp/pcr166. [DOI] [PubMed] [Google Scholar]
- 45.Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, Kawamoto T, et al. OsHMA3, a P1Btype of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011;189:190–199. doi: 10.1111/j.1469-8137.2010.03459.x. [DOI] [PubMed] [Google Scholar]
- 46.Kabir AH. Biochemical and molecular changes in rice seedlings (Oryza sativa L.) to cope with chromium stress. Plant Biol. 2016;18:710–719. doi: 10.1111/plb.12436. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki A, Yamaji N, Ma JF. Overexpression of OsHMA3 enhances cd tolerance and expression of Zn transporter genes in rice. J Exp Bot. 2014;65:6013–6021. doi: 10.1093/jxb/eru340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan J, Wang P, Wang P, Yang M, Lian XM, Tang Z, et al. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of Japonica rice cultivars. Plant Cell Environ. 2016;39:1941–1954. doi: 10.1111/pce.12747. [DOI] [PubMed] [Google Scholar]
- 49.Khan MI, Khan NA. Ethylene reverses photosynthetic inhibition by nickel and zinc in mustard through changes in PS II activity, photosynthetic nitrogen use efficiency, and antioxidant metabolism. Protoplasma. 2014;251:1007–1019. doi: 10.1007/s00709-014-0610-7. [DOI] [PubMed] [Google Scholar]
- 50.Khan M. Iqbal R., Nazir Faroza, Asgher Mohd., Per Tasir S., Khan Nafees A. Selenium and sulfur influence ethylene formation and alleviate cadmium-induced oxidative stress by improving proline and glutathione production in wheat. Journal of Plant Physiology. 2015;173:9–18. doi: 10.1016/j.jplph.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 51.Thao NP, Khan MI, Thu NB, Hoang XL, Asgher M, Khan NA, et al. Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress. Plant Physiol. 2015;169:73–84. doi: 10.1104/pp.15.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avramova Z. Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress response genes. Plant J. 2015;83:149–159. doi: 10.1111/tpj.12832. [DOI] [PubMed] [Google Scholar]
- 53.Liu R, How-Kit A, Srammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci U S A. 2015;112:10804–10809. doi: 10.1073/pnas.1503362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Correia B, Valledor L, Meijón M, Rodriguez JL, Dias MC, Santos C, et al. Is the interplay between epigenetic markers related to the acclimation of cork oak plants to high temperatures? PLoS One. 2013;8:e53543. doi: 10.1371/journal.pone.0053543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.González RM, Ricardi MM, Lusem ND. Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics. 2013;8:864–872. doi: 10.4161/epi.25524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diez CM, Roessler K, Gaut BS. Epigenetics and plant genome evolution. Curr Opin Plant Biol. 2014;18:1–8. doi: 10.1016/j.pbi.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Yan SH, Zhao L, Tan JJ, Zhang Q, Gao F, et al. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 2014;14:105. doi: 10.1186/1471-2229-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai LF, Chen YL, Luo XD, et al. Level and pattern of DNA methylation changes in rice cold tolerance introgression lines derived from Oryza rufipogon Griff. Euphytica. 2015;205:73–83. doi: 10.1007/s10681-015-1389-0. [DOI] [Google Scholar]
- 59.Yong-Villalobos L. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc Natl Acad Sci U S A. 2015;112:E7293–E7302. doi: 10.1073/pnas.1522301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawashima T, Berger F. Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet. 2014;15:613–624. doi: 10.1038/nrg3685. [DOI] [PubMed] [Google Scholar]
- 61.Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J, et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife. 2016;5:e13546. doi: 10.7554/eLife.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pecinka A, Mittelsten Scheid O. Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol. 2012;53:801–808. doi: 10.1093/pcp/pcs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saze H. Transgenerational inheritance of induced changes in the epigenetic state of chromatin in plants. Genes Genetic Syst. 2012;87:145–152. doi: 10.1266/ggs.87.145. [DOI] [PubMed] [Google Scholar]
- 64.Probst AV, Mittelsten SO. Stress-induced structural changes in plant chromatin. Curr Opin Plant Biol. 2015;27:8–16. doi: 10.1016/j.pbi.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 65.Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-ibarra J, et al. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015;11:e1004915. doi: 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rasmann S, Vos MD, Casteel CL, Tian DL, Halitschke R, Sun JY, et al. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 2012;158:854–863. doi: 10.1104/pp.111.187831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grossniklaus U, Kelly WG, Fergusonsmith AV, Pembrey M, Lindquist S. Transgenerational epigenetic inheritance: how important is it? Nat Rev Genet. 2013;14:228–235. doi: 10.1038/nrg3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baulcombe DC, Dean C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb Perspect Biol. 2004;6:a019471. doi: 10.1101/cshperspect.a019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kinoshita T, Seki M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2004;55:1859–1863. doi: 10.1093/pcp/pcu125. [DOI] [PubMed] [Google Scholar]
- 70.Liu Z. L., Han F. P., Tan M., Shan X. H., Dong Y. Z., Wang X. Z., Fedak G., Hao S., Liu Bao. Activation of a rice endogenous retrotransposon Tos17 in tissue culture is accompanied by cytosine demethylation and causes heritable alteration in methylation pattern of flanking genomic regions. Theoretical and Applied Genetics. 2004;109(1):200–209. doi: 10.1007/s00122-004-1618-8. [DOI] [PubMed] [Google Scholar]
- 71.Ngezahayo F, Xu C, Wang HY, Jiang LL, Pang JS, Liu B. Tissue culture induced transpositional activity of mPing is correlated with cytosine methylation in rice. BMC Plant Biol. 2009;9:91. doi: 10.1186/1471-2229-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kidwell Kimberlee K., Osborn Thomas C. Plant Genomes: Methods for Genetic and Physical Mapping. Dordrecht: Springer Netherlands; 1992. Simple plant DNA isolation procedures; pp. 1–13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of gene-specific primers used for RT-PCR analysis and amplification of probe used for Southern blotting. (DOC 50 kb)
Table S2. List of primers used for bisulfite sequencing of Tos17. (DOC 39 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].