Abstract
Background:
Early data showed that FOXP3 could induce epithelial-mesenchymal transition by stimulating the Wnt/β-catenin signaling pathway in non-small cell lung cancer (NSCLC). However, how the expression of FOXP3 is regulated in NSCLC remains unknown. We thus explored the impacts of the long noncoding RNA EGFR antisense RNA 1 (EGFR-AS1) and hypoxia-inducible factor-2A (HIF2A) on FOXP3 expression and the cancer stemness of NSCLC.
Methods:
Lung tissues samples from 87 patients with NSCLC and two NSCLC cell lines were used in this study. The regulation of FOXP3 and lung cancer cell stemness by EGFR-AS1 and HIF2A was determined at molecular levels in NSCLC tissue samples and cultured cells in the presence/absence of the smoking carcinogen, 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (also known as nicotine-derived nitrosamine ketone). The results were confirmed in tumor xenograft models.
Results:
We found that NNK decreased the expression of EGFR-AS1 in the long term, but increased the expression of HIF2A and FOXP3 to stimulate lung cancer cell stemness. EGFR-AS1 significantly inhibited FOXP3 expression and NSCLC cell stemness, whereas HIF2A obviously promoted both. The enhancement of lung cancer stemness by FOXP3 was, at least partially, via stimulating Notch1, as the inhibition of Notch1 could markedly diminish the effect of FOXP3.
Conclusions:
FOXP3, the expression of which is under the fine control of EGFR-AS1, is a critical molecule that promotes NSCLC cancer cell stemness through stimulating the Notch1 pathway.
Keywords: cancer cell stemness, EGFR antisense RNA 1, FOXP3, hypoxia-inducible factor-2 alpha, non-small cell lung cancer
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung malignances and median survival is only 6–12 months.1 It is well accepted that the development of all malignancies including metastasis and their low sensitivity to antitumor treatments could be attributed to the cancer stem cells (CSCs), which have the ability to initiate and colonize at distant secondary tissue sites.2–7 Among the various molecular signaling pathways in cells, Notch, WNT, and hedgehog are the three main pathways that are believed to be related to cancer stemness.2,4–7 Understanding the key signaling pathways of lung CSCs is of pivotal and clinical importance for new drug discovery and development.2
FOXP3, a transcription factor, belongs to the family of FOX proteins.8 FOXP3 was initially discovered in regulatory T (Treg) cells and it plays a significant role in the process and maintenance of Treg cells. Previous findings have indicated that FOXP3 can work to induce epithelial-mesenchymal transition (EMT) by stimulating the Wnt/β-catenin signaling pathway in cancer cells.9 How the expression of FOXP3 is regulated in NSCLC, however, remains unknown.
Noncoding RNAs are believed to have various impacts on the development, progression, and treatment of cancers. Among these cancer-related noncoding RNAs, numerous microRNAs (miRNAs) have been found to participate in the regulation of FOXP3 in several types of cancers. Qin and colleagues reported that miR-126 could enhance the expression of FOXP3 in Treg cells, resulting in an increase in the antitumor effect of CD8+ T cells in breast cancer.10 In T-cell acute lymphoblastic leukemia, miR146a could act as a tumor suppressor and increase the oncological immune response by upregulating the expression of FOXP3.11 In addition to miRNA, the expression of FOXP3 can also be regulated by some long noncoding RNAs (lncRNAs). It has been reported that lncEGFR upregulation in Treg cells correlates positively with tumor size and expression of FOXP3 in liver cancer.12
The FOXP3 long intergenic noncoding RNA is a negative regulator that could suppress the expression of FOXP3.13 EGFR antisense RNA 1 (EGFR-AS1) is located on human chromosome 7, in the direction opposite to that of EGFR gene transcription according to the database of the University of California Santa Cruz (http://genome.ucsc.edu/). Apparently, EGFR-AS1 is near lncEGFR, making it possible that EGFR-AS1 has similar functions to lncEGFR or plays a related role in the regulation of FOXP3.
Hypoxia-inducible factors, including hypoxia-inducible factor 1A (HIF1A) and hypoxia-inducible factor 2A (HIF2A), are transcription factors that are responsible for the induction of genes associated with cell survival under hypoxia.14 The overexpression of HIF1A or HIF2A is associated with solid cancers.15–17 According to the study of Clambey and colleagues, HIF1A could directly bind to the FOXP3 promoter, and then, with the cooperation of transforming growth factor beta, upregulate the expression of FOXP3 in the cutaneous T cells.15 However, whether HIF2A can function as HIF1A remains unclear.
Cigarette smoking is well known as the primary inducer of NSCLC.18–21 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (also known as nicotine-derived nitrosamine ketone), as the major component of cigarette smoking, greatly contributes to lung cancer tumorigenesis via multiple pathways including stimulating lung CSCs.18,22 However, its tumorigenesis mechanism, especially the pathway related to lung CSCs, is still not fully known.
In this study, we aimed to determine how EGFR-AS1 and HIF2A regulated FOXP3 expression in NSCLC cells, and its impact on lung cancer cell stemness. The results of this study have revealed some novel mechanisms on FOXP3 expression regulation in NSCLC cells and identified new potential therapeutic targets for this malignant disorder.
Materials and methods
Ethics statement
An informed consent for human tissues for research purposes only was obtained from all patients recruited in this study. The use of human samples in this study was approved (2014.649 and 2015.729) by the joint Chinese University of Hong Kong (CUHK) – New Territories East Cluster Clinical Research Ethics Committee. All animal experiments were conducted in accordance with the Animals (Control of Experiments) Ordinance Chapter 340, and approved (14/092/GRF-4-B) by the Animal Experimentation Ethics Committee of CUHK.
Tissue collection
A total of 87 pairs of NSCLC tissues and the corresponding adjacent nontumor lung tissues were obtained from patients who underwent surgery in the Prince of Wales Hospital between 2003 and 2016. All the patients were diagnosed with NSCLC based on laboratory tests and imaging examinations before surgery and histopathological evaluation after surgery. Clinical characteristics were available for all samples (Table 1). No patients had received any local or systemic treatment before surgery. All collected tissue samples were fixed in formalin for histological evaluation and snap-frozen in liquid nitrogen and stored at −80°C until experimentation.
Table 1.
Clinical characteristics of patients with NSCLC.
| Total | AS1 |
HIF2A |
|||||
|---|---|---|---|---|---|---|---|
| H | L | H | L | ||||
| Smoking status | |||||||
| Smoker | 60 | 19 | 41 | p = 0.006 | 55 | 5 | p > 0.05 |
| Nonsmoker | 27 | 17 | 10 | 25 | 2 | ||
| Sex | |||||||
| Male | 59 | 20 | 39 | p > 0.05 | 55 | 4 | p > 0.05 |
| Female | 28 | 16 | 12 | 25 | 3 | ||
| Age (years) | 66.16 ± 7.92 | 66.58 ± 1.4 | 65.86 ± 1.07 | p > 0.05 | 66.59 ± 0.89 | 61.29 ± 2.47 | p > 0.05 |
| Tumor diameter (cm) | 3.77 ± 1.82 | 3.28 ± 0.23 | 4.12 ± 0.28 | p = 0.033 | 3.78 ± 0.21 | 3.67 ± 0.49 | p > 0.05 |
| Tumor differentiation | |||||||
| Well differentiated | 65 | 28 | 37 | p > 0.05 | 59 | 6 | p > 0.05 |
| Poorly differentiated | 22 | 8 | 14 | 21 | 1 | ||
| Stage | |||||||
| IA | 26 | 14 | 12 | p > 0.05 | 26 | 0 | p > 0.05 |
| IB | 18 | 9 | 9 | 16 | 2 | ||
| IIA | 13 | 2 | 11 | 13 | 0 | ||
| IIB | 14 | 4 | 10 | 12 | 2 | ||
| IIIA | 11 | 5 | 6 | 9 | 2 | ||
| IIIB | 2 | 0 | 2 | 2 | 0 | ||
| IV | 3 | 2 | 1 | 2 | 1 | ||
| T stage | |||||||
| 1 | 33 | 17 | 16 | p > 0.05 | 33 | 0 | p = 0.012 |
| 2 | 38 | 12 | 26 | 35 | 3 | ||
| 3 | 14 | 7 | 7 | 11 | 3 | ||
| 4 | 2 | 0 | 2 | 1 | 1 | ||
| Lymph metastasis | |||||||
| Positive | 26 | 10 | 16 | p > 0.05 | 23 | 3 | p > 0.05 |
| Negative | 61 | 26 | 35 | 57 | 4 | ||
AS1 antisense RNA 1; H, high expression of EGFR-AS1; HIF2A, hypoxia-inducible factor-2A; L, lower expression of EGFR-AS1; NSCLC, non-small cell lung cancer.
Immunohistochemistry (IHC)
An immunohistochemical assay was performed according to standard protocol on formalin-fixed paraffin sections using a primary antibody to HIF2A (Santa Cruz, 1:50, Santa Cruz Biotechnology, Dallas, TX, USA). The staining intensities were scored using the immunoreactive score (IRS) method by a pathologist and an investigator separately. The IRS method is described in Supplementary Table 1.
Cell lines and culture conditions
Cell lines, including HEK293NT cell lines, and NSCLC cell lines of NCH-H460 and NCH-H23, were obtained from the American Type Culture Collection, and were characterized by mycoplasma detection, DNA fingerprinting, isozyme detection, and determination of cell viability. All the cells were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) and antibiotics at 37°C in humidified air with 5% CO2 or with 1% O2. NNK, protease inhibitor cocktail, and fluorescein isothiocyanate-conjugated immunoglobulins were supplied by Sigma (Saint Louis, MO, USA). NNK was resolved in dimethyl sulfoxide (DMSO) at a concentration of 2 M, and when applied to treat cells, it was diluted in a medium to a final concentration of 10 µM.21
The specific inhibitor of the Notch signaling pathway, N-[2S-(3,5-difluorophenyl) acetyl]-L-alanyl-2-phenyl-1,1-dimethylethyl ester-glycine (DAPT),23–25 was supplied by Cayman Chemical (Ann Arbor, MI, USA). DAPT was resolved in DMSO at a concentration of 5 mM, and when applied to treat cells, it was diluted in a medium to a final concentration of 25 µM.23–25 The specific incubation period was determined by the experiments that followed.
Transfection of cell lines and construction of stably transfected cell lines
The plasmid of HA-HIF2alpha-pcDNA3 was a gift from William Kaelin (Addgene plasmid #18950). The plasmid of pCDH-HIF2a-Puro was a gift from Eric Jonasch and Xiande Liu (Addgene plasmid #71708). Full-length EGFR-AS1 was generated by polymerase chain reaction (PCR) from cDNA with primers incorporating appropriate restriction enzyme sites: EGFR-AS1 FWD (EcoRV): 5’- GAGGATATCCCCATTTCTACACAGTGTCTGTTTCCTCAGA-3’, EGFR-AS1 REV (HindIII):5’- GCGAAGCTTCTATTGCATCGGTACTGAACATATACGGACTTT-3’. The PCR products were first cloned into pCR2.1 (Invitrogen), using TOPO cloning from which EGFR-AS1 was isolated using EcoRV/HindIII and cloned into similarly digested pcDNA3.1(-) (Invitrogen). The plasmid of pLKO.1 was used as the backbone for the construction of shRNAs. The overexpression plasmid of pHIV-FOXP3 and shRNAs for FOXP3 were constructed as described.11 The shRNAs for HIF2A were constructed according to the protocol of the Genetic Perturbation Platform (https://www.broadinstitute.org), and the sequence is shown in Supplementary Table 2.
The vectors and corresponding negative controls were transfected into cell lines with lipofectamine 2000 DNA transfection reagent (Invitrogen) according to the provided protocol. Lentivirus for the overexpression and shRNAs for the knockdown were constructed with auxiliary plasmids including pRSV-REV, pMDLg/pRRE, psPAX2, and pMD2.G in accordance with standard protocols. After transient transfection or lentivirus infection, the cell lines were screened by G418 or puromycin for different durations according to the respective instructions. The vectors were then sequenced to testify the construction by Biological Genomics Institute (Shenzhen, China).
Western blot analysis
A lysis buffer containing the mammalian protein extraction reagent radioimmunoprecipitation assay (Beyotime, Haimen, China), a protease inhibitor cocktail (Roche, Basel, Switzerland), and phenylmethylsulfonyl fluoride (Roche) were used to lyse the cells after treatments. The protein concentration was detected by the Bradford method using Bradford detergent compatible (DC). Samples containing 20 μg of protein from each cell line were electrophoresed. The protein was transferred on to 0.22 μm nitrocellulose membranes (Millipore, Bedford, MA, USA). After the protein was blocked in 5% skim milk for 1 h at room temperature, the membranes were incubated with primary antibodies at 4°C overnight. After incubation with the corresponding secondary antibodies conjugated to horseradish peroxidase, the enhanced chemiluminescence chromogenic substrate from Griffin Biotech (Hong Kong, China) and photoplates from Kodak (Rochester, NY, USA) were used to detect specific bands on the membranes. Antibodies for HIF2A (Santa Cruz Biotechnology), FOXP3 (Cell Signaling Technology, Danvers, MA, USA), OCT4A (Cell Signaling Technology), ALDH1A1 (Santa Cruz Biotechnology), Notch1 (Cell Signaling Technology), and HES1 (Cell Signaling Technology) were applied as primary antibodies. Protein expression was semi-quantified by the control β-tubulin (Santa Cruz Biotechnology) or GAPDH (Santa Cruz Biotechnology). Then the gray scale ratio was assessed by Adobe Photoshop CS6 (San Jose, CA, USA). The photographs were the representatives for the triple experiments.
RNA extraction and quantitative reverse-transcriptase-PCR (qRT-PCR) analyses
Total RNA was extracted from the frozen tissues or cultured cells using TRIZOL reagent (Invitrogen). For qRT-PCR, a reverse transcription kit (Takara, Dalian, China) was used to synthesize cDNA. SYRB Real-time PCR Master Mix (Takara) was used for real-time PCR. The results were normalized with the expression of GAPDH or actin as a reference. The primers are shown in Supplementary Table 3. Real-time PCR and data collection were performed using a Bio-Rad CFX Manager 3.0 instrument (Bio-Rad, Hercules, CA, USA).
Cell proliferation assays
A MTT kit (Sigma) was used to assay cell proliferation and conducted according to the manufacturer’s instruction. Transfected and negative controlled H460 cells or H23 cells were harvested 24 h after transfection, and then cultured on 96-well plates for different periods. MTT 10 µl was added to each well and the OD490 was assessed hourly after 4 h.
TUNEL analysis of apoptosis
A TUNEL kit (Roche) was used for the detection and quantification of apoptosis at single cell level, based on labeling if the DNA strand broke. The assay was performed in accordance with the provided instructions.
Cell invasion assay
At 24 h after transfection, approximately 5 × 10^5 transiently transfected or negative controlled cells in serum-free media were cultured in the upper chamber for migration assays, 8 μm pore size (Millipore), and invasion assays with 20% Matrigel (Sigma). The lower chambers were filled with 600 ml of RPMI 1640 media containing 20% FBS. After 24 h of incubation at 37°C, the cells that had migrated or invaded through the membrane were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet (Sigma). The cells below the surface were photographed, and the cells in three random fields were counted.
Colony formation
After 3000 stably transfected cells were cultured in each well of a 6-well plate for 10–14 days, colonies containing more than 50 cells were counted.
Tumor-sphere formation
A total of 5000 stably transfected cells were cultured in each well of a 6-well ultra-low attachment plate (Corning, Wiesbaden, Germany) for 10 days. All the cells were cultured in Cancer Stem Premium (ProMab Biotechnologies, Richmond, CA, USA) at 37°C in humidified air with 5% CO2. Tumor spheres containing more than 50 cells were counted. The efficiency of tumor-sphere forming was calculated by dividing the number of tumor spheres formed (50 mm) by the original number of single cells seeded and expressed as the mean percentage of efficiency.
Co-immunoprecipitation
After HEK293NT cells were cotransfected with plasmids as shown in the figures for 48 h, the nuclear protein was extracted for immunoprecipitation. The corresponding antibodies were added to the protein. Then the mixture was incubated with continuing slow rotation overnight at 4°C to capture the targeted protein. Protein A/G-agarose beads (Santa Cruz Biotechnology) were added to the protein mix for a 4-h incubation at 4°C. After centrifuging and washing, the samples were used for western blot analysis.
Dual-luciferase reporter assay
H460 and H23 cells were plated in 12-well plates and cotransfected with various plasmids as indicated in the figures. Cells were collected 48 h after transfection, and luciferase activities were analyzed by the dual-luciferase reporter assay kit (Promega, Fitchburg, WI, USA). Reporter activity was normalized to the control Renilla.
Chromatin immunoprecipitation (CHIP) assay
CHIP assay was performed with a Magna CHIP kit (Merck, Darmstadt, Germany) according to the manufacturer’s instructions. H460 cells were sonicated to shear the chromatin to a manageable size. The antibody for HIF2A (Novus, Centennial, CO, USA) was used for immunoprecipitation. Real- time PCR was conducted to detect targeted DNA. The primers used are shown in Supplementary Table 3.
Tumor xenograft assay
Female nude mice (6–8 weeks old, 20–22 g) were provided by Laboratory Animal Service Center of CUHK, and kept on a 12-h light, 12-h dark cycle with free access to food and water. The nude mice were randomly divided into three groups that were subcutaneously injected respectively with 2 × 106 H460-HIF2A, H460-EGFR-AS1, and H460-control cells into the left and right flank of the mice for tumor xenograft experiments. Tumor size was measured every 3 days for 18 days by a micrometer. Tumor volume was calculated by length × width × width/2. Tumors were harvested and fixed in formalin for histological evaluation or snap-frozen in liquid nitrogen for both mRNA and protein preparations. No adverse event of the animals was observed in this study.
Statistical analysis
In this study, when the gene expression in the tumor tissue was higher than in the corresponding normal tissue, the corresponding patient would be defined as a high expression sample, and in the opposite case, the patient would be considered as a low expression sample. The sample size for the study was estimated using Rollin Brant’s Sample Size Calculators (www.stat.ubc.ca/~rollin/). We set alpha level at 0.05 and the desired power at 0.95 to estimate the number of mice needed for each group. Student’s t-tests and analyses of variance were used to analyze data with GraphPad Prim 6.0 software (GraphPad, La Jolla, CA, USA). p values of less than 0.05 were considered statistically significant.
Results
Effects of NNK on the expression of EGFR-AS1, HIF2A and FOXP3, and lung cancer cell stemness
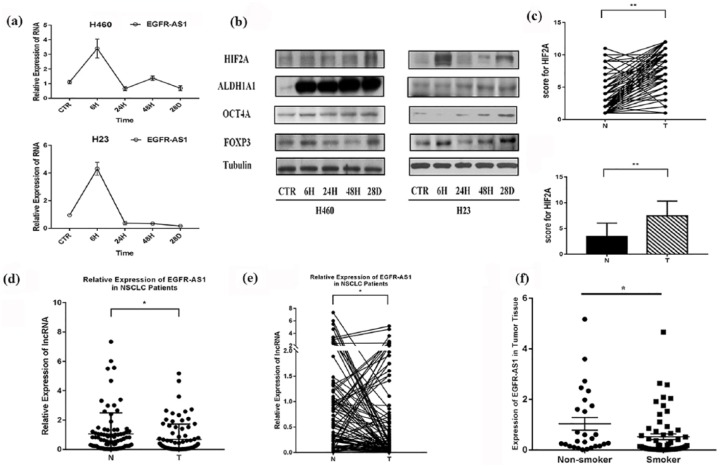
NSCLC cells were treated with NNK for different periods, and the expression of EGFR-AS1 was measured by real-time PCR, while the expression of FOXP3, HIF2A, ALDH1A1, and OCT4A, two lung cancer stem-cell markers,2,5,26,27 was evaluated by western blot analysis. It was found that at 6 h after NNK treatment, the expression of EGFR-AS1 began to decrease, whereas the expression of FOXP3, HIF2A, ALDH1A1, and OCT4A was increased along with the treatment (Figure 1(a) and (b)).
Figure 1.
Effects of smoking on the expression of EGFR-AS1, HIF2A, and FOXP3, and lung cancer cell stemness. (a) Two NSCLC cell lines were treated with NNK for different periods. The expression of EGFR-AS1 was increased significantly at first (p < 0.01), and then reduced from the elevated level (p < 0.01). (b) Two NSCLC cell lines were treated with NNK for different periods. The expression of FOXP3, HIF2A, and two lung cancer stem-cell markers (ALDH1A1 and OCT4A) was evaluated by western blot. (c) and (d) The expression of HIF2A in patient tissues was evaluated by immunohistochemistry (IHC). IRS was applied to quantify the expression of HIF2A in adjacent normal tissues (N) and tumor tissues (T), **p < 0.01. (e) and (f) Real-time PCR was used to evaluate the expression of long noncoding RNA EGFR-AS1 in samples from patients with NSCLC, *p < 0.05. (f) The correlation analysis showed that the expression of EGFR-AS1 was negatively associated with smoking status, **p < 0.01. The experiments were performed in triplicate at least. EGFR-ASI, EGFR antisense RNA 1; HIF2A, hypoxia-inducible factor-2A; IRS, immunoreactive score; NSCLC, non-small cell lung cancer; NNK, 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone; PCR, polymerase chain reaction.
Expression of EGFR-AS1 and HIF2A in NSCLC and their association with clinical characteristics
The expression of HIF2A in patient tissues was evaluated by IHC. It showed that the expression of HIF2A was upregulated in tumor tissues compared with adjacent nontumor tissues (Figure 1(c) and (d); Supplementary Figure 1). Real-time PCR was used to evaluate the expression of lncRNA EGFR-AS1 in patients with NSCLC, and its expression in tumor tissues was significantly downregulated compared with adjacent nontumor tissues (Figure 1(d) and (e)).
The correlation analysis showed that the expression of EGFR-AS1 was associated with smoking status and tumor diameter, and the expression of HIF2A with T stage (Table 1). The majority of the smoking patients were associated with the lower expression of EGFR-AS1, whereas most nonsmoking patients were associated with the higher expression of EGFR-AS1 (Figure 1(f)). The finding appeared to indicate that smoking was negatively related to the expression of EGFR-AS1 in patients with NSCLC (Figure 1(f)).
The influence of EGFR-AS1 upon FOXP3 expression and NSCLC cells stemness
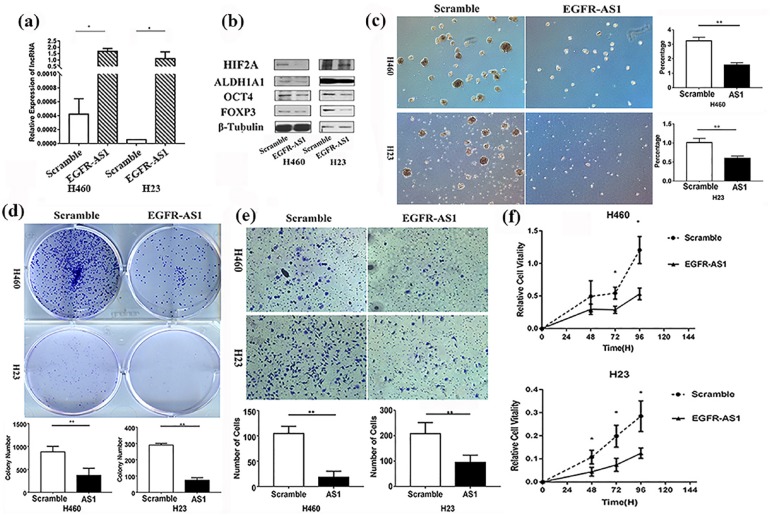
EGFR-AS1 expression plasmid was transfected into NSCLC cells to construct stable expression cell lines. The cell lines with EGFR-AS1 overexpression were successfully established, and the levels of HIF2A, FOXP3, and two CSC markers were significantly decreased in cells with EGFR-AS1 overexpression compared with the control (Figure 2(a) and (b)).
Figure 2.
The influence of EGFR-AS1 on FOXP3 expression and NSCLC cells stemness in vitro. (a) EGFR-AS1 plasmid was transfected into the NSCLC cells, and the expression of EGFR-AS1 was evaluated by qRT-PCR assay, *p < 0.05. (b) EGFR-AS1 plasmid was transfected into the NSCLC cells, and then the levels of HIF2A, FOXP3, and two CSC markers were evaluated by western blot. (c) EGFR-AS1 expression plasmid was transfected into NSCLC cells to construct stable expression cell lines, and then tumor-sphere formation was evaluated (left panel). Compared with scramble, EGFR-AS1 decreased the efficiency of tumor-sphere formation (right panel), **p < 0.01. (d) Colony-formation assay was conducted to evaluate the proliferative ability of the two cell lines **p < 0.01. (e) Transwell assay was conducted to evaluate the invasive ability of the two cell lines, **p < 0.01. (f) MTT assay was used to evaluate the proliferative ability of the two cell lines, *p < 0.05. The experiments were performed in triplicate at least. CSC, cancer stem cell; EGFR-ASI, EGFR antisense RNA 1; HIF2A, hypoxia-inducible factor-2A; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcriptase-polymerase chain reaction.
The tumor-sphere formation assay was conducted to evaluate the cancer stemness in these stable cell lines. Compared with control, the cell lines with EGFR-AS1 overexpression appeared to show lower cancer cell stemness (Figure 2(c)). We further performed colony-formation, transwell, and MTT assays to evaluate the cellular transformation, invasion, and proliferative ability, respectively, features of which are known to be closely related to the malignant ability of CSCs. The results showed that all these three features were significantly reduced in cells with EGFR-AS1 overexpression, compared with the control (Figure 2(d), (e) and (f)). Moreover, TUNEL analysis demonstrated that the cells with EGFR-AS1 overexpression had a higher apoptosis ratio than the control cells (Supplementary Figure 2).
Having established the inhibitory features of EGFR-AS1 in vitro, we used the mouse xenograft model to test its in vivo function. To this end, NSCLC H460 cells with stable EGFR-AS1 expression were subcutaneously injected into nude mice. The results indicated that the growth of the tumor formed by EGFR-AS1-overexpressed cells was significantly reduced compared with the tumor formed by the H460 cells transfected with the scramble (Supplementary Figure 3). In line with the reduction in tumor growth, the expression of HIF2A, FOXP3, ALDH1A1, and OCT4A in the EGFR-AS1-overexpresed tumor was much lower than in the tumor with the scramble (Supplementary Figure 3).
Effects of HIF2A on FOXP3 expression and NSCLC cell stemness
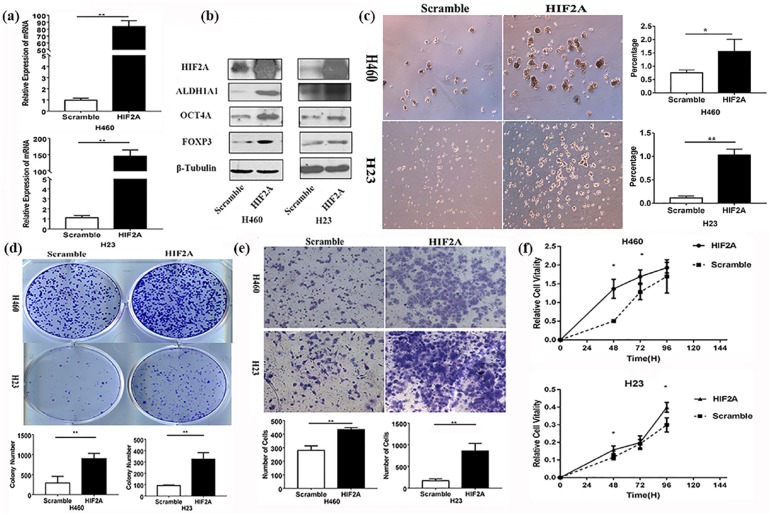
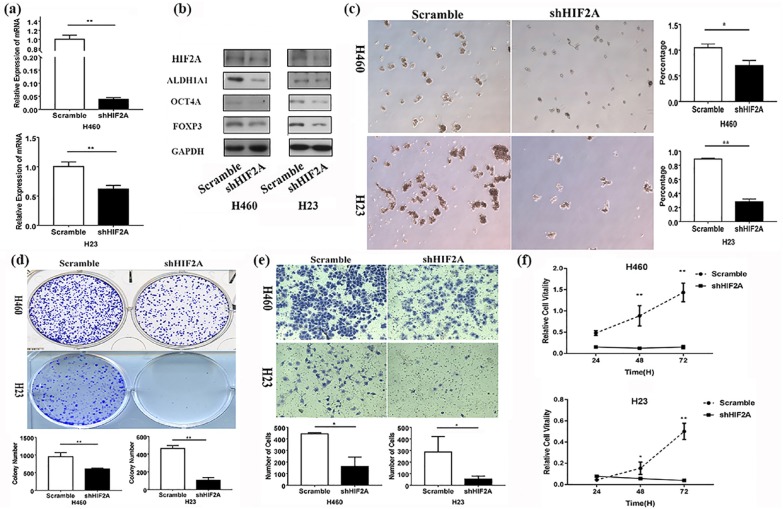
We found that the expression of FOXP3 and two CSC markers, ALDH1A1 and OCT4A, was significantly higher in NSCLC cells with HIF2A overexpression than those without (Figure 3(a) and (b)). The tumor-sphere formation assay showed that the HIF2A overexpression significantly enhanced sphere formation (Figure 3(c)). The cellular transformation, invasion, and proliferative ability were all markedly increased in NSCLC cells with HIF2A overexpression, compared with cells without HIF2A transfection, as was evident from the colony-formation, transwell, and MTT assays, respectively (Figure 3(d), (e) and (f)). Knockdown of HIF2A by its shRNA could reverse the above results (Figure 4). Further, TUNEL analysis revealed that apoptosis occurred less frequently in NSCLC cells with HIF2A overexpression compared with the control (Supplementary Figure 2). In our in vivo test, we found that the subcutaneous tumor formed by H460 cells with HIF2A overexpression was much larger than the tumor formed by cells without HIF2A overexpression, and the expression of FOXP3, ALDH1A1, and OCT4A in the former was much higher than the latter (Supplementary Figure 5).
Figure 3.
Effects of HIF2A on FOXP3 expression and NSCLC cell stemness in vitro. (a) After HIF2A plasmid was transfected into the NSCLC cell lines, the expression of HIF2A was evaluated by qRT-PCR assay, **p < 0.01. (b) After HIF2A plasmid was transfected into the NSCLC cell lines, the expression of FOXP3 and two CSC markers, ALDH1A1 and OCT4A, was evaluated by western blot. (c) After the stable expression of HIF2A in H23 and H460 cells, tumor-sphere formation was evaluated (left panel). Compared with scramble, HIF2A increased the efficiency of tumor-sphere formation (right panel) **p < 0.01, *p < 0.05. (d) Colony-formation assay was conducted to evaluate the proliferative ability of the four cell lines, **p < 0.01. (e) Transwell assay was conducted to evaluate the invasive ability of the two cell lines, **p < 0.01. (f) MTT assay was used to evaluate the proliferative ability of the four cell lines, **p < 0.01, *p < 0.05. The experiments were performed in triplicate at least. CSC, cancer stem cell; HIF2A, hypoxia-inducible factor-2A; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcriptase-polymerase chain reaction.
Figure 4.
Effects of shHIF2A on FOXP3 expression and NSCLC cell stemness in vitro. (a) After HIF2A shRNA was transfected into the NSCLC cell lines, the expression of HIF2A was evaluated by qRT-PCR assay **p < 0.01. (b) After HIF2A shRNA plasmid was transfected into the NSCLC cell lines, the expression of FOXP3 and two CSC markers, ALDH1A1 and OCT4A, was evaluated by western blot. (c) After the stable expression of HIF2A shRNA in H23 and H460 cells, tumor-sphere formation was evaluated (left panel). Compared with scramble, shHIF2A decreased the efficiency of tumor-sphere formation (right panel) **p < 0.01, *p < 0.05. (d) Colony-formation assay was conducted to evaluate the proliferative ability of the two cell lines, **p < 0.01. (e) Transwell assay was conducted to evaluate the invasive ability of the two cell lines, *p < 0.01. (f) MTT assay was used to evaluate the proliferative ability of the four cell lines, ** p < 0.05, *p < 0.01. The experiments were performed in triplicate at least. CSC, cancer stem cell; HIF2A, hypoxia-inducible factor-2A; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcriptase-polymerase chain reaction.
Effects of FOXP3 on lung cancer stemness
Regarding the impact of FOXP3 on lung cancer stemness, we found that the function of FOXP3 was similar to that of HIF2A. FOXP3 overexpression promoted CSC marker (ALDH1A1 and OCT4A) expression (Figure 5(a) and (b)) and tumor-sphere formation (Figure 5(c)), but inhibited apoptosis in NSCLC cells (Supplementary Figure 2). The effect of FOXP3 on lung cancer stemness was confirmed by the inhibitory test as the knockdown of FOXP3 by its shRNA reduced tumor-sphere formation and CSC marker (ALDH1A1 and OCT4A) expression (Figure 5(e), (f) and (g)).
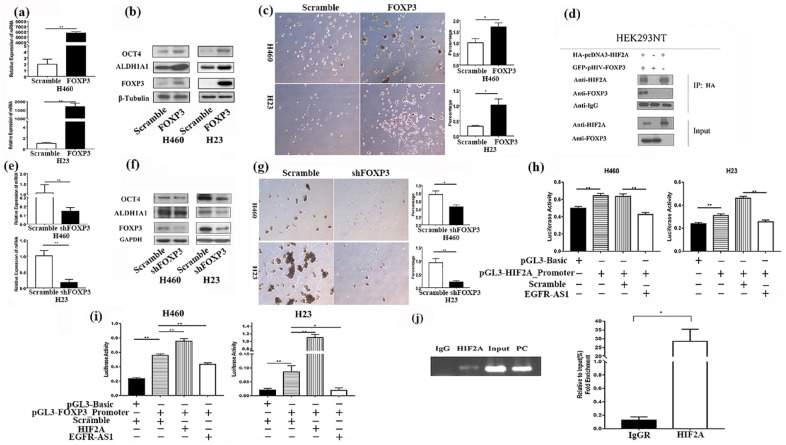
Figure 5.
The regulation of FOXP3 expression in NSCLC cells by EGFR-AS1/HIF2A and the effects of FOXP3 on lung cancer stemness through the Notch pathway. (a) After FOXP3 plasmid was transfected into the NSCLC cell lines, the expression of FOXP3 was evaluated by qRT-PCR assay, **p < 0.01. (b) After FOXP3 plasmid was transfected into the NSCLC cell lines, the expression of FOXP3 and two CSC markers, ALDH1A1 and OCT4A, was evaluated by western blot. (c) After the stable expression of FOXP3 in H23 and H460 cells, tumor-sphere formation was evaluated (left panel). Compared with scramble, FOXP3 increased the efficiency of tumor-sphere formation (right panel) **p < 0.01, *p < 0.05. (d) The immunoprecipitation experiment results demonstrated that HIF2A could bind with FOXP3. (e) After FOXP3 shRNA was transfected into the NSCLC cell lines, the expression of FOXP3 was evaluated by qRT-PCR assay, **p < 0.01. (f) After FOXP3 shRNA plasmid was transfected into the NSCLC cell lines, the expression of FOXP3 and two CSC markers, ALDH1A1 and OCT4A, was evaluated by western blot. (g) After the stable expression of FOXP3 shRNA in H23 and H460 cells, tumor-sphere formation was evaluated (left panel). Compared with scramble, shFOXP3 decreased the efficiency of tumor-sphere formation (right panel) **p < 0.01, *p < 0.05. (h) Luciferase report assay was conducted to evaluate the influence of EGFR-AS1 and HIF2A on the activity of HIF2A promoter, **p < 0.01. (i) Luciferase report assay was conducted to evaluate the influence of EGFR-AS1 and HIF2A on the activity of the FOXP3 promoter, **p < 0.01, *p < 0.05. (j) Chromatin immunoprecipitation assays were conducted to evaluate the influence of HIF2A on the FOXP3 promoter, *p < 0.05. The experiments were performed in triplicate at least. CSC, cancer stem cell; EGFR-ASI, EGFR antisense RNA 1; HIF2A, hypoxia-inducible factor-2A; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcriptase-polymerase chain reaction.
The regulation of FOXP3 expression in NSCLC cells by EGFR-AS1/HIF2A
The data above showed that EGFR-AS1 and HIF2A have opposite effects on FOXP3 expression (Figures 2 and 3). Here, we further investigated the possible mechanisms responsible. Our results demonstrated that HIF2A could bind with FOXP3, as was evident in the immunoprecipitation experiment (Figure 5(d)), and increase the activity of the FOXP3 promoter (Figure 5(i)). Furthermore, CHIP assays indicated that HIF2A was directly bound to the promoter of FOXP3 in NSCLC H460 cells (Figure 5(j)). In addition, we found that EGFR-AS1 could suppress the activities of both the HIF2A promoter and FOXP3 promoter (Figure 5(h) and (i)).
The Notch pathway in the regulation of NSCLC cell stemness by FOXP3
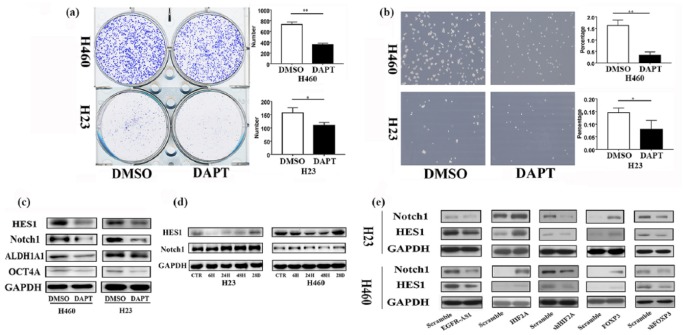
Our results showed that once the Notch signaling pathway was inhibited by the Notch inhibitor DAPT,23–25 it would be difficult for NSCLC cells to form colonies and tumor spheres (Figure 6(a) and (b)). Together with the reduced formation of colonies and tumor spheres, the expression of cancer stem biomarkers was significantly decreased (Figure 6(c)). These results support the pivotal role of the Notch signaling pathway in the regulation of NSCLC cell stemness.28 Our data further showed that after NNK could enhance the expression of Notch1 and HES1 (Figure 6(d)), the result of which was in agreement with the positive effect of NNK on the expression of NSCLC stem-cell markers ALDH1A1 and OCT4A (Figure 1). Furthermore, the overexpression of either HIF2A or FOXP3 significantly upregulated the levels of Notch1 and HES1 proteins, but the overexpression of EGFR-AS1 downregulated them; HIF2A or FOXP3 knockdown achieved opposite results (Figure 6(e)).
Figure 6.
The critical role of the Notch pathway in FOXP3-mediated NSCLC cell stemness. (a) Colony- formation assay was conducted to evaluate the impact of the Notch inhibitor DAPT on the proliferative ability of the NSCLC cell lines, **p < 0.01, *p < 0.05. (b) After the treatment of DAPT, tumor-sphere formation was assayed (left panel). Compared with DMSO, DAPT decreased tumor- sphere formation (right panel) **p < 0.01, *p < 0.05. (c) The expression of cancer stem markers was evaluated by western blot after DAPT treatment. (d) After NNK treatment, the expression of Notch1 and HES1 increased with the treatment time extended. (e) Western blot analysis was used to evaluate the influence of EGFR-AS1, HIF2A, and FOXP3 on the Notch pathway. The experiments were performed in triplicate at least. DAPT, N-[2S-(3,5-difluorophenyl) acetyl]-L-alanyl-2-phenyl-1,1-dimethylethyl ester-glycine; DMSO, dimethyl sulfoxide; EGFR-ASI, EGFR antisense RNA 1; HIF2A, hypoxia-inducible factor-2A; NNK, 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone; NSCLC, non-small cell lung cancer.
Discussion
In this study, we have identified a novel mechanism by which NNK promoted the expression of FOXP3 and HIF2A to enhance lung CSCs. Although NNK increased the expression of EGFR-AS1 in NSCLC cells at the early stage of treatment, it decreased the expression of EGFR-AS1 in the long term. According to our data, the transcription factor HIF2A could bind to the FOXP3 promoter to increase FOXP3 expression, promoting lung cancer cell stemness in the Notch1-dependent manner (Figure 7). In contrast to HIF2A, lncRNA EGFR-AS1 could suppress FOXP3 expression at least in part by reducing the activity of the FOXP3 promoter. Therefore, FOXP3 appears to be a key molecule in the promotion of the Notch1-dependent NSCLC cell stemness. The balance between HIF2A and lncRNA EGFR-AS1 may decide the level of FOXP3 and thus the fate of this FOPX3-mediated cancer stemness in NSCLC.
Figure 7.
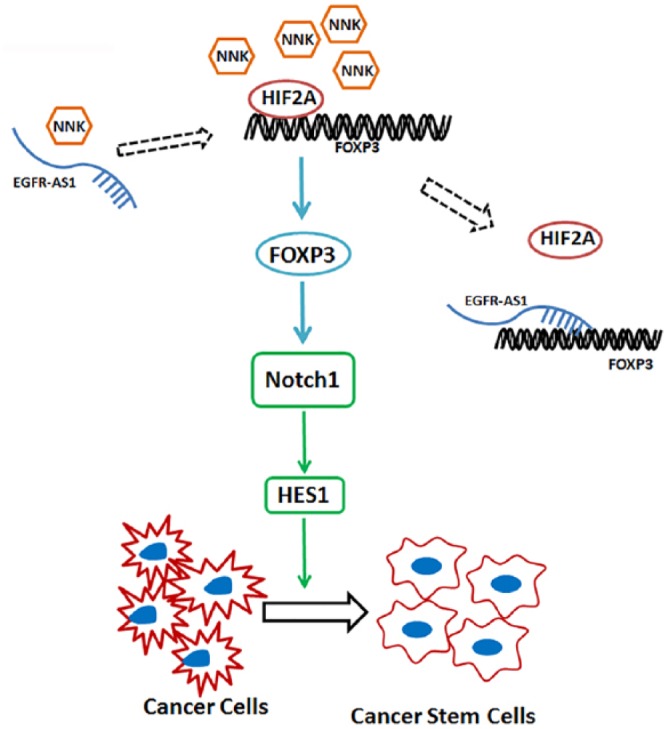
The potential mechanism of FOXP3-mediated regulation of cancer cell stemness. NNK could decrease EGFR-AS1 but increase HIF2A and FOXP3. The transcription factor HIF2A may bind to the FOXP3 promoter to increase the expression of FOXP3, promoting lung cancer cell stemness through the activation of the Notch pathway. In contract to HIF2A, the long noncoding RNA EGFR-AS1 may interact with the FOXP3 promoter to suppress the expression of FOXP3, thus inhibiting lung CSCs. CSC, cancer stem cells; EGFR-ASI, EGFR antisense RNA 1; HIF2A, hypoxia-inducible factor-2A; NNK, 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone.
NNK is known to upregulate the expression of CSC markers including ALDH and CD133 in NSCLC,29,30 indicating a positive role of NNK in the development or/and maintenance of lung CSCs. However, the molecular mechanism responsible remains unknown. Our current study has indicated that FOXP3 can be the key molecule in NNK-mediated lung CSCs. In this study, we first showed that NNK induced the expression of FOXP3. The increased FOXP3 was able to stimulate the expression of lung CSC markers, ALDH1A1 and OCT4A, and enhanced tumor-sphere formation. The positive role of FOXP3 in the enhancement of lung CSCs was confirmed by the inhibitory experiment in which FOXP3 siRNA was employed to block FOXP3. The involvement of FOXP3 in NSCLC cell stemness is also supported by our early result showing that FOXP3 induced NSCLC growth and metastasis by stimulating EMT and Wnt/β-catenin signaling,9 both of which are closely associated with lung CSCs.2,31
Having established the positive role of FOXP3 in lung CSCs, we explored how FOXP3 regulated lung CSCs. In this regard, the involvement of EGFR-AS1 and HIF2A was discovered. Our data showed that the expression of EGFR-AS1 was downregulated in cancer tissues of NSCLC compared with the adjacent normal tissues. EGFR-AS1 suppressed the expression of HIF2A and FOXP3, proliferation, metastasis, and cancer cell stemness. The correlation analysis revealed that the higher expression of EGFR-AS1 was associated with smaller tumor diameter, but negatively related to smoking status. Compared with nonsmoking patients, the percentage of smoking patients with a lower expression of EGFR-AS1 was much higher, suggesting that smoking may contribute to the lower expression of EGFR-AS1. In summary, the above findings appear to support an antitumor role of EGFR-AS1 in NSCLC. It is noted that our result is not in line with two other findings in which EGFR-AS1 was found to promote the growth or treatment resistance in hepatocellular carcinoma and head and neck squamous cell carcinoma.32,33
The oncogenic role of HIF2A in NSCLC has been documented.34 We confirmed this finding in our study as all NSCLC tumor tissues expressed HIF2A and 92% (80/87) of cases had a significant increase of HIF2A compared with the nontumor tissues. Significantly, we discovered that HIF2A can act on the FOXP3 promoter to stimulate its expression. Furthermore, HIF2A may bind to FOXP3 protein, which may enhance the oncogenic role of FOXP3. In contrast to HIF2A, our results have shown that EGFR-AS1 can suppress the activity of the FOXP3 promoter. Therefore, the balance between HIF2A and EGFR-AS1 may be critical for FOXP3 expression (Figure 6). Our findings have also revealed the involvement of Notch1 and HES1 in FOXP3-mediated stimulation of lung CSCs. This result is in line with the active roles of Notch1 and HES1 in the regulation of CSCs. It has been reported that HIF1A controls the expression of FOXP3 in Treg cells and some other cancer cells.35,36 According to our results, HIF2A could not only bind to the FOXP3 promoter in order to increase the expression of FOXP3, but also binds with FOXP3 protein, which may promote its oncogenic role in lung cancer cells. Therefore, HIF2A plays a positive role in the regulation of FOXP3 expression in NSCLC cells.
The Notch signaling pathway is highly conserved in cancer cells and plays a pivotal role in the linkages between angiogenesis and CSC self-renewal.37 As a result, it has received increased attention as a target for eliminating CSCs.38 Though the targeted therapy of inhibiting Notch can reduce tumor growth, the complete shutdown of the Notch pathway is unnecessary or even harmful for the therapeutic purpose.39 In this study, under the regulation of EGFR-AS1 and HIF2A, FOXP3 could activate the Notch signaling pathway by upregulating the expression of Notch1 and HES1, stimulating cancer cell stemness. Therefore, these findings have identified some downstream molecules in the Notch pathway, the targeting of which may control cancer cell stemness in NSCLC.
There were some limitations in this study. First, the sample size may not have been large enough and thus it may not have obtained some significant correlations between clinicopathological features and the biomarkers tested. Second, the mechanism study on EGFR-AS1/FOXP3 on lung CSCs has not been tested in vivo. Third, the relationship between smoking carcinogens and EGFR-AS1/FOXP3 needs further tests, for example, whether EGFR-AS1 overexpression can change the carcinogenic effect of NNK. Nevertheless, these limitations may point to the future direction along the lines of this study.
In conclusion, cigarette smoking carcinogen NNK may decrease the expression of EGFR-AS1, but enhance the expression of HIF2A and FOXP3 to promote lung CSCs by stimulating Notch1 and HES1 (Figure 7). The level of FOXP3 is positively regulated by HIF2A but negatively by EGFR-AS1 in NSCLC. Since lung cancer cell FOXP3 is important in the maintenance of lung CSCs, the downregulation of FOXP3 or its downstream molecules may offer a novel channel for the targeted therapy of NSCLC.
Supplemental Material
Supplemental material, Supplementary_materials_190228 for EGFR-AS1/HIF2A regulates the expression of FOXP3 to impact the cancer stemness of smoking-related non-small cell lung cancer by Haolong Qi, Shanshan Wang, Juekun Wu, Shucai Yang, Steven Gray, Calvin S.H. Ng, Jing Du, Malcolm J. Underwood, Ming-Yue Li and George G Chen in Therapeutic Advances in Medical Oncology
Acknowledgments
We thank Professors Jun Yu and Ling Qin for use of their facilities, and Dr Yujuan Dong, Dr Jianwei Ren, Dr Yi Liu, Dr Nuozhou Wang, Dr Jianqing Yu, Dr Hao Jia, Mr Rocky Ho, Mr Ernest Chak, and Miss Angel Kong for their technical assistance
Footnotes
Funding: This study was supported by a grant from the Research Grants Council of the Hong Kong SAR (No: CUHK462613), and the National Natural Science Foundation of China National (No: 81472742).
Conflict of interest statement: The authors declare no conflicts of interest in preparing this article.
Supplemental material: Supplemental material for this article is available online.
ORCID iD: George G Chen  https://orcid.org/0000-0001-7276-3830
https://orcid.org/0000-0001-7276-3830
Contributor Information
Haolong Qi, Department of Surgery, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong, China; Department of Hepatobiliary Surgery, Renmin Hospital of Wuhan University, Wuhan, China.
Shanshan Wang, Department of Otorhinolaryngology, Head and Neck Surgery, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong, China.
Juekun Wu, Department of Surgery, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, China.
Shucai Yang, Department of Clinical Laboratory, Pingshan District People’s Hospital of Shenzhen, Shenzhen, China.
Steven Gray, Thoracic Oncology Research Group, Trinity Centre for Health Sciences, St James’s Hospital, Dublin, Ireland.
Calvin S.H. Ng, Department of Surgery, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong, China
Jing Du, Peking University Shenzhen Hospital, Shenzhen, China.
Malcolm J. Underwood, Department of Surgery, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong, China
Ming-Yue Li, Department of Surgery, the Chinese University of Hong Kong, Prince of Wales Hospital, Shatin, NT, Hong Kong, China; Shenzhen Research Institute, The Chinese University of Hong Kong, Shenzhen, Guangdong, China.
George G Chen, Department of Surgery, the Chinese University of Hong Kong, Prince of Wales Hospital, Shatin, NT, Hong Kong, China; Shenzhen Research Institute, The Chinese University of Hong Kong, Shenzhen, Guangdong, China.
References
- 1. Brainard J, Farver C. The diagnosis of non-small cell lung cancer in the molecular era. Mod Pathol. Epub ahead of print 2 January 2019. DOI: 10.1038/s41379-018-0156-x. [DOI] [PubMed] [Google Scholar]
- 2. Heng WS, Gosens R, Kruyt FAE. Lung cancer stem cells: origin, features, maintenance mechanisms and therapeutic targeting. Biochem Pharmacol 2019; 160: 121–133. [DOI] [PubMed] [Google Scholar]
- 3. Liu Y, Yang SC, Li MY, et al. Tumorigenesis of smoking carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone is related to its ability to stimulate thromboxane synthase and enhance stemness of non-small cell lung cancer stem cells. Cancer Lett 2016; 370: 198–206. [DOI] [PubMed] [Google Scholar]
- 4. Huang WC, Kuo KT, Adebayo BO, et al. Garcinol inhibits cancer stem cell-like phenotype via suppression of the Wnt/Beta-catenin/STAT3 axis signalling pathway in human non-small cell lung carcinomas. J Nutr Biochem 2018; 54: 140–150. [DOI] [PubMed] [Google Scholar]
- 5. Liu T, Wu X, Chen T, et al. Downregulation of DNMT3a by MIR-708–5p inhibits lung cancer stem cell-like phenotypes through repressing Wnt/Beta-catenin signaling. Clin Cancer Res 2018; 24: 1748–1760. [DOI] [PubMed] [Google Scholar]
- 6. Giroux Leprieur E, Tolani B, Li H, et al. Membrane-bound full-length Sonic Hedgehog identifies cancer stem cells in human non-small cell lung cancer. Oncotarget 2017; 8: 103744–103757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sosa Iglesias V, Giuranno L, Dubois LJ, et al. Drug resistance in non-small cell lung cancer: a potential for notch targeting? Front Oncol 2018; 8: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jia H, Qi H, Gong Z, et al. The expression of Foxp3 and its role in human cancers. Biochim Biophys Acta Rev Cancer 2019; 1871: 170–178. [DOI] [PubMed] [Google Scholar]
- 9. Yang S, Liu Y, Li MY, et al. Foxp3 promotes tumor growth and metastasis by activating Wnt/Beta-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer 2017; 16: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qin AD, Wen ZK, Zhou Y, et al. Microrna-126 regulates the induction and function of Cd4(+) Foxp3(+) regulatory T cells through Pi3k/Akt pathway. J Cell Mol Med 2013; 17: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saki N, Abroun S, Soleimani M, et al. The roles of miR-146a in the differentiation of jurkat T-lymphoblasts. Hematol 2014; 19: 141–147. [DOI] [PubMed] [Google Scholar]
- 12. Jiang RQ, Tang JW, Chen Y, et al. The long noncoding RNA LNC-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nature Commun 2017; 8: 15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zemmour D, Pratama A, Loughhead SM, et al. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Natl Acad Sci U S A 2017; 114: E3472–E3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang GL, Jiang BH, Rue EA, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 1995; 92: 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clambey ET, McNamee EN, Westrich JA, et al. Hypoxia-inducible factor-1 alpha-dependent induction of Foxp3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci U S A 2012; 109: E2784–E2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu LP, Hu BG, Ye C, et al. HBx mutants differentially affect the activation of hypoxia-inducible factor-1α in hepatocellular carcinoma. Br J Cancer 2014; 110: 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsu HL, Liao PL, Cheng YW, et al. Chloramphenicol induces autophagy and inhibits the hypoxia inducible factor-1 alpha pathway in non-small cell lung cancer cells. Int J Mol Sci 2019; 20: E157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tu CY, Cheng FJ, Chen CM, et al. Cigarette smoke enhances oncogene addiction to C-met and desensitizes EGFR-expressing non-small cell lung cancer to EGFR TKIs. Mol Oncol 2018; 12: 705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du XM, Qi F, Lu SY, et al. Nicotine upregulates FGFR3 and RB1 expression and promotes non-small cell lung cancer cell proliferation and epithelial-to-mesenchymal transition via downregulation of MIR-99b and MIR-192. Biomed Pharmacother 2018; 101: 656–662. [DOI] [PubMed] [Google Scholar]
- 20. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018; 553: 446–454. [DOI] [PubMed] [Google Scholar]
- 21. Li MY, Liu Y, Liu LZ, et al. Estrogen receptor alpha promotes smoking-carcinogen-induced lung carcinogenesis via cytochrome P450 1B1. J Mol Med 2015; 93: 1221–1233. [DOI] [PubMed] [Google Scholar]
- 22. Guo J, Kim D, Gao J, et al. IKBKE is induced by stat3 and tobacco carcinogen and determines chemosensitivity in non-small cell lung cancer. Oncogene 2013; 32: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Hu BD, Guo J, Ye YZ, et al. Specific inhibitor of NOTCH3 enhances the sensitivity of NSCLC cells to gemcitabine. Oncol Rep 2018; 40: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gelsomino L, Panza S, Giordano C, et al. Mutations in the estrogen receptor alpha hormone binding domain promote stem cell phenotype through notch activation in breast cancer cell lines. Cancer Lett 2018; 428: 12–20. [DOI] [PubMed] [Google Scholar]
- 25. Dai G, Deng S, Guo WC, et al. Notch pathway inhibition using DAPT, a gamma-secretase inhibitor (GSI), enhances the antitumor effect of cisplatin in resistant osteosarcoma. Mol Carcinogenesis 2019; 58: 3–18. [DOI] [PubMed] [Google Scholar]
- 26. Po A, Silvano M, Miele E, et al. Noncanonical GLI1 signaling promotes stemness features and in vivo growth in lung adenocarcinoma. Oncogene 2017; 36: 4641–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prabavathy D, Swarnalatha Y, Ramadoss N. Lung cancer stem cells-origin, characteristics and therapy. Stem Cell Invest 2018; 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zou B, Zhou XL, Lai SQ, et al. Notch signaling and non-small cell lung cancer. Oncol Lett 2018; 15: 3415–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirata N, Yamada S, Sekino Y, et al. Tobacco nitrosamine NNK increases ALDH-positive cells via ROS-Wnt signaling pathway in A549 human lung cancer cells. J Toxicol Sci 2017; 42: 193–204. [DOI] [PubMed] [Google Scholar]
- 30. Balbo S, Upadhyaya P, Villalta PW, et al. DNA adducts in aldehyde dehydrogenase-positive lung stem cells of a/J Mice treated with the tobacco specific lung carcinogen 4-(Methylnitrosamino)-1-(3-Pyridyl)-1-Butanone (NNK). Chem Res Toxicol 2013; 26: 511–513. [DOI] [PubMed] [Google Scholar]
- 31. Choi SI, Kim SY, Lee JH, et al. Osteopontin production by TM4SF4 signaling drives a positive feedback autocrine loop with the STAT3 pathway to maintain cancer stem cell-like properties in lung cancer cells. Oncotarget 2017; 8: 101284–101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qi HL, Li CS, Qian CW, et al. The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. Tumour Biol 2016; 37: 1079–1089. [DOI] [PubMed] [Google Scholar]
- 33. Tan DSW, Chong FT, Leong HS, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med 2017; 23: 1167–1175. [DOI] [PubMed] [Google Scholar]
- 34. Giatromanolaki A, Koukourakis MI, Sivridis E, et al. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer 2001; 85: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feldhoff LM, Rueda CM, Moreno-Fernandez ME, et al. Il-1beta induced HIF-1alpha inhibits the differentiation of human Foxp3(+) T cells. Sci Rep 2017; 7: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alcantara-Hernandez M, Torres-Zarate C, Perez-Montesinos G, et al. Overexpression of hypoxia-inducible factor 1 alpha impacts Foxp3 levels in mycosis fungoides-cutaneous T-cell lymphoma: clinical implications. Int J Cancer 2014; 134: 2136–2145. [DOI] [PubMed] [Google Scholar]
- 37. Pannuti A, Foreman K, Rizzo P, et al. Targeting notch to target cancer stem cells. Clin Cancer Res 2010; 16: 3141–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Venkatesh V, Nataraj R, Thangaraj GS, et al. Targeting notch signalling pathway of cancer stem cells. Stem Cell Investig 2018; 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hayashi T, Gust KM, Wyatt AW, et al. Not all NOTCH is created equal: the oncogenic role of NOTCH2 in bladder cancer and its implications for targeted therapy. Clin Cancer Res 2016; 22: 2981–2992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_materials_190228 for EGFR-AS1/HIF2A regulates the expression of FOXP3 to impact the cancer stemness of smoking-related non-small cell lung cancer by Haolong Qi, Shanshan Wang, Juekun Wu, Shucai Yang, Steven Gray, Calvin S.H. Ng, Jing Du, Malcolm J. Underwood, Ming-Yue Li and George G Chen in Therapeutic Advances in Medical Oncology