Abstract
The increasing use of antibiotics is being driven by factors such as the aging of the population, increased occurrence of infections, and greater prevalence of chronic diseases that require antimicrobial treatment. The excessive and unnecessary use of antibiotics in humans has led to the emergence of bacteria resistant to the antibiotics currently available, as well as to the selective development of other microorganisms, hence contributing to the widespread dissemination of resistance genes at the environmental level. Due to this, attempts are being made to develop new techniques to combat resistant bacteria, among them the use of strictly lytic bacteriophage particles, CRISPR–Cas, and nanotechnology. The use of these technologies, alone or in combination, is promising for solving a problem that humanity faces today and that could lead to human extinction: the domination of pathogenic bacteria resistant to artificial drugs. This prospective paper discusses the potential of bacteriophage particles, CRISPR–Cas, and nanotechnology for use in combating human (bacterial) infections.
Keywords: multidrug-resistant bacteria, bacteriophage particles, phage therapy, CRISPR–Cas, nanotechnology
Bacterial Resistance
Since their discovery in 1929, antibiotics have been widely used in human and veterinary medicine, either for treatments or in attempts to prevent bacterial infections. The excessive use of antibiotics, whether for prevention or treatment, has significantly increased the level of bacterial resistance worldwide (Ali et al., 2018). The associated numbers of human deaths are alarming, reaching 50,000 per year in the United States and Europe (Simlai et al., 2016), with an estimated 10 million deaths per year by 2050, surpassing the current deaths resulting from all types of cancer (approximately 8.2 million) (Jansen et al., 2018).
The first list of antibiotic-resistant pathogens was published by the World Health Organization (WHO) in 2017. This list showed that out of the 12 resistant pathogens, seven were noted to be resistant to beta-lactam antibiotics. Consequently, there is renewed focus on the production of new antibiotics, establishing a goal for future research strategies (WHO, 2017).
The overuse and misuse of antibiotics in humans have led to the selective emergence of bacteria resistant to the currently available antibiotics, as well as resistant non-pathogenic microbiota, hence leading to the generalized dissemination of resistance genes at the environmental level (Nitsch-Osuch et al., 2016). There is greatest concern when this phenomenon occurs with Enterococcus spp., Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., together given the acronym ESKAPE, which highlights the ability of these microorganisms to escape the action of antimicrobial agents (Boucher et al., 2009).
Antimicrobial resistance has become globalized, following the first reports of its appearance in India, with its subsequent spread to Pakistan, the United States, Canada, Japan, and the United Kingdom (Rios et al., 2016). This resistance can occur in different ways, depending on the acquired and selective genetic changes or insertion of external genes, which leads to previously non-existent responses. Several mechanisms of resistance have emerged in recent times, including alteration of the target (by a DNA gyrase), increased efflux (export of a drug out of the microorganism), inactivation of fluoroquinolones (by an aminoglycoside N-acetyltransferase), inhibition of the 30S ribosomal subunit (by aminoglycosides), and protection of the target by DNA-binding proteins (the Qnr family) (Redgrave et al., 2014; Munita and Arias, 2016; Kapoor et al., 2017).
Some of these changes are already well known, such as alteration of the chemical structure of antimicrobial agents (Alekshun and Levy, 2007), decrease of the concentration of the antimicrobial at its site of action (Gonzalez-Bello, 2017; Willers et al., 2017), changes in the target of antimicrobial action (Sieradzki and Markiewicz, 2004), and alteration of membrane permeability (Hao et al., 2018). There are mechanisms of permeability reduction that do not involve porin expression, such as changes in the cell envelope of P. aeruginosa that are associated with resistance to polymyxin B (Falagas and Kasiakou, 2005). In addition to antibiotics that act on the cell wall, such as penicillins and glycopeptides, the activities of other antimicrobials that act on the bacterial ribosome may also decrease due to changes in their primary target. This phenomenon mainly affects macrolides and tetracyclines (Poehlsgaard and Douthwaite, 2005; Wu et al., 2005).
The presence of these mechanisms of resistance is increasingly common in large numbers of microorganisms, due to the selective pressure exerted by antimicrobials, leading to a natural selection that results in the dominance of certain groups of resistant bacteria, with concomitant death of sensitive microorganisms (Tello et al., 2012).
In a meta-analysis carried out by Bell et al. (2014), in which 243 studies were evaluated, it was concluded that “Increased consumption of antibiotics may not only produce greater resistance at the individual patient level but may also produce greater resistance at the community, country, and regional levels, which can harm individual patients.” Another study of the same year evaluated the consumption of antibiotics worldwide between 2000 and 2010. It was found that the consumption of antibiotics increased by around 36%, with the countries of the BRICS group (Brazil, Russia, India, China, and South Africa) accounting for approximately 76% of the increase (Van Boeckel et al., 2014).
Therefore, the data reflect a worrying trend regarding the treatment of infectious diseases, since not only are these drugs being increasingly used (Van Boeckel et al., 2014), but also their use is directly proportional to the increase in resistance indicators (Bell et al., 2014). In the absence of any significant discovery of new molecules for the control of resistant microorganisms (Hogberg et al., 2010), there is an urgent need for redefining the relationship of humans with infectious diseases.
In summary, the problem faced in relation to bacterial resistance is a concern that must be urgently addressed, since functional meta-genomic studies of soil microorganisms have revealed a wide range of genetic determinants that confer resistance to antibiotics, of which only one fraction has been described in human pathogens (Forsberg et al., 2014).
Hence, there is a pressing need for a new generation of antimicrobials able to mitigate the spread of antibiotic resistance and preserve beneficial microbiota. Among the possibilities for the solution of problems related to bacterial resistance, the use of nanotechnology, CRISPR–Cas9, and therapy with bacteriophage particles can be highlighted as potential future strategies. These techniques could be employed individually to directly combat microorganisms, as well as in combination in integrated strategies.
The scientific community has indicated that there are no perspectives for any significant clinical introduction of new antimicrobials in the short term. The main recommended approach is rational use of the classical antibiotics that have been used for the past 50 years, together with techniques that enhance their activity. This may be achieved using substances that increase antibiotic activity by reducing or blocking the resistance mechanism, such as beta-lactamase, efflux pump, and quorum sensing inhibitors, as well as bacteriophages and new drug delivery systems, among other techniques (Moo et al., 2019; Mulani et al., 2019; Pham et al., 2019; Vikesland et al., 2019).
CRISPRs
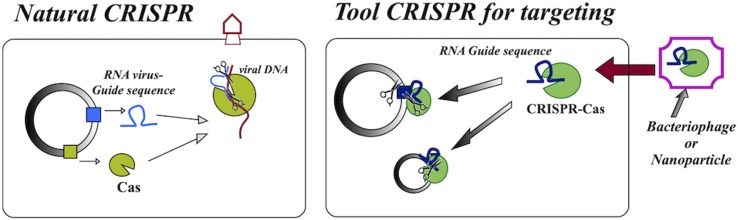
CRISPRs (clustered regularly interspaced short palindromic repeats) are adaptive immune systems derived from bacteria and archaea. CRISPR–Cas systems use RNA for target DNA recognition and the Cas enzyme for subsequent destruction of nucleic acids, so they require only one protein for binding and cleavage. Due to this simplicity, researchers have developed a new molecular tool based on natural CRISPRs ( Figure 1 ). This tool has different applications, one of them being the possibility of antimicrobial action, since they are cytotoxic systems that can be directed to kill bacteria, immunizing them against resistant plasmids (Sorek et al., 2013;Bikard et al., 2014; Hsu et al., 2014).
Figure 1.
Schematic drawing showing the natural CRISPR–Cas complex found in bacteria, which functions as an “immune system” against viruses, and the CRISPR–Cas tool used as an agent, based on the complex naturally present in bacteria.
For medical purposes, CRISPR–Cas systems can enable the selective and specific removal of microorganisms. Although there are other antimicrobial approaches, they offer only partial solutions, while CRISPR systems are generalized and programmable strategies (Gomaa et al., 2014) that can be employed to selectively and quantitatively remove individual bacterial strains, based purely on sequence information, hence creating opportunities in the treatment of multidrug-resistant infections.
In studies of the use of these systems as antimicrobials, Gomaa et al. (2014) reported that both heterologous and endogenous systems could selectively kill bacterial species and strains. It was shown that all sequences in the target genome led to cell death, suggesting that, theoretically, any genomic location could be a distinct target for antimicrobials based on CRISPRs. Another possibility would be the use of this technology for antimicrobial action using RNA-guided nucleases (RGNs), targeting specific resistance genes or undesirable polymorphisms, allowing programmable remodeling of the microbiota (Citorik et al., 2014).
In a study carried out by Fuente-Núñez and Lu (2017) concerning CRISPR–Cas constructs designed to function as precision antimicrobials, these were shown to be capable of eliminating drug-resistant microbes, with CRISPR–Cas selectively targeting genes involved in antibiotic resistance, biofilm formation, and virulence. However, although studies show that CRISPRs are effective, there are still problems to be overcome in relation to an efficient delivery vehicle, which is the next step for the implementation of CRISPR–Cas systems as antimicrobial agents (Beisel et al., 2014). Focusing on the problem of CRISPR transportation and delivery, Pan et al. (2017) were able to identify eight depolymerases in the multi-host bacteriophage K64-1, which, together with K64dep (S2-5), characterized elsewhere, gave a total of nine capsule depolymerases.
Currently, obtaining bacteriophages as carriers of CRISPRs is still a challenge. Shen et al. (2018) succeeded in obtaining positive results in studies aimed at obtaining a Klebsiella bacteriophage by genome alteration, which was suggested as a possibility for the use of targeted CRISPRs. One option is to use nanotechnology for the delivery of CRISPRs, which could provide surface modifications that ensure the desired specificity (Yan et al., 2015). As pointed out by Pursey et al., (2018), there is still a great deal to discover concerning the use of CRISPR–Cas in the fight against resistant bacteria, with further research especially needed in relation to its safe use.
Another concern is the possibility that bacteria could present resistance against CRISPR–Cas, since the original mechanisms are present in them. However, in a study by Chen et al. (2019), performed with multidrug-resistant Shigella, it was shown that the bacteria that presented resistance genes also presented a decrease in the activity of natural CRISPR–Cas.
If we consider the different possibilities of target genes for CRISPR–Cas, we can conclude that there is a need for an interdisciplinary study, where there is collaboration of researchers who study sequences, find a safe way of delivery, and evaluate the existence of resistance to technology. Different studies show that bacteria tend to store different genes, and different combinations between virulence and resistance are an alarming threat, as it suggests the feasibility of adaptability. A study carried out by Oliveira Santos et al. (2018), where they showed the possible adaptability of the KPC-2 gene to different mobile elements, is an example of the need to consider different possibilities for the application of CRISPR–Cas. Regarding the onset of carbapenem-resistant K. pneumoniae, recent publication showed the introduction of two new DNA editing systems. One is the plasmid pCasKP-pSGKP and the other is the plasmid system pBECKP, where both systems showed efficiency in genome editing, which will facilitate further investigations for treatment of resistance to carbapenems (Wang et al., 2018).
Although the CRISPR–Cas tool offers a new possibility of fighting multidrug bacteria, some studies show that they do not present activity in some strains, as demonstrated by Hullahalli et al. (2017, 2018) in studies with Enterococcus faecalis where they present a study that determines the genetic basis of phenotypes associated with CRISPR–Cas tolerance, showing the importance of having a better knowledge of the response of organisms and possible strategies for dealing with conflicts induced by the use of CRISPRS, which may lead to tolerant phenotypes to this tool. Therefore, these studies show that knowledge of the genome and the metabolic pathways of the different resistant multidrug bacteria should be investigated so that resistance problems will not occur in the future in relation to new strategies used to fight resistant bacteria.
Nanotechnology in the Fight Against Resistant Bacteria
Nanotechnology applied to the synthesis of new antibiotics is an important approach, since the use of nanometric size materials can result in greater contact between the compound and the bacteria, with improved bioavailability, increased absorption, faster passage of the drug into the cell, and enhanced mucoadhesion. There is also the possibility of producing controlled release systems for the targeted delivery of encapsulated or surface adsorbed drugs (Zaidi et al., 2017;Jamil and Imran, 2018). One new approach is to use nanoparticles (NPs) of a metal such as silver, which can affect the bacterial respiration system, inducing the generation of reactive oxygen species (ROS). This approach could be used synergistically with antimicrobials, with effects such as inhibition and alteration of the synthesis of the cell wall, as well as its rupture (Shahverdi et al., 2007; Kumar et al., 2018).
One of the concerns regarding the use of nanoparticles is in relation to the resistance that bacteria can present to them, or the possibility of stimulating the transmission of MultiDrug-Resistant (MDR) genes. An example is provided by the work of Ansari et al. (2014), where Al2O3 nanoparticles were observed to promote the horizontal conjugative transfer of MDR genes, hence increasing the resistance to antibiotics.
The use of NPs to eliminate microorganisms can involve microbicidal or microbiostatic effects. In the latter case, the growth of bacteria is interrupted and the metabolic activities are halted, with microbial death then induced by the immune cells of the host. Nanotechnology can also solve problems related to drug solubility, since encapsulation can improve permeation through the membrane, increase circulation times, and enhance efficiency, while there is also the possibility of directing the drug towards the desired site of action in the body (Rodzinski et al., 2016).
The use of nanoparticles appears to have potential for the treatment of infectious diseases, especially considering that NPs may be able to access locations where the pathogens are present. However, there are a number of issues to be resolved, such as the scarcity of toxicity data, few existing preclinical studies, and the need for regulation (Zaidi et al., 2017).
Polymeric Nanoparticles and Nanocrystals
The use of polymeric nanocapsules as carriers for antibiotics, or the use of drug nanocrystals that are stable during delivery, can be successfully applied to a range of commonly used drugs. Polylactide-co-glycolide (PLGA) is an especially useful substance that can be employed in nanotechnological drug delivery applications (Kalhapure et al., 2014; Hemeg, 2017;Boya et al., 2017; Shaaban et al., 2017).
Hong et al. (2017) used bacitracin A (BA) modified with PLGA for synthesis of nano-BA, resulting in a core–shell structure with an average diameter of 150 nm. It was found that the nanoparticles strongly increased the antibacterial activity, than does free BA, with effective inhibition of the growth of various types of Gram (+) and Gram (−) bacteria. The formulation provided improved wound healing in rats than did use of a commercial Polysporin® ointment.
Yu et al. (2016) reported the development of a multifunctional release system with encapsulation of gentamicin sulfate/zirconium bis(monohydrogen orthophosphate) (α-ZrP) using chitosan (CHI). The formulation (α-ZrP CHI) extended the release of the drug, than did unencapsulated α-ZrP, which was attributed to the unique lamellar structure and the CHI encapsulation. The methodology provided a model for the future development of new delivery vehicles.
Shaaban et al. (2017) reported that nanoantibiotics produced by incorporating imipenem in PLGA or PCL nanocapsules provided better results, than did classical imipenem. The nanoencapsulated formulations showed antimicrobial and anti-adherent activities in evaluations using clinical isolates of imipenem-resistant bacteria.
Other types of nanoparticles that have received attention are lipid nanoparticles (liposomes) (Derbali et al., 2019) and nanoceramics applied in orthopedic surgeries where systemic drug administration has limitations (Kumar and Madhumathi, 2016).
Gaspar et al. (2017) reported the use of solid lipid nanoparticles containing rifabutin (RFB) for pulmonary administration to treat tuberculosis. The nanoparticles increased the activity of the drug against M. tuberculosis infection, suggesting that RFB-solid lipid nanoparticles (SLN) encapsulation could be a promising approach for tuberculosis treatment. A major advantage of encapsulation is that it provides sustained release of the drug, resulting in greater efficiency of treatment, as well as easier absorption, enabling satisfactory results to be achieved with a smaller amount of the active agent.
Although the use of nanoparticles can be advantageous, some studies have shown that the microenvironment where they are released (such as blood and lung fluid) may alter the creation of the nanoparticle–pathogen complex, due to the formation of a corona around the nanoparticle. Siemer et al. (2019) exposed nanoparticles to different bacteria and showed that formation of the pathogen–nanoparticle complex was assisted by its small size and that the presence of a corona significantly inhibited formation of the complex. Therefore, in addition to in vitro analyses, new studies are needed that consider the microenvironment in which the nanoparticle will be released and exert its action.
Metallic Nanoparticles
The use of metallic nanoparticles can be a good option in the fight against resistant bacteria. Studies have reported the synthesis and use of different nanoparticulate metals, metal oxides, metal halides, and bimetallic materials showing antimicrobial activity. Nanoparticles have been synthesized consisting of Ag, Au, Zn, Cu, Ti, and Mg, among other metals (Zakharova et al., 2015; Hajipour et al., 2012;Sunitha et al., 2013; Dizaj et al., 2014; He et al., 2016; Senarathna et al., 2017;Eymard-Vernain et al., 2018). However, consideration should be given to their potential toxicity (Lima et al., 2012; Dakal et al., 2016; Durán et al., 2016a).
Eymard-Vernain et al. (2018) showed that MgO nanoparticles presented bactericidal action, mainly affecting the expression of genes related to oxidative stress, together with membrane alteration. Verma et al. (2018) reported excellent antibacterial activity of ZnO nanoparticles, with a size-dependent effect, since the use of smaller nanoparticles resulted in more ROS and increased cell membrane rupture.
Other studies have investigated the bactericidal potential of carbon nanotubes, either plain or functionalized, as well as their use to assist the transport and translocation of antibiotics (Cong et al., 2016; Mocan et al., 2017).
With the development of nanotechnology, many studies have been carried out concerning the application of nanoparticles as antimicrobials. These nanomaterials present different diameters, structures, and modes of action. Some of them have produced good results, showing that nanotechnology can be used as one of the strategies in the fight against multidrug-resistant bacteria in the future (Supplementary Table 1).
Silver nanoparticles are the most studied metallic nanoparticles, with their antimicrobial activity having been recognized by the United States Food and Drug Administration (FDA) since the year 1920. The mechanisms of action of silver nanoparticles (AgNP) on bacteria have been exhaustively investigated. There is a consensus that adhesion of the nanoparticles to the cell membrane can lead to electrostatic changes, porosity alteration, rupture, leakage of cytoplasmic content, interference in bacterial respiratory processes, blocking of enzyme activity, and DNA destruction. It has also been observed that there is the production of ROS, with consequent effects on the DNA (Choi and Hu, 2008; Durán et al., 2010; Prabhu and Poulose, 2012; Rai et al., 2012; Kon and Rai, 2013; Yuan et al., 2017).
The adhesion of nanoparticles to bacterial membranes mainly occurs due to the presence of proteoglycans (Kim et al., 2017) and results in rupture or increased porosity of the membrane. This enables access of the nanoparticles into the cell, where they can interact with enzymes and DNA (Grigor’eva et al., 2013; Kasithevar et al., 2017). AgNPs may also interact with membrane proteins, leading to cell stress, or may interact with the lipid part of the membrane, affecting its fluidity (Morones et al., 2005; Chwalibog et al., 2010). Some studies have suggested that the observed effects are actually caused by silver ions released from AgNPs (Jung et al., 2008; Xiu et al., 2011; Xiu et al., 2012; Chernousova and Epple, 2013). Accordingly, the AgNPs only act as vehicles for the delivery of ions that cause adverse effects in the respiratory chain and protein synthesis, as well as DNA alterations (Chen et al., 2011; Li et al., 2014).
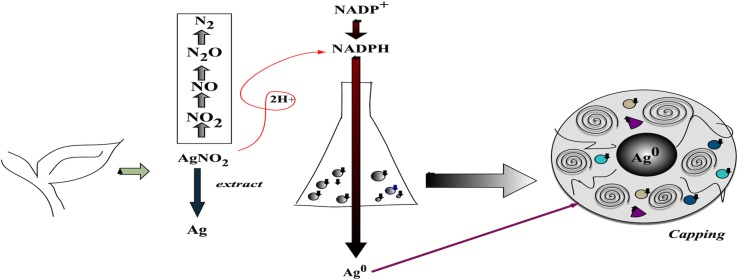
The biogenic synthesis of silver nanoparticles ( Figure 2 ) has received increasing attention in recent years. These nanoparticles present positive characteristics in terms of their improved stability and dispersion, due to the coating formed during the synthesis. In addition, there may be a positive effect of synergy between the nanoparticles and the compounds originating from the organism used. Biogenic synthesis is considered simple, low cost, and suitable for large-scale nanoparticle production (Lima et al., 2012; Kasithevar et al., 2017).
Figure 2.
Scheme, based on the literature, illustrating the synthesis of biogenic nanoparticles. The synthesis uses AgNO3 together with extract (or metabolites) and enzymes from the organism. These nanoparticles have a characteristic outer layer (coating) containing metabolites.
Biogenic nanoparticles have been found to present lower toxicity, while providing effective bactericidal activity against both Gram (−) and Gram (+) bacteria (Durán et al., 2016b; Kasithevar et al., 2017). These nanoparticles have also shown potential for use in the control of fungi (Balashanmugam and Kalaichelvan, 2015; Ahmad et al., 2016; Guilger et al., 2017).
Nanocages
Nanocages are hollow and porous nanometric structures that may be used for the transport and delivery of antibiotics. They can be synthesized from various substances, including metals, proteins, and polymers, and have been investigated in terms of their potential for combating multidrug-resistant bacteria. Reported advantages of these structures are that they provide greater adhesion, retention at the site of infection, increased systemic circulation, and good biocompatibility (Wang et al., 2016; Mekeer et al., 2018).
Wang et al. (2018) synthesized gold nanocages using membrane coating of macrophages pretreated with S. aureus. Clinical treatments performed with local or systemic injection showed that the system provided increased bactericidal effectiveness. Ruozi et al. (2017) synthesized apoferritin-based nanocages, which were used for the encapsulation of streptomycin. The system showed promise for the delivery of antimicrobials, although further characterization, biocompatibility, and efficacy studies were still required. A study by Wu et al. (2019), using silica, silver, and gold nanospheres, showed that the Au–Ag@SiO2 nanocage had broad-spectrum bactericidal properties. The nanocage could be used for antibiotic transport, as well as for infrared-induced hyperthermia therapy against bacterial infection.
Bacteriophages
Bacteriophages (or phages, for short), which are viruses that only infect bacterial cells, are among the most ubiquitous biological entities, with a total estimated abundance of at least 1,030 types (Chibani-Chennoufi et al., 2004). Despite being known for more than 100 years, only now is renewed interest in phages driving studies of them as potential alternatives or complements to current antibiotics, due to their unique affinities and ability to kill bacteria resistant to antibiotics (Hagens and Loessner, 2010; Hyman and Abedon, 2010; Summers, 2012). The interaction between phage particles and bacteria generally involves specific receptors located in the outer membranes of bacteria. Despite the great potential of phages for treating and/or controlling infections caused by antibiotic-resistant bacteria, only a few clinical trials have been performed in humans and are accepted by public health authorities such as the FDA and the European Medicines Agency (EMA) (Rios et al., 2016).
Phages are ubiquitous in the biosphere and are highly specific to particular bacteria species, acting as natural predators of bacteria. They exhibit high tissue permeability and do not affect the beneficial intestinal microflora (so they do not promote secondary infections). Their exponential growth results in their accumulation in extremely high concentrations where they are needed the most, as long as the bacterial host still exists (Hagens and Loessner, 2010; Wittebole et al., 2013; Rios et al., 2016;Harada et al., 2018). However, phage-based therapy requires that the bacterium responsible for the infection is firstly isolated, before the identification and isolation of a specific and strictly lytic phage can be achieved. In addition, due to their protein nature, plain phage particles may be recognized by the immune system, resulting in a drastic reduction of their therapeutic efficacy (Chan and Abedon, 2012; Wittebole et al., 2013).
Bacterial resistance to phage particles generally occurs due to non-adsorption, membrane coating due to mucilage production by bacteria, and destruction of viral genetic material by restriction endonucleases (Wittebole et al., 2013).
Following oral or intravenous administration, phage particles may affect the major body systems, namely, the cardiovascular, digestive, immune, and nervous systems (Moutinho et al., 2012). Furthermore, due to their protein nature, phage particles are prone to denaturation by conformational changes that may be either reversible or irreversible, or to destruction by the immune system. The solution lies in protecting them, either by encapsulation within nanocarriers (Rios et al., 2018) that are invisible towards the digestive and immune systems, or by binding them to a macroscopic support so that they become insoluble (Balcão et al., 2013; Balcão et al., 2014). The combination of these strategies can provide phages with structural and functional stabilization (Balcão and Vila, 2015), enabling them to be potentially used for the eradication of antibiotic-resistant bacteria.
Several studies have described phage-based CRISPR-driven techniques for the prevention of bacterial drug resistance (Barrangou, 2015; Bikard and Barrangou, 2017; Doss et al., 2017;Hatoum-Aslan, 2018; Pursey et al., 2018). In this approach, bacteriophages are designed to carry and deliver CRISPR–Cas in bacteria, in order to combat multidrug-resistant bacteria. Such systems are being developed by biotechnology companies such as Locus Biosciences (Morrisville, NC, USA) and Eligo Bioscience (Paris, France) (Reardon, 2017).
Recent biotechnological advances therefore open the door to the possibility of tailoring bacteriophage particles to improve their characteristics, including i) enhancing the ability of phages to penetrate bacterial biofilms; ii) increasing phage efficacy; iii) broadening the spectrum of phage lytic activities to infections caused by different bacteria; and iv) making phages more stable and specific (Maura and Debarbieux, 2011; Rios et al., 2016; Harada et al., 2018).
At the present time, due to the increase in bacterial resistance to antibiotics, together with the likely ineffectiveness of antibiotics within a few years, there is an urgent need to develop new antimicrobial strategies. This is a new era, in which the emergence of new solutions and discoveries will be crucial.
Future Trends and Possible Solutions
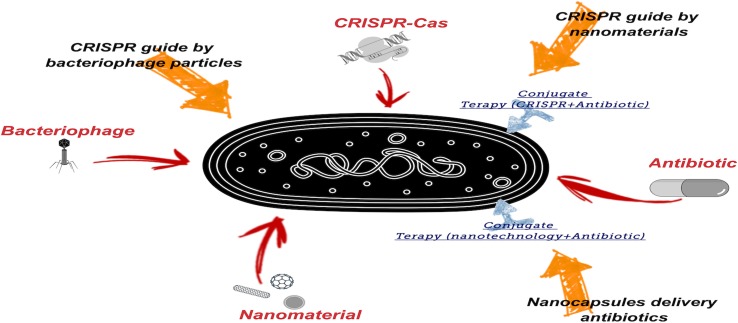
The use of new technologies to combat multidrug-resistant bacteria is ever more necessary, because although there are still effective antibiotics, resistance to them is constantly increasing. The strategies discussed in this paper may provide new ways of fighting multidrug-resistant bacteria. This could include associations between different strategies, as well as their use in combination with antibiotics, in order to combat this critical emerging problem ( Figure 3 ).
Figure 3.
Proposed new technological tools to combat multidrug-resistant bacteria. Emphasis is given to the need to use more than one tool.
The use of CRISPRs, a relatively new technology, may be one of the available solutions. Coupled with nanotechnological delivery methods, this technique could be sufficiently specific and provide the activity required to combat multidrug-resistant bacteria. For this, nanocapsules could be synthesized that are able to reach specific targets, which would facilitate the delivery of CRISPRs.
Biogenic metal nanoparticles, such as silver nanoparticles, may be an option in conjugated treatments to combat MDR bacteria. These nanoparticles offer the benefits of synergy between the effects of the metal and the metabolites of the organism used for their production. They present low toxicity and can act to disrupt existing mechanisms of resistance in bacteria.
Bacteriophages can be used successfully to fight multidrug-resistant bacteria, but although it is not difficult to find the correct virus for each specific bacterium host, the task is nevertheless not straightforward. Consequently, the use of bacteriophage particles as carriers for CRISPRs seems to be a faster and more efficient solution, although such delivery may not always be guaranteed. Recent studies show that CRISPR technology can assist in the modification of bacteriophages, making them more specific for the intended purpose.
To conclude, a deeper understanding of these new and innovative therapeutic strategies is of utmost importance. Until such new strategies have been mastered, structured, and made commercially available, it is imperative to control the use of the currently available chemical antibiotics. It is also essential that health professionals use wisely, and only as a last resort, new antibiotics that may become available in the near future, in order to prevent the emergence and spread of bacterial resistance to them.
Author Contributions
All authors participated in writing the manuscript, specifically RL with the themes nanotechnology and CRISPRs, FF with multidrug resistance, and VB with bacteriophage technology.
Funding
The funding for this work was provided by the São Paulo State Research Foundation [FAPESP, grants #2016/08884-3 (PneumoPhageColor project), #2016/12234-4 (TransAppIL project), #2018/05522-9 (PsaPhageKill project, BPE fellowship granted to VB), and #2017/13328-5 (Biogenic Metal Nanoparticles project)]. Support was provided by the National Council for Scientific and Technological Development (CNPq), in the form of Research Productivity (PQ) fellowships awarded to VB (grants #306113/2014-7 and #308208/2017-0) and RL (grant #303967/2015-3). Funding support was also provided by CESAM (UID/AMB/50017/2019) and FCT/MCTES.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Universidade of Sorocaba/UNISO for supporting the publication charges.
References
- Ahmad A., Wei Y., Syed F., Tahir K., Taj R., Khan A. U., et al. (2016). Amphotericin B-conjugated biogenic silver nanoparticles as an innovative strategy for fungal infections. Microb. Pathog. 99, 271–281. 10.1016/j.micpath.2016.08.031 [DOI] [PubMed] [Google Scholar]
- Alekshun M. N., Levy S. B. (2007). Molecular mechanisms of antibacterial multidrug resistance. Cell 128 (6), 1037–1050. 10.1016/j.cell.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Ali J., Rafiq Q. A., Ratcliffe E. (2018). Antimicrobial resistance mechanisms and potential synthetic treatments. Future Sci. OA. 4 (4), Fso290. 10.4155/fsoa-2017-0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M. A., Khan H. M., Khan A. A., Cameotra S. S., Saquib Q., Musarrat J. (2014). Interaction of Al2O3 nanoparticles with Escherichia coli and their cell envelope biomolecules. J. Appl. Microbiol. 116 (4), 772–783. 10.1111/jam.12423 [DOI] [PubMed] [Google Scholar]
- Azam A., Ahmed A. S., Oves M., Khan M. S., Habib S. S., Memic A. (2012). Antimicrobial activity of metal oxide nanoparticles against gram-positive and gram-negative bacteria: a comparative study. Int. J. Nanomedicine 7, 6003–6009. 10.2147/IJN.S35347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashanmugam P., Kalaichelvan P. T. (2015). Biosynthesis characterization of silver nanoparticles using Cassia roxburghii DC aqueous extract, and coated on cotton cloth for effective antibacterial activity. Int. J. Nanomedicine 10 (1), 87–97. 10.2147/IJN.S79984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcão V. M., Barreira S. V. P., Nunes T. M., Chaud M. V., Tubino M., Vila M. M. D. C. (2014). Carbohydrate hydrogels with stabilized phage particles for bacterial biosensing: bacterium diffusion studies. Appl . Biochem. Biotechnol. 172, 1194–1214. 10.1007/s12010-013-0579-2 [DOI] [PubMed] [Google Scholar]
- Balcão V. M., Moreira A. R., Moutinho C. G., Chaud M. V., Tubino M., Vila M. M. (2013). Structural and functional stabilization of phage particles in carbohydrate matrices for bacterial biosensing. . Technol. 53, 55–69. 10.1016/j.enzmictec.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Balcão V. M., Vila M. M. D. C. (2015). Structural and functional stabilization of protein entities: state-of-the-art. Adv. Drug Deliv. Rev. 93, 25–41. 10.1016/j.addr.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Barrangou R. (2015). The roles of CRISPR–Cas systems in adaptive immunity and beyond. Curr. Opin. Immunol. 32, 36–41. 10.1016/j.coi.2014.12.008 [DOI] [PubMed] [Google Scholar]
- Beisel C. L., Gomaa A. A., Barrangou R. (2014). A CRISPR design for next-generation antimicrobials. Genome Biol. 15, 516. 10.1186/s13059-014-0516-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B. G., Schellevis F., Stobberingh E., Goossens H., Pringle M. (2014). A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. Bioorg. Med. Chem. Lett. 14, 13. 10.1186/1471-2334-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Barrangou R. (2017). Using CRISPR–Cas systems as antimicrobials. Curr. Opin. Microbiol. 37, 155–160. 10.1016/j.mib.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Bikard D., Euler C. W., Jiang W., Nussenzweig P. M., Goldberg G. W., Duportet X., et al. (2014). Exploiting CRISPR–Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32 (11), 1146–1150. 10.1038/nbt.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! an update from the Infectious diseases society of America. Dis. Soc. Am. 48 (1), 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Boya V. N., Lovett R., Setua S., Gandhi V., Nagesh P. K. B., Khan S., et al. (2017). Probin mucin interaction behavior of magnetic nanoparticles. J. Coll. Inter. Sci. 488, 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet L., Lyon D. Y., Hotze E. M., Alvarez P. J., Wiesner M. R. (2009). Comparative photoactivity and antibacterial properties of C60 fullerenes and titanium dioxide nanoparticles. Environ. Sci. Technol. 43 (12), 4355–4360. 10.1021/es803093t [DOI] [PubMed] [Google Scholar]
- Chan B. K., Abedon S. T. (2012). “Phage therapy pharmacology phage cocktails,” in Advances in applied microbiology, vol. 78 Eds. Laskin A. I., Sariaslani S., Gadd G. M. (San Diego: Elsevier Academic Press Inc.), 1–23. 10.1016/B978-0-12-394805-2.00001-4 [DOI] [PubMed] [Google Scholar]
- Chen M., Yang Z., Wu H., Pan X., Xie X., Wu C. (2011). Antimicrobial activity and the mechanism of silver nanoparticle thermosensitive gel. Int. J. Nanomedicine. 6, 2873–2877. 10.2147/IJN.S23945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Liu H., Liang W., Hong L., Zhang B., Huang L., et al. (2019). Insertion sequences in the CRISPR–Cas system regulate horizontal antimicrobial resistance gene transfer in. Int. J. Antimicrob. Agents 53 (2), 109–115. 10.1016/j.ijantimicag.2018.09.020 [DOI] [PubMed] [Google Scholar]
- Chernousova S., Epple M. (2013). Silver as antibacterial agent: ion, nanoparticle, and metal. Angew. Chem. Int. Ed. Engl. 52 (6), 1636–1653. 10.1002/anie.201205923 [DOI] [PubMed] [Google Scholar]
- Chibani-Chennoufi S., Bruttin A., Dillmann M.-L., Brüssow H. (2004). Phage–host interaction: an ecological perspective. J. Bacteriol. 186 (12), 3677–3686. 10.1128/JB.186.12.3677-3686.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi O., Hu Z. (2008). Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ. Sci. Technol. 42 (12), 4583–4588. 10.1021/es703238h [DOI] [PubMed] [Google Scholar]
- Chwalibog A., Sawosz E., Hotowy A., Szeliga J., Mitura S., Mitura K., et al. (2010). Visualization of interaction between inorganic nanoparticles and bacteria or fungi. Int. J. Nanomedicine. 5, 1085–1094. 10.2147/IJN.S13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citorik R. J., Mimee M., Lu T. K. (2014). Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32 (11), 1141–1145. 10.1038/nbt.3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong S., Cao Y., Fang X., Wang Y., Liu Q., Gui H., et al. (2016). Carbon nanotube macroelectronics for active matrix polymer-dispersed liquid crystal displays. ACS Nano. 10 (11), 10068–10074. 10.1021/acsnano.6b04951 [DOI] [PubMed] [Google Scholar]
- Dakal T. C., Kumar A., Majumdar R. S., Yadav V. (2016). Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 7, 1831. 10.3389/fmicb.2016.01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbali R. M., Aoun V., Moussa G., Frei G., Tehrani S. F., Del’Orto J. C., et al. (2019). Tailored nanocarriers for the pulmonary delivery of levofloxacin against Pseudomonas aeruginosa: a comparative study. Mol. Pharm. 16 (5), 1906–1916. 10.1021/acs.molpharmaceut.8b01256 [DOI] [PubMed] [Google Scholar]
- Dizaj S. M., Lotfipour F., Barzegar-Jalali M., Zarrintan M. H., Adibkia K. (2014). Antimicrobial activity of the metals and metal oxide nanoparticles, mater. Sci. Eng. C: Mater. Biol. Appl. 44, 278–284. 10.1016/j.msec.2014.08.031 [DOI] [PubMed] [Google Scholar]
- Doss J., Culbertson K., Hahn D., Camacho J., Barekzi N. (2017). A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses 9, 50. 10.3390/v9030050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán N., Durán M., de Jesus M. B., Seabra A. B., Fávaro W. J., Nakazato G. (2016. a). Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomedicine 12 (3), 789–799. 10.1016/j.nano.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Durán N., Nakazato G., Seabra A. B. (2016. b). Antimicrobial activity of biogenic silver nanoparticles, and silver chloride nanoparticles: an overview and comments. Appl. Microbiol. Biotechnol. 100 (15), 6555–6570. 10.1007/s00253-016-7657-7 [DOI] [PubMed] [Google Scholar]
- Durán N., Marcato P. D., De Conti R., Alves O. L., Costa F. T. M., Brocchi M. (2010). Potential use of silver nanoparticles on pathogenic bacteria, their toxicity and possible mechanisms of action. J. Braz. Chem. Soc. 21 (6), 949–959. 10.1590/S0103-50532010000600002 [DOI] [Google Scholar]
- Eymard-Vernain E., Luche S., Rabilloud T., Lelong C. (2018). Impact of nanoparticles on the Bacillus subtilis (3610) competence. Sci. Rep. 8: 2978, Correction in: Sci. Rep. 2018 8, 6486. 10.1038/s41598-018-21402-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Kasiakou S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Infect. Dis. Soc. Am. 40 (9), 1333–1341. 10.1086/429323 [DOI] [PubMed] [Google Scholar]
- Forsberg K. J., Patel S., Gibson M. K., Lauber C. L., Knight R., Fierer N., et al. (2014). Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616. 10.1038/nature13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente-Núñez C., Lu T. K. (2017). CRISPR–Cas9 technology: applications in genome engineering, development of sequence-specific antimicrobials, and future prospects. Integr. Biol. 9, 109–122. 10.1039/c6ib00140h [DOI] [PubMed] [Google Scholar]
- Gaspar D. P., Gaspar M. M., Eleutério C. V., Grenha A., Blanco M., Gonçalves L. M. D., et al. (2017). Microencapsulated solid lipid nanoparticles as a hybrid platform for pulmonary antibiotic delivery. Mol. Pharm. 514 (9), 2977–2990. 10.1021/acs.molpharmaceut.7b00169 [DOI] [PubMed] [Google Scholar]
- Gomaa A. A., Klumpe H. E., Luo M. L., Selle K., Barrangou R., Beisel C. L. (2014). Programmable removal of bacterial strains by use of genome-targeting CRISPR–Cas systems. MBio. 5 (1), e00928–00913. 10.1128/mBio.00928-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Bello C. (2017). Antibiotic adjuvants—a strategy to unlock bacterial resistance to antibiotics. Bioorg. Med. Chem. Lett. 27 (18), 4221–4228. 10.1016/j.bmcl.2017.08.027 [DOI] [PubMed] [Google Scholar]
- Grigor’eva A., Saranina I., Tikunova N., Safonov A., Timoshenko N., Rebrov A., et al. (2013). Fine mechanisms of the interaction of silver nanoparticles with the cells of Salmonella typhimurium and staphylococcus aureus. Biometals 26 (3), 479–488. 10.1007/s10534-013-9633-3 [DOI] [PubMed] [Google Scholar]
- Guilger M., Pasquoto-Stigliani T., Bilesky-José N., Grillo R., Abhilash P. C., Fraceto L. F., et al. (2017). Biogenic silver nanoparticles based on Trichoderma harzianum: synthesis, characterization, toxicity evaluation and biological activity. Sci. Rep. 7, 44421. 10.1038/srep44421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S., Han J. W., Dayem A. A., Eppakayala V., Kim J. H. (2012). Oxidative stress-mediated antibacterial activity of graphene oxide and reduced graphene oxide in Pseudomonas aeruginosa. Int. J. Nanomedicine 7, 5901–5914. 10.2147/IJN.S37397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habash M. B., Park A. J., Vis E. C., Harris R. J., Khursigara C. M. (2014). Synergy of silver nanoparticles and aztreonam against Pseudomonas aeruginosa PAO1 biofilms. Antimicrob. Agents Chemother. 58 (10), 5818–5830. 10.1128/AAC.03170-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagens S., Loessner M. J. (2010). Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr. Pharm. Biotechnol. 11 (1), 58–68. 10.2174/138920110790725429 [DOI] [PubMed] [Google Scholar]
- Hajipour M. J., Fromm K. M., Ashkarran A. A., Jimenez de Aberasturi D., de Larramendi I. R., Rojo T., et al. (2012). Antibacterial properties of nanoparticles. Trends Biotechnol. 30 (10), 499–511. 10.1016/j.tibtech.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Hao M., Ye M., Shen Z., Hu F., Yang Y., Wu S., et al. (2018). Porin deficiency in carbapenem-resistant enterobacter aerogenes strains. Microb. Drug Resist. 24 (9), 1–7. 10.1089/mdr.2017.0379 [DOI] [PubMed] [Google Scholar]
- Harada L. K., Silva E. C., Campos W. F., Del Fiol F. S., Vila M., Dąbrowska K., et al. (2018). Biotechnological applications of bacteriophages: state of the art. Microbiol. Res. 212–213, 38–58. 10.1016/j.micres.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Hatoum-Aslan A. (2018). Phage genetic engineering using CRISPR–Cas systems. Viruses 10, 335. 10.3390/v10060335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Ingudam S., Reed S., Gehring A., Strobaugh T. P., Jr., Irwin P. (2016). Study on the mechanism of antibacterial action of magnesium oxide nanoparticles against foodborne pathogens. J. Nanobiotechnology 14, 54. 10.1186/s12951-016-0202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemeg H. A. (2017). Nanomaterials for alternative antibacterial therapy. Int. J. Nanomedicine 12, 8211–8225. 10.2147/IJN.S132163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogberg L. D., Heddini A., Cars O. (2010). The global need for effective antibiotics: challenges and recent advances. Trends Pharmacol. Sci. 31 (11), 509–515. 10.1016/j.tips.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Hong W., Gao X., Qiu P., Yang J., Qiao M., Shi H., et al. (2017). Synthesis, construction, and evaluation of self-assembled nano-bacitracin A as an efficient antibacterial agent in vitro and in vivo. Int. J. Nanomedicine 12, 4691–4708. 10.2147/IJN.S136998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F. (2014). Development and applications of CRISPR–Cas9 for genome engineering. Cell 157 (6), 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullahalli K., Rodrigues M., Palmer K. L. (2017). Exploiting CRISPR–cas to manipulate enterococcus faecalis populations. Elife 6, e26664. 10.7554/eLife.26664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullahalli K., Rodrigues M., Nguyen U. T., Palmer K. (2018). An attenuated CRISPR–Cas system in Enterococcus faecalis permits DNA acquisition. MBio. 9 (3), e00414–18. 10.1128/mBio.00414-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman P., Abedon S. T. (2010). “Bacteriophage host range and bacterial resistance,” in Advances in applied microbiology, vol. 70 Eds. Laskin A. I., Sariaslani S., Gadd G. M. (San Diego: Elsevier Academic Press Inc.), 217–248. 10.1016/S0065-2164(10)70007-1 [DOI] [PubMed] [Google Scholar]
- Jamil B., Imran M. (2018). Factors pivotal for designing of nanoantimicrobials: an exposition. Crit. Rev. Microbiol. 44 (1), 79–94. 10.1080/1040841X.2017.1313813 [DOI] [PubMed] [Google Scholar]
- Jansen K. U., Knirsch C., Anderson A. S. (2018). The role of vaccines in preventing bacterial antimicrobial resistance. Nat. Med. 24 (1), 10–19. 10.1038/nm.4465 [DOI] [PubMed] [Google Scholar]
- Jung W. K., Koo H. C., Kim K. W., Shin S., Kim S. H., Park Y. H. (2008). Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and. Appl. Environm. Microbiol. 74 (7), 2171–2178. 10.1128/AEM.02001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhapure R. S., Suleman N., Mocktar C., Seedat N., Govender T. (2014). Nanoengineered drug delivery systems for enhancing antibiotic therapy. J. Pharm. Sci. 104 (3), 872–905. 10.1002/jps.24298. [DOI] [PubMed] [Google Scholar]
- Kapoor G., Saigal S., Elongavan A. (2017). Action and resistance mechanisms of antibiotics: a guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 33 (3), 300–305. 10.4103/joacp.JOACP_349_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasithevar M., Periakaruppan P., Muthupandian S., Mohan M. (2017). Antibacterial efficacy of silver nanoparticles against multi-drug resistant clinical isolates from post-surgical wound infections. Microb. Pathog. 107, 327–334. 10.1016/j.micpath.2017.04.013 [DOI] [PubMed] [Google Scholar]
- Khan S., Alam F., Azam A., Khan A. U. (2012). Gold nanoparticles enhance methylene blue-induced photodynamic therapy: a novel therapeutic approach to inhibit Candida albicans biofilm. Int. J. Nanomedicine 7, 3245–3257. 10.2147/IJN.S31219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Li B., Linhardt R. J. (2017). Pathogenesis and inhibition of flaviviruses from a carbohydrate perspective. Pharmaceuticals 10, 44. 10.3390/ph10020044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon K., Rai M. (2013). Metallic nanoparticles: mechanism of antibacterial action and influencing factors. J. Comp. Clin. Pathol. Res. 2, 160–174. [Google Scholar]
- Kumar M., Curtis A., Hoskins C. (2018). Application of nanoparticle technologies in the combat against anti-microbial resistance. Pharmaceutics 10 (1), 11. 10.3390/pharmaceutics10010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar T. S., Madhumathi K. (2016). Antibiotic delivery by nanobioceramics. Ther. Deliv. 7 (8), 573–588. 10.4155/tde-2016-0025 [DOI] [PubMed] [Google Scholar]
- Li J., Rong K., Zhao H., Li F., Lu Z., Chen R. (2013). Highly selective antibacterial activities of silver nanoparticles against Bacillus subtilis. J. Nanosci. Nanotechnol. 13 (10), 6806–6813. 10.1166/jnn.2013.7781 [DOI] [PubMed] [Google Scholar]
- Li J., Qiao Y., Zhu H., Meng F., Liu X. (2014). Existence, release, and antibacterial actions of silver nanoparticles on Ag–PIII TiO2 films with different nanotopographies. Int. J. Nanomedicine 9, 3389–3402. 10.2147/IJN.S63807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima R., Seabra A. B., Durán N. (2012). Silver nanoparticles: a brief review of cytotoxicity and genotoxicity of chemically and biogenically synthesized nanoparticles. J. Appl. Toxicol. 32 (11), 867–879. 10.1002/jat.2780 [DOI] [PubMed] [Google Scholar]
- Maura D., Debarbieux L. (2011). Bacteriophages as twenty-first century antibacterial tools for food and medicine. Appl. Microbiol. Biotechnol. 90 (3), 851–859. 10.1007/s00253-011-3227-1 [DOI] [PubMed] [Google Scholar]
- Meeker D. G., Wang T., Harrington W. N., Zharov V. P., Johnson S. A., Jenkins S. V., et al. (2018). Versatility of targeted antibiotic-loaded gold nanoconstructs for the treatment of biofilm-associated bacterial infections. Int. J. Hyperthermia 34 (2), 209–219. 10.1080/02656736.2017.1392047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocan T., Matea C. T., Pop T., Mosteanu O., Buzoianu A. D., Suciu S., et al. (2017). Carbon nanotubes as anti-bacterial agents. Cell. Mol. Life Sci. 74 (19), 3467–3479. 10.1007/s00018-017-2532-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moo C. L., Yang S. K., Yusoff K., Ajat M., Thomas W., Abushelaibi A., et al. (2019). Mechanisms of antimicrobial resistance (AMR) and alternative approaches to overcome AMR. Curr. Drug Discov. Technol. 10.2174/1570163816666190304122219 [DOI] [PubMed] [Google Scholar]
- Morones J. R., Elechiguerra J. L., Camacho A., Holt K., Kouri J. B., Ramírez J. T., et al. (2005). The bactericidal effect of silver nanoparticles. Nanotechnology 16 (10), 2346–2353. 10.1088/0957-4484/16/10/059 [DOI] [PubMed] [Google Scholar]
- Moutinho C. G., Matos C. M., Teixeira J. A., Balcão V. M. (2012). Nanocarrier possibilities for functional targeting of bioactive peptides and proteins: state-of-the-art. J. Drug Target. 20, 114–141. 10.3109/1061186X.2011.628397 [DOI] [PubMed] [Google Scholar]
- Mulani M. S., Kamble E. E., Kumkar S. N., Tawre M. S., Pardesi K. R. (2019). Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front. Microbiol. 10, 539. 10.3389/fmicb.2019.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita J. M., Arias C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4 (2). 10.1128/microbiolspec.VMBF-0016-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira Santos I. C., Albano R. M., Asensi M. D., D’Alincourt Carvalho-Assef A. P. (2018). Draft genome sequence of KPC-2-producing Pseudomonas aeruginosa recovered from a bloodstream infection sample in Brazil. J. Glob. Antimicrob. Resist. 15, 99–100. 10.1016/j.jgar.2018.08.021 [DOI] [PubMed] [Google Scholar]
- Nitsch-Osuch A., Gyrczuk E., Wardyn A., Życinska K., Brydak L. (2016). Antibiotic prescription practices among children with influenza. Adv. Exp. Med. Biol. 905, 25–31. 10.1007/5584_2015_198 [DOI] [PubMed] [Google Scholar]
- Pan Y.-J., Lin T.-L., Chen C.-C., Tsai Y.-T., Cheng Y.-H., Chen Y.-Y., et al. (2017). Klebsiella phage ΦK64-1 encodes multiple depolymerases for multiple host capsular types. J. Virol. 91, e02457–02416. 10.1128/JVI.02457-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T. N., Loupias P., Dassonville-Klimpt A., Sonnet P. (2019). Drug delivery systems designed to overcome antimicrobial resistance. Med. Res. Rev. 2019, 1–54. 10.1002/med.21588 [DOI] [PubMed] [Google Scholar]
- Poehlsgaard J., Douthwaite S. (2005). The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3 (11), 870–881. 10.1038/nrmicro1265 [DOI] [PubMed] [Google Scholar]
- Prabhu S., Poulose E. K. (2012). Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int. Nano Lett. 2, 32. 10.1186/2228-5326-2-32 [DOI] [Google Scholar]
- Pursey E., Sünderhauf D., Gaze W. H., Westra E. R., van Houte S. (2018). CRISPR–Cas antimicrobials: challenges and future prospects. PLoS Pathog. 14 (6), e1006990. 10.1371/journal.ppat.1006990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M. K., Deshmukh S. D., Ingle A. P., Gade A. K. (2012). Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J. Appl. Microbiol. 112 (5), 841–852. 10.1111/j.1365-2672.2012.05253.x [DOI] [PubMed] [Google Scholar]
- Redgrave L. S., Sutton S. B., Webber M. A., Piddock L. J. V. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22 (8), 438–445. 10.1016/j.tim.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Rios A. C., Vila M. M. D. C., Lima R., Del Fiol F. S., Tubino M., Teixeira J. A., et al. (2018). Structural and functional stabilization of bacteriophage particles within the aqueous core of a W/O/W multiple emulsion: a potential biotherapeutic system for the inhalational treatment of bacterial pneumonia. Process Biochem. 64, 177–192. 10.1016/j.procbio.2017.09.022 [DOI] [Google Scholar]
- Rios A. C., Moutinho C. G., Pinto F. C., Del Fiol F. S., Jozala A., Chaud M. V., et al. (2016). Alternatives to overcoming bacterial resistances: state-of-the-art. Microbiol. Res. 191, 51–80. 10.1016/j.micres.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Rodzinski A., Guduru R., Liang P., Hadjikhani A., Stewart T., Stimphil E., et al. (2016). Targeted and controlled anticancer drug delivery and release with magnetoelectric nanoparticles. Sci. Rep. 6, 20867. 10.1038/srep20867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruozi B., Veratti P., Vandelli M. A., Tombesi A., Tonelli M., Forni F., et al. (2017). Apoferritin nanocage as streptomycin drug reservoir: technological optimization of a new drug delivery system. Int. J. Pharm. 518 (1-2), 281–288. 10.1016/j.ijpharm.2016.12.038 [DOI] [PubMed] [Google Scholar]
- Salem W., Leitner D. R., Zingl F. G., Schratter G., Prassl R., Goessler W., et al. (2015). Antibacterial activity of silver and zinc nanoparticles against Vibrio cholerae and enterotoxic Escherichia coli. Int. J. Med. Microbiol. 305 (1), 85–95. 10.1016/j.ijmm.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senarathna U. L. N. H., Fernando S. S. N., Gunasekara T. D. C. P., Weerasekera M. M., Hewageegana H. G. S. P., Arachchi N. D. H., et al. (2017). Enhanced antibacterial activity of TiO2 nanoparticle surface modified with Garcinia zeylanica extract. Chem. Cent. J. 11, 7. 10.1186/s13065-017-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban M. I., Shaker M. A., Mady F. M. (2017). Imipenem/cilastatin encapsulated polymeric nanoparticles for destroying carbapenem-resistant bacterial isolates. J. Nanobiotechnology 15 (1), 29. 10.1186/s12951-017-0262-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahverdi A., Fakhimi A., Shahverdi H., Minaian S. (2007). Synthesis and effect of silver nanoparticles on the anti-bacterial activity of different antibiotics against staphylococcus aureus and. escherichia coli. Nanomed. Nanotechnol. Biol. Med. 3, 168–171. 10.1016/j.nano.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Shen J., Zhou J., Chen G. Q., Xiu Z. L. (2018). Efficient genome engineering of a virulent Klebsiella bacteriophage using CRISPR–Cas9. J. Virol. 92 (17), e00534–00518. 10.1128/JVI.00534-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemer S., Westmeier D., Barz M., Eckrich J., Wünsch D., Seckert C., et al. (2019). Biomolecule-corona formation confers resistance of bacteria to nanoparticle-induced. Biomaterials 192, 551–559. 10.1016/j.biomaterials.2018.11.028 [DOI] [PubMed] [Google Scholar]
- Sieradzki K., Markiewicz Z. (2004). Mechanism of vancomycin resistance in methicillin resistant Staphylococcus aureus. J. Microbiol. 53 (4), 207–14. [PubMed] [Google Scholar]
- Simlai A., Mukherjee K., Mandal A., Bhattacharya K., Samanta A., Roy A. (2016). Partial purification and characterization of an antimicrobial activity from the wood extract of mangrove plant Ceriops decandra. EXCLI. J. 15, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Lawrence C. M., Wiedenheft B. (2013). CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266. 10.1146/annurev-biochem-072911-172315 [DOI] [PubMed] [Google Scholar]
- Summers W. C. (2012). The strange history of phage therapy. Bacteriophage 2 (2), 130–133. 10.4161/bact.20757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunitha A., Rimal I. R. S., Sweetly G., Sornalekshmi S., Arsula R., Praseetha P. K. (2013). Evaluation of antimicrobial activity of biosynthesized iron and silver nanoparticles using the fungi Fusarium oxysporum and Actinomycetes sp. Nano. Biomed. Eng. 5 (1), 39–45. 10.5101/nbe.v5i1.p39-45 [DOI] [Google Scholar]
- Tello A., Austin B., Telfer T. C. (2012). Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ. Health Perspect. 120 (8), 1100–1106. 10.1289/ehp.1104650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N., Mir A., Mallik D., Sinha A., Nayar S., Webster T. J. (2010). Bactericidal effect of iron oxide nanoparticles on Staphylococcus aureus. Int. J. Nanomedicine 5, 277–283. 10.2147/IJN.S9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel T. P., Gandra S., Ashok A., Caudron Q., Grenfell B. T., Levin S. A., et al. (2014). Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect. Dis. 14 (8), 742–750. 10.1016/S1473-3099(14)70780-7 [DOI] [PubMed] [Google Scholar]
- Verma S. K., Jha E., Panda P. K., Das J. K., Thirumurugan A., Suar M., et al. (2018). Molecular aspects of core–shell intrinsic defect induced enhanced antibacterial activity of ZnO nanocrystals. Nanomedicine (Lond) 13 (1), 43–68. 10.2217/nnm-2017-0237 [DOI] [PubMed] [Google Scholar]
- Vikesland P., Garner E., Gupta S., Kang S., Maile-Moskowitz A., Zhu N. (2019). Differential drivers of antimicrobial resistance across the world. Acc. Chem. Res. 52 (4), 916–924. 10.1021/acs.accounts.8b00643 [DOI] [PubMed] [Google Scholar]
- Wang C., Wang Y., Zhang L., Miron R. J., Liang J., Shi M., et al. (2018). Pretreated macrophage-membrane-coated gold nanocages for precise drug delivery for treatment of bacterial infections. Adv. Mater. 30 (46), e1804023. 10.1002/adma.201804023 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wan J., Miron R. J., Zhao Y., Zhang Y. (2016). Antibacterial properties and mechanisms of gold–silver nanocages. Nanoscale 8 (21), 11143–11152. 10.1039/C6NR01114D [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang S., Chen W., Song L., Zhang Y., Shen Z., et al. (2018). CRISPR–Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 84 (23), e01834–01818. 10.1128/AEM.01834-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz I. S., Maoz M., Panitz D., Eichler S., Segal E. (2015). Combination of CuO nanoparticles and fluconazole: preparation, characterization, and antifungal activity against Candida albicans. J. Nanopart. Res. 17 (8), 342. 10.1007/s11051-015-3149-4 [DOI] [Google Scholar]
- Willers C., Wentzel J. F., du Plessis L. H., Gouws C., Hamman J. H. (2017). Efflux as a mechanism of antimicrobial drug resistance in clinical relevant microorganisms: the role of efflux inhibitors. Expert Opin. Ther. Targets 21 (1), 23–36. 10.1080/14728222.2017.1265105 [DOI] [PubMed] [Google Scholar]
- Wittebole X., de Roock S., Opal S. M. (2013). A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 4 (8), 1–10. 10.4161/viru.25991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2017). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics.
- Wu J. Y., Kim J. J., Reddy R., Wang W. M., Graham D. Y., Kwon D. H. (2005). Tetracycline-resistant clinical Helicobacter pylori isolates with and without mutations in 16S rRNA-encoding genes. Antimicrob. Agents Chemother. 49 (2), 578–583. 10.1128/AAC.49.2.578-583.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Li A., Zhao X., Zhang C., Yu B., Zhao N., et al. (2019). Silica-coated gold–silver nanocages as photothermal antibacterial agents for combined anti-infective therapy. ACS. Appl. Mater. Interfaces. 11 (19), 17177–17183. 10.1021/acsami.9b01149 [DOI] [PubMed] [Google Scholar]
- Xiu Z. M., Ma J., Alvarez P. J. (2011). Differential effect of common ligands and molecular oxygen on antimicrobial activity of silver nanoparticles versus silver ions. Environ. Sci. Technol. 45 (20), 9003–9008. 10.1021/es201918f [DOI] [PubMed] [Google Scholar]
- Xiu Z. M., Zhang Q. B., Puppala H. L., Colvin V. L., Alvarez P. J. (2012). Negligible particle-specific antibacterial activity of silver nanoparticles. Nano. Lett. 12 (8), 4271–4275. 10.1021/nl301934w [DOI] [PubMed] [Google Scholar]
- Yan M., Wen J., Liang M., Lu Y., Kamata M., Chen I. S. Y. (2015). Modulation of gene expression by polymer nanocapsule delivery of DNA cassettes encoding small RNAs. PLoS ONE 10 (6), e0127986. 10.1371/journal.pone.0127986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Gao X., Zhang R., Li Z., Tan Z., Su H. (2016). Synthesis and characterization of α-ZrP@CHI drug deliver system. J. Nanosci. Nanotechnol. 16 (4), 3628–3631. 10.1166/jnn.2016.11859 [DOI] [PubMed] [Google Scholar]
- Yuan Y.-G., Peng Q.-L., Gurunathan S. (2017). Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: an alternative approach for antimicrobial therapy. Int. J. Mol. Sci. 18, 569. 10.3390/ijms18030569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharova O. V., Godymchuk A. Y., Gusev A. A., Gulchenko S. I., Vasyukova I. A., Kuznetsov D. V. (2015). Considerable variation of antibacterial activity of Cu nanoparticles suspensions depending on the storage time, dispersive medium, and particle sizes. BioMed Research International 2015, Article ID 412530, 11. 10.1155/2015/412530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S., Misba L., Khan A. U. (2017). Nano-therapeutics: a revolution in infection control in post antibiotic era. Nanomedicine 13 (7), 2281–2301. 10.1016/j.nano.2017.06.015 [DOI] [PubMed] [Google Scholar]