Abstract
Leeches (Hirudinida) comprise a charismatic, yet often maligned group of worms. Despite their ecological, economic, and medical importance, a general consensus on the phylogenetic relationships of major hirudinidan lineages is lacking. This absence of a consistent, robust phylogeny of early-diverging lineages has hindered our understanding of the underlying processes that enabled evolutionary diversification of this clade. Here, we used an anchored hybrid enrichment-based phylogenomic approach, capturing hundreds of loci to investigate phylogenetic relationships among major hirudinidan lineages and their closest living relatives. We recovered Branchiobdellida as sister to a clade that includes all major lineages of hirudinidans and Acanthobdella, casting doubt on the utility of Acanthobdella as a “missing link” between hirudinidans and the clitellate group formerly known as Oligochaeta. Further, our results corroborate the reciprocal monophyly of jawed and proboscis-bearing leeches. Our phylogenomic resolution of early-diverging leeches provides a useful framework for illuminating the evolution of key adaptations and host–symbiont associations that have allowed leeches to colonize a wide diversity of habitats worldwide.
Keywords: Acanthobdella, anchored hybrid enrichment, Hirudinida, phylogeny, symbiosis, sanguivory
Introduction
Leeches (Hirudinida) often evoke images of stealthy blood-feeders, triggering a negative visceral reaction that has been ingrained across human cultures for thousands of years. This stigma is not entirely undeserved. Leeches can have deleterious effects on wildlife, economically important fisheries, and human health due to their potential role as vectors of blood-borne pathogens (Slesak et al. 2015). Despite this, leeches have served positive roles throughout much of human history, applied—literally—to ease or treat a wide variety of ailments and diseases (Thearle 1998; Phillips and Siddall 2009). The value of these organisms expands beyond bloodletting in the medical realm, though; leeches are also used as ecological bioindicators and model organisms in developmental biology and neurobiology (Metcalfe et al. 1988; Bendell and McNicol 1991; Minelli and Fusco 2004; Weisblat and Kuo 2009; Le Marrec-Croq et al. 2013). However, despite the economic, scientific, and medical importance of leeches, a general consensus on the phylogenetic relationships of major hirudinidan lineages has been lacking. Absence of a consistent, robust phylogeny of major leech lineages has hindered our understanding of the underlying processes that enabled the evolutionary success of leeches and their allies.
Historically, studies of morphological characters and feeding behavior separated Hirudinida into two major groups: Rhynchobdellida, the proboscis-bearing leeches (e.g., the giant Amazonian leech Haementeria ghilianii, duck leech Theromyzon tessulatum, and fish leeches [family Piscicolidae]), and Arhynchobdellida, the jawed leeches (e.g., the horse leech Haemopis sanguisuga, stinging leech Haemadipsa picta, and the “medicinal” leeches [Macrobdella spp. and Hirudo spp. among others]) (Sawyer 1986). However, molecular data have failed to provide consistent support for this hypothesized scheme. Indeed, the monophyly of the proboscis-bearing Rhynchobdellida has been repeatedly challenged (Siddall and Burreson 1995, 1998; Trontelj et al. 1999; Martin et al. 2000; Siddall et al. 2001; Rousset et al. 2008) suggesting the possibility that either multiple evolutionary gains or independent losses underlie the diversification history of the protrusible proboscis used to feed on host body fluids (i.e., blood or hemolymph) across hirudinidans (Tessler et al. 2018).
Morphological and molecular studies also conflict in fundamental areas critical to understanding the evolution of Hirudinida and its allies, that is, Branchiobdellida (crayfish worms) and Acanthobdellida (the unusual Arctic fish “leech”)—these three groups collectively comprise Hirudinea (Sawyer 1986; Tessler et al. 2018). The evolutionary origin of a parasitic lifestyle and associated morphological adaptations for blood feeding (i.e., sanguivory) in Hirudinea are unclear. Resolution of the origins of parasitism and sanguivory in leeches hinges on the relationships among members of Rhynchobdellida and Arhynchobdellida as well as phylogenetic placement of Hirudinida relative to the ectocommensal Branchiobdellida and ectoparasitic Acanthobdellida. Acanthobdellida and Branchiobdellida exhibit a suite of characters consistent with both Hirudinida and the group of primarily free-living clitellate annelids formerly known as “Oligochaeta.” Acanthobdellida in particular has been heralded as a “missing link,” with behavior and morphology believed to represent a transitional form between Lumbriculidae (a clade of microdrile clitellate annelids) and the true leeches (Clark 1969; Sawyer 1986). Among the most striking morphological characters exhibited by Acanthobdellida is the series of paired anteroventral chetae used to facilitate feeding behavior (fig. 1); chetae, while found on lumbriculids and other families formerly considered oligochaetes, are absent from both hirudinidans and branchiobdellidans.
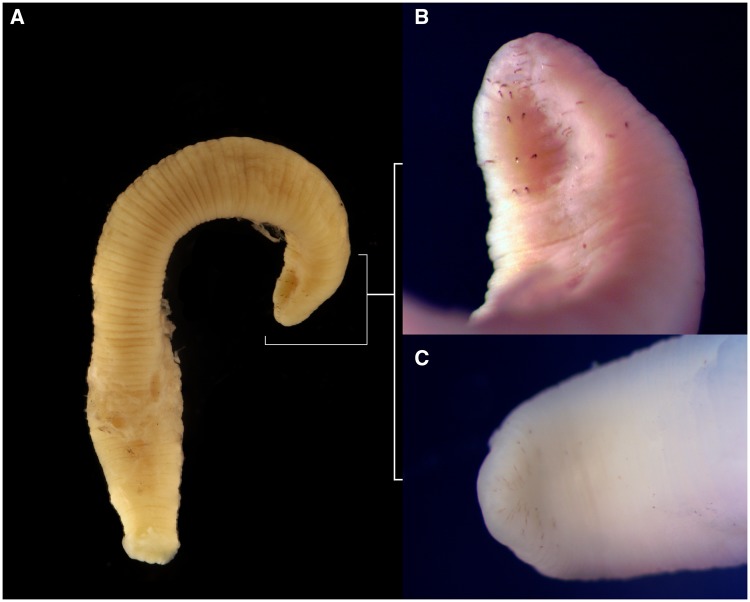
Fig. 1.
—Images of adult (NCSM 90081) and juvenile (NCSM 90080) Acanthobdella peledina specimens, the former from which we obtained one of the two sequences used in this study. (A) Adult, whole body; (B) Adult, close-up of anterior end, showing the diagnostic paired chaetae surrounding the oral region, and; (C) juvenile, close-up of anterior end, showing the diagnostic chaetae, albeit less distinct than in the adult.
Placement of Acanthobdellida as the link between Branchiobdellida and Hirudinida reinforced an Aristotelean concept, now discounted, that transitions in host associations within these groups mirror the rise of major vertebrate clades, from an initial switch from invertebrates to fishes, followed by successive transitions to amphibians, reptiles, birds, nonhuman mammals, and ultimately humans (Sawyer 1986). Members of the order Branchiobdellida are all obligate ectosymbionts of crustaceans, primarily crayfishes. Hirudinidans have host associations that include numerous invertebrate and vertebrate hosts (fig. 2). In contrast, acanthobdellidans are known to primarily parasitize only arctic salmonids though instances of parasitism on other teleosts such as turbot have been reported (Hauck et al. 1979). Both morphological and molecular data have been used to support Acanthobdellida as sister to Hirudinida (i.e., Branchiobdellida + [Acanthobdellida + Hirudinida]) (Purschke et al. 1993; Siddall and Burreson 1995; Trontelj et al. 1999; Martin 2001; Marotta et al. 2008; Tessler et al. 2018), yet have also supported Acanthobdellida as sister to Branchiobdellida + Hirudinida (Siddall et al. 2001; Gelder and Siddall 2001; Rousset et al. 2008). A primary challenge in overcoming these fundamental areas of uncertainty is the time scale which encompasses the early evolutionary history of leeches and their allies. Leech lineages date back to the late Paleozoic to early Mesozoic (Jansson et al. 2008; Parry et al. 2014). Consequently, the resolving power of some of the genetic markers that have been previously relied upon for these groups (e.g., cytochrome c oxidase I, 12S rDNA, 16S rDNA, 28S rDNA, 18S rDNA, and/or internal transcribed spacer 1 [ITS-1]); (Siddall and Burreson 1998; Martin et al. 2000; Siddall et al. 2001; Phillips and Siddall 2009; Williams et al. 2013; Tessler et al. 2018) is likely limited. As such, addressing these questions may require genomic-scale sequencing that spans the major lineages of Hirudinida, Branchiobdellida, and Acanthobdellida. However, no phylogenomic-scale data sets have yet been brought to bear on the relationships among or within these three groups.
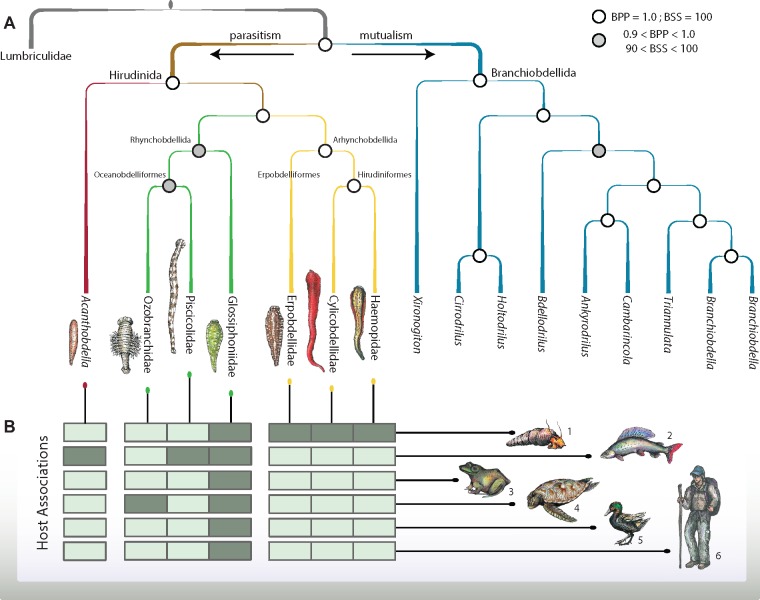
Fig. 2.
—Phylogenetic relationships of major leech and branchiobdellidan lineages and host associations. (A) Evolutionary relationships based on Bayesian and maximum likelihood analyses of DNA sequence data captured through anchored hybrid enrichment. Clade support values are indicated by circles at nodes (Bayesian posterior probability = BPP; Bootstrap support = BSS). Branches are color-coded to show historically accepted higher taxa: brown = Hirudinida, green = Rhynchobdellida (now Oceanobdelliformes and Glossiphoniiformes), yellow = Arhynchobdellida (represented in this study by members of what are now Erpobdelliformes and Hirudiniformes), red = Acanthobdellida, blue = Branchiobdellida. (B) Matrix indicates associations of leeches (columns aligned to phylogeny) with generalized hosts (rows) depicted graphically as 1) invertebrates, 2) fishes (marine, brackish, and freshwater), 3) amphibians and reptiles except turtles, 4) turtles (both marine and freshwater), 5) birds, and 6) mammals. Dark shaded cells in the matrix indicate leech-host affiliation. Upward lines stemming from the matrix link the matrix in A with dots color coded by respective branches, horizontal lines correspond with generalized hosts.
We investigated phylogenetic relationships among Acanthobdellida and major hirudinidan and branchiobdellidan lineages using DNA sequence data captured through anchored hybrid enrichment (AHE) (Lemmon et al. 2012; Faircloth et al. 2013). Phylogenomic analyses of 301 loci comprising 55,502 base pairs provided strongly supported topological resolution consistent across inference methods, and robust to assessments of phylogenetic information content and deviations from nucleotide frequency stationarity (e.g., bias). These results shed light on several long-held hypotheses of leech evolution, and provide the necessary framework for examining broader-reaching questions related to the origin of host–symbiont associations, sanguivory, and adaptations that have allowed these lineages to colonize a wide diversity of habitats.
Materials and Methods
Sequence Data Acquisition
A total of 23 samples from 22 taxa were included in this study, including 16 ingroup and six outgroup taxa (supplementary materials, Supplementary Material online). Ingroup taxa were strategically chosen to represent deep divergences within major hirudinidan and branchiobdellidan lineages. Outgroup taxa were selected to represent a wide taxonomic breadth within Lumbriculidae, acknowledged to be sister to the leeches and their allies. We included two specimens of A. peledina, representing one of two described species in order Acanthobdellida. This overall taxon sampling strategy follows a theory of phylogenetic experimental design, which posits a positive relationship between the proximity of taxa to an internode and the predicted power of resolution (Townsend and Lopez-Giraldez 2010; Dornburg, Townsend, and Wang 2017). DNA was isolated using a Qiagen DNEasy kit (Qiagen, Inc.), with DNA yields quantified using a Qubit 2.0 Fluorometer (Invitrogen). Given the importance of placement of Acanthobdellida relative to Hirudinida and Branchiobdellida, DNA extraction of each of the two A. peledina specimens was performed in a lab in which no annelids had ever been handled or sequenced.
AHE Data Collection and Analysis
AHE data (probe design, see supplementary materials, Supplementary Material online) were collected at Florida State University’s Center for Anchored Phylogenomics (www.anchoredphylogeny.com). After quantification using Qubit fluorometer, DNA extracts were sonicated using a Covaris ultrasonicator to a fragment size of 150–500 bp. From this DNA, indexed libraries were prepared following (Lemmon et al. 2012; Prum et al. 2015), using a Beckman Coulter FXp liquid-handling robot. Libraries were then Qubit-quantified and pooled in two pools of ∼12 samples for enrichment. The probe set Ann1a was produced by Agilent and used to enrich the libraries. Enriched libraries were quantified using KAPA qPCR, then sequenced on one Illumina HiSeq2500 lane with a paired-end 150 bp protocol with 8 bp indexing.
Paired Illumina reads were quality-filtered using the Casava high-chastity filter, then merged following (Rokyta et al. 2012) in order to produce longer (merged) reads and to identify adapters to be removed. Reads were assembled using Helobdella, Dendrobaena, and Mesenchytraeus as references, following the quasi-de novo assembly approach described by Hamilton et al. (2016). Assembly clusters comprising <15 mapped reads were discarded in order to avoid potential low-level contamination. For each locus, orthology of the consensus sequences derived from the remaining assembly clusters was determined using pairwise sequence divergences in a neighbor-joining approach (see Hamilton et al. 2016 for details). Orthologous sequences were aligned using MAFFT v7.023b, then auto-trimmed/masked following (Hamilton et al. 2016), with the following settings: MINGOODSITES = 7, MINPROP = 0.5, and MISSINGALLOWED = 12. Final alignments were inspected in Geneious R9 to ensure that no aberrant sequences were present.
The previous published version of this study was based upon DNA sequence data that unknowingly included some host contamination and changed the topology of the resulting phylogenies once removed. The following protocol was designed and implemented in this revised version. To detect and remove contaminated sequences, each locus was further inspected for potential vertebrate host contamination by blasting sequences from five leeches (both Acanthobdella individuals, Ozobranchus, Piscicola, and Haemopis) against the following genomes on ENSEMBL: Human, Chinese softshell turtle, spotted gar, midas cichlid, eastern happy, and northern pike. This strategy allowed us to capture lineages that span the ray-finned fish Tree of Life (Near et al. 2012), while also screening for turtle host and human contamination in other species. For each locus, all potential vertebrate matches were downloaded and aligned to the alignment of all sequenced taxa for each AHE locus. Phylogenetic searches were conducted in IQTREE (Nguyen et al. 2015) using the best-fit substitution model clade support quantified with 1,000 ultrafast phylogenetic bootstraps (Minh et al. 2013) and a SH-like approximate likelihood ratio test with 1,000 bootstraps (Guindon et al. 2010). Using this approach, we found strong evidence for localized contamination of fish DNA within both Acanthobdella specimens (12% and 43%, respectively) and the fish leech Piscicola geometra (50%), and turtle host DNA within the sea turtle leech Ozobranchus margoi (16%). We also identified human contamination in a few other instances (supplementary materials, Supplementary Material online). Taxon-locus pairs identified as contaminated were removed from the data set prior to any additional analyses.
Assessment of Phylogenetic Information Content and Data Compositional Patterns
Theory and empirical data have long supported a relationship between the rate of character evolution and the utility of a character in resolving a specific phylogenetic problem (Townsend 2007; Townsend et al. 2012) Although the median rate of evolution for loci targeted by AHE is low, substantial heterogeneity within individual loci has been shown to negatively impact inference (Dornburg, Townsend, Brooks, et al. 2017; Dornburg et al. 2018). To assess predicted levels of convergences in character state, or homoplasy, across our data, site-specific rates, λi…λj were quantified for each locus using IQTREE (Nguyen et al. 2015), and a guide chronogram generated using nonparametric rate smoothing with cross-validation of the rate smoothing parameter on the concatenated RAxML tree topology using the APE package in R (Paradis 2006). Site rates were used to assess variation in information content between loci by explicitly quantifying the predicted impact of homoplasy on the resolution of specific phylogenetic problems in the R package PhyInformR (Dornburg et al. 2016). To accomplish this we quantified quartet internode homoplasy probabilities (QIHP), which represents the predicted probability of having greater strength of support at a given internode for an incorrect rather than correct topology as a result of homoplasious site patterns (Townsend et al. 2012). These calculations are agnostic to the empirical topology, requiring only a temporal depth (T) and internode distance (t0) to be specified for resolution of the hypothetical topology. We quantified QIHP values for each locus based on resolving a hypothetical quartet with branch lengths that approximate the expected relative divergences between the most recent common ancestor of Acanthobdella, Branchiobdellida, and the “true” leeches. Loci were retained up to the locus-median change point of QIHP using the R package changepoint (Killick and Eckley 2014). Although this approach has been shown to be robust to deviations in guide tree topology and branch lengths (Dornburg, Townsend, Brooks, et al. 2017), we further quantified quartet internode resolution probabilities (QIRP), which represents the predicted probability of having greater strength of support at a given internode for a correct rather than incorrect topology based on a substitutions reflecting the evolutionary history of character change (Townsend 2007; Townsend et al. 2012), across a range of T and t0 values. This additional quantification allowed us to assess confidence in the predicted resolving power of our data across a range of hypothetical branch lengths. Additionally, as nucleotide compositional biases have been found in some AHE data sets (Dornburg, Townsend, Brooks, et al. 2017; Reddy et al. 2017), we used the software BaCoCa (Kück and Struck 2014) to identify and filter any loci identified as deviating from stationarity based on a chi-square homogeneity test with a threshold of 0.05.
We replicated the above quantifications of phylogenetic information content with five legacy markers (COI, 16S, 18S, 28S, ITS1) used in Tessler et al. (2018) and earlier studies of leech relationships. This allowed us to assess if differences in inferred tree topologies among legacy data sets and compared with AHE data would be predicted based on expectations of homoplasy. All data were accessioned using GenBank (supplementary materials, Supplementary Material online). For each gene we quantified expectations of phylogenetic informativeness (Townsend 2007) and phylogenetic signal versus noise (Townsend et al. 2012) using PhyInformR (Dornburg et al. 2016).
Phylogenomic Analyses
We assembled three data sets for maximum likelihood and Bayesian analyses: 1) a concatenated analysis containing data from all loci (301 loci; 55,502 bp); 2) a data set with loci identified as containing high levels of homoplasy removed (251 loci; 42,024 bp); and 3) a data set that also removed loci identified as containing significant levels of compositional biases from the second data set (245 loci; 41,029 bp). Each concatenated data set was first analyzed using a maximum likelihood approach in RAxML v8.2.8 (Stamatakis 2014) with 500 rapid bootstraps and a thorough ML search using a GTR+ model of nucleotide substitution. Although the GTR model represents the most complex time reversible nucleotide substitution model, the influence of potential nucleotide model overparameterization has been shown to be negligible for topological inference (Dornburg et al. 2008). In contrast, dividing multi-locus data sets into smaller partitions to more accurately model among-site rate variation can improve the accuracy of topological inference (Kainer and Lanfear 2015). For each of our data sets, we used PartitionFinder v2.0 to find the best-fit partitioning strategy for our data using a heuristically optimized search and the Bayesian Information Criterion (Lanfear et al. 2016). The candidate pool of partition strategies for each data set ranged from a single partition for all data, to a partition for each locus. Each data set was then analyzed using its best-fit partition strategy. All analyses were additionally repeated using the maximum likelihood algorithms available in IQTREE (Nguyen et al. 2015) to assess congruence between likelihood algorithms, using the option to identify the best-fit substitution model and quantifying clade support with 1,000 ultrafast phylogenetic bootstraps (Minh et al. 2013) and a SH-like approximate likelihood ratio test with 1000 bootstraps (Guindon et al. 2010).
All maximum likelihood analyses were also repeated in a Bayesian framework using the open MPI distribution of MrBayes v.3.2 (Ronquist et al. 2012). Each analysis was run for 30 million generations based on preliminary analyses that assessed sampling levels required for convergence and effective sampling of the state space for each parameter by the MCMC sampler. Following analysis convergence was assessed through: 1) visual inspection of state likelihoods, 2) quantification of potential scale reduction factors, ensuring an average deviation of clade splits below 0.001 between replicate runs, and 3) convergence diagnostics available in AWTY (Nylander et al. 2008). To ensure that all samples stemmed from the target distribution, burn-in was independently assessed for each run based on convergence diagnostics.
To assess the impact of independent gene histories on topological parameter estimates, we used ASTRAL-II (Mirarab and Warnow 2015) to estimate a species tree based on maximum likelihood inference of each individual locus. Each locus was analyzed in RAxML v8.2.8 (Stamatakis 2014) and independent gene trees were subject to a heuristic search that maximizes the number of quartets found across all trees in a species tree (Mirarab and Warnow 2015). Three searches were conducted to mirror the analyses above, allowing us to assess how robust inference of gene trees and the resulting species tree were to both homoplasy and compositional biases: 1) a search based on all loci; 2) a search based on loci below the QIHP threshold for filtration; and 3) a search based on loci that were below the QIHP threshold and not identified as possessing significant biases in base composition.
Results and Discussion
Relationships among Leeches and Their Allies
Both maximum likelihood and Bayesian analyses of our 301-locus data set resulted in complete and strongly supported resolution of relationships among leeches and their allies (bootstrap support [BSS] ≥ 90 and Bayesian posterior probability [BPP] > 0.90 across all nodes; fig. 2, supplementary figs. 1–3, Supplementary Material online). We recovered strong support (BSS = 100; BPP = 1.0; fig. 2) for a monophyletic Branchiobdellida sister to a clade comprising Hirudinida + the acanthobdellidan Acanthobdellapeledina (BSS = 100; BPP = 1.0; fig. 2). This placement of A. peledina is robust to data filtration, partitioning strategy, and inference method (supplementary materials, Supplementary Material online). All analyses support a close relationship among members of the taxa formerly considered Arhynchobdellida (i.e., Erpobdelliformes [represented here by Erpobdellidae] and Hirudiniformes [represented here by the nonblood feeding groups Cylicobdellidae and Haemopidae] (BSS = 100; BPP = 1.0; fig. 2) as well as among Rhynchobdellida (i.e., Glossiphoniidae, Piscicolidae, and Ozobranchidae, the latter two in combination forming Oceanobdelliformes sensu, Tessler et al. 2018; 90 < BSS < 100; 0.9 < BPP 1.0; fig. 2). Our results strongly support (i.e., corroborate; Borda and Siddall 2004; Tessler et al. 2018) a divergence between the arhynchobdellid clades Erpobdelliformes and Hirudiniformes (BSS = 100; BPP = 1.0; fig. 2).
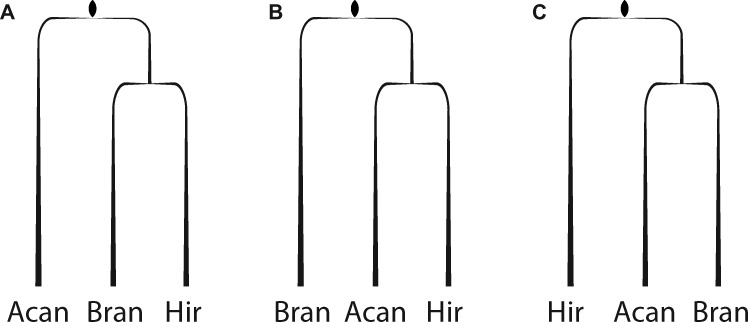
Phylogenomic studies have increasingly provided resolution to the annelid Tree of Life (Struck et al. 2011); however, resolution of evolutionary relationships among leeches and their close relatives has historically been lacking. Described in 1850, A. peledina has been considered a “living fossil,” or “missing link” in the evolutionary history of leeches and their allies. Yet, the placement of Acanthobdellida relative to Branchiobdellida and Hirudinida has been inconsistent, with molecular and morphological data supporting Acanthobdellida as sister to Branchiobdellida + Hirudinida (fig. 3A;Siddall et al. 2001; Gelder and Siddall 2001; Rousset et al. 2008), Branchiobdellida as sister to Acanthobdellida + Hirudinida (fig. 3B; Siddall and Burreson 1995; Martin 2001; Marotta et al. 2008; Tessler et al. 2018), or Hirudinida as sister to Branchiobdellida + Acanthobdellida (fig. 3C;Trontelj et al. 1999; Martin et al. 2000). Our results support the second hypothesis (fig. 3B), resolving Acanthobdella as sister to the leech representatives in our study and provides a foundation for increased taxon sampling within both ingroups and outgroups that can further refine relationships among these worms.
Fig. 3.
—Simplified cladograms displaying postulated relationships among Acanthobdellida (Acan), Branchiobdellida (Bran), and Hirudinida (Hir) based on prior molecular and/or morphological data: A. Acanthobdellida as sister to Branchiobdellida + Hirudinida; B. Branchiobdellida as sister to Acanthobdellida + Hirudinida, and; C. Hirudinida as sister to Acanthobdellida + Branchiobdellida.
All this considered, why have previous molecular-based studies that have inferred relationships among Branchiobdellida, Acanthobdellida, and Hirudinida, either directly or indirectly, differed in their results (Siddall and Burreson 1995; Trontelj et al. 1999; Martin et al. 2000; Siddall et al. 2001; Gelder and Siddall 2001; Martin 2001; Marotta et al. 2008; Rousset et al. 2008; Tessler et al. 2018)? Each of these prior molecular studies have relied on sequences of one or more genes or gene fragments (nuclear and mitochondrial), including COI, 12S rDNA, 16S rDNA, 18S rDNA, 28S rDNA, and ITS1. We quantified the predicted phylogenetic utility of five of these genes for topological resolution of a hypothetical internode at the temporal depth of early leech divergences. Our results strongly suggest that these legacy markers contain low levels of phylogenetic information content for a phylogenetic problem at this depth, even when concatenated (fig. 4).
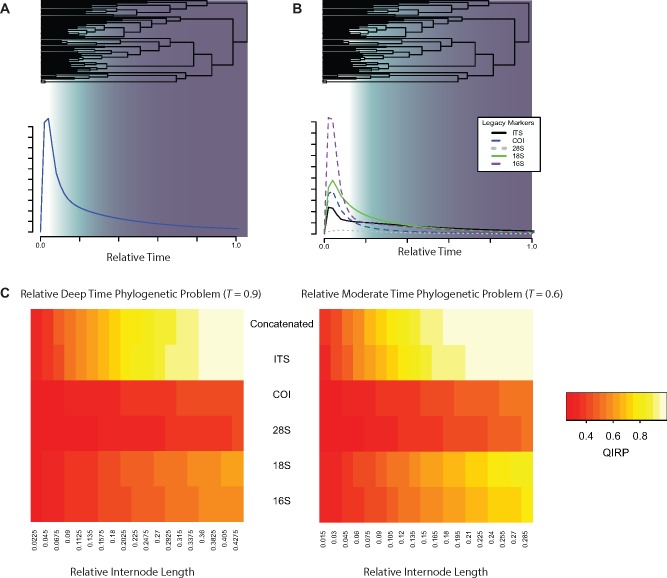
Fig. 4.
—Quantification of phylogenetic informativeness present in legacy markers. (A) Phylogenetic informativeness profile of the combined information from five legacy markers, COI, ITS-1, 28S rDNA, 18S rDNA, and 16S rDNA. (B) Phylogenetic informativeness profile of individual legacy markers. (C) Heatmaps indicating probabilities of a legacy marker (Y-axis) resolving a phylogenetic problem of a specific internode length (X-axis) for a relatively deep time internode (T = 0.9; left) and a relatively moderate time (T = 0.6). Colors correspond to the predicted probability of correct resolution of the hypothetical quartet, with white indicating higher predictions of loci contributing to correct topological inference. Shading in A & B corresponds to the decline of informativeness and the correlated onset of convergences in character state (i.e., “the rainshadow of noise”) (Dornburg, Townsend, and Wang 2017; Townsend and Leuenberger 2011).
Analysis of phylogenetic informativeness (PI) of both the concatenated data set (fig. 4A) and each individual legacy marker (fig. 4B), demonstrate a steep decline of PI prior to focal divergences between Acanthobdella and other major lineages. Such a decline in PI has been likened to a “rainshadow of noise” that indicates an increase in the saturation of character-state substitutions (Townsend and Leuenberger 2011; Dornburg et al. 2014; Dornburg, Townsend, and Wang 2017). Correspondingly, quantification of the predicted probability of each legacy marker or the entire matrix of concatenated legacy markers correctly resolving hypothetical internodes at moderate (fig. 4C) or deep levels of topological divergence (fig. 4D) was extremely low for the internode distances among major leech groups predicted in our study. In contrast to the legacy marker data set, conserved element approaches, such as those using AHE (e.g., Lemmon et al. 2012; Faircloth et al. 2013) or ultraconserved elements (UCE’s; e.g., Faircloth et al. 2012), have consistently shown great utility for resolving phylogenetic relationships at deep timescales (e.g., Faircloth et al. 2013; Brandley et al. 2015; Faircloth et al. 2015; Prum et al. 2015; Young et al. 2016; Dornburg, Townsend, Brooks, et al. 2017). For disentangling the evolutionary history of leeches and their allies, our quantification of phylogenetic information content demonstrates high levels of predicted phylogenetic information at deep timescales for this class of data. While individual loci did decline in PI and exhibit levels of noise (QIHP) similar to legacy markers (fig. 5A), results based on filtering these few loci (fig. 4B) were congruent with relationships based on the unfiltered data set (supplementary materials, Supplementary Material online). This is likely the result of high levels of signal overwhelming the total noise in the data set, as evident from the predicted probability of the concatenated alignment having the power to resolve a range of hypothetical internodes at both deep (fig. 5C) and moderate (fig. 5D) timescales. From a perspective of phylogenetic experimental design, our results suggest these markers represent a more appropriate data choice relative to legacy markers to address questions at these deep timescales, supporting the recognition that scrutiny of legacy marker data in tandem with phylogenomic scale data is essential to disentangling sources of topological incongruence (Parker et al. 2019). However, despite predictions of high utility for concatenated AHE loci, the predicted utility of individual loci for specific internodes was low for nodes at deep timescales characterized by short internodes (fig. 5C and D). This prediction is consistent with the lack of support and short branches in our species tree, both of which indicate gene tree uncertainty (supplementary materials, Supplementary Material online).
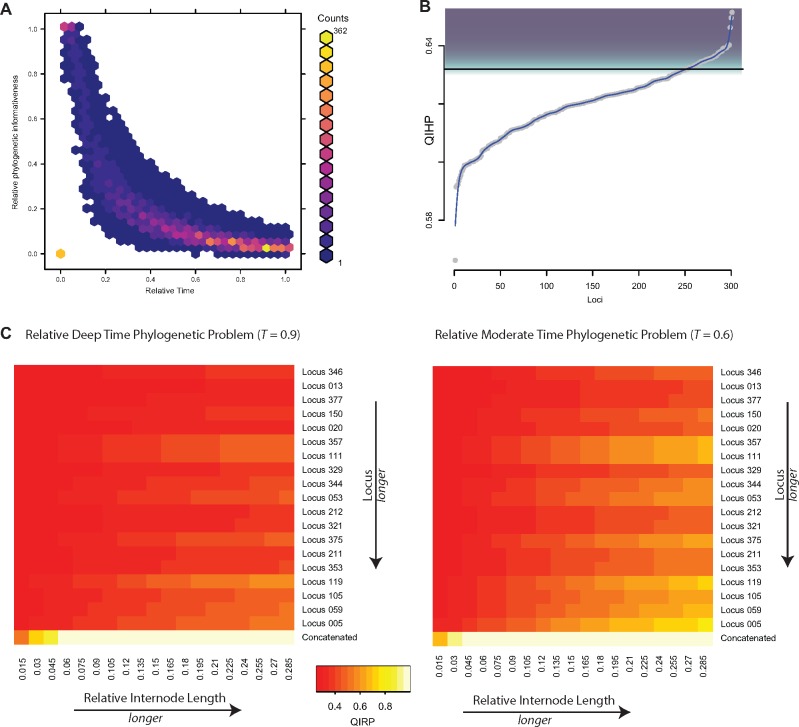
Fig. 5.
—Quantification of phylogenetic informativeness present in AHE data. (A) Phylogenetic informativeness (PI) profile of the information in each AHE locus represented through a hexbin plot of the relative phylogenetic informativeness (PI) over time. Colors correspond to the number of loci occupying each bin of informativeness through time following (Near et al. 2018). (B) Example of changepoint plot used as filtration criterion following (Dornburg, Townsend, Brooks, et al. 2017). (C) Heatmaps indicating probabilities of the combined AHE data set and representative loci (Y-axis) resolving a phylogenetic problem of a specific internode length (X-axis) for a relatively deep time internode (T = 0.9; left) and a relatively moderate time (T = 0.6). Colors correspond to the predicted probability of correct resolution of the hypothetical quartet. Loci are sorted by length. Counts in A represent the number of loci represented in a given hexbin. Shading in B corresponds to the loci selected possessing levels of phylogenetic noise that exceed the filtration threshold.
Placement of Acanthobdella as sister to Hirudinida is a hypothesis that has been repeatedly suggested over the past several decades (see Siddall and Burreson 1995; Martin 2001; Marotta et al. 2008; Tessler et al. 2018). The concept that Acanthobdella, Branchiobdellida, and Hirudinida are closely allied has rarely been in dispute. Indeed, Grube (1850) listed his newly described species, A. peledina, as the sole member of Acanthobdellea, within the “Discophora” (= Hirudinea), along with Branchiobdellea, Clepsinea, and Hirudinaèea, the latter two comprising the leeches as then known. However, placement of Acanthobdella relative to Branchiobdellida and Hirudinida, as well as Lumbriculidae, has been long debated. Our results are consistent with the most recent comprehensive analysis of these groups based on 5 legacy markers (Tessler et al. 2018) in recovering Acanthobdella sister to Hirudinida. This finding supports the more than a century old conclusions of Livanow (1906) who posited : “All of these facts provide us with sufficient evidence to establish for Acanthobdella a special group called Acanthobdellea that would be the equivalent of the other Hirudinean groups, the Rhynchobdellea and the Gnathobdellea [= the jawed leeches, Arhynchiobdellida]” (p. 837 in German; 215 in English translation). Livanow (1906) continues, stating “There are not enough reasons for combining the Acanthobdellids with the Discodrilida [= branchiobdellidans]…” (p. 837 in German; 215 in English translation).
Although Acanthobdella shares numerous morphological and ecological similarities with leeches (see, e.g., Livanow 1906; Brinkhurst and Gelder 1989), it possesses unusual characters similar to “oligochaetes” that have been used to justify keeping the taxon separate, including paired anterior chaetae presumed to aid in attachment to the host for feeding. Interestingly, Chapman and Brinkhurst (1987) demonstrated that variation in chetae, including gain, or reduction and loss, can be induced in tubifid clitellate annelids by varying salinity, pH, water hardness, and mercury. These findings perhaps add uncertainty in the relative weight of these characters for taxonomic inference about Acanthobdella in the absence of environmental information.
However, the alignment of molecular and morphological data supports the placement of Acanthobdella as sister to Hirudinida. Our inferred tree topology further provides a strong foundation from which to address questions regarding the evolution of host–symbiont associations and key adaptations that have allowed leeches to colonize a wide diversity of habitats globally. We resolve a deep divergence between rhynchobdellid (proboscis-bearing) leeches and the arhynchobdellid (jawed) leeches, (BSS = 100; BPP = 1.0; fig. 2), supporting an ancient divergence in leech feeding morphology. This result has been recovered with inconsistent support by other studies (Siddall and Burreson 1995, 1998; Trontelj et al. 1999; Martin et al. 2000; Siddall et al. 2001; Rousset et al. 2008).
Our results in toto illuminate the origins of sanguivory in the group, implied as a single transition to blood feeding (in the ancestor of Hirudinida inclusive of Acanthobdella), followed by a number of reversals (e.g., select taxa within Hirudiniformes and the ancestor of Erpobdelliformes and …). These results suggest a deep evolutionary shift from free-living to commensal association and in symbiotic strategy between the predominantly mutualist branchiobdellidans and parasitic hirudinidans + Acanthobdella. However, given that the feeding mode of Acanthobdella requires parasitization of fish lineages with Cenozoic origins (Near et al. 2012, 2013) in ecologically extreme high latitude environments, Acanthobdella may represent an early-diverged lineage that has only recently specialized by taking advantage of new ecological opportunities. Our analyses highlight the complexity in the evolution of host association and habitat colonization. Host association is rare within Clitellata (see Gelder 1980), with a remarkable exception being the clade that contains Branchiobdellida, Hirudinida, and Acanthobdella. These results demonstrate that the phylogeny of taxa within Hirudinea and their major transitions in host associations do not mirror the rise of major vertebrate clades (e.g., host specificity with Aristotelean progression, Sawyer 1986; fig. 2). Instead, we find a complex pattern of host switching and generalization with multiple shifts across three clades: Acanthobdellida Oceanobdelliformes + Glossiphoniiformes (=Rhynchobdellida), and Erpobdelliformes + Hirudiniformes (=Arhynchobdellida). Acanthobdellida contains A. peledina, which feeds primarily on the blood of salmonid and some other teleost fishes, such as turbot, in Arctic freshwaters (Hauck et al. 1979). Oceanobdelliformes comprises Piscicolidae, and Ozobranchidae, groups that feed primarily on the blood of marine and freshwater fishes, and marine turtles, respectively. Glossiphoniiform leeches feed on aquatic gastropods, amphibians, turtles, birds, and mammals, whereas the erpobdelliform and hirudiniform leeches comprises a number of families with a diversity of invertebrate and vertebrate host preferences. These results suggest evolutionary host switching or host generalization to be an important part of the diversification of these groups (Hauck et al. 1979).
The placement of Acanthobdella sister to leeches, together with the deep divergence between Oceanobdelliformes + Glossiphoniiformes and Erpobdelliformes + Hirudiniformes (BSS = 100; BPP = 1.0; fig. 2) supports, in part, previous studies that hypothesized a pattern of evolutionary habitat transition from freshwater (Branchiobdellida and Acanthobdellida) to marine environments (most Oceanobdelliformes) and a secondary return to freshwater and terrestrialism (Erpobdelliformes + Hirudiniformes); in these scenarios Acanthobdellida was the transitional form placed between Branchiobdellida and Hirudinida existing in freshwater but feeding on euryhaline hosts (Apakupakul et al. 1999; Borda and Siddall 2004). However, it is possible, given our results, that a freshwater existence is pervasive, and not secondarily obtained. Acanthobdella may not represent a link between lineages as much a uniquely specialized arctic parasite that has taken advantage of the rise of Cenozoic fishes including salmonids and other extant teleost fishes as a means of persisting in extreme habitats.
Conclusion
A conserved element approach has great potential for enhancing our understanding of the early evolutionary origins of hirudineans, a group of organisms with ecological, economical, and cultural importance. Our phylogenomic analyses based on the capture of hundreds of loci provide needed resolution and support to relationships among the of major hirudinean lineages. Most notably, we provide insight into the position of the Arctic fish leech, A. peledina, relative to Hirudinida (leeches) and Branchiobdellida (crayfish worms). The placement of this unusual taxon suggests a single origin of sanguivory and raises intriguing questions regarding the origin and persistence of the chetae of Acanthobdella. This taxonomic framework is consistent with well-conceived historic hypotheses (i.e., Livanow 1906), and also provides the foundation necessary to begin testing the significance of major adaptations and host associations in the successful diversification of this cross-culturally emblematic group of organisms.
Availability of Data and Materials
All raw sequence reads are available on NCBI (accession SAMN11310017–SAMN11310038).
This project - including alignments, phylogenetic software analysis input files, resulting output files, and associated R scripts - has been deposited on Zenodo under DOI:10.5281/ zenodo.2630841; https://zenodo.org/record/2630841#.XKe 78etKhE4. All versions of the data, including those used in the previous published version of this paper, are available.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The original publication of this study was based upon a data set with DNA sequence data unknowingly contaminated with host DNA that a reader brought to the attention of the authors and editors. Protocols to detect contaminated sequences were designed, implemented, and the analyses repeated and subsequently changed the topology of the phylogenetic trees and their interpretation, which led to this revised version. We thank this highly engaged reader for indicating the sequence contamination in our data set, and the journal editorial team for providing us with the opportunity to address this issue.
The authors thank Aleksander Bielicki, Heidi Golden, Michael Kendrick, Alex Huryn, Ivana Maguire, Goran Klobcar, Juhyung Lee, Carl Williams, and Mark Roberts for assistance obtaining specimens. Special thanks to Michelle Kortyna and Sean Holland at the Center for Anchored Phylogenomics for assistance with data collection and analysis, to Marianne Barrier for assistance in designing contamination detection protocols, and to E. Ferraro and F. Reed for their assistance conducting ENSEMBL-based searches.
This study was supported by the U.S. National Science Foundation WormNet II (Assembling the Annelid Tree of Life) grant (DEB-1136604 to S.W.J. and DEB-1036516 to F.E.A.).
Author Contributions
S.W.J., C.E., F.E.A., and B.W.W. conceived and designed the study. B.W.W. selected the taxa with contributions from A.J.P. and conducted the initial laboratory preparations. A.R.L. selected the loci and designed the probes. A.R.L. and E.M.L. collected the molecular data and conducted bioinformatics analyses. A.D., K.L.Z., F.E.A., and B.W.W. performed phylogenomic analyses and tests. A.D., B.W.W., and K.L.Z. designed figures and K.L.Z. created the illustrations. A.J.P., A.D., K.L.Z., and B.W.W. wrote the paper with contributions from all authors.
Data deposition: All sequence data has been archived in the short read archive as SAMN11310017–11310038.
Literature Cited
- Apakupakul K, Siddall ME, Burreson EM.. 1999. Higher level relationships of leeches (Annelida: Clitellata: Euhirudinea) based on morphology and gene sequences. Mol Phylogenet Evol. 12(3):350–359. [DOI] [PubMed] [Google Scholar]
- Bendell BE, McNicol DK.. 1991. An assessment of leeches (Hirudinea) as indicators of lake acidification. Can J Zool. 69(1):130–133. [Google Scholar]
- Borda E, Siddall ME.. 2004. Arhynchobdellida (Annelida: Oligochaeta: Hirudinida): phylogenetic relationships and evolution. Mol Phylogenet Evol. 30(1):213–225. [DOI] [PubMed] [Google Scholar]
- Brandley MC, et al. 2015. Evaluating the performance of anchored hybrid enrichment at the tips of the tree of life: a phylogenetic analysis of Australian Eugongylus Group Scincid Lizards. BMC Evol Biol. 15(1):62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhurst RO, Gelder SR.. 1989. Did the lumbriculids provide the ancestors of the Branchiobdellidans, Acanthobdellidans and Leeches? Hydrobiologia 180(1):7–15. [Google Scholar]
- Chapman PM, Brinkhurst RO.. 1987. Hair today, gone tomorrow: induced chaetal changes in tubificid oligochaetes Hydrobiologia 155(1):45–55. [Google Scholar]
- Clark RB. 1969. Systematics and phylogeny: annelida, echiura, sipuncula. Chem Zool. 1–68. [Google Scholar]
- Dornburg A, Fisk JN, Tamagnan J, Townsend JP.. 2016. PhyInformR: phylogenetic experimental design and phylogenomic data exploration in R. BMC Evol Biol. 16 (1):262.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornburg A, Santini F, Alfaro ME.. 2008. The influence of model averaging on clade posteriors: an example using the triggerfishes (family Balistidae). Syst Biol. 57(6):905–919. [DOI] [PubMed] [Google Scholar]
- Dornburg A, Su Z, Townsend JP.. 2018. Optimal rates for phylogenetic inference and experimental design in the era of genome-scale datasets. Syst Biol. 10.1093/sysbio/syy047. [DOI] [PubMed] [Google Scholar]
- Dornburg A, Townsend JP, Friedman M, Near TJ.. 2014. Phylogenetic informativeness reconciles ray-finned fish molecular divergence times. BMC Evol Biol. 14(1):169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornburg A, Townsend JP, Brooks W, et al. 2017. New insights on the sister lineage of percomorph fishes with an anchored hybrid enrichment dataset. Mol Phylogenet Evol. 110:27–38. [DOI] [PubMed] [Google Scholar]
- Dornburg A, Townsend JP, Wang Z.. 2017. Maximizing power in phylogenetics and phylogenomics: a perspective illuminated by fungal big data. Adv Genet. 100:1–47. [DOI] [PubMed] [Google Scholar]
- Faircloth BC, Branstetter MG, White ND, Brady SG.. 2015. Target enrichment of ultraconserved elements from arthropods provides a genomic perspective on relationships among hymenoptera. Mol Ecol Resour. 15(3):489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth BC, et al. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst Biol. 61(5):717–726. [DOI] [PubMed] [Google Scholar]
- Faircloth BC, Sorenson L, Santini F, Alfaro ME.. 2013. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS One 8(6):e65923.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelder SR. 1980. A review of the symbiotic oligochaeta (Annelida). Zoologischer Anzeiger Jena. 204(1/2):69–81. [Google Scholar]
- Gelder SR, Siddall ME.. 2001. Phylogenetic assessment of the branchiobdellidae (Annelida, Clitellata) using 18S rDNA, mitochondrial cytochrome c oxidase subunit I and morphological characters. Zool Scr. 30(3):215–222. [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hamilton CA, Lemmon AR, Lemmon EM, Bond JE.. 2016. Expanding anchored hybrid enrichment to resolve both deep and shallow relationships within the spider tree of life. BMC Evol Biol. 16(1): 10.1186/s12862-016-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck AK, Fallon MJ, Burger CV.. 1979. New host and geographical records for the leech Acanthobdella peledina Grube 1851 (Hirudinea, Acanthobdellidae). J Parasitol. 65(6):989. [Google Scholar]
- Jansson I-M, McLoughlin S, Vajda V.. 2008. Early Jurassic annelid cocoons from eastern Australia. Alcheringa 32(3):285–296. [Google Scholar]
- Kainer D, Lanfear R.. 2015. The effects of partitioning on phylogenetic inference. Mol Biol Evol. 32(6):1611–1627. [DOI] [PubMed] [Google Scholar]
- Killick R, Eckley IA.. 2014. Changepoint: an R package for changepoint analysis. J Stat Soft. 58(3): 10.18637/jss.v058.i03. [DOI] [Google Scholar]
- Kück P, Struck TH.. 2014. BaCoCa – a heuristic software tool for the parallel assessment of sequence biases in hundreds of gene and taxon partitions. Mol Phylogenet Evol. 70:94–98. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. msw260. [DOI] [PubMed] [Google Scholar]
- Le Marrec-Croq F, Drago F, Vizioli J, Sautière P-E, Lefebvre C.. 2013. The leech nervous system: a valuable model to study the microglia involvement in regenerative processes. Clin Dev Immunol. 2013:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon AR, Emme SA, Lemmon EM.. 2012. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst Biol. 61(5):727–744. [DOI] [PubMed] [Google Scholar]
- Livanow N. 1906. Acanthobdella peledina Grube, 1851. Zool Jahrb Anat. 22:637–866. [Google Scholar]
- Marotta R, Ferraguti M, Erséus C, Gustavsson LM.. 2008. Combined-data phylogenetics and character evolution of Clitellata (Annelida) using 18S rDNA and morphology. Zool J Linnean Soc. 154(1):1–26. [Google Scholar]
- Martin P. 2001. On the origin of the Hirudinea and the Demise of the Oligochaeta. Proc R Soc Lond B. 268(1471):1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Kaygorodova I, Sherbakov DY, Verheyen E.. 2000. Rapidly evolving lineages impede the resolution of phylogenetic relationships among Clitellata (Annelida). Mol Phylogenet Evol. 15(3):355–368. [DOI] [PubMed] [Google Scholar]
- Metcalfe JL, Fox ME, Carey JH.. 1988. Freshwater leeches (Hirudinea) as a screening tool for detecting organic contaminants in the environment. Environ Monit Assess. 11(2):147–169. [DOI] [PubMed] [Google Scholar]
- Minelli A, Fusco G.. 2004. Evo-Devo perspectives on segmentation: model organisms, and beyond. Trends Ecol Evol. 19(8):423–429. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Warnow T.. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31(12):i44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, et al. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA. 109(34):13698–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, et al. 2013. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci USA. 110(31):12738–12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, et al. 2018. Phylogenetic analysis of antarctic notothenioids illuminates the utility of RADseq for resolving cenozoic adaptive radiations. Mol Phylogenet Evol. 129:268–279. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL.. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24(4):581–583. [DOI] [PubMed] [Google Scholar]
- Paradis E. 2006. Analysis of phylogenetics and evolution with R. Springer-Verlag New York. [Google Scholar]
- Parker E, Dornburg A, Domínguez-Domínguez O, Piller KR.. 2019. Assessing phylogenetic information to reveal uncertainty in historical data: an example using Goodeinae (Teleostei: Cyprinodontiformes: Goodeidae). Mol Phylogenet Evol. 134:282–290. [DOI] [PubMed] [Google Scholar]
- Parry L, Tanner A, Vinther J.. 2014. The origin of Annelids. Palaeontology 57(6):1091–1103. [Google Scholar]
- Phillips AJ, Siddall ME.. 2009. Poly-paraphyly of Hirudinidae: many lineages of medicinal leeches. BMC Evol Biol. 9:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prum RO, et al. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526(7574):569–573. [DOI] [PubMed] [Google Scholar]
- Purschke G, Westheide W, Rohde D, Brinkhurst RO.. 1993. Morphological reinvestigation and phylogenetic relationship of Acanthobdella peledina (Annelida, Clitellata). Zoomorphology 113(2):91–101. [Google Scholar]
- Reddy S, et al. 2017. Why do phylogenomic data sets yield conflicting trees? Data type influences the avian tree of life more than taxon sampling. Syst Biol. 66(5):857–879. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Lemmon AR, Margres MJ, Aronow K.. 2012. The venom-gland transcriptome of the eastern Diamondback Rattlesnake (Crotalus Adamanteus). BMC Genomics 13(1):312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset V, Plaisance L, Erséus C, Siddall ME, Rouse GW.. 2008. Evolution of habitat preference in Clitellata (Annelida). Biol J Linnean Soc Linnean Soc Lond. 95(3):447–464. [Google Scholar]
- Sawyer RT. 1986. Leech biology and behaviour. New York: Oxford University Press. [Google Scholar]
- Siddall ME, et al. 2001. Validating Livanow: molecular data agree that Leeches, Branchiobdellidans, and Acanthobdella peledina form a monophyletic group of Oligochaetes. Mol Phylogenet Evol. 21(3):346–351. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Burreson EM.. 1995. Phylogeny of the Euhirudinea: independent evolution of blood feeding by leeches? Can J Zool. 73(6):1048–1064. [Google Scholar]
- Siddall ME, Burreson EM.. 1998. Phylogeny of leeches (Hirudinea) based on mitochondrial cytochromec oxidase subunit I. Mol Phylogenet Evol. 9(1):156–162. [DOI] [PubMed] [Google Scholar]
- Slesak G, Inthalath S, Dittrich S, Paris DH, Newton PN.. 2015. Leeches as further potential vectors for rickettsial infections. Proc Natl Acad Sci USA. 112(48):E6593.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck TH, et al. 2011. Phylogenomic analyses unravel annelid evolution. Nature 471(7336):95–98. [DOI] [PubMed] [Google Scholar]
- Tessler M, et al. 2018. Worms that suck: phylogenetic analysis of Hirudinea solidifies the position of Acanthobdellida and Necessitates the dissolution of Rhynchobdellida. Mol Phylogenet Evol. 127:129–134. [DOI] [PubMed] [Google Scholar]
- Thearle MJ. 1998. Leeches in medicine. ANZ J Surg. 68(4):292–295. [DOI] [PubMed] [Google Scholar]
- Townsend JP. 2007. Profiling phylogenetic informativeness. Syst Biol. 56(2):222–231. [DOI] [PubMed] [Google Scholar]
- Townsend JP, Leuenberger C.. 2011. Taxon sampling and the optimal rates of evolution for phylogenetic inference. Syst Biol. 60(3):358.. [DOI] [PubMed] [Google Scholar]
- Townsend JP, Lopez-Giraldez F.. 2010. Optimal selection of gene and ingroup taxon sampling for resolving phylogenetic relationships. Syst Biol. 59(4):446–457. [DOI] [PubMed] [Google Scholar]
- Townsend JP, Su Z, Tekle YI.. 2012. Phylogenetic signal and noise: predicting the power of a data set to resolve phylogeny. Syst Biol. 61(5):835–849. [DOI] [PubMed] [Google Scholar]
- Trontelj P, Sket B, Steinbrück G.. 1999. Molecular phylogeny of leeches: congruence of nuclear and mitochondrial rDNA data sets and the origin of bloodsucking. J Zool Syst Evol Res. 37(3):141–147. [Google Scholar]
- Weisblat DA, Kuo D-H.. 2009. Helobdella (leech): a model for developmental studies. Cold Spring Harb Protoc. 2009(4):pdb.emo121. [DOI] [PubMed] [Google Scholar]
- Williams BW, Gelder SR, Proctor HC, Coltman DW.. 2013. Molecular phylogeny of North American Branchiobdellida (Annelida: Clitellata). Mol Phylogenet Evol. 66(1):30–42. [DOI] [PubMed] [Google Scholar]
- Young AD, et al. 2016. Anchored enrichment dataset for true flies (order Diptera) reveals insights into the phylogeny of flower flies (family Syrphidae). BMC Evol Biol. 16(1):143.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence reads are available on NCBI (accession SAMN11310017–SAMN11310038).
This project - including alignments, phylogenetic software analysis input files, resulting output files, and associated R scripts - has been deposited on Zenodo under DOI:10.5281/ zenodo.2630841; https://zenodo.org/record/2630841#.XKe 78etKhE4. All versions of the data, including those used in the previous published version of this paper, are available.