Abstract
Background
Accumulating evidence suggests immunomodulatory and context-dependent effects of TP53 mutations in cancer. We performed an exploratory analysis of the transcriptional, immunobiological and prognostic associations of TP53 mutations within the gene expression-based consensus molecular subtypes (CMSs) of colorectal cancer (CRC).
Materials and methods
In a single-hospital series of 401 stage I–IV primary CRCs, we sequenced the whole coding region of TP53 and analysed CMS-dependent transcriptional consequences of the mutations by gene expression profiling. Immunomodulatory associations were validated by multiplex, fluorescence-based immunohistochemistry of immune cell markers. Prognostic associations of TP53 mutations were analysed in an aggregated series of 635 patients classified according to CMS, including publicly available data from a French multicentre cohort (GSE39582).
Results
TP53 mutations were found in 60% of the CRCs. However, gene set enrichment analyses indicated that their transcriptional consequences varied among the CMSs and were most pronounced in CMS1-immune and CMS4-mesenchymal. Subtype specificity was primarily seen as an upregulation of gene sets reflecting cell cycle progression in CMS4 and a downregulation of T cell activity in CMS1. The subtype-dependent immunomodulatory associations were reinforced by significant depletion of several immune cell populations in mutated tumours compared with wild-type (wt) tumours exclusively in CMS1, including cytotoxic lymphocytes (adjusted p value in CMS1=0.002 and CMS2−4>0.9, Microenvironment Cell Populations (MCP)-counter algorithm). This was validated by immunohistochemistry-based quantification of tumour infiltrating CD8+ cells. Within CMS1, the immunomodulatory association of TP53 mutations was strongest among microsatellite stable (MSS) tumours, and this translated into a propensity for metastatic disease and poor prognostic value of the mutations specifically in the CMS1/MSS subtype (both series overall survival: TP53 mutation vs wt: HR 5.52, p=0.028).
Conclusions
Integration of TP53 mutation status with the CMS framework in primary CRC suggested subtype-dependent immunobiological associations with prognostic and potentially immunotherapeutic implications, warranting independent validation.
Keywords: colorectal cancer, consensus molecular subtypes (cms), tp53 mutations, immunomodulation, prognosis
Key questions.
What is already known about this subject?
Tumour-infiltrating immune cells have impact on patient prognosis and efficacy of immunotherapy in colorectal cancer (CRC).
The prognostic value of TP53 mutations is debated.
Accumulating evidence suggests context-dependent effects of TP53 mutations, including a potential role in tumour immune responses.
The gene expression-based consensus molecular subtypes (CMSs) of CRC have demonstrated value as a framework for discovery of subtype-specific impact of clinically relevant biomarkers.
What does this study add?
The transcriptional effects of TP53 mutations varied among the CMSs, and the most distinct associations were increased cell cycle progression in CMS4 and downregulation of T cell activity in CMS1.
The CMS1-specific immunomodulatory association with TP53 mutations was of greatest magnitude in microsatellite stable (MSS) tumours, identified by transcriptome signature and validated by immunohistochemistry-based quantification of CD8+ T cells.
A CMS1–MSS-specific association between TP53 mutations and metastatic propensity and poor prognosis was found.
How might this impact on clinical practice?
TP53 mutations may have negative prognostic value in CMS1 MSS tumours, possibly mediated by a subtype-specific effect on tumour immune cell infiltration.
The results suggest that wild-type TP53 may be a positive predictive marker for response to immune checkpoint blockade in CMS1 MSS tumours, while TP53 mutations identify a non-immunogenic subgroup of CMS1 MSS tumours requiring immune-stimulatory adjuvants to maximise the therapeutic benefit of immune therapy.
Introduction
The tumour suppressor gene TP53 has long been known to be a central player in colorectal carcinogenesis1 2 and is among the most frequently mutated genes, both in colorectal cancer (CRC) and across tumour types.3 4 In some studies, TP53 mutated CRCs have been shown to be associated with inferior prognosis and resistance to chemotherapy and radiotherapy.5–7 Furthermore, differential predictive and prognostic impacts of different TP53 mutation types have been suggested.7 8 However, the current evidence for its prognostic and predictive values is not strong enough to justify implementation of TP53 mutation analysis in clinical practice.
An increasing body of evidence shows that TP53 function and mutational consequences are dependent on cell type, differentiation state and environmental signals.9 Cell cycle arrest and apoptosis are the best understood downstream effects of TP53 activity9; however, accumulating evidence suggests that TP53 contributes in the regulation of numerous cellular processes, including tumour immune responses.10 11 Microsatellite instability (MSI) defines a subtype of CRCs with strong anticancer immune responses, caused by a large number of frameshift mutations and corresponding expression of neoantigens,12 13 explaining both the superior prognosis in early-stage CRC14 15 and the great efficacy of immunotherapy in the metastatic setting.16 Furthermore, classification of CRCs according to the four gene expression-based consensus molecular subtypes (CMSs) also identifies a group of particularly immunogenic tumours, CMS1-immune, which comprises the majority of MSI CRCs and a small group of microsatellite stable (MSS) tumours.17 While TP53 mutations are most common in MSS3 and in the CMS2-epithelial/canonical subtype,17 the potentially differential biological effects and clinical relevance of the mutations among molecular subtypes of CRC are unknown. Recent studies have demonstrated the value of CMS as a framework for discovery of novel drug associations,18 as well as immunobiological and prognostic impacts of mutations in cancer-critical genes.19 20 This encouraged us to conduct an exploratory subtype-specific analysis also of TP53 in a population-based series of primary CRCs.
Materials and Methods
Patient material and sample preparation
A total of 401 primary tumour samples from a consecutive series of patients operated for CRC stages I–IV at Oslo University Hospital, Norway, between 2005 and 2014 were analysed. All samples were rapidly frozen in liquid nitrogen and stored at −80°C until further processing. DNA extraction was performed as previously described.21 22 Comprehensive clinical and pathological data were prospectively registered for all patients. To increase the statistical power of the prognostic analyses, publicly available data from a French multicentre cohort of stage I–IV colon cancers23 (n=319 with conclusive CMS classification, TP53 mutation status and clinical data) were included (accessed from National Center for Biotechnology Information’s Gene Expression Omnibus with accession number GSE39582). Clinical and molecular characteristics of both cohorts are summarised in table 1. The research conformed to the Helsinki Declaration and was approved by the Regional Committee for Medical and Health Research Ethics (REC number 1.2005.1629). Written consent was obtained from all patients. The research biobanks have been registered according to national legislation.
Table 1.
Clinical and molecular characteristics of the Oslo series and GSE39582 dataset
| Characteristic | Oslo series (n=401) | GSE39582 (n=319) |
| Age at diagnosis, mean (range) | 71 (27–97) | 68 (24–92) |
| Gender | ||
| Male | 195 (49%) | 179 (56%) |
| Female | 206 (51%) | 140 (44%) |
| Tumour localisation | ||
| Right | 171 (43%) | 119 (37%) |
| Left | 227 (57%) | 200 (63%) |
| Synchronous | 3 (0%) | 0 |
| Stage | ||
| I | 84 (21%) | 21 (7%) |
| II | 151 (38%) | 137 (43%) |
| III | 113 (28%) | 130 (41%) |
| IV | 53 (13%) | 31 (10%) |
| Chemotherapy | ||
| Overall | 87 (22%) | 125 (41%) |
| Stages I, II, III, IV | 0, 6 (4%), 53 (47%), 28 (53%) | 0, 34 (25%), 87 (67%), 4 (24%)* |
| TP53 | ||
| wt | 160 (40%) | 149 (47%) |
| mut | 241 (60%) | 170 (53%) |
| Samples scored (n) | 401 | 319 |
| MSI status† | ||
| MSI | 72 (18%) | 34 (11%) |
| MSS | 320 (82%) | 281 (89%) |
| Samples classified (n) | 392 | 315 |
| CMS† | ||
| 1 | 62 (20%) | 46 (14%) |
| 2 | 138 (44%) | 154 (48%) |
| 3 | 54 (17%) | 43 (14%) |
| 4 | 62 (20%) | 76 (24%) |
| Samples classified (n) | 316 | 319 |
| KRAS† | ||
| wt | 273 (68%) | 195 (62%) |
| mut | 128 (32%) | 122 (39%) |
| Number of samples scored | 401 | 317 |
| BRAF† | ||
| wt | 333 (83%) | 266 (92%) |
| mut | 68 (17%) | 23 (8%) |
| Number of samples scored | 401 | 289 |
*Percentage refers to the fraction of patients with available data on chemotherapy (n=17 among patients with stage IV disease).
†Percentages refer to the fraction of patients with conclusive status of the relevant molecular marker.
CMS, consensus molecular subtype; MSI, microsatellite instability; MSS, microsatellite stable; mut, mutation; wt, wild type.
MSI status and TP53 mutation analysis
MSI analysis was performed as previously described.15TP53 mutation status was assessed in all 401 samples by sequencing the entire coding region (exons 2–11), as well as the first ten and last 10 nucleotides of each intron. Sequencing was done by Sanger sequencing technology using BigDye Terminator V.1.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer’s procedure. The samples were run on the 3730 DNA Analyzer (Applied Biosystems). All electropherograms were scored manually and independently by two of the authors, assisted by the SeqScape V.2.5 and Sequencing Analysis V.5.3.1 software (both Applied Biosystems). All mutations were resequenced for validation. Splice mutations were defined as any mutation affecting any of the first or the last 10 nucleotides of an intron, based on the finding that intron mutations outside the invariant AG and GT dinucleotides in the consensus splice acceptor and splice donor site, respectively, may lead to mis-splicing.24 Tumours harbouring only synonymous mutations were classified as TP53 wild type (wt) in survival analysis. Mutation analyses had previously been performed for KRAS (exon 2: codons 12 and 13)14 and BRAF (codon 600)25 in all 401 samples. KRAS exon 3 codon 61 was analysed in a subset of samples (n=127).
Microarray gene expression analysis, CMS
All samples have previously been analysed for gene expression using Affymetrix GeneChip Human Exon V.1.0 ST Array (n=199)21 26 and can be accessed from GEO (GSE24550, GSE29638, GSE69182 and GSE79959) or Human Transcriptome Array V.2.0 (n=202) (GSE7995927 and GSE965218). The samples were classified according to CMS using the random forest classifier developed by Guinney et al.17 Confident CMS classification was obtained in 79% (n=316) of the tumours using default settings.18 The hallmark gene sets from the Molecular Signatures Database V.5.228 were analysed with the camera package29 to assess biological consequences of TP53 mutations within the individual CMS subgroups. The levels of infiltrating immune and stromal cell populations in tumours were estimated using the R package Microenvironment Cell Population (MCP)-counter V.1.130 on gene expression data. The resulting estimates were compared according to TP53 mutation status and CMS classification using independent samples t-tests and Bonferroni correction to account for multiple hypothesis testing.
Multiplex immunohistochemistry
For validation of immune cell infiltration estimated by gene expression, the majority of the tumours (n=294, 73%) were analysed by immunohistochemistry on tissue microarrays with a core diameter of 1.0 mm per sample. The arrays were constructed using the Minicore three-tissue arrayer from Mitogen. Multiplexed immunohistochemical staining of CD3, CD8 and CD56, together with epithelial and nuclear segmentation markers, was performed on 4 µm thick tissue sections following the Opal protocol (PerkinElmer), using a four-plex kit (NEL810001KT, PerkinElmer), together with Opal 620 reagent (FP1495001KT, PerkinElmer). The Opal protocol was followed except that slide deparaffination, antigen retrieval and antibody stripping were all performed using DAKO buffers (three-in-one, catalogue # K800521-2 and K800421-2) in the PT-link module (DAKO).
The five-plex staining was performed in the following order: anti-CD56 (1:400, Clone MRQ-42; Cell Marque) stained with Opal 620, anti-CD8 (1:800, Clone C8/144B; DAKO) stained with Opal 520, anti-CD3 (1:300, Clone F7.2.38; DAKO) stained with Opal 570, a cocktail of antibodies consisting of anti-E-cadherin (1:20.000, Clone 36; BD Biosciences), anti-pan Cytokeratin (1:4000, C-11; Abcam) and anti-pan Cytokeratin Type I/II (1:2000, AE1/AE3; Thermo Fisher) stained with Opal 690. All primary antibody solutions were incubated for 30 min at room temperature. Nuclei were stained with DAPI prior to mounting in Prolong Diamond Antifade Mountant (Thermo Fisher). The slides were multispectrally imaged using the Vectra Imaging platform V.3.0.4 (PerkinElmer) at ×20 magnification. Images were spectrally unmixed in Inform V.2.3.0 (PerkinElmer) using spectra derived from single-plex images of samples stained with each fluorophore individually, and tissue autofluorescence was removed by using a spectrum derived from unstained tissue but otherwise similarly treated. A training algorithm built in the same software was used to classify the tissue into epithelial and stromal regions based on the corresponding markers, and finally individual cells were segmented based on the DAPI signal. Areas with necrotic tissue or any technical artefacts were excluded from subsequent analyses.
The raw data file containing data on each individual cell in the entire cohort was analysed using R V.3.3.1, and cell positivity thresholds for each marker were determined based on evaluation of the expression distribution across all cells, as well as by careful manual inspection of images in Inform to evaluate the signal-to-noise level across individual samples. To assess how sensitive the analysis was to the threshold selected, we tested a high threshold and a low threshold for each marker, one above and one below what we determined manually. Cell positivity fractions based on the two thresholds were calculated by summing up the number of positive cells for each marker and dividing by the total number of cells in each tissue sample. Comparison of these data resulted in highly correlated cell positivity fractions across samples (Pearson correlation coefficient; CD3=0.95, CD8=0.98, CD56=0.94), indicating a strong agreement regardless of thresholds selected. The high thresholds for each marker were subsequently used for downstream analysis.
Statistical analyses
Statistical analyses were performed with the SPSS V.21.0 software. Fisher’s exact test or Spearman correlation test was used when appropriate to test for association between categorical variables. Five-year overall survival (OS) and 5-year relapse-free survival were defined according to the guidelines by Punt et al,31with date of surgery as starting point for both endpoints. Survival curves were generated using the Kaplan-Meier method, and differences between curves were assessed with the log-rank test. The Cox proportional hazards model was used for univariable and multivariable analyses to assess the independence of prognostic factors. All statistical tests were two-tailed and results with p<0.05 were considered significant.
Results
Distinct transcriptional associations of TP53 mutations among the CMSs
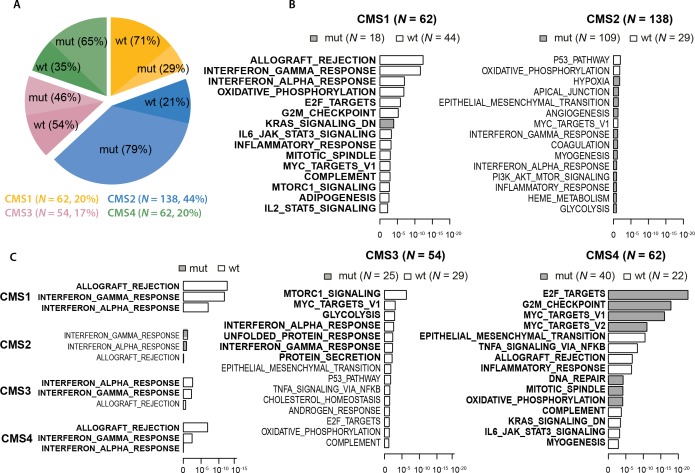
TP53 mutations were detected in 60.1% (n=241) of the 401 cases in the Oslo series. Consistent with the original CMS publication,17 the TP53 mutation rate varied across the CMS groups (p<0.001, Fisher’s exact test; figure 1A, online supplementary table S1A) and was highest in CMS2 (79%) and CMS4 (65%). CMS1 had the lowest mutation rate (29%), in accordance with the enrichment of MSI in this subtype. When analysing MSS tumours only, the mutation rate in CMS1 increased to 63%, while remaining similar in the three other subtypes (online supplementary table S1B).
Figure 1.
TP53 mut rates and their transcriptional and immunomodulatory effects according to the CMSs. (A) The distribution of CMS groups and their TP53 mut rates in the Oslo series (N=316 CMS-classified samples). Numbers to the right refer to the amount and proportion of tumours in each CMS group. (B) Gene set analysis of 50 ‘hallmark gene sets' between TP53 wt and mut tumours within the individual CMS groups. The 15 most differentially expressed gene sets in each CMS group are shown, ranked by unadjusted p values (x-axes, log10 scale). Grey and white indicate gene sets with upregulation in TP53 mutated and wt samples, respectively. Significantly differentially expressed gene sets (FDR values <0.05) are highlighted in bold. (C) Significance levels (x-axis) from expression enrichment analysis of gene sets reflecting T cell activity between TP53 wt and mutated tumours according to CMS group. Analysis was performed on 316 CMS-classified samples in the Oslo series. CMS, consensus molecular subtype; FDR, False Discovery Rate; IL, interleukin; mut, mutation; wt, wild type.
esmoopen-2019-000523supp001.pdf (1.2MB, pdf)
To investigate CMS-dependent phenotypical effects of TP53 mutations, gene set enrichment analysis of the 50 ‘'hallmark gene sets’, which represent various biological states and processes,28 was performed comparing TP53 wt and mutated tumours within each CMS group. This indicated that TP53 mutations had the most distinct effects in CMS1 and CMS4, and were associated with significant dysregulation of 20 (40%) and 24 (48%) of the gene sets, respectively, compared with 7 (14%) in CMS3 and none in CMS2 (online supplementary table S2). Comparing CMS1 and CMS4, most (16) of the differentially expressed gene sets overlapped between the two subtypes. However, in 50% of these, TP53 mutations were associated with dysregulation in the opposite direction in the two subtypes, indicating subgroup-specific transcriptomic consequences. This included upregulation of gene sets reflecting cell cycle progression (eg, E2F_TARGETS, G2M_CHECKPOINT and MYC_TARGETS_V1) in TP53 mutated tumours in CMS4 and downregulation in CMS1 (figure 1B, online supplementary table S2). Intriguingly, gene sets reflecting T cell activity (ie, ALLOGRAFT_REJECTION and INTERFERON_GAMMA/ALPHA_RESPONSE) were the most significantly downregulated gene sets in TP53 mutated tumours in CMS1. Indications of immunomodulatory effects of TP53 mutations could also be seen in CMS3 and CMS4, but of weaker statistical significance than in CMS1 (figure 1C).
CMS-dependent association between TP53 mutation status and immune cell infiltration
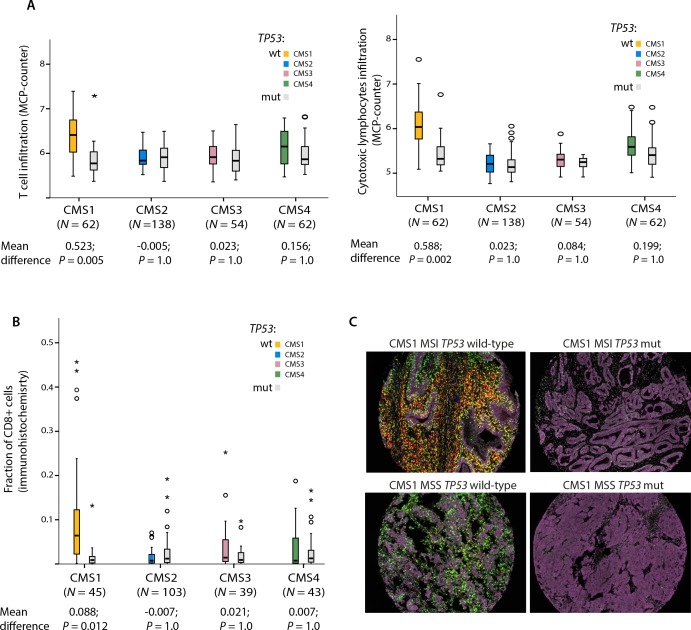
To scrutinise the potential immunomodulatory effect of TP53 mutations according to CMS in the Oslo series, we quantified the abundances of infiltrating immune and stromal cell populations in the tumours using the gene expression-based MCP-counter algorithm.30 Several cell populations of the innate and adaptive immune systems, including T cells and cytotoxic lymphocytes, were significantly depleted in TP53 mutated compared with wt tumours in CMS1. In contrast, no significant association between TP53 mutations and any kind of immune or stromal cell infiltration was seen in any of the other CMS groups (table 2 and figure 2A).
Table 2.
Tumour immune cell infiltration according to TP53 mutation status within the consensus molecular subtypes (Oslo series, N=316)
| Cell population | CMS1 (n=62) | CMS2 (n=138) | CMS3 (n=54) | CMS4 (n=62) | ||||
| Mean difference* | Adjusted P value† | Mean difference* | Adjusted P value† | Mean difference* | Adjusted p value† | Mean difference* | Adjusted P value | |
| T cells | 0.52 | 0.005 | −0.005 | 1.0 | 0.023 | 1.0 | 0.16 | 1.0 |
| CD8 T cells | 0.24 | 0.76 | −0.055 | 1.0 | 0.06 | 1.0 | 0.24 | 0.16 |
| Cytotoxic lymphocytes | 0.59 | 0.002 | 0.023 | 1.0 | 0.084 | 1.0 | 0.20 | 1.0 |
| NK cells | 0.22 | 0.12 | 0.025 | 1.0 | 0.034 | 1.0 | 0.21 | 0.12 |
| B lineage | 0.14 | 1.0 | −0.01 | 1.0 | −0.19 | 1.0 | 0.37 | 1.0 |
| Monocytic lineage | 0.66 | 0.006 | −0.098 | 1.0 | 0.082 | 1.0 | 0.32 | 1.0 |
| Myeloid dendritic cells | 0.31 | 0.03 | −0.011 | 1.0 | −0.04 | 1.0 | 0.21 | 1.0 |
| Neutrophils | 0.012 | 0.90 | 0.083 | 0.76 | 0.025 | 1.0 | 0.25 | 0.24 |
| Endothelial cells | 0.054 | 1.0 | −0.12 | 0.96 | 0.011 | 1.0 | 0.09 | 1.0 |
| Fibroblasts | 0.14 | 1.0 | −0.107 | 1.0 | 0.233 | 1.0 | 0.43 | 0.2 |
*Difference in abundances of eight immune and two stromal cell populations between TP53 wild-type and mutated tumours (positive values indicate higher infiltration in wild type), measured based on gene expression (Microenvironment Cell Populations-counter algorithm).
†Independent samples t-test. P values have been adjusted for multiple hypotheses testing according to Bonferroni (multiplied by 40), and adjusted p values <0.05 are highlighted in bold.
CMS, consensus molecular subtype.
Figure 2.
Tumour immune cell quantification by multiplex immunohistochemistry. (A) Infiltration levels of T cells and cytotoxic lymphocytes in the microenvironment of TP53 wild-type and mutated tumours in CMS1-4 in the Oslo series (N=316). Abundances of the various cell populations are estimated based on gene expression (MCP-counter algorithm). P values from independent samples t-test have been multiplied by 40 to account for multiple hypotheses testing according to Bonferroni; all test results are presented in table 1. (B) Comparison of differences in the abundance of CD8+ cells between TP53 wild-type and mutated tumours in CMS1-4. Analysis performed on 230 patients with confident CMS classification and available multiplex immunohistochemistry data in the Oslo series. P values from independent samples t-test have been multiplied by 12 to account for multiple hypotheses testing according to Bonferroni; all test results are presented in online supplementary table S4. (C) Illustration of immune cell infiltration according to MSI and TP53 mutation status in four CMS1 CRCs. Epithelial cells in purple, CD8+ cells in green, CD3+ cells in red and CD56+ cells in blue. Yellow represents colocalised signals from CD8 and CD3. CMS, consensus molecular subtype; CRC, colorectal cancer; MCP, Microenvironment Cell Population; MSI, microsatellite instability; mut, mutation; wt, wild type.
This CMS1-specific role of TP53 mutations was of the greatest magnitude and was statistically significant only among MSS tumours, while a similar non-significant trend was seen in MSI tumours (online supplementary figure S1). Performing the same analysis according to MSI status, but irrespective of CMS classification, did not reveal any clear differences in T cell infiltration (online supplementary table S3), suggesting that the immunomodulatory associations of TP53 mutations were more dependent on the transcriptomic subtypes than the mutator phenotype. Quantification of immune cell infiltration by multiplex immunohistochemistry validated the CMS-dependent immunomodulatory associations; TP53 mutated CMS1 tumours exhibited significantly fewer CD8+ T cells than its wt counterpart, while no similar trends were seen in any other CMS group (figure 2B–C and online supplementary table S4). However, we were not able to validate the CMS-dependent correlation between immune infiltration and TP53 mutation status in the gene expression data from the French multicentre cohort.
TP53 mutations have CMS-dependent prognostic impact in correspondence with the immunomodulatory association
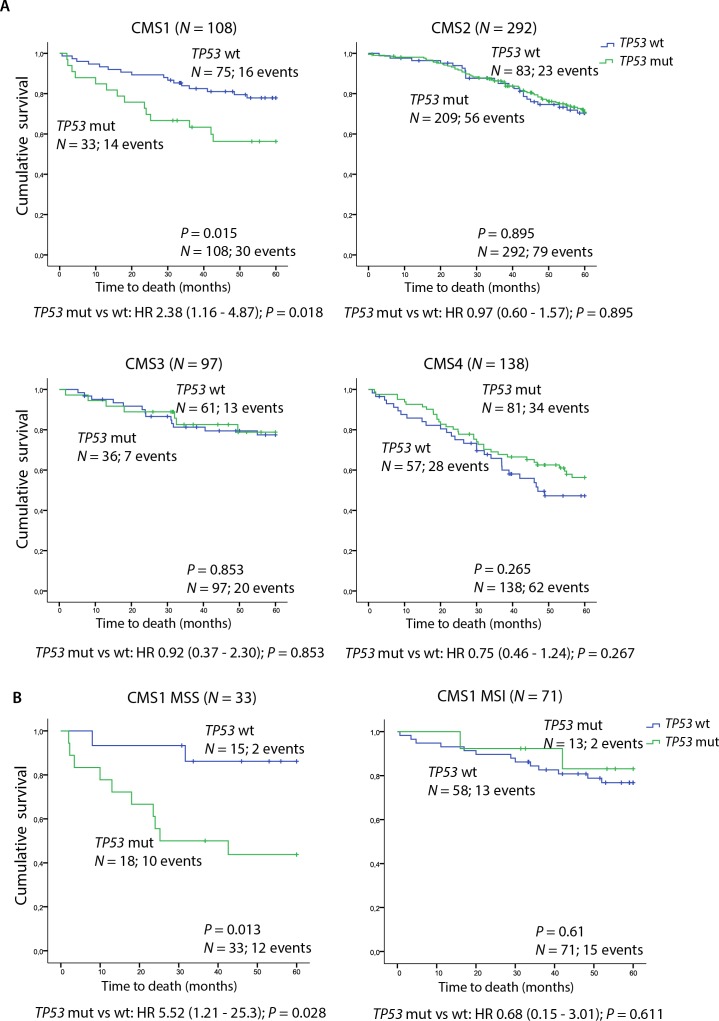
Further, we investigated whether the CMS-dependent correlation between TP53 mutations and immune profiles seen in the in-house series corresponded with patient prognosis. To increase the statistical power of subgroup analyses, public data from the French multicentre cohort23 were added, giving a total of 635 patients with confident CMS classification and TP53 mutation status. TP53 mutations were clearly associated with inferior OS in CMS1 (HR 2.38 (95% CI 1.16–4.87), p=0.018; figure 3A), whereas no significant prognostic effect was seen in any of the other subtypes. CMS1-specific correlation between TP53 mutations and poor prognosis was also seen in each patient series independently, although non-significant (online supplementary figure S2). Further stratification according to MSI status showed that the negative prognostic impact in CMS1 was specific to MSS tumours (MSS: HR 5.52 (95 % CI 1.21–25.3), p=0.028 vs MSI: HR 0.68 (95% CI 0.15–3.01), p=0.611, pinteraction=0.049; figure 3B), corresponding with the stronger association between TP53 mutations and immune infiltration in this subgroup. Here, TP53 mutations also had a negative prognostic effect in multivariable analysis incorporating age, gender, location, stage and BRAFV600E mutation status (HR 14.2 (95% CI 1.06–190), p=0.045). A negative prognostic value of TP53 mutations specific to CMS1 MSS tumours was found in each patient series independently, although not reaching statistical significance (online supplementary figure S3). Splitting the survival analysis into stage I–IV disease showed that the negative prognostic effect in CMS1 MSS was mainly explained by an enrichment of stage IV disease among TP53 mutated tumours in this subgroup (p=0.009, online supplementary figure S4 and table S5). Notably, an association between TP53 mutations and metastatic disease was not found in the other molecular subgroups (online supplementary table S5).
Figure 3.
Prognostic impact of TP53 mutations according to the consensus molecular subtypes. (A) Kaplan-Meier survival curves showing 5-year OS comparing TP53 wt and mutated tumours in patients with conclusive CMS classification in the combined Oslo series and French multicentre cohort (N=635). (B) Five-year OS comparing TP53 wt and mutated tumours according to MSI status in patients with CMS1 tumours (N=104) from the combined patient cohorts. CMS, consensus molecular subtype; MSI, microsatellite instability; MSS, microsatellite stable; mut, mutation; OS, overall survival; wt, wild type.
Discussion
In our population-based series of primary CRCs, TP53 mutations have subtype-dependent associations with metastatic propensity and patient prognosis, potentially mediated by a CMS1-specific immunomodulatory effect. Accumulating experimental and clinical data have demonstrated that TP53 dysfunction skews the tumour microenvironment towards a state of chronic inflammation, characterised by infiltration of immune-suppressing and tumour-promoting cells, such as regulatory T cells and myeloid-derived suppressor cells, in disfavour of cytotoxic T cells.11 Further, TP53 has been shown to be involved in an immune-stimulating positive feedback loop with interferon signalling, which can be activated by both viral infection and oncogenic stress.10 32 Our results may suggest that the loss of this TP53-dependent function in CRC is most pronounced in CMS1, where an association with metastatic disease and poor survival is seen. The positive prognostic impact of T cell infiltration in CRC is well established,33 34 and CMS1 tumours have gene expression profiles characterised by immune infiltration, primarily of TH1 and cytotoxic T cells.17 We therefore propose that a TP53 mutant-mediated transformation towards an immunosuppressive milieu could be of particular importance in CMS1, whose good prognosis in early-stage CRC is dependent on the presence of active, cytotoxic T cells. This highlights the relevance of investigating biomarkers within biologically homogeneous subgroups and conforms to comprehensive preclinical and clinical data showing that both wt TP53 activity and the consequences of TP53 mutations are highly dependent on the cellular and microenvironmental contexts.9 Recent studies have revealed associations between increased WNT35 and TGFβ signalling36 and immune evasion in CRC, and our results may suggest that additional canonical pathways in colorectal carcinogenesis have significant impact on tumour immune responses.
However, our findings must be interpreted with caution due to small sample sizes with increasing substratification. This is accentuated by the lack of biological validation in the French multicentre series. Yet, the inability to validate the CMS-dependent immunobiological associations of TP53 mutations using gene expression data may be due to technical limitations. The MCP-counter scores estimating the various cell population abundances are dependent on the gene expression platform used to produce the data,30 potentially confounding comparison between the patient series. Immunohistochemical staining is still considered the gold standard for quantification of immune cell populations and was used to validate the subtype-dependent immunobiological gene expression patterns found in the in-house series. Moreover, despite major advances in gene expression-based classifications of CRC, the specific transcriptomic contribution from and the interaction between stromal and cancer cells is still uncertain and these analyses may be influenced by factors such as tumour purity.37 The lack of external immunobiological validation could also partly be due to the lower TP53 mutation rate (53%) in the French cohort. The TP53 mutation rate of 60% in our in-house series is also higher than in previously published large materials8 and databases38 on CRC, with TP53 mutation rates of 42% and 43%, respectively. Similarly, the TP53 mutation rates were clearly higher across all CMS groups in our material compared with the results in Guinney et al.17 These discrepancies can largely be explained by the coverage of all exons and intronic splice regions in our material, highlighting the importance of non-restricted sequencing when examining the biological and clinical consequences of TP53 mutations. Moreover, the molecular underpinnings of the subtype-dependent immunomodulatory associations of TP53 mutations remain to be elucidated. This highlights the exploratory nature of our study, warranting both validation in independent clinical cohorts and functional studies to provide a mechanistic explanation of our findings.
In addition to MSI, CMS1 is a potential positive predictive marker for response to immune checkpoint inhibition. Of potential relevance to stratified treatment, our findings may suggest that wt TP53 pinpoints the most highly immunogenic CMS1 MSS tumours. The results may also point to a subgroup of non-immunogenic, TP53 mutated CMS1 MSS tumours in need of immune-stimulatory adjuvants to maximise the potential therapeutic benefit of checkpoint inhibitors. One potential strategy for this subgroup would be to reactivate the TP53 pathway, which has been shown to induce immunogenic cell death in preclinical models, likely through activation of innate immunity.39 This strategy is currently limited by the lack of TP53-reactivating drugs being approved for clinical use. However, pharmacological reactivators of mutant TP53, such as APR-246 have shown clinical efficacy40 and are currently under further evaluation in clinical trials. Although the immunomodulatory properties of such TP53-targeting drugs are yet to be explored, the increasing evidence of an interaction between TP53 function and antitumour immunity warrants investigations of their combinatorial use with immune-stimulatory drugs.
In conclusion and in agreement with the importance of combined biomarker analysis for precision cancer medicine, this work suggests that integrated analysis of TP53 mutations in the CMS framework may identify a subgroup of CRCs in which TP53 has strong immunomodulatory associations. A potential clinical relevance was reinforced by a poor prognostic value of the mutations specifically in this CMS1–MSS subgroup, also encouraging analyses of TP53 mutation status as a predictive marker for immunotherapy in CMS1.
Footnotes
Contributors: Conception and design: JS, AS and RAL. Development of methodology: AS, JB and RAL. Acquisition of data: JS, CHB, IAE, SAD, MH, MGG, JB and AN. Analysis and interpretation of data: JS, AS, CHB, IAE, SAD, PWE, AN, JB and RAL. Writing of the manuscript: JS, AS, MGG and RAL. Review and/or revision of the manuscript: all authors. Study supervision: AS and RAL.
Funding: This work was supported by the Norwegian Cancer Society (project numbers 6824048-2016 and 182759-2016); the foundation Stiftelsen Kristian Gerhard Jebsen; the South-Eastern Norway Regional Health Authority (project number 2016123) and the Research Council of Norway (FRIPRO Toppforsk, project number 250993).
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The research was approved by the Regional Committee for Medical and Health Research Ethics South-Eastern Norway (REC number 1.2005.1629).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available in a public, open access repository.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- 2.Lothe RA, Fossli T, Danielsen HE, et al. . Molecular genetic studies of tumor suppressor gene regions on chromosomes 13 and 17 in colorectal tumors. J Natl Cancer Inst 1992;84:1100–8. 10.1093/jnci/84.14.1100 [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kandoth C, McLellan MD, Vandin F, et al. . Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munro AJ, Lain S, Lane DP. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer 2005;92:434–44. 10.1038/sj.bjc.6602358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat 2003;21:271–6. 10.1002/humu.10175 [DOI] [PubMed] [Google Scholar]
- 7.Diep CB, Thorstensen L, Meling GI, et al. . Genetic tumor markers with prognostic impact in Dukes' stages B and C colorectal cancer patients. J Clin Oncol 2003;21:820–9. 10.1200/JCO.2003.05.190 [DOI] [PubMed] [Google Scholar]
- 8.Russo A, Bazan V, Iacopetta B, et al. . The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol 2005;23:7518–28. 10.1200/JCO.2005.00.471 [DOI] [PubMed] [Google Scholar]
- 9.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell 2017;170:1062–78. 10.1016/j.cell.2017.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muñoz-Fontela C, Mandinova A, Aaronson SA, et al. . Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat Rev Immunol 2016;16:741–50. 10.1038/nri.2016.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y, Guo G. Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int J Mol Sci 2016;17 10.3390/ijms17111942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turajlic S, Litchfield K, Xu H, et al. . Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 2017;18:1009–21. 10.1016/S1470-2045(17)30516-8 [DOI] [PubMed] [Google Scholar]
- 13.Germano G, Lamba S, Rospo G, et al. . Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 2017;552:116–20. 10.1038/nature24673 [DOI] [PubMed] [Google Scholar]
- 14.Dienstmann R, Mason MJ, Sinicrope FA, et al. . Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann Oncol 2017;28:1023–31. 10.1093/annonc/mdx052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merok MA, Ahlquist T, Røyrvik EC, et al. . Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol 2013;24:1274–82. 10.1093/annonc/mds614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le DT, Durham JN, Smith KN, et al. . Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guinney J, Dienstmann R, Wang X, et al. . The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sveen A, Bruun J, Eide PW, et al. . Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res 2018;24:794–806. 10.1158/1078-0432.CCR-17-1234 [DOI] [PubMed] [Google Scholar]
- 19.Lal N, White BS, Goussous G, et al. . KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin Cancer Res 2018;24:224–33. 10.1158/1078-0432.CCR-17-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smeby J, Sveen A, Merok MA, et al. . CMS-dependent prognostic impact of KRAS and BRAFV600E mutations in primary colorectal cancer. Ann Oncol 2018;29:1227–34. 10.1093/annonc/mdy085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ågesen TH, Sveen A, Merok MA, et al. . ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis. Gut 2012;61:1560–7. 10.1136/gutjnl-2011-301179 [DOI] [PubMed] [Google Scholar]
- 22.Berg M, Agesen TH, Thiis-Evensen E, et al. . Distinct high resolution genome profiles of early onset and late onset colorectal cancer integrated with gene expression data identify candidate susceptibility loci. Mol Cancer 2010;9 10.1186/1476-4598-9-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marisa L, de Reyniès A, Duval A, et al. . Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453 10.1371/journal.pmed.1001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson SL, Coli R, Daly IW, et al. . Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet 2001;68:753–8. 10.1086/318808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vedeld HM, Merok M, Jeanmougin M, et al. . CpG island methylator phenotype identifies high risk patients among microsatellite stable BRAF mutated colorectal cancers. Int J Cancer 2017;141:967–76. 10.1002/ijc.30796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sveen A, Ågesen TH, Nesbakken A, et al. . ColoGuidePro: a prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res 2012;18:6001–10. 10.1158/1078-0432.CCR-11-3302 [DOI] [PubMed] [Google Scholar]
- 27.Sveen A, Johannessen B, Tengs T, et al. . Multilevel genomics of colorectal cancers with microsatellite instability-clinical impact of JAK1 mutations and consensus molecular subtype 1. Genome Med 2017;9 10.1186/s13073-017-0434-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Subramanian A, Pinchback R, et al. . Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res 2012;40:e133 10.1093/nar/gks461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becht E, Giraldo NA, Lacroix L, et al. . Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17 10.1186/s13059-016-1070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Punt CJA, Buyse M, Köhne C-H, et al. . Endpoints in adjuvant treatment trials: a systematic review of the literature in colon cancer and proposed definitions for future trials. J Natl Cancer Inst 2007;99:998–1003. 10.1093/jnci/djm024 [DOI] [PubMed] [Google Scholar]
- 32.Takaoka A, Hayakawa S, Yanai H, et al. . Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003;424:516–23. 10.1038/nature01850 [DOI] [PubMed] [Google Scholar]
- 33.Galon J, Costes A, Sanchez-Cabo F, et al. . Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–4. 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 34.Pagès F, Berger A, Camus M, et al. . Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005;353:2654–66. 10.1056/NEJMoa051424 [DOI] [PubMed] [Google Scholar]
- 35.Grasso CS, Giannakis M, Wells DK, et al. . Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov 2018;8:730–49. 10.1158/2159-8290.CD-17-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tauriello DVF, Palomo-Ponce S, Stork D, et al. . TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- 37.Isella C, Terrasi A, Bellomo SE, et al. . Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015;47:312–9. 10.1038/ng.3224 [DOI] [PubMed] [Google Scholar]
- 38.Bouaoun L, Sonkin D, Ardin M, et al. . TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat 2016;37:865–76. 10.1002/humu.23035 [DOI] [PubMed] [Google Scholar]
- 39.Xue W, Zender L, Miething C, et al. . Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007;445:656–60. 10.1038/nature05529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehmann S, Bykov VJN, Ali D, et al. . Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol 2012;30:3633–9. 10.1200/JCO.2011.40.7783 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2019-000523supp001.pdf (1.2MB, pdf)