A genome-wide association study recently identified an MSH3/DHFR locus associated with Huntington’s disease progression. Flower, Lomeikaite et al. identify tandem repeat variants at this locus, and show that a three-repeat allele is associated with reduced somatic expansion, delayed onset and slower progression in Huntington’s disease and myotonic dystrophy type 1.
Keywords: Huntington’s disease, myotonic dystrophy, transcriptomics, movement disorders, association study
Abstract
The mismatch repair gene MSH3 has been implicated as a genetic modifier of the CAG·CTG repeat expansion disorders Huntington’s disease and myotonic dystrophy type 1. A recent Huntington’s disease genome-wide association study found rs557874766, an imputed single nucleotide polymorphism located within a polymorphic 9 bp tandem repeat in MSH3/DHFR, as the variant most significantly associated with progression in Huntington’s disease. Using Illumina sequencing in Huntington’s disease and myotonic dystrophy type 1 subjects, we show that rs557874766 is an alignment artefact, the minor allele for which corresponds to a three-repeat allele in MSH3 exon 1 that is associated with a reduced rate of somatic CAG·CTG expansion (P = 0.004) and delayed disease onset (P = 0.003) in both Huntington’s disease and myotonic dystrophy type 1, and slower progression (P = 3.86 × 10−7) in Huntington’s disease. RNA-Seq of whole blood in the Huntington’s disease subjects found that repeat variants are associated with MSH3 and DHFR expression. A transcriptome-wide association study in the Huntington’s disease cohort found increased MSH3 and DHFR expression are associated with disease progression. These results suggest that variation in the MSH3 exon 1 repeat region influences somatic expansion and disease phenotype in Huntington’s disease and myotonic dystrophy type 1, and suggests a common DNA repair mechanism operates in both repeat expansion diseases.
Introduction
Huntington’s disease and myotonic dystrophy type 1 (DM1) are autosomal dominant disorders caused by CAG·CTG trinucleotide repeat expansions. Huntington’s disease is characterized by a progressive movement disorder, cognitive impairment and psychiatric symptoms (Bates et al., 2014), and DM1 by myotonia, muscular dystrophy, cognitive impairment, cardiac conduction defects and endocrine dysfunction (Harper, 2001). No disease-modifying treatments are available for either (Bates et al., 2015; Meola and Cardani, 2015).
Huntington’s disease is caused by a (CAG)n repeat expansion in HTT exon 1 and DM1 by a (CTG)n expansion in the 3′ untranslated region (UTR) of DMPK (Brook et al., 1992; Bates et al., 2014). In both, inherited repeat length is the major determinant of disease course, correlating inversely with the age at onset and positively with disease severity. The repeat is unstable, and expansion during germline transmission results in genetic anticipation (Hunter et al., 1992; Bates et al., 2014). Repeat tracts are also unstable in somatic cells, tending to expand over time, particularly in Huntington’s disease striatum (Kennedy et al., 2003) and DM1 muscle (Ashizawa et al., 1993), the most prominently affected tissues in each disease. Such expansion-biased, age-dependent and tissue-specific somatic instability is thought to contribute to disease onset and progression (Kennedy et al., 2003; Shelbourne et al., 2007; Swami et al., 2009; Morales et al., 2012).
In mouse models, the DNA mismatch repair proteins MSH2 and MSH3 are essential for CAG·CTG repeat expansion, and their inactivation limits expansion events and improves disease phenotype (van den Broek et al., 2002; Foiry et al., 2006; Dragileva et al., 2009; Pinto et al., 2013; Tome et al., 2013). In patients with DM1, a candidate gene association study reported a coding single nucleotide polymorphism (SNP) (rs26279, p.A1045T) in MSH3 exon 23 that was associated with the rate of somatic expansion (Morales et al., 2016). Genome-wide association studies (GWAS) in patients with Huntington’s disease identified variation in DNA repair genes that modify disease course, and pathway analyses in each study further highlighted DNA repair (GeM-HD, 2015; Moss et al., 2017; Lee et al., 2017). Such variants also influence onset in other CAG expansion diseases, suggesting a common mechanism operates in conditions caused by repeat expansion (Bettencourt et al., 2016). The lead variant in a recent GWAS linking MSH3 with Huntington’s disease progression was the imputed SNP rs557874766, which nominally results in Pro67Ala at the N-terminus (Moss et al., 2017).
However, rs557874766 is located within a 9 bp tandem repeat in exon 1 of MSH3 and the 5′ UTR of the dihydrofolate reductase gene (DHFR) on the opposite strand. This repeat is polymorphic in copy number (Nakajima et al., 1995; Morales et al., 2016) and sequence (Morales, 2006), which led us to hypothesize that rs557874766 could be an alignment artefact. Additionally, the 500-bp region flanking the MSH3 repeat is highly polymorphic, containing six SNPs and a 1-bp indel. We conducted targeted Illumina sequencing of the MSH3 exon 1 region in 218 Huntington’s disease and 247 DM1 subjects, which allowed us to obtain accurate haplotype information for the region. Using whole blood RNA-Seq in Huntington’s disease, we investigated whether sequence variation at the MSH3/DHFR locus influences their expression.
Materials and methods
Cohorts
The 218 Huntington’s disease subjects were from TRACK-HD (Tabrizi et al., 2009). The DM1OPTIMISTIC cohort of 247 subjects was from OPTIMISTIC (van Engelen and Consortium, 2015) and the independent DM1CostaRica cohort of 199 subjects was previously reported in Morales et al. (2016).
Progenitor allele length
Progenitor pure CAG length for Huntington’s disease was determined by MiSeq sequencing (Ciosi et al., 2018). Five subjects were excluded because they were part of a twin pair (n = 1) or the progenitor CAG length could not be unambiguously identified (n = 4) (Ciosi et al., unpublished results). DM1 progenitor allele length was determined by small pool PCR (van Engelen and Consortium, 2015; Cumming et al., in press). DM1 patients were tested for CCG repeat interruptions, known cis-modifiers of CTG repeat stability and disease phenotype (Cumming et al., 2018, in press).
Phenotypes
Two phenotypes were common to both cohorts: age at onset and rate of somatic expansion of the pathogenic CAG·CTG repeat. Huntington’s disease age at onset represents onset of motor symptoms (Tabrizi et al., 2009). DM1 age at onset was subject self-assessment of the first occurrence of symptoms likely related to DM1 (Cumming et al., in press). Somatic CAG·CTG expansion in blood was previously quantified in both cohorts (Ciosi et al., unpublished results; Cumming et al., in press). For Huntington’s disease MiSeq data, the measure of somatic expansion was the proportion of reads in the sample that correspond to somatic expansions (reads with more CAG repeats than the progenitor allele) relative to the number of reads obtained for the progenitor allele (Ciosi et al., unpublished results). For DM1, it was the difference in number of repeats between the modal allele and the estimated progenitor allele length (Cumming et al., 2018). In both cohorts, relative rate of somatic expansion corresponds to the variation in the measures of somatic expansion that is not explained by age and CAG·CTG repeat length. Positive values reflect a faster rate of somatic expansion.
Two phenotypes were only available for Huntington’s disease; progression score (Moss et al., 2017) and gene expression. Progression score was derived for 213 TRACK-HD subjects in Ciosi et al. (unpublished results), as described in Moss et al. (2017). It measures typical Huntington’s disease progression that is not explained by age and pure CAG repeat length, with positive scores reflecting faster progression. Blood MSH3 and DHFR expression levels were available for 108 Huntington’s disease subjects (Moss et al., 2017).
Illumina sequencing of MSH3 exon 1
MiSeq amplicon sequencing, adapted from Ciosi et al. (2018), was used to genotype the MSH3 exon 1 repeat and flanking variants (Supplementary Fig. 1). The region was amplified using locus-specific primers incorporating Illumina indexed adaptors (Supplementary Table 1) (Ciosi et al., 2018). PCR was carried out using 10 ng of blood genomic DNA, 10% DMSO, 1 µM of each primer, 1× Custom PCR master mix (Thermo Scientific, SM0005), 0.048% (v/v) 2-mercaptoethanol and 0.5 U of Taq polymerase (Sigma) in a total volume of 10 µl. Thermal cycling conditions were: an initial denaturation at 96°C for 5 min, followed by 30 cycles of (96°C for 45 s), (60°C for 45 s) and (70°C for 2 min), with a final extension at 65°C for 1 min followed by 70°C for 10 min. Six hundred sequencing cycles were run 400 nt forward, 200 nt reverse. Quality control confirmed >80% of bases had Phred quality >30.
Bioinformatic analyses
Genotyping was conducted on the University of Glasgow Galaxy platform (heighliner.cvr.gla.ac.uk). Paired-end reads were merged and aligned to multiple references corresponding to potential 9 bp repeat alleles (Supplementary material), followed by variant calling. For repeat homozygotes, haplotypes were confirmed from .sam files using Tablet (Milne et al., 2013). The Galaxy workflow is available at https://www.myexperiment.org/workflows/5087.html. Conservation analysis used PhastCons and PhyloP (UCSC), with species sequence alignment in Clustal Omega.
Transcriptome-wide association study
The transcriptome-wide association study (TWAS) method of Gusev et al. (2016) was used to impute cortical gene expression from 452 dorsolateral prefrontal cortex samples from the CommonMind Consortium (CMC, 2017) into the TRACK-HD GWAS of Huntington’s disease progression (n = 243) (Moss et al., 2017). Following the Gusev et al. (2016) approach, we tested association between imputed cortical gene expression and Huntington’s disease progression.
Statistical analyses
Linear regression modelling of genotype-phenotype correlation was conducted in R (R Core Team, 2013). An additive genetic model was used to score genotypes. For age at onset analysis, we controlled for CAG·CTG repeat length in Huntington’s disease and DM1, and for repeat interruptions in DM1 (Supplementary Table 4). Meta-analysis of somatic expansion and age at onset in Huntington’s disease and DM1 was conducted with METAL (Willer et al., 2010). PLINK 1.07 (Purcell et al., 2007) was used to derive allele frequencies, Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium. Haplotype relationships were visualized as a network using median joining on NETWORK (Bandelt et al., 1999).
Data availability
Data are available from the corresponding author on request.
Results
Rs557874766 is an alignment artefact
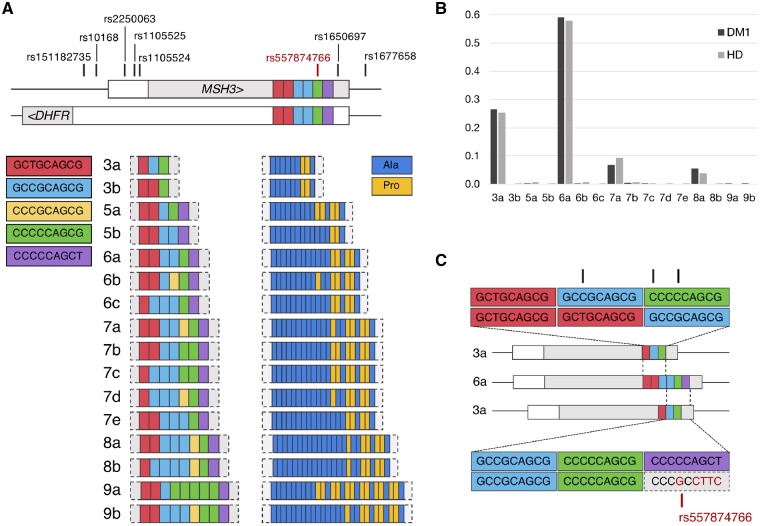
We observed 16 MSH3 repeat alleles, differing in sequence and length from three to nine repeats (Fig. 1A and Supplementary Table 2). Alleles contained combinations of five types of repeat units, with coding potential for proline or alanine (Fig. 1A). They were numbered by repeat length, suffixed alphabetically by frequency i.e. ‘3a’ represents the most common three-repeat allele.
Figure 1.
MSH3/DHFR 9bp tandem repeat allele structure and frequency observed in Huntington’s disease and DM1 cohorts. (A) Schematic representation of the 9 bp tandem repeat alleles observed in this study and their coding potential. Repeat units are colour-coded by DNA and amino acid sequence. Location of the repeat and flanking variants in relation to MSH3/DHFR locus are shown in the top panel. This locus contains overlapping MSH3 exon 1 and DHFR promoter regions. For both MSH3 and DHFR, the 5’-untranslated region is shown in white and coding sequence in light grey. The direction of transcription is indicated by arrows for each gene. (B) Repeat allele frequencies observed in Huntington’s disease (HD) and DM1. Four common alleles, 3a, 6a, 7a and 8a, are observed in Huntington’s disease and DM1 cohorts at similar frequencies. (C) Schematic showing potential misalignments of 3a and 6a alleles, resulting in the apparent SNP rs557874766, shown in red on the lower alignment. Black marks in the top alignment represent mismatches that could be created in a similar manner as rs557874766, by misalignment of the 3a and 6a repeat alleles.
The most common allele in both cohorts, 6a (Fig. 1B), corresponds to the human reference sequence (NC_000005.10, GRCh38.p12). Illumina sequencing revealed that rs557874766 (Moss et al., 2017) was not a SNP, but an alignment artefact resulting from the complex 9-bp repeat sequence (Fig. 1C). Individuals with the rs557874766 minor allele instead carry a three-repeat allele, 3a, the second most common allele observed in both cohorts. Two subjects with Huntington’s disease imputed as homozygous for the rs557874766 major allele were determined to be heterozygous for the 3a repeat allele by both Illumina and Sanger sequencing (Supplementary Fig. 2), highlighting the importance of directly genotyping such complex loci. We conclude that rs557874766 does not exist in the form of an SNP and results from incorrect alignment of the 3a allele to the reference 6a allele (Fig. 1C).
The MSH3 exon 1 repeat region is poorly conserved between species, with mean scores of 0.29 [standard deviation (SD) 0.41] and 0.25 (SD 0.91) in PhastCons and PhyloP, respectively (Supplementary Table 3). Sequence alignment of 20 mammalian reference genomes showed most have two repeats (Supplementary Fig. 3). Together with a four- and a five-repeat allele, the 3a allele has been observed in gorillas and chimpanzees, suggesting 3a is an ancestral allele in humans (Morales, 2006).
MSH3/DHFR variants are associated with rate of somatic expansion and disease phenotypes in Huntington’s disease and DM1
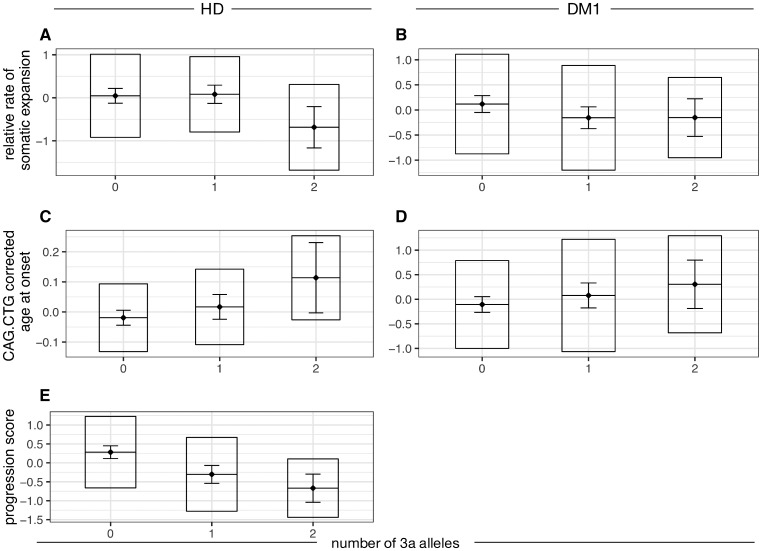
The 3a allele correlated negatively with relative rate of somatic expansion in subjects with Huntington’s disease (P = 0.032) and showed similar effect direction, though above nominal significance, in DM1 (P = 0.053) (Fig. 2 and Supplementary Table 2). Additionally, 3a was associated with delayed age at onset by 1.05 years (P = 0.0029) and slower progression in Huntington’s disease by 0.52 units (P = 3.86 × 10−7), which corresponds to 0.37 and 0.10 units per year on the UHDRS total motor score and total functional capacity, respectively. In DM1, the association between 3a and age at onset showed a consistent effect direction, approaching significance (P = 0.061). In meta-analysis, 3a was significantly associated with relative rate of somatic expansion (P = 0.004) and age at onset (P = 0.003) in Huntington’s disease and DM1. Detailed analysis of the relationship between repeat alleles and phenotypes (Supplementary Table 5) shows that the 3a allele accounts for the reduced somatic expansion rate, delayed onset and slower progression observed in Huntington’s disease. The association with somatic expansion appears to be driven by 3a homozygotes, whereas that with progression seems to follow an additive pattern with the number of 3a alleles. For onset, the pattern of association is unclear. In DM1, the number of seven-repeat alleles was associated with reduced expansion rate (Supplementary Table 5).
Figure 2.
The number of MSH3 3a repeat alleles is associated with Huntington’s disease and DM1 phenotypes. Boxplots for three measures of disease phenotype are shown: rate of somatic expansion corrected for the inherited CAG·CTG length in Huntington’s disease (A) and for the inherited CAG·CTG length and variant repeats in DM1 (B); age at onset corrected for the inherited CAG·CTG length in Huntington’s disease (C) and DM1 (D); progression score in Huntington’s disease (E). For each dataset, the diamond and horizontal line spanning the diamond indicate the mean, the box the standard deviation and the whiskers the 95% confidence intervals of the mean. HD = Huntington’s disease.
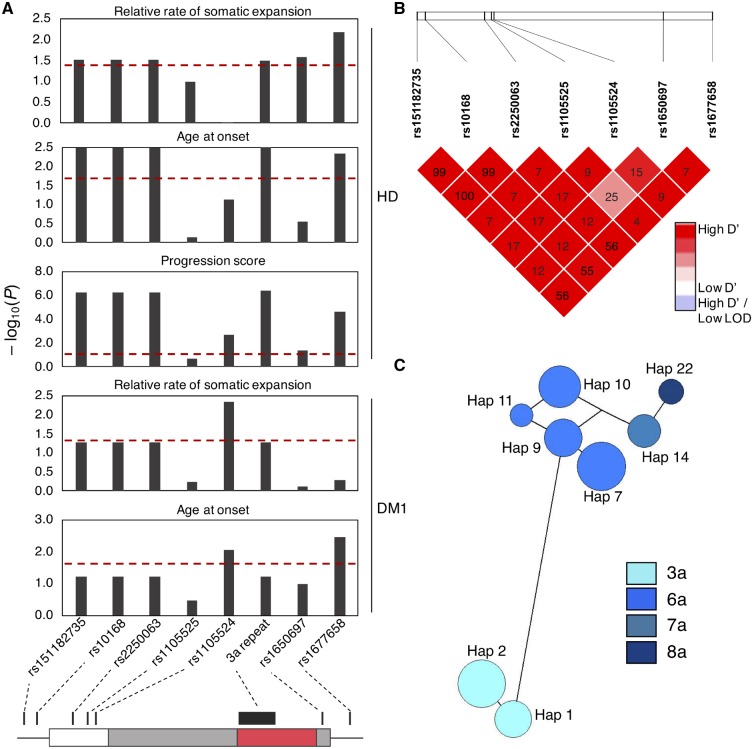
In addition to testing repeat allele effects, we also assessed correlation between flanking SNP genotypes and disease phenotypes. All the flanking variants were in HWE (Supplementary Table 6) and in strong linkage disequilibrium with each other (Fig. 3B). Three variants (rs151182735, rs10168 and rs2250063) were in nearly complete linkage disequilibrium with the 3a allele, and as such were as significantly associated with phenotypes (Fig. 3A and Supplementary Table 6). All three are non-coding variants 5’ to the repeat and their alternative alleles are associated with reduced MSH3 and DHFR expression in the prefrontal cortex (CMC, 2017) and in multiple tissues in GTEx (GTEx, 2015) (Supplementary Table 7). Three SNPs, rs1105524, rs1650697 and rs1677658, also correlated with some phenotypes, though not uniformly (Fig. 3A and Supplementary Table 6). Rs1105524 and rs1677658 are non-coding variants, whereas rs1650697 corresponds to Ile79Val. All three are expression quantitative trait loci (eQTL) for MSH3 and DHFR in the prefrontal cortex (CMC, 2017) and in multiple tissues in GTEx (Supplementary Table 7). Previously, in a separate DM1 cohort (DM1CostaRica), Morales et al. (2016) reported association between both rs1677658 (P = 0.009) and rs10168 (P = 0.031) and somatic expansion, though neither survived correction for multiple testing for the candidate SNPs analysed. However, the direction of effect for both SNPs was the same as in the present study, and a significant association in meta-analyses with the two DM1 cohorts (rs1677658 P = 0.03, rs10168 P = 0.004) and all three DM1 and Huntington’s disease cohorts (rs1677658 P = 8.85 × 10−4, rs10168 P = 3.37 × 10−4) suggests these variants influence somatic expansion (Supplementary Table 6). Morales et al. (2016) reported an association between somatic expansion and age at onset, though the direct effect of MSH3 genotype on age at onset was not found to be significant. In the present study, meta-analyses of the two DM1 cohorts (rs1677658 P = 0.009, rs10168 P = 0.04) and all three DM1 and Huntington’s disease cohorts (rs1677658 P = 8 × 10−4, rs10168 P = 0.003) found the MSH3 genotype was significantly associated with age at onset (Supplementary Table 6). Meta-analyses of the three-repeat allele with all three DM1 and Huntington’s disease cohorts provide further support for its protective effect on somatic expansion (DM1OPTIMISTIC + DM1CostaRicaP = 0.004, DM1OPTIMISTIC + DM1CostaRica + Huntington’s disease P = 3.46 × 10−4) and age at onset (DM1OPTIMISTIC + DM1CostaRicaP = 0.04, DM1OPTIMISTIC + DM1CostaRica + Huntington’s disease P = 0.003) (Supplementary Table 2).
Figure 3.
Variants at the MSH3/DHFR locus are associated with phenotypes in Huntington’s disease and DM1. (A) Bar charts showing associations between variant genotypes and disease phenotypes: relative rate of somatic expansion and age at onset corrected for the CAG·CTG length and progression score for Huntington’s disease, and rate of somatic expansion and age at onset corrected for the CAG·CTG length and repeat interruptions for DM1. Each bar represents association for a single variant. Red dotted line represents the P = 0.05 significance threshold. Variant location in relation to the MSH3 exon 1 region is shown in the bottom panel. White box = 5’ untranslated region; grey = coding sequence; red = MSH3 repeat region; intron is shown by a black line. (B) Linkage disequilibrium heat map for the seven variants flanking the MSH3 repeat. Colour intensity represents the D’ value for each SNP pair. R2 values are indicated in text for each variant pair. (C) Haplotype network for eight haplotypes with frequency > 0.035 observed at the MSH3 exon 1 region. Circles represent different haplotypes. The size of the circle is proportional to the number of individuals with a particular haplotype. Each haplotype is connected with the most similar haplotype by a line. Length of the line represents the number of genotypes that are different between each two haplotypes. Circles are colour coded according to the repeat allele found on the haplotype.
The associations of SNPs with phenotypes were conditioned on the effects of MSH3 repeat alleles (Supplementary Table 8). As rs151182735, rs10168 and rs2250063 perfectly correlated with 3a, their independent effects could not be determined (Supplementary Table 6). With the exception of rs1677658 (linkage disequilibrium with 3a: r2 = 0.610) and rs1650697 (linkage disequilibrium with 3a: r2 = 0.143), whose alternative alleles were associated with delayed and early age at onset, respectively in the combined Huntington’s disease and DM1 meta-analysis (P = 0.015 and P = 0.029; Supplementary Table 8), there was no significant evidence for association between SNPs and expansion rate, onset or progression independent of repeat alleles.
Considering variants with minor allele frequency >0.1 and all of the repeat alleles, we observed 25 haplotypes in the region, named Hap1 to Hap25 (Supplementary Table 9). The 3a repeat allele occurs on both Hap1 and Hap2, which differ only in the presence of the rs1677658 alternative allele on the more common Hap2. Hap1 was associated with reduced somatic expansion in DM1 (P = 0.032) and slower progression in Huntington’s disease (P = 0.020), whereas Hap2 was associated with reduced somatic expansion (P = 0.021) and delayed onset (P = 4.03 × 10−5) in both Huntington’s disease and DM1, and with slower progression (P = 1.64 × 10−5) and reduced expression of MSH3 (P = 0.024) and DHFR (P = 1.12 × 10−3) in Huntington’s disease (Supplementary Table 9).
Overall, this analysis clarifies the sequence and variants present in MSH3 exon 1 and demonstrates that MSH3 repeat variants are associated with disease phenotypes in both Huntington’s disease and DM1.
MSH3 and DHFR expression in blood is associated with repeat alleles
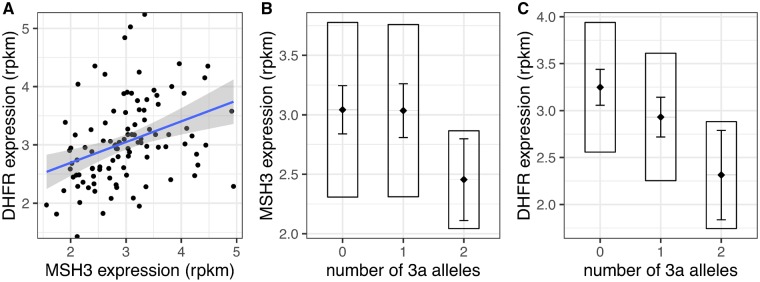
Each 3a allele was associated with reduced DHFR expression (P = 2.48 × 10−4; Fig. 4C) and homozygosity for 3a was associated with reduced MSH3 expression (P = 0.0273; Fig. 4B), whereas each 7a or 8a allele was associated with increased MSH3 expression (P = 8.55 × 10−4 and P = 8.26 × 10−3, respectively). The sum of MSH3 repeat lengths on both alleles appeared to correlate with MSH3 (P = 7.00 × 10−3) and DHFR expression (P = 1.76 × 10−3), which would suggest increasing repeat length increases expression of both (Supplementary Fig. 4). However, a more detailed analysis of MSH3 repeat alleles (Supplementary Table 5) shows the number of seven- or eight-repeat alleles is associated with increased expression of MSH3 (P = 4.53 × 10−6), and that this explains the apparent association with the sum of repeat lengths. In this relatively small cohort, MSH3 (age at onset P = 0.446, progression P = 0.440) and DHFR (age at onset P = 0.911, progression P = 0.284) expression in blood were not themselves directly associated with disease phenotype. MSH3 expression was not significantly associated with somatic expansion (P = 0.625), whereas the association of DHFR expression, while nominally significant (P = 0.049), did not survive correction for the number of phenotypes tested.
Figure 4.
Association of the MSH3 3a allele with MSH3 and DHFR expression in Huntington’s disease whole blood. Whole blood RNA-Seq in a subset of 108 Huntington’s disease subjects. (A) Significant correlation between MSH3 and DHFR expression levels (r2 = 0.120, P = 2.06 × 10−4). Grey area around the blue regression line represents 95% confidence interval of the model. (B) Homozygosity for MSH3 3a repeat allele is associated with lower MSH3 expression in blood (P = 0.028). (C) MSH3 3a repeat allele is associated with lower DHFR expression (P = 2.33 × 10−4). Rpkm = reads per kilobase of transcript per million mapped reads. In boxplots, the diamond and horizontal line spanning the diamond indicate the mean, the box indicates the standard deviation and the whiskers indicate the 95% confidence intervals of the mean.
In the detailed analysis, the number of three-repeat alleles was associated with reduced DHFR expression (P = 2.33 × 10−4; Fig. 4C), and this was sufficient to explain the apparent association of DHFR expression with other repeat alleles (Supplementary Table 5), including that observed with increasing total repeat length. DHFR and MSH3 expression are correlated (r2 = 0.120, P = 2.06 × 10−4; Fig. 4A). However, association between DHFR and three-repeat alleles remains significant after correcting for MSH3 expression (P = 7.51 × 10−4), and association between MSH3 and seven- or eight-repeat alleles remains significant after correcting for DHFR expression (P = 1.30 × 10−7). In the best-fitting model for DHFR expression, the alternative allele at rs1105524 (linkage disequilibrium with 3a: r2 = 0.192) increases and rs1650697 decreases DHFR expression independently of the three-repeat alleles (Supplementary Table 8). Otherwise, the repeat allele is the major determinant of MSH3 and DHFR expression, and there is no evidence of independent SNP effects.
MSH3 expression in cortex is associated with onset and progression in Huntington’s disease
In a TWAS, increased expression of both MSH3 and DHFR in prefrontal cortex (CMC, 2017) was associated with faster progression in TRACK-HD (Moss et al., 2017) at similar levels of significance (P = 2.52 × 10−6 and P = 4.08 × 10−6, respectively; Supplementary Table 10), making it difficult to distinguish which is more functionally relevant. This ties in with the observation that SNPs significantly associated with somatic expansion, age at onset and progression (Supplementary Table 6) were eQTLs for both MSH3 and DHFR in CMC data. Notably, however, increased MSH3 expression was significantly associated with early onset (P = 1.71 × 10−3) in a TWAS of the GeM dataset (GeM-HD, 2015), while DHFR expression was not significantly associated with onset (Supplementary Table 10). This favours MSH3 over DHFR expression as a modifier of Huntington’s disease course.
Discussion
MSH3 has recently been identified as a genetic modifier of somatic instability in DM1 (Morales et al., 2016), and progression in Huntington’s disease (Moss et al., 2017). The MSH3 signal in the GWAS of Huntington’s disease progression was driven by an imputed SNP, rs557874766, located within a 9 bp tandem repeat sequence in exon 1 of MSH3, which is also in the 5′ UTR of DHFR on the opposite strand. MSH3 and DHFR are organized head-to-head, transcribed in opposite directions and are regulated by the same promoter. Here we demonstrate that rs557874766 is an alignment artefact and corresponds to a three-repeat allele, 3a, which was the shortest repeat allele observed and is likely ancestral. At the protein level, in silico modelling predicts that 6a results in the gain of a surface α-helix (Kallberg et al., 2012) at the N-terminus of MSH3.
A total of 16 MSH3 repeat alleles were observed, varying in sequence and length from three to nine repeats. Repeat alleles 6a and 3a are the first and second most common in this European cohort, though previous studies suggest a seven-repeat allele may be second most common in East Asian populations (Nakajima et al., 1995). In Huntington’s disease, 3a was associated with reduced somatic expansion, delayed onset and slower progression. In DM1, each 3a allele showed a trend towards reduced somatic expansion and delayed onset but was significantly associated with both measures in meta-analysis of Huntington’s disease and DM1. Longer seven-repeat alleles were associated with reduced somatic expansion only in DM1. Whether this reflects a subtle difference in MSH3 biology between the two disorders, or simply a sampling error, remains undetermined.
The MSH3 repeat lies between binding domains for PCNA (Clark et al., 2000) and EXO1 (Schmutte et al., 2001), both of which are involved in mismatch repair (MMR) (Kleczkowska et al., 2001). PCNA is a sliding clamp that participates in DNA replication, but in MMR it delivers MSH proteins to mismatches and increases binding specificity (Flores-Rozas et al., 2000). EXO1 excises the daughter strand after mismatch recognition, as well as being involved in end resection during homologous recombination (Goellner et al., 2015). The MSH3 repeat region is poorly conserved between species, with other mammals having between zero and five repeats. This lack of evolutionary constraint suggests functional redundancy in the MMR pathway and a lack of a major effect of N-terminal MSH3 variation outside the context of repeat expansion disease. Unlike other MMR components, germline heterozygous MSH3 mutations are not associated with increased risk of cancer, most likely because MSH2/MSH6 can also initiate repair at replication errors (Edelmann et al., 2000; Jiricny, 2006; Haugen et al., 2008).
Three non-coding variants 5’ of the repeat were in near complete linkage disequilibrium with 3a, so it is not possible to determine their independent effects on disease phenotypes. All three are associated with reduced MSH3 expression in multiple tissues, including cortex (CMC and GTEx). Controlling for repeat alleles, no SNPs were significantly associated with phenotypes, except the intronic rs1677658 and the exon 1 rs1650697 variants, which contributed to delayed or early onset, respectively in the combined Huntington’s disease and DM1 dataset. Rs1677658 was associated with reduced MSH3 and DHFR expression (CMC and GTEx), whereas rs1650697 was associated with increased DHFR in Huntington’s disease blood, as well as multiple tissues in GTEx. Hap2, the MSH3 haplotype most significantly linked with reduced somatic expansion and delayed onset in Huntington’s disease and DM1, and with slower progression in Huntington’s disease, contains the 3a allele, along with alternative alleles of non-coding variants rs151182735, rs10168 and rs2250063, which are in complete linkage disequilibrium with it, and rs1677658. It is thus difficult to assess which (if any) MSH3 variants (repeats or SNPs) are driving associations with disease phenotypes, and further investigation in a larger sample is warranted.
Whole blood transcriptomic analysis in a subset of the Huntington’s disease patients found the 3a allele was associated with reduced expression of MSH3 and DHFR, and seven- or eight-repeat alleles with increased MSH3 expression. DHFR, which shares a promoter with MSH3 (Drummond, 1999), is a ubiquitously expressed enzyme involved in purine, thymidylic acid and amino acid synthesis, but has not previously been implicated in Huntington’s disease pathogenesis. Our TWAS found that increased expression of MSH3 and DHFR in cortex are associated with faster Huntington’s disease progression (Moss et al., 2017). While MSH3 expression was significantly associated with early onset in our GeM TWAS (P = 1.71 × 10−3) (GeM-HD, 2015), DHFR expression was not associated with disease course. This is consistent with Huntington’s disease mouse brain, in which expression of MSH3, but not DHFR, correlates with somatic expansion (Tome et al., 2013). Neither MSH3 nor DHFR expression in blood was significantly associated with somatic expansion, onset or progression in this sample. However, investigation in a larger sample, or in a more relevant tissue, such as striatum, would be of interest.
Collectively, our results suggest the MSH3 3a repeat allele reduces somatic expansion and improves phenotype in both Huntington’s disease and DM1, potentially through altering MSH3 expression levels. However, given the proximity of the repeat region to MMR protein binding domains, the 3a allele could also alter MSH3 function in the recognition and repair of insertion-deletion loops, double-strand breaks or single-strand annealing (Lyndaker and Alani, 2009; Schmidt and Pearson, 2016). Repetitive DNA sequences form unusual secondary structures such as slipped strands, hairpin loops, G-quadruplexes and R-loops (Mirkin, 2007; Neil et al., 2017), the stability of which correlates with expansion (Gacy et al., 1995). MSH3 may recognize these structures (Owen et al., 2005) and initiate repair, during which out of register synthesis could result in repeat expansion (Khan et al., 2015; Neil et al., 2017). This preliminary study elucidates variation in MSH3 that modifies Huntington’s disease and identifies the same signal in an independent trinucleotide repeat disease. Though beyond the scope of the present study, in the future it will be important to replicate these findings in additional independent cohorts for each disease. Together, these results suggest a common mechanism, involving somatic expansion, operates in vivo in distinct trinucleotide repeat diseases to influence disease course. Therefore, modulation of MSH3 has significant therapeutic potential in a range of diseases caused by repeat expansions.
Supplementary Material
Acknowledgements
We thank the people who have enabled this work through their participation in the TRACK-HD and OPTIMISTIC studies.
Glossary
Abbreviations
- DM1
myotonic dystrophy type 1
- SNP
single nucleotide polymorphism
- TWAS
transcriptome-wide association study
Appendix 1
TRACK-HD investigators and OPTIMISTIC consortium
See Supplementary material for full details.
TRACK-HD investigators
Peter Kraus, Rainer Hoffman, Alan Tobin, Beth Borowsky, S. Keenan, Kathryn B. Whitlock, Sarah Queller, Colin Campbell, Chiachi Wang, Doug Langbehn, Eric Axelson, Hans Johnson, Tanka Acharya, Dave M. Cash, Chris Frost, Rebecca Jones, Caroline Jurgens, Ellen P. ‘t Hart, Jeroen van der Grond, Marie-Noelle N. Witjes-Ane, Raymund A.C. Roos, Eve M. Dumas, Simon J.A. van den Bogaard, Cheryl Stopford, David Craufurd, Jenny Callaghan, Natalie Arran, Diana D. Rosas, S. Lee, W Monaco, Alison O’Regan, Cassie Milchman, E. Frajman, Izelle Labuschagne, Julie Stout, Melissa Campbell, Sophie C. Andrews, Natalie Bechtel, Ralf Reilmann, Stefan Bohlen, Chris Kennard, Claire Berna, Stephen Hicks, Alexandra Durr, C Pourchot, Eric Bardinet, Kevin Nigaud, Romain Valabrègue, Stephane Lehericy, Cecilia Marelli, Celine Jauffret, Damian Justo, Blair Leavitt, Joji Decolongon, Aaron Sturrock, Alison Coleman, Rachelle Dar Santos, A. Patel, Claire Gibbard, Daisy Whitehead, Ed Wild, Gail Owen, Helen Crawford, Ian Malone, Nayana Lahiri, Nick C. Fox, Nicola Z. Hobbs, Rachael I. Scahill, Roger Ordidge, Tracey Pepple, Joy Read, Miranda J. Say, Bernhard Landwehrmeyer.
OPTIMISTIC consortium
Ferroudja Daidj, Guillaume Bassez, Baptiste Lignier, Florence Couppey, Stéphanie Delmas, Jean-François Deux, Karolina Hankiewicz, Celine Dogan, Lisa Minier, Pascale Chevalier, Amira Hamadouche, Michael Catt, Vincent van Hees, Sharon Catt, Ameli Schwalber, Juliane Dittrich, Marie Kierkegaard, Stephan Wenninger, Benedikt Schoser, Angela Schüller, Kristina Stahl, Heike Künzel, Martin Wolff, Anna Jellinek, Cecilia Jimenez Moreno, Grainne Gorman, Hanns Lochmüller, Michael Trenell, Sandra van Laar, Libby Wood, Sophie Cassidy, Jane Newman, Sarah Charman, Renae Steffaneti, Louise Taylor, Allan Brownrigg, Sharon Day, Antonio Atalaia, Joost Raaphorst, Kees Okkersen, Baziel van Engelen, Stephanie Nikolaus, Yvonne Cornelissen, Marlies van Nimwegen, Daphne Maas, Ellen Klerks, Sacha Bouman, Hans Knoop, Linda Heskamp, Arend Heerschap, Ridho Rahmadi, Perry Groot, Tom Heskes, Katarzyna Kapusta, Jeffrey Glennon, Shaghayegh Abghari, Armaz Aschrafi, Geert Poelmans, Shaun Treweek, Fiona Hogarth, Roberta Littleford, Peter Donnan, Adrian Hapca, Michael Hannah, Emma McKenzie, Petra Rauchhaus, Sarah A. Cumming, Darren G. Monckton, Berit Adam, Catharina Faber, Ingemar Merkies.
Contributor Information
TRACK-HD Investigators:
Peter Kraus, Rainer Hoffman, Alan Tobin, Beth Borowsky, S Keenan, Kathryn B Whitlock, Sarah Queller, Colin Campbell, Chiachi Wang, Doug Langbehn, Eric Axelson, Hans Johnson, Tanka Acharya, Dave M Cash, Chris Frost, Rebecca Jones, Caroline Jurgens, Ellen P ‘t Hart, Jeroen van der Grond, Marie-Noelle N Witjes- Ane, Raymund A C Roos, Eve M Dumas, Simon J A van den Bogaard, Cheryl Stopford, David Craufurd, Jenny Callaghan, Natalie Arran, Diana D Rosas, S Lee, W Monaco, Alison O’Regan, Cassie Milchman, E Frajman, Izelle Labuschagne, Julie Stout, Melissa Campbell, Sophie C Andrews, Natalie Bechtel, Ralf Reilmann, Stefan Bohlen, Chris Kennard, Claire Berna, Stephen Hicks, Alexandra Durr, C Pourchot, Eric Bardinet, Kevin Nigaud, Romain Valabre, ` gue, Stephane Lehericy, Cecilia Marelli, Celine Jauffret, Damian Justo, Blair Leavitt, Joji Decolongon, Aaron Sturrock, Alison Coleman, Rachelle Dar Santos, A Patel, Claire Gibbard, Daisy Whitehead, Ed Wild, Gail Owen, Helen Crawford, Ian Malone, Nayana Lahiri, Nick C Fox, Nicola Z Hobbs, Rachael I Scahill, Roger Ordidge, Tracey Pepple, Joy Read, Miranda J Say, and Bernhard Landwehrmeyer
OPTIMISTIC Consortium:
Ferroudja Daidj, Guillaume Bassez, Baptiste Lignier, Florence Couppey, Stéphanie Delmas, Jean-François Deux, Karolina Hankiewicz, Celine Dogan, Lisa Minier, Pascale Chevalier, Amira Hamadouche, Michael Catt, Vincent van Hees, Sharon Catt, Ameli Schwalber, Juliane Dittrich, Marie Kierkegaard, Stephan Wenninger, Benedikt Schoser, Angela Schüller, Kristina Stahl, Heike Künzel, Martin Wolff, Anna Jellinek, Cecilia Jimenez Moreno, Grainne Gorman, Hanns Lochmüller, Michael Trenell, Sandra van Laar, Libby Wood, Sophie Cassidy, Jane Newman, Sarah Charman, Renae Steffaneti, Louise Taylor, Allan Brownrigg, Sharon Day, Antonio Atalaia, Joost Raaphorst, Kees Okkersen, Baziel van Engelen, Stephanie Nikolaus, Yvonne Cornelissen, Marlies van Nimwegen, Daphne Maas, Ellen Klerks, Sacha Bouman, Hans Knoop, Linda Heskamp, Arend Heerschap, Ridho Rahmadi, Perry Groot, Tom Heskes, Katarzyna Kapusta, Jeffrey Glennon, Shaghayegh Abghari, Armaz Aschrafi, Geert Poelmans, Shaun Treweek, Fiona Hogarth, Roberta Littleford, Peter Donnan, Adrian Hapca, Michael Hannah, Emma McKenzie, Petra Rauchhaus, Sarah A Cumming, Darren G Monckton, Berit Adam, Catharina Faber, and Ingemar Merkies
Funding
This work was in part supported by the UK Dementia Research Institute, European Union, Medical Research Council (UK), Rosetrees Trust, CHDI, and Myotonic Dystrophy Support Group. We thank the CHDI Foundation, a non-profit biomedical research organization exclusively dedicated to developing therapeutics that will substantially improve the lives of Huntington’s disease-affected individuals, who funded the TRACK-HD study; the European Union’s Seventh Framework Programme (FP7/2007–2013) under grant agreement n° 2012–305121 “Integrated European –omics research project for diagnosis and therapy in rare neuromuscular and neurodegenerative diseases (NEUROMICS)” [305121] and no 305697 (the OPTIMISTIC project); the Medical Research Council (MRC) for their support of the MRC Centre for Neuropsychiatric Genetics and Genomics, MR/L010305/1, and PhD studentship to MF (1477284); the Rosetrees Trust, a private, family funded charity that supports medical research (JS16/M574); College of Medical, Veterinary and Life Sciences of University of Glasgow for PhD studentship to V.L. (200526–01). This work was supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK.
Competing interests
D.G.M. has been a scientific consultant and/or received an honoraria or stock options from Biogen Idec, AMO Pharma, Charles River, Vertex Pharmaceuticals, Triplet Therapeutics, LoQus23, BridgeBio, Small Molecule RNA and Lion Therapeutics. D.G.M. also had a research contract with AMO Pharma. D.G.M. is on the Scientific Advisory Board of the Myotonic Dystrophy Foundation and is a scientific advisor to the Myotonic Dystrophy Support Group. In the past 2 years, S.J.T. has undertaken consultancy services, including advisory boards, with F. Hoffmann-La Roche Ltd., Ixitech Technologies, Takeda Pharmaceuticals International, Teva Pharmaceuticals, Alnylam Pharmaceuticals Inc., GSK, Heptares Therapeutics, UCB Pharma S.A., University College Irvine, Triplet Therapeutics, LoQus therapeutics and Vertex Pharmaceuticals Inc. All honoraria for these consultancies were paid through the offices of UCL Consultants Ltd., a wholly owned subsidiary of University College London.
References
- Ashizawa T, Dubel JR, Harati Y. Somatic instability of CTG repeat in myotonic dystrophy. Neurology 1993; 43: 2674–8. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bates G, Tabrizi S, Jones L. Huntington’s disease. Oxford: Oxford University Press; 2014. [Google Scholar]
- Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR, et al. Huntington disease. Nat Rev Dis Primers 2015; 1: 15005. [DOI] [PubMed] [Google Scholar]
- Bettencourt C, Hensman-Moss D, Flower M, Wiethoff S, Brice A, Goizet C, et al. DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann Neurol 2016; 79: 983–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell 1992; 68: 799–808. [DOI] [PubMed] [Google Scholar]
- Ciosi M, Cumming SA, Alshammari AM, Symeonidi E, Herzyk P, McGuinness D, et al. Library preparation and MiSeq sequencing for the genotyping-by-sequencing of the Huntington disease HTT exon one trinucleotide repeat and the quantification of somatic mosaicism. Protocol Exchange 2018. PROTOCOL (Version 1). doi: 10.1038/protex.2018.089. [Google Scholar]
- Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem 2000; 275: 36498–501. [DOI] [PubMed] [Google Scholar]
- CMC. CommonMind Consortium Knowledge Portal. 2017. Available from: https://www.synapse.org//#!Synapse:syn2759792/wiki/ (10 August 2017, date last accessed).
- Cumming SA, Hamilton MJ, Robb Y, Gregory H, McWilliam C, Cooper A, et al. De novo repeat interruptions are associated with reduced somatic instability and mild or absent clinical features in myotonic dystrophy type 1. Eur J Hum Genet 2018; 26: 1635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, et al. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis 2009; 33: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JT. Genomic amplification of the human DHFR/MSH3 locus remodels mismatch recognition and repair activities. Adv Enzyme Regul 1999; 39: 129–41. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Umar A, Yang K, Heyer J, Kucherlapati M, Lia M, et al. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res 2000; 60: 803–7. [PubMed] [Google Scholar]
- Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet 2000; 26: 375–8. [DOI] [PubMed] [Google Scholar]
- Foiry L, Dong L, Savouret C, Hubert L, te Riele H, Junien C, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet 2006; 119: 520–6. [DOI] [PubMed] [Google Scholar]
- Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 1995; 81: 533–40. [DOI] [PubMed] [Google Scholar]
- GeM-HD GMoHsDG-HC. Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell 2015; 162: 516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goellner EM, Putnam CD, Kolodner RD. Exonuclease 1-dependent and independent mismatch repair. DNA Repair 2015; 32: 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx GTC. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York, NY) 2015; 348: 648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 2016; 48: 245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper PS. Myotonic dystrophy. 3rd edn. London: Saunders WB; 2001. [Google Scholar]
- Haugen AC, Goel A, Yamada K, Marra G, Nguyen TP, Nagasaka T, et al. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res 2008; 68: 8465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A, Tsilfidis C, Mettler G, Jacob P, Mahadevan M, Surh L, et al. The correlation of age of onset with CTG trinucleotide repeat amplification in myotonic dystrophy. J Med Genet 1992; 29: 774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 2006; 7: 335–46. [DOI] [PubMed] [Google Scholar]
- Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 2012; 7: 1511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, Ennis M, et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet 2003; 12: 3359–67. [DOI] [PubMed] [Google Scholar]
- Khan N, Kolimi N, Rathinavelan T. Twisting right to left: A…A mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation. PLoS Comput Biol 2015; 11: e1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev 2001; 15: 724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Chao MJ, Harold D, Abu Elneel K, Gillis T, Holmans P, et al. A modifier of Huntington’s disease onset at the MLH1 locus. Hum Mol Genet 2017; 26: 3859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndaker AM, Alani E. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. Bioessays 2009; 31: 315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola G, Cardani R. Myotonic dystrophies: an update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim Biophys Acta 2015; 1852: 594–606. [DOI] [PubMed] [Google Scholar]
- Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, et al. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform 2013; 14: 193–202. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature 2007; 447: 932–40. [DOI] [PubMed] [Google Scholar]
- Morales F, Couto JM, Higham CF, Hogg G, Cuenca P, Braida C, et al. Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum Mol Genet 2012; 21: 3558–67. [DOI] [PubMed] [Google Scholar]
- Morales F, Vasquez M, Santamaria C, Cuenca P, Corrales E, Monckton DG. A polymorphism in the MSH3 mismatch repair gene is associated with the levels of somatic instability of the expanded CTG repeat in the blood DNA of myotonic dystrophy type 1 patients. DNA Repair 2016; 40: 57–66. [DOI] [PubMed] [Google Scholar]
- Morales FA. Somatic mosaicism and genotype-phenotype correlations in myotonic dystrophy type 1. PhD thesis. University of Glasgow; 2006. [Google Scholar]
- Moss DJH, Pardinas AF, Langbehn D, Lo K, Leavitt BR, Roos R, et al. Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol 2017; 16: 701–11. [DOI] [PubMed] [Google Scholar]
- Nakajima E, Orimo H, Ikejima M, Shimada T. Nine-bp repeat polymorphism in exon 1 of the hMSH3 gene. Jpn J Hum Genet 1995; 40: 343–5. [DOI] [PubMed] [Google Scholar]
- Neil AJ, Kim JC, Mirkin SM. Precarious maintenance of simple DNA repeats in eukaryotes. BioEssays 2017; 39: 1700077. doi:10.1002/bies.201700077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen BA, Yang Z, Lai M, Gajec M, Badger JD 2nd, Hayes JJ, et al. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol 2005; 12: 663–70. [DOI] [PubMed] [Google Scholar]
- Pinto RM, Dragileva E, Kirby A, Lloret A, Lopez E, St Claire J, et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genet 2013; 9: e1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. http://www.R-project.org/.
- Schmidt MH, Pearson CE. Disease-associated repeat instability and mismatch repair. DNA Repair 2016; 38: 117–26. [DOI] [PubMed] [Google Scholar]
- Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem 2001; 276: 33011–8. [DOI] [PubMed] [Google Scholar]
- Shelbourne PF, Keller-McGandy C, Bi WL, Yoon SR, Dubeau L, Veitch NJ, et al. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Gen 2007; 16: 1133–42. [DOI] [PubMed] [Google Scholar]
- Swami M, Hendricks AE, Gillis T, Massood T, Mysore J, Myers RH, et al. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet 2009; 18: 3039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Durr A, Craufurd D, et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol 2009; 8: 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC. R: a language and environment for statistical computing. 2013. Available from: http://www.R-project.org/ (27 August 2018, date last accessed).
- Tome S, Manley K, Simard JP, Clark GW, Slean MM, Swami M, et al. MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington’s disease mice. PLoS Genet 2013; 9: e1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet 2002; 11: 191–8. [DOI] [PubMed] [Google Scholar]
- van Engelen B, Consortium O. Cognitive behaviour therapy plus aerobic exercise training to increase activity in patients with myotonic dystrophy type 1 (DM1) compared to usual care (OPTIMISTIC): study protocol for randomised controlled trial. Trials 2015; 16: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding author on request.