Abstract
Pandemics of vector-borne human and plant diseases often depend on the behaviors of their arthropod vectors. Arboviruses, including many bunyaviruses, manipulate vector behavior to accelerate their own transmission to vertebrates, birds, insects, and plants. However, the molecular mechanism underlying this manipulation remains elusive. Here, we report that the non-structural protein NSs of Tomato spotted wilt orthotospovirus, a prototype of the Tospoviridae family and the Orthotospovirus genus, is a key viral factor that indirectly modifies vector preference and increases vector performance. NSs suppresses the biosynthesis of plant volatile monoterpenes, which serve as repellents of the vector western flower thrips (WFT, Frankliniella occidentalis). NSs directly interacts with MYC2, the jasmonate (JA) signaling master regulator and its two close homologs MYC3 and MYC4, to disable JA-mediated activation of terpene synthase genes. The dysfunction of the MYCs subsequently attenuates host defenses, increases the attraction of thrips, and improves thrips fitness. Moreover, MYC2 associated with NSs of Tomato zonate spot orthotospovirus, another Euro/Asian-type orthotospovirus, suggesting that MYC2 is an evolutionarily conserved target of Orthotospovirus species for suppression of terpene-based resistance to promote vector performance. These findings elucidate the molecular mechanism through which an orthotospovirus indirectly manipulates vector behaviors and therefore facilitates pathogen transmission. Our results provide insights into the molecular mechanisms by which Orthotospovirus NSs counteracts plant immunity for pathogen transmission.
Author summary
Most bunyaviruses are transmitted by arthropod vectors, and some of them can modify the behaviors of their arthropod vectors to increase transmission to mammals, birds, and plants. NSs is a non-structural bunyavirus protein with multiple functions that acts as an avirulence determinant and silencing suppressor. In this study, we identified a new function of NSs as a conserved manipulator of vector behavior via plant. NSs suppresses jasmonate-mediated plant immunity against thrips by directly interacting with several homologs of MYC transcription factors, the core regulators of the jasmonate-signaling pathway. This hijacking by NSs enhances thrips preference and performance. Therefore, our data support the hypothesis that MYC2 is a convergent target that plant pathogens manipulate to promote their survival in plants.
Introduction
Arthropod-borne viruses (arboviruses) are virulent causal agents of diseases in humans, animals, and plants. Vector behaviors have critical ecological and evolutionary consequences for arboviruses, which rely exclusively on their arthropod vectors for dispersal to new hosts. Therefore, it is of evolutionary significance for an arbovirus to alter its vector’s behavior to facilitate its own transmission. For plant viruses, such influence of vectors by viruses can include plant-mediated indirect effects or direct manipulation within the vector after acquisition. Among the indirect effects, infected plants tend to be more attractive to vectors [1]. For example, Geminiviridae and Luteoviridae viruses almost universally induce preferred settling of the vectors onto infected plants [2–5], and this phenomenon also exists among the Potyviridae and Bunyaviridae [6–9]. Moreover, viruses can positively or negatively affect the performance or fitness of arthropod vectors on the host. Persistently transmitted viruses, which need a sustained feeding of insect vectors to be acquired or transmitted, in particular, have positive effects on vector performance. For example, insect vectors perform better on Geminiviridae- and Tospoviridae-infected plants [9–12]. For nonpersistently transmitted viruses, vectors acquire or transmit the viruses in seconds through probing or feeding, such as Potyviridae, Caulimoviridae and Bromoviridae, also can positively or negatively affect their vectors for efficient virus spread [1, 6, 13–15].
Bunyavirales encompasses nine families of viruses with single-stranded negative-sense RNA genomes. As a prototype of the plant-infected Tospoviridae family, Tomato spotted wilt orthotospovirus (TSWV) is transmitted mainly by Frankliniella occidentalis Pergande (Western flower thrips, WFT) in a persistent and propagative manner [16,17]. Plant infection with TSWV influences several vector behaviors, such as biting and host choice to increase virus transmission, similar to the animal-infecting members of Bunyavirales [18–20]. For instance, non-viruliferous F. occidentalis prefers to settle on TSWV-infected pepper (Capsicum annuum L.) and Datura stramonium plants over noninfected controls [9]. However, the underlying molecular mechanism of this conserved indirect manipulation of vector behaviors by Orthotospovirus and Bunyavirales species is still unclear, although this plant immunity suppression is thought to occur in TSWV-infected Arabidopsis thaliana [21]. The bunyavirus families are divided based on their different coding strategies for the additional non-structural proteins, NSm and NSs, which are often involved in host-pathogen interactions. Orthotospovirus NSm protein facilitates the movement of viral ribonucleoproteins from cell to cell within the plant host. NSm of TSWV has recently been identified as the avirulence factor recognized by the product of resistance gene Sw-5b from tomato (Solanum lycopersicum L.) [22]. The NSs proteins of many bunyaviruses modulate host innate immune responses, and NSs in Orthotospovirus functions as a silencing suppressor in both plants and insects [23,24]. These proteins are responsible for establishing systemic infection in plants and for virus transmission by insect vectors [25,26].
Many plant species emit herbivore-induced plant volatiles (HIPVs), as an indirect anti-herbivore defense strategy [27–30]. HIPVs can repel insects such as aphids and caterpillars or deter lepidopteran oviposition [31–33], and are a common induced defense mechanism among plants including cotton and tomato [34,35]. Phytohormones such as jasmonate (JA) play vital roles in regulating HIPV production upon insect attack [36,37]. Several viruses have been shown to modify this JA-regulated volatile biosynthesis to affect the communication between plant and insect vector. For instance, begomoviruses inhibit the JA pathway and modify volatile terpene-mediated defense responses against whitefly [38]. The JA-mediated biosynthesis of secondary metabolites is believed to be associated with thrips resistance [39]. However, whether and how TSWV influence JA signaling remains elusive, although this virus is thought to hijack the antagonistic relation between JA and salicylic acid signaling [40].
In this study, we showed that TSWV benefits to thrips vector by suppressing a JA-regulated defense pathway of plants against herbivores. We identified the NSs protein from thrip-borne TSWV as a viral genetic factor induced attraction of its insect vector. Various NSs from orthotospovirus suppress the JA signaling pathway in the host plant by directly interacting with MYCs, key regulators of the JA signaling pathway, to reduce host defense responses against thrips. Our results establish a molecular mechanism underlying how TSWV attracts and benefits to its thrips vector by targeting plant MYC proteins.
Results
TSWV infection enhances plant attractiveness to the thrips vector and suppresses plant terpene synthesis
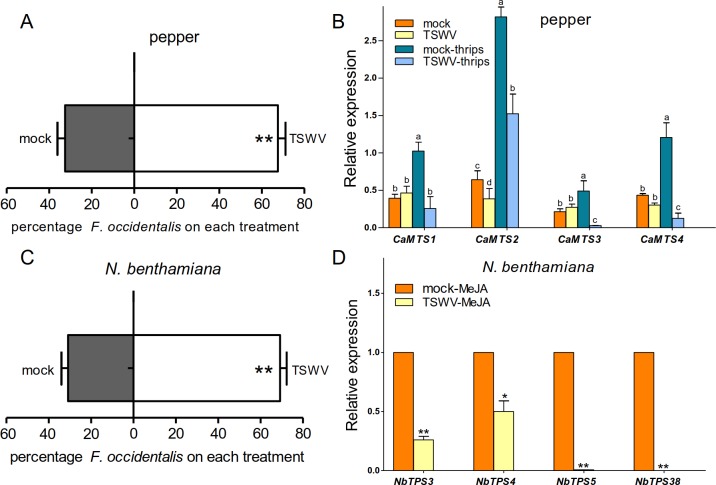
We first investigated the indirect effect of TSWV infection on the behavioral responses of the vector Frankliniella occidentalis Pergande (Western flower thrips, WFT). We conducted a two-choice assay between infected and non-infected plants. Pepper (Capsicum annuum L.), a natural host of TSWV and an important crop worldwide, was first tested in the tripartite thrip–orthotospovirus–plant interaction. A group of 50 non-viruliferous WFT was released from the center of the two-choice arena between two types of pepper plants. Consistent with previous results from Maris et al. [9], ~68% of thrips approached TSWV-infected plants, whereas the remaining approached non-infected plants (Fig 1A), suggesting that TSWV infection indirectly increases the attractiveness of peppers to the thrips vector.
Fig 1. TSWV infection enhances plant attractiveness to the thrips vector and suppresses plant terpene synthesis.
(A) Thrips preference (as percentage recaptured WFT out of 50 released) on different pepper plants. Four-week-old pepper plants were infected with TSWV (TSWV) or inoculated with buffer (mock). Plants of a similar size were used for thrips bioassay at 14 days post viral infection (dpi). Data are mean + SE, n = 6. **P < 0.01, Wilcoxon matched pairs tests. (B) Relative expression levels of TPS genes in pepper with or without thrips infestation for 6 h. Values are means + SE, n = 3. P < 0.05, one-way ANOVA plus Duncan’s multiple range tests. (C) Thrips preference (as percentage recaptured WFT out of 50 released) on N. benthamiana plants. Three-week-old N. benthamiana plants were infected with TSWV (TSWV) or inoculated with buffer (mock). Leaves of a similar size were used for the thrips bioassay at 14 dpi. Data are mean + SE, n = 6. **P < 0.01, Wilcoxon matched pairs tests. (D) Relative expression levels of various TPS genes in mock or TSWV-infected N. benthamiana after MeJA treatment. N. benthamiana plants were sprayed with 100 μM MeJA (Sigma-Aldrich) containing 0.01% (v/v) Tween 20. Values are means + SE, n = 3. *P < 0.05 **P < 0.01, Student’s t-test.
The attraction of insect vectors induced by the infection of other viruses is dependent on plant volatiles [38,41]. We therefore measured the expression levels of terpene synthase (TPS) genes in pepper leaves based on our previous functional analysis of TPS genes [38]. Reverse-transcription quantitative PCR (RT-qPCR) analysis showed that the expression of four pepper monoterpene synthase genes (CaMTS1, CaMTS2, CaMTS3, and CaMTS4), which are related to monoterpene synthesis, were upregulated after thrips infestation (Fig 1B). However, the terpene biosynthesis gene expression activated by thrips was significantly lower in TSWV-infected plants compared with the control (Fig 1B).
Another model (host) plant for tripartite interaction research, Nicotiana benthamiana, was also tested. Similar to the observations in pepper, TSWV-infected N. benthamiana leaves also were more attractive to thrips than non-infected leaves (Fig 1C). Moreover, RT-qPCR analysis indicated that the terpene synthase genes NbTPS5 and NbTPS38 responded to thrips infestation in N. benthamiana (S1A Fig). Consistent with the above results, NbTPS5 and NbTPS38 expression was notably induced by methyl jasmonate (MeJA) treatment, reflecting the same trends as during thrips infestation (S1B Fig). JA signaling is normally rapidly activated by thrips feeding [40]. Considering that N. benthamiana is not a good host for thrips as indicated by their poor survival rate, and the finding that MeJA induces similar expression of TPS genes in N. benthamiana as thrips infestation (S1 Fig), we used MeJA to mimic WFT infestation in further tripartite interaction experiments. The expression of NbTPS3, NbTPS4, NbTPS5, and NbTPS38 was less changed in TSWV-infected plants compared to the control plants when induced by methyl jasmonate (MeJA) (Fig 1D).
TSWV infection induces a terpene-dependent preference in the thrips vector
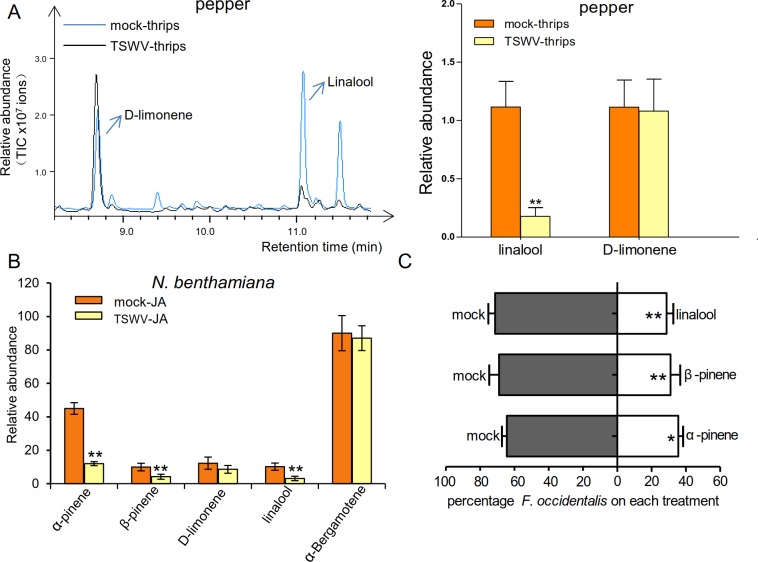
To explore the metabolic consequences of the altered TPS gene expression, we investigated changes in the emission of plant volatile compounds after TSWV infection. Plants have evolved a blend of HIPVs that are emitted in response to, and directly repel, herbivores [27–33]. We measured the volatile emission collected in the headspace of peppers with or without thrips infestation. When infested by thrips, damaged plants emitted more volatiles than control plants (S2 Fig). It is noteworthy that TSWV-infected plants emitted significantly less linalool, which is the main monoterpene collected from peppers after herbivory, compared to non-infected plants, consistent with their lower expression of TPS genes. In addition, there was no significant difference in the emissions of the monoterpene D-limonene (Fig 2A). We also monitored the emission of volatile compounds in the headspace of N. benthamiana, after applying MeJA to mimic WFT infestation; this plant hormone is known to elicit the production of various terpenes [42]. Among the five detected terpenes, the levels of three volatile monoterpenes, linalool, α-pinene, and β-pinene, were significantly lower in TSWV-infected plants compared to non-infected plants (Fig 2B). To examine whether linalool, α-pinene and β-pinene play a role in plant–WFT interactions, we performed a two-choice assay in which non-viruliferous WFT had the choice between the changed monoterpenes and the solvent control hexane. The α-pinene and β-pinene directly repelled thrips similarly to linalool (Fig 2C). These consistent results on pepper and N. benthamiana revealed that TSWV infection induces a terpene-dependent preference in the thrips vector and that this feature is common among various TSWV hosts.
Fig 2. TSWV infection increases attractiveness to the thrips vector in a terpene-dependent manner.
(A) Representative GC/MS ion chromatograms of the headspace volatile compounds of control (mock-thrips) and TSWV-infected peppers (TSWV-thrips) after thrips infestation for 6 h. The peaks of specific products are marked with arrows in the left panel. Relative abundance of terpenes emitted after thrips infestation are showed in the right panel. Values are means + SE, n = 4. **P < 0.01, Student’s t-test. (B) Terpenes emitted by N. benthamiana after TSWV infection (under MeJA treatment). Values are mean relative amounts (percentage of internal standard peak area) ± SE, n = 4. **P < 0.01, Student’s t-test. (C) Thrips preference (as percentage recaptured WFT out of 50 released) on the pure monoterpenes (linalool, α-pinene, β-pinene) and solvent control (n-hexane) in a two-choice assay. Data are mean percentages + SE, n = 6. *P < 0.05 **P < 0.01, Wilcoxon matched pairs tests.
NSs manipulates the preference behavior of WFT on plants
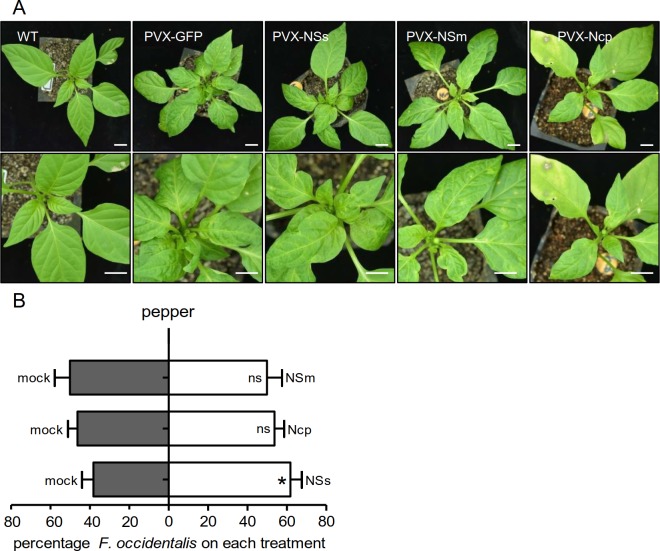
Our data demonstrated that the orthotospovirus TSWV increases the attraction of insect vector WFT to its host plant by inhibiting terpene synthase expression in the host. Next, to explore which viral protein(s) in TSWV manipulate vector host choice, we selected three of the five viral proteins in TSWV, including a structural protein nucleocapsid protein (Ncp) and two non-structural proteins, NSm and NSs [24]. We used the heterologous Potato virus X (PVX) model system for systemic ectopic expression of individual genes for TSWV NSs, NSm or Ncp [43]. PVX-GFP, used to express green fluorescent protein (GFP) in the plant, was served as the control. There were no obvious morphological differences between these recombinant PVX vector-infected peppers (Fig 3A). We performed a WFT two-choice assay to determine whether the expression of a single viral protein is sufficient to attract WFT. PVX-NSs-infected plants but not PVX-NSm- or PVX-Ncp-infected plants were significantly more attractive to WFT than PVX-GFP-infected plants (Fig 3B), indicating the expression of NSs alone is sufficient to attract WFT in peppers.
Fig 3. NSs from TSWV is a vector behavior manipulator.
(A) Phenotype of pepper leaves inoculated with recombinant Potato virus X (PVX) vectors. PVX-NSs, PVX-NSm or PVX-Ncp was transformed into peppers via agroinfiltration. PVX-GFP was used as the control. Bar = 2 cm. (B) Attractiveness of different infiltrated peppers. Agrobacteria carrying individual recombinant PVX vectors were infiltrated into peppers. Plants of a similar size were used for thrips two-choice assays at 10 dpi. Data are mean choice percentages + SE, n = 6. *P < 0.05, Wilcoxon matched pairs tests.
TSWV NSs interacts with MYC2 and its homologs MYC3 and MYC4
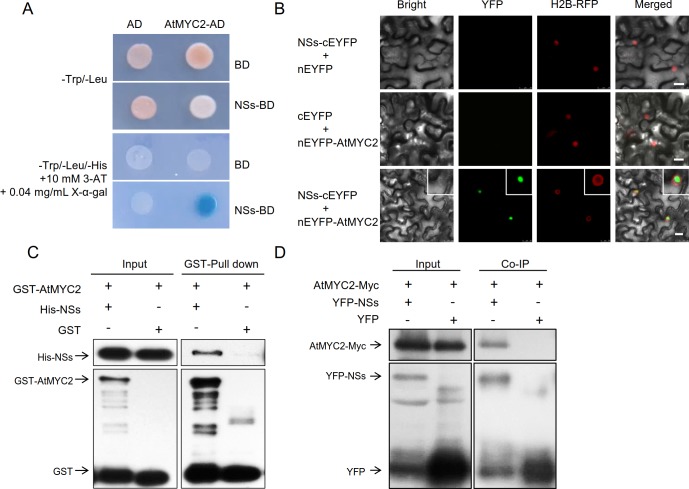
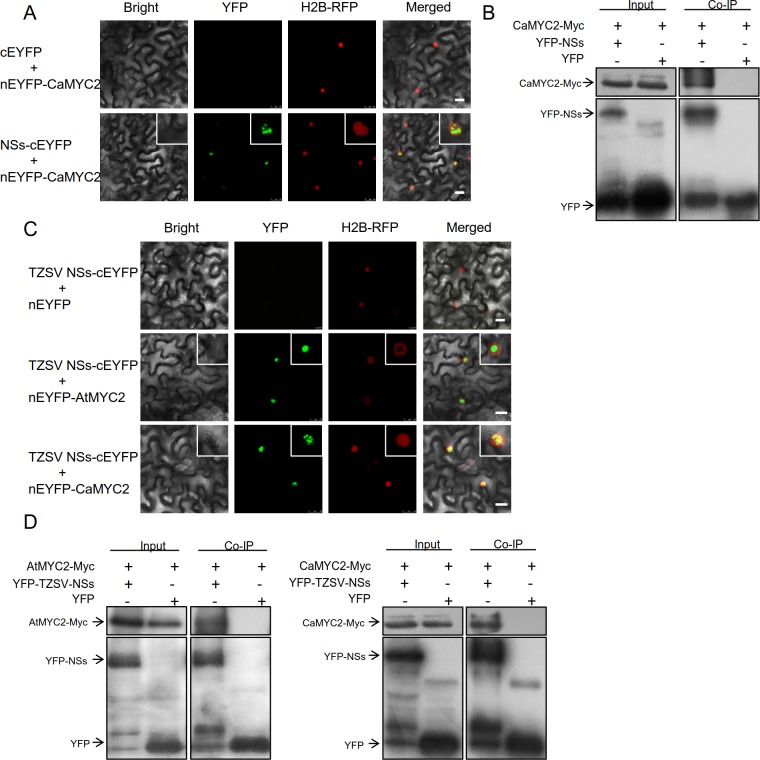
To explore the host protein targets of NSs, we screened an Arabidopsis cDNA library by yeast two-hybrid analysis and identified AtMYC2, a key components of the JA signaling pathway [44–46]. Based on the importance of the JA signaling pathway to plant–herbivore interactions, we further confirmed the interaction between AtMYC2 and NSs. In a yeast two-hybrid assay, the yeast transformants harboring AD-AtMYC2 and BD-NSs could grow on SD-Leu-Trp-His medium with 0.04 mg/mL X-α-gal and turned blue, while the negative control transformants did not (Fig 4A). A bimolecular fluorescence complementation (BiFC) assay confirmed the AtMYC2 and NSs interaction in plants. NSs-cEYFP and nEYFP-AtMYC2 constructs were co-expressed in transgenic N. benthamiana lines that expressed a nucleus-localized histone H2B-red fluorescent protein (H2B-RFP) fusion marker protein. A strong interaction (represented by fluorescence) was observed in the nucleus (Fig 4B), while no fluorescence was observed in the negative controls (Fig 4B). GST pull-down assay was used to verify the direct physical interaction between NSs and AtMYC2 in vitro. His-NSs was pulled down by GST-AtMYC2, but not by GST alone (Fig 4C). Moreover, in a co-immunoprecipitation (Co-IP) assay, AtMYC2-Myc was coimmunoprecipitated by YFP-NSs, but not the YFP control (Fig 4D). Taken together, these results demonstrate that NSs directly interacts with MYC2 in vitro and in vivo.
Fig 4. TSWV NSs interacts with MYC2.
(A) Yeast two-hybrid assay between NSs and AtMYC2. Yeast cotransformed with the indicated plasmids was spotted onto synthetic medium (SD-Leu-Trp-His) containing 0.04 mg/mL X-α-gal and 10 mM 3-amino-1,2,4-triazole (3-AT). The empty vectors pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. (B) Bimolecular fluorescence complementation (BiFC) assay. NSs-cEYFP and nEYFP-AtMYC2 were transiently expressed in H2B-RFP transgenic N. benthamiana leaf epidermal cells via agroinfiltration. Bars = 15 μm. (C) GST pull-down assays between NSs and AtMYC2. (D) Interaction between NSs and AtMYC2 in Co-immunoprecipitation (Co-IP) assay. Total protein was extracted from N. benthamiana leaves transiently expressing 35S:MYC2-Myc together with 35S:YFP-NSs or 35S:YFP alone. GFP-trap beads were used to precipitate the interaction complex, Anti-GFP and Anti-Myc antibodies were using to detect the immunoprecipitates.
MYC3 and MYC4 are two closely related bHLH transcription factors that function partially redundantly with MYC2 to activate JA responses in Arabidopsis [47]. To determine whether TSWV NSs targets MCY3 and MYC4 as well, we performed a yeast two-hybrid assay and a BiFC assay. MYC2 relatives MYC3 and MYC4 were also found to interact with TSWV NSs as indicated by AD-AtMYCs (MYC3 and MYC4) and BD-NSs yeast transformants turned blue when grown on SD-Leu-Trp-His medium with 0.04 mg/mL X-α-gal (S3A Fig). In the BiFC assay, N. benthamiana coexpressing MYC3 and NSs exhibited fluorescence in the cytoplasm and nucleus, while coexpression of MYC4 and NSs led to fluorescence only in the cytoplasm (S3B Fig). These results indicate that MYC family transcription factors are targeted by NSs protein.
MYCs positively regulate volatile-dependent immunity against WFT in Arabidopsis
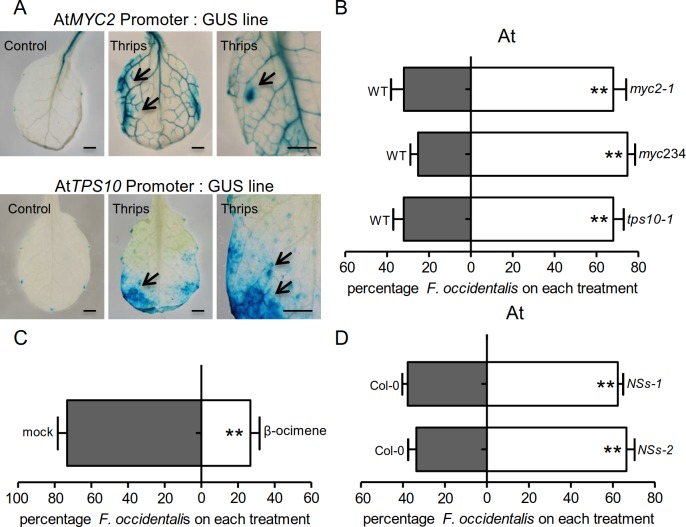
We previously showed that Arabidopsis MYC2 plays important roles in JA-regulated plant defense responses, e.g. directly regulates TPS10 transcript levels to promote plant volatile biosynthesis [38]. Thus, we hypothesized that AtMYC2, which interacts with virulence factor NSs, is involved in the viral-induced, volatile-dependent attraction of WFT to the host plant. To validate this hypothesis, we performed a GUS staining assay using two transgenic Arabidopsis lines expressing an AtMYC2 or AtTPS10 promoter: GUS reporter gene. As shown in Fig 5A, high GUS expression was detected after 24 h of WFT infestation. This expression pattern suggests that AtMYC2 and AtTPS10 both function in defense responses against WFT in Arabidopsis.
Fig 5. MYC2 and its homologs in Arabidopsis are essential regulators of host immunity responses against WFT.
(A) GUS staining of AtMYC2p-GUS and AtTPS10p-GUS seedlings after 24 h of thrips feeding. An untreated line was used as a control. Arrows indicate thrips feeding sites. Bars = 2 mm. (B) Thrips preference (as percentage recaptured WFT out of 50 released) between the mutants and WT control in a two-choice assay. Three-week-old Arabidopsis plants cultured in MS medium were used for the thrips two-choice assay. Data are mean percentages + SE, n = 6. **P < 0.01, Wilcoxon matched pairs tests. (C) β-ocimene is less attractive to thrips than mock treatment in a two-choice assay. Data are mean percentages + SE, n = 6. **P < 0.01, Wilcoxon matched pairs tests. (D) Thrips preference (as percentage recaptured WFT out of 50 released) between the 35S:YFP-NSs transgenic Arabidopsis lines (NSs-1; NSs-2) and mock control in a two-choice assay. Three-week-old transgenic Arabidopsis plants cultured in MS medium were used. Data are mean percentages + SE, n = 6. **P < 0.01, Wilcoxon matched pairs tests.
To analyze the effects of AtMYC2 and AtTPS10 on the feeding preferences of thrips, we performed two-choice assays using myc2-1, tps10-1, and wild-type Col-0 Arabidopsis. As shown in Fig 5B, the myc2-1 and tps10-1 mutants were more attractive to WFT than wild type. We also tested the effect of triple mutant myc234 on host preference, finding that WFT strongly preferred myc234 plants over the wild type (Fig 5B). AtTPS10 encodes a monoterpene synthase that produces β-ocimene [48]. We therefore carried out a two-choice assay of β-ocimene to examine whether the attraction of tps10 is terpene-dependent. β-ocimene had a strong repellent effect on WFT (Fig 5C). These results indicate that AtMYC2 is essential for terpene-dependent immunity against the thrips vector. We further examined if the TSWV NSs contributes to the preference of thrips on Arabidopsis. In two-choice assays, two transgenic Arabidopsis 35S:YFP-NSs (NSs-1; NSs-2) lines were significantly more attractive to thrips compared to controls (Fig 5D), supporting the conclusion that NSs protein can modify vector feeding behavior in a terpene-dependent manner.
NSs promotes thrips vector performance by targeting MYCs
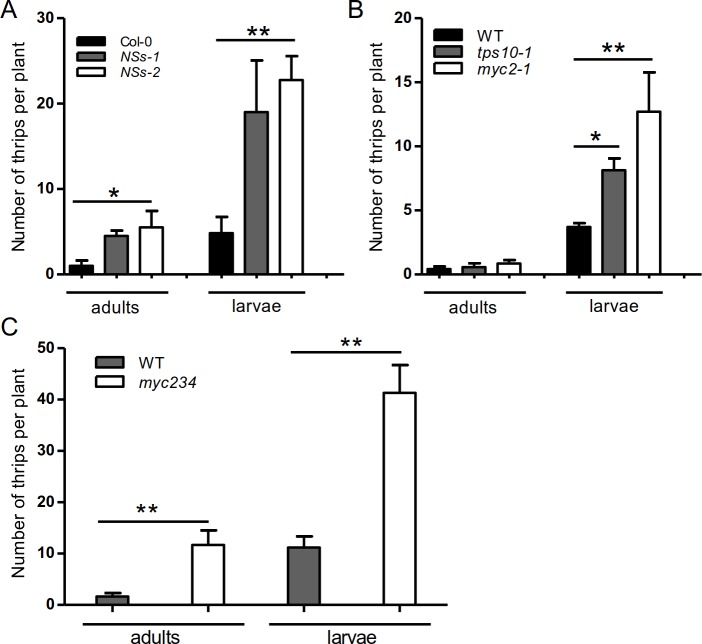
The viral transmission cycle can be roughly divided into two phases. In the first phase, the TSWV-infected plants attract non-viruliferous thrips to feed, with volatiles playing a key role in this early process (Figs 1–5). In the second phase, a (viruliferous) thrips population is established on TSWV-infected plants to facilitate virus transmission. To investigate whether NSs influences thrips population establishment, we performed a thrips spawning experiment with a slight modification [40]. Seven female adult thrips were allowed to feed on 35S:YFP-NSs (NSs-1; NSs-2) or wild-type Arabidopsis for two weeks. We counted the number of new adults and larvae to analyze the effect of NSs on the thrips population. Plants expressing NSs were more suitable for WFT population growth than wild type (Fig 6A). We reasoned that NSs targets MYCs to disable the activation of terpene synthase genes, thereby attenuating the defense of the host plant against thrips. To investigate this hypothesis, we conducted another spawning experiment using myc2-1, tps10-1, and myc234 mutants. More WFT were found on the mutants compared with wild type; these lines were equally suitable for WFT growth compared to the lines expressing NSs, confirming the important role for NSs in the tripartite WFT–TSWV–plant interaction (Fig 6B and 6C).
Fig 6. NSs promotes WFT performance by targeting MYC-mediated host defense.
(A-C) Effects of different genes on the number of WFT offspring. Seven adult females fed on each three-week-old Arabidopsis line. After 2 weeks, new larvae and adults were counted. Values are means ± SE, n = 8. *P < 0.05, **P < 0.01, Student’s t-test.
A conserved protein interaction between Orthotospovirus NSs and plant MYC2
TSWV-infected pepper plants were more attractive to the thrips vector than healthy plants (Fig 1A). Therefore, we asked whether NSs could interact with AtMYC2 orthologs in pepper. We examined the interaction between NSs and the homologous protein of AtMYC2 in pepper (CaMYC2). Our BiFC assay results showed interaction fluorescence of NSs–CaMYC2 in the nucleus, while there was no fluorescence of control (Fig 7A). In Co-IP assays, CaMYC2-Myc protein was coimmunoprecipitated by YFP-NSs, but not by YFP alone (Fig 7B). Taken together, our results suggest that NSs–MYC2 interaction is relatively conserved in pepper.
Fig 7. A conserved protein interaction between Orthotospovirus NSs and plant MYC2 proteins.
(A) BiFC assays of the interaction between TSWV NSs and CaMYC2. H2B-RFP transgenic N. benthamiana plants, which express a nucleus marker were used in this assay. Bars = 15 μm. (B) Co-immunoprecipitation (Co-IP) assay of the interaction between TSWV NSs and CaMYC2. GFP-trap beads were used to precipitate the interaction complex. (C) BiFC assays of the interaction between TZSV NSs–AtMYC2 and TZSV NSs–CaMYC2. H2B-RFP transgenic N. benthamiana plants were used in this assay. Bars = 15 μm. (D) Co-immunoprecipitation (Co-IP) assay of the interaction between TZSV NSs–AtMYC2 (left panel) and TZSV NSs–CaMYC2 (right panel). GFP-trap beads were used to precipitate the interaction complex.
Other orthotospoviruses also encode a NSs protein and might similarly manipulate vector behavior to accelerate their own transmission [49]. To explore whether the interaction between NSs–MYC2 is conserved among orthotospoviruses, we used Tomato zonate spot orthotospovirus (TZSV), a new species of genus Orthotospovirus that threatens food security in Southwest China [50]. The evolutionary relationship of TSWV and TZSV is not very close [51], as TSWV represents the American- and TZSV represents the Euro/Asian-type orthotospoviruses (S4 Fig). We examined TZSV NSs–AtMYC2/CaMYC2 interactions by BiFC and Co-IP assays. Notably, BiFC showed interaction fluorescence of TZSV NSs–AtMYC2 and TZSV NSs–CaMYC2 aggregated in the nucleus, while the Co-IP assays again confirmed the interaction between TZSV NSs–AtMYC2 (left panel) and TZSV NSs–CaMYC2 (right panel) (Fig 7D), providing evidence that TZSV NSs interacts with both AtMYC2 and CaMYC2 in vivo, consistent with NSs–MYC2 interaction in TSWV (Figs 4 and 7A). These results indicated that the interaction between NSs and MYC2 may be conserved in orthotospoviruses.
In summary, our results suggest that NSs targets MYCs to attenuate host defense responses to thrips, thereby manipulating terpene-dependent chemical communication between the plant and the thrips vector.
Discussion
TSWV suppresses host terpene biosynthesis and promotes the performance of its thrips vector
Vector-borne virus-infected plants often attract the pathogens’ vectors [1]. Here, we demonstrate a possible molecular mechanism of this virus-induced indirect manipulation through the shared host plant. Non-viruliferous thrips feeding was reported to induce a negative change in plant quality for their survival [10]. Consistent with this, we showed that the expression of various TPSs were induced strongly by herbivory (Fig 1B) and repellent terpenes were produced as a consequence (S2 Fig). Orthotospoviruses depend on the vector thrips for transmission, and enhanced performance of WFT on virus-infected plants would be beneficial to the virus and the vector. We found that the induction of plant defense was greatly decreased in TSWV-infected plants, thus promoting the performance of WFT vector (Figs 1, 5 and 6). Our results establish the existence of an indirect mutualistic relationship between Orthotospoviruses and the thrips vector. This indirect mutualism refers to a positive effect of virus on its insect vector. Virus suppresses plant defense against the insect vector leading to enhanced vector performance and population, which in turn promote virus transmission.
Among the monoterpenes manipulated by TSWV in various plants, linalool functions as a repellent to WFT both in pepper and N. benthamiana (Fig 2). It is one of the most common defensive monoterpene compounds in the HIPVs released from plant species in response to herbivore attacks [52]. Linalool has been shown to affect the feeding behavior of insects, as well as to attract pollinators, repel herbivores, and affect insect spawning decisions [38,52]. It also inhibits the growth of WFT [53], in agreement with the conclusion that linalool is an anti-WFT secondary metabolite hijacked by TSWV (Fig 2A). Since volatiles are essential to herbivory responses, exogenous application of monoterpenes such as linalool may be a promising approach to avoid herbivore feeding damage and even plant pathogen transmission under field conditions, without the need for engineering in plants.
NSs represses MYC2-mediated JA signaling pathway to achieve indirect tospovirus–WFT mutualism
Behavioral manipulation has been observed in animal-infecting bunyaviruses for many years. As early as 1980, La Crosse virus (LACV) was reported to modify the feeding behavior of mosquito vectors [18]. Rift Valley fever virus (RVFV) was found to affect mosquito vector morbidity and mortality [19]. However, the molecular mechanism underlying this manipulation was unclear, and no specific information was available regarding viral determinants of the virus–host–vector interaction in bunyaviruses.
Our study identifies NSs of TSWV as an indirect vector behavior manipulator that suppresses host plant defense responses to attract and benefit the fitness of WFT, which in turn facilitates disease dispersal from plant to plant. Notably, NSs is conserved in bunyaviruses, and TSWV NSs is an avirulence determinant that triggers a hypersensitive response in resistant plants [54]. NSs is also a well-known viral suppressor of host RNA interference in both plants and insects and is essential for TSWV transmission by WFT [16,23–26]. Here, we showed that the expression of NSs is sufficient to control the behavior of WFT (Figs 3, 5D and 6A) by suppressing the host defense against insects through MYC proteins (Fig 4). Additionally, the non-viruliferous female thrips were reported to produce more offspring on virus-infected plants, which is in agreement with their preference for TSWV-infected plants [9,10,21]. Taken together, the infection of TSWV could counter plant defense to benefit its vector, thus promoting its spread through the NSs protein.
Effectors target the plant MYC immunity hub
Earlier studies showed that effectors from bacterial, fungal and oomycete pathogens converge onto common host proteins in Arabidopsis [55]. Our results suggest that viral effectors also share the same plant targets. JA signaling is essential for plant defense against pathogen and insect attack in several phytopathological systems [56,57]. However, plant arboviruses target JA signaling to increase the suitability of host plants for their vectors [38,58]. JA-dependent plant defenses affect WFT performance and preference, and TSWV infection reduces the levels of these responses. In JA-insensitive coi1-1 mutants, WFT do not show a preference for TSWV-infected plants [21]. Our results suggest that the MYC proteins involved in the JA pathway are responsible for plant terpene immunity against WFT (Fig 5A–5C). MYCs are downstream genes of the JA receptor COI1, and MYC2-orchestrated transcriptional reprogramming occurs during JA signaling [48].
Functional blocking of MYCs increases WFT preference and promotes WFT performance, including developmental duration and fecundity in Arabidopsis (Fig 6). We hypothesize that several MYC-regulated indole and aliphatic glucosinolates that function as defensive chemicals against herbivores might be repressed. Alternatively, the levels of nutrients (such as amino acids) are likely altered in the host, which could affect the feeding behavior and preference of thrips, as previously reported [8]. In addition, the interaction between TZSV NSs and MYC2 indicates that TZSV infection of plants may also benefit its insect vector like TSWV infection does (Fig 7B). Therefore it seems like NSs of Orthotospovirus conservatively interacted with MYC2 and its homologs in plant host (Fig 7).
By interrupting MYC-regulated plant defense via NSs, Orthotospovirus species appear to indirectly manipulate the preference and performance of WFT, as is the case for βC1 in Begomovirus. We previously demonstrated that βC1 of Tomato yellow leaf curl China virus (TYLCCNV) interacts with MYC2 to subvert plant resistance and to promote vector performance [38]. Notably, Begomovirus and Orthotospovirus species are persistently transmitted, which tend to induce attraction and promote performance of vectors on infected plants for increased transmission efficiency, indicating that viruses with same transmission mechanisms can have common manipulation tactics. Interestingly, the silencing suppressor 2b of the nonpersistently transmitted virus Cucumber mosaic virus (CMV, Bromoviridae) also suppresses JA signaling, and myc234 triple mutant plants were observed to attract the CMV aphid vector [58], although CMV appears to attract vectors deceptively [15]. These similar results on evolutionarily different viruses and plant hosts suggest that manipulation of the JA pathway could be a general feature in tripartite virus–vector–plant interactions. Notably, these independently evolved virulence proteins were known as silencing suppressors that convergently targeted the host RNA silencing machinery, and our studies establish that the same occurs for the manipulation of plant–insect vector interactions.
These similar effects and pathogen manipulation tactics indicate that the mechanistic and evolutionary principle for diverse pathogens seems to be convergent, even in human pathogens. For instance, CCR5, which is the first described cellular receptor of human immunodeficiency virus (HIV), is necessary and sufficient for the pathogenesis of many pathogens [59]. The HIV, Toxoplasma gondii, poxviruses (vaccinia and myxoma), and Staphylococcus aureus exploit CCR5 to target and kill mammalian immune cells [60–63]. Why pathogens from different kingdoms tend to keep finding the same host targets to disrupt their defenses, and whether this is a consequence of selective pressure in evolution remain to be further determined.
In summary, we have demonstrated that the emission of several monoterpenes is greatly decreased by the TSWV infection, which in turn promotes WFT preference and performance, uncovering a molecular mechanism underpinning the virus-induced manipulation through the shared host plant of the WFT vector. This work presents a mechanism by which a pathogen regulates host-derived olfactory cues for vector attraction. These results will also help to address similar tripartite interaction systems in plants, animals and humans and will allow innovative control methods through interference of vector transmission.
Materials and methods
Plant materials
Pepper accession Lingfeng (Capsicum annuum L.), Nicotiana benthamiana and Arabidopsis thaliana (Col-0) plants were grown in insect-free growth chambers following standard procedures [38]. The Arabidopsis myc2-1, tps10-1, and myc234 mutants (Col-0 background) were described previously [38]. The 35S:YFP-NSs transgenic lines NSs-1 and NSs-2 were generated using the Agrobacterium-mediated floral-dip method [64].
Naive Western flower thrips colony and mechanical inoculation of TSWV
A starting colony of Western flower thrips (WFT, Frankliniella occidentalis Pergande) (Thysanoptera: Thripidae) was kindly provided by Prof. Youjun Zhang (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences). The thrips were maintained on green bean pods (Phaseolus vulgaris L.) in a climate chamber as described previously [65]. Tomato spotted wilt orthotospovirus (isolate TSWV-YN) obtained from Prof. Xiaorong Tao (Nanjing Agriculture University) was mechanically inoculated onto pepper and N. benthamiana as described by Mandal et al. [66]. Infected leaves were ground in 0.05 M phosphate buffer (pH 7.0) and applied to the host plant using a soft finger-rubbing technique. Infected plants were tested at 10–14 dpi by RT-qPCR prior to the thrip two-choice assays.
Thrip two-choice assay
The two-choice assays on plants or leaves were performed as described previously [8,9]. Peppers inoculated with TSWV or buffer was used for the assay at 10–14 days post inoculation. A TSWV-infested and a control plant were confined in a pot covered with a fine mesh. For N. benthamiana, detached leaves of TSWV-infected plants and non-infected plants were separately placed in a 16 cm-Petri dish, which was covered with a moist filter paper. For Arabidopsis, plants were cultivated on solid Murashige and Skoog medium for 3–5 weeks, and whole plants were used for the two-choice assay. Fifty F. occidentalis adults were released to the center of the two tested plants or the leaves of N. benthamiana, the number of thrips that settled on each plant or leaf was counted at 12h (pepper) or 24 h (N. benthamiana, Arabidopsis) after release. For two-choice assays with individual monoterpene, 2 cm × 2 cm filter paper containing 40 μL of a 1:100 (v/v) solution of standard chemical substance from Sigma dissolved in n-hexane or n-hexane alone (as a control) was placed in a 16cm-Petri dish. Thrips were released between the two tested samples, and the thrips were counted 5 min after release. The Petri dishes were contained in a thrip culture chamber throughout the experiment to maintain consistent environmental conditions.
Thrip infestation assay
Plants were infested with non-viruliferous thrips as described previously [56]. Twenty adult thrips (7–14 d after eclosion) were grouped and starved for 3 h before the plant infestation assay. Arabidopsis plants grown on solid MS medium or soil-grown pepper and N. benthamiana plants were infested with adult thrips for the indicated time period. The thrips were gently removed and the leaf samples collected in liquid nitrogen for further analysis. For the GUS-reporter line expression assays, transgenic Arabidopsis plants were infested with thrips for 24 h, followed by GUS activity analysis. The experiment was repeated at least twice with similar results.
Volatile analysis
For volatile analysis on pepper plants, plants were infested with thirty adults in a nylon mesh cage for 6 h before volatile collection. The volatiles emitted from insect‐exposed TSWV-infected and control plants were collected with a solid phase microextraction (SPME; Supelco, Belafonte, PA, USA) fiber consisting of 100 μm polydimethylsiloxane (Supelco). Chemical analysis was performed by gas chromatography-mass spectrometry (GC-MS) (Shimadzu, QP2010) coupled with a DB5MS column (Agilent, Santa Clara, CA, USA, 30 m x 0.25 mm x 0.25 μm). The SPME fiber was thermally desorbed in the injector at 250°C for 1 min. The initial oven temperature was held at 40°C for 3 min, increased to 240°C with a gradient of 5°C/min, and maintained at 240°C for 5 min. The inlet temperature was 250°C. The collection of volatiles for each treatment was repeated 4–6 times.
The collection, isolation, and identification of volatiles from N. benthamiana plants were performed as described previously [38,67]. Plant volatiles were collected for 12 h at a gas flow rate of 300 mL/min and analyzed by GC-MS. At least four plants per group were used.
Plasmid construction
For PVX heterologous virus protein expression in pepper, the TSWV virus genes NSs, NSm, and Ncp were cloned into the PVX vector pGR208 by using gene-specific primers in S1 Table. For agroinfiltration transient expression vectors construction, the indicated DNA fragments were PCR cloning into pENTR-3C entry vector, then transformed into the agroinfiltration destination vector under the control of a CaMV 35S promoter. All constructs used for protein expression in plants were transformed into Agrobacterium tumefaciens strain EHA105. Agrobacterium carrying the binary vectors were infiltrated into the abaxial sides of pepper and N. benthamiana leaves [49].
Yeast two-hybrid analysis
The Arabidopsis Mate and Plate Library was screened using yeast mating method according to the Matchmaker Gold Yeast Two-Hybrid System manufacturer’s protocol (Clontech). Briefly, full-length NSs was amplified and inserted into the pGBKT7 vector by Gateway recombination, then the constructs was transformed into yeast strain Y2HGold and testing for autoactivation by using the Yeastmaker Yeast Transformation System (Clontech). Then the Arabidopsis Mate and Plate Library and BD-NSs yeast clones were mated in YPDA medium. After incubation, isolated destination clones were selected from diploid-selection medium (SD/-Leu/-Trp). These primary positive interactors were secondary screened on medium plates (SD/-Leu/-Trp/-His) and third time screened on medium plates (SD/-Leu/-Trp/ -His/X-a-Gal). PCR and BLAST searches were used to obtain sequence information on corresponding AD- and BD-clones per colony.
The interaction between TSWV NSs and AtMYCs were confirmed according to the manufacturer’s protocol (Clontech). The pGBKT7-NSs and pGAD424-MYCs constructs were co-transformed into yeast strain Y2HGold. Yeast cotransformed with the indicated plasmids was spotted onto synthetic medium (SD-Leu-Trp-His) containing 10 mM 3-amino-1,2,4-triazole and 0.04 mg/mL X-α-gal. The empty vectors pGBKT7 (BD) and pGADT7 (AD) were used as negative controls [38].
Bimolecular fluorescence complementation (BiFC)
BiFC was performed as described previously [38]. The indicated constructs were fused with the N-or C- terminal of YFP and transformed into A. tumefaciens strain EHA105. The recombinant constructs of A. tumefaciens were infiltrated in 4–6 week old transgenic N. benthamiana (expressing a nuclear marker-H2B-RFP)[68] leaves via agroinfiltration. The fluorescent signals were detected at 2 dpi via confocal microscopy.
In Vitro pull-down assay
His and GST tag fusion proteins were purified using His- and GST-Trap (GE Healthcare) according to the manufacturer’s instructions [38]. GST-AtMYC2 (2 μg) and His-NSs (2 μg) fusion proteins were mixed and incubated with 25 μL GST-Trap for 2 h at 4°C in a binding buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.25% Triton X-100, and 35 mM b-mercaptoethanol). After six washes with binding buffer, pulled-down proteins were resuspended in 2xSDS buffer and detected by immunoblot using Anti-GST and Anti-His antibodys.
Co-immunoprecipitation (Co-IP)
A.tumefaciens carrying the 35S:MYC2-Myc or 35S:YFP-NSs constructs were infiltrated into N. benthamiana leaves. About 1g leaf tissue was collected and ground to powder in liquid nitrogen. Proteins were extracted in a cold extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 0.1% Triton, 0.5% NP-40, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), one protease inhibitor cocktail/100 mL (Sigma-Aldrich, USA)). Then the protein extracts were incubated with 25 μL GFP-trap beads for 3 h at 4°C. After that, the beads were washed three times with extraction buffer and resuspended in 2xSDS buffer before used for immunoblot analysis.
Quantitative RT-PCR
Total RNA was extracted from leaf and plant samples using an RNeasy Plant Mini Kit (Qiagen) with column DNase treatment. RNA was reverse transcribed using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, China). Four to six independent biological samples were collected and analyzed. RT-qPCR was performed using SYBR Green Real-Time PCR Master Mix (Toyobo, China) on the CFX 96 system (Bio-Rad). Pepper Ca-ACT1 and N. benthamiana Nb-EF1α were used as the internal controls (Listed in S1 Table).
Thrip spawning assay
The thrip spawning assay was performed as described previously with some modifications [40]. Arabidopsis plants were grown in soil covered with Parafilm (Bemis, USA) to prevent any thrips from escaping and to facilitate counting. Three-week-old plants were placed in an acryl cylinder chamber (7 cm × 5 cm) and covered with a fine mesh. Seven female adults (7–14 d after eclosion) were allowed to infest a single plant for two weeks, and new larvae and adult thrips were counted. Eight plants of each genotype were used per experiment. The experiment was repeated at least twice with similar results.
GUS staining
Transgenic Arabidopsis plants expressing the AtMYC2 or AtTPS10 promoter:GUS reporter gene were infested with thrips for 24 h and incubated in GUS staining buffer (0.5 mg/mL X-glucuronide, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 10 mM EDTA, 0.1% Triton X-100, 0.1 M pH 7.0 phosphate buffer) at 37°C overnight. The stained seedlings were cleared by washing with 70% ethanol. Untreated plants were used as a negative control. The experiment was repeated at least twice with similar results.
Data analysis
Significant differences in gene expression and volatile organic compound levels were determined by Student’s t tests or one-way ANOVA; if the ANOVA result was significant (P < 0.05), Duncan’s multiple range tests were used to detect significant differences between groups. Thrip choices between different treatments were analyzed by nonparametric Wilcoxon matched pairs tests. All statistical tests were carried out with GraphPad Prism.
Accession numbers
Sequence data in this study can be found in Sol Genomics Network (https://solgenomics.net), TAIR (www.Arabidopsis.org) or GenBank/EMBL under the following accession numbers: CaMYC2 (CA00g50270), CaMTS1 (CA08g16370), CaMTS2 (CA08g16380), CaMTS3 (CA08g16410), CaMTS4 (CA08g16420), AtMYC2 (AT1G32640), AtMYC3 (AT5G46760), AtMYC4 (AT4G17880), AtTPS10 (AT2G24210), TSWV NSs (JF960235.1), TSWV NSm (JF960236.1), and TSWV Ncp (JF960235.1), TZSV(EF552433.1).
Supporting information
(A) Relative expression levels of various TPS genes in N. benthamiana after thrips infestation. Four-week-old N. benthamiana plants were infested with twenty thrips adults in a confined pot for 48h. Total RNA was prepared from treated plants for RT-qPCR analysis. Values are means + SE, n = 3. **P < 0.01, Student’s t-test. (B) Relative expression levels of various TPS genes in N. benthamiana after 100 μM MeJA treatment for 24h. Values are means + SE, n = 3. **P < 0.01, Student’s t-test.
(TIF)
Representative extracted ion chromatograms of GC/MS headspace volatile compounds of peppers. Plants under the same growth condition were infested with (pepper-thrips) or without (pepper) thrips for 6 h.
(TIF)
(A) Interaction between TSWV NSs–AtMYC3 and TSWV NSs–AtMYC4 in a yeast two-hybrid assay. Yeast cotransformed with the indicated plasmids was spotted onto synthetic medium (SD-Leu-Trp-His) containing 0.04 mg/mL X-α-gal and 10mM 3-amino-1,2,4-triazole (3-AT). The empty vectors pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. (B) Interaction between TSWV NSs–AtMYC3 and TSWV NSs–AtMYC4 in a BiFC assay. Indicated construsts were transiently expressed in H2B-RFP transgenic N. benthamiana leaf epidermal cells by agroinfiltration. Bars = 15 μm.
(TIF)
ClustalW was used to construct the phylogenetic tree. It was constructed based on the amino acid sequences of the NSs protein from 23 orthotospoviruses.
(TIF)
(DOCX)
Acknowledgments
We thank Profs. Youjun Zhang (Chinese Academy of Agricultural Sciences, China) and Xiaorong Tao (Nanjing Agriculture University, China) for providing the WFT clone and TSWV-YN strain. We also thank the three anonymous reviewers whose helpful suggestions have led to an improvement of our manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
JY was supported by the Chinese Academy of Sciences (Strategic Priority Research Program Grant NO. XDB11040300) and National Natural Science Foundation of China (31830073,31522046, 31672001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Eigenbrode SD, Bosque-Pérez NA, Davis TS. Insect-borne plant pathogens and their vectors: ecology, evolution, and complex interactions. Annu Rev Entomol. 2018; 63:169–191. 10.1146/annurev-ento-020117-043119 [DOI] [PubMed] [Google Scholar]
- 2.Legarrea S, Barman A, Marchant W, Diffie S, Srinivasan R. Temporal effects of a Begomovirus infection and host plant resistance on the preference and development of an insect vector, Bemisia tabaci,and implications for epidemics. PLoS One. 2015;10(11):e0142114 10.1371/journal.pone.0142114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiménez-Martínez ES, Bosque-Pérez NA, Berger PH, Zemetra RS, Ding H, Eigenbrode SD. Volatile cues influence the response of Rhopalosiphum padi (Homoptera: Aphididae) to barley yellow dwarf virus-infected transgenic and untransformed wheat. Environ Entomol. 2004;33:1207–1216. 10.1603/0046-225x-33.5.1207. [DOI] [Google Scholar]
- 4.Medina-Ortega KJ, Bosque-Pérez NA, Ngumbi E, Jiménez-Martínez ES, Eigenbrode SD. Rhopalosiphum padi (Hemiptera: Aphididae) responses to volatile cues from Barley yellow dwarf virus-infected wheat. Environ Entomol. 2009;38(3):836–845. 10.1603/022.038.0337 [DOI] [PubMed] [Google Scholar]
- 5.Eigenbrode SD, Ding H, Shiel P, Berger PH. Volatiles from potato plants infected with potato leafroll virus attract and arrest the virus vector, Myzus persicae (Homoptera: Aphididae). Proc Biol Sci. 2002;269(1490):455–460. 10.1098/rspb.2001.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casteel CL, Yang C, Nanduri AC, De Jong HN, Whitham SA, Jander G. The NIa-Pro protein of Turnip mosaic virus improves growth and reproduction of the aphid vector, Myzus persicae (green peach aphid). Plant J. 2014;77(4):653–663. 10.1111/tpj.12417 [DOI] [PubMed] [Google Scholar]
- 7.Hodge S, Powell G. Do plant viruses facilitate their aphid vectors by inducing symptoms that alter behavior and performance? Environ Entomol. 2008;37(6):1573–1581. 10.1603/0046-225x-37.6.1573 [DOI] [PubMed] [Google Scholar]
- 8.Shalileh S, Ogada PA, Moualeu DP, Poehling HM. Manipulation of Frankliniella occidentalis (Thysanoptera: Thripidae) by Tomato Spotted Wilt Virus (Tospovirus) via the host plant nutrients to enhance its transmission and spread. Environ Entomol. 2016;45(5):1235–1342. 10.1093/ee/nvw102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maris PC, Joosten NN, Goldbach RW, Peters D. Tomato spotted wilt virus infection improves host suitability for its vector Frankliniella occidentalis. Phytopathology. 2004;94(7):706–711. 10.1094/PHYTO.2004.94.7.706 [DOI] [PubMed] [Google Scholar]
- 10.Belén B, Janssen A, Maris PC, Peters D, Sabelis MW. Herbivore arthropods benefit from vectoring plant viruses. Ecol Lett. 2005;8(1):70–79. 10.1111/j.1461-0248.2004.00699.x. [DOI] [Google Scholar]
- 11.Colvin J, Omongo CA, Govindappa MR, Stevenson PC, Maruthi MN, Gibson G, et al. Host-plant viral infection effects on arthropod-vector population growth, development and behaviour: management and epidemiological implications. Adv Virus Res. 2006;67:419–452. 10.1016/S0065-3527(06)67011-5 [DOI] [PubMed] [Google Scholar]
- 12.Costa HS, Brown JK, Byrne DN. Life history traits of the whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) on six virus-infected or healthy plant species. Environ Entomol. 1991;20:1102–1107. 10.1093/ee/20.4.1102. [DOI] [Google Scholar]
- 13.Donaldson JR, Gratton C. Antagonistic effects of soybean viruses on soybean aphid performance. Environ Entomol. 2007;36(4):918–925. 10.1603/0046-225x(2007)36[918:aeosvo]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 14.Khan ZR, Saxena RC. Behavior and biology of Nephotettix virescens (Homoptera: Cicadellidae) on Tungro virus-infected rice plants: epidemiology implications. Environ Entomol. 1985;14:297–304. 10.1093/ee/14.3.297. [DOI] [Google Scholar]
- 15.Mauck KE, De Moraes CM, Mescher MC. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc Natl Acad Sci U S A. 2010;107(8):3600–3605. 10.1073/pnas.0907191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wijkamp I, Almarza N, Goldbach R, Peters D. Distinct levels of specificity in thrips transmission of tospoviruses. Phytopathology. 1995;85(10):1069–1074. 10.1094/Phyto-85-1069. [DOI] [Google Scholar]
- 17.Roselló S, Díez MJ, Nuez F. Viral diseases causing the greatest economic losses to the tomato crop. I. The Tomato spotted wilt virus—a review. Sci Hortic. 1996;67(3–4):117–150. 10.1016/S0304-4238(96)00946-6. [DOI] [Google Scholar]
- 18.Grimstad PR, Ross QE, Craig GB Jr. Aedes triseriatus (Diptera: Culicidae) and La Crosse virus. II. Modification of mosquito feeding behavior by virus infection. J Med Entomol. 1980;17(1):1–7. 10.1093/jmedent/17.1.1 [DOI] [PubMed] [Google Scholar]
- 19.Turell MJ, Gargan TP 2nd, Bailey CL. Culex pipiens (Diptera: Culicidae) morbidity and mortality associated with Rift valley fever virus infection. J Med Entomol. 1985;22(3):332–337. 10.1093/jmedent/22.3.332 [DOI] [PubMed] [Google Scholar]
- 20.Stafford CA, Walker GP, Ullman DE. Infection with a plant virus modifies vector feeding behavior. Proc Natl Acad Sci U S A. 2011;108(23):9350–9355. 10.1073/pnas.1100773108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe H, Shimoda T, Ohnishi J, Kugimiya S, Narusaka M, Seo S, et al. Jasmonate-dependent plant defense restricts thrips performance and preference. BMC Plant Biol. 2009;9:97 10.1186/1471-2229-9-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leastro MO, De Oliveira AS, Pallás V, Sánchez-Navarro JA, Kormelink R, Resende RO. The NSm proteins of phylogenetically related tospoviruses trigger Sw-5b-mediated resistance dissociated of their cell-to-cell movement function. Virus Res. 2017;240:25–34. 10.1016/j.virusres.2017.07.019 [DOI] [PubMed] [Google Scholar]
- 23.Ocampo Ocampo T, Gabriel Peralta SM, Bacheller N, Uiterwaal S, Knapp A, Hennen A, et al. Antiviral RNA silencing suppression activity of Tomato spotted wilt virus NSs protein. Genet Mol Res. 2016;15(2). 10.4238/gmr.15028625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver JE, Whitfield AE. The genus Tospovirus: emerging bunyaviruses that threaten food security. Annu Rev Virol. 2016;3(1):101–124. 10.1146/annurev-virology-100114-055036 [DOI] [PubMed] [Google Scholar]
- 25.Whitfield AE, Ullman DE, German TL. Tospovirus-thrips interactions. Annu Rev Phytopathol. 2005;43:459–489. 10.1146/annurev.phyto.43.040204.140017 [DOI] [PubMed] [Google Scholar]
- 26.Margaria P, Bosco L, Vallino M, Ciuffo M, Mautino GC, Tavella L, et al. The NSs protein of tomato spotted wilt virus is required for persistent infection and transmission by Frankliniella occidentalis. J Virol. 2014;88(10):5788–5802. 10.1128/JVI.00079-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontana A, Held M, Fantaye CA, Turlings TC, Degenhardt J, Gershenzon J. Attractiveness of constitutive and herbivore-induced sesquiterpene blends of maize to the parasitic wasp Cotesia marginiventris (Cresson). J Chem Ecol. 2011;37(6):582–591. 10.1007/s10886-011-9967-7 [DOI] [PubMed] [Google Scholar]
- 28.Frost CJ, Mescher MC, Dervinis C, Davis JM, Carlson JE, De Moraes CM. Priming defense genes and metabolites in hybrid poplar by the green leaf volatile cis-3-hexenyl acetate. New Phytol. 2008;180(3):722–734. 10.1111/j.1469-8137.2008.02599.x [DOI] [PubMed] [Google Scholar]
- 29.Erb M, Veyrat N, Robert CA, Xu H, Frey M, Ton J, et al. Indole is an essential herbivore-induced volatile priming signal in maize. Nat Commun. 2015;16(6):62–73. 10.1038/ncomms7273 pmid: 25683900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauch-Mani B, Baccelli I, Luna E, Flors V. Defense priming: An adaptive part of induced resistance. Annu Rev Plant Biol. 2017;68:485–512. 10.1146/annurev-arplant-042916-041132 [DOI] [PubMed] [Google Scholar]
- 31.Hegde M, Oliveira JN, da Costa JG, Loza-Reyes E, Bleicher E, Santana AE, et al. Aphid antixenosis in cotton is activated by the natural plant defence elicitor cis-jasmone. Phytochemistry. 2012;78(8):81–88. 10.1016/j.phytochem.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 32.De Moraes CM, Mescher MC, Tumlinson JH. Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature. 2001;410(6828):577–580. 10.1038/35069058 [DOI] [PubMed] [Google Scholar]
- 33.Zakir A, Sadek M.M, Bengtsson M, Hansson B.S, Witzgall P, Anderson P, et al. Herbivore‐induced plant volatiles provide associa-tional resistance against an ovipositing herbivore. Journal of Ecology. 2013;101(2):410–417. 10.1111/1365-2745.12041. [DOI] [Google Scholar]
- 34.Sánchez-Hernández C, López MG, Délano-Frier JP. Reduced levels of volatile emissions in jasmonate‐deficient spr2 tomato mutants favour oviposition by insect herbivores. Plant Cell Environ. 2006;29(4):546–557. [DOI] [PubMed] [Google Scholar]
- 35.Huang XZ, Xiao YT, Köllner TG, Jing WX, Kou JF, Chen JY, et al. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 2018;41(1):261–274. 10.1111/pce.13088 [DOI] [PubMed] [Google Scholar]
- 36.Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- 37.Howe GA, Major IT, Koo AJ. Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol. 2018;69:387–415. 10.1146/annurev-arplant-042817-040047 [DOI] [PubMed] [Google Scholar]
- 38.Li R, Weldegergis BT, Li J, Jung C, Qu J, Sun Y, et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell. 2014;26(12):4991–5008. 10.1105/tpc.114.133181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G, Klinkhamer PGL, Escobar-Bravo R, Leiss KA. Type VI glandular trichome density and their derived volatiles are differently induced by jasmonic acid in developing and fully developed tomato leaves: Implications for thrips resistance. Plant Sci. 2018;276:87–98. 10.1016/j.plantsci.2018.08.007 [DOI] [PubMed] [Google Scholar]
- 40.Abe H, Tomitaka Y, Shimoda T, Seo S, Sakurai T, Kugimiya S, et al. Antagonistic plant defense system regulated by phytohormones assists interactions among vector insect, thrips and a tospovirus. Plant Cell Physiol. 2011;53(1):204–212. 10.1093/pcp/pcr173 [DOI] [PubMed] [Google Scholar]
- 41.Fereres A, Peñaflor MF, Favaro CF, Azevedo KE, Landi CH, Maluta NK, et al. Tomato infection by whitefly-transmitted circulative and non-circulative viruses induce contrasting changes in plant volatiles and vector behaviour. Viruses. 2016; 8(8):225–240. 10.3390/v8080225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessler A, Baldwin IT, Defensive function of herbivore-induced plant volatile emissions in nature. Science. 2001;291(5511):2141–2144. 10.1126/science.291.5511.2141 [DOI] [PubMed] [Google Scholar]
- 43.Thomas CL, Jones L, Baulcombe DC, Maule AJ. Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 2001;25(4):417–425. [DOI] [PubMed] [Google Scholar]
- 44.Kazan K, Manners JM. MYC2: the master in action. Mol Plant. 2013; 6(3):686–703. 10.1093/mp/sss128 [DOI] [PubMed] [Google Scholar]
- 45.Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012;24(6):2635–2648. 10.1105/tpc.112.098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111(6):1021–1058. 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao C, Qi S, Liu K, Li D, Jin C, Li Z, et al. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol Biochem. 2016;108:63–70. 10.1016/j.plaphy.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 48.Böhlmann J, Martin D, Oldham NJ, Gershenzon J. Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-β-ocimene synthase. Arch Biochem Biophys. 2000; 375(2):261–269. 10.1006/abbi.1999.1669 [DOI] [PubMed] [Google Scholar]
- 49.Hedil M, Sterken MG, de Ronde D, Lohuis D, Kormelink R. Analysis of tospovirus NSs proteins in suppression of systemic silencing. PLoS One. 2015; 10(8):e0134517 10.1371/journal.pone.0134517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niu Y, Wang D, Cui L, Wang B, Pang X, Yu P. Monoclonal antibody-based colloid gold immunochromatographic strip for the rapid detection of Tomato zonate spot tospovirus. Virol J. 2018;15(1):15 10.1186/s12985-018-0919-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turina M, Kormelink R, Resende RO. Resistance to Tospoviruses in vegetable crops: epidemiological and molecular aspects. Annu Rev Phytopathol. 2016; 54(1):347–371. 10.1146/annurev-phyto-080615-095843 [DOI] [PubMed] [Google Scholar]
- 52.Boachon B, Junker RR, Miesch L, Bassard JE, Höfer R, Caillieaudeaux R, et al. CYP76C1(cytochrome P450)‐mediated linalool metabolism and the formation of volatile and soluble linalool oxides in Arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell, 2015;27(10):2972–2990. 10.1105/tpc.15.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang T, Stoopen G, Thoen M, Wiegers G, Jongsma MA. Chrysanthemum expressing a linalool synthase gene 'smells good', but 'tastes bad' to western flower thrips. Plant Biotechnol J. 2013;11(7):875–882. 10.1111/pbi.12080 [DOI] [PubMed] [Google Scholar]
- 54.De RD, Butterbach P, Lohuis D, Hedil M, van Lent JW, Kormelink R. Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol Plant Pathol. 2013;14(4):405–415. 10.1111/mpp.12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe. 2014;16(3):364–375. 10.1016/j.chom.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abe H, Ohnishi J, Narusaka M, Seo S, Narusaka Y, Tsuda S, et al. Function of jasmonate in response and tolerance of Arabidopsis to thrip feeding. Plant Cell Physiol. 2008;49(1):68–80. 10.1093/pcp/pcm168 [DOI] [PubMed] [Google Scholar]
- 57.Pineda A, Soler R, Weldegergis BT, Shimwela MM, Van Loon JJ, Dicke M. Non-pathogenic rhizobacteria interfere with the attraction of parasitoids to aphid-induced plant volatiles via jasmonic acid signalling. Plant Cell Environ. 2013; 36(2):393–404. 10.1111/j.1365-3040.2012.02581.x [DOI] [PubMed] [Google Scholar]
- 58.Wu D, Qi T, Li WX, Tian H, Gao H, Wang J, et al. Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 2017; 27(3):402–415. 10.1038/cr.2017.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alonzo F 3rd, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, et al. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature. 2013;493(7430):51–55. 10.1038/nature11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381(6584):661–666. 10.1038/381661a0 [DOI] [PubMed] [Google Scholar]
- 61.Golding H, Aliberti J, King LR, Manischewitz J, Andersen J, Valenzuela J, et al. Inhibition of HIV-1 infection by a CCR5-binding cyclophilin from Toxoplasma gondii. Blood. 2003;102(9):3280–3286. 10.1182/blood-2003-04-1096 [DOI] [PubMed] [Google Scholar]
- 62.Aliberti J, Valenzuela JG, Carruthers VB, Hieny S, Andersen J, Charest H, et al. Molecular mimicry of a CCR5 binding-domain in the microbial activation of dendritic cells. Nat Immunol. 2003;4(5):485–490. 10.1038/ni915 [DOI] [PubMed] [Google Scholar]
- 63.Lalani AS, Masters J, Zeng W, Barrett J, Pannu R, Everett H, et al. Use of chemokine receptors by poxviruses. Science. 1999;286(5446):1968–1971. [DOI] [PubMed] [Google Scholar]
- 64.Yang J, Iwasaki M, C, Machida Y, Zhou X, Chua N. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008;22(18):2564–2577. 10.1101/gad.1682208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murai T, Loomans AJ. Evaluation of an improved method for mass-rearing of thrips and a thrips parasitoid. Entomol Exp Appl. 2001;101(3):281–289. 10.1023/A:1019252708938. [DOI] [Google Scholar]
- 66.Mandal B, Csinos AS, Martinezochoa N, Pappu HR. A rapid and efficient inoculation method for Tomato spotted wilt tospovirus. J Virol Methods. 2008; 149(1):195–198. 10.1016/j.jviromet.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 67.Li R, Afsheen S, Xin Z, Han X, Lou Y. OsNPR1 negatively regulates herbivore-induced JA and ethylene signaling and plant resistance to a chewing herbivore in rice. Physiol Plant. 2013;147(3):340–351. 10.1111/j.1399-3054.2012.01666.x [DOI] [PubMed] [Google Scholar]
- 68.Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009;59(1):150–162. 10.1111/j.1365-313X.2009.03850.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Relative expression levels of various TPS genes in N. benthamiana after thrips infestation. Four-week-old N. benthamiana plants were infested with twenty thrips adults in a confined pot for 48h. Total RNA was prepared from treated plants for RT-qPCR analysis. Values are means + SE, n = 3. **P < 0.01, Student’s t-test. (B) Relative expression levels of various TPS genes in N. benthamiana after 100 μM MeJA treatment for 24h. Values are means + SE, n = 3. **P < 0.01, Student’s t-test.
(TIF)
Representative extracted ion chromatograms of GC/MS headspace volatile compounds of peppers. Plants under the same growth condition were infested with (pepper-thrips) or without (pepper) thrips for 6 h.
(TIF)
(A) Interaction between TSWV NSs–AtMYC3 and TSWV NSs–AtMYC4 in a yeast two-hybrid assay. Yeast cotransformed with the indicated plasmids was spotted onto synthetic medium (SD-Leu-Trp-His) containing 0.04 mg/mL X-α-gal and 10mM 3-amino-1,2,4-triazole (3-AT). The empty vectors pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. (B) Interaction between TSWV NSs–AtMYC3 and TSWV NSs–AtMYC4 in a BiFC assay. Indicated construsts were transiently expressed in H2B-RFP transgenic N. benthamiana leaf epidermal cells by agroinfiltration. Bars = 15 μm.
(TIF)
ClustalW was used to construct the phylogenetic tree. It was constructed based on the amino acid sequences of the NSs protein from 23 orthotospoviruses.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.