Abstract
Mitogen-activated protein kinase (MAPK) cascades play central roles in response to biotic and abiotic stresses. However, the mechanisms by which various MAPK members regulate the plant immune response in rice remain elusive. In this article, to characterize the mechanisms, the knock-out and overexpression mutants of OsMPK15 were constructed and the disease resistance was investigated under the various fungal and bacterial inoculations. The knock-out mutant of OsMPK15 resulted in the constitutive expression of pathogenesis-related (PR) genes, increased accumulation of reactive oxygen species (ROS) triggered by the pathogen-associated molecular pattern (PAMP) elicitor chitin, and significantly enhanced the disease resistance to different races of Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae (Xoo), which cause the rice blast and bacterial blight diseases, respectively. On contrary, the expression of PR genes and ROS were down-regulated in the OsMPK15-overexpressing (OsMPK15-OE) lines. Meanwhile, phytohormones such as salicylic acid (SA) and jasmonic acid (JA) were accumulated in the mpk15 mutant lines but decreased in the OsMPK15-OE lines. The expression of SA- and JA-pathway associated genes were significantly upregulated in the mpk15 mutant, whereas it was down regulated in the OsMPK15-OE lines. We conclude that OsMPK15 may negatively regulate the disease resistance through modulating SA- and JA-mediated signaling pathway.
Keywords: OsMPK15, PRs, SA/JA, ROS, M. oryzae, Xoo, rice
Introduction
The plant has evolved two-layered immune systems to fight off the pathogen attacks, including pattern-recognition receptors (PRRs) triggered immunity (PTI) and specific pathogen-derived effectors triggered immunity (ETI) that is activated by recognition of cytoplasmic resistance proteins (Monaghan and Zipfel, 2012). Perception of extracellular signals, transmission into a cell, and the subsequent activation of defense response via phosphorylation play a central role in defense signaling (Yang et al., 1997). Among them, the mitogen-activated protein kinase (MAPK) cascade is one of the well-characterized and conserved signaling pathways that play vital importance in plant growth and development as well as abiotic and biotic stress response (Hirt, 1997; Yang et al., 1997; Zhang and Klessig, 2001).
Plant MAPK cascade signaling modules consist of three functionally intertwined protein kinases including MAPK kinase kinase (MPKKK), MAPK kinase (MPKK), and MAPK. The basic process is that MPKKK phosphorylate and activate MPKKs, which in turn phosphorylate and activate MAPKs. The activated MAPKs are commonly imported into nucleus, and further interact with specific downstream components such as transcription factors (Tena et al., 2001; Asai et al., 2002; Koo et al., 2009; Jalmi and Sinha, 2016). Until now, at least 17 rice MAPKs have been identified according to in silico search of rice genome database but still a large number of members remain yet to be characterized (Reyna and Yang, 2006).
With increase in number of MAPKs characterized, rice, as a monocotyledonous model plant, is subjected to study the roles of MAPKs in signaling pathway. The first characterized rice MAPK is OsBWMK1 (OsMPK12), which was induced by Magnaporthe oryzae and mechanical wounding. OsBWMK1 protein contains TDY motif and has a kinase activity that mediates salicylic acid (SA)-dependent defense responses by phosphorylating and activating the target transcription factor OsWRKY33 and OsEREBP1, resulting in high levels of pathogenesis-related (PR) transcript (He et al., 1999; Cheong et al., 2003; Koo et al., 2009). Furthermore, overexpression of OsMAPK12-1 (a transcript of OsBWMK1) inhibited seed germination and seedling growth, and enhanced disease resistance to Xoo. PXO099 and Xanthomonas oryzae pv. oryzicola (Xoc) RS105; whereas the knockdown lines displayed the opposite phenotype (Xiao et al., 2017). Transcript of OsBIMK1 will be accumulated upon treatment with benzothiadiazole (BTH) and jasmonic acid (JA), mechanical wounding, or rice-M. oryzae interaction (Song and Goodman, 2002). Suppression of abscisic acid (ABA) and pathogen infection induced by OsMAPK5 may result in a constitutive expression of PR genes such as PR1 and PR10, indicating that OsMAPK5 negatively regulates fungal and bacterial disease resistance (Xiong and Yang, 2003; Xie et al., 2014; Bertini et al., 2018). OsMAPK6 was characterized to be post-translationally activated in a cell culture by a sphingolipid elicitor and regulated by the small GTPase OsRac1, heterotrimeric G-protein, and OsMPKK10.2. OsMPKK10.2 phosphorylating and activating OsMAPK3/6 in vitro, resulting in enhanced resistance to both Xoc resistance and drought tolerance (Ma et al., 2017). By contrast, knockdown (RNAi) of OsMAPK6 resulted in a reduction of OsPAL mRNA transcript (Liebertherr et al., 2005; Yuan et al., 2007). OsMAPK6 was also essential for the chitin elicitor-induced biosynthesis of diterpenoid phytoalexins from glycolysis to secondary metabolite biosynthesis (Kishi-Kaboshi et al., 2010). Recently, group C member OsMPK7 is regulated by OsMKK3, which interacts with and phosphorylates the target protein OsWRKY33, mediating the resistance against Xoo via up-regulation of PR genes, cell wall structure maintenance, and cell metabolism (Jalmi and Sinha, 2016).
In addition to its important role in biotic stresses, plant MAPK cascade is also involved in grain size through control cell division (Bent, 2001). Map-based cloning suggested that rice small grain 1 (smg1) mutant that encodes OsMKK4, could promote grain size by influencing cell proliferation (Duan et al., 2014). Other examples include the dwarf and small grain 1 (dsg1), which exhibits small grains, dwarfism, and erect leaves. DSG1 encodes a MAPK 6 (OsMAPK6), plants of which have larger grain and sparser panicles than the wild type (WT) plants due to cell differentiation and proliferation. OsMAPK6 strongly interacts with OsMKK4, which promotes cell proliferation and grain size via brassinoseroids (BR) signaling pathway (Liu et al., 2015). GSN1 encodes the MAPK phosphatase OsMKP1, which interacts with and inactivates the MAPK OsMAPK6 via dephosphorylation. Meanwhile, the suppression of OsMAPK6, OsMKK4, and OsMKKK10 resulted in smaller grains and dense panicles (Guo et al., 2018).
Although many studies on the MAPK cascades in immune response have been conducted in Arabidopsis and rice, those work are mainly focused on the TEX motif members such as the OsMAPK3/6, OsMPK4, and OsMPK5 mediated signaling pathway. There are 17 identified MAPKs members in rice genome, of which 11 members contain the TDY phosphorylation site and six members have the TEY motif. They could be divided into six groups by phylogenetic analysis, of which OsMPK15 belongs to group F (TDY type) with a length of 498 amino acid. Previous studies found that the transcript of OsMPK15 was suppressed by virulent blast race earlier than 24 h while it was accumulated 48 h after inoculated with the avirulent M. oryzae race (Reyna and Yang, 2006). Little is known about the mechanisms of OsMPK15 in the defense response against M. oryzae and Xoo. In our study, the Crispr/Cas9-edited mutagenesis and overexpression approaches were performed to characterize the function of OsMPK15 in disease resistance. We found that OsMPK15 acted as a negative regulator in defense response to the M. oryzae and Xoo pathogens. We also found that the knock-out of OsMPK15 significantly increased grain length.
Materials and Methods
Plant Materials Generation and Growth Conditions
A mpk15 mutant was generated using Crispr/Cas9 editing system by Biogle company (Hangzhou, China). The target sequence of OsMPK15 (LOC_Os11g17080) was designed TTCCTCTATCAGTTGCTTCGAGG. The mpk15 mutant lines were homozygous through sequence confirmation. The full length ORF of OsMPK15 was cloned into a modified pCAMBIA 1301 vector under the control of maize ubiquitin promoter, yielding the pCAMBIA1301-Ubi::OsMPK15 plasmid. The OsMPK15-overexpressing (OsMPK15-OE) lines were generated by introducing the yielded plasmid to the rice cultivar ZH11. Transgenic rice plants were grown in a controlled field under natural conditions.
Pathogen Inoculation
To evaluate the rice blast disease resistance at the seedling stage, we used punch inoculation on 30-day-old seedling rice leaves (Park et al., 2012). M. oryzae isolates were grown on oat agar medium for 10 days. Spores were collected and the concentration was adjusted to 105 conidia /ml supplemented with 0.02% tween-20 before inoculation. Punch inoculation of detached leaves was carried out as follows: dip 5 μl spore suspension for each drop using pipette tip at three spots on each leaf. The inoculated leaves were kept in the dark for 24 h at 25°C with 100% humidity and then moved to the growth chamber under the 25°C, 12 h light/12 h dark cycle normal condition. Lesion length was measured at 6 days post-inoculation (dpi). Relative quantification of fungal DNA amount was calculated using qRT-PCR method by assessing the genomic DNA level of a M. oryzae 28S rDNA and relative fungal growth was presented as ratios obtained by comparison of the genomics fungal 28S rDNA levels with a rice OsEF1 genomic DNA levels according to the previous report (Hong et al., 2017).
To evaluate resistance of rice bacterial blight disease, Philippine Xoo strain PXO96, and Chinese Xoo strain Zhe817 were grown in NA media. Rice plants at the booting stage were inoculated using the leaf-clipping method (Sun et al., 2004; Hong et al., 2017) and the inoculated plants were moved to a greenhouse under controlled conditions at 30°C in daytime and 25°C in the night and maintained suitable humidity. Disease phenotype was recorded by measuring the lesion length at 15 dpi.
Measurement of ROS
Leaf disks from 3-month-old plants were cut and preincubated in sterile distilled water overnight. ROS generation after chitin treatment was measured using the luminol chemiluminescence assay (Park et al., 2012). Briefly, three-leaf disks per sample were placed in a 1.5 ml microcentrifuge tube containing 100 μl luminol (L-012 from Wako Pure Chemical Corporation instead, CAS. No. 143556-24-5), 1.0 μl horseradish peroxidase (Sigma), and 1.0 μl elicitor (800 nM Chitin, water as a mock control) and the luminescence readout was recorded every 10 s for 20 min in Glomax 20/20 Luminometer (Promega). At least three biological replications were performed for each sample.
SA and JA Quantification
Quantification of SA and JA was carried out according to the method described previously (Fu et al., 2012; Hong et al., 2017). Quantification of SA and JA was performed by an HPLC-MS system (Model 1290/6460, Agilent) with stable isotope-labeled SA and JA as an internal standard. Each measurement was replicated three times.
Gene Expression Analysis by Real-Time PCR
Total RNA of the wild-type, mpk15 mutant, and OsMPK15-OE lines at the same developmental stage were extracted using the TRIzol reagent (Invitrogen, Shanghai, China). First-strand cDNA was synthesized using PrimeScript RT reagent Kit with gDNA eraser (TaKaRa, Dalian, China) according to the manufacturer’s instructions. 0.5 μl cDNA was used for a template of qRT-PCR with a LightCycler 480 II real-time PCR system (Roche, United States). The OsActin gene (LOC_Os03g50885) was used as an internal control. Gene-specific primers are listed in Supplementary Table S1. Three independent biological and technical replicates were run for each expression assessment.
Protein-Protein Interaction Prediction
Protein-protein interaction could be mined from the online resources such as STRING 10.5 database1. Briefly, the full amino acid sequence and organism of OsMPK15 were submitted for a query, and the predicted computationally scored protein and protein interaction network could be obtained.
Evaluation of Agronomic Traits
Plant height and tiller number for different rice lines were examined under the field conditions. Harvested grains were air-dried and stored at room temperature for 1 month. Twenty random main panicles from each line were chosen for measuring the panicle weight, the total number of grains per panicle, seed setting rate, and grain yield per plant using the conventional methods. 1,000-grain weight and grain length and width from the WT, mpk15 mutant, OsMPK15-OE plants were calculated using an automatic seed analysis system (WanShen SC-G, Hangzhou, China). The measurements were repeated at least three times and the repeated data were averaged for analysis.
Statistical Analysis
The two-sample student’s t-test was used to compare the difference in mean and a p-value of ≤ 0.05 was considered significant.
Results
Generation of CRISPR/Cas9 Edited mpk15 Mutant and OsMPK15-OE Lines
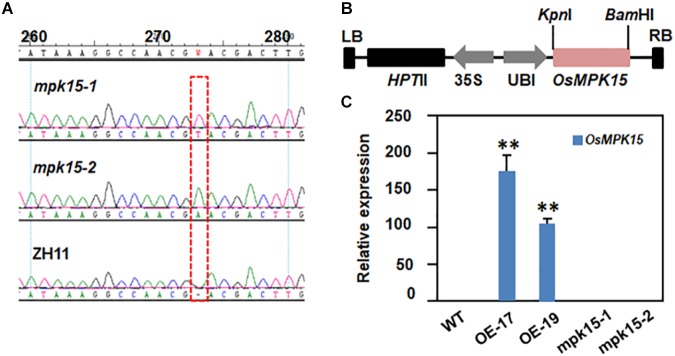
To better understand the molecular mechanism of rice MAPK family underlying plant-microbe interaction and immune response, we explored the expression pattern of OsMPK15 in response to M. oryzae infection and phytohormone treatment. Since previous studies have pointed to the conclusions that the transcript of OsMPK15 was suppressed by virulent blast race earlier than 24 h while it was accumulated 48 h after inoculated with the avirulent race (Reyna and Yang, 2006) and that the transcript of OsMPK15 was also accumulated by JA treatment (Reyna and Yang, 2006), we therefore hypothesized that OsMPK15 may be involved in the disease resistance. To test this hypothesis, we generated both the mpk15 mutant and OsMPK15-OE lines using the reverse-genetic approach. Sequencing analysis demonstrated that the two mpk15 mutant lines were homozygous, suitable for the proposed investigation, which inserted “A” or “T” resulting in a shift mutation, respectively (Figure 1A). For overexpression of OsMPK15, the full coding sequence of OsMPK15 was constructed into a modified binary vector under the control of maize ubiquitin promoter (Figure 1B) and transformed into the rice cultivar ZH11. We selected two over-expressing lines, the transcript of OsMPK15 in OE-17 and OE-19 was 173.1 and 104.5 times higher than the WT by qRT-PCR (Figure 1C). During our study, we did not observe any significant changes in morphological and developmental phenotypes between the OsMPK15-OE and WT plants at the seedling and adult stages (Figure 2A). On the flag, top second, and top third leaves, we found that slightly lesion mimic was found in the WT while there was no lesion mimic in the mpk15 mutant under the field condition. However, increased susceptibility to bacterial blight was observed in the top third leaf of OsMPK15-OE line (Figure 2B).
FIGURE 1.
Generation of Crispr/Cas9 edited mpk15 mutant and OsMPK15-OE lines. (A) Sequence confirmation of the homogenous mpk15-1 and mpk15-2 mutant lines., (B) Schematic diagram of the overexpression construct for rice transformation. HPT II, Hygromycin phosphotransferase II; LB, left border; RB, right border; Ubi, maize ubiquitin promoter; 35S, CaMV 35S promoter. (C) OsMPK15 expression levels in the WT, mpk15 mutant, and OsMPK15-OE lines at 30-days-old seedling. Data presented are the means ± SD from three independent experiments. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
FIGURE 2.
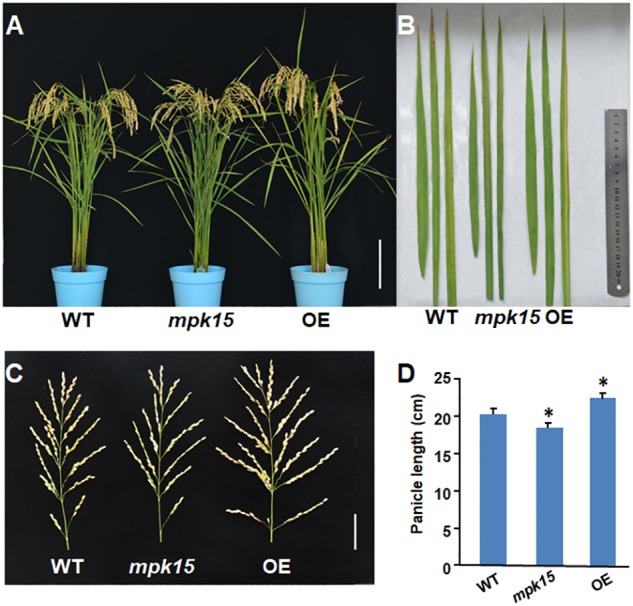
Phenotypic comparison of the WT, mpk15 mutant, and OsMPK15-OE lines. (A) The phenotype of the WT, mpk15 mutant, and OsMPK15-OE lines at 102 days post-sowing (dps). Bars indicate a length of 15 cm. (B) Magnified view of the flag leaf, top second, and top third leaves in the WT, mpk15 mutant, and OsMPK15-OE line at 102 dps. Among them, the top third of OsMPK15-OE has obvious bacterial blight lesion. (C,D) panicle phenotype and length in the WT, mpk15 mutant, and OsMPK15-OE lines. Bars indicate 5 cm. Data presented are the mean ± SD from three independent experiments. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
The Role of OsMPK15 in Disease Resistance to M. oryzae
To investigate the role of OsMPK15 in rice innate immunity, we first examined whether OsMPK15 is responsible for resistance to rice blast at the seedling stage. Two M. oryzae compatible strains, 46-2 and RB22, were used to inoculate the 30-days-old seedling of mpk15 mutant, OsMPK15-OE and non-transgenic WT seedlings by the punch inoculation and disease levels were evaluated by the lesion diameter and the in planta fungal growth. As shown in Figure 3A,B, the overall blast disease phenotype on the OsMPK15-OE lines OE-17 and OE-19 was more severe than those in the non-transgenic WT plants after inoculation with strains 46-2 and RB22. Typical blast lesions were seen on leaves of OsMPK15-OE and non-transgenic WT plants, whereas almost no typical blast lesion was observed on the leaves of mpk15 mutant lines (Figure 3A,B). At 6 dpi of M. oryzae, the average blast lesion sizes on the inoculated leaves of OsMPK15-OE plants were increased by 2.14 to 2.41 folds while the lesion size on the inoculated leaves of the mpk15 mutant plants were decreased by 60.5 to 68.9%, as compared with that in the WT plants (Figure 3D). Next the in planta fungal growth, measured by the genomic DNA level of the 28S rDNA gene of M. oryzae, indicated that OsMPK15-OE plants had more growth of M. oryzae in the inoculated leaves, increased by 7.60 to 9.92-folds, whereas the mpk15 mutant plants decreased by 81.71 to 89.15%, as compared with that in the WT plants (Figure 3E). Taken together, these results demonstrated that the mpk15 mutant plants exhibited an increased resistance while OsMPK15-OE displayed increased susceptibility to M. oryzae.
FIGURE 3.
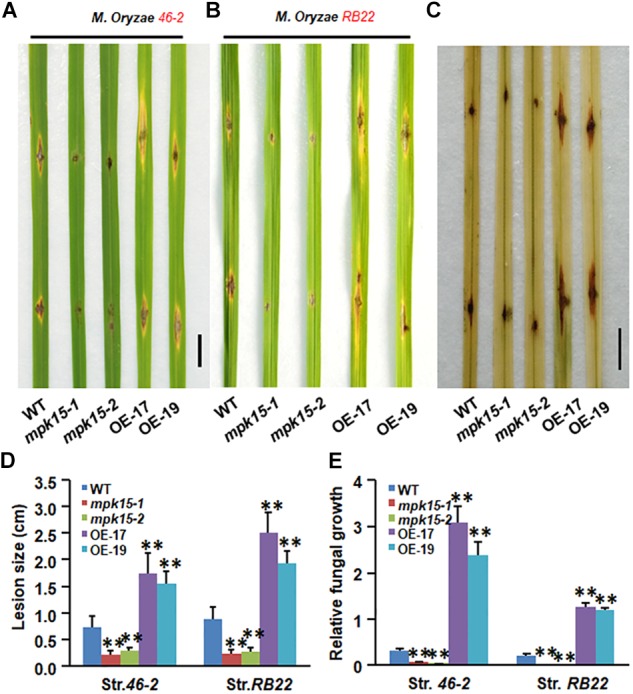
OsMPK15 negatively regulates M. oryzae disease resistance in detached leaves. (A,B) Phenotype of the at 30-days-old WT, mpk15 mutant, and OsMPK15-OE plants inoculated by the punch method with spore suspension (1 × 105 per ml) of M. oryzae strain 46-2 or race RB22 at 6 dpi. Bars indicate 1 cm. (C) DAB staining in M. oryzae strain RB22 inoculated leaves of WT, mpk15 mutant, and OsMPK15-OE plants at 6 dpi. Bars indicate 1 cm. (D) Disease lesion size statistics of M. oryzae strain 46-2 inoculated WT, mpk15 mutant, and OsMPK15-OE plants. At least 30 plants in each experiment were evaluated for lesion size with three replicates. (E) Quantification of M. oryzae growth in strain 46-2 and RB22-inoculated leaves of the WT, mpk15 mutant, and OsMPK15-OE plants at 6 dpi. Amounts of M. oryzae 28S rDNA and rice OsEF1 genomic DNA were estimated by qRT-PCR and relative fungal growth were shown as ratios of Mo28S/OsEF1. Three independent experiments were performed with similar results in (A–E) Data presented in (D,E) are the mean ± SD from three independent experiments. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
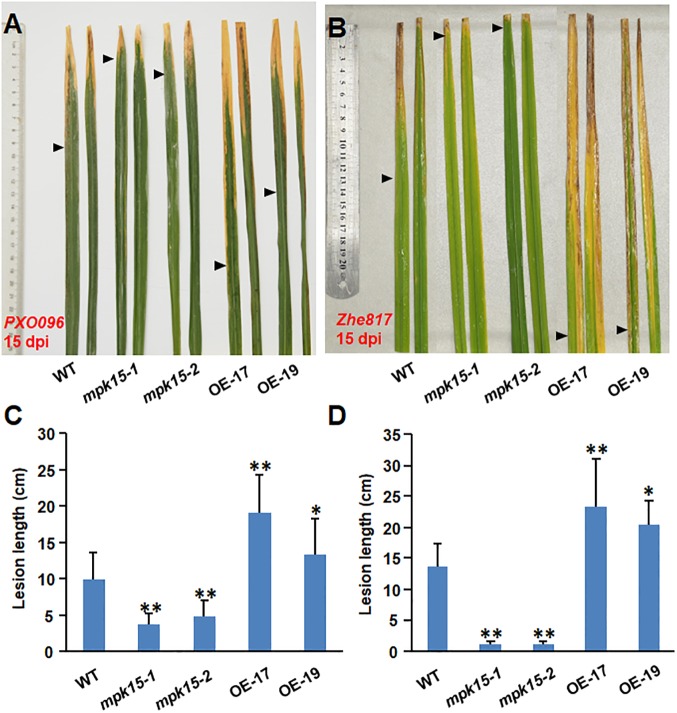
OsMPK15 Negatively Regulates Resistance to X. oryzae pv. oryzae
Rice bacterial leaf blight, caused by Xoo, is also one of the most devastating diseases threating rice production. We then evaluated the resistance of the OsMPK15-OE and mpk15 mutant lines against Xoo by leaf-clipping inoculating of adult plants at 3-month-old plants with two Xoo strains, PXO96 and Zhe817, and the disease severity was evaluated by measuring lesion length at 15 dpi. As shown in Figure 4A,B, the overall Xoo-caused blight disease on the mpk15 mutant lines was less severe while the OsMPK15-OE plants were more severe, as compared with those in WT plants. The average length of the blight lesions on the PXO96 strain inoculated flag leaves of the mpk15 mutant plants were 3.68 and 4.81 cm, representing the reductions of 51.47 and 62.82%, while those of the OsMPK15-OE plants were 18.98 and 13.32 cm, indicating the increases of 34.45 and 91.58%, as compared with that (9.90 cm) in the WT plants (Figure 4C). The similar, even more evident, a pattern was observed in Figure 4D, as inoculated with Zhe817 strain. These results indicate that the mpk15 mutant lines exhibited an increased resistance while the OsMPK15-OE lines displayed increased susceptibility to Xoo.
FIGURE 4.
OsMPK15 negatively regulates the bacterial disease resistance to X. oryzae pv. oryzae. The WT, mpk15 mutant, and OsMPK15-OE plants were inoculated with Xoo strains PXO96 and Zhe817 using the leaf clipping method at the 90-days-old (booting stage). (A,B) Disease symptom on the PXO96 and Zhe817-inoculated leaves at 15 dpi. Rulers were indicated on the left. (C,D) Lesion length on the inoculated leaves at 15 dpi. At least 12 plants in each experiment were used for measurement of the lesion length. Data presented in (C,D) are the mean ± SD from three independent experiments. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
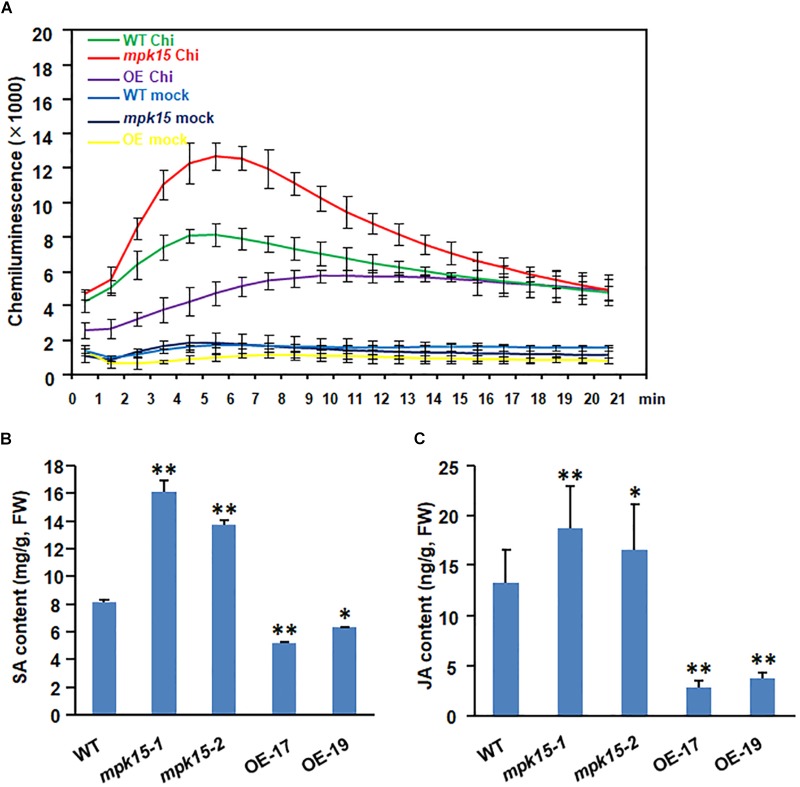
Chitin-Induced ROS Accumulation in the mpk15 Mutant Lines
To investigate whether OsMPK15 can suppress the plant PTI response, we used chemical luminescence to monitor the dynamics of ROS bursts in the mpk15 mutant, OsMPK15-OE, and WT plants treated with chitin, which is recognized by OsCEBiP/OsCERK1 in rice (Kaku et al., 2006; Shimizu et al., 2010; Park et al., 2012). The data revealed that the ROS accumulation in the mpk15 mutant lines was significantly higher than that in WT in response to chitin treatments while the accumulation in the OsMPK15-OE lines was significantly lower than that in WT. The peak ROS level occurred about 5 and 6 min after the chitin treatment (Figure 5A). The chitin treated ROS accumulation in the mpk15 mutant lines was 1.56 times greater than that in WT, whereas the OsMPK15-OE lines had only 58.3% of the WT plants at the peak time (Figure 5A). In contrast, the water controls of different lines all were kept the basal ROS level. This finding was also confirmed by quantification of the endogenous H2O2 using 3,-3′-diaminobenzidine (DAB) staining (Figure 3C). These results suggested that the chitin-triggered ROS accumulation is suppressed in OsMPK15-OE lines and increases in the mpk15 mutant lines.
FIGURE 5.
Increase in the ROS accumulation and disease-related hormone levels in the mpk15 mutants. (A) ROS accumulation dynamics in the WT, mpk15 mutant, and OsMPK15-OE plants at 90 dps after treatment of chitin and water (mock) treatment served as a control. Error bars are computed from three replicates (n = 3). (B,C) The 30-day-old seedling leaf samples of the WT, mpk15 mutant and OsMPK15-OE lines under normal conditions were used for analyses of SA and JA contents. FW: fresh weight. Data presented are the mean ± SD from three independent experiments. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
SA and JA Accumulation in the mpk15 Mutant Lines
Salicylic acid and jasmonic acid are the most important plant hormones that play vital roles in regulating immune responses (Qiu et al., 2007; Koo et al., 2009; Pieterse et al., 2012; Yang et al., 2012; Yang et al., 2013; Liu et al., 2016). To further investigate whether these pathways are involved in the OsMPK15-mediated defense signaling pathway, we then checked the endogenous SA and JA levels in the 4-week-old OsMPK15-OE, mpk15 mutant, and WT plants under normal conditions. The SA levels in the mpk15 lines were 1.69 to 1.98-folds higher than those in the non-transgenic WT plants while the OsMPK15-OE lines had reduced 36.69 and 22.06% as compared with the WT plants, respectively (Figure 5B). Similarly, the endogenous JA levels in the mpk15 mutant lines were increased by 41.47and 24.06% while the OsMPK15-OE lines were reduced by 78.87 and 71.91% as compared with WT plants, respectively (Figure 5C). These results suggested that the SA and JA accumulation was reduced in the OsMPK15-OE lines and increased in the mpk15 lines under normal conditions.
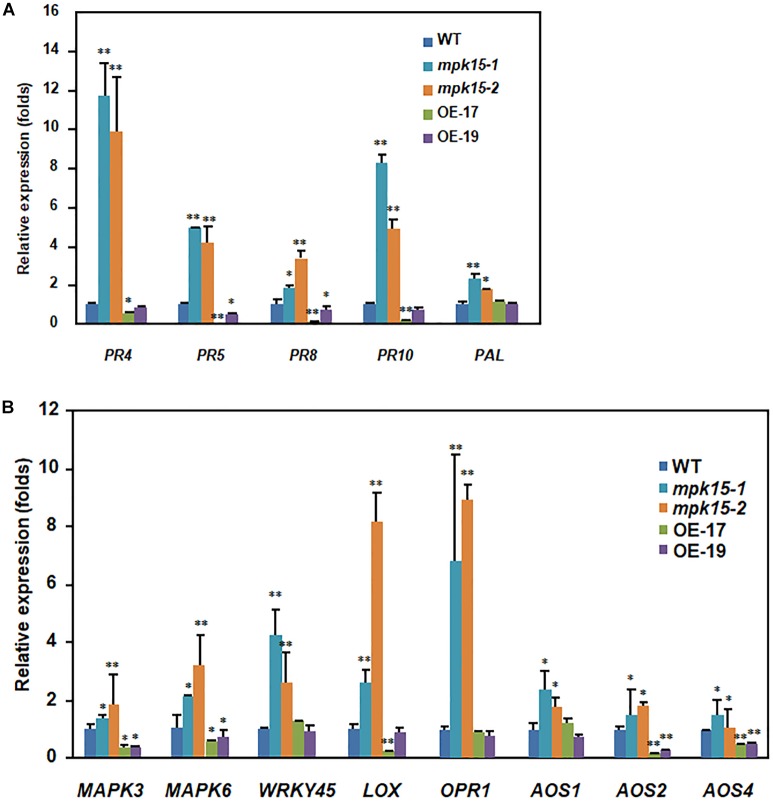
Improved resistance to M. oryzae and Xoo, increased the accumulation of chitin-triggered ROS, and elevated SA and JA levels in the mpk15 mutant lines motivated us to examine whether the mpk15 mutant alters the expression of defense genes. We analyzed the expression patterns of PR4, PR5, PR8, PR10, and PAL in the 4-week-old mpk15 mutant, OsMPK15-OE, and WT plants under normal conditions. As shown in Figure 6A, the expression levels of PR4, PR5, PR8, PR10, and PAL, of which PAL is a SA biosynthesis gene, were markedly upregulated in the mpk15 mutant lines, showing increases of 9.85, 4.15, 3.41, 4.94, and 2.23-folds as compared with the WT plants while keeping at a very lower levels (8.9 to 89% of WT) in the OsMPK15-OE lines. In addition, we also investigated the expression of MAPK central genes including MAPK3 and MAPK6, the SA signaling marker gene WRKY45 (Shimomo et al., 2007), and the JA biosynthesis genes LOX, OPR1, AOS1, AOS2, and AOS4 (Lee et al., 2001; Feussner and Wasternack, 2002). The MAPK3, MAPK6, and WRKY45 were significantly up-regulated in the mpk15 mutant lines and suppressed in the OsMPK15-OE lines (Figure 6B). In the mpk15 mutant lines, the JA biosynthesis genes including LOX and OPR1 were markedly up-regulated, showing increases of 2.61 and 6.78-folds higher than those in the WT plants while the other JA biosynthesis genes such as AOS1, AOS2, and AOS4 were slightly upregulated, showing increases of 1.46 to 2.36 times higher than those of the WT plants; whereas they were suppressed the in OsMPK15-OE lines (Figure 6B), in good agreement with the elevated SA and JA content in the mpk15 mutant as shown in Figure 5C,D. These results demonstrated OsMPK15 might play a negative role in the regulation of SA- and JA-signaling pathway against M. oryzae and Xoo.
FIGURE 6.
Expression pattern of defense signaling genes in the WT, mpk15 mutant, and OsMPK15-OE lines. The 30-day-old seedlings samples of different lines under normal conditions were used for qRT-PCR analyses. (A) The expression pattern of pathogenesis-related (PR) genes includes PR4, PR5, PR8, PR10, and PAL. (B) Expression pattern of the MPK3, MPK6, SA-mediated signaling marker gene WRKY45, and JA biosynthesis genes include LOX, OPR1, AOS1, AOS2, and AOS4. Data presented are the mean ± SD from three independent experiments. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
Protein-Protein Interaction Prediction of OsMPK15
To further search the upstream regulator of OsMPK15 and the downstream substrate with M. oryzae and Xoo interaction, we used STRING 10.5 database2 for screening possible protein-protein interaction. Bioinformatics prediction suggested that there were 10 proteins to interact with the OsMPK15 protein (Supplementary Figure S1 and Supplementary Table S2). One protein named OsMPKK5 (LOC_Os06g09190) may be the upstream regulator of OsMPK15; the other five members were protein phosphatase. We also speculate that PP2C or protein phosphatase may interact with the OsMPK15 protein to dephosphorylate The putative role of MAPK to suppress the defense response (Schweighofer et al., 2007; Sidonskaya et al., 2016), needs to be further confirmed by the yeast two-hybrid or Co-IP methods in the follow-up study.
Decreased Grain Yield and Increased Grain Length in the mpk15 Mutant
Interestingly, in addition to the involvement in disease resistance, we also compared the agronomic traits between the mpk15 mutant, OsMPK15-OE, and WT plants. The plant height of the OsMPK15-OE lines was higher than that in WT (Figure 2A, 7A). Furthermore, the grain number per panicle and panicle length of the OsMPK15-OE line were more than those of WT while the mpk15 mutant had less grain than WT (Figure 2C,D). We speculated that enhanced disease resistance in the mpk15 mutant might consume much more energy, leading to less biomass and grain number panicle.
FIGURE 7.
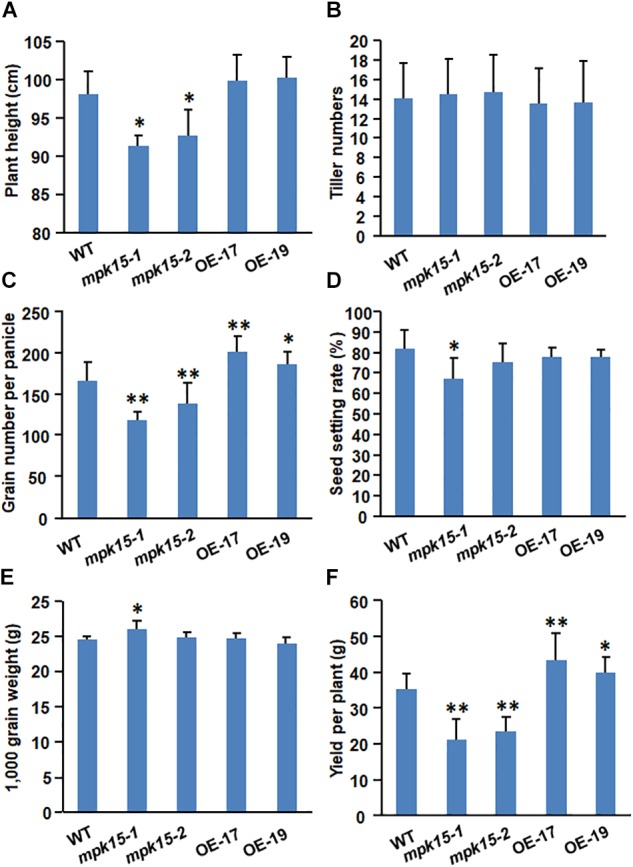
Agronomic phenotypes of the WT, mpk15 mutant, and OsMPK15-OE lines. (A,B) Comparison of plant height and tiller numbers among the WT, mpk15 mutant, and OsMPK15-OE plants (n = 10). (C–F) Comparison of grain number per panicle, seed setting rate, 1,000-grain weight, and yield per plant among the WT, mpk15 mutant, and OsMPK15-OE lines (n = 10). Data presented are the mean ± SD from 2 years of independent experiments with a similar result. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
Compared with the wild-type plants, the mpk15 mutant lines displayed a marked decrease of grain number per panicle and seed setting rate and increased 1,000-grain weight, respectively; whereas, the OsMPK15-OE lines significantly increased the grain number per panicle (Figure 7C). The tiller numbers, seed setting rate, and 1,000-grain weight were not significantly different between OsMPK15-OE and WT (Figure 7B,D,E). Our field assessments of grain yield per plant showed that the OsMPK15-OE lines had an increased grain yield per plant of 14.02 and 23.23% compared with the wild-type plant whereas the mpk15 mutant lines were only 60.2 to 66.93% of the WT plants, respectively (Figure 7F). In addition, the knock-out of OsMPK15 produces larger seeds while no significant differences were observed in grain length, width, and seedling development between the OsMPK15-OE and WT plants (Figure 8A–D). These results demonstrated that OsMPK15 may positively regulate grain yield production by sacrificing disease resistance.
FIGURE 8.
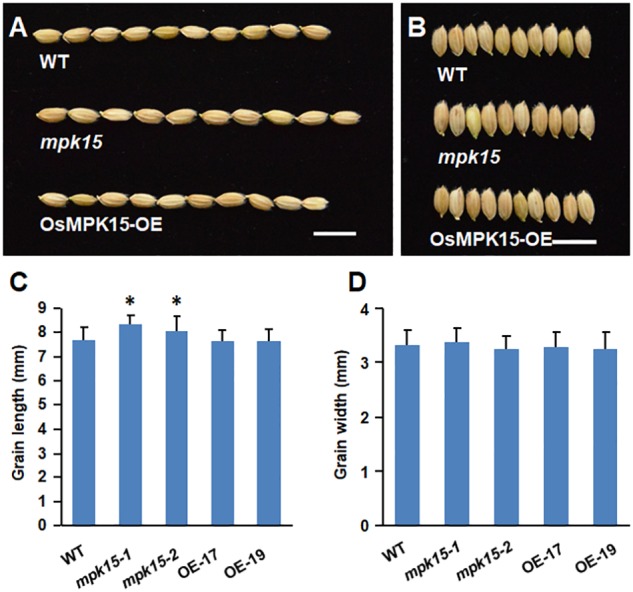
Grain length and width of the WT, mpk15 mutant, and OsMPK15-OE lines. (A,B) The phenotype of grain length and width from the WT, mpk15 mutant, and OsMPK15-OE lines. Bars indicate 10 mm. (C,D) Statistics of grain length and width (n = 300), respectively. ∗P ≤ 0.05; ∗∗P ≤ 0.01 (Student’s t-test).
Discussion
Rice durable and broad-spectrum disease resistance is a trait of agronomic importance for stable high yield variety. Rice MAPK cascade is one of the evolutionarily conserved signaling pathways in regulating defense response (Zhang and Klessig, 2001). Although substantial progress has been made, however, most of the rice MAPKs studies were focused on the “TEY” motif group such as OsMAPK3/6, whereas the “TDY” group such as group E remains uncharacterized. In the present study, we generated the mpk15 mutants and OsMPK15-OE lines and further provided the evidence of group E member of MAPK, OsMPK15 negatively regulating M. oryzae and Xoo disease resistance.
OsMPK15 Is a Switch in Response to M. oryzae and Xoo Infection
Knock-out of OsMPK15 lines enhanced disease resistance to different races of M. oryzae and Xoo while OsMPK15-OE lines significantly increased susceptibility as compared to WT plants (Figure 3, 4). We also found that the transcript of PR genes, OsMAPK3/6 and SA/JA signaling pathway associated genes were elevated in the mpk15 lines, while those were suppressed significantly in the OsMPK15-OE lines (Figure 6), leading to a relatively high level of SA and JA in the mpk15 lines (Figure 5C,D), implying that OsMPK15 negatively regulates disease resistance. These results demonstrate that OsMPK15 functions as an inhibitor to suppress the OsMAPK3/6 cascade signaling pathway in the normal growth condition, which is essential to activate the defense response (Liebertherr et al., 2005; Kishi-Kaboshi et al., 2010; Ma et al., 2017). Allocation of the resources and energy from growth and development to defense response in case of pathogen infection may cause less biomass and low yield (Figure 7F).
Salicylic acid and JA are important hormones in response to biotic stress response. Activation of ETI and systemic acquired resistance often induces the biosynthesis of SA (Century et al., 1997; Shapiro and Zhang, 2001). SA/BTH-induced OsWRKY45 was regulated by the MAPK-dependent phosphorylation in one of the two branches of the rice SA pathway (Nakayama et al., 2013; Ueno et al., 2013). The involvement of defense responses of the mpk15 mutant is partially supported by the up-regulation of PRs such as OsPR10 (Figure 6A), which was modulated by the SA signaling pathway (Mitsuhara et al., 2008), indicating that the knock-out of OsMPK15 could activate the SA signaling pathway, which is in agreement with the previous report that AtMPK4 and OsMAPK5 negatively modulate the PR gene expression and broad-spectrum resistance (Petersen et al., 2000; Xiong and Yang, 2003). We also observed a high level of JA in the mpk15 mutant accompanied by an increased expression of the JA biosynthesis genes such as LOX, OPR1, and AOS 1/2/4 (Figure 6B). In addition to induced expression of PR genes such as OsPR5 (Figure 6A), the knock-out of OsMPK15 could activate JA signaling pathway by upregulating the expression of JA biosynthesis and signaling genes (Mitsuhara et al., 2008). Generally, a high level of JA may repress vegetative growth and affect male fertility. The relatively lower seed setting rate (Figure 7D) in the mpk15 lines may be attributed to a high level of JA, which is consistent with the previous report (Hou et al., 2010; Yang et al., 2012).
OsMPK15 Positively Regulates Yield
There is a general consensus that improved plant disease resistance is usually accompanied by a decrease in growth and yield (Ning et al., 2017). Together with the increased broad-spectrum resistance in the mpk15 lines, the yield per plant was dramatically reduced compared with the WT plants (Figure 7F). These results indicate that the mpk15 lines will allocate the energy into defense against different pathogens, resulting in low yield and biomass. By contrast, the OsMPK15-OE plants will transfer more flow from defense response to growth and development. These results suggest that the increased broad-spectrum disease resistance in the mpk15 mutant came at the penalty of yield losses, which is supported by the previous reports (Jorgensen, 1992; Brown, 2002; Sharp et al., 2002).
We also found that the grain length was dramatically increased and reduced in the mpk15 and OsMPK15-OE lines, respectively (Figure 8C). In the previous study, activation of OsMAPK6 and its upstream regulator OsMKK4 positively influence rice grain size via cell proliferation of brassinosteroids (BRs) signaling and homeostasis (Duan et al., 2014; Liu et al., 2015). By contrast, the deactivation of OsMAPK6 by the MAPK phosphatase 1 (OsMKP1) also leads to smaller grains (Xu et al., 2018). In our study, the transcript of OsMAPK6 was accumulated in the knock-out of OsMPK15 lines (Figure 6B), leading to longer grain length in the mpk15 lines, which was consistent with the former studies (Liu et al., 2015), indicating OsMPK15 acting as an inhibitor for grain length.
Conclusion
In conclusion, we demonstrate that OsMPK15 negatively regulates the disease resistance to different races of M. oryzae and Xoo through modulating expression of PRs and SA/JA-associated genes as well as ROS burst. Furthermore, the knock-out of OsMPK15 also results in long grain size.
Gene Accession Numbers
Sequence data from this study can be found in the GenBank/EMBL data libraries with the following accession numbers: OsMPK15 (LOC_Os11g17080), OsPR4 (LOC_Os11g37970), OsPR5 (LOC_Os12g43380), OsPR8 (LOC_Os10g28080), OsPR10 (LOC_Os12g36880), OsPAL (LOC_Os02g41680), OsMAPK3 (LOC_Os03g17700), OsMAPK6 (LOC_Os06g06090), OsWRKY45 (LOC_Os05g25770), OsLOX (LOC_Os08g39840), OsOPR1 (LOC_Os06g11290), OsAOS1 (LOC_Os03g55800), OsAOS2 (LOC_Os03g12500), OsAOS4 (LOC_Os02g12690), OsActin (LOC_Os03g50885).
Author Contributions
YH conceived, designed and performed the experiments and drafted the manuscript. YH and QL analyzed the data. YC and YZ help my plant materials in the field condition, XL has a critical reading to the manuscript and made important suggestions. All authors discussed the results, provided critical feedback, and contributed to the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to thank the Rice Product Quality Supervision and Inspection Center in the Ministry of Agriculture, for determining the SA and JA content of rice leaves.
Funding. This project was supported by grants from the National Key Transgenic Research Projects (2016ZX08001002), the Natural Science Foundation of Innovation Research Group (31521064), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-CNRRI), the Zhejiang Provincial Key New Varieties Breeding effort (2016C02050-1), and Natural Science Foundation of China (31701338 and 31571944).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00752/full#supplementary-material
References
- Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., et al. (2002). MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Bent A. F. (2001). Plant mitogen-activated protein kinase cascades: negative regulatory roles turn out positive. Proc. Natl. Acad. Sci. U.S.A. 98 784–786. 10.1073/pnas.98.3.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini L., Proietti S., Focaracci F., Benedetta S., Caruso C. (2018). Epigenetic control of defense genes following MeJA-induced priming in rice (O. sativa). J. Plant Physiol. 228 166–177. 10.1016/j.jplph.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Brown J. K. (2002). Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 5 339–344. [DOI] [PubMed] [Google Scholar]
- Century K. S., Shapiro A. D., Repetti P. P., Dahlbeck D., Holub E., Staskawicz B. J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278 1963–1965. [DOI] [PubMed] [Google Scholar]
- Cheong Y. H., Moon B. C., Kim J. K., Kim C. Y., Kim M. C., Kim I. H., et al. (2003). BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 132 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P., Rao Y., Zeng D., Yang Y., Xu R., Zhang B., et al. (2014). SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77 547–557. 10.1111/tpj.12405 [DOI] [PubMed] [Google Scholar]
- Feussner I., Wasternack C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53 275–297. [DOI] [PubMed] [Google Scholar]
- Fu J., Chu J., Sun X., Wang J., Yan C. (2012). Simple, rapid and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28 1081–1087. 10.2116/analsci.28.1081 [DOI] [PubMed] [Google Scholar]
- Guo T., Chen K., Dong N. Q., Shi C. L., Ye W. W., Gao J. P., et al. (2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMAPK6 cascades to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30 871–888. 10.1105/tpc.17.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Fong S., Yang D., Wang G. (1999). BWMK1, a novel MAPK kinase induced by fungal infection and mechanical wounding in rice. Mol. Plant Microbe Interact. 12 1064–1073. [DOI] [PubMed] [Google Scholar]
- Hirt H. (1997). Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci. 2 11–15. [Google Scholar]
- Hong Y., Yang Y., Zhang H., Huang L., Li D., Song F. (2017). Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial disease. Sci. Rep. 7:41037. 10.1038/srep41037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L. Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19 884–894. 10.1016/j.devcel.2010.10.024 [DOI] [PubMed] [Google Scholar]
- Jalmi S. K., Sinha A. K. (2016). Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRKY30 in mediating resistance against Xanthomonas oryzae in rice. Sci. Rep. 6:37974. 10.1038/srep37974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen J. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63 141–152. [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Nat. Acad. Sci. U.S.A. 103 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M., Okada K., Kurimoto L., Murakami S., Umezawa T., Shibuya N., et al. (2010). A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63 599–612. 10.1111/j.1365-313X.2010.04264.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo S. C., Moon B. C., Kim J. K., Kim C. Y., Sung S. J., Kim M. C., et al. (2009). OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem. Biophy. Res. Commun. 387 365–370. 10.1016/j.bbrc.2009.07.026 [DOI] [PubMed] [Google Scholar]
- Lee M., Qi M., Yang Y. (2001). A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol. Plant Microbe Interact. 14 527–535. [DOI] [PubMed] [Google Scholar]
- Liebertherr D., Thao N. P., Nakashima A., Umemura K., Kawasaki T., Shimamoto K. (2005). A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol. 138 1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Sonbol F., Huot B., Gu Y., Withers J., Mwimba M., et al. (2016). Salicylic acid receptors activate jasmonic acid signaling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7:13099. 10.1038/ncomms13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Hua L., Dong S., Chen H., Zhu X., Jiang J., et al. (2015). OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 84 672–681. 10.1111/tpj.13025 [DOI] [PubMed] [Google Scholar]
- Ma H., Chen J., Zhang Z., Ma L., Yang Z., Zhang Q., et al. (2017). MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 92 557–570. 10.1111/tpj.13674 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I., Iwai T., Seo S., Yanagawa Y., Kawahigasi H., Hirose S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds. Mol. Genet. Genom. 279 415–427. 10.1007/s00438-008-0322-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15 349–357. 10.1016/j.pbi.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Nakayama A., Fukushima S., Goto S., Matsushita A., Shimono M., Sugano S., et al. (2013). Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 13:150. 10.1186/1471-2229-13-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y., Liu W., Wang G. (2017). Balancing immunity and yield in crop plants. Trends Plant Sci. 22 1069–1079. 10.1016/j.tplants.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Park C. H., Chen S., Shirsekar G., Zhou B., Khang C. H., Songkumarn P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant cell 24 4748–4762. 10.1105/tpc.112.105429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., Brodersen P., Naested H., Andreasson E., Lindhart U., Johansen B., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Qiu D., Xiao J., Ding X., Xiong M., Cai M., Cao Y., et al. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 5492–499. [DOI] [PubMed] [Google Scholar]
- Reyna N. S., Yang Y. N. (2006). Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 19 530–540. [DOI] [PubMed] [Google Scholar]
- Schweighofer A., Kazanaviciute V., Scheikl E., Teige M., Doczi R., Hirt H., et al. (2007). The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro A. D., Zhang C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Sharp G. L., Martin J. M., Lanning S. P., Blake N. K., Brey C. W., Sivamani E., et al. (2002). Field evaluation of transgenic and classical sources of wheat streak mosaic virus resistance. Crop Sci. 42 105–110. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64 204–214. 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomo M., Sugano S., Nakayama A., Jiang C., Ono K., Toki S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidonskaya E., Schweighofer A., Shubchynskyy V., Kammerhofer N., Hofmann J., Wieczorek K., et al. (2016). Plant resistance against the parasitic nematode heterodera schachtii is mediated by MPK3 and MPK6 kinases, which are controlled by the MAPK phosphatase AP2C1 in Arabidopsis. J. Exp. Bot. 67 107–118. 10.1093/jxb/erv440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F. M., Goodman R. M. (2002). OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta 215 997–1005. [DOI] [PubMed] [Google Scholar]
- Sun X., Cao Y., Yang Z., Xu C., Li X., Wang S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37 517–527. [DOI] [PubMed] [Google Scholar]
- Tena G., Asai T., Chiu W. L., Sheen J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4 392–400. [DOI] [PubMed] [Google Scholar]
- Ueno Y., Yoshida R., Kishi-Kaboshi M., Matsushita A., Jiang C., Goto S., et al. (2013). MAP kinases phosphorylate rice WRKY45. Plant Signal. Behav. 8:e24510. 10.4161/psb.24510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Tang Z., Li X., Hong Y., Li B., Xiao W., et al. (2017). Overexpressing OsMAPK12-1 inhibits plant growth and enhances resistance to bacterial disease in rice. Funct. Plant Biol. 44 694–704. [DOI] [PubMed] [Google Scholar]
- Xie K., Chen J., Wang Q., Yang Y. (2014). Direct phosphorylation and activation of a mitogen-activated protein kinase by a Calcium-dependent protein kinase in rice. Plant Cell 26 3077–3089. 10.1105/tpc.114.126441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L. Z., Yang Y. N. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Yu H., Wang J., Duan P., Zhang B., Li J., et al. (2018). A mitogen-activated protein kinase phosphatase influences grain size and weight in rice. Plant J. 95 937–946. 10.1111/tpj.13971 [DOI] [PubMed] [Google Scholar]
- Yang D. L., Yang Y., He Z. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant. 6 675–685. 10.1093/mp/sst056 [DOI] [PubMed] [Google Scholar]
- Yang D. L., Yao J., Mei C. S., Tong X. H., Zeng L. J., Li Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109 e1192–e1200. 10.1073/pnas.1201616109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shah J., Klessig D. F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11 1621–1639. [DOI] [PubMed] [Google Scholar]
- Yuan B., Shen X. L., Li X. H., Xu C. G., Wang S. P. (2007). Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 226 953–960. [DOI] [PubMed] [Google Scholar]
- Zhang S., Klessig D. F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6 520–527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.