Abstract
Changes in soil bacterial communities, which are crucial for the assessment of ecological restoration in Chinese plantations, have never been studied in the “Three North Shelterbelt” project in the semi-arid areas. We used high-throughput sequencing of the 16S rDNA gene to investigate the soil bacterial community diversity, structure, and functional characteristics in three plantation forests, including Populus × canadensis Moench (PC), Pinus sylvestris var. mongolica (PS), and Pinus tabuliformis (PT). In addition, soil environment factors were measured. There were distinct differences in soil characteristics among different plantation forests. Compared to PS and PT, PC had a higher soil pH, dissolved organic carbon (DOC), and available P, as well as a lower C/N ratio. Furthermore, afforestation with different tree species significantly altered the abundance of Proteobacteria, and Chloroflexi in the soil, and its influence on the bacterial diversity indices. The bacterial community compositions and functional groups related to C and N cycling from PS, and PT were grouped tightly, indicating that the soil bacterial phylogenetic distance of PS and PT were closer than that between PS plus PT and PC. Our results implied that the soil characteristics, as well as the diversity, compositions and functions related to C and N cycling of soil bacterial community obviously differed from the following afforestation, especially between PC and PS plus PT, which in turn enormously established the correlation between the soil microbial community characteristics and the afforestation tree species.
Keywords: Soil bacterial community, Bacterial functional characteristics, Afforestation, Thesemi-arid areas
Introduction
Desertification has always been an important global ecological environmental problem in the arid and semi-arid regions (Li et al., 2004; Torres et al., 2015; Becerril-Piña et al., 2015), that is mainly caused by climate change and human activities in arid, semi-arid and some sub-humid regions (Chasek et al., 2015; Salvati et al., 2015; Wijitkosum, 2016), impacting 25% of the total terrestrial area (Reynolds et al., 2007; Allington & Valone, 2010). Desertification has caused a loss of soil nutrients, a decline in land productivity and environmental degradation (Li et al., 2018). This leads to a decline or degradation in sand-stabilization, soil conservation, water resource regulation, carbon sequestration and other desert ecosystem services, and it endangers both regional and national economic, social, and environmental security (Martã-Nez-Valderrama et al., 2016; Sutton et al., 2016). Research sponsored by the United Nations Environment Programme (UNEP) shows that the global economic losses caused by desertification and drought are as high as $4. 2 × 1010 USD per year, which is equivalent to all official aid given to Africa in 2009 (United Nations Convention to Combat Desertification (UNCCD), 2011) (Furtado & Macedo, 2008). With rapid economic growth, China is also confronted with various environmental problems, including sandstorms, severe desertification, and land degradation in dry northern regions (Liu & Diamond, 2005).
Numerous researches in arid and semi-arid areas have suggested that afforestation is one of the most commonly used techniques and effective sand-fixing methods to reduce the harm of desertification (Gao et al., 2002; Nunezmir et al., 2015). In addition, afforestation exhibits a significantly positive feedback to the regional environment (Verón & Paruelo, 2010; Zhang et al., 2014; Peng et al., 2014), successfully combating the desertification and dust storms (Fan et al., 2014; Tan & Li, 2015). In order to prevent ecosystem degradation, artificial shrubs and trees have been widely planted in many degraded areas of China (Zhao et al., 2017). Since the 1950s, large desertification control projects have been implemented (Zou et al., 2002; Piao et al., 2005; Qadir, Qureshi & Cheraghi, 2010). Since 1978, a series of ecological restoration programs have been initiated in China to alleviate these increasingly serious environment problems and to restore degraded land (Zhang et al., 2016), primarily through afforestation and reforestation (Zhang et al., 2014), of which the ‘Three North Shelterbelt Development Program (TNSDP)’ is the largest afforestation program in the world (Li et al., 2012). This special program in China gives us the advantage of studying the benefits of afforestation in China.
Currently, planted forests in China account for about 31.8% of the forested area, which is the most of any country in the world (State Forest Administration, 2010). By 2008, afforestation of 1,511,700 hm2 of land had been completed in the TNSDP of Liaoning. In this area, 212,200 hm2 of degraded forest was covered, accounting for 13.97% of the preserved area. Great achievement has been made in the afforestation of the Horqin Sandy Land in northern China (Zuo et al., 2012; Zhao et al., 2014; Ge et al., 2015), and the ecological environment has been considerably improved. Furthermore, afforestation has increased soil organic carbon sequestration in soil (Deng et al., 2006; Zhou et al., 2014). During vegetation recovery, soil characteristics have also been identified as the main factors driving plant growth, plant production, and dune ecosystem function (Zuo et al., 2008; Zuo et al., 2009; Qiu et al., 2018). A variety of researchers have focused on the influences of vegetation on the soil water content (Yang et al., 2018) and soil characteristics (Deng et al., 2017).
Compared with the physical and chemical properties of soil, soil microorganisms are more efficient and dynamic indicators of soil quality (Van der Heijen, Bardgett & Straalen, 2010; Bridge & Spooner, 2001). Soil microorganisms play a vital role in soil processes, which can profoundly impact the main biogeochemical cycles of C and N, as well as provide protection to larger organisms through the formation of biofilms (Chatterjee et al., 2008; Ritz et al., 2009; Burton et al., 2010). Previous research suggests that the soil microbial distribution is regulated by the different revegetation types (Deng et al., 2019). In addition, soil characteristics play important roles in shaping the soil bacterial community diversity and structure in the terrestrial ecosystems (Liu et al., 2018). Extensive studies have indicated that tree species and soil characteristics influence the microbial community structure and compositions (Ding et al., 2017). Within the fragile local ecological environment, there is also a close correlation between the soil biological properties and vegetation types. Thus, it is of great significance to study the characteristics of soil microorganisms in different plantation forests used for sand fixation forest (Wang et al., 2016). This information will allow for a better understanding of how soil improvement effects sand fixation forests and prevents land desertification. Previous findings have reported that grassland afforestation changes the chemical properties and composition of the soil, as well as the ecological functions of the soil bacterial communities. And these effects of afforestation on the microorganisms have been modulated by changes in soil chemical characteristics (Wu et al., 2019). Moreover, the forest soil habitat is an efficient means to restore local vegetation and studying this area can shed new light on the distribution of local soil eukaryotic microorganisms in semi-arid areas (Zhao et al., 2018).
Northwest Liaoning has an arid climate, frequent gales, water shortage and low vegetation cover. It is also the key governance area of the TNSDP. In recent decades, Populus × canadensis Moench (PC), Pinus sylvestris var. mongolica (PS), and Pinus tabuliformis (PT) have become the main afforestation tree species and widely used for afforestation in the “Three North Shelterbelt” due to their strong stress resistance, ability to accelerate ecological rehabilitation and improve ecological stability. Previously, numerous studies in shelterbelt have focused primarily on the aboveground ecosystem, and the soil physicochemical characteristics (Wang et al., 2014; Zhou et al., 2016). However, variations in the soil bacterial communities, which are crucial for the assessment of ecological restoration in plantations in this area, have never been studied. Therefore, we selected a typical artificial shelterbelt in the semi-arid area of northwestern Liaoning as the study area. The soil bacterial community diversity, compositions and functional characteristics were analyzed for different plantation forests composed of Populus × canadensis Moench, Pinus sylvestris var. mongolica, and Pinus tabuliformis. We propose the following hypotheses: (1) the three different plantation forests harbor different soil bacterial diversity and community structures; (2) the functional groups related to C and N cycling differ between PC and PS plus PT; and (3) consistent differences occur in the soil bacterial community and functional characteristics following afforestation. This research provides a reference for vegetation restoration and sustainable management of artificial forests in this area. Furthermore, this study has important theoretical and practical significance for the selection of tree species used for sand fixation in semi-arid regions. Our results provide a scientific basis for the recovery of degraded soils.
Material and Methods
Sites description
Research was conducted at Fujia forest farm, Changtu County, northwest of Liaoning province (123°32′E∼123°55′E∼42°53′N∼43°21′N). This area is located at the southern edge of Horqin Sandy Land and belongs to the Liaohe alluvial plain. The soil texture is characterized by Arenosols (Lv et al., 2018). The study area is located at an attitude of 91.1∼173.4 m, with the relatively flat topography and a small amount of elliptic or circular dune distribution. The climate in this region is classified as a temperate semi-humid semi-arid continental climate (Lu et al., 2017), with long and cold winters, and hot summers. There is little rainfall, with an average annual precipitation of 400 to 550 mm that is concentrated in July to August. The average annual evaporation is approximately 1,843 mm. The average temperature is 7 °C with extreme maximum and minimum temperature of 35.6 °C and −31.5 °C. In recent decades, Populus × canadensis Moench, Pinus sylvestris var. mongolica, and Pinus tabuliformis were the main plantation forests in the “Three North Shelterbelt” (Fig. 1). Prior to afforestation, the vegetation type was Arachis hypogaea (peanut) farmland, and the site information was shown in Table 1.
Figure 1. Location of three forest type sites.
(A) Stusy site; (B) Populus × canadensis Moench; (C) Pinus sylvestris var. mongolica; (D) Pinus tabuliformis..
Table 1. Site information.
| Different samples | Age of stand | Stand density (plant hm−1) | Height (m) | Diameter at breast height (cm) | Crown density (%) |
|---|---|---|---|---|---|
| PC | 18 | 773 | 15.34 | 14.32 | 65% |
| PS | 33 | 642 | 14.52 | 22.43 | 70% |
| PT | 33 | 575 | 13.56 | 20.51 | 65% |
Notes.
- PC
- Populus × canadensis Moench
- PS
- Pinus sylvestris var. mongolica
- PT
- Pinus tabuliformis
Soil sampling
Three independent replicate plots (20 m ×20 m) within the same climate were established in August 2018 for each forest type. The distance between each sampling plot was greater than 50 m but less than 200 m. To ensure the representativeness of soil samples, 10–15 soil cores of topsoil (0∼10 cm) were collected for each triplicate plot using soil auger with an “S” shape. After removing the litter layer, the soil cores were combined to one composite sample, giving a total of nine samples. All soil samples were stored on ice box after being sealed in plastic bags for transport to the laboratory. In the laboratory, the samples were sieved through a two mm mesh to remove plant roots, stones, litter, and other debris. Samples were subsequently divided into three parts. One part was air-dried for analysis of soil characteristics, including soil pH, the contents of total carbon (C), total nitrogen (N), and available phosphorus (P). The second part was stored at 4 °C for DOC analysis, and the third part was stored at −80 °C for DNA extraction.
Measurement of soil characteristics
The soil pH was analyzed using an electrode pH meter in the soil-water (1:5 w/v) suspension (Bao, 2000; Ren et al., 2016). The contents of soil total C and total N were determined using an elemental analyzer (Elementar, Hesse, Germany) (Schrumpf et al., 2011). The concentrations of available P was determined using the extraction-flame photometry with a 0.5 M NaHCO3 extraction (Emteryd, 1989). Additionally, the content of dissolved organic carbon (DOC) was extracted from fresh soil using deionized water (1:5 w/v) (Gong et al., 2009) and determined via a TOC analyzer (Multi N/C 3100, Analytik Jena AG).
Soil DNA Extraction and 16S rDNA Sequencing
DNA was extracted from 0.5 g of soil using the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA), following the manufacturer’s instructions. A NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to determine the quantity and quality of the extracted DNA. PCR was performed to target V3-V4 hypervariable region of the bacterial 16S rRNA genes was constructed to amplify, with the forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Deng et al., 2018). PCR amplifications were carried out in two steps. Firstly, each of three independent 25 µl reactions per DNA sample contained five µl of Q5 High-Fidelity GC buffer (5 ×); five µl of Q5 reaction buffer (5 ×); one µl (10 uM) of forward primer, one µl (10 uM) of reverse primer; 0.25 µl (5 U/µl) of Q5 High-Fidelity DNA Polymerase; two µl of dNTPs (2.5 mM); two µl of DNA Template (40-50 ng); and 8.75 µl of ddH2O (Deng et al., 2018). Cycling conditions were as follows: one cycle of denaturation at 98 °C for 5 min; then denaturation at 98 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s (25 cycles); and with a final extension at 72 °C for 5 min. Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN, USA) and PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA) were used to purify and quantify PCR amplicons. Amplicons were then pooled at equal concentrations after the individual quantification step, and sequencing of pair-end 2 × 300 bp was performed using the Illlumina MiSeq platform with the MiSeq Reagent Kit v3 (Shanghai Personal Biotechnology Co., Ltd, Shanghai, China).
Functional prediction using FAPROTAX
The database of Functional Annotation of Prokaryotic Taxa (FAPROTAX), which is based on the available functional information in the existing microbiology literature, summarize the names of related species from functional classification and annotation information. FAPROTAX can be used to extrapolate functions of cultured prokaryotes to estimate metabolic or other ecological relevant functions, which is more suitable for the functional annotation and prediction of the biogeochemical cycle of environmental samples. The annotated operational taxonomic unit (OTU) table from the Silva database was read, and the annotated OTU information was matched with the species information in the database using a python program and the predicted functions were outputted. The details of this approach are provided by Louca, Parfrey & Doebeli (2016), Louca et al. (2017). The relative abundances of the functional groups in each sample was calculated as the cumulative abundance of OTUs assigned to each functional group, which was obtained by normalizing the cumulative abundance of OTUs correlated with at least one function. Thus, functional annotation of the OTUs was established based on FAPROTAX. We then investigated potential functions involved in geographical location and environmental conditions.
Bioinformatics and processing of sequencing data
The QIIME software (v1.9.0) and the UPARSE pipeline (Zhong, Yan & Shang, 2015) were used to analyze the raw data obtained from Illumina sequencing. The bacterial raw data was submitted to the NCBI Sequence Read Archive (SRA) under accession number PRJNA495735. The operational taxonomic assignment of OTUs with similarities >97% was conducted using the UPARSE pipeline (Edgar, 2013). Then the operational taxonomic classification and identity of OTUs were determined using a BLAST algorithm against sequences within the Silva Database via QIIME software (Kõljalg et al., 2013). OTU-level alpha diversity indices, such as Simpson index, Chao1 index, Shannon index, and ACE index were computed using the OTU table in QIIME (Caporaso et al., 2010).
Statistical analysis
Among samples, the unique and shared OTUs of the soil bacterial community were used to create Venn diagrams using the R (R v.3.4.4) with the “VennDiagram” package (Zaura et al., 2009). The heatmap representation of the top 50 classified bacterial genera in each sample was established using R (R v.3.4.4) with the “gplot” and “pheatmap” packages (R Development Core Team, 2009). The relationships between soil characteristics and the bacterial community functions related to C and N cycling based on Spearman’s rank correlation analysis were built using R (v.3.4.4) with the “psych” and “corrplot” packages (R Development Core Team, 2009). The differences in bacterial community structure across samples were established through beta diversity analysis and visualized via nonmetric multidimensional scaling (NMDS) based on unweighted UniFrac distance metrics (Lozupone et al., 2007).
Multifactorial ANOVA (MANOVA) and Canonical correspondence analysis (CCA) were applied to analyze the effects of tree species on all the measured soil characteristics. Multiple comparisons of means at a 95% confidence interval were performed using Tukey’s honest significance difference (HSD) post-hoc test. Soil bacterial diversity and relative abundances were analyzed in SPSS (v. 19.0) using a one-way analysis of variation (ANOVA) and least significant difference (LSD) multiple comparison tests (Banerjee, 2016). Spearman’s rank correlation was used to estimate the relationships between soil characteristics and soil bacterial community diversity. Similar patterns of functional groups of bacterial community were analyzed with Principal component analysis via the STAMP software (Parks et al., 2014). The linkages between soil environmental factors and bacterial community composition at the phylum level were performed by canonical correspondence analysis (CCA) via Canoco 4.5 (Braak & Smilauer, 2002).
Results
Soil characteristics in different plantation forests
Tree species had a strong significant effect on all the soil pH (F = 6.58, P = 0.031), total C (F = 30.54, P = 0.001), DOC (F = 6.02, P = 0.037), total N (F = 6.47, P = 0.032), C/N (F = 45.49, P < 0.001), and available P (F = 78.06, P < 0.001) (Table 2). There were distinct differences in the soil chemical characteristics among the three plantation forests. The soil pH ranged from 5.53 to 5.92. The highest pH value occurred in PC with 5.92, and significantly higher than PS and PT (P < 0.05). The soil DOC was highest in PC with 105.46 mg kg−1, and significantly higher than in PS and PT (P < 0.05). Soil total C and total N concentrations in PS were the highest with 12.00 g kg−1, and 1.05 g kg−1, respectively, followed by PT. And both of them occurred significantly higher than PC (P < 0.05). The maximum value of the C/N ratio occurred in PT with 11.79, followed by PS with 11.41, and lowest in PC with 9.15. No significant differences were observed in the soil total C content and C/N ratio between PS and PT (P > 0.05). Soil available P was highest in PC with 16.00 mg kg−1, which was significantly higher than those of PS and PT (P < 0.05) (Table 2). Compared to PS and PT, the PC had the highest soil pH value, DOC, and available P, as well as the lowest C/N ratio.
Table 2. Results of MANOVA and post-hoc analyses on the effects of tree species and their interactions on soil properties.
| Factors | pH | Total C (g kg−1) | DOC (mg kg−1) | Total N (g kg−1) | C/N ratio | Available P (mg kg−1) |
|---|---|---|---|---|---|---|
| Tree species | – | – | – | – | – | – |
| F-value | 6.58 | 30.54 | 6.02 | 6.47 | 45.49 | 78.06 |
| P-value | 0.031 | 0.001 | 0.037 | 0.032 | <0.001 | <0.001 |
| PC | 5.92a | 7.82b | 105.46a | 0.85b | 9.15b | 16.00a |
| PS | 5.57b | 12.00a | 83.42ab | 1.05a | 11.41a | 3.77b |
| PT | 5.53b | 11.46a | 80.10b | 0.97a | 11.79a | 3.09b |
Notes.
- PC
- Populus × canadensis Moench
- PS
- Pinus sylvestris var. mongolica
- PT
- Pinus tabuliformis
Different small letters meant significant difference at 0.05 level.
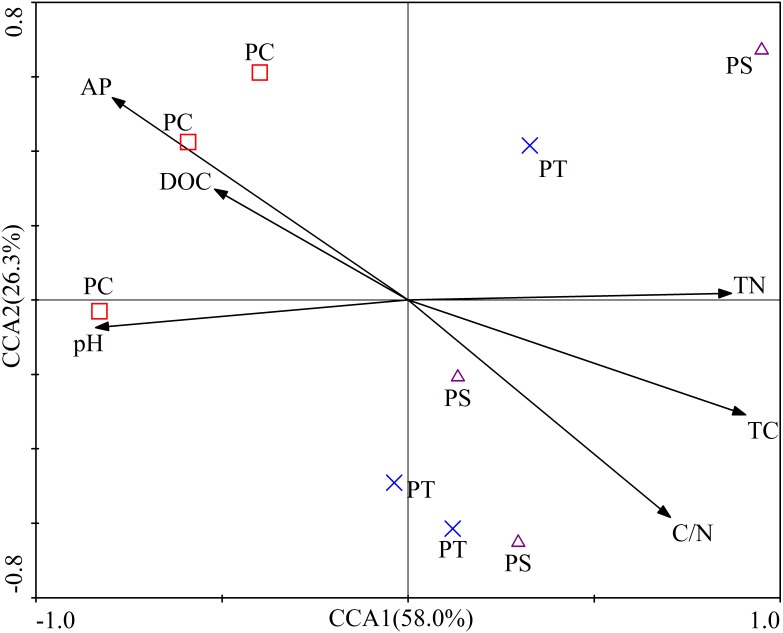
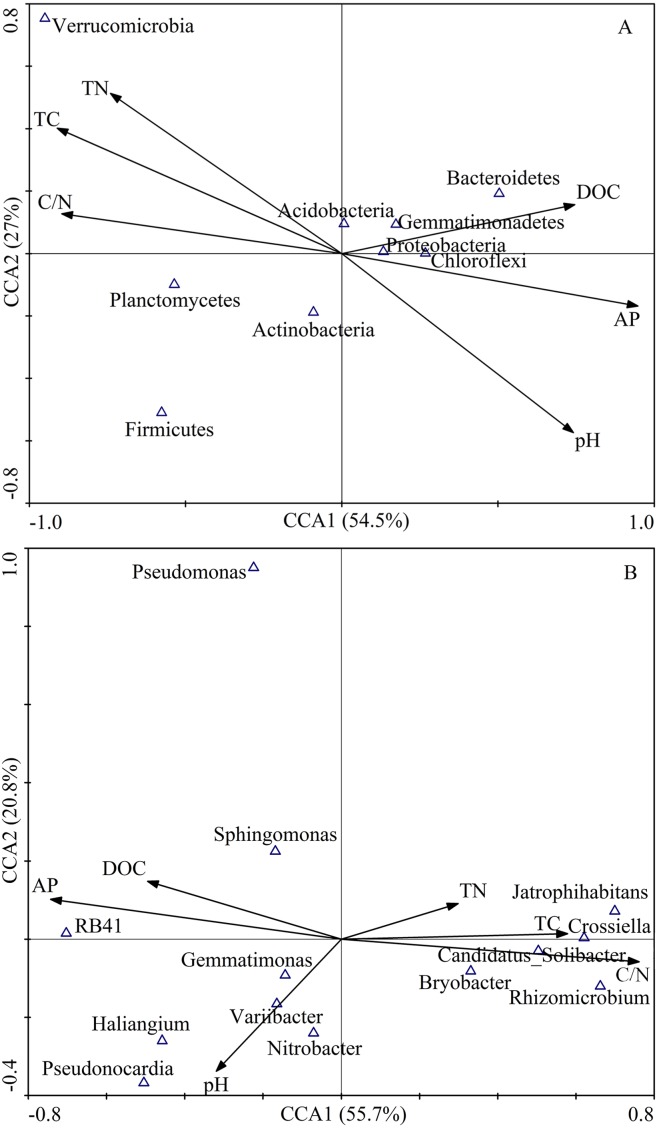
The first two axes of the CCA accounted for 84.3% of the total variance. The CCA plot showed a clear separation in the space among the three plantation forests. In fact, the PC distinctly separated from PS, and PT, especially along CCA1 (Fig. 2). The investigated soil characteristics also clearly separated into the quadrants. Soil total N was situated in the first quadrant; available P and soil pH were in the second quadrant; soil pH was in the third quadrant; soil total C and C/N ratio were in the fourth one (Fig. 2). The results illustrated that the different forest types had different soil characteristics, significantly different between PC and PT plus PS.
Figure 2. Results of canonical correspondence analysis-plot of all the measured soil environment factors.
PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis. TC: total C; DOC: Dissolved organic carbon; TN: total N; AP: available P.
Soil bacterial community diversity under different plantation forests
There were significant differences among different plantation forests regarding soil total C and total N contents ACE index and Chao1 index (F = 7.64, P = 0.02; F = 7.92, P = 0.02; Table 3). The maximum values of the ACE index and Chao1 index occurred in PS with 4,074.80, and 3,952.39, respectively, followed by PT and PC. The soil bacterial Shannon index and Simpson index among three plantation forests significantly differed (F = 6.89, P = 0.03; F = 5.68, P = 0.04; Table 3). The Shannon index and Simpson index were lowest in PT with 10.09 and 0.996, respectively. The Spearman’s rank correlations indicated that soil total N significantly positively correlated with the Chao 1 index (r = 0.78, P < 0.05) and ACE index (r = 0.83, P < 0.01). The Simpson index was significantly positively related with soil available P (r = 0.77, P < 0.05), and DOC contents (r = 0.67, P < 0.05), while, the Simpson index was highly negatively correlated with the C/N ratio (r = − 0.73, P < 0.05) (Table 4).
Table 3. Soil bacterial diversity indices in different plantation forests.
| Diversity indices | PC | PS | PT | F test | P value |
|---|---|---|---|---|---|
| No. of sequences | 48,848 ± 3,650aA | 51,532 ± 2,892aA | 52,395 ± 2,529aA | 1.10 | 0.39 |
| OTUs number(Phylum) | 2,750 ± 349bA | 3,378 ± 150aA | 2,837 ± 141bA | 6.36 | 0.03 |
| Shannon index | 10.46 ± 0.11aA | 10.48 ± 0.16aA | 10.09 ± 0.16bA | 6.89 | 0.03 |
| ACE index | 2,898.70 ± 537.94bB | 4,074.80 ± 181.79aA | 3,192.86 ± 344.77bAB | 7.64 | 0.02 |
| Chao1 index | 2,876.13 ± 518.02bB | 3,952.39 ± 67.89aA | 3,140.32 ± 290.58bAB | 7.92 | 0.02 |
| Simpson index | 0.998 ± 0.000aA | 0.997 ± 0.001abA | 0.996 ± 0.001bA | 5.68 | 0.04 |
Notes.
Data are means ± standard error (n = 3).
- PC
- Populus × canadensis Moench
- PS
- Pinus sylvestris var. mongolica
- PT
- Pinus tabuliformis
Different small letters meant significant difference at 0.05 level. Different capital letters meant significant difference at 0.01 level.
Table 4. Spearman’s rank correlations between the soil bacterial diversity indices and available edaphic factors.
| pH | Total C | DOC | Total N | C/N ratio | Available P | |
|---|---|---|---|---|---|---|
| Simpson index | 0.65 | −0.65 | 0.67* | −0.55 | −0.73* | 0.77* |
| Chao1 index | −0.48 | 0.55 | −0.35 | 0.78* | 0.25 | −0.43 |
| ACE index | −0.58 | 0.62 | −0.43 | 0.83** | 0.30 | −0.45 |
| Shannon index | 0.22 | −0.18 | 0.27 | −0.09 | −0.35 | 0.20 |
Notes.
correlation significant at 0.05 level.
correlation significant at 0.01 level (two-tailed).
Soil bacterial community structure in different plantation forests
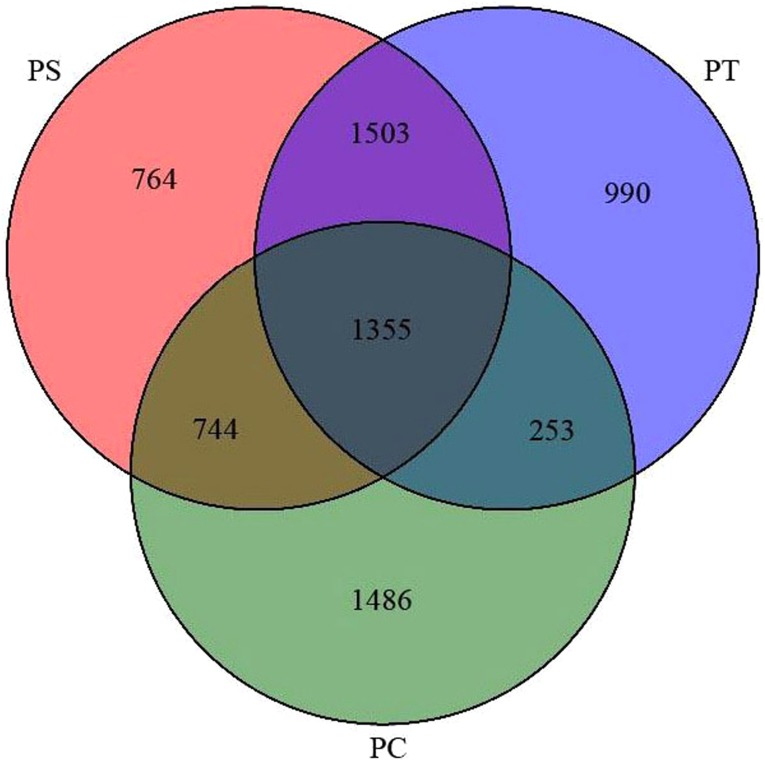
After quality trimming and chimera removal, 48,848, 51,532, and 52,395 high-quality sequences were generated from the PC, PS, and PT sites, respectively. Rarefaction curves for all the soil samples were shown in Fig. S1. As shown in the Venn diagram (Fig. 3), the total number of shared bacterial OTUs in PC, PS, and PT was 1,355. The number of bacterial OTUs shared between two sites was 1,503 for PT and PS, 744 for PS and PC, and 253 for PT and PC. The unique OTUs harbored in PC, PS and PT were 1486, 764 and 990, respectively.
Figure 3. Venn diagram representation of shared and unique OTUs of soil bacteria across three different plantation forests.
PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
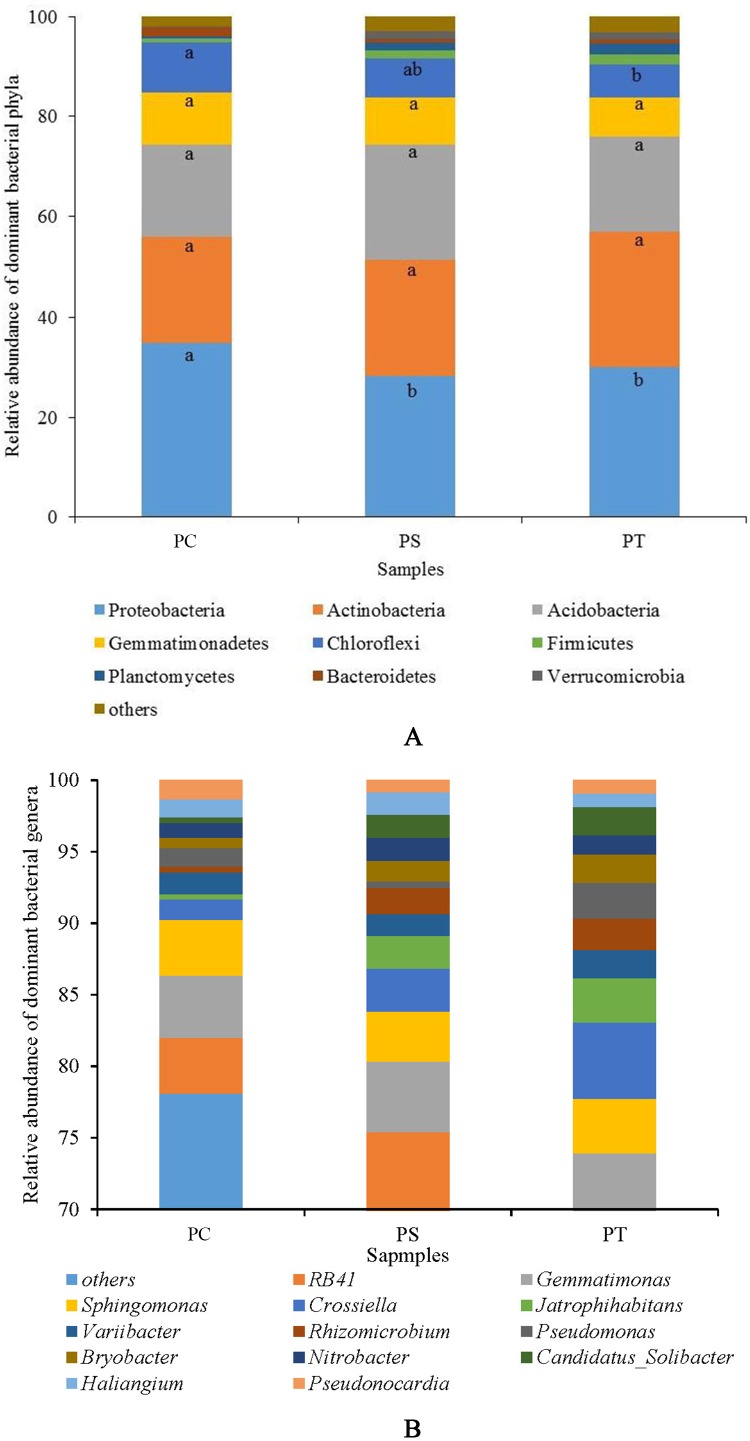
Sequences analysis showed a total of 29 phyla, and 793 genera within the three plantation forest samples. Nine dominant phyla (relative abundance >1%) were observed, of which the total average relative abundances represented more than 95%. Proteobacteria was the most dominant bacterial phylum (31.01%), followed by Actinobacteria (23.76%), Acidobacteria (20.08%), Gemmatimonadetes (9.17%), Chloroflexi (8.07%), Firmicutes (1.52%), Planctomycetes (1.45%), Bacteroidetes (1.18%), and Verrucomicrobia (1.07%) (Fig. 4). The average relative abundances of the Proteobacteria subgroups (Alpha-, Beta-, Gamma-, and Delta-Proteobacteria) were 18.90%, 5.45%, 3.10%, and 3.56%, respectively (Fig. S2). The relative abundances of Proteobacteria and Chloroflexi varied significantly (P < 0.05) among the different forest types, with the highest abundances in PC with 34.78% and 10.02%, respectively (Fig. 4A). No significant differences were observed for other phyla among the different plantation forests (P > 0.05).
Figure 4. The relative abundance of dominant bacterial phyla (A) and genera (B) in different plantation forests.
PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis. Different small letters meant significant difference at 0.05 level.
At the genus level, 13 dominant bacterial genera (relative abundances >1%) were observed, namely RB41, Gemmatimonas, Sphingomonas, Crossiella, Jatrophihabitans, Variibacter, Rhizomicrobium, Pseudomonas, Bryobacter, Nitrobacter, Candidatus-Solibacter, Haliangium, and Pseudonocardia, accounting for more than 20% of the total relative abundances (Fig. 4B). The average relative abundances of Jatrophihabitans, Rhizomicrobium, Bryobacter, Candidatus-Solibacter, and Haliangium were significantly different among the three plantation forests (P < 0.05).
Effects of tree species on the compositions of the soil bacterial community
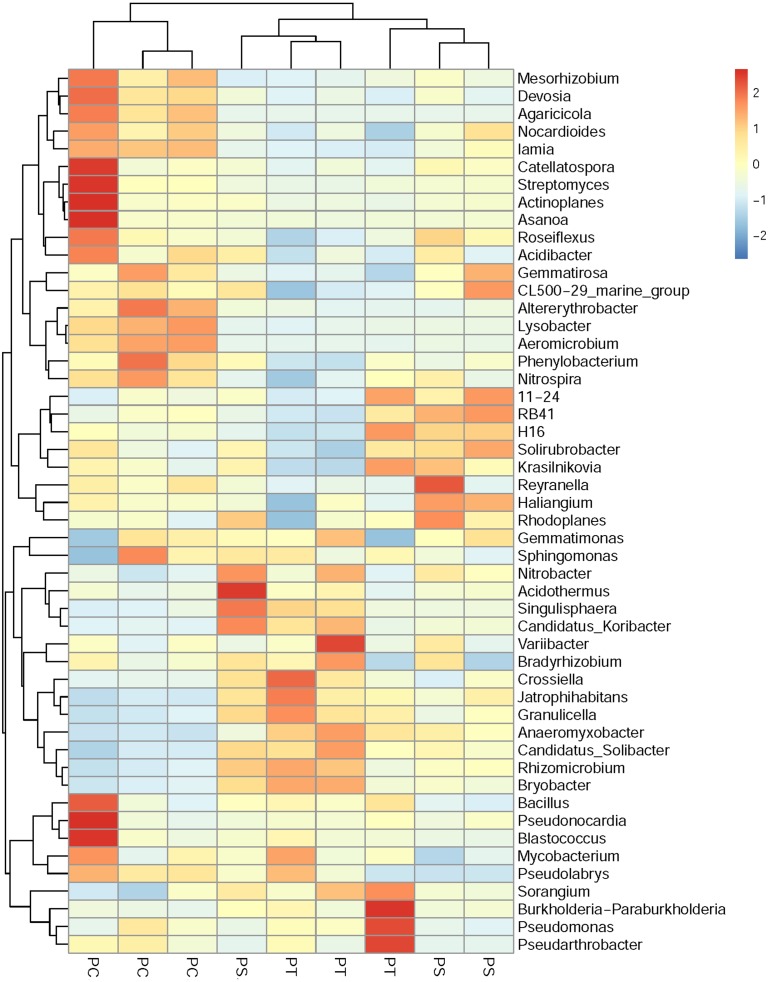
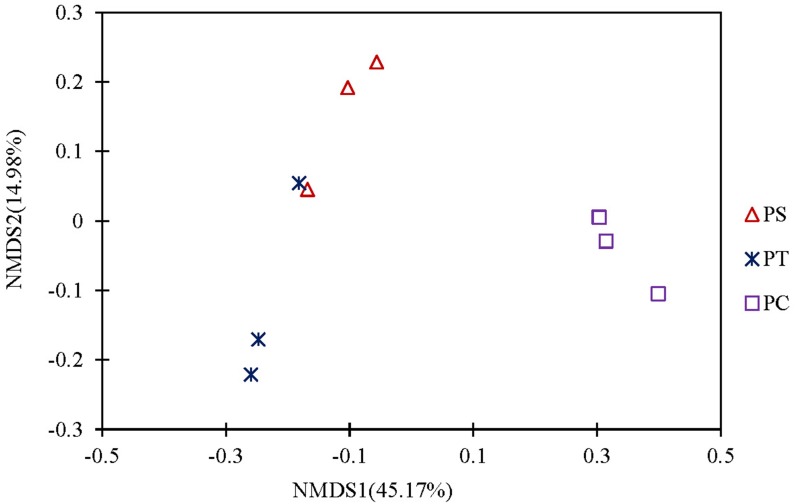
A cluster heatmap analysis was used to analyze the differences in the bacterial community compositions among the three plantation forests at the genus level (Fig. 5). The relative abundance and distribution of soil bacteria in different plantation forests changed significantly. Results showed that the soil samples were divided into two groups: one group contained the PT and PS, and the other group was PC. In order to show the bacterial community structures of PC, PS and PT, NMDS plot based on the unweighted uniFrac metric was calculated (Fig. 6). The NMDS plot also showed that the samples were classified into two large groups, one group corresponding to the communities from the PS, and PT, and the other group from PC. The samples from the PS and PT were grouped tightly, indicating that they shared a high similarity in their bacterial compositions. Furthermore, the PS and PT tended to be separate from the PC, especially along NMDS1, which contributed to 45.17% of the variance. Both analyses demonstrated that different plantation forests had different influences on the soil bacterial communities. Moreover, the phylogenetic relationship of the PT and PS was closer than those between the PT plus PS and PC.
Figure 5. Heatmap and hierarchical cluster analysis based on the relative abundances of the top 50 genera identified in the soil bacterial communities.
The samples are grouped according to the similarity of each other, and the clustering results are arranged horizontally according to the clustering results. In the figure, red represents the genus with higher abundance in the corresponding sample, and blue represents the genus with lower abundance. PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
Figure 6. Non-metric Multidimensional scaling analysis (NMDS) based on unweighted Unifrac metric illustrating the soil bacterial community structure among different plantation forests.
PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
Effects of soil environment factors on the soil bacterial community compositions
Results of the CCA showed that soil bacterial community structure had significant correlations between and soil characteristics (Fig. 7). At the phylum level, the first ordination CCA axis (CCA1) was strongly correlated with pH (r = 0.71), total C (r = − 0.87), DOC (r = 0.71), total N (r = − 0.71), C/N (r = − 0.85), and available P (r = 0.91), explaining 54.5% of the total variability of the bacterial community structures. Both axes together explained 81.5% of the variation (Fig. 7A). At the genus level, the first ordination CCA axis (CCA1) was strongly correlated with total C (r = 0.58), C/N (r = 0.76), and available P (r = − 0.74), explaining 55.7% of the total variability of the bacterial community structures. Both axes together explained 76.5% of the variation (Fig. 7B). Thus, soil DOC, C/N, and available P were important variables that played vital roles in the shaping of the bacterial communities.
Figure 7. CCA of abundant bacterial communities at the phylum (A) and genus (B) level and soil chemical characteristics for soil samples from different plantation forests.
TC: total C; DOC: dissolved organic carbon; TN: total N; AP: available P.
The relative abundances of Proteobacteria (r = 0.83, P < 0.01) and Bacteroidetes (r = 0.78, P < 0.05) had significantly positive correlations with the DOC content. The relative abundances of Proteobacteria (r = − 0.67, P < 0.05), Chloroflexi (r = − 0.68, P < 0.05), and Bacteroidetes (r = − 0.77, P < 0.05) were significantly negatively correlated with C/N. In contrast, the relative abundances of Proteobacteria (r = 0.68, P < 0.05), Chloroflexi (r = 0.78, P < 0.05), and Bacteroidetes (r = − 0.70, P < 0.05) were positively correlated with available P. The relative abundance of Verrucomicrobia was dramatically negatively correlated with soil pH (r = − 0.83, P < 0.01), while, Verrucomicrobia was significantly positively correlated with total C (r = 0.90, P < 0.01), and total N (r = 0.83, P < 0.01). The relative abundance of Planctomycetes showed a significantly positive correlation with C/N (r = 0.70, P < 0.05), and negative correlation with available P (r = 0.88, P < 0.01) (Table 5).
Table 5. Spearman’s rank correlations between the relative abundances of dominant bacterial groups and available edaphic factors.
| pH | Total C | DOC | Total N | C/N ratio | Available P | |
|---|---|---|---|---|---|---|
| Proteobacteria | 0.60 | −0.55 | 0.83** | −0.58 | −0.67* | 0.68* |
| Actinobacteria | −0.25 | 0.15 | −0.55 | 0.18 | 0.23 | −0.15 |
| Acidobacteria | −0.17 | 0.27 | −0.05 | 0.28 | 0.23 | −0.35 |
| Gemmatimonadetes | 0.35 | −0.25 | 0.55 | −0.17 | −0.32 | 0.20 |
| Chloroflexi | 0.52 | −0.63 | 0.55 | −0.52 | −0.68* | 0.78* |
| Firmicutes | −0.33 | 0.40 | −0.53 | 0.33 | 0.38 | −0.43 |
| Planctomycetes | −0.35 | 0.60 | −0.40 | 0.33 | 0.70* | −0.88** |
| Bacteroidetes | 0.40 | −0.32 | 0.78* | −0.28 | −0.77* | 0.70* |
| Verrucomicrobia | −0.83** | 0.90** | −0.50 | 0.83** | 0.53 | −0.58 |
Notes.
correlation significant at 0.05 level.
correlation significant at 0.01 level (two-tailed).
Bacterial functional annotation and distribution in different plantation forests
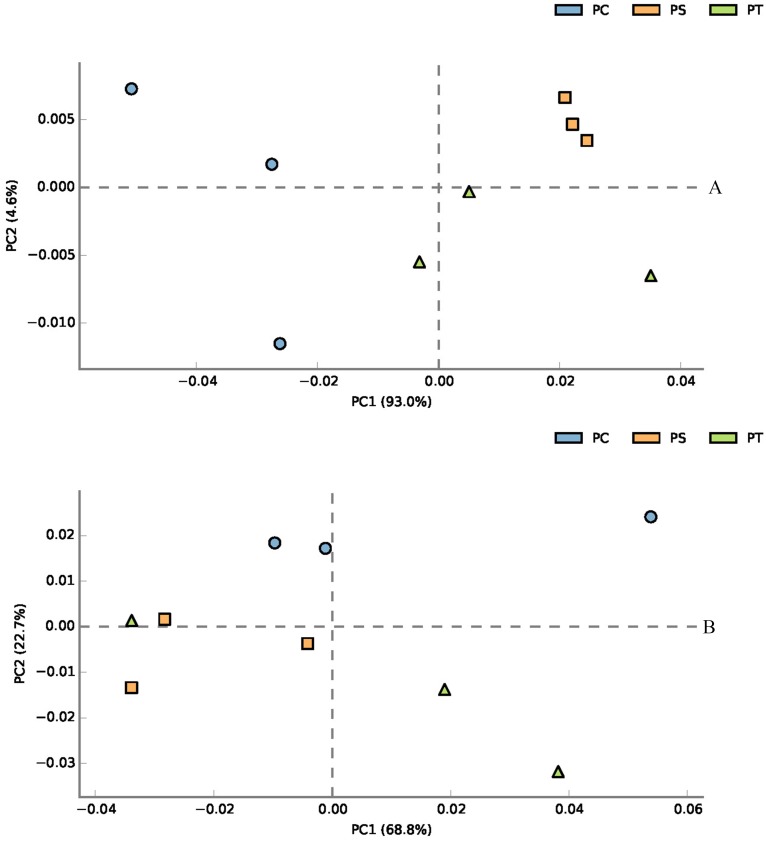
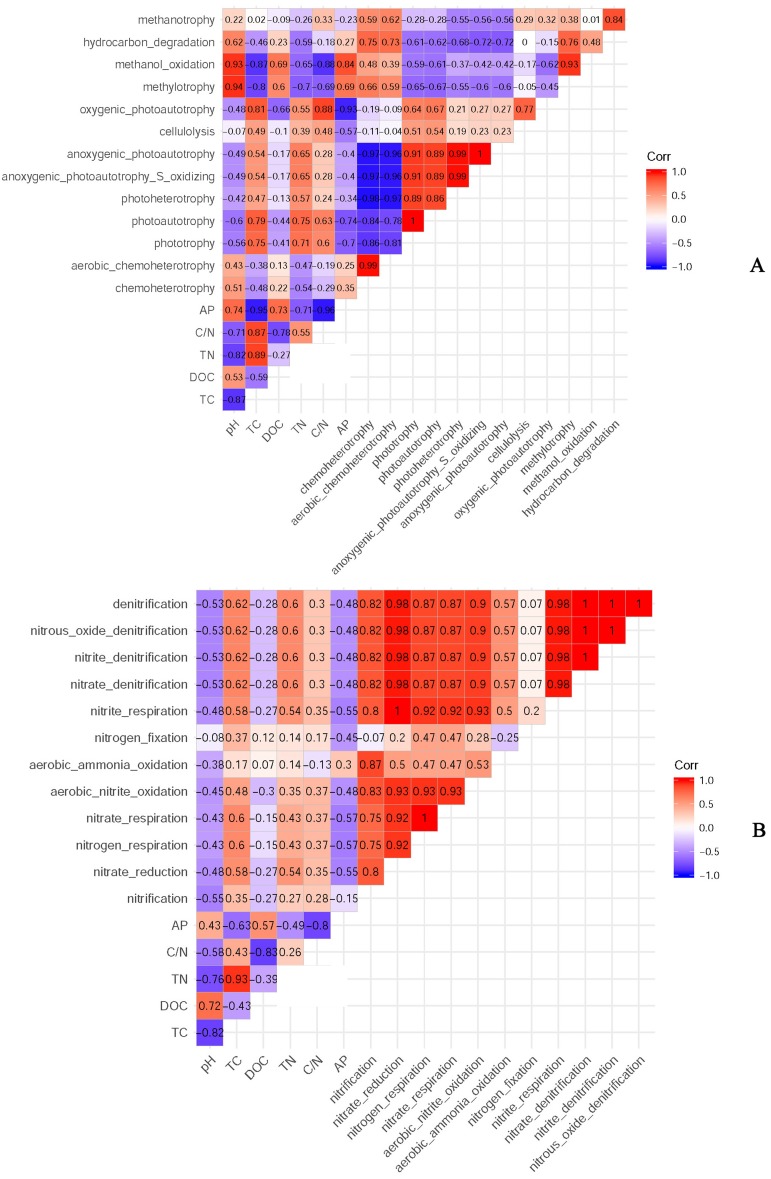
According to the classification annotation results of the 16S rDNA sequences, a total of 51 functional groups were identified using FAPROTAX. These functional groups contained 5063 OTUs, and OTUs per functional group were listed in Table S1. We examined 13 ecological functional groups related to the C cycling, including chemoheterotrophy, aerobic chemoheterotrophy, phototrophy, photoautotrophy, photoheterotrophy, anoxygenic photoautotrophy S oxidizing, anoxygenic photoautotrophy, cellulolysis, oxygenic photoautotrophy, methylotrophy, methanol oxidation, hydrocarbon degradation, and methanotrophy (Table S2). The PCA plot showed that the functional groups related to C cycling in PT and PS were separate from those of PC, especially along PCA1 (Fig. 8A). Additionally, we examined 12 ecological functional groups connected to the N cycling, including nitrification, nitrate reduction, nitrogen respiration, nitrate respiration, aerobic nitrite oxidation, aerobic ammonia oxidation, nitrogen fixation, nitrite respiration, nitrate denitrification, nitrite denitrification, nitrous oxide denitrification, and denitrification (Table S3). The PCA plot showed that the functional groups related to N cycling in PT and PS were separated from those of PC, especially along PCA2 (Fig. 8B), indicating that the functional groups of PC differed from those of PS plus PT. We performed the Spearman’s rank correlation analysis to explore the relationships between the microbial functional groups and the six key environmental variables (Fig. 9). Soil pH value, total C, total N, C/N, and available P were the main factors influencing the functional groups related to C cycling (Fig. 9A). Whereas, total C was the main factor influencing the functional groups related to N cycling (Fig. 9B).
Figure 8. PCA plot of functional groups related to C cycling (A) and N cycling (B).
PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
Figure 9. The relationships between soil bacterial functional groups related to C (A) and N (B) cycles and soil environmental factors.
TC: total C; DOC: dissolved organic carbon; TN: total N; AP: available P.
Discussion
Soil chemical characteristics following afforestation with different tree species
Afforestation with different plantation forests had significant difference in soil conditions. PC had the highest pH value, when compared to PS and PT (Table 2), which was similar to the study demonstrating that the soil in pine stands had a lower pH than the oak and birch tree stands (Yoshimura et al., 2008). This difference might be the result of higher litter acidity in coniferous forests (Augusto et al., 2002). We observed that soil DOC in PC was higher than that in PS and PT. Our results were consistent with a previous study stating that soil organic matter and nitrogen were higher in broadleaf forests than those in coniferous forests (Jiang et al., 2012). Previous findings had established that both tree species and afforestation time dramatically influenced soil characteristics (Kim et al., 2018; Kang et al., 2018). In our study, the C/N values decreased in the order of PT >PS >PC, which was consistent with a previous finding that coniferous forests (pine) soil contained more carbon and had a higher C/N ratio than broadleaf forests (Yoshimura et al., 2008). The potential role of different forest types in variation of soil C/N ratio was also supported by previous finding (Mcgroddy, Daufresne & Hedin, 2004). In summary, the soil characteristics following afforestation with different tree species in the same area exhibited obvious differences, especially between PC and PS plus PT.
The bacterial community diversity response to different plantation forests
Similar to the soil characteristics, the Chao 1 index, and ACE index in PC were significantly lower than those in PS and PT (P < 0.01). Simultaneously, here we found that the Simpson index and Shannon index existed significant differences among different plantation forests (F = 6.89, P = 0.03; F = 5.68, P = 0.04; Table 3). This result might be due to the differences in the chemical compositions and decomposition rate of the litter (Kang et al., 2018). The Spearman’s rank correlations illustrated the Chao 1 index and ACE index existed significantly positively correlated with soil total N, which was similar to the research from northeast China that reported that the H’ value was positively correlated with the total N (Hui et al., 2014). The Simpson index was significantly positively related to soil available P, while, a previous study suggested that there was no significant correlation between diversity indices and P content (Wang et al., 2018). These results verified that there were significant differences in soil bacterial diversity among different forest lands.
The bacterial community compositions response to different plantation forests
The abundances of dominant bacterial phyla varied among the different plantation forests. In our study, Proteobacteria was the most dominant group, which was similar to the findings from Chinese pine plantations on the Loess Plateau (Dang et al., 2017), while, the research from Wulai forest reported Acidobacteria was the dominant member (Lin et al., 2014). Owing to differences in the lifestyles of Proteobacteria and Acidobacteria, they can be used as indicators of nutritional status (Hartman et al., 2008). In our study, the relative abundance of Proteobacteria in PC was significantly higher than that in PS and PT. In addition, the relative abundances of Proteobacteria had significantly positive correlations with DOC content. Our results concur with previous findings establishing that the availability of carbon was positively related with the abundance Proteobacteria (Fierer, Bradford & Jackson, 2007). In our study, Alphaproteobacteria was the dominant taxa at the class level. This is in agreement with the results obtained from a boreal peatland in Central Finland (Sun et al., 2014).
The phylum Acidobacteria is abundant in various soil environments (Zimmermann et al., 2005; Araujo et al., 2012; Meng et al., 2013). In our study, the abundance of Acidobacteria was relatively higher in the soil communities of PS and PT than that in PC, which was consistent with a previous investigation that the relative abundance of Acidobacteria was relatively higher in the coniferous forest than those of broadleaf forest (Christianl et al., 2008). Soil pH is generally considered as a key factor in shaping bacterial community structures (Preem et al., 2012). Research has established that the relative abundance of the Acidobacteria is dramatically associated with acidic soils (Jones et al., 2009), and more specifically, when the pH is lower than 5.5, Acidobacteria abundance increases (Lauber et al., 2009). However, our results found that the soil pH had no relation to the relative abundance of Acidobacteria, which might be due to the narrow range of pH from 5.53 to 5.92 (Table 2). PC stand had the highest abundance of Proteobacteria and lowest abundance of Acidobacteria. In consideration of the comparatively higher relative abundance of Proteobacteria observed in the copiotrophic soils and the relatively higher Acidobacteria abundance obtained in the oligotrophic soils (Fierer, Bradford & Jackson, 2007), we suggested that the PC plantation improved the soil nutrient conditions with lower C/N value. Gemmatimonadetes was the dominant bacterial community in our research, previous research illustrated that Gemmatimonadetes has been found in arid soils, such as grassland, prairie, and pasture soil, as well as pine soils (Debruyn et al., 2011). In our study, the relative abundances of Chloroflexi, and Bacteroidetes were positively correlated with available P, and available P might be one of the important factors influencing the bacterial community. Identically, previous findings have indicated that the phosphorus content has an effect on community structures (Fierer, Bradford & Jackson, 2007; Bergkemper et al., 2016).
The results of clear differentiation provided by the heatmap (Fig. 5) and NMDS (Fig. 6) plots illustrated that significant differences in the bacterial community compositions were observed among PS, PT and PC. The soil bacterial communities of the PT and PS sites were similar to each other, indicating that the hierarchical clustering distance between two coniferous forests was shorter than the distance between the coniferous and broadleaf forests. Our results were agreement with previous study which have established that the compositions of the soil bacterial community in hardwood forest differed from those in conifer forests (Lin et al., 2011; Ushio et al., 2008), which could release different quality and quantity of litter and root exudates (Sauheitl et al., 2010). In addition, the compositions of soil bacteria between PS and PT were also different. These results confirmed our hypothesis that the three different plantation forests harbored different soil bacterial community diversity and structure, suggesting that afforestation tree species had correlation with the soil bacterial community, which was consistent with previous findings (Ren et al., 2016; Gunina et al., 2017).
The bacterial functional groups response to different plantation forests
The C and N cycle in the terrestrial ecosystem and its regulatory mechanism are the hot topics in the science research of soil ecology and global change ecology (Maia et al., 2010). It is known that soil bacterial communities play an important role in biogeochemical cycles (Jenkins et al., 2017). In our study, we examined 13 functional groups related to C cycling. In contrast, a previous study from a temperate deciduous broadleaved forest and a tropical mountain rainforest detected eight ecological functional groups connected with the carbon cycle (Wei et al., 2018). Soil nitrogen fixation, nitrification, denitrification, ammonification, and other major nitrogen transformation processes are mainly mediated by soil bacteria (Yoon et al., 2015). And the soil nitrogen cycle, especially the biological nitrification and denitrification processes, can affect the production and emission of greenhouse gases, such as CO2, CH4, and N2O (Gregorich et al., 2005). To some extent, the denitrifying community of bacteria plays a vital role in the soil nitrogen cycle, and the relative abundances of specific OTUs are more valuable in predicting community function (Bent et al., 2016). In our study, the functional groups of denitrification were significantly higher in the PS than PT and PC (P < 0.05). For soil bacterial function, the functional groups related to C and N cycling in PT and PS were distinctly separate from those of PC, indicating that the functional groups of the broadleaf forest differed from those of the coniferous forests. Different plantation tree species could distinctly affect the community compositions of decomposers (Kubartová et al., 2007). Due to the existence of functional gene redundancy, these functional profiles are observed among bacterial communities (Fierer et al., 2012). Different plantation forests affect soil characteristics (Bhatia, 2008), thereby causing the change in the soil microbial diversity (Nair & Ngouajio, 2012), and functional diversity (Zhang et al., 2007). As a result, we believe our work has broad implications for reforestation in the semi-arid areas.
Conclusions
Our results revealed that soil characteristics after afforestation with different tree species under the same climatic conditions showed dramatic differences, especially between Populus × canadensis Moench and Pinus sylvestris var. mongolica and Pinus tabuliformis. Compared to Pinus sylvestris var. mongolica and Pinus tabuliformis, the plantation of Populus × canadensis Moench increased the soil pH value, DOC content, and soil available P content, while the C/N ratio decreased. Furthermore, the soil bacterial community compositions, diversity, and functions are different among plantation types, especially for Populus × canadensis Moench and Pinus sylvestris var. mongolica plus Pinus tabuliformis.
The bacterial diversity indices and the relative abundances of Proteobacteria, and Chloroflexi in the soil significantly differed among plantation types. The bacterial community compositions and functional groups related to C and N cycling from Pinus sylvestris var. mongolica, and Pinus tabuliformis were grouped tightly, indicating that the phylogenetic distance for microorganisms under different plantation types could be divided into two groups, including Pinus sylvestris var. mongolica, plus Pinus tabuliformis, and Populus × canadensis Moench. Our results highlighted that the soil bacterial community compositions and functions obviously differed following afforestation, especially between Populus × canadensis Moench and Pinus sylvestris var. mongolica and Pinus tabuliformis, which in turn enormously established the correlation between the soil microbial community characteristics and the afforestation tree species. Moreover, the bacterial community structure and functions related to C and N cycling showed consistent differences among different plantation forests following afforestation in the semi-arid areas.
Supplemental Information
The length of the curve reflects the size of the sample sequencing. The longer the curve, the higher the depth of the sequencing, and the greater the possibility of observing higher diversity. The flatness of the curve reflects the impact of the sequencing depth on the diversity of the observed samples. The flatter the curve indicates that the sequencing result is sufficient to reflect the diversity of the current samples, and the further increase of the sequencing depth is unable to detect a large number of new OTU that have not been discovered. On the contrary, it indicates that the diversity is not close to saturation, and further increasing the sequencing depth will help to observe more new OTU. PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
Data are means ± standard error (n = 3). PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis. Different small letters meant significant difference at 0.05 level. Different big letters meant significant difference at 0.01 level.
Data are means ± standard error (n = 3). PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis. Different small letters meant significant difference at 0.05 level. Different big letters meant significant difference at 0.01 level.
Funding Statement
This research was financially supported by the special fund for Forestry Scientific Research in the Public Interest (No.201404303-05), the National Science and Technology Support Program of China (2015BAD07B30103, the Sub-project of the National Key Research and Develepment Program (2017YFC050410501), the Special Fund for Forest Scientific Research in the Public Welfare (201304216), and Cfern & Beijing Techno Solutions Award Funds on Excellent Academic Achievements. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Wenxu Zhu, Email: zhuwx@syau.edu.cn.
Yongbin Zhou, Email: 13998160246@163.com, yyzyb@163.com.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Jiaojiao Deng conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables.
Yan Zhang contributed reagents/materials/analysis tools.
You Yin performed the experiments, prepared figures and/or tables.
Xu Zhu conceived and designed the experiments.
Wenxu Zhu conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Yongbin Zhou performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper.
Data Availability
The following information was supplied regarding data availability:
The bacterial raw data are available at the NCBI: PRJNA495735.
References
- Allington & Valone (2010).Allington GRH, Valone TJ. Reversal of desertification: the role of physical and chemical soil properties. Journal of Arid Environments. 2010;74(8):973–977. doi: 10.1016/j.jaridenv.2009.12.005. [DOI] [Google Scholar]
- Araujo et al. (2012).Araujo JF, De Castro APD, Costa MM, Togawa RC, Júnior GJ, Quirino BF, Bustamante MM, Williamson L, Handelsman J, Krüger RH. Characterization of soil bacterial assemblies in Brazilian Savanna-Like vegetation reveals Acidobacteria dominance. Microbial Ecology. 2012;64(3):760–770. doi: 10.1007/s00248-012-0057-3. [DOI] [PubMed] [Google Scholar]
- Augusto et al. (2002).Augusto L, Ranger J, Dan B, Rothe A. Impact of several common tree species of European temperate forests on soil fertility. Annals of Forest Science. 2002;59(3):233–253. doi: 10.1051/forest:2002020. [DOI] [Google Scholar]
- Banerjee (2016).Banerjee S. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biology Biochemistry. 2016;97:188–198. doi: 10.1016/j.soilbio.2016.03.017. [DOI] [Google Scholar]
- Bao (2000).Bao SD. Soil and agricultural chemistry analysis. China Agriculture Press; BeiJing: 2000. [Google Scholar]
- Becerril-Piña et al. (2015).Becerril-Piña R, Mastachi-Loza CA, González-Sosa E, Díaz-Delgado C, Bâ KM. Assessing desertification risk in the semi-arid highlands of central Mexico. Journal of Arid Environments. 2015;120:4–13. doi: 10.1016/j.jaridenv.2015.04.006. [DOI] [Google Scholar]
- Bent et al. (2016).Bent E, Németh D, Wagner-Riddle C, Dunfield KE. Residue management leading to higher field-scale N2O flux is associated with different soil bacterial nitrifier and denitrifier gene community structures. Applied Soil Ecology. 2016;108:288–299. doi: 10.1016/j.apsoil.2016.09.008. [DOI] [Google Scholar]
- Bergkemper et al. (2016).Bergkemper F, Welzl G, Lang F, Krüger J, Schloter M, Schulz S. The importance of C, N and P as driver for bacterial community structure in German beech dominated forest soils. Journal of Plant Nutrition and Soil Science. 2016;179(4):472–480. doi: 10.1002/jpln.201600077. [DOI] [Google Scholar]
- Bhatia (2008).Bhatia CR. Role of microbial diversity for soil, health and plant nutrition. In: Nautiyal CS, Dion P, editors. Molecular mechanisms of plant and microbe coexistence. vol. 15. Springer; Berlin Heidelberg: 2008. pp. 53–74. (Soil biology). [DOI] [Google Scholar]
- Braak & Smilauer (2002).Braak CJFT, Smilauer P. Ithaca: 2002. http://www.canoco.com . (Microcomputer Power). Biometris (WU MAT) PRI Biometris. [Google Scholar]
- Bridge & Spooner (2001).Bridge P, Spooner B. Soil fungi: diversity and detection. Plant and Soil. 2001;232(1–2):147–154. doi: 10.1023/a:1010346305799. [DOI] [Google Scholar]
- Burton et al. (2010).Burton J, Chen C, Xu Z, Ghadiri H. Soil microbial biomass, activity and community composition in adjacent native and plantation forests of subtropical Australia. Journal of Arid Environments. 2010;10(7):1267–1277. doi: 10.1007/s11368-010-0238-y. [DOI] [Google Scholar]
- Caporaso et al. (2010).Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK. QIIME allows analysis of highthroughput community sequencing data. Nature Methods. 2010;75:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasek et al. (2015).Chasek P, Safriel U, Shikongo S, Vivian FF. Operationalizing Zero Net Land Degradation: the next stage in international efforts to combat desertification? Journal of Arid Environments. 2015;112(Part A):5–13. doi: 10.1016/j.jaridenv.2014.05.020. [DOI] [Google Scholar]
- Chatterjee et al. (2008).Chatterjee A, Vance GF, Pendall E, Stahl PD. Timber harvesting alters soil carbon mineralization and microbial community structure in coniferous forests. Soil Biology Biochemistry. 2008;40(7):1901–1907. doi: 10.1016/j.soilbio.2008.03.018. [DOI] [Google Scholar]
- Christianl et al. (2008).Christianl L, Michaels S, Marka B, Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biology Biochemistry. 2008;40(9):2407–2415. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- Dang et al. (2017).Dang P, Yu X, Le H, Liu J, Shen Z, Zhao Z. Effects of stand age and soil properties on soil bacterial and fungal community composition in Chinese pine plantations on the Loess Plateau. PLOS ONE. 2017;12(10):e0186501. doi: 10.1371/journal.pone.0186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debruyn et al. (2011).Debruyn JM, Nixon LT, Fawaz MN, Johnson AM, Radosevich M. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Applied and Environmental Microbiology. 2011;77(17):6295–6300. doi: 10.1128/AEM.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng et al. (2019).Deng JJ, Yin Y, Luo JY, Zhu WX, Zhou YB. Different revegetation types alter soil physical-chemical characteristics and fungal community in the Baishilazi Nature Reserve. PeerJ. 2019;6:e6251. doi: 10.7717/peerj.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng et al. (2018).Deng JJ, Yin Y, Zhu WX, Zhou YB. Variations in soil bacterial community diversity and structures among different revegetation types. Frontiers in Microbiology. 2018;9:2874. doi: 10.3389/fmicb.2018.02874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng et al. (2017).Deng Q, Mcmahon DE, Xiang Y, Yu CL, Jackson RB, Hui D. A global meta-analysis of soil phosphorus dynamics after afforestation. New Phytologist. 2017;213(1):181–192. doi: 10.1111/nph.14119. [DOI] [PubMed] [Google Scholar]
- Deng et al. (2006).Deng XP, Shan L, Zhang H, Turner NC. Improving agricultural water use efficiency in arid and semiarid areas of China. Agricultural Water Management. 2006;80(1–3):23–40. doi: 10.1016/j.agwat.2005.07.021. [DOI] [Google Scholar]
- Ding et al. (2017).Ding X, Zhang B, Lü X, Wang J, Horwath WR. Parent material and conifer biome influence microbial residue accumulation in forest soils. Soil Biology Biochemistry. 2017;107:1–9. doi: 10.1016/j.soilbio.2016.12.020. [DOI] [Google Scholar]
- Edgar (2013).Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods. 2013;10:996–998. doi: 10.1038/NMETH.2604. [DOI] [PubMed] [Google Scholar]
- Emteryd (1989).Emteryd O. Swedish University of Agricultural Sciences. Department of Forest Site Research; Uppsala: 1989. Chemical and physical analysis of inorganic nutrients in plant, soil, water and air. [Google Scholar]
- Fan et al. (2014).Fan B, Guo L, Li N, Chen J, Lin H, Zhang X, Shen M, Rao Y, Wang C, Ma L. Earlier vegetation green-up has reduced spring dust storms. Scientific Reports. 2014;4:6749. doi: 10.1038/srep06749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer, Bradford & Jackson (2007).Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88(6):1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Fierer et al. (2012).Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. Isme Journal. 2012;6(5):1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado & Macedo (2008).Furtado C, Macedo TBS. UN convention to combat desertification: recent developments. Max Planck Yearbook of United Nations Law Online. 2008;19(4):814–818. doi: 10.1016/S1003-6326(08)60356-8. [DOI] [Google Scholar]
- Gao et al. (2002).Gao Y, Qiu GY, Shimizu H, Tobe K, Sun B, Wang J. A 10-year study on techniques for vegetation restoration in a desertified Salt Lake area. Journal of Arid Environments. 2002;524(4):483–497. doi: 10.1006/jare.2002.1013. [DOI] [Google Scholar]
- Ge et al. (2015).Ge X, Li Y, Luloff AE, Dong K, Xiao J. Effect of agricultural economic growth on sandy desertification in Horqin Sandy Land. Ecological Economic. 2015;119:53–63. doi: 10.1016/j.ecolecon.2015.08.006. [DOI] [Google Scholar]
- Gong et al. (2009).Gong W, Yan XY, Wang JY, Hu TX, Gong YB. Long-term manuring and fertilization effects on soil organic carbon pools under a wheat–maize cropping system in North China Plain. Plant and Soil. 2009;314(1–2):67–76. doi: 10.1007/s11104-008-9705-2. [DOI] [Google Scholar]
- Gregorich et al. (2005).Gregorich EG, Rochette P, Vandenbygaart AJ, Angers D. Greenhouse gas contributions of agricultural soils and potential mitigation practices in Eastern Canada. Soil and Tillage Research. 2005;83(1):53–72. doi: 10.1016/j.still.2005.02.009. [DOI] [Google Scholar]
- Gunina et al. (2017).Gunina A, Smith AR, Godbold DL, Jones DL, Kuzyakov Y. Response of soil microbial community to afforestation with pure and mixed species. Plant and Soil. 2017;412(1–2):357–368. doi: 10.1007/s11104-016-3073-0. [DOI] [Google Scholar]
- Hartman et al. (2008).Hartman WH, Richardson CJ, Vilgalys R, Bruland GL. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(46):17842–17847. doi: 10.1073/pnas.0808254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui et al. (2014).Hui L, Ye D, Wang X, Settles ML, Wang J, Hao ZQ, Zhou LS, Dong P, Jiang Y, Ma ZS. Soil bacterial communities of different natural forest types in Northeast China. Plant and Soil. 2014;383(1–2):203–216. doi: 10.1007/s11104-014-2165-y. [DOI] [Google Scholar]
- Jenkins et al. (2017).Jenkins JR, Viger M, Arnold EC, Harris ZM, Ventura M, Miglietta F, Cyrii G, Edwards RJ, Rumpel C, Fornasier F, Zavalloni C, Tonon G, Alberti G, Taylor G. Biochar alters the soil microbiome and soil function: results of next-generation amplicon sequencing across Europe. GCB Bioenergy. 2017;9(3):519–621. doi: 10.1111/gcbb.12371. [DOI] [Google Scholar]
- Jiang et al. (2012).Jiang Y, Chen C, Xu Z, Liu Y. Effects of single and mixed species forest ecosystems on diversity and function of soil microbial community in subtropical China. Journal of Soils and Sediments. 2012;12(2):228–240. doi: 10.1007/s11368-011-0442-4. [DOI] [Google Scholar]
- Jones et al. (2009).Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. The ISME Journal. 2009;3(4):442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg et al. (2013).Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 2013;22(21):5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Kang et al. (2018).Kang H, Gao H, Yu W, Yi Y, Wang Y, Ning M. Changes in soil microbial community structure and function after afforestation depend on species and age: case study in a subtropical alluvial island. Science of The Total Environment. 2018;625:1423–1432. doi: 10.1016/j.scitotenv.2017.12.180. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2018).Kim S, Zang H, Mortimer P, Shi LL, Li YJ, Xu JC, Ostermann A. Tree species and recovery time drives soil restoration after mining: a Chronosequence Study. Land Degradation and Development. 2018;29(6):1738–1747. doi: 10.1002/ldr.2951. [DOI] [Google Scholar]
- Kubartová et al. (2007).Kubartová A, Moukoumi J, Béguiristain T, Ranger J, Berthelin J. Microbial diversity during cellulose decomposition in different forest stands: I. microbial communities and environmental conditions. Microbial Ecology. 2007;54(3):393–405. doi: 10.1007/s00248-007-9286-2. [DOI] [PubMed] [Google Scholar]
- Lauber et al. (2009).Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-Based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology. 2009;75(15):5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2018).Li D, Xu D, Wang Z, Ding X, Song A. Ecological compensation for desertification control: a review. Journal of Geographical Sciences. 2018;28(3):367–384. doi: 10.1007/s11442-018-1478-9. [DOI] [Google Scholar]
- Li et al. (2012).Li MM, Liu A, Zou CJ, Xu WD, Shimizu H, Wang KR. An overview of the “Three-North” Shelterbelt project in China. Forest Ecosystems. 2012;14(1):70–79. doi: 10.1007/s11632-012-0108-3. [DOI] [Google Scholar]
- Li et al. (2004).Li XR, Ma FY, Xiao HL, Wang XP, Kim KC. Long-term effects of revegetation on soil water content of sand dunes in arid region of Northern China. Journal of Arid Environments. 2004;57(1):1–16. doi: 10.1016/S0140-1963(03)00089-2. [DOI] [Google Scholar]
- Lin et al. (2014).Lin YT, Hu HW, Whitman WB, Coleman DC, Chiu CY. Comparison of soil bacterial communities in a natural hardwood forest and coniferous plantations in perhumid subtropical low mountains. Botanical Studies. 2014;55:31–39. doi: 10.1186/1999-3110-55-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin et al. (2011).Lin YT, Jangid K, Whitman WB, Coleman DC, Chiu CY. Change in bacterial community structure in response to disturbance of natural hardwood and secondary coniferous forest soils in Central Taiwan. Microbial Ecology. 2011;61(2):429–437. doi: 10.1007/s00248-010-9748-9. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2018).Liu J, Dang P, Gao Y, Zhu HL, Zhu HN, Zhao F, Zhao Z. Effects of tree species and soil properties on the composition and diversity of the soil bacterial community following afforestation. Forest Ecology and Management. 2018;427(1):342–349. doi: 10.1016/j.foreco.2018.06.017. [DOI] [Google Scholar]
- Liu & Diamond (2005).Liu J, Diamond J. China’s environment in a globalizing world. Nature. 2005;435(7046):1179–1186. doi: 10.1038/4351179a. [DOI] [PubMed] [Google Scholar]
- Louca et al. (2017).Louca S, Jacques SMS, Pires APF, Leal JS, González AL, Doebeli M, Farjalla VF. Functional structure of the bromeliad tank microbiome is strongly shaped by local geochemical conditions. Environmental Microbiology. 2017;19(8):3132–3151. doi: 10.1111/1462-2920.13788. [DOI] [PubMed] [Google Scholar]
- Louca, Parfrey & Doebeli (2016).Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353(6305):1272–1277. doi: 10.1126/science.aaf4507. [DOI] [PubMed] [Google Scholar]
- Lozupone et al. (2007).Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Applied and Environmental Microbiology. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2017).Lu Z, Wei Y, Li Z, Guo X, Zhou Y. Characteristics of sap flow and its influencing factors of Pinus sylvestris var. mongolica insandy land of Northwest Liaoning. Chinese Journal of Ecology. 2017;36(11):3182–3189. doi: 10.13292/j.1000-4890.201711.020. [DOI] [Google Scholar]
- Lv et al. (2018).Lv G, Zhai JX, Li YX, Wang L, Wang YC. Soil infiltration characteristics of different plant community in sandy land of northwestern Liaoning. Agricultural Research in the Arid Areas. 2018;36(4):133–139. [Google Scholar]
- Maia et al. (2010).Maia SMF, Ogle SM, Cerri CEP, Cerri CC. Soil organic carbon stock change due to land use activity along the agricultural frontier of the southwestern Amazon, Brazil, between 1970 and 2002. Global Change Biology. 2010;16(10):2775–2788. doi: 10.1111/j.1365-2486.2009.02105.x. [DOI] [Google Scholar]
- Martã-Nez-Valderrama et al. (2016).Martã-Nez-Valderrama J, Ibáñez J, Del Barrio G, Sanjuán ME, Alcalá FJ, Martínez-Vicente S, Ruiz A, Puigdefábregas J. Present and future of desertification in Spain: implementation of a surveillance system to prevent land degradation. Science of the Total Environment. 2016;563-564:169–178. doi: 10.1016/j.scitotenv.2016.04.065. [DOI] [PubMed] [Google Scholar]
- Mcgroddy, Daufresne & Hedin (2004).Mcgroddy ME, Daufresne T, Hedin LO. Scaling of C:N:P stoichiometry in forests worldwide: implications of terrestrial redfield-type ratios. Ecology. 2004;85(9):2390–2401. doi: 10.1890/03-0351. [DOI] [Google Scholar]
- Meng et al. (2013).Meng H, Li K, Nie M, Wan JR, Quan ZX, Fang CM, Chen JK, Gu LD, Li B. Responses of bacterial and fungal communities to an elevation gradient in a subtropical montane forest of China. Applied Microbiology and Biotechnology. 2013;97(5):2219–2230. doi: 10.1007/s00253-012-4063-7. [DOI] [PubMed] [Google Scholar]
- Nair & Ngouajio (2012).Nair A, Ngouajio M. Soil microbial biomass, functional microbial diversity, and nematode community structure as affected by cover crops and compost in an organic vegetable production system. Applied Soil Ecology. 2012;58:45–55. doi: 10.1016/j.apsoil.2012.03.008. [DOI] [Google Scholar]
- Nunezmir et al. (2015).Nunezmir GC, Iannone BVI, CurtisK, Fei SL, Oliet JA. Evaluating the evolution of forest restoration research in a changing world: a big literature review. New Forest. 2015;46(5–6):669–682. doi: 10.1007/s11056-015-9503-7. [DOI] [Google Scholar]
- Parks et al. (2014).Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng et al. (2014).Peng SS, Piao S, Zeng Z, Ciais P, Zhou L, Li LZ, Li LZX, Myneni RB, Yin Y, Zeng H. Afforestation in China cools local land surface temperature. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(8):2915–2919. doi: 10.1073/pnas.1315126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao et al. (2005).Piao S, Fang J, Liu H, Zhu B. NDVI-indicated decline in desertification in China in the past two decades. Geophysical Research Letters. 2005;32(6):347–354. doi: 10.1029/2004gl021764. [DOI] [Google Scholar]
- Preem et al. (2012).Preem JK, Truu J, Truu M, Ülo M, Oopkaup K, Lõhmus K, Helmiisaari HS, Uri V, Zobel M. Bacterial community structure and its relationship to soil physico-chemical characteristics in alder stands with different management histories. Ecological Engineering. 2012;49(4):10–17. doi: 10.1016/j.ecoleng.2012.08.034. [DOI] [Google Scholar]
- Qadir, Qureshi & Cheraghi (2010).Qadir M, Qureshi AS, Cheraghi SAM. Extent and characterisation of salt-affected soils in Iran and strategies for their amelioration and management. Land Degradation and Development. 2010;19(2):214–227. doi: 10.1002/ldr.818. [DOI] [Google Scholar]
- Qiu et al. (2018).Qiu K, Xie Y, Xu D, Pott R. Ecosystem functions including soil organic carbon, total nitrogen and available potassium are crucial for vegetation recovery. Scientific Reports. 2018;8(1):7607. doi: 10.1038/s41598-018-25875-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009).R Development Core Team . R Foundation for Statistical Computing; Vienna: 2009. pp. 12–21. [Google Scholar]
- Ren et al. (2016).Ren C, Zhao F, Kang D, Yang G, Han X, Tong X, Feng Y, Ren GX. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. Forest Ecology and Management. 2016;376:59–66. doi: 10.1016/j.foreco.2016.06.004. [DOI] [Google Scholar]
- Reynolds et al. (2007).Reynolds JF, Smith DM, Lambin EF, Turner BL, Mortimore M, Batterbury SP, Downing TE, Dowlatabadi H, Fernández RJ, Herrick JE, Huber-Sannwald E, Jiang H, Leemans R, Lynam T, Maestre FT, Ayarza M, Walker B. Global Desertification: building a science for dryland development. Science. 2007;316(5826):847–851. doi: 10.1126/science.1131634. [DOI] [PubMed] [Google Scholar]
- Ritz et al. (2009).Ritz K, Black HIJ, Campbell CD, Harris JA, Wood C. Selecting biological indicators for monitoring soils: a framework for balancing scientific and technical opinion to assist policy development. Ecological Indicators. 2009;9(6):1212–1221. doi: 10.1016/j.ecolind.2009.02.009. [DOI] [Google Scholar]
- Salvati et al. (2015).Salvati L, Mavrakis A, Colantoni A, Colantoni A, Mancino G, Ferrara A. Complex Adaptive Systems, soil degradation and land sensitivity to desertification: a multivariate assessment of Italian agro-forest landscape. Science of The Total Environment. 2015;521–522:235–245. doi: 10.1016/j.scitotenv.2015.03.094. [DOI] [PubMed] [Google Scholar]
- Sauheitl et al. (2010).Sauheitl L, Glaser B, Dippold M, Leiber K, Weigelt A. Amino acid fingerprint of a grassland soil reflects changes in plant species richness. Plant and Soil. 2010;334(1–2):353–363. doi: 10.1007/s11104-010-0387-1. [DOI] [Google Scholar]
- Schrumpf et al. (2011).Schrumpf M, Schulze ED, Kaiser K, Schumacher J. How accurately can soil organic carbon stocks and stock changes be quantified by soil inventories? Biogeosciences. 2011;8:1193–1212. doi: 10.5194/bg-8-1193-2011. [DOI] [Google Scholar]
- State Forest Administration (2010).State Forest Administration The seventh national forest resources inventory and the forest resources conditions, China. Forest Resources Management. 2010;(1):1–8. [Google Scholar]
- Sun et al. (2014).Sun H, Terhonen E, Koskinen K, Paulin L, Kasanen R, Asiegbua FO. Bacterial diversity and community structure along different peat soils in boreal forest. Applied Soil Ecology. 2014;74:37–45. doi: 10.1016/j.apsoil.2013.09.010. [DOI] [Google Scholar]
- Sutton et al. (2016).Sutton PC, Anderson SJ, Costanza R, Kubiszewski I. The ecological economics of land degradation: impacts on ecosystem service values. Ecological Economics. 2016;129:182–192. doi: 10.1016/j.ecolecon.2016.06.016. [DOI] [Google Scholar]
- Tan & Li (2015).Tan M, Li X. Does the Green Great Wall effectively decrease dust storm intensity in China? A study based on NOAA NDVI and weather station data. Land Use Policy. 2015;43:42–47. doi: 10.1016/j.landusepol.2014.10.017. [DOI] [Google Scholar]
- Torres et al. (2015).Torres L, Abraham EM, Rubio C, Barbero-Sierra C, Ruiz-Pérez M. Desertification research in Argentina. Land Degradation and Development. 2015;26(5):433–440. doi: 10.1002/ldr.2392. [DOI] [Google Scholar]
- Ushio et al. (2008).Ushio M, Wagai R, Balser TC, Kitayama K. Variations in the soil microbial community composition of a tropical montane forest ecosystem: does tree species matter? Soil Biology Biochemistry. 2008;40(10):2699–2702. doi: 10.1016/j.soilbio.2008.06.023. [DOI] [Google Scholar]
- Van der Heijen, Bardgett & Straalen (2010).Van der Heijen MGA, Bardgett RD, Van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecology Letters. 2010;11(3):296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- Verón & Paruelo (2010).Verón SR, Paruelo JM. Desertification alters the response of vegetation to changes in precipitation. Journal of Applied Ecology. 2010;47(6):1233–1241. doi: 10.1111/j.1365-2664.2010.01883.x. [DOI] [Google Scholar]
- Wang et al. (2014).Wang K, Wang D, Zhang C, Chen X, Song L. Effects of shelterbelt on soil physical and chemical properties in northwestern Liaoning Province. Journal of Northeast Forestry University. 2014;42:77–79+99. [Google Scholar]
- Wang et al. (2016).Wang YW, Chai Q, Ouyang XZ, Luo L. Soil microbial populations and enzyme activities under different sand-fixation forests in oasis-desert ecotone of Minqin Desert. Arid Land Geography. 2016;39:104–111. [Google Scholar]
- Wang et al. (2018).Wang Q, Wang C, Yu WW, Turak A, Chen D, Huang Y, Ao JH, Jiang Y, Huang ZR. Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese Fir Plantations. Frontiers in Microbiology. 2018;9:1543. doi: 10.3389/fmicb.2018.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2018).Wei H, Peng C, Yang B, Song H, Li Q, Jiang L, Wei G, Wang K, Wang H, Liu S, Chen D, Li Y, Wang M. Contrasting soil bacterial community, diversity, and function in two forests in China. Frontiers in Microbiology. 2018;9:1693. doi: 10.3389/fmicb.2018.01693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijitkosum (2016).Wijitkosum S. The impact of land use and spatial changes on desertification risk in degraded areas in Thailand. Sustainable Environment Research. 2016;26(2):84–92. doi: 10.1016/j.serj.2015.11.004. [DOI] [Google Scholar]
- Wu et al. (2019).Wu SH, Huang BH, Gao J, Wang SQ, Liao PC. The effects of afforestation on soil bacterial communities in temperate grassland are modulated by soil chemical properties. PeerJ. 2019;7:e6147. doi: 10.7717/peerj.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2018).Yang T, Ala M, Zhang Y, Wu J, Wang A. Characteristics of soil moisture under different vegetation coverage in Horqin Sandy Land, northern China. PLOS ONE. 2018;13(6):e019880. doi: 10.1371/journal.pone.0198805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon et al. (2015).Yoon S, Cruz-García C, Sanford R, Ritalahti KM, Löffler FE. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3(-)/NO2(-) reduction pathways in Shewanella loihica strain PV-4. The ISME Journal. 2015;9(5):1093–1104. doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura et al. (2008).Yoshimura C, Gessner M, Tockner K, Furumai H. Chemical properties, microbial respiration, and decomposition of coarse and fine particulate organic matter. Journal of the North American Benthological Society. 2008;27(3):664–673. doi: 10.1899/07-106.1. [DOI] [Google Scholar]
- Zaura et al. (2009).Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy core microbiome of oral microbial communities. BMC Microbiology. 2009;9(1):259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang F, Xing Z, Rees HW, Dong Y, Li S, Meng FR. Assessment of effects of two runoff control engineering practices on soil water and plant growth for afforestation in a semi-arid area after 10 years. Ecological Engineering. 2014;64:430–442. doi: 10.1016/j.ecoleng.2013.12.024. [DOI] [Google Scholar]
- Zhang et al. (2016).Zhang Y, Peng C, Li W, Tian L, Zhu Q, Chen H, Fang X, Zhang L, Mu X, Li Z, Li S, Yang Y, Wang J, Xiao X. Multiple afforestation programs accelerate the greenness in the ‘Three North’ region of China from 1982 to 2013. Ecological Indicators. 2016;61(Part 2):404–412. doi: 10.1016/j.ecolind.2015.09.041. [DOI] [Google Scholar]
- Zhang et al. (2007).Zhang Y, Zhang X, Liu X, Xiao Y, Qu L, Wu L, Zhou J. Microarray-based analysis of changes in diversity of microbial genes involved in organic carbon decomposition following land use/cover changes. FEMS Microbiology Letters. 2007;266(2):144–151. doi: 10.1111/j.1574-6968.2006.00511.x. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2014).Zhao HL, Li J, Liu RT, Zhou RL, Qu H. Effects of desertification on temporal and spatial distribution of soil macro-arthropods in Horqin sandy grassland, Inner Mongolia. Geoderma. 2014;223–225:62–67. doi: 10.1016/j.geoderma.2014.01.026. [DOI] [Google Scholar]
- Zhao et al. (2018).Zhao H, Li X, Zhang Z, Zhao Y, Chen P, Zhu YW. Drivers and assemblies of soil eukaryotic microbes among different soil habitat types in a semi-arid mountain in China. PeerJ. 2018;6:e6042. doi: 10.7717/peerj.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2017).Zhao H, Li X, Zhang Z, Zhao Y, Yang J, Zhu Y. Species diversity and drivers of arbuscular mycorrhizal fungal communities in a semi-arid mountain in China. PeerJ. 2017;5:e4155. doi: 10.7717/peerj.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Yan & Shang (2015).Zhong Y, Yan W, Shang GZ. Impact of long-term N additions upon coupling between soil microbial community structure and activity. and nutrient-use efficiencies. Soil Biology and Biochemistry. 2015;91:151–159. doi: 10.1016/j.soilbio.2015.08.030. [DOI] [Google Scholar]
- Zhou et al. (2014).Zhou W, Lewis BJ, Wu S, Yu D, Zhou L, Wei YW, Dai LM. Biomass carbon storage and its sequestration potential of afforestation under natural forest protection program in China. Chinese Geographical Science. 2014;24(4):406–413. doi: 10.1007/s11769-014-0702-5. [DOI] [Google Scholar]
- Zhou et al. (2016).Zhou YB, Guo XW, Wei YW, Wang X, Li J, Li ZY, Zheng ZR. The vertical distribution of soil carbon, nitrogen and phosphorus for typical plantations in the Semi-arid Areas of Northwest Liaoning. Journal of Shenyang Agricultural University. 2016;47:418–424. [Google Scholar]
- Zimmermann et al. (2005).Zimmermann J, Gonzalez JM, Saiz-Jimenez C, Ludwig W. Detection and phylogenetic relationships of highly diverse uncultured Acidobacterial communities in Altamira Cave using 23S rRNA sequence analyses. Geomicrobiology Journal. 2005;22(7–8):379–388. doi: 10.1080/01490450500248986. [DOI] [Google Scholar]
- Zou et al. (2002).Zou XY, Li S, Zhang CL, Dong GR, Dong YX, Yan P. Desertification and control plan in the Tibet Autonomous Region of China. Journal of Arid Environments. 2002;51(2):183–198. doi: 10.1006/jare.2001.0943. [DOI] [Google Scholar]
- Zuo et al. (2008).Zuo X, Zhao H, Zhao X, Guo Y, Li Y, Luo Y. Plant distribution at the mobile dune scale and its relevance to soil properties and topographic features. Environmental Geology. 2008;54(5):1111–1120. doi: 10.1007/s00254-007-1104-0. [DOI] [Google Scholar]
- Zuo et al. (2009).Zuo X, Zhao X, Zhao H, Zhang T, Guo Y, Li YQ, Huang YX. Spatial heterogeneity of soil properties and vegetation—soil relationships following vegetation restoration of mobile dunes in Horqin Sandy Land, Northern China. Plant and Soil. 2009;318(1–2):53–167. doi: 10.1007/s11104-008-9826-7. [DOI] [Google Scholar]
- Zuo et al. (2012).Zuo X, Zhao X, Zhao H, Zhang T, Li Y, Li YQ, Wang S, Li W, Powers R. Scale dependent effects of environmental factors on vegetation pattern and composition in Horqin Sandy Land Northern China. Geoderma. 2012;173–174(1–9):1–9. doi: 10.1016/j.geoderma.2011.10.003. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The length of the curve reflects the size of the sample sequencing. The longer the curve, the higher the depth of the sequencing, and the greater the possibility of observing higher diversity. The flatness of the curve reflects the impact of the sequencing depth on the diversity of the observed samples. The flatter the curve indicates that the sequencing result is sufficient to reflect the diversity of the current samples, and the further increase of the sequencing depth is unable to detect a large number of new OTU that have not been discovered. On the contrary, it indicates that the diversity is not close to saturation, and further increasing the sequencing depth will help to observe more new OTU. PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis.
Data are means ± standard error (n = 3). PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis. Different small letters meant significant difference at 0.05 level. Different big letters meant significant difference at 0.01 level.
Data are means ± standard error (n = 3). PC: Populus × canadensis Moench; PS: Pinus sylvestris var. mongolica; PT: Pinus tabuliformis. Different small letters meant significant difference at 0.05 level. Different big letters meant significant difference at 0.01 level.
Data Availability Statement
The following information was supplied regarding data availability:
The bacterial raw data are available at the NCBI: PRJNA495735.