Abstract
The hMRI toolbox is an open-source toolbox for the calculation of quantitative MRI parameter maps from a series of weighted imaging data, and optionally additional calibration data. The multi-parameter mapping (MPM) protocol, incorporating calibration data to correct for spatial variation in the scanner's transmit and receive fields, is the most complete protocol that can be handled by the toolbox. Here we present a dataset acquired with such a full MPM protocol, which is made freely available to be used as a tutorial by following instructions provided on the associated toolbox wiki pages, which can be found at http://hMRI.info, and following the theory described in: hMRI – A toolbox for quantitative MRI in neuroscience and clinical research [1].
Keywords: hMRI, hMRI toolbox, qMRI, Quantitative MRI, Analysis tools, qMRI software
Specifications Table
| Subject area | Neuroimaging |
| More specific subject area | Quantitative MRI, Multi-Parameter Mapping (MPM) |
| Type of data | In vivo MRI data (Fig. 1, Fig. 2, Table 1) |
| How data was acquired | 3T MRI, Siemens Prisma |
| Data format | NIfTI image volumes after defacing for anonymization (only) |
| Experimental factors | A deliberate motion was performed during the experiment after acquisition 9 and returned to approximately the original position after acquisition 12. |
| Experimental features | No special treatment was performed except to anonymise via SPM∖Util∖De-face Images. |
| Data source location | Wellcome Centre for Human Neuroimaging, London, UK |
| Data accessibility | Data is available at http://hMRI.info |
| Related research article | Tabelow K, Balteau E, Ashburner J, Callaghan MF, Draganski B, Helms G, Kherif F, Leutritz T, Lutti A, Phillips C, Reimer E, Ruthotto L, Seif M, Weiskopf N, Ziegler G, Mohammadi S (2019). hMRI - A toolbox for quantitative MRI in neuroscience and clinical research. NeuroImage, in press [1]. |
Value of the data
|
1. Data
This dataset is comprised of imaging volumes (full list in Table 1) acquired with the multi-parameter mapping quantitative MRI protocol. It consists of calibration data to map the transmit field (series 4), main B0 field (series 5 and 6) and the net receive field (series 7, 8, 10, 11, 13 and 14). It also consists of multi-echo volumes with variable flip angle (series 9 and 15) and additional magnetisation transfer (MT) weighting (series 12).
Table 1.
Image series number, showing the chronology of the acquisition, together with the sequence name and the data description.
| Image Series No. | Sequence Name | Description |
|---|---|---|
| 4 | mfc_seste_b1map_v1e | B1+ Mapping Data |
| 5 | gre_field_mapping_1acq_rl | B0 Mapping Magnitude |
| 6 | gre_field_mapping_1acq_rl | B0 Mapping Phase Difference |
| 7 | mfc_smaps_v1a_Array | Net Receive Sensitivity Mapping of Array |
| 8 | mfc_smaps_v1a_QBC | Net Receive Sensitivity Mapping of Body Coil |
| 9 | pdw_mfc_3dflash_v1i_R4 | Lower flip angle multi-echo FLASH |
| Participant moved to new position via primary rotation about z | ||
| 10 | mfc_smaps_v1a_Array | Net Receive Sensitivity Mapping of Array |
| 11 | mfc_smaps_v1a_QBC | Net Receive Sensitivity Mapping of Body Coil |
| 12 | mtw_mfc_3dflash_v1i_R4 | FLASH acquisition with MT pre-pulse |
| Participant returned to approximate alignment with the original position | ||
| 13 | mfc_smaps_v1a_Array | Net Receive Sensitivity Mapping of Array |
| 14 | mfc_smaps_v1a_QBC | Net Receive Sensitivity Mapping of Body Coil |
| 15 | t1w_mfc_3dflash_v1i_R4 | Higher flip angle multi-echo FLASH |
A tutorial describing how to process the dataset with the hMRI toolbox is available at https://github.com/hMRI-toolbox/wiki/MapCreation#example. From these data maps of the effective transverse relaxation rate (R2*), the longitudinal relaxation rate (R1), proton density (PD) and magnetisation transfer saturation (MT) can be generated.
The acquisition of these data was approved by the local ethics committee and informed written consent was obtained from the participant prior to scanning. All data were acquired on a whole body 3T Prisma system (Siemens Healthineers, Erlangen, Germany). The data were acquired using the body coil for signal transmission and a 64 channel coil for signal reception.
2. Experimental design, materials, and methods
A summary of the data acquisition and experimental design is given in Table 1. The participant was centred within the head coil at the outset of the exam.
2.1. B1+ mapping data (image series No.4)
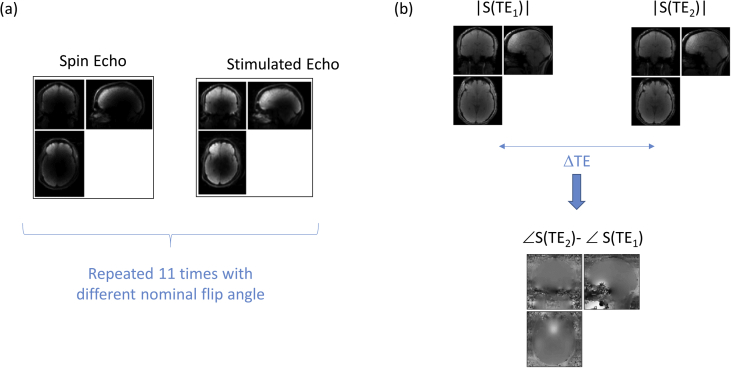
The scanning session began by acquiring calibration data (Fig. 1a, mfc_seste_b1map_v1e_004) to measure B1+ following a previously published method [2], [3]. Eleven spin-echo and stimulated-echo pairs were acquired with the nominal flip angle (i.e. α in an α-2α-α sequence) varying from 115° to 65° in 5° decrements (Fig. 1a). These data were acquired with 4mm isotropic resolution using a 3D-EPI readout with a 0.5 ms echo spacing. The sequence had an echo time of 39.06 ms, a mixing time of 33.8 ms and a repetition time (TR) of 500 ms. The field of view (FoV) was 256 (anterior-posterior, AP) x 192 (right-left, RL) x 192 (head-foot, HF) mm3. Partially parallel imaging with a speed up factor of 2 was used in each phase-encoded direction. A fully sampled volume was acquired at the outset to serve as the auto-calibrating lines for subsequent reconstruction of the aliased data using the GRAPPA algorithm [4] as implemented in the vendor's software. The primary phase-encoded direction of the EPI readout was right-left. The total acquisition time was 3 minutes.
Fig. 1.
Calibration data to map the transmit field, B1+. Eleven spin echo and stimulated echo pairs were acquired (for robustness to low SNR regions) with different nominal flip angle (a). These data were acquired with an EPI readout. To correct for resulting geometric image distortions, additional calibration data mapping spatial inhomogeneity in the main magnetic field were also acquired (b).
2.2. B0 mapping (image series No.5–6)
To correct for geometric distortions in the B1+ mapping data caused by the low bandwidth in the phase-encoded direction of the EPI readout (RL), additional data were acquired to map the inhomogeneity of the B0 field (gre_field_mapping_1acq_rl) and subsequently used to apply distortion correction on the B1+ calibration data. These data were acquired with 3mm effective isotropic resolution using a multi-echo gradient echo sequence with an excitation flip angle of 90°, a TR of 1.02s and a bandwidth of 260 Hz/pixel. Magnitude images with echo times of 10.00 and 12.46 ms respectively (gre_field_mapping_1acq_rl_0005) were reconstructed together with their phase difference (gre_field_mapping_1acq_rl_0006) by the vendor's software (Fig. 1b). The acquisition time was 2 minutes 14 seconds.
2.3. FLASH acquisitions (image series No 9, 12 and 15)
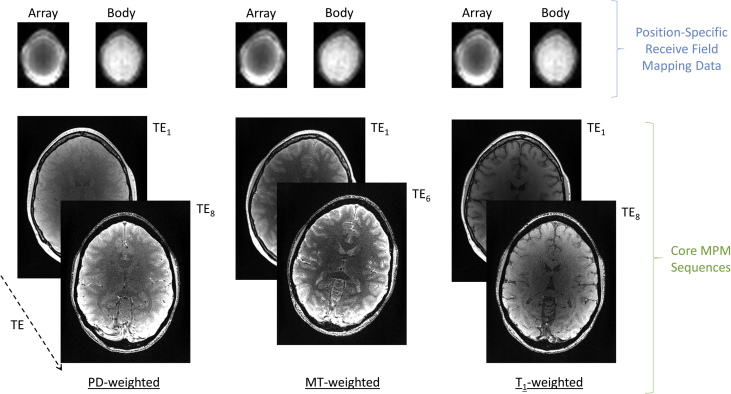
These were followed by the acquisition of spoiled multi-echo 3D fast low angle shot (FLASH) acquisitions with predominantly PD, MT or T1 weighting (Fig. 2, pdw_mfc_3dflash_v1i_R4_0009, mtw_mfc_3dflash_v1i_R4_0012 and t1w_mfc_3dflash_v1i_R4_0015 respectively). Each multi-echo FLASH volume had a TR of 25 ms.
Fig. 2.
The main MPM protocol consists of three high resolution, multi-echo, 3D FLASH volumes with proton density (PD), magnetisation transfer (MT) and T1 weighting. Prior to each high resolution acquisition shorter, low resolution, single echo acquisitions were acquired to map the net receive field sensitivity of the array coil, which will vary if inter-scan motion occurs (c.f. PD-weighted v's MT-weighted).
The PD-weighting was achieved with an excitation flip angle of 6°. The flip angle was increased to 21° to achieve T1-weighting. MT-weighting was achieved by applying a Gaussian RF pulse 2 kHz off resonance prior to excitation with a flip angle of 6°. The off-resonance MT saturation pulse was 4 ms in duration and had a nominal flip angle of 220°.
Each of these volumes were acquired with whole-brain coverage using a FoV of 256 (HF) x 224 (AP) x 179 (RL) mm3. Gradient echoes were acquired with alternating readout gradient polarity at eight equidistant echo times ranging from 2.30 to 18.40 ms in steps of 2.30 ms using a readout bandwidth of 488Hz/pixel. Only six echoes were acquired for the MT-weighted acquisition in order to maintain a TR of 25 ms for all FLASH volumes. To maximise spoiling of the transverse magnetisation, each FLASH volume was acquired with RF spoiling using a linear phase increment of 137°. In addition, a spoiling gradient moment, which imposed a 6π dephasing moment across a voxel dimension, was applied along the readout direction after the last echo had been acquired. To accelerate the data acquisition, partially parallel imaging was employed in each phase-encoded direction (AP and RL) with a speed up factor of 2 and forty integrated auto-calibrating lines in each direction for subsequent reconstruction with GRAPPA [4]. The acquisition time for each FLASH volume was 7 minutes 8 seconds. Each dataset was acquired with a 30° rotation of the sagittal plane so that any eye-related motion artefact propagated to the neck/inferior cerebellum rather than the cortex.
2.4. Net receive sensitivity mapping of array (image series no. 7, 10 and 13) and body coil (image series no. 8, 11 and 14)
Prior to the acquisition of each FLASH volume described above, two additional unaccelerated, low resolution (8 mm isotropic) volumes were acquired with the same FoV (Fig. 2). A single echo, with a TE of 2.20 ms, was acquired in each case using a 6° flip angle and a TR of 6.00 ms. The acquisition time for each of these calibration volumes was 5.90 seconds. The first was obtained using the 64 channel coil for signal reception (mfc_smaps_v1a_Array*), while the second was acquired using the body coil for signal reception (mfc_smaps_v1a_QBC*). These data were acquired to correct for the relative receive field sensitivity of the array coil, which will be position-specific [5].
2.5. Deliberate inter-scan motion
After the PD-weighted acquisition had been acquired, the participant performed a yaw rotation (i.e. about the z-axis) while the scanner was not running (see Table 1). The amplitude of the motion aimed to be as large as possible within the confines of the 64 channel coil.
This was done to be able to test the performance of the inter-scan motion correction scheme that accounts for position-specific modulation by the receiving coil. After the participant had moved sensitivity mapping data (image series 10–11) were acquired followed by acquisition of the MT-weighted dataset (image series 12).
The participant then returned to approximately the original position (i.e. that of the PD-weighted acquisition, centred within the head coil), again while the scanner was not running. Then, sensitivity mapping data (image series 13–14) were acquired followed by acquisition of the T1-weighted dataset (image series 15).
2.6. Processing
Prior to sharing, the DICOM images produced by the scanner were converted to NIfTI format using the DICOMImport utility as implemented in the hMRI toolbox ([1], http://hMRI.info). The NIfTI data were subsequently anonymised using the defacing utility as implemented in SPM12 (https://www.fil.ion.ucl.ac.uk/spm/) in order to comply with General Data Protection Regulation (GDPR) regulations.
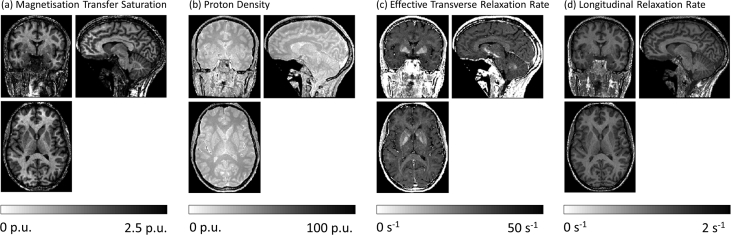
Following anonymization, the data were processed using the “Create hMRI maps” module of the hMRI toolbox. This module is accessed via the SPM batch menu via: SPM-Tools-hMRI Tools-Create hMRI maps. The processing included correction for receive field modulation as described in Papp et al. [5], transmit field inhomogeneity [2], [3] and imperfect RF spoiling correction [6]. The batch module, together with the toolbox configuration file are also supplied. Orthogonal views of the resulting maps are shown in Fig. 3. All outputs from this module, including the supplementary results (B1+ and B1− maps, and signal intensities for each contrast extrapolated to TE = 0 ms) and all meta-data (processing log, json files, and quality assurance (QA) metrics), are provided.
Fig. 3.
Orthogonal views of the quantitative multi-parameter maps magnetisation transfer saturation (a), proton density (b), effective transverse relaxation rate (c) and longitudinal relaxation rate (d), derived from this dataset using the hMRI toolbox.
Acknowledgments
The acquisition of this data was supported by The Wellcome Centre for Human Neuroimaging via core funding from Wellcome [203147/Z/16/Z]. AL is supported by the Swiss National Science Foundation (project grant Nr. 320030_184784) and the ROGER DE SPOELBERCH foundation. EB received funding from the European Structural and Investment Fund / European Regional Development Fund & the Belgian Walloon Government, project BIOMED-HUB (programme 2014-2020). BD is supported by the Swiss National Science Foundation (NCCR Synapsy, project grant Nr. 32003B_135679, 32003B_159780) and the Leenaards Foundation. LREN is very grateful to the Roger De Spoelberch and Partridge Foundations for their generous financial support. NW has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° 616905; and from the European Union's Horizon 2020 research and innovation programme under the grant agreement No 681094. SM was supported by the German Research Foundation (DFG Emmy Noether Stipend: MO 2397/4-1 and DFG Priority Program 2041 “Computational Connectomics”, AL 1156/2-1; GE 2967/1-1; MO 2397/5-1; MO 2249/3- 1). NW and SM received funding from the BMBF (01EW1711A and B) in the framework of ERA-NET NEURON. CP is supported by the F.R.S.-FNRS (Belgium). MFC is supported by the MRC and Spinal Research Charity through the ERA-NET Neuron joint call (MR/R000050/1). The Wellcome Centre for Human Neuroimaging is supported by core funding from the Wellcome [203147/Z/16/Z].
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Tabelow K., Balteau E., Ashburner J., Callaghan M.F., Draganski B., Helms G., Kherif F., Leutritz T., Lutti A., Phillips C., Reimer E., Ruthotto L., Seif M., Weiskopf N., Ziegler G., Mohammadi S. hMRI - a toolbox for using quantitative MRI in neuroscience and clinical research. Neuroimage. 2019;194:191–210. doi: 10.1016/j.neuroimage.2019.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutti A., Stadler J., Josephs O., Windischberger C., Speck O., Bernarding J., Hutton C., Weiskopf N. Robust and fast whole brain mapping of the RF transmit field B1 at 7T. PLoS One. 2012;7:e32379. doi: 10.1371/journal.pone.0032379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutti A., Hutton C., Finsterbusch J., Helms G., Weiskopf N. Optimization and validation of methods for mapping of the radiofrequency transmit field at 3T. Magn. Reson. Med. 2010;64:229–238. doi: 10.1002/mrm.22421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griswold M.a., Jakob P.M., Heidemann R.M., Nittka M., Jellus V., Wang J., Kiefer B., Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn. Reson. Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 5.Papp D., Callaghan M.F., Meyer H., Buckley C., Weiskopf N. Correction of inter-scan motion artifacts in quantitative R1 mapping by accounting for receive coil sensitivity effects. Magn. Reson. Med. 2016;76:1478–1485. doi: 10.1002/mrm.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preibisch C., Deichmann R. Influence of RF spoiling on the stability and accuracy of T1 mapping based on spoiled FLASH with varying flip angles. Magn. Reson. Med. 2009;61:125–135. doi: 10.1002/mrm.21776. [DOI] [PubMed] [Google Scholar]