Abstract
Chikungunya virus (CHIKV) is a mosquito-borne virus that causes epidemics widely in the world especially in the tropical and subtropical regions. Phylogenetic analysis has found that the CHIKV lineages were associated with the spatial and temporal distributions, which were related to the virus adaption to the major mosquito species and their distributions. In this study, we reported the complete genome sequences of eight CHIKV isolates from the outbreak in Pakistan last year. Then we reviewed the evolutionary history using extensive phylogenetic analysis, analyzed lineage-specific substitutions in viral proteins, and characterized the spreading pathway of CHIKV strains including the Pakistani strains. The results showed that the Pakistani stains belonged to the ECSA.IOL sub-lineage and derived from India. The genetic properties of the Pakistani strains including the adaptive substitution to vectors were further characterized, and the potential risks from the occurrence of CHIKV infection in Pakistan were discussed. These results provided better understanding of CHIKV evolution and transmission in the world and revealed the possible origination of the CHIKV outbreak and epidemic in Pakistan, which would promote the disease prevention and control in the identified countries and territories with the history of CHIKV infections as well as new regions with potential risk of CHIKV outbreaks.
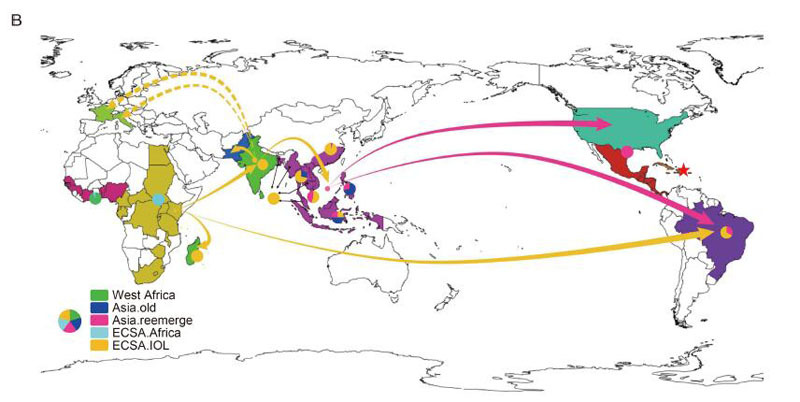
Electronic Supplementary Material
Supplementary material is available for this article at 10.1007/s12250-017-4077-5 and is accessible for authorized users.
Keywords: chikungunya virus (CHIKV), Pakistan, phylogenetic analysis
Electronic supplementary material
Extensive evolution analysis of the global chikungunya virus strains revealed the origination of CHIKV epidemics in Pakistan in 2016
Acknowledgment
This work was supported by the Science and Technology Basic Work Program (2013FY113500) from the Ministry of Science and Technology of China, the International Cooperation on key Technologies of Biosafety along the China-Pakistan Economic Corridor (153B42KYSB2017 0004), the Strategic Bio-resource Service Network Plan and Building the Biogenetic Resource Preserving Capacity Program from the Chinese Academy of Sciences (ZSSB-002), and was funded by the National Basic Scientific Data Sharing-Service Platform (XXH12504-3-15).
Contributor Information
Shu Shen, Email: shenshu@wh.iov.cn.
Fei Deng, Email: df@wh.iov.cn.
References
- Aamir UB, Badar N, Salman M, Ahmed M, Alam MM. Outbreaks of chikungunya in Pakistan. Lancet Infect Dis. 2017;17:483. doi: 10.1016/S1473-3099(17)30191-3. [DOI] [PubMed] [Google Scholar]
- Abraham R, Manakkadan A, Mudaliar P, Joseph I, Sivakumar KC, Nair RR, Sreekumar E. Correlation of phylogenetic clade diversification and in vitro infectivity differences among Cos-mopolitan genotype strains of Chikungunya virus. Infect Genet Evol. 2016;37:174–184. doi: 10.1016/j.meegid.2015.11.019. [DOI] [PubMed] [Google Scholar]
- AbuBakar S, Sam IC, Wong PF, MatRahim N, Hooi PS, Roslan N. Reemergence of endemic Chikungunya, Malaysia. Emerg Infect Dis. 2007;13:147–149. doi: 10.3201/eid1301.060617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal MF, Naqvi SQ, Sultan MA, Hanif A. Chikungunya fever among children presenting with nonspecific febrile illness during an epidemic of dengue fever in Lahore, Pakistan. Merit Res J Med Scis. 2015;3:69–73. [Google Scholar]
- Arankalle VA, Shrivastava S, Cherian S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, Sudeep AB, Mishra AC. Genetic divergence of Chikungunya viruses in India (1963-2006) with special reference to the 2005-2006 explosive epidemic. J Gen Virol. 2007;88:1967–1976. doi: 10.1099/vir.0.82714-0. [DOI] [PubMed] [Google Scholar]
- Casal PE, Chouhy D, Bolatti EM, Perez GR, Stella EJ, Giri AA. Evidence for homologous recombination in Chikungunya Virus. Mol Phylogenet Evol. 2015;85:68–75. doi: 10.1016/j.ympev.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Cauchemez S, Ledrans M, Poletto C, Quenel P d, Valk H, Colizza V, Boëlle PY. Local andregional spread of Chikungunya in the Americas. Euro Surveill. 2014;19:20854. doi: 10.2807/1560-7917.ES2014.19.28.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). 2006. Chikungunya fever diagnosed among international travelers–United States, 2005-2006. MMWR Morb Mortal Wkly Rep, 55: 1040–1042. [PubMed]
- Charrel R d, Lamballerie X, Raoult D. Chikungunya outbreaks–the globalization of vectorborne diseases. N Engl J Med. 2007;356:769–771. doi: 10.1056/NEJMp078013. [DOI] [PubMed] [Google Scholar]
- Darwish MA, Hoogstraal H, Roberts TJ, Ahmed IP, Omar F. A sero-epidemiological survey for certain arboviruses (Togaviridae) in Pakistan. Trans R Soc Trop Med Hyg. 1983;77:442–445. doi: 10.1016/0035-9203(83)90106-2. [DOI] [PubMed] [Google Scholar]
- Delisle E, Rousseau C, Broche B, Leparc-Goffart I, L'Ambert G, Cochet A, Prat C, Foulongne V, Ferre JB, Catelinois O, Flusin O, Tchernonog E, Moussion IE, Wiegandt A, Septfons A, Mendy A, Moyano MB, Laporte L, Maurel J, Jourdain F, Reynes J, Paty MC, Golliot F. Chikungunya outbreak in Montpellier, France, September to October 2014. Euro Surveill. 2015;pii:21108. doi: 10.2807/1560-7917.es2015.20.17.21108. [DOI] [PubMed] [Google Scholar]
- Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D. Vectors of Chikungunya virus in Senegal: current data and transmission cycles. Am J Trop Med Hyg. 1999;60:281–286. doi: 10.4269/ajtmh.1999.60.281. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima SH, Atif S, Rasheed SB, Zaidi F, Hussain E. Species Distribution Modelling of Aedes aegypti in two dengue-endemic regions of Pakistan. Trop Med Int Health. 2016;21:427–436. doi: 10.1111/tmi.12664. [DOI] [PubMed] [Google Scholar]
- Haroon MZ, Wazir MS. And yet one more adds to the sorrow. J Ayub Med Coll Abbottabad. 2016;28:637–638. [PubMed] [Google Scholar]
- Kariuki Njenga M, Nderitu L, Ledermann JP, Ndirangu A, Logue CH, Kelly CH, Sang R, Sergon K, Breiman R, Powers AM. Tracking epidemic Chikungunya virus into the Indian Ocean from East Africa. J Gen Virol. 2008;89:2754–2760. doi: 10.1099/vir.0.2008/005413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikungunya virus emergence is constrained in Asia by lineagespecific adaptive landscapes. Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform, 9: 299-306. [DOI] [PMC free article] [PubMed]
- Lam SK, Chua KB, Hooi PS, Rahimah MA, Kumari S, Tharmaratnam M, Chuah SK, Smith DW, Sampson IA. Chikungunya infection–an emerging disease in Malaysia. Southeast Asian J Trop Med Public Health. 2001;32:447–451. [PubMed] [Google Scholar]
- Laras K, Sukri NC, Larasati RP, Bangs MJ, Kosim R D, Wandra T, Master J, Kosasih H, Hartati S, Beckett C, Sedyaningsih ER, Beecham H 3, Corwin AL. Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans R Soc Trop Med Hyg. 2005;99:128–141. doi: 10.1016/j.trstmh.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Leparc-Goffart I, Nougairede A, Cassadou S, Prat C d, Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383:514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- Liu SQ, Li X, Zhang YN, Gao AL, Deng CL, Li JH, Jehan S, Jamil N, Deng F, Wei HP, Zhang B. 2017. Detection, isolation and characterization of chikungunya virus associated with chikungunya outbreak in Pakistan in 2016-17. Virol Sin, 32. (In Press) [DOI] [PMC free article] [PubMed]
- Lumsden WH. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53. II. General description and epidemiology. Trans R Soc Trop Med Hyg. 1955;49:33–57. doi: 10.1016/0035-9203(55)90081-X. [DOI] [PubMed] [Google Scholar]
- Mallhi TH, Khan YH, Khan AH, Tanveer N, Qadir MI. First chikungunya outbreak in Pakistan: a trail of viral attacks. New Microbes New Infect. 2017;19:13–14. doi: 10.1016/j.nmni.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoagouni C, Kamgang B, Kazanji M, Paupy C, Nakoune E. Potential of Aedes aegypti and Aedes albopictus populations in the Central African Republic to transmit enzootic chikungunya virus strains. Parasit Vectors. 2017;10:164. doi: 10.1186/s13071-017-2101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola P d, Lamballerie X, Jourdan J, Rovery C, Vaillant V, Minodier P, Brouqui P, Flahault A, Raoult D, Charrel RN. Novel Chikungunya Virus Variant in Travelers Returning from Indian Ocean Islands. Emerg Infect Dis. 2006;12:1493–1499. doi: 10.3201/eid1210.060610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A g Cs. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- Robinson MC. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53. I. Clinical features. Trans R Soc Trop Med Hyg. 1955;49:28–32. doi: 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci U S A. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, Chen R, Sherman MB, Weaver SC. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol. 2011;1:310–317. doi: 10.1016/j.coviro.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, Chen R, Yun R, Rossi SL, Plante KS, Guerbois M, Forrester N, Perng GC, Sreekumar E, Leal G, Huang J, Mukhopadhyay S, Weaver SC. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014;5:4084. doi: 10.1038/ncomms5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One. 2009;4:e6835. doi: 10.1371/journal.pone.0006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandingham D H, Klingler K, Tsetsarkin K, McElroy KL, McElroy KL, Powers AM, Lehane MJ, Higgs S. Differential infectivities of o'nyong-nyong and chikungunya virus isolates in Anopheles gambiae and Aedes aegypti mosquitoes. Am J Trop Med Hyg. 2005;72:616–621. doi: 10.4269/ajtmh.2005.72.616. [DOI] [PubMed] [Google Scholar]
- Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, Nasar F, Schuh AJ, Holmes EC, Higgs S, Maharaj PD, Brault AC, Weaver SC. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol. 2010;84:6497–6504. doi: 10.1128/JVI.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC. Urbanization and geographic expansion of zoonotic arboviral diseases: mechanisms and potential strategies for prevention. Trends Microbiol. 2013;21:360–363. doi: 10.1016/j.tim.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Forrester NL. Chikungunya: Evolutionary history and recent epidemic spread. Antiviral Res. 2015;120:32–39. doi: 10.1016/j.antiviral.2015.04.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extensive evolution analysis of the global chikungunya virus strains revealed the origination of CHIKV epidemics in Pakistan in 2016


