Abstract
Intrinsically disordered regions (IDRs) of proteins are involved in many diseases. The rational drug design against disease-mediating proteins is often based on the 3D structure; however, the flexible structure of IDRs hinders the use of such structure-based design methods. Here, we developed a rational design method to obtain a peptide that can bind an IDR using only sequence information based on the statistical contact energy of amino acid pairs. We applied the method to the disordered C-terminal domain of the tumor suppressor p53. Titration experiments revealed that one of the designed peptides, DP6, has a druggable affinity of ~1 μM to the p53 C-terminal domain. NMR spectroscopy and molecular dynamics simulation revealed that DP6 selectively binds to the vicinity of the target sequence in the C-terminal domain of p53. DP6 inhibits the nonspecific DNA binding of a tetrameric form of the p53 C-terminal domain, but does not significantly affect the specific DNA binding of a tetrameric form of the p53 core domain. Single-molecule measurements revealed that DP6 retards the 1D sliding of p53 along DNA, implying modulation of the target searching of p53. Statistical potential-based design may be useful in designing peptides that target IDRs for therapeutic purposes.
Subject terms: Intrinsically disordered proteins, Fluorescence imaging, DNA-binding proteins, Virtual screening
Introduction
Intrinsically disordered regions (IDRs) of proteins lack a defined 3D structure. Approximately 10–35% of prokaryotic and 15–45% of eukaryotic proteins are estimated to contain IDRs1. IDRs function as hubs in protein-protein interaction networks, the regulation of transcription and signaling pathways, and phase separation. IDRs are involved in many diseases and are considered drug targets2–4. The major drug design targeting IDR is based on the experimental screening of chemicals4. By contrast, structure-based drug design, which is one of the most commonly used methods to design drugs targeting proteins, is difficult to apply to IDRs due to their flexible structure4. A unique successful method of the rational drug design targeting IDRs is virtual screening of chemicals that bind pockets formed in IDR conformations5,6. Before the screening, pockets of the IDR need to be estimated by MD simulation or NMR. Accordingly, a rational design approach without 3D structure information is required for targeting IDRs.
p53, an intrinsically disordered protein containing IDRs, is a multifunctional transcription factor that can suppress cell tumorigenesis and is a desirable drug target7,8. Fifty percent of the gene mutations identified in tumor cells occur in p53 gene9. p53 is composed of an N-terminal (NT) domain, a core domain, a linker, a tetramerization domain, and a C-terminal (CT) domain. The core and tetramerization domains form a specific tertiary structure, while the other two domains and the linker are IDRs. The core and CT domains are involved in the specific and nonspecific DNA binding, respectively. The 1D sliding of p53 along DNA is essential to searching for the target site among tremendous amounts of nonspecific DNA10. p53 possesses two sliding modes with a different conformation11–13. The CT domain facilitates the 1D sliding of p5311,14 and regulates the transcription of downstream genes15–18. Several studies focused on drug design or function modification for p538,17,19–27 have mainly targeted the folded domains, and successful drug design for the IDRs of p53 has been limited to two studies28,29. Gabizon et al. identified several peptides that bind to the tetrameric form of the disordered CT domain of p53 by screening peptides derived from natural proteins bound to p5329.
Peptides are promising drug candidates30,31 and may work for targeting IDRs29. The combination of 20 amino acids with different characteristics has the potential to generate peptides with high affinity for the target IDR. Also, the flexible peptide can fit any conformation of the IDR. We need to search for peptides with a high affinity for the target IDR among an enormous number of candidates; for example, for a 16-residue peptide, 2016 candidates are possible. The theoretical pool of peptides is very large, while using peptides from natural proteins limits the number of peptides that can be tested, hence it is needed to develop a computer-based method for drug design.
Here, we aimed to develop the design method of peptides that can bind IDRs and then apply the method to the p53 IDR. The method uses only the IDR’s sequence information without considering its structure. For the design, we used the Miyazawa-Jernigan (MJ) potential, which reflects the residue-residue potential for favorable and unfavorable contacts based on the analysis of known protein crystal structures in protein crystal structures32. MJ potential has been used in protein folding simulation and structural prediction33–37. As a target IDR, we chose the CT domain of p53. We designed six peptides, and found that three peptides bound to the CT domain tighter than the peptides identified by Gabizon et al.29. A series of experiments including NMR, molecular dynamics (MD) simulations, ensemble titration, and single-molecule fluorescence demonstrated that one designed peptide with the highest affinity can modulate the nonspecific DNA binding of p53 and the 1D sliding of p53 along DNA. This is the first approach to target IDRs using only their sequence information and is potentially applicable to many disease-related proteins containing IDRs.
Results
MJ potential-based peptide design identifies a peptide with micromolar affinity for the p53 CT domain
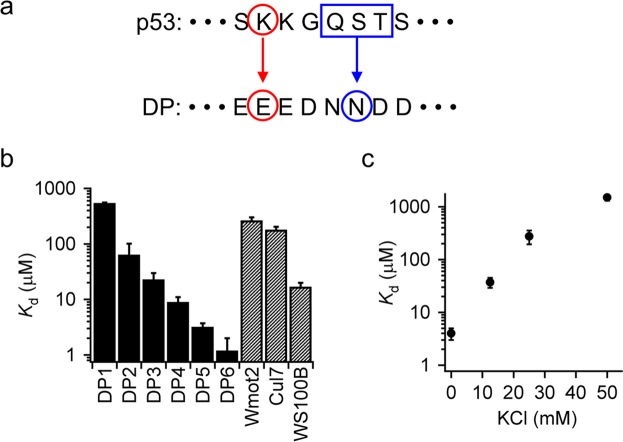
Using MJ potential32, we designed peptide sequences to bind the disordered CT domain of p53. Specifically, six peptides with 13 or 16 residues were designed to minimize the total statistical binding energy for one by one residue (designed peptides DP1–4) or one by three residues including two adjacent residues (designed peptides DP5–6) (Supplementary Figure S1 and Table 1). One by one assumes the contacts of two residues facing in the two sequences, while one by three assumes the contacts to three sequential residues (Fig. 1a). The binding energy between the ith residue of the CT domain and the jth residue of a designed peptide was calculated as eij + err − eir − ejr, as defined by Miyazawa and Jernigan32,38, where eij denotes the energy difference between the formation of contacts between the ith and jth residues, and the same residues exposed to solvent. The notation r represents averaging over all amino acids. To test how well our method worked, we titrated the designed peptides against the CT peptide (residues 367–393) labeled with a fluorescent dye, FAM, using a fluorometer with fluorescence anisotropy11 (Supplementary Fig. S2a). All titration curves were well fitted with equations 1 and 2 (see Methods) based on one by one binding. The apparent dissociation constant of the designed peptides (KD) with the CT peptide ranged from 1.2 ± 0.8 μM to 550 ± 10 μM in the absence of salt, and DP6 showed the strongest binding to the CT domain (Fig. 1b and Table 1). One by three design (DP5–6) improved the binding ability of the designed peptide to the CT peptide by at least six-fold compared to one by one design (DP1–4). The affinity of the designed peptides for the CT peptide correlated with the estimated binding energy (Supplementary Fig. S3). These results suggest that interactions between residues that are favorable in natural folded proteins can stabilize complexes involving disordered proteins.
Table 1.
Length, sequence, and dissociation constant of peptides used in this study.
| Peptide | L* | Sequence | Kd (μM)† |
|---|---|---|---|
| DP1 | 13 | EHEEIFMESKGWR | 60 ± 40 |
| DP2 | 13 | EDEEGNDSDEHEE | 23 ± 7 |
| DP3 | 16 | DEHEEIFMESKGWRDR | 550 ± 10 |
| DP4 | 16 | DHIEDEEGNDSDEHEE | 9 ± 2 |
| DP5 | 13 | EEEEDNNDDDEEE | 3.2 ± 0.5 |
| DP6 | 16 | EEEEEDNNDDDEEEWF | 1.2 ± 0.8 |
| Wmot2 (266–280)‡ | 16 | WSTNGDTFLGGEDGDQ | 270 ± 40 |
| Cul7 (386–400)‡ | 15 | LDDYEEISAGDEGEF | 180 ± 20 |
| WS100B (61–75)‡ | 16 | WLDNDGDGECDFQEFM | 17 ± 3 |
*Residue length. †Kd was determined by titrating peptides against the CT peptide of p53 in the absence of salt. The error of Kd is the SEM of the fitting. ‡The peptides were identified from natural proteins29.
Figure 1.
Binding of designed peptides to the monomeric CT domain of p53. (a) Schematic diagram of peptide design. Red and blue indicate one by one residue and one by three residue design, respectively. DP, designed peptide. (b) Dissociation constants of designed peptides (DP1–DP6) or peptides from natural proteins (Wmot2, Cul7, and WS100B) for the CT peptide of p53 in the absence of salt. (c) Salt dependence of the dissociation constant of DP6 with the CT peptide. In panels (b,c), the error is the SEM of the fitting.
Next, we compared the affinity of our designed peptides to that of peptides derived from natural p53-binding proteins and confirmed to bind to the CT peptide in an earlier report29. To this end, we titrated three natural peptides against the CT peptide under the same conditions and determined the KD (Fig. 1b and Supplementary Fig. S2b). The KD of DP6 with respect to the CT domain was at least 12 times stronger than the KD of the natural peptides (Table 1).
To elucidate the mechanism of DP6 binding to the CT domain, we examined the dependence of KD on salt concentration (Fig. 1c). KD decreased as the K+ concentration increased, suggesting that the binding was mainly governed by electrostatic interactions between the +6 net charge of DP6 and the −4 net charge of the CT peptide. The circular dichroism spectra of the DP6–CT peptide complex and of the DP6 and CT peptides alone showed no significant secondary structure, implying disordered-disordered contacts in the complex (Supplementary Fig. S4).
DP6 binds selectively to the target region and its vicinity via electrostatic and hydrophobic interactions
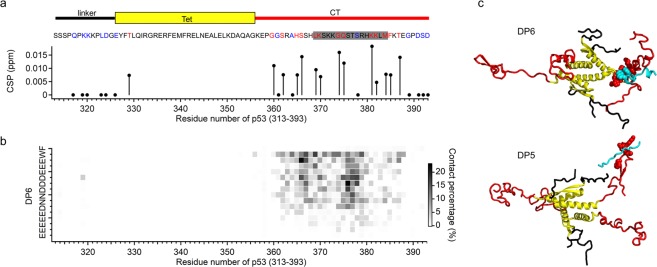
To examine whether DP6 binds specifically to the designed target position of p53, we conducted NMR analysis of a 15N-labeled p53 (residues 313–393) tetramer including linker, Tet, and CT domains in the absence and presence of DP6 (Fig. 2a and Supplementary Fig. S5). Clear chemical shift perturbations in the 1H-15N HSQC spectrum upon binding to DP6 were observed in residues 329, 360, 362, 365, 366, 369, 370, 374, 375, 381, 382, 384, 385, and 387 of p53. These residues correspond to the designed target of the CT domain (residues 369–384) for DP6 and its vicinity. To further investigate the complex structure, we conducted MD simulations of the p53 (313–393) tetramer and DP6. The contact map showed significant interaction between residues 360–385 of p53 and DP6, which is consistent with the chemical shift perturbations (Fig. 2a,b). Furthermore, the identified binding site of DP6 agreed with the p53 sequence region having lower MJ binding energy for DP6 (Supplementary Fig. S6). In the MD simulation trajectory, the DP6 and CT domains showed flexibility, and no specific tertiary structure was formed (Supplemental Movie). In these structures, contacts between the acidic residues of DP6 and the basic residues of the CT domain were always observed. In addition, W15 and F16 in DP6 and hydrophobic regions in the CT domain (especially, residues 361–369, 371, 373–379, 385, 387, 388) always formed a hydrophobic cluster (Fig. 2c). These results suggest that electrostatic and hydrophobic interactions strongly stabilized the DP6–CT domain complex compared with other peptides. In fact, such hydrophobic interactions were not observed between the CT domain and DP5, which may explain the relatively weak affinity compared with that of DP6 (Fig. 2c). Accordingly, we conclude that DP6 binds selectively to the designed target region and its vicinity in the CT domain of p53.
Figure 2.
Binding of the designed peptide DP6 to tetrameric p53 (313–393). (a) Chemical shift perturbations (CSP) of the 1H-15N HSQC spectrum of p53 (313–393) in the absence/presence of DP6. Red and blue characters represent residues showing positive or no CSP, respectively. Gray highlighting indicates the target sequence of p53 used for the design of DP6. (b) Contact map between residues of DP6 and p53 in MD simulation. (c) Typical snapshots of DP6 and p53 (313–393) tetramer complex in MD simulation. Cyan, black, yellow, and red denote designed peptide, linker, the Tet domain, and the CT domain of p53, respectively. Hydrophobic amino acids of the CT domain, DP5, and DP6 in contacts are depicted in space-filling representation.
DP6 interferes with nonspecific DNA binding of p53
To examine the effect of DP6 on the DNA binding of p53, we prepared three p53 constructs: full-length p53 (FL-p53), and two mutants each containing one of its two DNA binding domains, a tetrameric form of the CT domain (TetCT mutant), and a tetrameric form of the core domain (CoreTet mutant) as reported previously12 (Fig. 3a). For the p53 mutants, we used a thermostabilized form of p53 with a single exposed cysteine, which is suitable for in vitro ensemble and single-molecule studies11.
Figure 3.
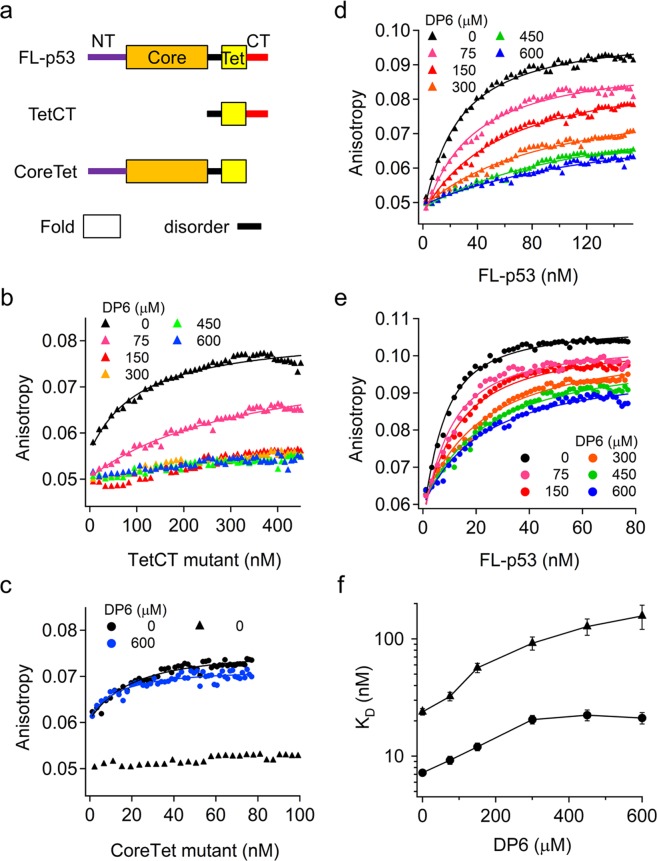
Effect of the designed peptide DP6 on the DNA binding of the p53 tetramer. (a) p53 constructs used in this study. NT, core, Tet, and CT represent the N-terminal, core, tetramerization, and C-terminal domains of p53, respectively. Thick and thin primary structures correspond to folded and disordered regions, respectively. (b) Titration of the TetCT mutant against nspDNA at various DP6 concentrations. (c) Titration of the CoreTet mutant against spDNA in the presence (blue circles) and absence (black circles) of 600 μM DP6 and against nspDNA in the absence of DP6 (triangles). (d) Titration of FL-p53 against nspDNA at various DP6 concentrations. (e) Titration of FL-p53 against spDNAat various DP6 concentrations. (f) Affinity of FL-p53 for nspDNA (triangles) and spDNA (circles) at various DP6 concentrations. The errors were the SEM of the fitting. In panels (b–e), tetramer concentrations are used for the p53 mutants, and the solid curves are the best-fitted curves using Equations (1) and (2) described in the Methods.
To examine the effect of DP6 on the affinity of the TetCT mutant for DNA, we titrated TetCT mutant against nonspecific DNA (nspDNA) labeled with 6-FAM at 0–600 μM DP6 based on fluorescence anisotropy11. The solution used here included 100 mM KCl to mimic physiological conditions. In the absence of DP6, the TetCT mutant bound to nspDNA (KD = 110 ± 20 nM for tetramer). The anisotropy change for nspDNA was repressed as the DP6 concentration increased (Fig. 3b). However, no significant change in anisotropy was observed at more than 150 μM DP6, suggesting that DP6 inhibits the binding of the TetCT mutant to nspDNA by competitive binding to the CT domain of the TetCT mutant. For control experiments investigating the effect of DP6 on the binding of the core domain to specific DNA (spDNA), we titrated the CoreTet mutant against spDNA in the presence/absence of 600 μM DP6 (Fig. 3c, circles). DP6 did not weaken the binding of the CoreTet mutant to spDNA (Supplementary Table S1). Nonspecific binding of the CoreTet mutant was not observed, confirming that DNA binding of the CoreTet mutant in the presence of DP6 was specific (Fig. 3c, triangles). Accordingly, the results suggest that DP6 binds specifically to the CT domain of p53 and prevents nonspecific binding to DNA, thereby competitively maintaining the association of the core domain with spDNA.
We next examined the effect of DP6 on the affinity of FL-p53 to DNA by titrating FL-p53 against nspDNA and spDNA at 0–600 μM DP6. The anisotropy change for nspDNA was repressed as the DP6 concentration increased, indicating that the nonspecific affinity of FL-p53 was weakened by DP6 (Fig. 3d). The anisotropy change for spDNA was also repressed by DP6 (Fig. 3e), but the repression was more effective for nspDNA than for spDNA. The KD of FL-p53 for nspDNA gradually increased with the addition of DP6 and was 7-fold higher at 600 μM than at 0 μM DP6 (Fig. 3d and Supplementary Table S1). By contrast, the KD for spDNA increased to 3-fold and was saturated at 300 μM DP6 (Fig. 3e and Supplementary Table S1). In contrast to DP6, DP5 did not affect the affinity of FL-p53, suggesting that W and/or F of DP6 strengthens the binding of the designed peptide to FL-p53 (Supplementary Fig. S7 and Supplementary Table S2). These results demonstrate that DP6 weakens the nonspecific DNA binding of FL-p53 more efficiently than the specific DNA binding.
DP6 retards 1D sliding of p53 along DNA
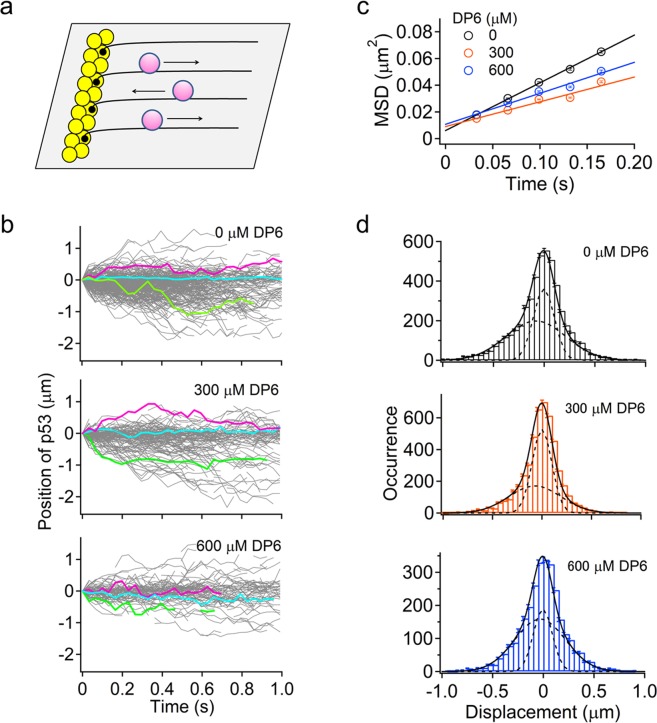
To test whether DP6 affects the 1D sliding of p53 along DNA, we visualized the 1D sliding of FL-p53 labeled with a fluorescent dye, Atto532, along nspDNA at 0, 300, and 600 μM DP6 using single-molecule fluorescence microscopy coupled with the DNA array “DNA garden”10,11,39 (Fig. 4a). We conducted the measurements within 50 min after the dilution of stock solution under conditions in which p53 maintains a tetrameric form40 and obtained 158–290 trajectories in various DP6 concentrations (Fig. 4b). The mean square displacement (MSD) plots of FL-p53 were linear in all conditions, indicating the diffusional motion of p53 along DNA at various DP6 concentrations (Fig. 4c). The average diffusion coefficient, calculated from the slope of MSD, decreased to 0.5-fold at 300 μM DP6 and 0.6-fold at 600 μM (Supplementary Table S3). These results demonstrate that DP6 retards the 1D sliding of p53 along DNA.
Figure 4.
Effect of the designed peptide DP6 on 1D sliding of the p53 tetramer along DNA. (a) Schematic diagram of the single-molecule detection of p53 labeled with Atto532 on a DNA array. Yellow circles, pink circles, and solid lines represent NeutrAvidin, p53, and DNA, respectively. (b) Typical single-molecule trajectories of FL-p53 along nonspecific DNA at various DP6 concentrations. Several traces are colored for clarity. (c) Time courses of the averaged MSD of FL-p53 at various DP6 concentrations. (d) Displacement distributions for the sliding dynamics of FL-p53 at various DP6 concentrations. Bars represent the displacement distributions observed at time intervals of 165 ms. Errors of the bar were estimated by a bootstrap method with 1,000 iterations. Solid and dashed curves are best-fitted curves obtained using Equation (3) described in Methods and the distributions of each mode, respectively.
To examine how DP6 affects the two sliding modes of p53, we analyzed the displacement distributions of p53 at the time interval of 165 ms in different DP6 concentrations. All displacement distributions were well fitted with the sum of two Gaussian functions, which assumes two diffusional motions (Eq. 3, see Methods), indicating that FL-p53 possesses two sliding modes in the presence of DP6. The diffusion coefficient of the fast mode decreased to 0.8-fold at 300 μM DP6 and to 0.7-fold at 600 μM, thereby reducing the average diffusion coefficient (Supplementary Table S3). By contrast, no significant change in the slow mode is consistent with the tight interaction of the core domains with DNA identified in the slow mode12. Accordingly, the fast mode of FL-p53 sliding, rather than the slow mode, is retarded upon the association of DP6 with FL-p53.
Discussion
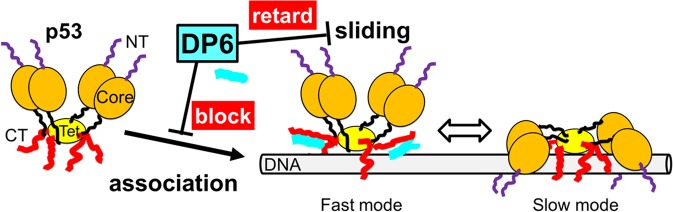
In this study, we developed the design method of peptides that can bind IDRs using only the IDR’s sequence information without its structural information. To test this method, we targeted the disordered CT domain of p53. We found that DP6 has the high affinity for the CT domain, and modulates the affinity of p53 for DNA and the target search. These results suggest that our design method has a potential to generate druggable peptide candidates for IDRs. Here, we discuss the mechanism for action of DP6 and the comparison of drug design methods for IDRs.
We propose a mechanism for how DP6 affects p53 (Fig. 5). One of the actions of DP6 is to competitively block the association of the CT domain of p53 with nonspecific DNA. Previous spectroscopic studies including chemical shift perturbations in NMR and the effects of mutation on the affinity revealed that residues 365−382, including lysine residues 372, 373, 381, and 382 in the CT domain, interact with DNA41,42. The NMR data in this study demonstrate that DP6 interacts with residues 360–387 of p53 (Fig. 2a). Therefore, it is likely that DP6 sterically prevents the association of the CT domain with DNA, reducing the affinity of p53 for nonspecific DNA (Fig. 3b). This finding is supported by the fact that additives such as small nucleotides and antibodies strengthen specific affinity by blocking the CT domain43. Before the formation of the p53–DNA complex, the CT domain is blocked by DP6 from association with nonspecific DNA.
Figure 5.
Model for regulation of the affinity and 1D sliding of p53 by the designed peptide DP6. DP6 (cyan) weakens the association of p53 with DNA (gray bar) and retards the diffusion of the fast mode of p53 along DNA upon association with the CT domain of p53 (red). Purple, orange, and yellow represent the NT, core, and Tet domains of p53, respectively. The structure of the p53–DNA complex is described based on the results of our previous study12.
The other action of DP6 is to retard 1D sliding along DNA. Once the CT domain binds to DNA instead of DP6, the p53–DP6 complex slides along DNA (Fig. 5). DP6 mainly affects the fast mode in which the CT domain of p53 interacts with nonspecific DNA without core domain–DNA interaction12 (Fig. 4d). The observed reduction in the diffusion coefficient in the fast mode may be attributed to the larger molecular size of the p53–DP6 complex and/or intercalation with DNA by DP6. The larger size of p53 may be caused by the detachment of the CT domain from DNA and the association of DP6 with p53 and should slow the 1D sliding along DNA because of the dependence of the 1D sliding on molecular size44,45. As observed in DNA glycosylase46, tryptophan in DP6 may insert between DNA bases during the sliding of the p53–DP6 complex and thereby retard the sliding. By contrast, DP6 does not affect the slow mode, in which p53 reads the DNA sequence and recognizes the target, because in the slow mode, the core domain and CT domain interact with DNA12. This result confirms the specific DNA binding ability of p53. Thus, DP6 may act as a potential inhibitor of p53 by preventing the target search kinetically but not the target binding itself.
The MJ-potential-based approach has unique characteristics comparing with other approaches in the drug design targeting IDRs. Successful approach in the drug design for IDRs is the experimental or virtual screening of chemicals4. The experimental screening does not use any information of the IDR sequence and structure, but requires a high cost and consumes time. As in the case of p53, a small chemical RITA was discovered to bind disordered NT domain of p53 by using the experimental screening28. In the virtual screening, the IDR pockets are identified using MD or NMR and then the chemicals that can bind the pockets are screened4. The virtual screening may not work in the case of no pockets identified in the IDR conformation ensemble. By contrast, our method may be applicable to the IDRs without pocket structures, because the structural information is not used. The MJ-based approach can be applied to the IDRs with known primary sequence. In fact, the designed peptide DP6 identified in this study can bind the CT domain of p53 much stronger than the peptides from natural proteins (Table 1) and the affinity of DP6 is comparable to that of drug candidates discovered in other disordered proteins4. This suggests the successful searching for the druggable peptide among an enormous number of candidates. The virtual screening of peptides based on the MJ contact energy is simple and does not need a supercomputer. Accordingly, the MJ-based approach provides an alternative strategy of drug design targeting IDRs and may apply to many disease-related proteins containing IDRs.
Materials and Methods
Calculation of contact energy
The binding energy between the ith residue of the CT domain and the jth residue of the designed peptide, E, for one by one residue design was calculated as eij + err − eir − ejr, as defined by Miyazawa and Jernigan32. The notation r represents averaging over all amino acids. For eij, err, eir, and ejr, we used the values reported by Miyazawa and Jernigan32. We calculated the binding energy by replacing the jth residue of a designed peptide with each of the 20 amino acids, and then determined the amino acid that gave the lowest binding energy. This procedure was repeated to obtain the sequence of a 13-residue or 16-residue peptide, and the total binding energy was calculated by summing each binding energy. A designed peptide with minimal total energy was selected among peptides designed for different initial residues of the CT domain. In addition, we selected peptides designed for different regions of the CT domain, but not with the minimum total energy. For the calculation involving one by three residues, including two adjacent residues, we added the binding energy between adjacent residues of the CT peptide and each residue of a designed peptide.
p53 mutants and peptides
We prepared FL-p53, TetCT mutant, and CoreTet mutant as described previously11. For FL-p53, the thermostable and cysteine-modified mutant of human p53 (C124A, C135V, C141V, W146Y, C182S, V203A, R209P, C229Y, H233Y, Y234F, N235K, Y236F, T253V, N268D, C275A, C277A, K292C) was used11. The TetCT mutant corresponds to residues 293−393 of human p53 with an additional N-terminal cysteine11. The CoreTet mutant corresponds to residues 1–363 of FL-p5312. Expression and purification of the three mutants was conducted as previously described11,12. Briefly, all mutants with a GST tag were expressed in Escherichia coli and were collected from a GST column after cleavage of the GST tag and further purified by using a heparin column. The DNA binding ability of all mutants was confirmed by titration experiments as described elsewhere10–12. For NMR analysis, the p53 gene corresponding to residues 313–393 of human p53 in pGEX-6P-1 was generated using a KOD-Plus-Mutagenesis Kit (TOYOBO, Osaka, Japan). 15N-labeled p53 (313–393) was expressed in BL21 (DE3) plysS in 15N M9 media at 16 °C for 18 h after the addition of 0.5 mM IPTG and purified as described above12. For titration experiments, CT peptide (residues 367–393 of human p53) labeled with FAM at the N-terminus, designed peptides, and peptides from natural proteins were synthesized without caps and obtained with at least 95% purity (Toray Research Center Inc., Tokyo, Japan).
NMR spectroscopy
1H/15N HSQC experiments were performed at 5 °C using a 1H 600 MHz NMR spectrometer (DRX-600; Bruker, Billerica, MA, USA). The solution contained 0.5 mM 15N-labeled p53 (313–393), 0 or 20 μM DP6, 10 mM HEPES, and 10% 2H2O at pH 7.0. HSQC cross-peaks were assigned to individual amide groups with reference to the assignments of p53 (313–393)41. Spectral analysis was performed using the software Topspin 1.1 (Bruker, Billerica, MA, USA) and NMRViewJ47.
MD simulation
A tetramer of p53 (313–393) and the DP6 peptide were simulated using Amber16 simulator48 with the AMBER ff99SB force field49 and Generalized Born energy for solvation50. For the initial structure of p53 (313–393), the tetramerization domain and the missing disordered region were generated using PDB code 1OLH and modeled in PyMol software, respectively. The initial structure of DP6 was generated in extended form. Initially, DP6 was located at six positions on the x-, y-, and z-axes ± 100 Å from the center of mass of the p53 (313–393) tetramer as described in51. For system relaxation, 20 ns simulations were conducted with constraint of the p53 tetrameric domains while decreasing the temperature from 1000 to 280 K. Then, four 100 ns simulations without any constraint were conducted starting from each initial conformation with randomized initial velocity at 280 K. The temperature was controlled using a Langevin thermostat. The solvent viscosity of water was set to 1.0 ps−1. Noncovalent interactions were used without cutoffs. The covalent bonds of hydrogen atoms in p53 and DP6 were constrained using the SHAKE method52, and the integration time step was 2 fs. For the contact map, 80–100 ns conformations were used. A contact was defined as a distance of less than 6.5 Å between the centers of two side chains except for Gly, where Cα was used38.
Titration experiments
The fluorescence anisotropy of fluorescent CT peptide or DNA was measured at 25 °C using a fluorescence spectrometer (FP-6500, JASCO Co., Tokyo, Japan) with an automatic titrator and home-build autorotating polarizer11. To measure the affinity of the designed peptides to the CT peptide, nonlabeled designed peptides or peptides from natural proteins29 were titrated into a solution containing 5 nM FAM-labeled CT peptide, 20 mM HEPES, 0.5 mM EDTA, 1 mM DTT, and 0.2 mg/mL BSA (pH 7.9). To examine the salt-dependence of the binding of DP6 to CT peptide, KCl was also added. To measure the affinity of p53 mutants to DNA, nonlabeled p53 mutants were titrated into a solution including 5 nM 6-FAM-DNA, 20 mM HEPES, 0.5 mM EDTA, 1 mM DTT, 0.2 mg/mL BSA, 100 mM KCl, 2 mM MgCl2, and various concentrations of DP6 (pH 7.9). spDNA and nspDNA were 30-bp sequences of the p21 gene and a random sequence, respectively, as described elsewhere11 (Sigma-Aldrich Co., Tokyo, Japan). The titration curves were fitted by the following equations:
| 1 |
| 2 |
where robs, rA, rAB, KD, cA, and cB are the observed anisotropy, anisotropy of free molecule A, anisotropy of the complex between molecules A and B, dissociation constant, total concentration of molecule A and total concentration of molecule B, respectively. For p53 mutants, the concentration was calculated for the tetramer.
Single-molecule measurements of p53 mutants using the DNA garden method
The 1D sliding of FL-p53 labeled with Atto532 along DNA was measured by a custom-built TIRF microscope, as described previously11,39. The λDNA array tethered on the coverslip of the flow cell was constructed using microcontact printing as described previously39. FL-p53 was labeled with Atto532 as described elsewhere11. FL-p53 with Atto532 at 2–4 nM was introduced into the flow cell with the DNA array by a syringe pump and measured at a flow rate of 500 μL/min, corresponding to 90% extension of λDNA. The solution contained 20 mM HEPES, 0.5 mM EDTA, 1 mM DTT, 0.2 mg/mL BSA, 1 mM DTT, 2 mM Trolox, 100 mM KCl, 2 mM MgCl2, and 0–600 μM DP6 at pH7.9. The experiments were conducted immediately after the dilution of FL-p53 from the stock solution at more than 10 μM and finished within 50 min to prevent the dissociation of the tetramer40. The analysis was described previously11. For distribution analysis, the following fitting equation was used:
| 3 |
where P(δx), δt, δx, Ai, vi, and Di are the occurrence of δx, time interval, displacement in the time interval, amplitude of the ith mode, drift velocity of the ith mode, and diffusion coefficient of the ith mode, respectively.
Supplementary information
Acknowledgements
This work was supported by MEXT/JSPS KAKENHI, JP15H01625 (to K.K.) and JP16K07313 (to K.K.), and the Research Program of “Dynamic Alliance for Open Innovation Bridging Human, Environment and Materials” in “Network Joint Research Center for Materials and Devices” (to T.K.). We thank Dr. Soichiro Kitazawa (Ritsumeikan University) for an earlier trial of the NMR experiments.
Author Contributions
K.K. contributed design of work and the development of the design strategy, and wrote the manuscript. E.M. contributed the acquisition of titration data. T.W. and R.K. contributed the acquisition of NMR data. H.T. contributed the preparation of a p53 mutant for NMR. S.K. contributed the preparation of p53 mutants and the acquisition of titration data. Y.I. and A.M. contributed the acquisition of single-molecule data. T.K. contributed the development of the design strategy and MD simulations, and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kiyoto Kamagata, Email: kiyoto.kamagata.e8@tohoku.ac.jp.
Tomoshi Kameda, Email: kameda-tomoshi@aist.go.jp.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44688-0.
References
- 1.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem. Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Uversky VN. Drugs for ‘protein clouds’: targeting intrinsically disordered transcription factors. Curr. Opin. Pharmacol. 2010;10:782–788. doi: 10.1016/j.coph.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Sammak S, Zinzalla G. Targeting protein-protein interactions (PPIs) of transcription factors: Challenges of intrinsically disordered proteins (IDPs) and regions (IDRs) Prog. Biophys. Mol. Biol. 2015;119:41–46. doi: 10.1016/j.pbiomolbio.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Ruan H, Sun Q, Zhang W, Liu Y, Lai L. Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov Today. 2019;1:217–227. doi: 10.1016/j.drudis.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Jin F, Yu C, Lai L, Liu Z. Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins. PLoS Comput. Biol. 2013;9:e1003249. doi: 10.1371/journal.pcbi.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu C, et al. Structure-based Inhibitor Design for the Intrinsically Disordered Protein c-Myc. Sci. Rep. 2016;6:22298. doi: 10.1038/srep22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joerger AC, Fersht AR. The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2010;2:a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown CJ, Cheok CF, Verma CS, Lane DP. Reactivation of p53: from peptides to small molecules. Trends Pharmacol. Sci. 2011;32:53–62. doi: 10.1016/j.tips.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Hainaut P, Hollstein M. p53 and human cancer: The first ten thousand mutations. Adv. Cancer Res. 2000;77:81–137. doi: 10.1016/S0065-230X(08)60785-X. [DOI] [PubMed] [Google Scholar]
- 10.Itoh Y, et al. Activation of p53 facilitates the target search in DNA by enhancing the target recognition probability. J. Mol. Biol. 2016;428:2916–2930. doi: 10.1016/j.jmb.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Murata A, et al. One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca(2+) or Mg(2+) at millimolar concentrations. J. Mol. Biol. 2015;427:2663–2678. doi: 10.1016/j.jmb.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Murata A, et al. One-dimensional search dynamics of tumor suppressor p53 regulated by a disordered C-terminal domain. Biophys. J. 2017;112:2301–2314. doi: 10.1016/j.bpj.2017.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subekti DRG, et al. The disordered linker in p53 participates in nonspecific binding to and one-dimensional sliding along DNA revealed by single-molecule fluorescence measurements. Biochemistry. 2017;56:4134–4144. doi: 10.1021/acs.biochem.7b00292. [DOI] [PubMed] [Google Scholar]
- 14.Tafvizi, A., Huang, F., Fersht, A. R., Mirny, L. A. & van Oijen, A. M. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. USA108, 563–568 (2011). [DOI] [PMC free article] [PubMed]
- 15.Hupp TR, Sparks A, Lane DP. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 16.Kim AL, et al. Conformational and molecular basis for induction of apoptosis by a p53 C-terminal peptide in human cancer cells. J. Biol. Chem. 1999;274:34924–34931. doi: 10.1074/jbc.274.49.34924. [DOI] [PubMed] [Google Scholar]
- 17.Selivanova G, Ryabchenko L, Jansson E, Iotsova V, Wiman KG. Reactivation of mutant p53 through interaction of a C-terminal peptide with the core domain. Mol. Cell. Biol. 1999;19:3395–3402. doi: 10.1128/MCB.19.5.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yakovleva T, et al. p53 Latency. C-terminal domain prevents binding of p53 core to target but not to nonspecific DNA sequences. J. Biol. Chem. 2001;276:15650–15658. doi: 10.1074/jbc.M100482200. [DOI] [PubMed] [Google Scholar]
- 19.Issaeva N, et al. Rescue of mutants of the tumor suppressor p53 in cancer cells by a designed peptide. Proc. Natl. Acad. Sci. USA. 2003;100:13303–13307. doi: 10.1073/pnas.1835733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane DP, Hupp TR. Drug discovery and p53. Drug Discov Today. 2003;8:347–355. doi: 10.1016/S1359-6446(03)02669-2. [DOI] [PubMed] [Google Scholar]
- 21.Joerger AC, Ang HC, Fersht AR. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl. Acad. Sci. USA. 2006;103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boeckler FM, et al. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad. Sci. USA. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert JM, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Basse N, et al. Toward the rational design of p53-stabilizing drugs: probing the surface of the oncogenic Y220C mutant. Chem. Biol. 2010;17:46–56. doi: 10.1016/j.chembiol.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Kamada R, et al. Enhancement of transcriptional activity of mutant p53 tumor suppressor protein through stabilization of tetramer formation by calix[6]arene derivatives. Bioorg. Med. Chem. Lett. 2010;20:4412–4415. doi: 10.1016/j.bmcl.2010.06.053. [DOI] [PubMed] [Google Scholar]
- 26.Gureev MA, Davidovich PB, Tribulovich VG, Garabadzhiu AV. Natural compounds as a basis for the design of modulators of p53 activity. Russian Chemical Bulletin. 2014;63:1963–1975. doi: 10.1007/s11172-014-0687-3. [DOI] [Google Scholar]
- 27.Bauer MR, et al. Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of p53 Mutant Y220C Rescue Drugs. ACS Chem. Biol. 2016;11:2265–2274. doi: 10.1021/acschembio.6b00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Issaeva N, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat. Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 29.Gabizon R, et al. Specific Recognition of p53 Tetramers by Peptides Derived from p53 Interacting Proteins. PLoS One. 2012;7:e38060. doi: 10.1371/journal.pone.0038060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Audie J, Swanson J. Advances in the prediction of protein-peptide binding affinities: implications for peptide-based drug discovery. Chem. Biol. Drug Des. 2013;81:50–60. doi: 10.1111/cbdd.12076. [DOI] [PubMed] [Google Scholar]
- 31.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Miyazawa S, Jernigan RL. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J. Mol. Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 33.Kapsokalivas L, Gan X, Albrecht AA, Steinhofel K. Population-based local search for protein folding simulation in the MJ energy model and cubic lattices. Comput. Biol. Chem. 2009;33:283–294. doi: 10.1016/j.compbiolchem.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Rakshit S, Ananthasuresh GK. A novel approach for large-scale polypeptide folding based on elastic networks using continuous optimization. J. Theor. Biol. 2010;262:488–497. doi: 10.1016/j.jtbi.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Yun S, Guy HR. Stability tests on known and misfolded structures with discrete and all atom molecular dynamics simulations. J. Mol. Graph. Model. 2011;29:663–675. doi: 10.1016/j.jmgm.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perdomo-Ortiz A, Dickson N, Drew-Brook M, Rose G, Aspuru-Guzik A. Finding low-energy conformations of lattice protein models by quantum annealing. Sci. Rep. 2012;2:571. doi: 10.1038/srep00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maher B, Albrecht AA, Loomes M, Yang XS, Steinhofel K. A firefly-inspired method for protein structure prediction in lattice models. Biomolecules. 2014;4:56–75. doi: 10.3390/biom4010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal-structures - quasi-chemical approximation. Macromolecules. 1985;18:534–552. doi: 10.1021/ma00145a039. [DOI] [Google Scholar]
- 39.Igarashi C, et al. DNA garden: a simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA binding proteins. Bull. Chem. Soc. Jpn. 2017;90:34–43. doi: 10.1246/bcsj.20160298. [DOI] [Google Scholar]
- 40.Rajagopalan S, Huang F, Fersht AR. Single-Molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res. 2011;39:2294–2303. doi: 10.1093/nar/gkq800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, Fersht AR. Regulation of DNA binding of p53 by its C-terminal domain. J. Mol. Biol. 2004;342:801–811. doi: 10.1016/j.jmb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 42.Friedler A, Veprintsev DB, Freund SM, von Glos KI, Fersht AR. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure. 2005;13:629–636. doi: 10.1016/j.str.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 43.Anderson ME, Woelker B, Reed M, Wang P, Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: Implications for regulation. Mol. Cell. Biol. 1997;17:6255–6264. doi: 10.1128/MCB.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blainey PC, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamagata K, Mano E, Ouchi K, Kanbayashi S, Johnson RC. High Free-Energy Barrier of 1D Diffusion Along DNA by Architectural DNA-Binding Proteins. J. Mol. Biol. 2018;430:655–667. doi: 10.1016/j.jmb.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 48.Case, D. A. et al. AMBER 2016. University of California, San Francisco (2016).
- 49.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mongan J, Simmerling C, McCammon JA, Case DA, Onufriev A. Generalized Born model with a simple, robust molecular volume correction. J. Chem. Theory Comput. 2007;3:156–169. doi: 10.1021/ct600085e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurinomaru T, Kameda T, Shiraki K. Effects of multivalency and hydrophobicity of polyamines on enzyme hyperactivation of alpha-chymotrypsin. J. Mol. Catal. B-Enzym. 2015;115:135–139. doi: 10.1016/j.molcatb.2015.02.010. [DOI] [Google Scholar]
- 52.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.