Abstract
Sepsis is a life-threating condition with dysregulated systemic host response to microbial pathogens leading to disproportionate inflammatory response and multi-organ failure. Various biomarkers are available for the diagnosis and prognosis of sepsis; however, these laboratory parameters may show limitations in these severe clinical conditions. MicroRNAs (miRNA) are single-stranded non-coding RNAs with the function of post-transcriptional gene silencing. They normally control numerous intracellular events, such as signaling cascade downstream of Toll-like receptors (TLRs) to avoid excessive inflammation after infection. In contrast, abnormal miRNA expression contributes to the development of sepsis correlating with its clinical features and outcomes. Based on recent clinical studies altered levels of circulating miRNAs can act as potential diagnostic and prognostic biomarkers in sepsis. In this review, we summarized the available data about TLR-mediated inflammatory signaling with its intracellular response in immune cells and platelets upon sepsis, which are, at least in part, under the regulation of miRNAs. Furthermore, the role of circulating miRNAs is also described as potential laboratory biomarkers in sepsis.
Key words: microRNA, sepsis, monocyte, platelet, TLR, inflammation
INTRODUCTION
Sepsis is a life-threating condition with dysregulated systemic host response to microbial pathogens defined by the Third International Consensus Task Force (Sepsis-3), while septic shock is a subset of sepsis with circulatory, metabolic and cellular abnormalities (1). Based on the etiology of insults of sepsis, we distinguish pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns (DAMP) (2). Since similar mediators are released in both conditions that react with Tolllike receptors (TLRs), they inflict similar excessive inflammatory response (2). Sepsis still results in about 17% mortality due to multiorgan failure (3), which is caused by delayed diagnosis and treatment of patients as well as the inappropriate administration of broad spectrum antibiotics that contributes to the development of antibiotic resistance (4). Several clinical trials have aimed to test a large number of biomarkers for the early diagnosis of sepsis (5). Currently, FDA-(Food and Drug Administration, USA) approved procalcitonin (PCT) can effectively differentiate culture-negative and culture-positive sepsis from non-infectious systemic inflammatory response syndrome (SIRS) (6). In addition, several other diagnostic and prognostic biomarkers are available in sepsis, such as C-reactive protein (CRP) (7), serum lactate (8), and interleukin-6 (IL-6) (9). However, these parameters may be elevated in non-septic conditions as well (10).
MicroRNAs (miRNA) are evolutionarily conserved, single-stranded, non-coding RNAs of 20-25 nucleotides in length with the function of post-transcriptional gene silencing via decreasing messenger RNA (mRNA) levels to fine tune protein expression or via degradation of mRNA to inhibit translation (11). Each miRNA target hundreds of mRNAs, and each mRNA is under the control of several miRNAs. Among physiological conditions, miRNAs with a dynamic nature, tightly control intracellular processes to maintain homeostasis (11). For example, miRNAs are the fine-tuners of signaling downstream of TLRs to avoid excessive inflammation after infection (12). On the other hand, they have been implicated in the development of various human diseases, such as cardiovascular, autoimmune and malignant disorders (13-15). miRNAs secreted from the cells and their presence in plasma/serum denote the role of circulating (cell-free) miRNAs in pathogenesis (16). Although immune response is predominantly controlled at the transcriptional level, miRNA-mediated RNA interference operates at the translation level (17). Consequently, dysregulated intracellular and circulating miRNA expression has been correlated with the clinical features of SIRS (18), critically ill polytrauma (19), and sepsis (20-22). The function of miRNAs in the regulation of immune response and development of sepsis seems to be critical (23, 24), thus a better understanding of these mechanisms may result in improved diagnostic and therapeutic strategies in sepsis. In this review, we focus on TLR4-mediated inflammatory signaling with subsequent cellular events in immune cells and platelets upon sepsis, which are, at least in part, under the regulation of miRNAs. Furthermore, the role of circulating miRNAs is also summarized as potential diagnostic and prognostic biomarkers in sepsis.
BIOGENESIS AND RELEASE OF miRNAS
As the first step, primary miRNAs (pri-miRNAs) are usually transcribed by the function of RNA polymerase II in the cell nucleus (25). Pri-miRNAs are then processed into 70 bp “hairpin” miRNA precursors (pre-miRNAs) by to the endonuclease activity of Drosha (26). This reaction is catalyzed by the DiGeorge Syndrome Critical Region 8 (DGCR8) complex. Pre-miRNAs bind to the Exportin-5 transporter protein, which shifts them from the nucleus into the cytoplasm. Ribonuclease (RNAse) III called Dicer further processes pre-miRNAs into mature miRNA duplexes (27). The RNA-induced silencing complex (RISC) is formed by TRBP (transactivation-responsive RNA-binding protein), Dicer and Ago2 (Argonaute-2) proteins, which guides miRNAs to post-transcriptionally regulate mRNAs by binding to their 3’ untranslated region (UTR). In humans, they fine tune protein expression rather than inhibit it (11). The main steps of miRNA maturation are depicted in Figure 1.
Figure 1.
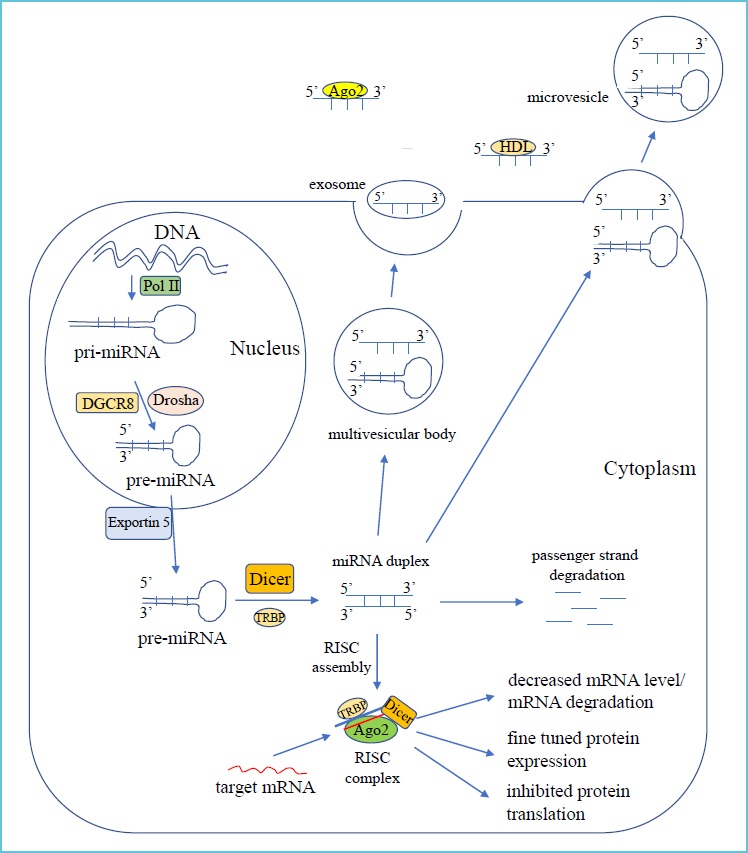
Key steps in miRNA biogenesis*
miRNAs can be released in several ways from parent cells into the plasma and remain stable in the circulation (28). They are highly resistant to endo- and exogenous RNase activity, excessive pH and temperature conditions (29). These characteristics are partly achieved by transport within exosomes and microvesicles, or being carried by RNA-binding protein, such as Ago2 protein or high-density lipoprotein (28). Microvesicles are generated by the budding of the plasma membrane and transfer functional miRNAs and also pre-miRNAs to target cells (Figure 1). These “RNA vectors” can alter cellular functions and induce biological responses (30). Cell-free miRNAs have been detected in various body fluids, such as plasma, serum, urine or saliva (31).
SIGNALING PATHWAY INVOLVED IN TLR4 ACTIVATION IN IMMUNE CELLS
Stimulation of TLRs induces the activation of NF-κB (nuclear factor kappa B) and MAPK (mitogen activated protein kinase) pathway causing the production of proinflammatory cytokines in macrophages and monocytes during the development of sepsis (32). Ten human subtypes of TLRs (TLR1-TLR10) are known to exist (33). These receptors become functional by diverse stimuli (e.g. TLR2 is activated by peptidoglycan of Gram-positive bacteria), and are localized in various cellular compartments, such as TLR2 and TLR4 are present on the cell surface, while TLR3 and TLR7-TLR9 sensing nucleotide derivatives, are located in the membrane of intracellular vesicles (33). One of the most characterized receptor in sepsis is TLR4 with agonist of lipopolysaccharide (LPS) of Gram-negative bacteria. Signaling pathway downstream of TLR4 has been investigated in detail (34). Briefly, after recognition of LPS, TLR4 recruits the myeloid differentiation primary response protein 88 (MyD88). MyD88 then recruits IL-1R-associated kinases (IRAK4, IRAK1 and IRAK2) that activate and ubiquitinylate TNFR-associated factor 6 (TRAF6). Due to the subsequent ubiquitination of TAK1-binding protein 2 (TAB2), TAK1 becomes activated. These events lead to the activation of the inhibitor of NF-κB kinase (IKK) complex consisting of IKK-α, IKK-β and NF-κB essential modulator (NEMO, or IKK-γ), which phosphorylates IΚBα and thereby releases NF-ΚB transcription factor containing p50 and p65 for translocation to the nucleus. This allows the transcription of proinflammatory genes, such as IL6 and TNF-α (34). Figure 2 depicts the major signaling events of TLR4-induced MyD88-dependent signaling with the inhibitory effect of miRNAs against different components of this pathway (described below) (Figure 2).
Figure 2.
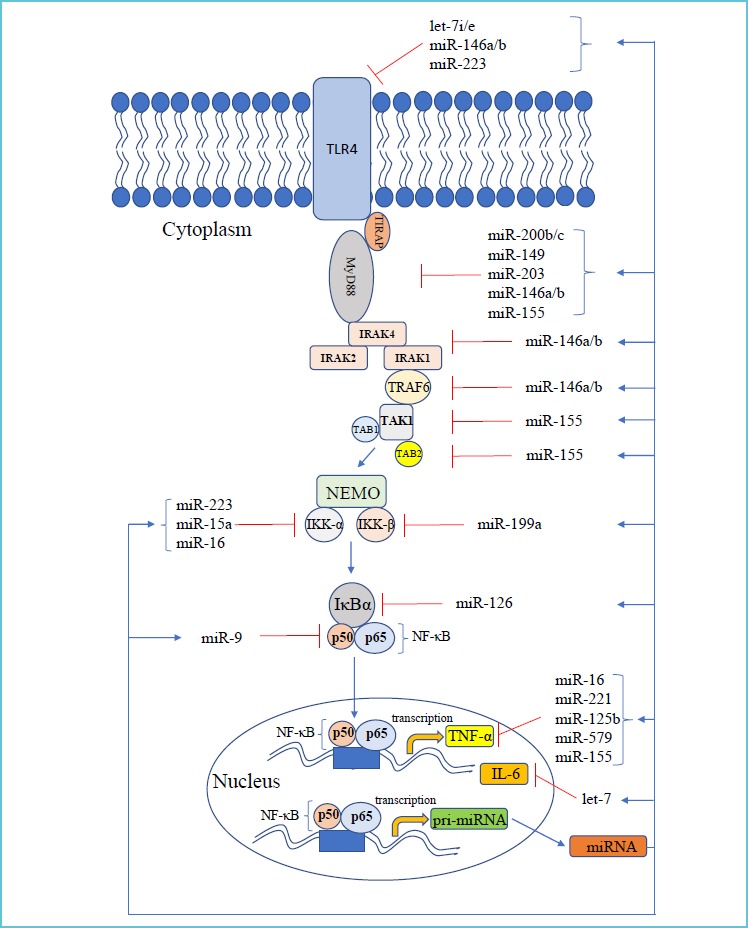
TLR4-mediated signaling with the normal inhibitory function of miRNAs*
In parallel, MyD88-independent TLR4 signaling is also induced upon infection causing production of type I interferons (35). This pathway is also under a broad regulation of miRNAs (36). Although it is evident that deregulated TLR4-induced NF-κB inflammatory response is predominantly involved in sepsis, administration of different anti-inflammatory drugs, such as TNF-α antagonist (37) or corticosteroids (38) resulted in only a moderate improvement in sepsis therapy suggesting that other regulatory factors may be also associated with sepsis.
TLR4-MEDIATED PLATELET ACTIVATION AND MEGAKARYOCYTE FUNCTION
Most TLR members are expressed on platelets and megakaryocytes (39). Hence, platelets participate in amplified inflammatory and immune response, and TLR2 and TLR4 can vastly contribute to sepsis related thrombotic complications. TLR-induced platelet activation causes platelet adhesion, aggregation, heterotypic aggregates formation, expression and release of proinflammatory cytokines and thrombin generation (39). Of note, platelets express TLR4, but not CD14, thus a low amount of soluble CD14 is required to initiate LPS-mediated platelet response (40). Upon infection, platelet activation is not directly induced by LPS via TLR4, but it is primed after stimulation elicited by other platelet agonists (41). As a result, release of soluble CD40L and platelet factor-4 is increased without higher P-selectin expression (42). Our group previously demonstrated that the rough form of LPS from S. minnesota induced platelet CD40L expression with elevated microparticle levels and potentiated platelet aggregation at low concentration of thrombin receptor activating peptide, however, P-selectin positivity was not enhanced (43). Septic patients frequently show increased platelet activation that may turn into severe thrombocytopenia because of neutrophil-dependent sequestration of activated platelets into lungs in a TLR4-dependent fashion (44). Moreover, platelet TLR4 is involved in TNF-α production after LPS administration (45), induces platelet binding to neutrophils causing formation of neutrophil extracellular nets (46), and propagates the splicing of unprocessed IL-1β and tissue factor to be translated in platelets (47, 48). Interestingly, low concentration of LPS (without induced systemic TNF-α production) caused platelet activation with enhanced platelet clearance and production increasing the thrombotic risk, while high LPS levels resulted in altered platelet reactivity not merely due to TLR4 function (49). Hence, platelets with TLR4 act at the crossroads of sepsis linking inflammation with coagulation abnormalities via propagation of thrombin generation (50) and expression and secretion of proinflammatory cytokine (47).
The molecular machinery of TLR4-mediated signaling in platelets is not fully elucidated as yet. TLR4 can trigger platelet activation via different signaling molecules. TLR4 interacts with MyD88 and TIR domain containing adaptor protein (TIRAP), and downstream proteins IRAK1, IRAK4 and TRAF6 are activated resulting in JUN N-terminal kinase (JNK) and PI3K/Akt pathway activation (51). MyD88 also forms complex with guanylyl cyclase (GC) causing cGMP protein kinase (PKG) activation and ERK phosphorylation (52). Additionally, platelets contain an intact, functional, and complete NF-κB pathway with non-genomic functions that becomes phosphorylated upon platelet stimulation (53, 54). Recently, the TLR4-PI3K-Akt-ERK-cPLA2-TXA2 pathway has been described during platelet activation (55). Figure 3 demonstrates the key intracellular elements of TLR4-induced signaling in platelets (Figure 3).
Figure 3.
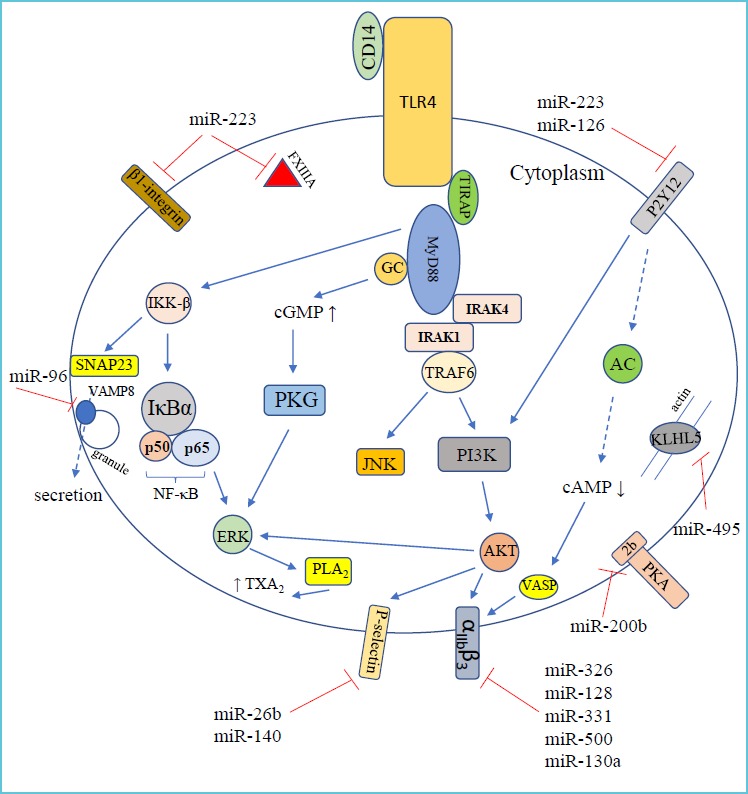
Signaling pathway downstream of TLR4 receptor in platelets with related cellular events and their regulatory miRNAs*
TLR4 is also involved in the regulation of platelet production. Expression of TLR4 was observed in human megakaryoblastic leukemia cell line MEG-01 (56), and also in human megakaryocytes in patients with myelodysplasia (57). TLR4 was elevated during maturation of murine megakaryocytes showing higher CD41 expression (44). TLR4-deficient mice showed decreased platelet count, turnover with lower RNA content and less responsiveness to thrombin-activated expression of P-selectin compared to wild type littermates (58).
In low-grade endotoxemia, platelet turnover often increases, which causes a larger number of newly formed, more active platelets (59). These platelets carry higher and altered RNA levels being more prone to produce proteins and to participate in thrombus formation compared to older platelets (60). This phenomenon was supported by a former animal model when megakaryocytes produced platelets with altered mRNA profile within 24 hours in septic mice and these platelets mediated lymphotoxicity via granzyme B (61). Accordingly, TLR4-induced signaling can modulate thrombopoiesis as well.
ALTERED LEVEL OF miRNAs BY TLR4 SIGNALING IN IMMUNE CELLS
TLR4-mediated signaling enables to modulate miRNA expression. Several miRNAs are up-regulated following LPS stimulation, such as miR-155, miR-146a, miR-21, miR-223, miR-9 and let-7e, etc. in monocytes and macrophages. Among them, there are subtypes with early (e.g. miR-155) and late (e.g. miR-21) response in expression, but these all are NF-κB-dependent.
On the other hand, some miRNAs are down-regulated (e.g. miR-125b and let-7i) due to TLR signaling via transcriptional repression or destabilization of miRNA transcripts (12).
REGULATION OF TLR4-MEDIATED SIGNALING CASCADE BY miRNAs IN SEPSIS
miRNAs have been implicated as an important link between the innate and adaptive immune systems, and their dysregulation might have a role in the pathogenesis of inflammatory diseases (12). miRNAs directly target signaling proteins and control NF-κB activity in immune cells, thus have been identified as novel regulators of immune system (12, 62). Figure 2 depicts those key cellular miRNAs, which negatively regulate the components of TLR4-induced NF-κB signaling pathway (Figure 2). TLR4 expression is highly restricted to immune cell types, such as macrophages, dendritic cells, T- and B-cells. The expression of receptor is regulated by let-7 family. As such, overexpression of let-7i resulted in reduced level of TLR4 in human cholangiocytes (63), while transfection of antisense miRNA to let-7e in macrophages caused enhanced LPS-induced cytokine response via higher expression of TLR4 (64). MiR-146b also targets TLR4 based on luciferase reporter assays of HEK293 cells when there was decreased activity of reporter genes containing the 3’-UTR of TLR4 when miR-146b was transfected (65). In addition, myeloid-specific miR-223, which has an important role in granulopoiesis (66), is also a regulator of TLR4 (67) (Figure 2). Based on these results, miRNAs can tone down TLR4 expression.
Signaling molecules downstream of TLR4 function under the control of miRNAs. Expression of MyD88 protein is affected by miR-200b and miR-200c (68). Furthermore, miR-149 negatively regulated MyD88 protein levels in RAW264.7 cells when lentiviral vector expressing miR-149 was transfected (69). Overexpression of miR-203 resulted in significantly repressed translation of MyD88 in macrophages (70). Finally, miR-155 decreased MyD88 expression at protein but not mRNA level suggesting that the miR-155-mediated inhibition is a post-transcriptional event in HEK293 cells (71). MiR-146 family negatively regulates the translation of IRAK1 and TRAF6. Taganov et al. reported for the first time that there are miRNAs which are up-regulated by TLR signaling, and in turn, miR-146a and miR-146b were found to suppress IRAK1 and TRAF6 (72). This role of miR-146b was later confirmed by others (65). The IKK complex is essential for NF-κB activation. Decreased levels of miR-15a, miR-16 and miR-223 were detected with elevated IKK-α levels when human monocytes were differentiated with granulocyte-monocyte colony stimulating factor (73). IKK-3 has complementary sequences for miR-199a and the transfection of this miRNA caused reduced IKK-3 level studied in ovarian cancer cells (74).
Similarly, a direct regulatory association between miR-126 and IκBα was described in HT29 cells (75) (Figure 2).
Via targeting transcription factors along TLR4 pathway, miRNAs can have a global impact on TLR4-induced gene expression (76). miRNAs induced by a particular signaling can inhibit the transcription factor, which is necessary for its expression. For example, elevated level of miR-155 feeds back and targets FOXP3 to decrease its expression (77). NFKB1 that is cleaved to form the NF-κB subunit p50, was shown to be targeted by miR-9 in human monocytes and neutrophils (78) (Figure 2).
TLR4-mediated NF-κB activation causes excessive production of proinflammatory cytokines, e.g. TNF-α and IL-6. Expression of these cytokines is directly targeted by miRNAs as well. The repressive effect of miR-579, miR-221, and miR-125 was studied on TNF-α during LPS tolerance in THP-1 cells (79). In addition, TNF-α translation could be also influenced by miR-16 (80) and miR-155 (81). Inflammatory response mediated by NF-κB rapidly reduced let-7 levels resulting in higher levels of IL-6 in cancer cells. As IL-6 activates NF-κB, thereby completes a positive feedback loop that maintains the epigenetic transformed state in the absence of the inducing signal (82) (Figure 2).
Among TLR4 signaling regulators, acetylcholinesterase that blocks NF-κB translocation, is regulated by miR-132 (83), translation inhibitor PDCD4 is targeted by miR-21 (84), while negative regulator SHIP1 is a primary target of miR-155 (85) in macrophages. These two later miRNAs with these effects can indeed fine tune TLR4 signaling that can be important for LPS tolerance or in the resolution of TLR4-induced responses (12). Overall, accumulating data above clearly demonstrate that miRNAs highly control each level of this very complex machinery of TLR-mediated signaling in immune cells.
IMPACT OF miRNAs ON PLATELET FUNCTION
Platelets play an important role in vascular integrity. They circulate in a resting state and become activated after vessel injury by exposed collagen and von Willebrand factor to adhere and aggregate for prevention of bleeding (86). miRNAs are also carried by platelets due to the delivery of miRNAs with mRNAs from megakaryocyte (87). The fact about functional miRNAs in platelets without nucleus was questioned for a long time, thus it was also obscure whether platelets are able to produce proteins de novo when being exposed to different challenges. Platelet miRNAs have been studied in relation to the expression of platelet receptors or other activation-related intracellular proteins (88). As yet, only a few miRNAs have been proved as regulator of platelet proteins, such miR-223 regulates ADP receptor P2Y12, intracellular FXIII-A, and integrin β1 expression (89). P2Y12 receptor is targeted by miR-126 as well (90). VAMP8/endobrevin, a critical v-SNARE protein involved in platelet granule secretion, is regulated by miR-96 (91), while the expression of αIIβ3 receptor is controlled by miR-326, miR-128, miR-331, miR-500 (92) and miR-130a (93).
Our group has recently reported that miR-26b and miR-140 could affect SELP (P-selectin) mRNA level in MEG-01 cells (94). In addition, Nagalla et al. described miR-200b:PRKAR2B (encoding the β-regulatory chain of cAMP-dependent protein kinase type II, PKA) and miR-495:KLHL5 (encoding a Kelch-like protein that binds actin) interactions in platelets (95) (Figure 3). Of note, these functions of platelet miRNAs above were not investigated specifically among septic conditions as yet.
CIRCULATING miRNAs AS LABORATORY BIOMARKERS IN SEPSIS
Besides their normal function in the regulation of gene expression, altered miRNA levels in plasma/serum have been intensively investigated with regards to their role as possible biomarkers in different human diseases, also in sepsis (20, 23, 24). Abnormal circulating miRNA levels reflected the pathophysiological processes during severe inflammation and bacterial infection, which was profiled in sepsis mice model using cecal ligation and puncture (CLP) (96). Cell-free miRNAs are stable in the circulation, and can be quantified more rapidly compared to time consuming microbial cultures. Hence, plasma or serum miRNAs may serve as potential biomarkers in the differential diagnosis of sepsis from SIRS, and may act as prognostic parameters under treatment (23). We have summarized the results of previous clinical studies profiling serum or plasma miRNAs (Table 1), and some key miRNAs are further described in detail below.
Table 1.
Circulating miRNA alterations in sepsis vs. SIRS subjects or healthy controls
| miRNAs | Body fluid/sample | Study populations | Methods | Results | Reference |
|---|---|---|---|---|---|
| miR-150 | plasma/blood leukocytes | 24 sepsis vs. 32 healthy controls | microarray | ↓ (sepsis vs. controls) | 98 |
| serum | 138 sepsis, 85 ICU w/o sepsis vs. 76 healthy controls | qRT-PCR | ↓ (non-survival vs. survival) | 99 | |
| plasma/blood leukocytes | 23 sepsis, 22 SIRS, 21 healthy controls | HiSeq Sequencing | ↓ (sepsis vs. SIRS) (sepsis vs. controls) | 100 | |
| plasma | 120 sepsis, 50 healthy controls | qRT-PCR | ↓ (sepsis vs. controls) | 101 | |
| miR-146a | serum | 50 sepsis, 30 SIRS, 20 healthy controls | qRT-PCR | ↓ (sepsis vs. SIRS) (sepsis vs. controls) | 105 |
| plasma | 14 sepsis, 14 SIRS | qRT-PCR | ↓ (sepsis vs. SIRS) | 106 | |
| blood leukocytes | 226 sepsis, 206 healthy controls | qRT-PCR | ↓ (sepsis vs. controls) | 108 | |
| blood leukocytes | 32 sepsis, 38 healthy controls | microarray | ↓ (sepsis vs. controls) | 107 | |
| miR-223 | serum | 117 sepsis survivor, 97 sepsis non-survivor | Solexa Sequencing | ↓ (non-survival vs. survival) | 111 |
| serum | 50 sepsis, 30 SIRS, 20 healthy controls | qRT-PCR | ↓ (sepsis vs. SIRS) (sepsis vs. controls) | 105 | |
| serum | 137 sepsis, 84 healthy controls | qRT-PCR | ↔ (sepsis vs. controls) | 110 | |
| plasma | 25 neonatal sepsis, 25 non-sepsis controls | qRT-PCR | ↓ (sepsis vs. controls) | 109 | |
| miR-15a/16 | serum | 166 sepsis, 32 SIRS, 24 healthy controls | qRT-PCR | ↑ (sepsis vs. SIRS) (sepsis vs. controls) | 113 |
| serum | 46 neonatal sepsis, 41 non-sepsis controls | qRT-PCR | ↑ (sepsis vs. controls) | 114 | |
| serum | 117 sepsis survivor, 97 sepsis non-survivor | Solexa Sequencing | ↑*/↓** (non-survival vs. survival) | 111 | |
| plasma | 29 sepsis w/shock, 33 sepsis w/o shock, 32 controls | qRT-PCR | ↑ (septic shock vs. sepsis) | 115 | |
| miR-122 | serum | 117 sepsis survivor, 97 sepsis non-survivor | Solexa Sequencing | ↑ (non-survival vs. survival) | 111 |
| serum | 54 sepsis w/coagulation disorder (CD), 69 sepsis w/o CD | qRT-PCR | ↑ (CD sepsis vs. non-CD sepsis) | 117 | |
| serum | 108 sepsis, 20 healthy controls | qRT-PCR | ↑ (sepsis vs. controls) | 118 |
* denotes alteration of miR-15a
** denotes change in miR-16 levels.
miR-150
This miRNA was formerly identified as a key regulator of immune cell differentiation and activation (17). During the maturation of B- and T-cells, miR-150 expression is down-regulated. When LPS was injected into humans, miR-150 levels went down in leukocytes (92). Since then, several trails reported that plasma/serum miR-150 was decreased in patients with sepsis at different degree (98, 99). Furthermore, the reduction of miR-150 showed a strong correlation with the severity of sepsis and the concentrations of plasma IL-18 (98). On the other hand, higher miR-150 correlated with the survival of septic patients suggesting its reliable prognostic value (99). Based on a genome-wide sequencing of cellular miRNAs, miR-150 was able to discriminate between patients who had SIRS and those with sepsis (100). Recently, plasma miR-150 was found to be lower than normal that correlated with renal dysfunction and 28-day survival as well as plasma levels of IL-6 and TNF-α (101). MiR-150 expression was significantly decreased in human umbilical vein endothelial cells (HUVECs) in vitro after LPS treatment, and over-expression of miR-150 could protect HUVECs from LPS-induced inflammatory response and apoptosis targeting NF-κB1 (101). Finally, reduced miR-150 level in peripheral leukocytes correlated with Gram-negative bacterial sepsis in the urogenital tract (102).
miR-146a and miR-223
MiR-146a has been widely studied in connection with immune response (72) and chronic inflammatory disorders (103). In parallel, miR-223 has been considered to play a key role in hematopoietic lineage differentiation (66), and was found to be up-regulated in the mucosa of colon in inflammatory bowel disease (104). Wang et al. described for the first time that there were significantly reduced serum levels of these miRNAs in septic patients compared to SIRS individuals and healthy controls (105). Additionally, the areas under the receiver operating characteristic curve (AUC-ROC) value was much higher in case of miR-223 (0.858) and miR-146a (0.804) vs. traditional biomarkers IL-6 (0.785) and CRP (0.589) (105). Similarly, septic subjects demonstrated lower plasma miR-146a levels than patients with SIRS (106). The expression of miR-146a was also down-regulated in blood leukocytes from sepsis (107, 108). Similarly, plasma miR-223 was decreased in neonatal sepsis vs. non-sepsis controls (109), while Benz et al. did not find significant difference in this miRNA level (110). However, when miR-223 was correlated with the outcome of sepsis, miR-223 was significantly lower in non-survivors in contrast to survivors of sepsis (111). Taken together, these studies suggested that decreased miR-223 and miR-146a levels were optimal for diagnosis of sepsis.
miR-15a and miR-16
These miRNAs were originally identified as tumor suppressors and dysregulated levels of miR-15a and miR-16 was found in certain solid tumors, such as in ovarian cancer (112). Since then, miR-15a and miR-16 were associated with innate immune system by targeting IκBα mRNA upstream of NF-κB pathway (73). Both miRNAs were found to be higher in both sepsis and SIRS subjects vs. normal controls (113). In addition, miR-15a had substantial AUC value (0.858) for the diagnosis of sepsis compared CRP (0.572) and PCT (0.605) and showed 94.4% specificity (113). Up-regulated miR-15a/16 was reported from the serum of neonatal sepsis patients, while transfection of miR15a/16 mimics into RAW264.7 macrophages down-regulated TLR4 and IRAK-1 in LPS-treated cells (114). Interestingly, diverse expression was seen in these miRNAs among survivor and non-survivors, i.e. serum miR-15a was significantly higher in non-survivors, while miR-16 was lower in this subgroup (111). Finally, higher level of miR-15a was associated with the development of septic shock in contrast to those with sepsis only (115).
miR-122
Although miR-122 is a liver-related miRNA, and thus showed elevated levels in liver injury earlier (116), its potential role in the diagnosis and prognosis of sepsis has been revealed as well. The level of miR-122 was elevated in sepsis, and even higher expression was found in those who did not survive sepsis (111). Moreover, increased serum miR-122 was correlated to coagulation disorders in sepsis (117). Very recently, multivariate regression analysis showed that serum miR-122 was an independent prognostic factor for 30 day-mortality based on Sepsis-3 criteria (118).
Other miRNAs
In addition to the outlined miRNAs, a large number of other miRNAs have been investigated in relation to sepsis. MiR-486 and miR-182 were overexpressed, while miR-342 was down-regulated in peripheral blood leukocytes of septic individuals (98). Altered levels of miRNAs predicted the outcome of sepsis. Serum miR-483 (111) and miR-297 (119) were reduced, while miR-574 (119) was increased in survivors than non-survivors. Tacke et al. found that miR-133a levels were elevated in sepsis that correlated with sepsis severity, SOFA (Sequential Organ Failure Assessment) scores and CRP/PCT concentrations (120). The members of miR-4772 family were up-regulated in both ex vivo blood leukocytes and in vitro stimulated monocytes by LPS, however it was not able to differentiate sepsis from SIRS (100). A miRNA regulatory network with pathway analysis, disease ontology analysis and protein-protein interaction network analysis were applied to test miRNA reliability. Huang et al. identified 7 miRNAs, which have the potential to be diagnostic (miR-15a, miR-16, miR-122, miR-146a, miR-223, miR-499, miR-150) and 3 prognostic sepsis biomarkers (miR-483, miR-574, miR-193) (121).
CELL-FREE miRNAs AS THROMBOSIS SENTINELS
Although the role of miRNAs in the regulation of hemostasis and in the development of coagulation disorders has not been totally clarified as yet, panels of plasma miRNAs may aid to diagnose and monitor coagulation-related diseases (122). For example, cell-free miRNAs alterations were directly associated with coronary artery disease, acute ischemic stroke, antiphospholipid syndrome reviewed in Ref. 122. Since platelets are major source of circulating miRNAs, change in their plasma levels can effectively indicate platelet activation and related vascular disorders (123). Further clinical studies are also required to evaluate the potential of circulating miRNAs for time-course detection of sepsis-induced platelet activation with or without disseminated intravascular coagulopathy, as it was performed in rats after non-lethal endotoxin injection (124).
CONCLUSIONS
The properties of miRNAs hold potentials for analyzing them as novel diagnostic and prognostic biomarkers. Although there are several circumstances that may challenge the analysis of circulating miRNAs, they may become routinely accessible, non-invasive molecular biomarkers in the near future based on the results of recent clinical trials (14). In addition, circulating miRNAs seem to be critical components of the pathogenesis of diseases like other already established biomarkers, such as BCR-ABL or HER2 in malignancy, thus they are not only the molecular remnants of different cell types, but rather of functional importance (125).
Consequently, miRNAs represent not only a new diagnostic repertoire, but targeted drugs can be developed to inhibit diseases with altered miRNA profile.
Acknowledgements
This publication was supported by OTKA Bridging Fund for Bela Nagy Jr. (Faculty of General Medicine, University of Debrecen). Béla Nagy Jr. was also supported by Szodoray Lajos Fellowship of the University of Debrecen.
REFERENCES
- 1.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajaee A, Barnett R, Cheadle WG. Pathogen- and Danger-Associated Molecular Patterns and the Cytokine Response in Sepsis. Surg Infect (Larchmt). 2018;19(2):107-116. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, et al. ; International Forum of Acute Care Trialists. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med. 2016;193(3):259-272. [DOI] [PubMed] [Google Scholar]
- 4.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(9):840-851. [DOI] [PubMed] [Google Scholar]
- 5.Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care. 2010;14(1):R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand D, Das S, Bhargava S, Srivastava LM, Garg A, Tyagi N, et al. Procalcitonin as a rapid diagnostic biomarker to differentiate between culture-negative bacterial sepsis and systemic inflammatory response syndrome: a prospective, observational, cohort study. J Crit Care. 2015;30(1):218.e7-12. [DOI] [PubMed] [Google Scholar]
- 7.Póvoa P, Coelho L, Almeida E, Fernandes A, Mealha R, Moreira P, et al. Early identification of intensive care unit-acquired infections with daily monitoring of C-reactive protein: a prospective observational study. Crit Care. 2006;10(2):R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro NI, Howell MD, Talmor D, Nathanson LA, Lisbon A, Wolfe RE, et al. Serum lactate as a predictor of mortality in emergency department patients with infection. Ann Emerg Med. 2005;45(5):524-528. [DOI] [PubMed] [Google Scholar]
- 9.Du B, Pan J, Chen D, Li Y. Serum procalcitonin and interleukin-6 levels may help to differentiate systemic inflammatory response of infectious and non-infectious origin. Chin Med J (Engl). 2003;116(4):538-542. [PubMed] [Google Scholar]
- 10.Sankar V, Webster NR. Clinical application of sepsis biomarkers. J Anesth. 2013;27(2):269-283. [DOI] [PubMed] [Google Scholar]
- 11.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the finetuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11(3):163-175. [DOI] [PubMed] [Google Scholar]
- 13.Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol. 2010;2(4):161-179. [PMC free article] [PubMed] [Google Scholar]
- 14.Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80(2):193-208. [DOI] [PubMed] [Google Scholar]
- 15.Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One. 2014;9(2):e89565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ojha R, Nandani R, Pandey RK, Mishra A, Prajapati VK. Emerging role of circulating microRNA in the diagnosis of human infectious diseases. J Cell Physiol. 2019;234(2):1030-1043. [DOI] [PubMed] [Google Scholar]
- 17.Tsitsiou E, Lindsay MA. MicroRNAs and the immune response. Curr Opin Pharmacol. 2009;9(4):514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caserta S, Mengozzi M, Kern F, Newbury SF, Ghezzi P, Llewelyn MJ. Severity of Systemic Inflammatory Response Syndrome Affects the Blood Levels of Circulating Inflammatory-Relevant MicroRNAs. Front Immunol. 2018;8:1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogobete AF, Sandesc D, Bedreag OH, Papurica M, Popovici SE, Bratu T, et al. MicroRNA Expression is Associated with Sepsis Disorders in Critically Ill Polytrauma Patients. Cells. 2018;7(12). pii: E271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benz F, Roy S, Trautwein C, Roderburg C, Luedde T. Circulating MicroRNAs as Biomarkers for Sepsis. Int J Mol Sci. 2016;17(1). pii: E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumache R, Rogobete AF, Bedreag OH, Sarandan M, Cradigati AC, Papurica M, et al. Use of miRNAs as biomarkers in sepsis. Anal Cell Pathol (Amst). 2015;2015:186716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reithmair M, Buschmann D, Marte M, Kirchner B, Hagl D, Kaufmann I, et al. Cellular and extracellular miRNAs are blood-compartment-specific diagnostic targets in sepsis. J Cell Mol Med. 2017;21(10):2403-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Essandoh K, Fan GC. Role of extracellular and intracellular microRNAs in sepsis. Biochim Biophys Acta. 2014;1842(11):2155-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingsley SMK, Bhat BV. Role of microRNAs in sepsis. Inflamm Res. 2017;66(7):553-569. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415-419. [DOI] [PubMed] [Google Scholar]
- 27.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293(5538):2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: A Collision of Two Paradigms. Trends Biochem Sci. 2016;41(10):883-892. [DOI] [PubMed] [Google Scholar]
- 29.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581-593. [DOI] [PubMed] [Google Scholar]
- 30.Nomura S. Extracellular vesicles and blood diseases. Int J Hematol. 2017;105(4):392-405. [DOI] [PubMed] [Google Scholar]
- 31.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujimoto H, Ono S, Efron PA, Scumpia PO, Moldawer LL, Mochizuki H. Role of Toll-like receptors in the development of sepsis. Shock. 2008;29(3):315-321. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805-820. [DOI] [PubMed] [Google Scholar]
- 34.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145-151. [DOI] [PubMed] [Google Scholar]
- 35.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185(10):6226-6233. [DOI] [PubMed] [Google Scholar]
- 36.Forster SC, Tate MD, Hertzog PJ. MicroRNA as Type I Interferon-Regulated Transcripts and Modulators of the Innate Immune Response. Front Immunol. 2015;6:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinhart K, Menges T, Gardlund B, Harm Zwaveling J, Smithes M, Vincent JL, et al. Randomized, placebo-controlled trial of the anti-tumor necrosis factor antibody fragment afelimomab in hyperinflammatory response during severe sepsis: The RAMSES Study. Crit Care Med. 2001;29(4):765-769. [DOI] [PubMed] [Google Scholar]
- 38.Rochwerg B, Oczkowski SJ, Siemieniuk RAC, Agoritsas T, Belley-Cote E, D’Aragon F, et al. Corticosteroids in Sepsis: An Updated Systematic Review and Meta-Analysis. Crit Care Med. 2018;46(9):1411-1420. [DOI] [PubMed] [Google Scholar]
- 39.D’ Atri LP, Schattner M. Platelet toll-like receptors in thromboinflammation. Front Biosci (Landmark Ed). 2017;22:1867-1883. [DOI] [PubMed] [Google Scholar]
- 40.Damien P, Cognasse F, Eyraud MA, Arthaud CA, Pozzetto B, Garraud O, et al. LPS stimulation of purified human platelets is partly dependent on plasma soluble CD14 to secrete their main secreted product, soluble-CD40-Ligand. BMC Immunol. 2015;16:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montrucchio G, Bosco O, Del Sorbo L, Fascio Pecetto P, Lupia E, Goffi A, et al. Mechanisms of the priming effect of low doses of lipopoly-saccharides on leukocyte-dependent platelet aggregation in whole blood. Thromb Haemost. 2003;90(5):872-881. [DOI] [PubMed] [Google Scholar]
- 42.Cognasse F, Hamzeh-Cognasse H, Lafarge S, Delezay O, Pozzetto B, McNicol A, et al. Toll-like receptor 4 ligand can differentially modulate the release of cytokines by human platelets. Br J Haematol. 2008;141:84-91. [DOI] [PubMed] [Google Scholar]
- 43.Kappelmayer J, Beke Debreceni I, Vida A, Antal-Szalmas P, Clemetson KJ, Nagy B., Jr Distinct effects of Reand S-forms of LPS on modulating platelet activation. J Thromb Haemost. 2013;11:775-778. [DOI] [PubMed] [Google Scholar]
- 44.Andonegui G, Kerfoot SM, McNagny K, Ebbert KV, Patel KD, Kubes P. Platelets express functional Toll-like receptor-4. Blood. 2005;106:2417-2423. [DOI] [PubMed] [Google Scholar]
- 45.Aslam R, Speck ER, Kim M, Crow AR, Bang KW, Nestel FP, et al. Platelet Toll-like receptor expression modulates lipopolysaccharide-induced thrombocytopenia and tumor necrosis factor-alpha production in vivo. Blood. 2006;107(2):637-641. [DOI] [PubMed] [Google Scholar]
- 46.Brinkmann V. Neutrophil Extracellular Traps in the Second Decade. J Innate Immun. 2018;10(5-6):414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shashkin PN, Brown GT, Ghosh A, Marathe GK, McIntyre TM. Lipopolysaccharide is a direct agonist for platelet RNA splicing. J Immunol. 2008;181:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rondina MT, Schwertz H, Harris ES, Kraemer BF, Campbell RA, Mackman N, et al. The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J Thromb Haemost. 2011;9:748-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jayachandran M, Miller VM, Brunn GJ, Owen WG. Platelet response as a sentinel marker of toll-like receptor 4 activation in mice. Thromb Res. 2010;126(5):414-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tóth J, Debreceni IB, Deak A, Peto K, Berhes M, Hajdu E, et al. Characteristics of thrombin generation in a fulminant porcine sepsis model. Thromb Res. 2017;158:25-34. [DOI] [PubMed] [Google Scholar]
- 51.Berthet J, Damien P, Hamzeh-Cognasse H, Pozzetto B, Garraud O, Cognasse F. Toll-like receptor 4 signal transduction in platelets: novel pathways. Br J Haematol. 2010;151(1):89-92. [DOI] [PubMed] [Google Scholar]
- 52.Zhang G, Han J, Welch EJ, Ye RD, Voyno-Yasenetskaya TA, Malik AB, et al. Lipopolysaccharide stimulates platelet secretion and potentiates platelet aggregation via TLR4/MyD88 and the cGMP-dependent protein kinase pathway. J Immunol. 2009;182(12):7997-8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spinelli SL, Casey AE, Pollock SJ, Gertz JM, McMillan DH, Narasipura SD, et al. Platelets and megakaryocytes contain functional nuclear factor-kappaB. Arterioscler Thromb Vasc Biol. 2010;30(3):591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivadeneyra L, Carestia A, Etulain J, Pozner RG, Fondevila C, Negrotto S, et al. Regulation of platelet responses triggered by Toll-like receptor 2 and 4 ligands is another non-genomic role of nuclear factor-kappaB. Thromb Res. 2014;133(2):235-243. [DOI] [PubMed] [Google Scholar]
- 55.Lopes Pires ME, Clarke SR, Marcondes S, Gibbins JM. Lipopolysaccharide potentiates platelet responses via toll-like receptor 4-stimulated Akt-Erk-PLA2 signalling. PLoS One. 2017;12(11):e0186981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward JR, Bingle L, Judge HM, Brown SB, Storey RF, Whyte MK, et al. Agonists of toll-like receptor (TLR)2 and TLR4 are unable to modulate platelet activation by adenosine diphosphate and platelet activating factor. Thromb Haemost. 2005;94(4):831-838. [PubMed] [Google Scholar]
- 57.Maratheftis Cl, Andreakos E, Moutsopoulos HM, Voulgarelis M. Toll-like receptor-4 is up-regulated in hematopoietic progenitor cells and contributes to increased apoptosis in myelodysplastic syndromes. Clin Cancer Res. 2007;13(4):1154-1160. [DOI] [PubMed] [Google Scholar]
- 58.Jayachandran M, Brunn GJ, Karnicki K, Miller RS, Owen WG, Miller VM. In vivo effects of lipopolysaccharide and TLR4 on platelet production and activity: implications for thrombotic risk. J Appl Physiol (1985). 2007;102:429-433. [DOI] [PubMed] [Google Scholar]
- 59.Stohlawetz P, Folman CC, von dem Borne AE, Pernerstorfer T, Eichler HG, Panzer S, et al. Effects of endotoxemia on thrombopoiesis in men. Thromb Haemost. 1999;81:613-617. [PubMed] [Google Scholar]
- 60.Harrison P, Goodall AH. “Message in the platelet”- more than just vestigial mRNA! Platelets. 2008;19:395-404. [DOI] [PubMed] [Google Scholar]
- 61.Freishtat RJ, Natale J, Benton AS, Cohen J, Sharron M, Wiles AA, et al. Sepsis alters the megakaryocyte-platelet transcriptional axis resulting in granzyme B-mediated lymphotoxicity. Am J Respir Crit Care Med. 2009;179:467-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 2011;3(3):159-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282(39):28929-28938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31(2):220-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci U S A. 2013;110(28):11499-11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. 2013;274(3):215-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83-86. [DOI] [PubMed] [Google Scholar]
- 68.Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME. The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-κB activation. Innate Immun. 2012;18(6):846-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu G, Zhang Z, Xing Y, Wei J, Ge Z, Liu X, et al. MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. J Cell Biochem. 2014;115(5):919-927. [DOI] [PubMed] [Google Scholar]
- 70.Wei J, Huang X, Zhang Z, Jia W, Zhao Z, Zhang Y, et al. MyD88 as a target of microRNA-203 in regulation of lipopolysaccharide or Bacille Calmette-Guerin induced inflammatory response of macrophage RAW264.7 cells. Mol Immunol. 2013;55(3-4):303-309. [DOI] [PubMed] [Google Scholar]
- 71.Tang B, Xiao B, Liu Z, Li N, Zhu ED, Li BS, et al. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett. 2010;584(8):1481-1486. [DOI] [PubMed] [Google Scholar]
- 72.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481-12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11(9):799-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27(34):4712-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng X, Wang H, Ye S, Guan J, Tan W, Cheng S, et al. Up-regulation of microRNA-126 may contribute to pathogenesis of ulcerative colitis via regulating NF-kappaB inhibitor IKBα. PLoS One. 2012;7(12):e52782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31(4):435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182(5):2578-2582. [DOI] [PubMed] [Google Scholar]
- 78.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106(13):5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem. 2010;285(27):20940-20951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120(5):623-634. [DOI] [PubMed] [Google Scholar]
- 81.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082-5089. [DOI] [PubMed] [Google Scholar]
- 82.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, et al. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31(6):965-973. [DOI] [PubMed] [Google Scholar]
- 84.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11(2):141-147. [DOI] [PubMed] [Google Scholar]
- 85.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106(17):7113-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ruggeri ZM. Platelets in atherothrombosis. Nat Med. 2002;8:1227-1234. [DOI] [PubMed] [Google Scholar]
- 87.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16:961-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edelstein LC, McKenzie SE, Shaw C, Holinstat MA, Kunapuli SP, Bray PF. MicroRNAs in platelet production and activation. J Thromb Haemost. 2013;11 Suppl 1:340-350. [DOI] [PubMed] [Google Scholar]
- 89.Elgheznawy A, Shi L, Hu J, Wittig I, Laban H, Pircher J, et al. Dicer cleavage by calpain determines platelet microRNA levels and function in diabetes. Circ Res. 2015;117(2):157-165. [DOI] [PubMed] [Google Scholar]
- 90.Kaudewitz D, Skroblin P, Bender LH, Barwari T, Willeit P, Pechlaner R, et al. Association of MicroRNAs and YRNAs With Platelet Function. Circ Res. 2016;118(3):420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kondkar AA, Bray MS, Leal SM, et al. VAMP8/endobrevin is overexpressed in hyperreactive human platelets: suggested role for platelet microRNA. J Thromb Haemost. 2010;8: 369-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rowley JW, Chappaz S, Corduan A, et al. Dicer1-mediated miRNA processing shapes the mRNA profile and function of murine platelets. Blood. 2016;127: 1743-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103(13):5078-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fejes Z, Poliska S, Czimmerer Z, Kaplar M, Penyige A, Gal Szabó G, et al. Hyperglycemia suppresses microRNA expression in platelets to increase P2RY12 and SELP levels in type 2 diabetes mellitus. Thromb Haemost. 2017;117:529-542. [DOI] [PubMed] [Google Scholar]
- 95.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117(19):5189-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu SC, Yang JC, Rau CS, Chen YC, Lu TH, Lin MW, et al. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS One. 2013;8(10):e77936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schmidt WM, Spiel AO, Jilma B, Wolzt M, Müller M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun. 2009;380(3):437-441. [DOI] [PubMed] [Google Scholar]
- 98.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009;4(10):e7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roderburg C, Luedde M, Vargas Cardenas D, Vucur M, Scholten D, Frey N, et al. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One. 2013;8(1):e54612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ma Y, Vilanova D, Atalar K, Delfour O, Edgeworth J, Ostermann M, et al. Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PLoS One. 2013;8(10):e75918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma Y, Liu Y, Hou H, Yao Y, Meng H. MiR-150 predicts survival in patients with sepsis and inhibits LPS-induced inflammatory factors and apoptosis by targeting NF-κB1 in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2018;500(3):828-837. [DOI] [PubMed] [Google Scholar]
- 102.How CK, Hou SK, Shih HC, Huang MS, Chiou SH, Lee CH, Juan CC. Expression profile of MicroRNAs in gram-negative bacterial sepsis. Shock. 2015;43(2):121-127. [DOI] [PubMed] [Google Scholar]
- 103.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EKL. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10(4):R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fasseu M, Treton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5(10). pii: e13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, et al. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394(1):184-188. [DOI] [PubMed] [Google Scholar]
- 106.Wang L, Wang HC, Chen C, Zeng J, Wang Q, Zheng L, et al. Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Exp Ther Med. 2013;5(4):1101-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou J, Chaudhry H, Zhong Y, Ali MM, Perkins LA, Owens WB, et al. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine. 2015;71(1):89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shao Y, Li J, Cai Y, Xie Y, Ma G, Li Y, et al. The functional polymorphisms of miR-146a are associated with susceptibility to severe sepsis in the Chinese population. Mediators Inflamm. 2014;2014:916202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dhas BB, Dirisala VR, Bhat BV. Expression Levels of Candidate Circulating microRNAs in Early-Onset Neonatal Sepsis Compared With Healthy Newborns. Genomics Insights. 2018;11:1178631018797079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Benz F, Tacke F, Luedde M, Trautwein C, Luedde T, Koch A, et al. Circulating microRNA-223 serum levels do not predict sepsis or survival in patients with critical illness. Dis Markers. 2015;2015:384208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients’ mortality: a prospective observational study. PLoS One. 2012;7(6):e38885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, et al. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69(23):9090-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie LX. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin Chem Lab Med. 2012;50(8):1423-1428. [DOI] [PubMed] [Google Scholar]
- 114.Wang X, Wang X, Liu X, Wang X, Xu J, Hou S, et al. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int J Clin Exp Med. 2015;8(4):5683-5690. [PMC free article] [PubMed] [Google Scholar]
- 115.Goodwin AJ, Guo C, Cook JA, Wolf B, Halushka PV, Fan H. Plasma levels of microRNA are altered with the development of shock in human sepsis: an observational study. Crit Care. 2015;19:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roderburg C, Benz F, Vargas Cardenas D, Koch A, Janssen J, Vucur M, et al. Elevated miR-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int. 2015;35(4):1172-1184. [DOI] [PubMed] [Google Scholar]
- 117.Wang HJ, Deng J, Wang JY, Zhang PJ, Xin Z, Xiao K, et al. Serum miR-122 levels are related to coagulation disorders in sepsis patients. Clin Chem Lab Med. 2014;52(6):927-933. [DOI] [PubMed] [Google Scholar]
- 118.Rahmel T, Schafer ST, Frey UH, Adamzik M, Peters J. Increased circulating microRNA-122 is a biomarker for discrimination and risk stratification in patients defined by sepsis-3 criteria. PLoS One. 2018;13(5):e0197637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang H, Meng K, Chen Wj, Feng D, Jia Y, Xie L. Serum miR-574-5p: a prognostic predictor of sepsis patients. Shock. 2012;37(3):263-267. [DOI] [PubMed] [Google Scholar]
- 120.Tacke F, Roderburg C, Benz F, Cardenas DV, Luedde M, Hippe HJ, et al. Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically ill patients. Crit Care Med. 2014;42(5):1096-1104. [DOI] [PubMed] [Google Scholar]
- 121.Huang J, Sun Z, Yan W, Zhu Y, Lin Y, Chen J, et al. Identification of microRNA as sepsis biomarker based on miRNAs regulatory network analysis. Biomed Res Int. 2014;2014:594350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tay J, Tiao J, Hughes Q, Jorritsma J, Gilmore G, Baker R. Circulating MicroRNA as Thrombosis Sentinels: Caveats and Considerations. Semin Thromb Hemost. 2018;44(3):206-215. [DOI] [PubMed] [Google Scholar]
- 123.Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res. 2013;112(4):595-600. [DOI] [PubMed] [Google Scholar]
- 124.Brooks MB, Turk JR, Guerrero A, Narayanan PK, Nolan JP, Besteman EG, et al. Non-Lethal Endotoxin Injection: A Rat Model of Hypercoagulability. PLoS One. 2017;12(1):e0169976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jackson DB. Serum-based microRNAs: are we blinded by potential? Proc Natl Acad Sci U S A. 2009;106(1):E5. [DOI] [PMC free article] [PubMed] [Google Scholar]


