Abstract
Background and Aims
Inflammatory bowel diseases are linked to an increased risk of developing colorectal cancer [CRC]. Previous studies suggested that the H2B ubiquitin ligase RING finger protein-20 [RNF20] inhibited inflammatory signaling mediated by the nuclear factor kappa-light-chain-enhancer of activated B cells [NF-κB]. However, the role of RNF40, the obligate heterodimeric partner of RNF20, in the context of inflammation and CRC has not been addressed. Here, we examined the effect of RNF40 loss on CRC cells in vitro and on inflammation and inflammatory signaling in vitro and in vivo.
Methods
We evaluated H2Bub1 levels in human and murine colorectal tumors by immunohistochemistry. Moreover, we correlated H2Bub1 and RNF40 levels in vivo and assessed the consequences of RNF40 depletion on cellular phenotype and gene expression in CRC cells in vitro. Finally, we examined the effect of a colon-specific loss of Rnf40 in a murine model of colitis, and assessed both local and systemic inflammation-associated consequences.
Results
In vitro studies revealed that the tumorigenic phenotype of CRC cells decreased after RNF40 depletion and displayed gene expression changes related to chromosome segregation and DNA replication, as well as decreased induction of several NF-κB-associated cytokines. This effect was associated with decreased nuclear localization of NF-κB following tumor necrosis factor alpha treatment. Consistently, the colon-specific loss of Rnf40 exerted a protective local, as well as systemic, effect following acute colitis.
Conclusions
Our findings suggest that RNF40 plays a central role in the maintenance of tumorigenic features and inflammatory signaling by promoting nuclear NF-κB activity.
Keywords: RNF40, H2Bub1, NF-κB
1. Introduction
Although inflammation is generally a protective mechanism, an ineffective resolution and, therefore, a potentially chronic inflammation can promote colorectal cancer formation.1 Recently, it was demonstrated that 15–20% of cancer-related deaths are linked to pre-existing inflammatory conditions.2 For instance, patients with inflammatory bowel diseases [IBDs], including ulcerative colitis, have a 3-fold increased risk of developing colitis-associated colorectal cancer [CRC].3,4 Given the increasing prevalence of IBDs over the past few years, numerous studies have attempted to elucidate underlying risk factors of this group of diseases. Besides environmental and lifestyle-related factors, a genome-wide association study analysis by Jostins et al. [2012] identified 163 genetic loci linked to IBDs.5 Among others, these loci were especially associated with immuno-regulatory/modulatory genes associated with immunodeficiencies, T cell function, and cytokine production.
One of the most well-known mediators of inflammatory signaling is the nuclear factor kappa-light-chain-enhancer of activated B cells [NF-κB] transcription factor family, consisting of five members: NF-κB1, NF-κB2, RelA, RelB, and c-Rel, which can act as homo- or heterodimers. Whereas RelA, RelB, and c-Rel regulate gene transcription by binding to DNA via their carboxy-terminal transactivation domains, NF-κB1 and NF-κB2 do not possess transactivation domains, and thus differentially control the expression of NF-κB target genes.6 The nuclear translocation of NF-κB dimers can be initiated by a variety of stimuli, including exposure to tumor necrosis factor alpha [TNFα], and facilitates the transcriptional activation of their target genes.6–8 These targets can be broadly categorized into positive cell-cycle regulators, anti-apoptotic factors, immunoregulatory cytokines, and negative feedback regulators of NF-κB.6,9,10 Depending on the transcription kinetics, these genes are referred to as immediate-early response genes [IEGs] or secondary/delayed response genes [SRGs]. IEGs, including FOS, JUN, and Early Growth Response 1 [EGR1], are transcribed within minutes,10 but the expression of SRGs is delayed, suggesting that their transcription is partially regulated by IEGs.11,12
Notably, epigenetic mechanisms are able to coordinate and regulate gene expression, including expression of the inflammatory machinery. Indeed, a recent study by Tarcic et al. linked the loss of the epigenetic modifier RNF20 with inflammation and inflammation-associated colorectal cancer in vitro and in vivo.13 More specifically, mice with a heterozygous global loss of Rnf20 were predisposed to colonic inflammation as well as inflammation-associated colorectal tumor growth. In addition, the authors observed highly increased NF-κB signaling in response to TNFα stimulation and RNF20 loss in mammary epithelial cells as well as in their mouse model. Generally, the RING finger protein RNF20 forms an obligate heterodimer with RNF40, which together are recruited by the adapter protein WW domain-containing adapter protein with coiled-coil [WAC] to elongating RNA Polymerase II during gene transcription. By virtue of its E3 ubiquitin ligase activity, the RNF20/RNF40 heterodimer monoubiquitinates H2B at lysine 120 [H2Bub1] and thereby promotes transcription.14 It was proposed that monoubiquitinated H2B facilitates transcriptional elongation by promoting the recruitment of the Facilitates Chromatin Transcription [FACT] complex, which enables histone H2A/H2B dimer displacement to facilitate RNA Polymerase II passage through chromatin across the gene body.15,16 Perhaps in part through its role in promoting the transcription of a subset of genes, H2Bub1 was described as a tumor-suppressive mark, since its loss was observed during tumor progression and correlated with poor patient survival in several cancer types, including colorectal cancer.17 However, the role of RNF40 in the context of inflammation and inflammation-associated CRC remains to be elucidated. Based on previously published studies and the role of H2Bub1 in cancer, we hypothesized that the depletion of RNF40 and therefore, the loss of H2Bub1, would increase inflammatory signaling as well as tumorigenic potential of colorectal cancer cells.
Unexpectedly, functional characterization assays as well as mRNA-seq analyses suggest a potential pro-tumorigenic role for RNF40 in human CRC cells in vitro. Moreover, after TNFα treatment, the induction of several cytokines and further NF-κB target genes was blocked by the knockdown of RNF40. Consistently, a mouse model with a colon-specific loss of Rnf40 displayed a milder inflammatory response compared with wild-type litter mates when challenged with an acute colitis. Together, these findings suggest that RNF40, and H2Bub1 in extension, may play an unexpected oncogenic role in CRC by promoting NF-κB-dependent inflammatory signaling.
2. Materials and Methods
2.1. Cell culture and siRNA transfection
Human colorectal cancer cell lines were grown in phenol-red free Dulbecco’s Modified Eagle’s Medium/F12 [DMEM/F12; for LS174T cells], RPMI [for COLO320DM, DLD1, SW480 cells], or McCoy’s [HCT116, HCT116 p53-/- cells] medium [Life Technologies] supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2. siRNA [GE Dharmacon siGENOME; Table S1, available as Supplementary data at ECCO-JCC online] transfections were performed using Lipofectamine® RNAiMAX [Invitrogen] according to the manufacturer’s instructions. Then 48 h post-transfection, cells were treated with 10 ng/mL of human recombinant TNFα [R&D Systems] dissolved in 0.2% bovine serum albumin [BSA] in culture medium for 0.5, and 6 h. As controls, cells were treated with 0.2% BSA in culture medium.
2.2. Cell characterization assays
Cells were seeded for characterization assays 24 h after siRNA transfection. To assess proliferation rates, 2000–5000 cells were seeded onto 96-well assay plates [Corning Life Sciences] and confluence was measured daily using a Celigo® S cell imaging cytometer [Nexcelom Bioscience LLC]. Clonogenic potential was evaluated by seeding 500 cells onto a six-well plate and waiting until macroscopically visible colonies formed. Soft agar colony formation assays were performed to test the ability to grow in an anchorage-independent manner. A base layer of 0.8% agarose in complete cell culture medium was poured into each well of a six-well plate; 10000 cells were resuspended in 0.48% agarose in medium and added into each well. Once solidified, agarose layers were covered by complete culture medium and plates were maintained at 37°C with 5% CO2 for about 1–2 weeks or until colonies were visible macroscopically. To stain colonies, cells were fixed in 4% PFA in PBS for 20 min, washed with PBS, and stained with 1% crystal violet in 20% ethanol for 20 min. Excess dye was removed by washing the plates with water. Upon drying, plates were scanned and the well area covered by cells, or the number of colonies, respectively, was quantified using FIJI.
2.3. Cell fractionation
To separate the nuclear and cytoplasmic contents, HCT116 cells were washed twice with PBS and 250 µl Buffer A [10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol, protease inhibitors; pH 7.9] were added per well of a six-well plate. Cells were incubated on ice for 5 min, scraped and centrifuged at 1300 g at 4°C for 5 min. The supernatant containing the cytoplasmic fraction was stored on ice and the pellet was washed with Buffer A. Upon centrifugation, the supernatant was discarded and the nuclear pellet was resuspended in 200 µl lysis buffer [3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease inhibitors]. The cytoplasmic and nuclear fractions were sonicated and stored at -20°C.
2.4. Western blotting and quantitative real-time polymerase chain reaction
Protein isolation, western blot analysis, RNA extraction, reverse transcription, and quantitative real-time polymerase chain reaction [qRT-PCR] were performed as previously described.21,24 Primary antibodies for western blot and primers for qRT-PCR are listed in Tables S2 and S3, available as Supplementary data at ECCO-JCC online, respectively.
2.5. Library preparation and next-generation sequencing
For mRNA-seq, HCT116 cells were transfected in triplicate with control and RNF40 siRNAs and either left untreated or treated with TNFα for 30 min. Libraries were prepared using TruSeq RNA Library Preparation Kit v2 [Illumina®] after verifying the RNA integrity on an agarose gel. Library quality was assessed using an Agilent Bioanalyzer 2100, and mRNA-seq was performed at the Transcriptome and Genome Analysis Laboratory [TAL], Göttingen, using HiSeq 4000 [Illumina®]. Sequence images were converted into bcl files [BaseCaller software, Illumina®] which were demultiplexed to fastq files [CASAVA v1.8.2]. Quality control of fastq files was carried out via FastQC and reads were mapped to the human reference genome [GRCh38.p10] using HISAT2 [version 2.0.3.3] on Galaxy. Read count files were generated using the featureCounts tool [version 1.4.6.p5] and analyzed for differential gene expression using DESeq2 [version 2.11.39]. All mRNA-seq data have been deposited at ArrayExpress [http://www.ebi.ac.uk/arrayexpress, accession number: E-MTAB-7197]. Significantly differentially regulated genes were selected by a Benjamini–Hochberg adjusted p-value <0.05 and log2-fold changes above 0.5 or below -0.5. Gene Set Enrichment Analysis [GSEA28,29] was performed as described previously19,25 with default settings [1000 permutations for gene sets, Signal2Noise metric for ranking genes]. Own GSEA gene sets were generated by combining all genes significantly upregulated upon TNFα treatment in control HCT116 cells in our experiments, or by summarizing upregulated genes in healthy and inflamed colorectal segments of IBD patients.31
2.6. Immunofluorescence
HCT116 cells were seeded onto coverslips 24 h post siRNA transfection. On the next day, cells were washed with PBS and fixed with 4% PFA in PBS for 20 min. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 15 min, washed and blocked with 10% FBS in PBS for 1 h. The primary RelA antibody [Table S2] was added in 5% FBS in PBS and incubated in a humidified chamber at 4°C overnight. Cells were washed and incubated with a secondary antibody in PBS [Thermo Scientific, Cat no. A11008] mixed with DAPI 1:1,000 for 1 h. Finally, the cells were washed and coverslips were mounted on microscope slides. Pictures were taken with a Zeiss LSM 510 Meta confocal microscope and fluorescence intensity was quantified using FIJI.
2.7. Generation of mice and genotyping
All animal work was performed in agreement with the institutional regulations for care and use of laboratory animals, and approved by the Lower Saxony State Office for Consumer Protection and Food Safety [LAVES; registration number: 15/2039]. The mouse line containing a mutated Apc1638N/+ allele was a kind gift from F. Bosman [Erasmus University Medical Center Rotterdam]. Apc1638N/+ were genotyped as described earlier18 and sacrificed at 6 months of age. The generation of mice harboring a conditional Rnf40 allele was described previously by our group.19 The Rnf40loxP mice were next crossed to a transgenic line expressing a constitutively active Cre recombinase under the control of the carbonic anhydrase 1 [Car1] promoter [CAC-Cre; kindly provided by J. Fleet, Purdue University].20 Mice were genotyped by pre-heating PCR samples to 95°C for 3 min. The respective Rnf40 DNA fragments were amplified for 35 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s. For the Cre recombinase, 35 cycles with 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s were used. Final elongation occurred at 72°C for 5 min [Rnf40 Forward: 5′-GCGAAAGTCACATTTGGCCT-3′, Rnf40 Reverse: 5′-CTTCACATTCCGTTCCTGCC-3′; Cre Forward: 5′-CGCGGTCTGGCAGTAAAAACTAT-3′, Cre Reverse: 5′-CCACCGTCAGTACGTGAGATAT-3′].
2.8. DSS treatment and determination of the Disease Activity Index
Mice were treated with 0.75% [w/v] dextran sodium sulphate [DSS; MO Biomedicals, LLC] in drinking water for 14 consecutive days and were injected intraperitoneally with Temgesic® [RB Pharmaceuticals] three times daily. During this treatment period, the severity of colitis was determined by calculating the disease activity index [DAI] on a daily basis. The DAI includes scores for weight loss, stool consistency, and intestinal bleeding intensity as follows: weight loss: 0–1% [0], 1–5% [1], 5–10% [2], 10–15% [3], >15% [4]; stool consistency: normal [0], soft [1], very soft [2], diarrhea [3]. To evaluate intestinal bleeding intensity, the stool guaiac test was performed. For this purpose, guaiac resin was dissolved in 70% EtOH until the solution was saturated. A few drops were applied to a Whatman filter paper and left to dry at room temperature. Feces were applied and distributed on the dried guaiac solution. Upon H2O2 addition, blue staining was observed in the presence of blood. Scoring of the stool guaiac test was: no blue staining [0], weak/sporadic staining [1], medium staining [2], strong blue staining [3], and bloody anus [4]. After adding the scores per tested aspect, a maximum score of 11 could be reached per day.
2.9. Tissue microarray [TMA]
The patient cohort consisted of 137 colorectal cancer patients, and the H2Bub1 staining intensity was scored based on the proportion of stained tumor cells [0%: score 0, ≤1%: 1, ≤10%: 2; ≤33%: 3, ≤66%: 4, >66%: 5] and staining intensity [no staining: score 0, weak: 1, moderate: 2, strong: 3]. Cases were subdivided based on the total score [sum of proportion and intensity scores] as low [0–2], medium [3–5], or high [6–8]. This research was performed according to the code of conduct for responsible use and was approved by a local ethics committee [Ref. 2006-154-N-MA].
2.10. Hematoxylin and eosin staining and immunohistochemistry
Histological analyses were performed as previously described.21 Briefly, organ sections were de-paraffinized in xylol for 20 min and rehydrated by storing them in decreasing concentrations of ethanol [100%, 90%, 70%] for 5 min each. For hematoxylin and eosin [H&E] staining, slides were washed with water and nuclei were stained in Mayer’s hematoxylin solution for 1 min. Excess dye was removed by rinsing slides under running tapwater for 5 min. Counterstaining was performed using eosin for 5–10 min. For immunohistochemistry [IHC], upon rehydration, antigen retrieval was carried out by boiling slides in 10 mM citric acid buffer or 1 mM EDTA for 15 min. Sections were quenched for endogenous peroxidases with 5% hydrogen peroxide in PBS and blocked with 10% fetal bovine serum [FBS] in PBS. Primary antibodies [Table S2] were diluted in 10% FBS in PBS and incubated overnight at 4°C. Biotinylated secondary antibodies [1:200] and ExtrAvidin-Peroxidase [1:1,000] were added each for 1 h. Staining was developed using 3,3’-diaminobenzidin-tetrahydrochloride [DAB] and counterstaining was carried out using hematoxylin. Slides were washed, dehydrated in increasing concentrations of ethanol, incubated in xylol and mounted.
2.11. Determination of histo-score
Inflammation intensity was scored based on the intactness of the intestinal epithelium and the rate of lymphocyte infiltration, in a range from 0 to 3 with increasing disruption of crypt structure as previously described.22 We scored as follows: normal intestinal epithelium [0], mild lymphocyte infiltration pushing crypts apart from each other [1], lymphocyte accumulations destroying crypt structure [2], no epithelial lining on top of the lymphocyte accumulation [3]. The percentage of damaged tissue was multiplied by the respective score [0–3] and the sum was divided by the maximum histo [H]-score, which was 30.
2.12. Mechanical bone testing
Tibiae were placed onto a Zwick device [145 660 Z020/TND] while a stamper moved towards the bone at 50 mm/min. To fix the bone on the plate, a primary force of 1N was applied. Measurements with an accuracy of 0.2–0.4% using 2–500 N were performed while the applied strength was linear, and were aborted once the curve declined by 10 N. Using the testXpert software [Zwick GmbH KG] the applied strength when the deformation was induced [yield load], during fracture [Fmax], and breaking the bone [failure load], and the bone stiffness, were determined.23
2.13. Statistical analyses
All graphs in this study were generated with GraphPad Prism version 5.04 [GraphPad Software]. Statistical analysis was performed using one-way analysis of variants [ANOVA] and Tukey post-hoc test [α = 0.05].
3. Results
3.1. H2Bub1 is distributed heterogeneously and independently from RNF40 in CRC
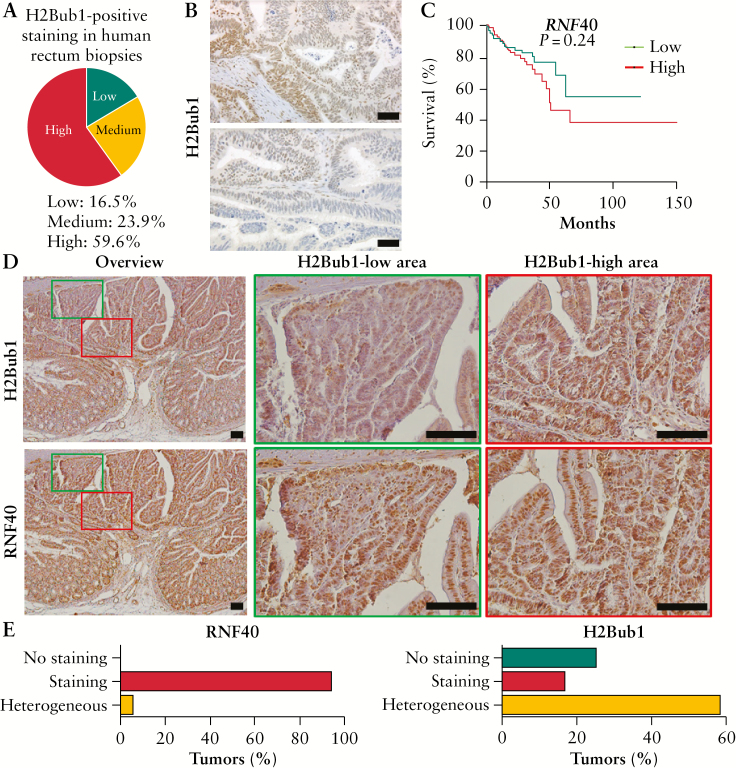
Previous studies suggested that a decrease in monoubiquitinated H2B at lysine 120 was associated with advanced tumor grade and stage in breast26 and colorectal cancer17 and therefore, H2Bub1 was proposed to largely have a tumor-suppressive function.27 Therefore, we performed immunohistochemical staining of H2Bub1 in a panel of human colorectal cancer samples with corresponding normal adjacent tissue as a control. Surprisingly, the majority of tumors [59.6%] were characterized by high H2Bub1 levels, and 23.9% showed medium and 16.5% low staining [Figure 1A]. Intriguingly, across patients we noticed intratumoral heterogeneity in H2Bub1-positive cells [Figure 1B]. Since H2Bub1 is mediated by the WAC-RNF20/RNF40 axis, the levels of RNF20, RNF40, and WAC could determine the clinical outcome of CRC patients as well. When stratifying patients according to their RNF40, RNF20, and WAC levels, using publicly available data generated by the TCGA Research Network [http://cancergenome.nih.gov/], no significant correlation was seen between low expression levels and poor prognosis [Figure 1C; Figure S1A, B, available as Supplementary data at ECCO-JCC online]. Although TCGA data suggest that RNF40 expression in colon and rectal tumors is similar to healthy tissue [Figure S1C, available as Supplementary data at ECCO-JCC online], a trend [albeit not statistically significant] that high, rather than low, RNF40 expression is associated with decreased survival was observed. The expression levels of RNF20 and WAC, on the other hand, did not correlate with survival.
Figure 1.
Colorectal tumors are frequently characterized by heterogeneous H2Bub1 levels. [A] Human rectal cancer biopsies were stained for H2Bub1, and the majority of cancer lesions showed high H2Bub1 levels. However, [B] several cancer lesions were characterized by heterogeneous H2Bub1 staining, as shown for two representative tumors. Scale bar: 100 µm. [C] Publicly available [TCGA] data were analyzed for RNF40-associated survival of CRC patients, indicating that tumors displaying high RNF40 expression show a tendency towards poorer survival. [D] Colorectal tumors of Apc1638N/+ mice were stained for H2Bub1 and RNF40 using IHC [n = 9]. Although areas with low and high H2Bub1 levels were detected within the same tumors, H2Bub1 levels were more variable in a manner independent of RNF40 amounts. Scale bar: 100 µm. [E] The percentage of colorectal cancer lesions [n = 23] with no, positive, or heterogeneous RNF40 and H2Bub1 staining was evaluated in Apc1638N/+ mice [n = 9]. Whereas the majority of tumors were characterized by high RNF40 abundance, H2Bub1 levels were frequently heterogeneous. CRC, colorectal cancer; IHC, immunohistochemistry.
To investigate the correlation between H2Bub1 and RNF40 in more detail in an in vivo system, Apc1638N/+ mice with a heterozygous truncating mutation in the Adenomatous Polyposis Coli [Apc] tumor suppressor gene were utilized. Using immunohistochemical staining, H2Bub1 and RNF40 protein levels were evaluated in colorectal tumors from these mice. We frequently detected colorectal tumors containing areas with high and low H2Bub1 staining [Figure 1D], and could thereby confirm our observations about intratumoral heterogeneity seen in human colorectal cancer samples. Notably, RNF40 levels were not affected at either the protein or the gene expression level [Figure S1D, available as Supplementary data at ECCO-JCC online]. In fact, although strong RNF40 staining was detected in almost all cancer lesions, more than half of these tumors were characterized by heterogeneous H2Bub1 levels [Figure 1E]. These findings demonstrate that colorectal tumors are frequently characterized by heterogeneous H2Bub1 levels, which appear to be independent of RNF40 levels in mice.
3.2. The tumorigenic properties of human CRC cells are reduced by low RNF40 levels
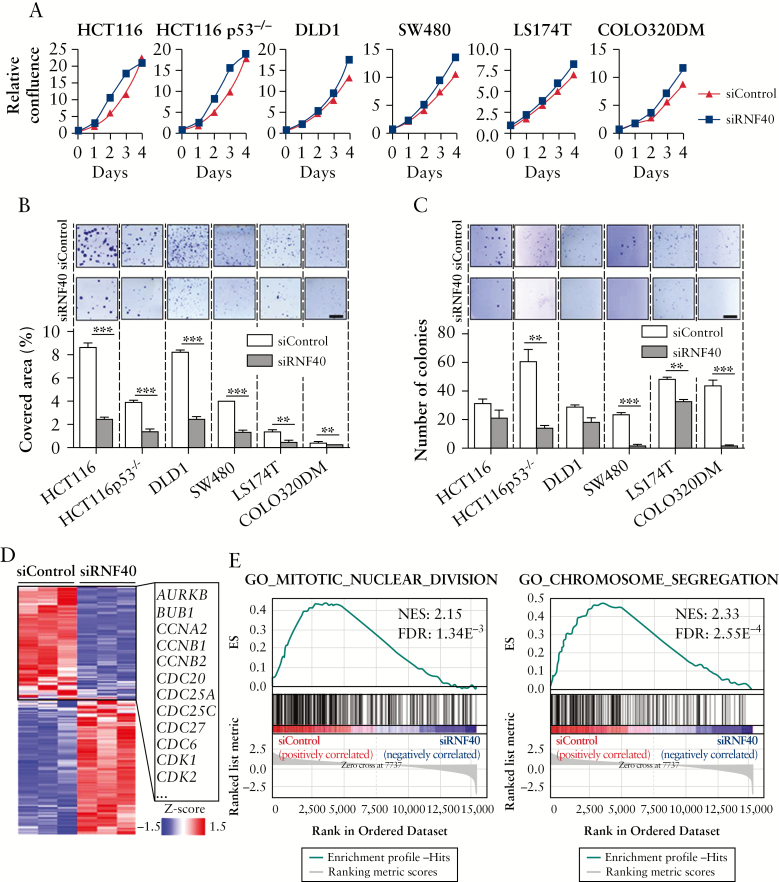
Since previous studies suggested a link between the loss of H2Bub1 levels and advanced tumor stages, indicating a potential tumor-suppressive role of RNF40, we aimed to test the effects of RNF40 loss on the proliferation and tumorigenic potential of six different human colorectal cancer cell lines. No morphological changes were observed 48 h after knockdown of RNF40 [Figure S1E, available as Supplementary data at ECCO-JCC online]. Cell confluence was measured on a daily basis and, notably, all cell lines showed a reduction in proliferation [Figure 2A]. Moreover, the clonogenic potential [Figure 2B] and anchorage-independent growth [Figure 2C] were reduced upon the loss of RNF40 in all cell lines tested.
Figure 2.
RNF40 knockdown reduces tumorigenic properties of CRC cells in vitro. RNF40 levels were reduced by siRNA transfection in HCT116, HCT116 p53-/-, DLD1, SW480, LS174T, and COLO320DM cells in four independent experiments. [A] Cellular confluence was measured daily, and decreased proliferation was observed in all cell lines following RNF40 depletion [siRNF40; n = 6]. Mean ± SEM. [B] The clonogenic potential of cells was determined by allowing single cells to form colonies and quantifying the well area covered by cells [n = 3]. Upon loss of RNF40, the cells were less likely to form colonies. Scale bar: 0.5 mm. Mean ± SEM. [C] The ability of cells to grow in an anchorage-independent manner was tested using a soft agar colony formation assay [n = 3]. A reduced number of colonies was detected upon the knockdown of RNF40. Scale bar: 0.5 mm. Mean ± SEM. [D] mRNA-seq was performed to evaluate the transcriptome-wide effect of RNF40 depletion in HCT116 cells, and revealed the downregulation of several cell cycle-associated genes [n = 3]. [E] GSEA indicates that pathways related to cell division are perturbed in RNF40-depleted cells. CRC, colorectal cancer; SEM, standard error of the mean; GSEA, Gene Set Enrichment Analysis.
To investigate the underlying transcriptome-wide effects of RNF40 loss on gene expression, we performed mRNA-seq studies in HCT116 cells transfected with control [siControl] or RNF40 [siRNF40] siRNAs [Figure S1F, available as Supplementary data at ECCO-JCC online]. Surprisingly, we found several cell cycle-associated genes to be downregulated [Figure 2D] and, accordingly, Gene Set Enrichment Analysis [GSEA28,29] revealed that cell division processes were impaired following RNF40 depletion [Figure 2E; Figure S1G, available as Supplementary data at ECCO-JCC online]. In summary, loss of RNF40 reduced the proliferation, clonogenic potential, and ability to grow in an anchorage-independent manner in all tested colorectal cancer cell lines. Moreover, RNF40-depleted HCT116 cells were characterized by proliferation defects likely caused by deregulation of cell cycle genes. These findings suggest that, in contrast to the effects observed in previous studies following RNF20 depletion, RNF40 is actually required for tumorigenic properties of colorectal cancer cells in vitro and, therefore, may have oncogenic functions in certain contexts.
3.3. RNF40 loss delays the nuclear translocation of RelA
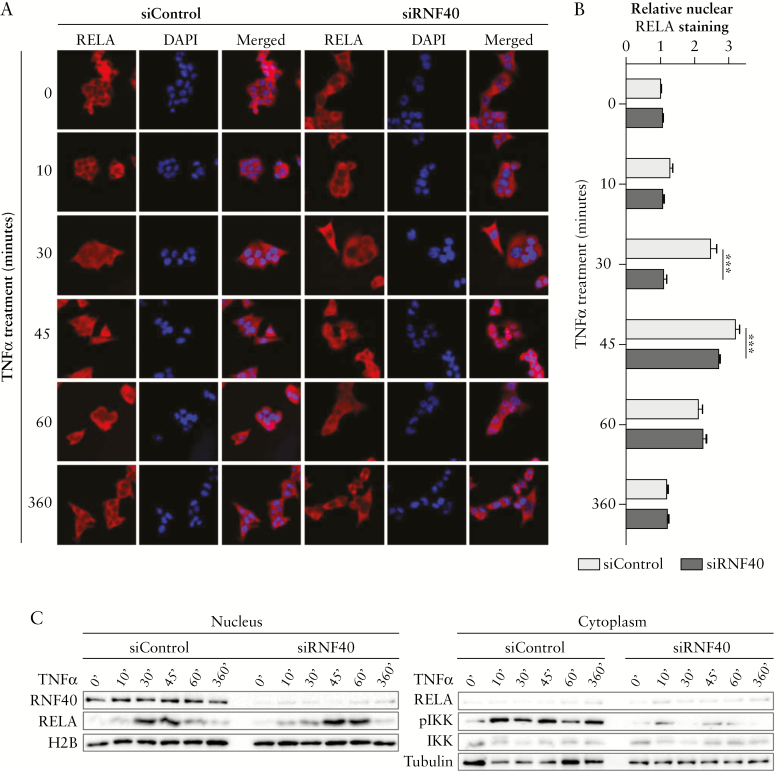
Inflammation is one of the major risk factors underlying the development of colorectal cancer.3,4 Based on our findings suggesting a potential pro-tumorigenic function of RNF40 in CRC and on the previous studies that described a suppressive function of RNF20 in inflammation,13 we sought to examine the influence of RNF40 on inflammatory processes. Therefore, we explored whether and how NF-κB activity might be affected by RNF40 loss following activation of inflammatory signaling, by treating HCT116 cells with TNFα following transfection with control [siControl] or RNF40-specific [siRNF40] siRNAs. In control cells, the NF-κB transcription factor RelA was readily detectable in the nucleus after 30, 45, and 60 min of TNFα treatment; the signal disappeared almost completely after 6 h [Figure 3A, B]. Surprisingly, in RNF40-depleted cells no staining was detected in the nucleus after 30 min. Instead, nuclear localization was delayed and observed only after 45 min. Generally, the translocation of NF-κB to the nucleus depends on the activation of IKK via phosphorylation, which then releases NF-κB members through the phosphorylation and targeted degradation of IκB proteins. Consistent with the observed defects in RelA translocation to the nucleus, we observed decreased levels of phosphorylated IKK after the depletion of RNF40 in cells treated with TNFα [Figure 3C]. These results suggest that the nuclear translocation of RelA is delayed in RNF40-deficient cells, probably due to decreased IKK activation and subsequent release from inhibitory factors.
Figure 3.
RNF40 depletion delays the nuclear translocation of RelA upon TNFα treatment. [A] Control and RNF40-depleted HCT116 cells were treated with TNFα [n = 3] 48 h after siRNA transfection, and stained for total RelA [red] and DAPI [blue]. Nuclear RelA was detected after 30 min of TNFα treatment in control cells, but the nuclei of RNF40-depleted cells were unstained at this time point. [B] Relative nuclear RelA staining was quantified in at least 50 cells using FIJI.30 Strong nuclear staining was detected after 30 and 45 min under RNF40 wild-type conditions, but the nuclear localization of RelA was delayed after RNF40 loss and only visible after 45 min of TNFα treatment. Mean ± SEM. [C] Nuclear and cytoplasmic protein was isolated from TNFα-treated HCT116 cells in two independent experiments. Western blot analysis revealed reduced levels of nuclear RelA and phosphorylated IKK [pIKK] in siRNF40 cells, which is needed to release NF-κB from its inhibitory complex [n = 2]. SEM, standard error of the mean.
3.4. RNF40 knockdown reduces inflammatory signaling
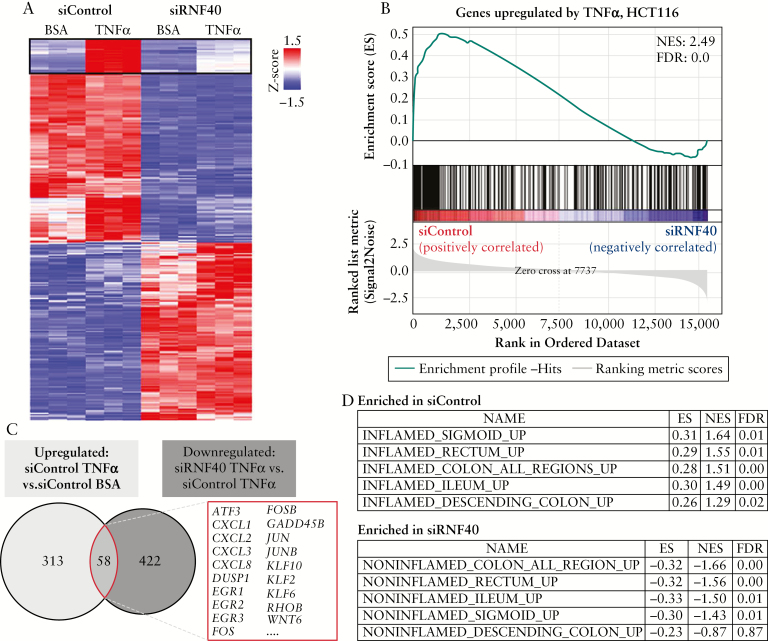
Reduced nuclear levels of RelA suggest that the NF-κB transcription factor activity is decreased, thereby affecting the transcription of NF-κB target genes. To gain an overview of the transcriptome-wide effects of RNF40 depletion, we assessed gene expression patterns in vehicle [BSA, negative control] and TNFα-treated HCT116 cells using mRNA-seq analysis. Principal component analysis confirmed the reproducibility of our results [Figure S1F, available as Supplementary data at ECCO-JCC online]. Notably, RNF40 depletion results in the differential regulation of a number of genes [Figure 4A]. Interestingly, the induced expression of several genes after TNFα treatment was severely impaired in the absence of RNF40. Gene set enrichment analyses [GSEA] confirmed this central role for RNF40 in controlling TNFα-induced transcription in HCT116 cells [Figure 4B]. When comparing the overlap of genes which are upregulated after TNFα treatment in wild-type cells with genes whose induction is impaired by RNF40 loss [downregulated in siRNF40 vs upregulated in siControl cells after TNFα treatment], we identified 58 genes differentially regulated following RNF40 reduction under inflammatory conditions [Figure 4C]. In particular, the induction of NF-κB target genes was blocked after TNFα treatment and RNF40 knockdown in HCT116 cells. For instance, this gene subset includes several cytokines [e.g. CXCL1, 2, 3, and 8], the EGR family of zinc finger transcription factors [EGR1, 2, and 3] as well as further transcription factors of the AP1 family including FOS, FOSB, JUN, and JUNB.
Figure 4.
RNF40 loss interferes with the induction of TNFα-responsive genes. [A] Heatmap depicting the transcriptome-wide effects of RNF40 depletion and TNFα treatment [n = 3] by showing significantly [padj ≤0.05] upregulated [log2 FC ≥0.75, red] or downregulated [log2 FC ≤-0.75, blue] genes. The expression of a subset of genes was induced by TNFα, but this induction was blocked by RNF40 loss. [B] All genes upregulated upon TNFα treatment in HCT116 wild-type cells were summarized as a gene set and compared with the expression pattern of control and RNF40-depleted cells using GSEA. This analysis confirms that the subgroup of genes normally upregulated under inflammatory conditions is downregulated following RNF40 depletion. [C] Venn diagram comparing the overlapping genes which are normally upregulated after TNFα treatment with genes which are upregulated by TNFα in control, but not in RNF40-depleted, cells. These 58 genes whose induction is blocked by RNF40 loss include several NF-κB target genes such as the cytokines CXCL1, 2, 3, and 8, as well as the transcription factors FOS and JUN. [D] Gene lists were generated from previously published expression data from IBD patients31 and compared with our mRNA-seq results from TNFα-treated HCT116 cells. Gene signatures of TNFα-treated wild-type cells and inflamed tissues were related to each other, whereas gene sets associated with non-inflamed were enriched in RNF40-depleted cells. GSEA, Gene Set Enrichment Analysis; IBD, inflammatory bowel disease.
We next sought to determine whether gene expression patterns altered following RNF40 depletion may be relevant for human IBD. Thus, we used previously published expression data from intestine biopsies isolated from IBD patients and compared these with our mRNA-seq data following RNF40 perturbation.31 When gene sets associated with inflamed or non-inflamed colon regions were compared with changes elicited by RNF40 depletion in TNFα-treated HCT116 cells, we found that genes related to inflamed tissue were enriched in control cells, whereas signatures associated with non-inflamed colon biopsies were enriched in RNF40-depleted cells [Figure 4D]. Together these findings demonstrate that inflammation-induced activation of NF-κB target genes requires RNF40 function, and that RNF40-dependent genes are dysregulated in IBD patient biopsies.
3.5. RNF40 is required for expression of NF-κB target genes after TNFα treatment
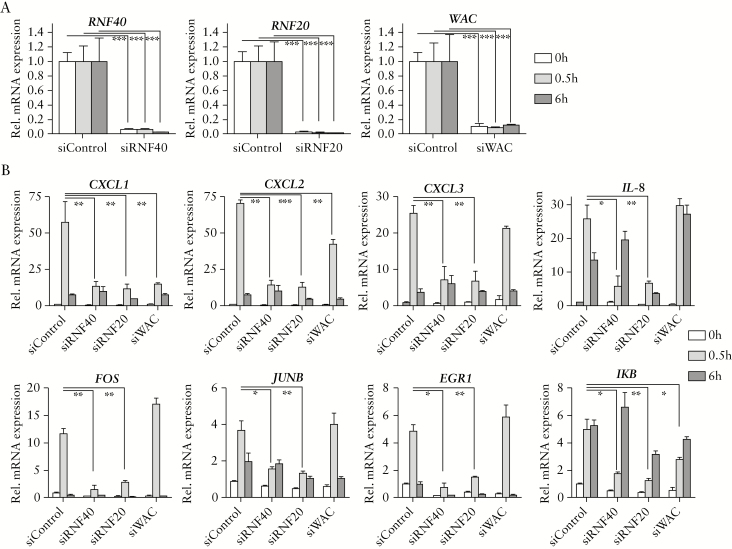
After mRNA-seq analysis, we aimed to confirm the effect of RNF40 loss on inflammation and, more specifically on NF-κB-associated inflammatory signaling, in more detail. Moreover, besides monitoring the consequences of depleting RNF40, we performed siRNA-mediated knockdowns for the other major components of the H2B ubiquitin ligase complex, RNF20 and WAC. We were able to confirm at the protein level that the loss of each of these proteins resulted in a loss of H2Bub1 [Figure S2A, available as Supplementary data at ECCO-JCC online], and therefore sought to investigate their potential roles in controlling inflammatory signaling. HCT116 cells were treated with TNFα 48 h after transfection, to monitor the early induction of NF-κB signaling after 30 min [IEGs] as well as the late induction after 6 h [SRGs]. After verifying the knockdown and the fact that RNF40, RNF20, and WAC expression were not affected by TNFα treatment [Figure 5A], the relative mRNA levels of CXCL1, CXCL2, CXCL3, IL-8, FOS, JUNB, EGR1, and IκB were tested. In agreement with our mRNA-seq data, the expression of several NF-κB target genes was induced upon addition of TNFα in control cells [Figure 5B]. In contrast, the induction of these factors was reduced by RNF40 depletion after 30 min as well as 6 h of incubation with TNFα. Whereas a similar dampening of NF-κB-mediated transcription can be observed in RNF20 knockdown cells, the loss of WAC showed only mild effects. Notably, these effects were not mediated by altered expression levels of upstream factors [Figure S2B, available as Supplementary data at ECCO-JCC online].
Figure 5.
Loss of RNF40 and RNF20 reduces induction of NF-κB target gene expression upon TNFα treatment. HCT116 siControl, siRNF40, siRNF20, and siWAC cells were stimulated with 10ng/mL of TNFα for 0, 0.5, and 6 h in three independent experiments, 48 h after siRNA transfection. [A] The siRNA-mediated knockdowns were verified, and relative mRNA expression levels of RNF40, RNF20, and WAC were not affected by TNFα treatment [n = 3]. Mean ± SEM. [B] Relative gene expression of several NF-κB target genes was assessed using qRT-PCR [n = 3]. Generally, the knockdown of RNF40 or RNF20 resulted in a reduced induction of these genes upon TNFα treatment, especially after 30 min, whereas WAC depletion only showed mild effects compared with the control siRNA. Mean ± SEM. SEM, standard error of the mean; qRT-PCR, quantitative real-time polymerase chain reaction.
3.6. Heterogeneous colorectal Rnf40 deletion reduces inflammation burden in vivo
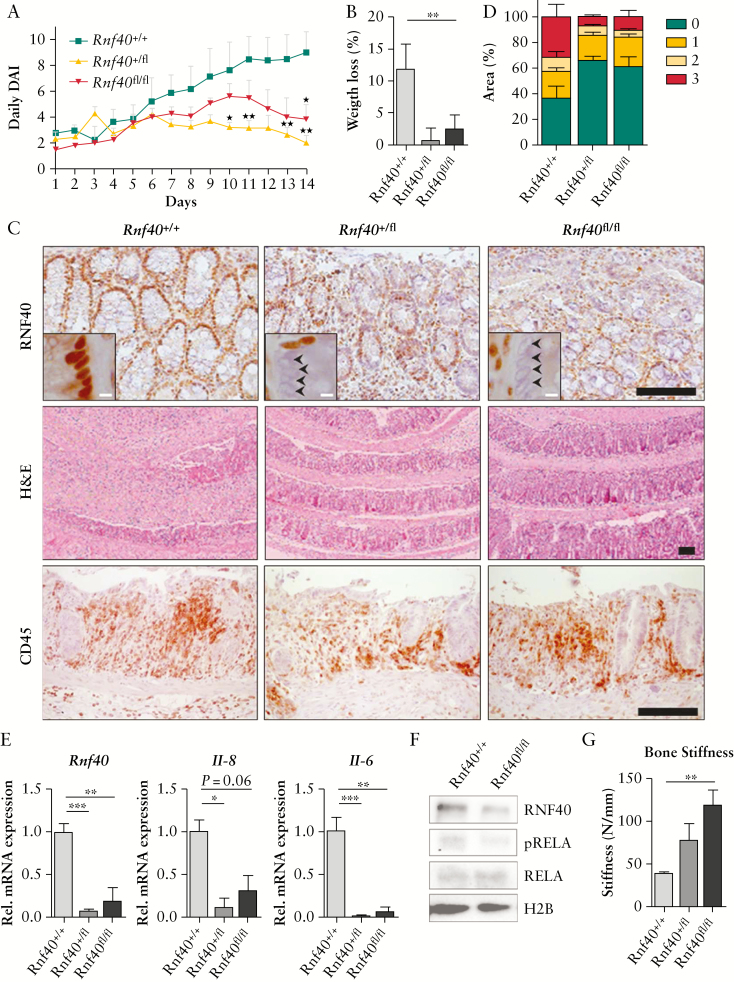
Whereas previous studies demonstrated that a global reduction of Rnf20 levels increased colitis burden in mice, our in vitro data suggested that Rnf40 deletion may, in fact, lower inflammation-associated symptoms in vivo. To test this hypothesis, we generated a mouse model with a colon-specific deletion of Rnf40 mediated by the CAC-Cre recombinase.20 We induced colorectal inflammation by treating mice with DSS, an approach frequently used for modelling acute colitis.34 During the entire treatment period, the disease activity index [DAI] was determined on a daily basis including scores for weight loss, stool consistency, and occult blood. In support of our in vitro findings, we observed that the DAI was lower after reducing Rnf40 levels in mice, especially at the end of the DSS treatment period [Figure 6A]. This difference in DAI among genotypes was mainly mediated by the minimal weight loss measured in CAC-Cre, Rnf40+/fl and Rnf40fl/fl animals [Figure 6B], whereas the stool consistency and the frequency of occult blood were not significantly affected by Rnf40 loss [Figure S2C, available as Supplementary data at ECCO-JCC online]. Indeed, we were able to confirm the reduction of RNF40 in the intestinal epithelium using IHC [Figure 6C]. Notably, H&E staining of the colon implicated only mild epithelial damage in Rnf40 heterozygous and knockout animals compared with wild types, whereas the infiltration of immunoregulatory cells was similar among genotypes. We next investigated the inflammatory response upon DSS treatment in more detail, using the Histo-score/H-score. Generally, this scoring system is based on the intactness of the colonic epithelium and lymphocyte infiltration. It ranges from 0 to 3, considering normal epithelium [0], as well as segments with mild [1], medium [2], or severe [3] damage. As expected, all mice were characterized by colorectal inflammation due to the DSS treatment. Intriguingly, in contrast to wild-type animals, Rnf40+/fl and Rnf40fl/fl mice showed only low proportions of epithelium with medium to severe signs of inflammation [Figure 6D]. We aimed to evaluate whether this effect is, similar to our in vitro outcomes, mediated by decreased inflammatory signaling and cytokine expression. For this purpose, we isolated RNA from the colons of the experimental animals and performed qRT-PCR. Indeed, reduced levels of Rnf40 were associated with decreased expression of interleukins 6 and 8 [Figure 6E]. Similar to our in vitro results [Figure 3C], levels of phosphorylated RelA [pRelA] were reduced proportionately to the reduction in Rnf40 expression [Figure 6F].
Figure 6.
The colorectal knockout of Rnf40 reduces symptoms of acute colitis in mice. [A] CAC-Cre; Rnf40flox mice were treated with 0.75% DSS for 14 days. The disease activity index [DAI] reflecting weight loss, stool consistency, and occult blood presence was determined daily. Colitis-associated symptoms were reduced by low colorectal Rnf40 levels [Rnf40+/+: n = 5; Rnf40+/fl: n = 6; Rnf40fl/fl: n = 6]. Mean ± SEM. [B] The low DAI observed in Rnf40+/fl and Rnf40fl/fl animals was mainly mediated by decreased weight loss. Mean ± SEM. [C] The reduction of RNF40 levels was confirmed using IHC, and the epithelial morphology was assessed using H&E staining. Especially in wild-type animals, the DSS treatment was associated with highly inflamed intestinal epithelium, but the reduction of Rnf40 rescued most crypt structures. Infiltration of CD45-positive immunoregulatory cells was similar among genotypes. Scale bars: Black: 100 µm, white: 5 µm. [D] When analysing the epithelial damage as a consequence of DSS treatment, the percentage of segments with no [0], mild [1], medium [2], or severe [3] damage was determined. Wild-type mice showed a high percentage of severely destroyed epithelium. but reduced Rnf40 expression exerted a protective effect. Mean ± SEM. [E] qRT-PCR using colon material revealed that reduced Rnf40 levels are associated with decreased cytokine expression. Mean ± SEM. [F] Protein lysates isolated from colons of CAC-Cre, Rnf40+/+, and Rnf40fl/fl mice indicated slightly reduced phosphorylated-RelA [pRelA] levels as assessed by western blot analysis. [G] To determine bone fragility, and therefore systemic effects of inflammation, tibiae were placed in a Zwick device applying pressure onto the bone, leading to fracture. The colorectal reduction of Rnf40 resulted in decreased bone fragility. Mean ± SEM. DSS, deextran sodium sulphate; SEM, standard error of the mean; qRT-PCR, quantitative real-time polymerase chain reaction; IHC, immunohistochemistry; H&E, hematoxylin and eosin.
Interestingly, in patients with inflammatory bowel diseases a decreased bone mineral density and increased fracture risk has been reported.32 To elucidate whether the dampened colorectal inflammation in CAC-Cre, Rnf40+/fl, and Rnf40fl/fl mice would also have systemic effects, bone biomechanical properties were tested. Using a Zwick device, the applied strength during bone deformation and bone fracture was determined. Moreover, the maximum forces applied before breaking the tibia and the bone stiffness were quantified. Indeed, the bone fragility was significantly decreased in animals with reduced Rnf40 levels compared with their wild-type littermates [Figure 6G; Figure S2D, available as Supplementary data at ECCO-JCC online]. These outcomes suggest that the colorectal depletion of Rnf40 exerts a protective effect against colorectal inflammation locally as well as systemically.
4. Discussion
Several studies have linked the loss of H2Bub1 levels to advanced tumorigenesis and, therefore, we and others have suggested that RNF40 and RNF20 may represent potential tumor suppressors.27,33 In line with this hypothesis, Tarcic and co-workers observed that a global reduction of Rnf20 caused increased colorectal inflammation and inflammation-associated tumorigenesis in mice, mainly due to accelerated NF-κB activity.13 Since the role of RNF40 was only insufficiently addressed in previous studies, and due to the high similarity between RNF20 and RNF40, we sought to elucidate the role of RNF40 in colorectal inflammation and the tumorigenic potential of CRC cells.
To obtain an impression of the levels of RNF40/H2Bub1 in CRC, we analyzed human colorectal cancer samples as well as colorectal tumor sections prepared from APC1638N/+ mice. We detected that intratumoral H2Bub1 amounts are highly heterogeneous in human and murine colorectal cancer lesions. This observation is in line with the general concept of the presence of epigenetically diverse cellular subpopulations within a single tumor. Differences in mutations, epigenetic regulation, morphology, and differentiation status within one disease site can be reasons underlying therapy resistance and, consequently, can also affect prognosis.35 Therefore, we were not surprised by our finding that also H2Bub1 levels can vary within a single tumor. Interestingly, in mouse tumors we detected that tumor regions with low H2Bub1 staining nevertheless maintained expression of RNF40. This suggests that tumor-associated loss of H2Bub1 is likely triggered by other means, which still remain to be elucidated. Although publicly available TGCA expression data indicate that RNF20 and WAC levels do not influence CRC patient survival, higher RNF40 mRNA levels showed a tendency towards being associated with poor prognosis. These findings suggest that the roles of RNF20, RNF40, and WAC in colorectal cancer are likely to be context-specific and cannot be strictly viewed as tumor-suppressive. Consistent with a potential tumorigenic function of RNF40, independent of the mutational profile, RNF40 reduction in six CRC cell lines was associated with decreased proliferation, clonogenic potential, and anchorage-independent growth. These findings were supported by mRNA-seq studies identifying RNF40-dependent genes as being associated with cell division processes. These observations support a potential pro-tumorigenic function of RNF40 whereby it might enhance the aggressiveness of CRC cells in vitro, supporting the tendency observed in the TGCA survival data.
Previously published data implied that RNF20 suppresses NF-κB signaling.13 However, our findings demonstrate that RNF40 loss delays the nuclear localization of the NF-κB member RelA. Accordingly, the transcription of NF-κB targets was reduced in RNF40 knockdown cells, whereas the expression of upstream factors was not affected. Due to their similarity in the contribution to H2B monoubiquitination, the effects of RNF20 and WAC reduction were also studied. RNF40 and RNF20 knockdown both decreased expression levels of various genes, but WAC reduction only negatively affected the transcription of a smaller subset of NF-κB targets. Therefore, since depletion of RNF20, RNF40, or WAC similarly resulted in a global loss of H2Bub1 levels, it appears that the effects observed on NF-κB-associated genes are likely independent of the H2B ubiquitin ligase function of RNF20 and RNF40. In support of our in vitro data demonstrating a role for RNF40 in mediating the effects of inflammatory signaling, the heterogeneous loss of Rnf40 decreased inflammation burden in a mouse model of acute colitis. Notably, this effect was not mediated by a decreased infiltration of immunoregulatory CD45-positive cells, suggesting that reduced NF-κB signaling underlies the protective effect of the Rnf40 knockout. These findings raise the question whether RNF40 may catalyze the ubiquitination of other protein targets in the cell, possibly thereby directly influencing NF-κB activity independent of epigenetic regulation via H2B monoubiquitination.
Surprisingly, our in vitro as well as in vivo data were not in agreement with the findings published by Tarcic et al.13 This apparent discrepancy may potentially be due to the possibility that RNF20 and RNF40 have opposing functions, or due to differences in the experimental setups. In particular, whereas Tarcic et al. used mice with a global heterozygous reduction of Rnf20, we generated mice with a colon-specific knockout of Rnf40 mediated by the colon-specific CAC-Cre recombinase. As discussed by Tarcic et al., to what extent the inflammation-associated effects were mediated by the reduction of Rnf20 in the colon or by secondary global effects modulating the immune response or microbiota, could not be ascertained in their experimental system. In addition, their mice underwent a protocol for chronic colitis in which mice were treated with 2% DSS for three cycles of 5 days in a row, each DSS cycle followed by 14 days’ treatment pause. In contrast, we administered a lower dose of DSS [0.75%] for 14 consecutive days to investigate the effects during acute colitis. Notably, we were able to demonstrate that our DSS treatment scheme has systemic consequences by increasing bone fragility in wild-type mice. To elucidate whether the differences between data from our study and the study on RNF20 are due to opposing functions of RNF20 and RNF40 in inflammation, or whether RNF20 and RNF40 play different roles in controlling inflammation in intestinal epithelial cells and immune cells, future studies exploring conditional loss of RNF20 or RNF40 in both immune and epithelial cells will be essential.
Our results demonstrate that RNF40 exerts oncogenic functions in vitro, thereby not only affecting proliferation, clonogenicity, and anchorage-independent growth, but also promoting the NF-κB-associated inflammatory response. This occurs through a reduction of the levels of IKK phosphorylation upon RNF40 loss, which results in delayed entry of RelA into the nucleus. The protective effect of RNF40 loss on inflammatory burden was confirmed in a DSS-based mouse model of acute colitis, and confirmed when comparing our mRNA-seq results with expression studies performed with human IBD biopsies. Further experimental approaches will help to unravel the exact molecular mechanisms that RNF40 uses to dampen the inflammatory response, and whether or how these mechanisms are different from RNF20-mediated processes. Additional studies will help to elucidate the effects of Rnf40 loss on inflammation-associated colorectal tumor formation and determine whether RNF40 may serve as a therapeutic target for reducing the devastating effects of inflammatory bowel diseases.
Funding
This work was supported by institutional funding provided to the Department of General, Visceral and Pediatric Surgery by the University Medical Center Göttingen. RLK is supported by the Dorothea Schlözer programme [University of Göttingen].
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
The authors would like to thank N. Molitor for her technical assistance, and the staff of the animal facility at the European Neuroscience Institute Göttingen [ENI-G]. Furthermore, we thank G. Salinas and F. Ludewig for performing next-generation sequencing at the Transcriptome and Genome Analysis Laboratory Göttingen.
Author Contributions
SAJ and RLK designed the study with input from WAF, RLC, and FW HUS, and PS analyzed tissue microarray data. RLK, RLC, MQ, and DM performed all cell culture experiments. RLK and RLC generated next-generation sequencing data. FW provided support with bioinformatic analyses. SAJ generated the targeting construct for the Rnf40 conditional knockout mouse. Mouse experiments were performed by RLK and FW with the help of RLC, MQ, and DS. Moreover, DS determined bone biomechanical parameters. SAJ and RLK wrote the manuscript. All authors have read and approved the final version of the article, including the authorship list. All authors give consent for the publication of the manuscript in the Journal of Crohn’s and Colitis.
References
- 1. Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol 2005;6:1191–7. [DOI] [PubMed] [Google Scholar]
- 2. Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest 2015;125:3347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 2001;48:526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. von Roon AC, Reese G, Teare J, Constantinides V, Darzi AW, Tekkis PP. The risk of cancer in patients with Crohn’s disease. Dis Colon Rectum 2007;50:839–55. [DOI] [PubMed] [Google Scholar]
- 5. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC] Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jost PJ, Ruland J. Aberrant NF-kappaB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood 2007;109:2700–7. [DOI] [PubMed] [Google Scholar]
- 7. Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 2012;26:203–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol 2010;2:a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2002;2:301–10. [DOI] [PubMed] [Google Scholar]
- 10. Bahrami S, Drabløs F. Gene regulation in the immediate-early response process. Adv Biol Regul 2016;62:37–49. [DOI] [PubMed] [Google Scholar]
- 11. Holzman LB, Marks RM, Dixit VM. A novel immediate-early response gene of endothelium is induced by cytokines and encodes a secreted protein. Mol Cell Biol 1990;10:5830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayashi M, Ueyama T, Nemoto K, Tamaki T, Senba E. Sequential mRNA expression for immediate early genes, cytokines, and neurotrophins in spinal cord injury. J Neurotrauma 2000;17:203–18. [DOI] [PubMed] [Google Scholar]
- 13. Tarcic O, Pateras IS, Cooks T, et al. . RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep 2016;14:1462–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu B, Zheng Y, Pham AD, et al. . Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 2005;20:601–11. [DOI] [PubMed] [Google Scholar]
- 15. Pavri R, Zhu B, Li G, et al. . Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006;125:703–17. [DOI] [PubMed] [Google Scholar]
- 16. Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 1998;92:105–16. [DOI] [PubMed] [Google Scholar]
- 17. Melling N, Grimm N, Simon R, et al. . Loss of H2Bub1 expression is linked to poor prognosis in nodal negative colorectal cancers. Pathol Oncol Res 2016;22:95–102. [DOI] [PubMed] [Google Scholar]
- 18. Cremers N, Neeb A, Uhle T, et al. . CD24 is not required for tumor initiation and growth in murine breast and prostate cancer models. PLoS One 2016;11:e0151468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie W, Nagarajan S, Baumgart SJ, et al. . RNF40 regulates gene expression in an epigenetic context-dependent manner. Genome Biol 2017;18:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xue Y, Johnson R, Desmet M, Snyder PW, Fleet JC. Generation of a transgenic mouse for colorectal cancer research with intestinal cre expression limited to the large intestine. Mol Cancer Res 2010;8:1095–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kosinsky RL, Wegwitz F, Hellbach N, et al. . Usp22 deficiency impairs intestinal epithelial lineage specification in vivo. Oncotarget 2015;6:37906–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meers GK, Bohnenberger H, Reichardt HM, Lühder F, Reichardt SD. Impaired resolution of DSS-induced colitis in mice lacking the glucocorticoid receptor in myeloid cells. PLoS One 2018;13:e0190846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komrakova M, Stuermer EK, Werner C, et al. . Effect of human parathyroid hormone hPTH [1-34] applied at different regimes on fracture healing and muscle in ovariectomized and healthy rats. Bone 2010;47:480–92. [DOI] [PubMed] [Google Scholar]
- 24. Bedi U, Scheel AH, Hennion M, Begus-Nahrmann Y, Rüschoff J, Johnsen SA. SUPT6H controls estrogen receptor activity and cellular differentiation by multiple epigenomic mechanisms. Oncogene 2015;34:465–73. [DOI] [PubMed] [Google Scholar]
- 25. Najafova Z, Tirado-Magallanes R, Subramaniam M, et al. . BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. Nucleic Acids Res 2017;45:127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prenzel T, Begus-Nahrmann Y, Kramer F, et al. . Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res 2011;71:5739–53. [DOI] [PubMed] [Google Scholar]
- 27. Johnsen SA. The enigmatic role of H2Bub1 in cancer. FEBS Lett 2012;586:1592–601. [DOI] [PubMed] [Google Scholar]
- 28. Mootha VK, Lindgren CM, Eriksson KF, et al. . PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schindelin J, Arganda-Carreras I, Frise E, et al. . Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peters LA, Perrigoue J, Mortha A, et al. . A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 2017;49:1437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ali T, Lam D, Bronze MS, Humphrey MB. Osteoporosis in inflammatory bowel disease. Am J Med 2009;122:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cole AJ, Clifton-Bligh R, Marsh DJ. Histone H2B monoubiquitination: roles to play in human malignancy. Endocr Relat Cancer 2015;22:T19–33. [DOI] [PubMed] [Google Scholar]
- 34. Hamdani G, Gabet Y, Rachmilewitz D, Karmeli F, Bab I, Dresner-Pollak R. Dextran sodium sulfate-induced colitis causes rapid bone loss in mice. Bone 2008;43:945–50. [DOI] [PubMed] [Google Scholar]
- 35. Dagogo-Jack I, Shaw AT. Tumor heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.