Abstract
The genetic basis combined with the sporadic occurrence of amyotrophic lateral sclerosis (ALS) suggests a role of de novo mutations in disease pathogenesis. Previous studies provided some evidence for this hypothesis; however, results were conflicting: no genes with recurrent occurring de novo mutations were identified and different pathways were postulated. In this study, we analyzed whole-exome data from 82 new patient-parents trios and combined it with the datasets of all previously published ALS trios (173 trios in total). The per patient de novo rate was not higher than expected based on the general population (P = 0.40). We showed that these mutations are not part of the previously postulated pathways, and gene-gene interaction analysis found no enrichment of interacting genes in this group (P = 0.57). Also, we were able to show that the de novo mutations in ALS patients are located in genes already prone for de novo mutations (P < 1 × 10−15). Although the individual effect of rare de novo mutations in specific genes could not be assessed, our results indicate that, in contrast to previous hypothesis, de novo mutations in general do not impose a major burden on ALS risk.
Keywords: ALS, amyotrophic lateral sclerosis, de novo mutations, disease pathway, motor neuron disease, trios
1 |. INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a heterogeneous disease in which many of the contributing genetic factors are still unknown. The identification of the underlying genetic factors is not only important for the understanding the disease pathogenesis, but also for individualized genetic counseling of patients and their families.
In recent years, de novo mutations have been implicated in the pathogenesis of a growing number of different diseases, for example, type 1 Charcot-Marie-Tooth (Motley et al., 2016), specific congenital syndromic disorders (Hoischen et al., 2011), nonsyndromic intellectual disability (de Ligt et al., 2012), and autism (Neale et al., 2012; O’Roak et al., 2012). In contrast to these disorders, ALS manifests later in life with peak incidence in the seventh decade (Huisman et al., 2011). Therefore, it is often not possible to obtain DNA from both parents, and thus it is difficult to acquire a large cohort of patient-parent trios. Recently, two studies (Chesi et al., 2013; Steinberg, Yu, Koboldt, Mardis, & Pamphlett, 2015) suggested a possible role of de novo mutations in the pathogenesis of ALS and hypothesized that genes with a function in either chromatin or transcription regulation might be mutated. In this study, we aimed to verify this role of de novo mutations in ALS, using next-generation sequencing data from, to our knowledge, the largest international cohort of ALS patients with healthy parents (patient-parent trios) and combining this data with the previously published data.
2 |. MATERIALS AND METHODS
2.1 |. Patients
All patients fulfilled the revised El Escorial criteria for possible, probable laboratory-supported, probable or definite ALS (Brooks, Miller, Swash, Munsat, & World Federation of Neurology Research Group on Motor Neuron Diseases, 2000). The patients had no first- or second-degree relatives with motor neuron disease or dementia.
The patients were recruited in The Netherlands from an ongoing prospective, population-based study (Huisman et al., 2011) (21 trios), in Germany by the MND-NET (18 trios), in Italy by the IRCCS Istituto Auxologico Italiano (39 trios), and in Sweden by the Department of Pharmacology and Clinical Neuroscience, Umea University (four trios). All participants gave written informed consent and local ethical committees have prospectively reviewed and approved this research at the participating centers.
Genetic prescreening encompassed at least testing of the C9orf72 repeat expansion. For the Dutch, Italian, German, and Swedish trios mutations in SOD1 and FUS were excluded. In Italian trios, mutations in TARDBP were excluded as well. In one patient from a Dutch trio (trio NL21), a C9orf72 repeat expansion was found, which was inherited by the mother. No first- or second-degree family member of this patient was diagnosed with ALS or frontotemporal dementia. The mother was over 60 years old, and her medical history reported only gastric acid- related problems and thus contained no neurological or psychiatric diseases. She reported no use of medication. Because the C9orf72 repeat expansion is known to co-occur with other mutations in ALS genes (van Blitterswijk et al., 2012; van Blitterswijk et al., 2013), we included this trio for further analysis.
2.2 |. Whole-genome/exome sequencing and variant detection
Genomic DNA was obtained from whole blood using standard procedures. In the Dutch samples, whole-genome sequencing was performed at Complete Genomics (Mountain View, CA) using DNA nanoball arrays and combinatorial probe-anchor ligation reads. Sequence reads were mapped to the National Center for Biotechnology Information (NCBI) reference genome 36. The mean coverage was 57×, and 97.7% of the genome was assessed by >10 individual reads per base. Variant calling confidence scores were provided by Complete Genomics, based on a likelihood model for true variant calls. Variants not passing the standard confidence score threshold were discarded. CGAtools 1.6.0 (Complete Genomics) was used to call de novo variants. Variants that were present in other nonrelated parental genomes or in 69 publicly available Complete Genomics genomes were considered to be platform-specific variants and also discarded. Passing variants were annotated using Annovar software 2013–08-24 (http://www.openbioinformatics.org/annovar) and nonsynonymous exonic SNVs were selected. SNVs had to be called in more than 10% of the reads at that base position. Due to platform-specific false-positive variant calling in simple tandem repeat regions and near indels (Reumers et al., 2012), variants in simple tandem repeat regions (UCSC table browser, http://genome.ucsc.edu) and within five bases of an indel were excluded. Liftover from NCBI build 36 to human reference genome hg19 was performed by the NCBI remap tool (https://www.ncbi.nlm.nih.gov/genome/tools/remap).
In the Italian samples, whole-exome sequencing was performed using the SeqCap EZ Human Exome Library 2.0 (Roche Nimblegen, Madison, WI) on a HiSeq2000 platform (Illumina, San Diego, CA). Reads were aligned to a human reference (hg19) using Burrows-Wheeler Aligner (BWA) and processed using the Genome Analysis ToolKit (GATK). After removal of duplicate reads, indel realignment (GATK IndelRealigner), and base quality recalibration (GATKTableRe-calibration) was performed. The average percentage of target bases with 10x coverage per sample was 93.11%. Variant detection and genotyping were performed using the UnifiedGenotyper (UG) tool from GATK. Variants not passing quality control criteria were eliminated (QD3, MQ60.0, HaplotypeScore> 13.0, MQRankSum<−12.5, ReadPosRankSum<−8.0). Outlying heterozygosity analyses were performed using PLINKv1.0734. This was a two-step procedure where we first established an LD pruned (R2<0.5, window size = 50, step = 5) set of autosomal markers with minor allele frequency (MAF) > 0.01 and P > 0.001 for deviation from Hardy-Weinberg equilibrium. These markers were then used to calculate per sample coefficients of an inbreeding (F), and samples were excluded where these fell outside the expected range (F < −0.1 or F > 0.1) (Cirulli et al., 2015). Samples were also discarded in case of genotypes with low quality (GQ 0.2 or clinically reported) or if the gender determined by genotypes did not match the clinically reported gender.
In the German and Swedish samples, whole-exome sequencing was performed using the enrichment kits SeqCap EZ Human Exome v.2.0 or v3.0 and an Illumina HiSeq2000 sequencer with a paired-end 100 bp protocol or for the last batch SureSelect Human All Exon V6 and an Illumina Hiseq4000 sequencer with a paired-end 75 bp protocol. Data analysis was performed with VARBANK exome pipeline of the Cologne Center for Genomics. In detail, reads were aligned to the human reference genome (hg19) with BWA-aln (version 0.6.2) followed by duplicate marking (Picard version 1.64), base quality score recalibration, and local indel realignment (both GATK version 1.6–11). Mean average coverage was 91.9 and at least 92.9% of the target sequences were covered 10×. For the de novo calling, BCF files were generated from the resulting BAM files for each trio using SAMtools mpileup (version 0.1.18), and variants were called with the open source program DeNovoGear (version 0.5.1; Ramu et al., 2013). The detection filters were set to a posterior probability >0.5. All de novo variants, compound heterozygous rare variants, and homozygous rare variants were uploaded to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
2.3 |. Sanger sequencing
All possible nonsynonymous de novo SNVs were validated by Sanger sequencing. Primer sequences and reaction conditions are available upon request. Sanger sequencing was performed using BigDye Terminator 3.1 sequencing kit (Applied Biosystems, Foster City, CA), DNA Analyzer 3700 or 3730XL (Applied Biosystems). PolyPhred (Nickerson, Tobe, & Taylor, 1997) version 6.18 or the DNASTAR Lasergene software (DNASTAR, Madison, WI) were used for data analysis.
2.4 |. Publicly available datasets
The Exome Aggregation Consortium (ExAC) dataset version 0.3 was downloaded from the Website (exac.broadinstitute.org); European (non-Finnish) subjects were included. The Genome of The Netherlands (GoNL) dataset release 4, and the de novo mutation rate map was downloaded from the Genome of The Netherlands Website (www.nlgenome.nl). A list of human genes with coding region size was obtained from the CCDS project (release 18, http://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi).
2.5 |. Functional enrichment analysis
The functional annotation chart of the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (Huang, Sherman, & Lempicki, 2009) was used for functional annotation, with homo sapiens as background. Pathways with Bonferroni-adjusted P values below 0.05 and false discovery rates below 0.1 were considered significant.
2.6 |. Protein-protein interaction networks
For network analysis between proteins, the Disease Association Protein-Protein Link Evaluator v0.17 (Rossin et al., 2011) (DAPPLE, http://genepattern.broadinstitute.org) was used with genes of interest as input and 1,000 permutations. The ALS Online Database (ALSoD, http://alsod.iop.kcl.ac.uk) was consulted on May 3, 2015.
2.7 |. Statistical analysis
Statistical analysis was performed using the statistical analysis program R version 3.1.0 (http://www.r-project.org/). Independent sample t-tests were used for comparison of de novo mutation rates between studies; a one-sample t-test was used for comparison with the general population. Wilcoxon rank sum tests were used to compare distributions of de novo rate, gene size, and de novo probability. All tests were two-tailed. Power calculations were performed using G*Power (V.3.1; Heinrich Heine Universität, Düsseldorf, Germany).
3 |. RESULTS
3.1 |. De novo mutations
We collected a total of 82 patient-parent trios from four different European populations for whole-exome or whole-genome sequencing. The rate of successful Sanger sequencing validation after computational de novo mutation-filtering steps was 52% for the Dutch cohort, 59% for the German/Swedish cohort, and 61% for the Italian cohort. In total, 69 nonsynonymous de novo mutations were found in the index patients; however, all of them in different genes (Table 1).
TABLE 1.
De novo mutations (DNM)
| Trio | DNM | Gene | Position | Transcript | Reference | Variant | Protein change | PolyPhen | ExAC MAF |
|---|---|---|---|---|---|---|---|---|---|
| NL2 | 1 | PACS1 | 11:66002865 | NM_018026.3 | G | A | p.R733Q | Possibly damaging | 0.000406 |
| NL3 | 1 | PTPN1 | 20:49195087 | NM_002827.3 | A | G | p.H208R | Benign | 0 |
| NL4 | 1 | ZNF618 | 9:116811441 | NM_133374.2 | G | A | p.R527Q | Probably damaging | 0.000017 |
| NL6 | 1 | SNRNP200 | 2:96955598 | NM_014014.4 | G | A | p.A960V | Probably damaging | 0.000033 |
| NL8 | 1 | PIK3C2B | 1:204415166 | NM_002646.3 | C | T | p.A866T | Probably damaging | 0 |
| NL11 | 2 | MAP3K6 | 1:27688646 | NM_004672.3 | C | T | p.A451T | Benign | 0.000008 |
| ZNF292 | 6:87970762 | NM_015021.1 | A | C | p.E2472A | Benign | 0 | ||
| NL12 | 1 | COG5 | 7:107013177 | NM_006348.3 | A | C | p.Q264P | Probably damaging | 0 |
| NL13 | 1 | MKI67 | 10:129901675 | NM_002417.4 | T | A | p.E2472A | Benign | 0 |
| NL14 | 1 | FBXL13 | 7:102553612 | NM_145032.3 | G | C | p.T310R | Probably damaging | 0 |
| NL15 | 2 | PXDNL | 8:52321174 | NM_144651.4 | T | C | p.I1004V | Benign | 0 |
| TLE2 | 19:3002420 | NM_001144761.1 | G | A | p.R674C | Probably damaging | 0.000076 | ||
| NL19 | 2 | LRIG2 | 1:113616050 | NM_014813.1 | G | A | p.V8L | Probably damaging | 0.000041 |
| OR2T11 | 1:248789610 | NM_001001964.1 | A | T | p.F274L | Possibly damaging | 0 | ||
| NL20 | 1 | SREBF1 | 17:17723001 | NM_004176.4 | G | C | p.L188V | Possibly damaging | 0 |
| G1 | 1 | PLD1 | 3:171392324 | NM_002662.4 | A | G | p.Y732C | Probably damaging | 0 |
| G2 | 1 | SYT15 | 10:46963852 | NM_181519.2 | A | G | p.M371V | Benign | 0.001571 |
| G3 | 2 | DOPEY2 | 21:37618674 | NM_005128.2 | C | A | p.P1466T | Benign | 0 |
| AHNAK2 | 14:105416174 | NM_138420.2 | T | A | p.C1872S | Benign | 0.008125 | ||
| G4 | 1 | HYAL4 | 7:123508339 | NM_012269.2 | A | C | p.L4F | Possibly damaging | 0 |
| G6 | 1 | SRP72 | 4:57368013 | NM_006947.3 | A | G | p.K668E | Probably damaging | 0 |
| G8 | 1 | ZNF793 | 19:38028548 | NM_001013659.2 | C | T | p.R330X | NA | 0 |
| G9 | 1 | C6orf89 | 6:36887382 | NM_152734.3 | T | C | p.I292T | Benign | 0.000231 |
| G13 | 1 | PRKD3 | 2:37501677 | NM_005813.3 | C | G | p.T513S | Benign | 0 |
| G14 | 1 | RTEL1 | 20:62324513 | NM_016434.3 | C | T | p.R957W | Probably damaging | 0.00005 |
| G16 | 2 | CAB39 | 2:231682556 | NM_016289.3 | A | G | p.N261D | Possibly damaging | 0 |
| ABL2 | 1:179086640 | NM_005158.4 | C | T | p.A397V | Probably damaging | 0 | ||
| G17 | 1 | TMEM131 | 2:98378517 | NM_015348.1 | G | C | p.V1624L | Benign | 0.000009 |
| G18 | 1 | OR1N1 | 9:125289536 | NM_012363.1 | C | T | p.R13G | Benign | 0.000018 |
| S2 | 1 | TECTA | 11:121016433 | NM_005422.2 | A | G | p.Y1238C | Benign | 0.000016 |
| S4 | 4 | OAS3 | 12:113379385 | NM_006187.2 | C | T | p.S63L | Benign | 0.000083 |
| BAHCC1 | 17:79409915 | NM_001080519.2 | G | A | p.G514R | Benign | 0.000026 | ||
| ZNF24 | 18:32917633 | NM_006965.2 | C | G | p.P224A | Benign | 0 | ||
| FBXW8 | 12:117383291 | NM_012174.1 | A | C | p.Q116H | Probably damaging | 0 | ||
| IT1 | 2 | NPTXR | 22:39222573 | NM_014293.3 | G | A | p.G344R | Possibly damaging | 0.000030 |
| RBM33 | 7:155530234 | NM_053043.2 | A | C | p.E195A | Possibly damaging | 0 | ||
| IT2 | 1 | C10orf90 | 10:128192697 | NM_001004298.2 | G | A | p.G455S | Probably damaging | 0.000015 |
| IT3 | 2 | ADAM33 | 20:3651693 | NM_025220.2 | G | A | p.R734X | NA | 0.000563 |
| PTCRA | 6:42893368 | NM_138296.2 | G | T | p.G265V | Probably damaging | 0 | ||
| IT4 | 2 | LTV1 | 6:144181653 | NM_032860.3 | G | A | p.E296K | Possibly damaging | 0 |
| SARDH | 9:136535790 | NM_007101.3 | G | A | p.P804L | Benign | 0.000034 | ||
| IT5 | 3 | ASCC3 | 6:100988080 | NM_006828.2 | G | T | p.D1912Y | Probably damaging | 0 |
| C8B | 1:57422504 | NM_000066.3 | G | T | p.C110F | Probably damaging | 0 | ||
| NEK10 | 3:27173451 | NM_001031741.3 | A | G | p.R370X | NA | 0 | ||
| IT6 | 1 | TRPM4 | 19:49671909 | NM_017636.3 | G | A | p.G123S | Probably damaging | 0 |
| IT7 | 2 | SLC6A17 | 1:110714820 | NM_001010898.2 | T | C | p.I142T | Probably damaging | 0 |
| VPS11 | 11:118949581 | NM_021729.4 | G | A | p.E714K | Benign | 0 | ||
| IT8 | 1 | CACNA1B | 9:141014708 | NM_000718.3 | G | T | p.R2042L | Probably damaging | 0 |
| IT9 | 1 | COL15A1 | 9:101748312 | NM_001855.4 | G | T | p.R189L | Probably damaging | 0 |
| IT10 | 1 | MFSD12 | 19:3557319 | NM_021731.2 | C | T | p.G28D | Probablydamaging | 0 |
| IT11 | 1 | ZFP64 | 20:50704964 | NM_199427.2 | A | C | p.S399R | Probably damaging | 0 |
| IT12 | 1 | MRGPRG | 11:3239743 | NM_001164377.1 | G | C | p.V101L | Benign | 0 |
| IT13 | 2 | ARHGEF18 | 19:7533837 | NM_001130955.1 | G | A | p.A1015T | Benign | 0.000053 |
| TJP3 | 19:3730576 | NM_001267560.1 | G | A | p.R162H | Possibly damaging | 0.000083 | ||
| IT14 | 1 | RRP1B | 21:45095000 | NM_015056.2 | A | G | p.I169V | Benign | 0 |
| IT15 | 4 | CLCN2 | 3:184075179 | NM_001171087.2 | G | A | p.R290Q | Probably damaging | 0 |
| LTBP4 | 19:41131902 | NM_001042544.1 | G | A | p.A1392T | Probably damaging | 0 | ||
| PDCD1 | 2:242794902 | NM_005018.2 | G | A | p.G103R | Benign | 0.000030 | ||
| SLC9A5 | 16:67289730 | NM_004594.2 | C | T | p.R270C | Probably damaging | 0 | ||
| IT16 | 2 | EPHA2 | 1:16464557 | NM_004431.3 | A | G | p.Q368R | Benign | 0 |
| PCK1 | 20:56140150 | NM_002591.3 | C | T | p.A458V | Probably damaging | 0.00003 | ||
| IT17 | 1 | PKD1 | 16:2158656 | NM_000296.3 | C | A | p.A2171D | Probably damaging | 0.00019 |
| IT18 | 2 | ODZ3 | 4:183522220 | NM_001080477.1 | C | G | p.P219A | Benign | 0 |
| PAPPA | 9:118950074 | NM_002581.3 | G | A | p.V353M | Possibly damaging | 0 | ||
| IT19 | 1 | CEL | 9:135939864 | NM_001807.4 | T | A | p.V50E | Probably damaging | 0 |
| IT20 | 1 | PROS1 | 3:93598107 | NM_000313.3 | G | A | p.R515H | Benign | 0 |
| IT21 | 1 | ZWILCH | 15:66821226 | NM_017975.3 | G | A | p.V336I | Benign | 0 |
| IT22 | 1 | SHD | 19:4290547 | NM_020209.3 | C | T | p.L314F | Benign | 0 |
| IT23 | 1 | SH2D3A | 19:6755290 | NM_005490.2 | C | T | p.T178M | Benign | 0 |
Note: Trios without DNM are not included.
This resulted in an average of 0.84 coding de novo mutations per trio, which is higher compared with previous studies in ALS with de novo mutation rates of 0.53 (Chesi et al., 2013) and 0.39 (Steinberg et al., 2015) (mean difference with previous studies combined 0.38, 95% confidence interval [CI] 0.14–0.62, P = 0.003), but consistent with the expected distribution in the general population (Samocha et al., 2014) (mean difference 0.14,95% CI −0.06 to 0.34, P = 0.16).
The number of de novo mutations per patient follows a Poisson distribution (Supp. Fig. S1; P = 0.17) indicating that the occurrence of multiple de novo mutations in certain individuals does not differ from what one would expect based on a de novo mutation rate of 0.84. There was no gene in which homozygous mutations were identified more than once when filtering our datasets for exonic variants with a MAF < 0.01 (Supp. Table S1). Compound heterozygous alterations were identified in OBSCN and VCAN in two trios each; however, no other gene had biallelic alterations in more than one trio (Supp. Table S2).
When we combined our trios with the previously published ALS trios (Chesi et al., 2013; Steinberg et al., 2015) (173 trios in total), the overall de novo mutation rate in ALS was 0.64. This also does not differ from the mutation rate in the general population (mean difference −0.058, 95% CI −0.18 to 0.067, P = 0.40).
3.2 |. Pathway enrichment analysis
Next, we investigated whether genes containing de novo mutations were enriched for certain functional pathways as has been shown in previous studies, using DAVID functional annotation analysis (Huang et al., 2009). Our 69 de novo mutation genes showed enrichment for phosphoproteins (P = 0.0086; Supp. Table S3).
We next combined our gene list with the previously published genes with de novo mutations in ALS (Chesi et al., 2013; Steinberg et al., 2015) to determine whether the total set of genes was enriched for specific pathways (111 de novo mutations in 110 genes; Supp. Table S4). The combined genes showed enrichment for phosphoproteins (P = 0.0004); however, the “chromatin,” “transcription regulation,” and “cell cycle” pathways showed no significant association in our or in the combined dataset.
3.3 |. Protein-protein interaction analysis
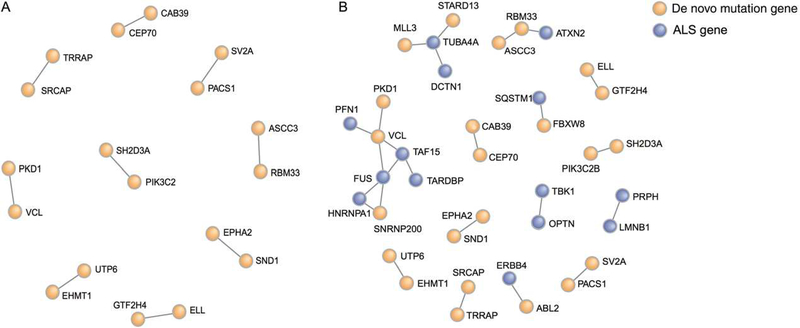
To analyze whether the de novo mutations have occurred randomly or in genes that are interconnected, we used the Disease Association Protein-Protein Link Evaluator (DAPPLE) (Rossin et al., 2011). This program analyzes the connectivity between genes using known protein interaction networks, and determines by permutation the likelihood of such interaction. Our total set of 110 genes with denovo mutations showed no more direct interactions than one would expect by chance (1,000 permutations, P = 0.57; Fig. 1A). Subsequently, we added major ALS genes from the ALS Online Database (ALSoD) and three recently discovered ALS genes (TUBA4A [Smith et al., 2014], TBK1 [Cirulli et al., 2015; Freischmidt et al., 2015], and NEK1 [Brenner et al., 2016; Kenna et al. 2016]) to the de novo mutation dataset (Supp. Fig. S2A). Although the known major ALS genes do show a network with higher connectivity than expected by chance (P = 0.03; Supp. Fig. S2B), combining these genes with our de novo mutation genes did not lead to an increase in connectivity (P = 0.41; Fig. 1B). This indicated that there is no overall increased interaction between the genes harboring de novo mutations in our study and known ALS genes.
FIGURE 1.
Protein interaction network of genes with de novo mutations by DAPPLE. Interacting genes are connected by a line, color represents the origin of the genes, genes found to harbor de novo mutations in this and previous studies are represented in orange, genes with a known association with development of ALS are represented in blue. A: The connectivity of the de novo mutation genes in the total network is not higher than one would expect by chance (P = 0.57). B: Protein interaction network after addition of the 34 known ALS genes. Two larger networks can be identified: one centered around FUS and one around TUBA4A.The connectivity in the total network is not significant (P = 0.41)
3.4 |. Comparison with de novo mutations in a control population
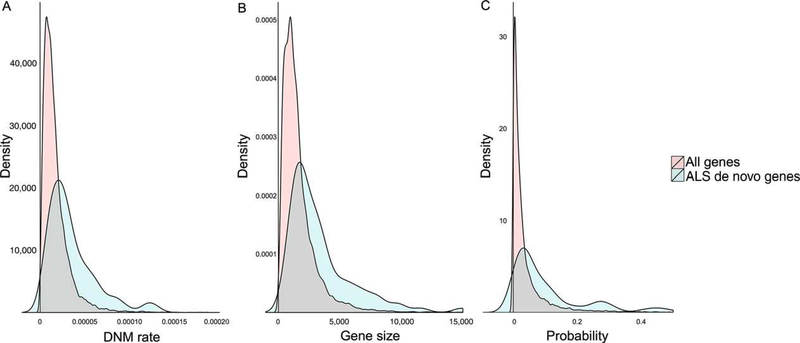
To further investigate whether the observed de novo mutations in ALS are likely to contribute to the disease, we used de novo mutation data from the GoNL, a whole-genome genotyping database of 769 healthy individuals including 250 child-parents trios from The Netherlands (Genome of the Netherlands Consortium, 2014). Using this database, we examined whether the de novo mutations identified in our study have occurred in genes that are relatively prone to de novo mutations or not. The GoNL mutation rate map (Francioli et al., 2015) was used to calculate the likelihood for each gene of having a de novo mutation in respect to the per-base mutation rate and gene size. We compared our de novo mutation genes to this population-based null distribution using the nonparametric Wilcoxon rank sum test, and found that the 69 genes identified by us have a higher overall probability for the occurrence of de novo mutations (P = 1.7 × 10−11). In addition, the previously reported genes with de novo mutations in ALS showed a similar pattern (Chesi et al., 2013; P = 1.4 × 10−4; Steinberg et al., 2015; P = 3.0 × 10−4), with both studies implicating genes with intrinsically elevated de novo mutation rates. The three studies combined also showed a clear shift toward genes with a higher de novo mutation rate (P < 1.0 × 10−15; Fig. 2). This demonstrates that the identified de novo mutations in ALS patients are within genes that in general are more likely to harbor de novo mutations, thus challenging the view that these de novo mutations are disease-specific or disease-causing.
FIGURE 2.
Distribution of the genes with de novo mutations compared with all genes in the genome. Genes with de novo mutations are from the combined data from this study, the study by Chesi et al. (2013) and the study by Steinberg et al. (2015). A: Average de novo mutation rate (DNM rate) per base of the genes with de novo mutations (blue) compared with the GoNL de novo database distribution (red); in general, the genes with de novo mutations have a higher per-base mutation rate (P < 1.0 × 10−15). B: Gene size (in base pairs) of the genes with de novo mutations (blue) compared with gene size of all genes from the genome (red); the genes with de novo mutations are on the whole larger (P < 1 × 10−15). C: Probability of de novo mutations per gene (the product of de novo mutation rate per base and gene size, blue) compared with the GoNL de novo database distribution (red); the genes with de novo mutations have, in general, a higher probability of de novo mutations (P < 1.0 × 10−15)
4 |. DISCUSSION
In this large international cohort of ALS patient-parent trios, we discovered that pattern and rates of de novo mutations in ALS patients in general do not differ from the general population. We could not find evidence for the previously reported role for de novo mutations in the development of ALS.
Due to the high mean age of symptom onset in ALS patients, it is more difficult to acquire DNA from unaffected parents compared with disorders with an earlier onset, for example, autism and schizophrenia. Therefore, this study included multiple international cohorts and combined this with previously published ALS trios. With these 173 trios, we could detect a de novo rate increase of 0.15 with a power of 0.8. A limitation of this study is that a potential smaller effect size could be missed due to sample size, although the clinical consequence of such a small effect can be questioned. Therefore, we also used DAVID, DAPPLE, and the GoNL de novo mutation data to further look into these mutations but found no differences between de novo patterns between patients and controls.
Using DAVID pathway analysis, we found no evidence for the previous reported association of de novo mutations with ALS via chromatin regulation, transcription regulation, or cell cycle pathways. This indicates again that the results of DAVID pathway analyses should be interpreted with caution if only a small number of genes are analyzed (Elbers et al., 2009).
De novo mutation rates increase with paternal age (Kong et al., 2012) and indeed a higher mean paternal age could be observed in patients with intellectual disability (Hehir-Kwa et al., 2011) and autism spectrum disorders (Neale et al., 2012; O’Roak et al. 2012). The average paternal age in our 82 trios was 28.8 years (data missing in seven trios), which is lower than in autism and intellectual disability where de novo mutations do play a role. A previous study also showed that paternal age of ALS patients is not higher than the paternal age of controls (de Jong et al., 2013), which is in line with our findings.
The trios in this research are sequenced on either Complete Genomics (cohort from The Netherlands) or Illumina (other cohorts) sequencing platforms. Although these platforms have different methods to assess the genome, comparative research showed these methods to have a large overlap in called SNVs, with a high false-positive rate in the platform-specific calls (Lam et al., 2012). Therefore, SNVs found with these two methods can be combined.
This study does not analyze de novo structural variants. Although structural variants could be important for explaining the missing heritability in ALS, analysis of structural variants from exome data is still challenging. Also, compared with SNVs, structural variants have a larger variation between different sequencing platforms (Lam et al., 2012). Therefore, these structural variants were not included in this study and further research is needed to assess their role on ALS development.
Our results do not rule out that individual de novo mutations in specific genes, as is reported for de novo mutations in FUS (Hübers et al., 2015), can be pathogenic in specific patients. However, these seem only to represent a minority of cases. In general, our results thus challenge a major role of de novo mutations in the development of ALS.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to all patients and their families for their participation in this study. The technical assistance of Marika Pusch, Ingrid Goebel, and Antje Knehr (Institute of Human Genetics, Medical Centre Hamburg-Eppendorf, Hamburg, Germany) is gratefully acknowledged. We furthermore thank the Regional Computing Center of the University of Cologne (RRZK) for providing computing time on the DFG-funded High Performance Computing (HPC) system CHEOPS as well as support for analyzing the German and Swedish samples.
Contract grant sponsors: ALS Foundation Netherlands; European Community’s Health Seventh Framework Programme (FP7/2007-2013); ZonMW under the frame of E-Rare-2; the ERA Net for Research on Rare Diseases (PYRAMID); EU Joint Programme-Neurodegenerative Disease Research (JPND) project (STRENGTH, SOPHIA); Medical Research Council and Economic and Social Research Council (UK); Health Research Board (Ireland); ZonMw (The Netherlands); Ministry of Health and Ministry of Education, University and Research (Italy); L’Agence nationale pour la recherché (France);The Netherlands Organisation for Health Research and Development (Vici scheme);AriSLA - Fondazione Italiana di Ricerca per la SLA (NOVALS 2012); ‘5 × 1000’ Healthcare Research of the Italian Ministry of Health; The Italian Ministry of Health (GR-2011-02347820’IRisALS’); Deutsche Forschungsgemeinschaft (DFG) (VO2028/1-1); BMBF-funded German Network for motor neuron diseases (MND-NET); Charcot Foundation for ALS Research; Virtual Helmholtz Association; International Graduate School in Molecular Medicine Ulm (IGradU).
Footnotes
DISCLOSURE STATEMENT
The authors declare that they have no conflict of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- Brenner D, Müller K, Wieland T, Weydt P, Böhm S, Lulé D, … Weishaupt JH (2016). NEK1 mutations in familial amyotrophic lateral sclerosis. Brain, 139, e28–e28. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Miller RG, Swash M, Munsat TL, & World Federation of Neurology Research Group on Motor Neuron Diseases. (2000). El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis and Other Motor Neuron Disorders, 1, 293–299. [DOI] [PubMed] [Google Scholar]
- Chesi A, Staahl BT, Jovičić A, Couthouis J, Fasolino M, Raphael AR, … Gitler AD (2013). Exome sequencing to identify de novo mutations in sporadic ALS trios. Nature Neuroscience 16, 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, … Goldstein DB (2015). Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science, 347, 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong SW, Huisman MHB, Hennekam EAM, Sutedja NA, van der Kooi AJ, de Visser M, … van den Berg LH (2013). Parental age and the risk of amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, 14, 224–227. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BWM, Kleefstra T, Yntema HG, Kroes T, … Vissers LE (2012). Diagnostic exome sequencing in persons with severe intellectual disability. The New England Journal of Medicine, 367, 1921–1929. [DOI] [PubMed] [Google Scholar]
- Elbers CC, van Eijk KR, Franke L, Mulder F, van der Schouw YT, Wijmenga C, & Onland-Moret NC (2009). Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genetic Epidemiology, 33, 419–431. [DOI] [PubMed] [Google Scholar]
- Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I, … Sunyaev SR (2015). Genome-wide patterns and properties of de novo mutations in humans. Nature Genetics, 47, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, … Weishaupt JH (2015). Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nature Neuroscience, 18, 631–636. [DOI] [PubMed] [Google Scholar]
- Genome of the Netherlands Consortium. (2014). Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nature Genetics, 46, 818–825. [DOI] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Rodríguez-Santiago B, Vissers LE, de Leeuw N, Pfundt R, Buitelaar JK, … Veltman JA (2011). De novo copy number variants associated with intellectual disability have a paternal origin and age bias. Journal of Medical Genetics, 48, 776–778. [DOI] [PubMed] [Google Scholar]
- Hoischen A, van Bon BWM, Rodríguez-Santiago B, Gilissen C, Vissers LELM, de Vries P, … de Vries BB (2011). De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nature Genetics, 43, 729–731. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, & Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hübers A, Just W, Rosenbohm A, Müller K, Marroquin N, Goebel I, … Volk AE (2015). De novo FUS mutations are the most frequent genetic cause in early-onset German ALS patients. Neurobiology of Aging, 36(3117), e1–e6. [DOI] [PubMed] [Google Scholar]
- Huisman MHB, de Jong SW, van Doormaal PTC, Weinreich SS, Schelhaas HJ, van der Kooi AJ, … van den Berg LH (2011). Population based epidemiology of amyotrophic lateral sclerosis using capture- recapture methodology. Journal of Neurology, Neurosurgery, and Psychiatry, 82, 1165–1170. [DOI] [PubMed] [Google Scholar]
- Kenna KP, van Doormaal PTC, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, … Landers JE (2016). NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nature Genetics, 48, 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, … Stefansson K (2012). Rate of de novo mutations and the importance of father’s age to disease risk. Nature, 488, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HYK, Clark MJ, Chen R, Chen R, Natsoulis G, O’Huallachain M, … Snyder M (2012). Performance comparison of whole-genome sequencing platforms. Nature Biotechnology, 30, 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Palaima P, Yum SW, Gonzalez MA, Tao F, Wanschitz JV, … Scherer SS (2016). De novo PMP2 mutations in families with type 1 Charcot-Marie-Tooth disease. Brain, 139, 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, … Daly MJ (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature, 485, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, & Taylor SL (1997). PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Research, 25, 27452751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, … Eichler EE (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature, 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu A, Noordam MJ, Schwartz RS, Wuster A, Hurles ME, Cartwright RA, & Conrad DF (2013). DeNovoGear: De novo indel and point mutation discovery and phasing. Nature Methods, 10, 985–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, Cleary J, … Del-Favero J (2012). Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nature Biotechnology, 30, 61–68. [DOI] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, … Daly MJ (2011). Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology PLoS Genetics, 7, e1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, … Daly MJ (2014). A framework for the interpretation of de novo mutation in human disease. Nature Genetics, 46, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BN, Ticozzi N, Fallini C, Gkazi A-S,Topp S, Kenna KP, … Landers JE (2014). Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron, 84, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg KM, Yu B, Koboldt DC, Mardis ER, & Pamphlett R (2015). Exome sequencing of case-unaffected-parents trios reveals recessive and de novo genetic variants in sporadic ALS. Scientific Reports, 5, 9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Baker MC, DeJesus-Hernandez M, Ghidoni R, Benussi L, Finger E, … Rademakers R (2013). C9ORF72 repeat expansions in cases with previously identified pathogenic mutations. Neurology, 81, 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, van Es MA, Hennekam EAM, Dooijes D, van Rheenen W, Medic J, … van den Berg LH (2012). Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Human Molecular Genetics, 21, 3776–3784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.