Abstract
Background
Long non-coding RNA differentiation antagonizing nonprotein coding RNA (lncRNA DANCR) has been reported to act as an oncogene in various human cancers. The role of DANCR in development of pancreatic cancer (PC) is unknown. The aim of our research was to investigate the biological role of DANCR in PC.
Material/Methods
Expressions of DANCR, miR-214-5p, and E2F2 mRNA in PC tissues and cell lines were examined by qRT-PCR. Western blotting was carried out for detection of E2F2 protein expression in PC cells. Transwell assays were used to examine the metastatic ability of PC cells, while CCK-8 and colony formation assay were applied to evaluate cell proliferation. The effects of DANCR on PC cells were assessed by knockdown in vitro and in vivo. The regulatory mechanism of competitive endogenous RNAs were obtained from bioinformatics prediction and luciferase reporter assay.
Results
DANCR was markedly upregulated in clinical tissues and cell lines of PC. High DANCR expression exhibited a significant correlation with poor prognosis. DANCR knockdown inhibited growth and metastasis of PC cells. Furthermore, DANCR acted as sponge to regulate miR-214-5p, and miR-214-5p inhibitor reversed the effects of DANCR knockdown on PC cells. Moreover, DANCR positively modulated E2F2 expression through miR-214-5p in PC cells.
Conclusions
Collectively, our findings demonstrated that lncRNA DANCR/miR-214-5p/E2F2 axis acts as an oncogene in PC development, which might provide a potential target for PC therapy.
MeSH Keywords: MicroRNAs; Pancreatic Neoplasms; RNA, Long Noncoding
Background
Pancreatic cancer (PC) is among the most aggressive and lethal of all cancers, and the 5-year survival rate is under 5% [1]. A majority of patients miss the optimal period for surgery after diagnosis due to the lack of obvious clinical symptoms and the occurrence of early distant metastasis [2,3]. In spite of great improvement in diagnosis and treatment in the last several decades, the mortality rate of PC is still dismal [4]. The molecular mechanisms of PC germination and development remain unclear. It is extremely important to explore novel effective biomarkers and therapeutic targets for the early diagnosis and therapy of PC.
Long non-coding RNAs (lncRNAs) are defined as regulatory noncoding transcripts of longer than 200 nucleotides in length [5]. Increasing research has reported that lncRNAs participate in multiple tumor biological processes, including cell proliferation and metastasis [6,7]. The newly discovered lncRNA differentiation antagonizing nonprotein coding RNA (DANCR) has been demonstrated to act as an oncogene in many human tumors [8–10]. In gastric cancer, knockdown of DANCR inhibits cell migration and invasion, and overexpression of DANCR shows the opposite influence [11]. In colorectal cancer, DANCR is highly expressed, and upregulation of DANCR facilitates cell growth and metastasis of colorectal cancer, both in vitro and in vivo [12]. However, the functional roles of DANCR in PC are unknown, and thus need to be further elucidated.
MicroRNAs (miRNAs), a group of short non-coding RNAs of less than 25 nucleotides in length, play a critical role in many biological processes [13]. Accumulating evidence has revealed that dysfunction of miRNAs plays a major role in tumorigenesis of cancers, including PC [14,15]. Recently, it was reported that there is a novel regulatory mechanism by which lncRNA acts as a competing endogenous RNA (ceRNA) by sponging miRNA to regulate the miRNA expression [16]. For instance, Liu et al. [17] indicated lncRNA NEAT1 facilitates ROCK1-mediated metastasis of ovarian cancer via working as a ceRNA of miR-382-3p.
This research showed that DANCR is upregulated in PC tissues and cell lines. DANCR can promote cell growth and metastasis in vitro and in vivo. In addition, DANCR functions as a molecular sponge of miR-214-5p, thus affecting E2F2 expression. In conclusion, our results demonstrate that the newly identified DANCR/miR-214-5p/E2F2 pathway modulates the development of PC, which provides a potential therapeutic target for PC.
Material and Methods
Tissue samples
We obtained 50 paired samples of PC and adjacent normal tissues from patients who underwent pancreatic resection surgery at the Second Affiliated Hospital of Wenzhou Medical University from 2013 to 2015. None of patients had previously undergone radiotherapy or chemotherapy before the operation. The study was approved by the Ethics Committee of the Second Affiliated Hospital of Wenzhou Medical University (2015 RES-130), and all patients signed the written informed consent. Clinicopathological data of these patients was documented.
Cell culture
PC cell lines (PANC-1, SW1990, CAPAN-1, BxPC-3, and AsPC-1), normal human pancreatic duct epithelial cells (HPDE6-C7), and HEK293T cells were obtained from American Type Culture Collection. Cells were cultured in RPMI 1640 or DMEM medium supplemented with 10% FBS.
Cell transfection
Two small interference RNAs targeting DANCR (si-DANCR) or negative control siRNAs (si-NC) were used, as reported previously [8]. The sequences were: 5′-UCGGAGGUGGAUUCUGUUA-3′ and 5′-AGCCAACTATCCCTTCAGT-3′. miR-214-5p mimics and inhibitor were synthesized by GenePharma (China). Lipofectamine 2000 reagent (Invitrogen, USA) was used in the cell transfection assay.
Quantitative real-time PCR
RNAs were isolated and then reverse-transcribed into cDNA. QRT-PCR was performed using the SYBR Green PCR Kit on the ABI 7500 Fast RT PCR system (Applied Biosystems, USA) according to the manufacturer’s protocols. Relative miRNA expression was normalized to U6. mRNA and lncRNA expression were normalized to GAPDH. The results were analyzed by 2−ΔΔCt method.
Cell viability assay
The cell viability was detected with CCK-8 assay. Cells were plated onto a 96-well plate. The solution was then added to each well at 24 h, 48 h, 72 h, and 96 h after transfection. Absorbance at the wavelength of 450 nm was examined with a microplate reader.
Cell cycle assay
The transfected cells were collected for cell cycle analysis 48 h later. Cells were fixed with 70% cold anhydrous ethanol, and then stained with propidium iodide. The cell cycle distribution was finally determined by flow cytometry.
Cell apoptosis assay
The transfected cells were harvested for cell apoptosis analysis by using the Annexin V-FITC apoptosis detection kit. After double staining with Annexin V-FITC and propidium iodide, cell apoptosis was detected by flow cytometry.
Colony formation assay
The PC cells were added to 6-well plates, then incubated in DMEM medium. After 2 weeks, cells were stained with crystal violet and the numbers of colonies were counted using a microscope.
Migration and invasion assay
Transwell assays were conducted to evaluate the metastatic ability of PC cells. After transfection for 48 h, cells in medium without serum were seeded into uncoated or Matrigel-coated upper chambers (BD Bioscience, USA), while medium supplemented with FBS was added into the lower chambers as a chemoattractant. The cells on the upper surface were carefully removed after 24-h incubation. Next, the migrating or invading cells were fixed by methanol, followed by staining with crystal violet. Five random fields of each well were counted.
Western blotting
The proteins were extracted by RIPA cell lysis buffer. Equal amounts of protein samples were separated by 10% SDS-PAGE, transferred onto the PVDF membrane, and then incubated with specific primary antibody for E2F2 or GAPDH (CST, USA). After incubation with HRP-conjugated secondary antibody, the results were analyzed Using the ECL detection system.
Luciferase reporter assay
DANCR fragments that had the putative or mutant binding sites of miR-214-5p were synthesized and cloned into pmirGLO vector (Promega, USA) in order to generate wild-type or mutant DANCR reporter plasmid (DANCR-wt and DANCR-mut). The wild-type E2F2 reporter plasmid (E2F2-wt) and mutant E2F2 reporter plasmid (E2F2-mut) were constructed using the same method. Luciferase reporter plasmid and miR-214-5p mimics or negative control were then co-transfected into the HEK293T cells. Luciferase activity was evaluated by the dual-luciferase reporter assay system.
Animal study
Male athymic BALB/c nude mice (n=20) were used in this study. PANC-1 cells transfected with sh-DANCR or sh-NC were subcutaneously injected into nude mice. The tumor volume was measured every week, and the tumor weight was measured 5 weeks after inoculation. Tumor volumes in nude mice were calculated according to the following formula: tumor volume (mm3)=0.5×length×width2. The animal study was approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University (approval no. 2016-0048).
Statistical analysis
Results are shown as mean ± standard deviation. Statistical analysis was done with the SPSS 20.0 software. We used the chi-square test, t test, and one-way ANOVA to estimate differences between groups. Overall survival of PC patients was evaluated with Kaplan-Meier method. P<0.05 was considered statistically significant.
Results
DANCR was upregulated in PC tissues and cell lines
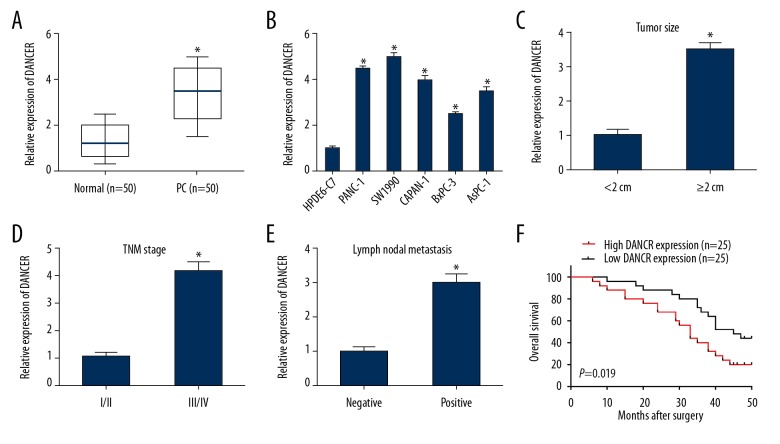
DANCR expression in 50 paired PC and adjacent normal tissues were measured by qRT-PCR assay. Significant upregulation of DANCR was observed in PC tissues in comparison to normal tissues (Figure 1A). We also examined DANCR expression in different PC cell lines. DANCR expression in PC cells (PANC-1, SW1990, CAPAN-1, BxPC-3, and AsPC-1) appeared to be higher than that in HPDE6-C7 cells (Figure 1B).
Figure 1.
DANCR was overexpressed in PC tissues and cell lines. (A) DANCR expression in PC tissues and normal tissues was determined by qRT-PCR. (B) DANCR expression in PC cells (PANC-1, SW1990, CAPAN-1, BxPC-3, and AsPC-1) and HPDE6-C7 cells was determined by qRT-PCR. (C–E) DANCR expression in PC patients was stratified by tumor size, TNM stage, or lymph nodal metastasis. (F) High DANCR expression was associated with a shorter overall survival of PC patients. * P<0.05.
Next, correlations between DANCR expression and clinicopathological characteristics of PC patients were evaluated. High DANCR expression exhibited significant correlations with tumor size, TNM stage, and lymph nodal metastasis (Table 1). DANCR expression was also overexpressed in PC patients with larger tumor size, advanced tumor stage, and positive lymph nodal metastasis (Figure 1C–1E). Moreover, the result showed high DANCR expression was markedly correlated with poor overall survival in patients with PC, as determined using Kaplan-Meier method (Figure 1F).
Table 1.
Correlation between lncRNA DANCR expression and clinicopathological characteristics of pancreatic cancer patients.
| Parameters | Total | DANCR expression | P value | |
|---|---|---|---|---|
| High (n=25) | Low (n=25) | |||
| Age (years) | 0.526 | |||
| <60 | 23 | 12 | 11 | |
| ≥60 | 27 | 13 | 14 | |
| Gender | 0.232 | |||
| Male | 24 | 14 | 10 | |
| Female | 26 | 11 | 15 | |
| TNM stage | 0.025* | |||
| I/II | 35 | 15 | 20 | |
| III/IV | 15 | 10 | 5 | |
| Tumor size (cm) | 0.032* | |||
| <2 | 24 | 8 | 16 | |
| ≥2 | 26 | 17 | 9 | |
| Distant metastasis | 0.248 | |||
| Negative | 23 | 10 | 13 | |
| Positive | 27 | 15 | 12 | |
| Lymph node metastasis | 0.020* | |||
| Negative | 30 | 11 | 19 | |
| Positive | 20 | 14 | 6 | |
| Tumor differentiation | 0.385 | |||
| Well/moderate | 33 | 16 | 17 | |
| Poor | 17 | 9 | 8 | |
P<0.05 was considered significant.
Knockdown of DANCR inhibited cell growth and metastasis of PC in vitro, and suppressed cell growth in vivo
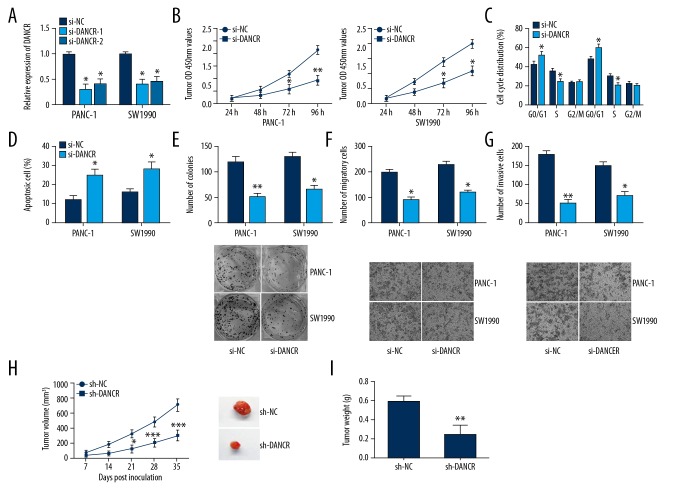
To explore effects of lncRNA DANCR on cell proliferation, migration, and invasion of PC, we silenced DANCR expression in PANC-1 and SW1990 cells through si-DANCR (Figure 2A). CCK-8 assay demonstrated that cell viability of PC was obviously inhibited in the si-DANCR group in comparison to the si-NC group (Figure 2B). Next, flow cytometry was performed to detect the cell cycle distribution and cell apoptosis. We found that DANCR knockdown led to cell cycle arrest at G0/G1 phase (Figure 2C) and promoted cell apoptosis (Figure 2D) in PANC-1 and SW1990 cells. In addition, DANCR knockdown markedly decreased the number of PC cell colonies using colony formation assay (Figure 2E). Furthermore, si-DANCR suppressed cell migration and invasion of PC, as shown by the results from Transwell assays (Figure 2F, 2G).
Figure 2.
DANCR inhibition suppressed proliferation, migration, and invasion of PC cells. (A) Transfection efficiency was evaluated by qRT-PCR. (B) Cell proliferation was detected by CCK-8 assay. (C) DANCR knockdown led to cell cycle arrest at G0/G1 phase in PANC-1 and SW1990 cells. (D) DANCR knockdown promoted cell apoptosis. (E) The colony-forming ability was examined by colony formation assay. (F, G) The migration and invasion abilities were examined through Transwell assays. (H) The tumor volume was measured every week to construct tumor growth curves. (I) Tumor weight was measured when the tumors were harvested. * P<0.05, ** P<0.01, *** P<0.001.
We also explored inhibition of DANCR on cell growth in vivo. Five weeks after injection, tumor volumes and weights of nude mice in the sh-DANCR group were remarkably decreased in comparison to these in the sh-NC group (Figure 2H, 2I). In conclusion, these results prove that DANCR inhibition suppresses cell growth of PC in vivo.
DANCR directly interacted with miR-214-5p in PC
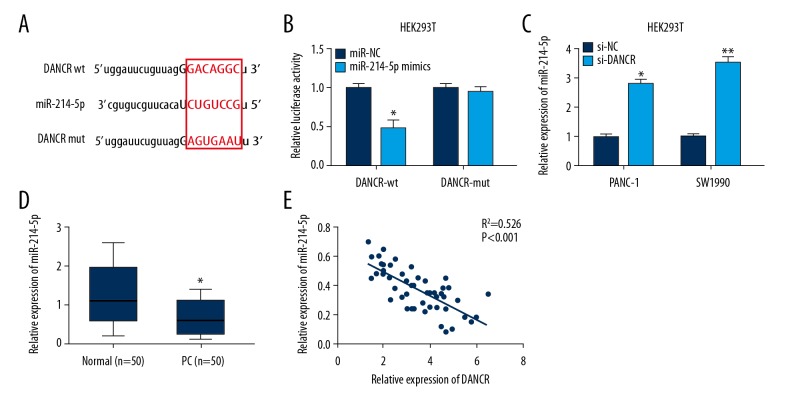
By using bioinformatics software (Starbase v3.0), miR-214-5p was found to be a potential target of DANCR (Figure 3A). miR-214-5p mimics dramatically reduced the relative luciferase activity of DANCR-wt by luciferase reporter assay, but did not affect the DANCR-mut reporter (Figure 3B). Moreover, miR-214-5p expression was markedly upregulated in PANC-1 and SW1990 cells transfected with si-DANCR compared with si-NC (Figure 3C).
Figure 3.
DANCR directly interacted with miR-214-5p in PC. (A) Bioinformatics prediction of miR-214-5p binding sites in DANCR 3′UTR. (B) Luciferase activity of DANCR-wt was decreased by miR-214-5p mimics. (C) miR-214-5p expression in PANC-1 and SW1990 cells transfected with si-DANCR or si-NC. (D) miR-214-5p expression in 50 paired PC and normal tissues was determined by qRT-PCR. (E) miR-214-5p expression exhibited a negative correlation with DANCR expression. * P<0.05, ** P<0.01.
miR-214-5p expression in 50 PC tissue samples was then detected. miR-214-5p was remarkably downregulated in PC tissue samples in comparison to adjacent normal tissues (Figure 3D). Furthermore, result from Pearson’s correlation analysis showed a negative correlation between miR-214-5p and DANCR expression in tissue samples (Figure 3E). These findings indicate that DANCR directly targets miR-214-5p in PC.
miR-214-5p mediates inhibition of DANCR knockdown in cell growth and metastasis of PC
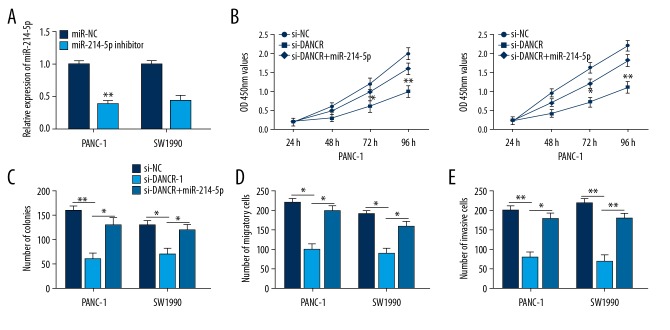
To further study whether DANCR exerts biological effects in PC by targeting miR-214-5p, we reduced miR-214-5p expression with miRNA inhibitor in PANC-1 and SW1990 cells (Figure 4A). CCK-8 and colony formation assay revealed DANCR inhibition dramatically suppressed cell growth of PC, and miR-214-5p inhibitor greatly restored the effects (Figure 4B, 4C). In addition, the repression of cell migration and invasion induced by DANCR knockdown was reversed by miR-214-5p inhibitor, as determined using Transwell assays (Figure 4D, 4E). In brief, knockdown of DANCR suppressed PC cell proliferation, migration, and invasion by miR-214-5p.
Figure 4.
miR-214-5p inhibitor reversed the effects of DANCR knockdown on PC cells. (A) Transfection efficiency was evaluated by qRT-PCR. (B, C) CCK-8 and colony formation assay revealed DANCR inhibition suppressed the growth of PC cells, while miR-214-5p inhibitor reversed the effects. (D, E) Transwell assays revealed the repression in cell migration and invasion induced by DANCR knockdown were rescued by miR-214-5p inhibitor. * P<0.05, ** P<0.01.
DANCR upregulated E2F2 expression by sponging miR-214-5p
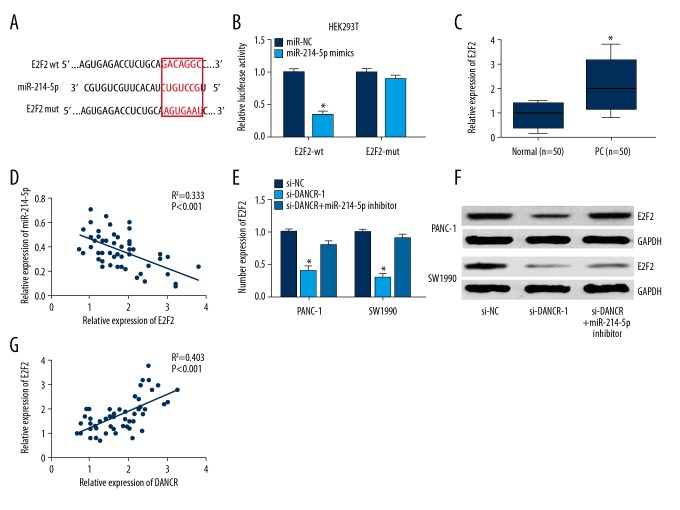
miRNAs can function as negative regulators of target genes at the post-transcriptional level. E2F2, an important regulator of cell proliferation and invasion in tumor progression, was identified as a potential target of miR-214-5p using bioinformatics software (TargetScan, Figure 5A). Luciferase reporter assay showed that miR-214-5p overexpression caused a remarkable decline in luciferase activity of E2F2-wt (Figure 5B). Expression of E2F2 mRNA in clinical tissues was measured with qRT-PCR assay. As shown in Figure 5C, E2F2 was obviously overexpressed in tumor tissues of PC patients in comparison to normal tissues. In addition, miR-214-5p expression exhibited a negative correlation with E2F2 expression (Figure 5D).
Figure 5.
DANCR upregulated E2F2 expression via miR-214-5p. (A) Bioinformatics prediction of miR-214-5p binding sites in E2F2 3′UTR. (B) Luciferase activity of E2F2-wt was reduced by miR-214-5p mimics in HEK293T cells. (C) Relative expression of E2F2 in clinical tissues was examined with qRT-PCR. (D) miR-214-5p expression exhibited a negative correlation with E2F2 expression. (E, F) DANCR knockdown inhibited mRNA and protein levels of E2F2 in PANC-1 and SW1990 cells, this repression was restored by miR-214-5p inhibitor. (G) Positive correlation between E2F2 and DANCR expression was found in PC tissues. * P<0.05.
Next, we investigated whether E2F2 expression was regulated by lncRNA DANCR in PC cells. We found that DANCR knockdown significantly inhibited E2F2 expression in PANC-1 and SW1990 cells, while this repression was restored by treatment with miR-214-5p inhibitor (Figure 5E, 5F). Furthermore, DANCR expression had a positive correlation with E2F2 expression in PC tissues (Figure 5G). These results show that DANCR upregulates E2F2 expression via sponging miR-214-5p in PC.
Discussion
PC is known as a very aggressive human disease with a poor prognosis. More and more studies demonstrate that the non-coding RNAs, including lincRNAs and miRNAs, have important roles in the regulation of tumorigenesis and development. Among them, lncRNA DANCR was found to be upregulated and to participate in various human malignancies. For instance, DANCR is overexpressed in hepatocellular carcinoma tissues, and inhibition of DANCR represses proliferation and metastasis of hepatocellular carcinoma in vitro and in vivo [9]. Moreover, Pan et al. reported that DANCR promotes cell growth and metastasis of gastric cancer, and might be a new diagnostic target [18]. Nonetheless, the functional role of DANCR in PC is still unclear.
Our findings showed that DANCR was overexpressed in PC tissues in comparison to normal tissues. High DANCR expression exhibited significant correlations with tumor size, TNM stage, lymph nodal metastasis, and shorter overall survival of PC patients. Furthermore, DANCR inhibition repressed cell growth and invasion of PC. These results indicate that DANCR can act as an oncogene in PC. However, the molecular mechanism of DANCR in PC progression is unclear.
In recent years, accumulating reports have shown that lncRNA can act as an endogenous sponge to specific targeted miRNA, then modulate its expression and function. The lncRNA ROR promotes radio-resistance in hepatocellular carcinoma cells through functioning as a competing endogenous RNA of miR-145 [19]. Wu et al. demonstrated the lncRNA UCA1 has an oncogenic effect in lung cancer, upregulating HMGB1 expression through sponging miR-193a [20]. DANCR has also been reported to act as a ceRNA and promote cell growth and metastasis of osteosarcoma by targeting miR-335-5p and miR-1972 [21]. In our study, DANCR directly interacted with miR-214-5p in PC, as shown by use of bioinformatics software and a luciferase reporter experiment. Moreover, DANCR depletion inhibited proliferation, migration, and invasion of PC cells, whereas addition of miR-214-5p inhibitor reversed these effects. Therefore, our data prove that DANCR regulates cell proliferation and invasion of PC via sponging miR-214-5p.
miR-214-5p is regarded as a tumor-suppressive miRNA and is known to play important regulative roles in cancer development, including osteosarcoma [22], hepatocellular carcinoma [23], and pancreatic cancer [24]. In accord with previous research, this study also showed miR-214-5p expression was obviously decreased in PC tissues and in cells. miRNAs participate in many cellular processes through targeting specific mRNAs. For example, miR-214-5p knockdown promotes survival and extracellular matrix formation through regulating COL4A1 in osteoblastic MC3T3-E1 cells [25]. Our study revealed E2F transcription factor 2 [E2F2) is a direct target of miR-214-5p in PC, as determined with the use of bioinformatics software and a luciferase reporter experiment.
Notably, E2F2 has been well characterized as a crucial regulator of physiological processes, such as cell cycle, proliferation, DNA damage repair, and autophagy [26]. Previous findings have indicated that E2F2 exhibits oncogenic activity in several types of cancer [27]. For instance, Xie et al. reported E2F2 is a significant upregulated transcription factor in ovarian cancer (OC) cells, and high E2F2 expression is associated with worse overall survival in OC patients [28]. In addition, E2F2 is overexpressed in renal clear cell carcinoma tissues compared to matched normal tissues, and high E2F2 expression is associated with worse prognosis [29]. In the present study, E2F2 was also obviously upregulated in PC tissue samples. Furthermore, miR-214-5p expression had a negative correlation with E2F2 expression, while DANCR expression had a positive correlation with E2F2 expression. Most importantly, DANCR upregulated E2F2 expression by miR-214-5p in PC.
Conclusions
lncRNA DANCR was observably upregulated in tissues and cell lines of PC, and was associated with poor overall survival of PC patients. Moreover, DANCR promoted PC cell growth and metastasis through working as a ceRNA of miR-214-5p, and positively regulated E2F2 expression in PC cells. These findings indicate that a novel axis of DANCR/miR-214-5p/E2F2 is involved in PC development, which might offer a target for PC treatment.
Footnotes
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ansari D, Tingstedt B, Andersson B, et al. Pancreatic cancer: Yesterday, today and tomorrow. Future Oncol. 2016;12(16):1929–46. doi: 10.2217/fon-2016-0010. [DOI] [PubMed] [Google Scholar]
- 3.Moniri MR, Dai LJ, Warnock GL. The challenge of pancreatic cancer therapy and novel treatment strategy using engineered mesenchymal stem cells. Cancer Gene Ther. 2014;21(1):12–23. doi: 10.1038/cgt.2013.83. [DOI] [PubMed] [Google Scholar]
- 4.Heestand GM, Kurzrock R. Molecular landscape of pancreatic cancer: Implications for current clinical trials. Oncotarget. 2015;6(7):4553–61. doi: 10.18632/oncotarget.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017;33(8):540–52. doi: 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495(2):1594–600. doi: 10.1016/j.bbrc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 8.Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 9.Ma X, Wang X, Yang C, et al. DANCR acts as a diagnostic biomarker and promotes tumor growth and metastasis in hepatocellular carcinoma. Anticancer Res. 2016;36(12):6389–98. doi: 10.21873/anticanres.11236. [DOI] [PubMed] [Google Scholar]
- 10.Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Mao Z, Li H, Du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37(6) doi: 10.1042/BSR20171070. pii: BSR20171070. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50(5):57. doi: 10.1038/s12276-018-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbato S, Solaini G, Fabbri M. MicroRNAs in oncogenesis and tumor suppression. Int Rev Cell Mol Biol. 2017;333:229–68. doi: 10.1016/bs.ircmb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Davis-Dusenbery BN, Hata A. MicroRNA in cancer: The involvement of aberrant microRNA biogenesis regulatory pathways. Genes Cancer. 2010;1(11):1100–14. doi: 10.1177/1947601910396213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qadir MI, Faheem A. miRNA: A diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27(3):197–204. doi: 10.1615/CritRevEukaryotGeneExpr.2017019494. [DOI] [PubMed] [Google Scholar]
- 16.Militello G, Weirick T, John D, et al. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–88. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Wang Y, Fu X, Lu Z. Long non-coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of miR-382-3p/ROCK1 axial. Cancer Sci. 2018;109(7):2188–98. doi: 10.1111/cas.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan L, Liang W, Gu J, et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 2018;9(2):1915–30. doi: 10.18632/oncotarget.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Shen Z, Zhi Y, et al. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 2018;645:117–25. doi: 10.1016/j.abb.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Wu H, Zhou C. Long non-coding RNA UCA1 promotes lung cancer cell proliferation and migration via microRNA-193a/HMGB1 axis. Biochem Biophys Res Commun. 2018;496(2):738–45. doi: 10.1016/j.bbrc.2018.01.097. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Wang D, Zhu T, Yin R. miR-214-5p targets ROCK1 and suppresses proliferation and invasion of human osteosarcoma cells. Oncol Res. 2017;25(1):75–81. doi: 10.3727/096504016X14719078133401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Wang H, Ren Z. MicroRNA-214-5p inhibits the invasion and migration of hepatocellular carcinoma cells by targeting Wiskott-Aldrich syndrome like. Cell Physiol Biochem. 2018;46(2):757–64. doi: 10.1159/000488734. [DOI] [PubMed] [Google Scholar]
- 24.Cao TH, Ling X, Chen C, et al. Role of miR-214-5p in the migration and invasion of pancreatic cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(21):7214–21. doi: 10.26355/eurrev_201811_16255. [DOI] [PubMed] [Google Scholar]
- 25.Li QS, Meng FY, Zhao YH, et al. Inhibition of microRNA-214-5p promotes cell survival and extracellular matrix formation by targeting collagen type IV alpha 1 in osteoblastic MC3T3-E1 cells. Bone Joint Res. 2017;6(8):464–71. doi: 10.1302/2046-3758.68.BJR-2016-0208.R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Huang J, Yang D, et al. Expression patterns of E2F transcription factors and their potential prognostic roles in breast cancer. Oncol Lett. 2018;15(6):9216–30. doi: 10.3892/ol.2018.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Tao H. Overexpression of microRNA-936 suppresses non-small cell lung cancer cell proliferation and invasion via targeting E2F2. Exp Ther Med. 2018;16(3):2696–702. doi: 10.3892/etm.2018.6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie L, Li T, Yang LH. E2F2 induces MCM4, CCNE2 and WHSC1 upregulation in ovarian cancer and predicts poor overall survival. Eur Rev Med Pharmacol Sci. 2017;21(9):2150–56. [PubMed] [Google Scholar]
- 29.Liang B, Zhao J, Wang X. Clinical performance of E2Fs 1-3 in kidney clear cell renal cancer, evidence from bioinformatics analysis. Genes Cancer. 2017;8(5–6):600–7. doi: 10.18632/genesandcancer.143. [DOI] [PMC free article] [PubMed] [Google Scholar]