Abstract
Context
The two major forms of circulating thyroid hormones (THs) are T3 and T4. T3 is regarded as the biologically active hormone because it binds to TH receptors (TRs) with greater affinity than T4. However, it is currently unclear what structural mechanisms underlie this difference in affinity.
Objective
Prompted by the identification of a novel M256T mutation in a resistance to TH (RTH)α patient, we investigated Met256 in TRα1 and the corresponding residue (Met310) in TRβ1, residues previously predicted by crystallographic studies in discrimination of T3 vs T4.
Methods
Clinical characterization of the RTHα patient and molecular studies (in silico protein modeling, radioligand binding, transactivation, and receptor–cofactor studies) were performed.
Results
Structural modeling of the TRα1-M256T mutant showed that distortion of the hydrophobic niche to accommodate the outer ring of ligand was more pronounced for T3 than T4, suggesting that this substitution has little impact on the affinity for T4. In agreement with the model, TRα1-M256T selectively reduced the affinity for T3. Also, unlike other naturally occurring TRα mutations, TRα1-M256T had a differential impact on T3- vs T4-dependent transcriptional activation. TRα1-M256A and TRβ1-M310T mutants exhibited similar discordance for T3 vs T4.
Conclusions
Met256-TRα1/Met310-TRβ1 strongly potentiates the affinity of TRs for T3, thereby largely determining that T3 is the bioactive hormone rather than T4. These observations provide insight into the molecular basis for underlying the different affinity of TRs for T3 vs T4, delineating a fundamental principle of TH signaling.
Met256-TRα1/Met310-TRβ1 determine the differential bioactivity of T3 vs T4, providing the molecular basis for the clinical concept that T4 functions as prohormone and T3 as bioactive hormone.
Thyroid hormones (THs) are indispensable for normal growth, development, and metabolism. T3 and T4 are the two major forms of TH. In 1952, it was recognized that T3 has greater biological potency than T4 (1–4). This fundamental discovery led to the clinical concept that T4, despite being the most abundant circulating iodothyronine, functions as a prohormone, with T3 being the biologically active hormone. Since then, this paradigm has remained unchanged, although the molecular and structural mechanisms underlying this have not been investigated in detail.
The genomic actions of THs are exerted through binding to the three functional isoforms of TH receptors (TRs), namely TRα1, TRβ1, and TRβ2, which are highly homologous but have distinctive expression patterns (5–7). Mutations in TRα and TRβ give rise to clinically distinct syndromes in humans, termed resistance to TH (RTH)α and RTHβ, respectively (8–14). RTHβ patients commonly present with goiter and tachycardia with abnormal thyroid function tests, including high serum free T3 (FT3) and free T4 (FT4) concentrations with normal or slightly increased TSH concentrations. The clinical phenotype of RTHα is distinct from RTHβ and includes growth retardation, macrocephaly, constipation, intellectual disability, and anemia. In RTHα, thyroid function tests are typically characterized by high to high-normal FT3, low to low-normal FT4, low reverse T3, and normal TSH concentrations.
The greater biological activity of T3 vs T4 is explained by differences in affinity for the functional isoforms of TH receptors (TRs). The binding affinity of T4 to the TRs is 10- to 30-fold less compared with T3 (15–17). Previous crystallographic studies revealed that the ligand-binding pocket of TRβ1 is able to accommodate both T3 and T4, although the helix (H)11-H12 loop is more loosely packed in the presence of T4 than T3 (16). These structural adaptations of TRβ1, which are required to accommodate the larger T4 molecule, have been attributed to possible steric hindrance of its bulky 5′-iodine moiety with the surrounding amino acids, especially the Met residue located at position 310 in TRβ1. Although no cocrystallization studies of TRα with T4 are available, a similar role for Met256 in TRα (the equivalent position of Met310 in TRβ) has been suggested (18). However, no functional studies to support the relevance of these residues for the differences in affinity for T3 and T4 have been performed.
Therefore, we combined structural modeling and in vitro approaches to determine the differential role of these Met residues in T3 vs T4 binding by TRs and characterized a newly identified TRα1-M256T and previously published TRβ1-M310T mutations, which naturally occur in patients with RTH (19–21). We showed that these Met residues are of particular importance for the binding of T3 but not T4. This observation provides the underlying molecular and structural basis for the role of T4 as prohormone and T3 as bioactive hormone in a paradigm for TH physiology and daily clinical practice.
Materials and Methods
TRα-M256T identification
The TRα-M256T mutation in an RTHα patient was identified by exome sequencing and was confirmed by Sanger sequencing as previously described (12) after obtaining informed consent. This study was conducted following the Declaration of Helsinki principles and was approved by the Medical Ethical Committee of the Erasmus Medical Center, Rotterdam, Netherlands (MEC-2015-362).
In silico prediction of TRα1-M256T function
The TRα1-M256T mutation bound to T3 and T4 was modeled into the wild-type (WT) TRα1 crystal structure (PDB-ID: 2H77) (22), and the M256T and M256A mutations were introduced using the side-chain substitution tool of the YASARA Structure Software (YASARA Bioscience GmbH, Vienna, Austria) (23) and processed as previously described (24).
DNA constructs and mutagenesis
The pcDNA3 FLAG-TRα1 and TRβ1 expression vectors containing full-length human TRα1 and TRβ1 with 5′ FLAG tagged (11, 24) and the pCMX VP16-TRα1 expression vector containing full-length human TRα1 fused with VP16 (25) have been described previously. TRα1-M256T, TRβ1-M310T, as well as the other TRα1 mutations (M256A, A263S, D211G, and R384H) were introduced using the QuickChange II Mutagenesis kit (Agilent Technologies, Amstelveen, Netherlands) according to the manufacturer’s protocol. The introduced mutations were confirmed by Sanger sequencing.
Radioligand competitive binding assays
FLAG-TRα1 WT, M256T, and M256A receptor proteins were synthesized using the TnT® T7 Quick Coupled Transcription/Translation System (Promega, Leiden, Netherlands). The affinity for T3 and T4 of the receptors was determined by competitive binding assays as previously described (24) using [125I]T3 and [125I]T4, respectively. The dissociation constant (Kd) was analyzed by GraphPad Prism program version 5.0 (GraphPad, La Jolla, CA) and shown as the mean ± SEM of three independent experiments performed in duplicate.
Cell culture and transfection
JEG-3 cells (ECACC Cat. no. 92120308, RRID:CVCL_0363; Sigma-Aldrich, Munich, Germany) were cultured and transfected as previously described (24, 26). Given the absence of 5′-deiodinating activity in this cell type (27), there is no intracellular deiodination of T4 to T3, which allowed us to study the direct effect of T3 and T4 on transactivation. For transcriptional activity assays, WT or mutant receptors were coexpressed with luciferase reporter constructs containing direct repeat TH response elements (DR4-TRE) as well as pMaxGFP as a transfection control. We also coexpressed WT and TRα1-M256T in a 1:1 equimolar ratio to determine the effect of the mutant on WT function (i.e., the dominant-negative effect). For receptor–cofactor interaction (two-hybrid) assays, VP16-fused WT or TRα1-M256T were coexpressed with a luciferase reporter construct containing the Gal4 binding site (UAStkLuc), together with pSG424 expression vectors containing the Gal4DBD fused to the interacting domains of NCoR1 or SRC1 (11). After transfection for 24 hours, cells were stimulated with 0 to 10,000 nM T3 (Cat. no. T2877; Sigma-Aldrich) or T4 (Cat. no. T2376; Sigma-Aldrich) in DMEM/F12 medium supplemented with 0.1% bovine serum albumin for 24 hours.
Immunoblotting
The expression of FLAG-tagged and VP16-fused receptors in JEG-3 cells was verified by immunoblotting nuclear extracts as previously described (24, 26). FLAG-tagged TRα1 and VP16-TRα1 were detected with a 1:1000 dilution of FLAG-M2 (#F1804; Sigma-Aldrich) and VP16 (sc-7545; Santa Cruz Biotechnology, Heidelberg, Germany) antibodies. The Histone 3 protein was detected as loading control with a 1:1000 dilution of a Histone 3 antibody (1B1B2) (#14269 Cell Signaling Technology, Leiden, Netherlands).
Luciferase assays
Luciferase activity was measured as previously described (12, 24). Data were expressed as percentage maximal response of WT stimulated by T3. EC50, IC50, and maximal response were calculated using GraphPad Prism program version 5.0 (GraphPad, La Jolla, CA). The results are shown as the mean ± SEM of at least three independent experiments performed in triplicate.
Statistical analysis
Statistical differences of logKd, logIC50, and logEC50 values between groups were analyzed by Student t test or one-way ANOVA with Tukey post test. The percentage maximal response of mutants was compared with WT by one-sample t test. Statistical significance was considered at P < 0.05.
Results
Clinical characterization
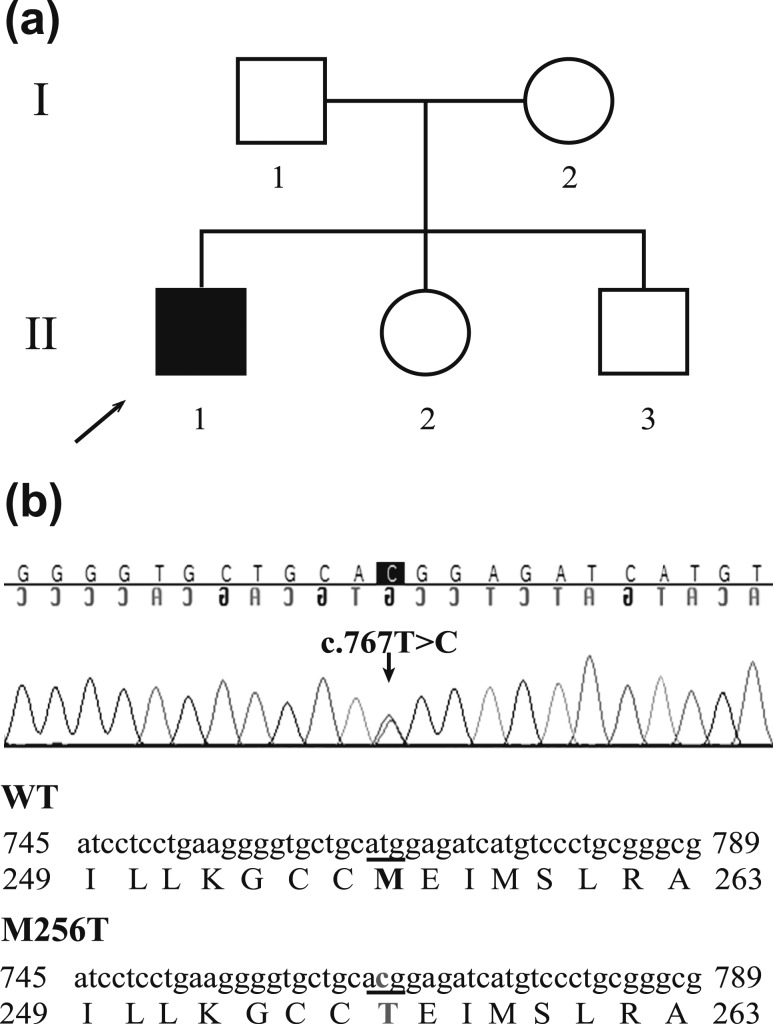
A de novo heterozygous missense mutation in the THRA gene (c.767T>C), resulting in substitution of Thr for Met at codon 256 (p.M256T), was identified in a 19-year-old male patient presenting with features similar to previously reported RTHα patients, including disproportionate ischial leg length (sitting height to height ratio +2.5 SD score), mild neurodevelopmental delay, coarse facies, macrocephaly (head circumference 60 cm, +2.5 SD score), and high serum T3/T4 ratio with normal TSH concentrations [FT4, 10.6 pmol/L (normal range, 11 to 25 pmol/L); total T4, 67 nmol/L (normal range, 58 to 128 nmol/L); total T3, 2.9 nmol/L (normal range, 1.4 to 2.5 nmol/L); reverse T3, 0.18 nmol/L (normal range, 0.22 to 0.54 nmol/L); T3/T4 ratio, 0.043 (normal range, 0.01 to 0.03); and TSH, 1.83 mU/L (normal range, 0.4 to 4.3 mU/L)] (Fig. 1). This mutation is not present in public databases (dbSNP, 1000Genome, and Exome Aggregation Consortium).
Figure 1.
(a) Pedigree chart demonstrating that only the index patient (II.1) has the clinical phenotype of RTHα. (b) Sequence analysis of exon 8 of the THRA gene shows a de novo heterozygous missense mutation (c.767T>C) in the index patient, resulting in a Met to Thr substitution at codon 256 (p.M256T).
Protein modeling
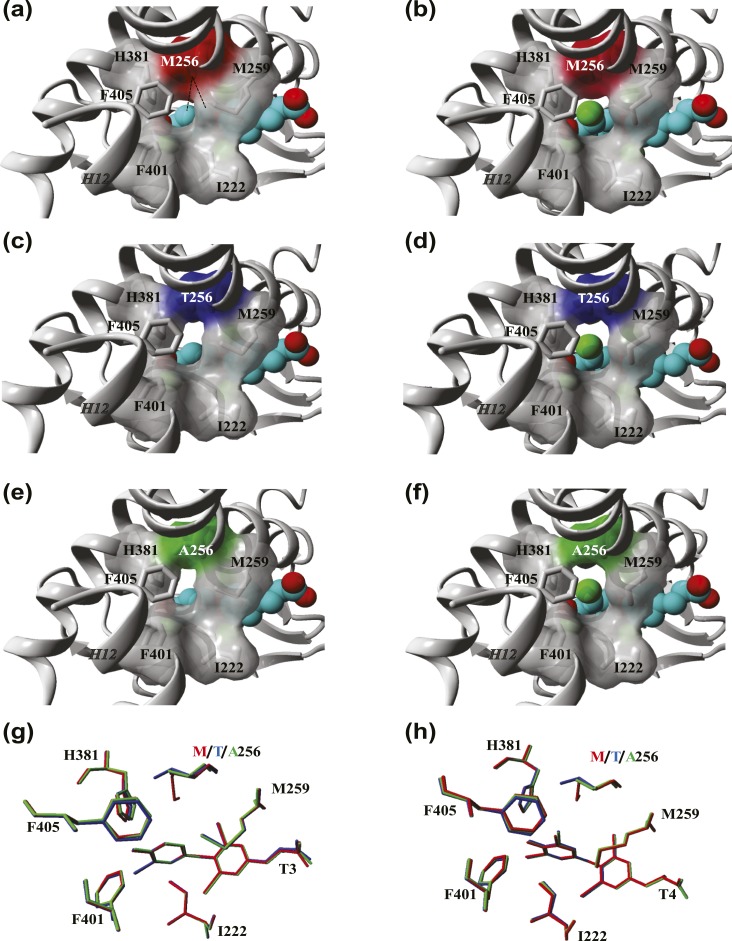
The role of the Met256 in TRα1 function and the potential effect of this mutation on the affinity of T3 and T4 were predicted by in silico modeling. Given the absence of a T4-bound TRα crystal structure, we studied the structural organization of the domains surrounding the outer ring of TH in the available T3- (PDB ID: 1xzx) and T4-liganded (PDB ID: 1y0x) crystal structures of TRβ1. In line with a previous report (16), we observed that the 5′ position of the outer ring of T3 and T4 is flanked by Ile276 (H3), Met310 and Met313 (H6), His435 (H11), and Phe455 and Phe459 (H12) of TRβ1. Together, these residues form a niche that allows the accommodation of T4 despite the presence of its bulky 5′-iodine. The same niche is present within the T3-liganded TRβ1 crystal but is considerably smaller in the absence of the 5′-iodine. Met310 (corresponding to Met256 of TRα1) is in the closest structural proximity to the 5′-carbon of the outer ring and forms an extensive network of (hydrophobic) interactions that link H6, H11, and H12.
We next modeled a T4 molecule into the ligand binding pocket of the available T3-liganded TRα1 crystal structure (PDB-ID: 2H77) [Fig. 2(b)]. Compared with the T3-liganded TRα1 structure [Fig. 2(a)], a slight outward shift of H11 and H12 was observed in the T4-liganded model, which was accompanied by reorientation of side-chains of residues surrounding the 5′-iodine. This resulted in a loss of the direct hydrophobic interactions between Met256 and the outer ring and a less tightly packed structural organization of the ligand binding pocket. These changes were similar to those observed in the corresponding TRβ1 crystal structures, validating the accuracy of the modeling procedure.
Figure 2.
Comparison of the architecture of the TRα1 ligand binding pocket in the presence of T3 and T4. (a and b) Close-up view of the ligand-binding pocket of the TRα1 crystal structure in complex with (a) T3 (PDB ID: 2h77) and (b) T4. The residue side-chains lining the niche that accommodates the outer ring of T3 and T4 are highlighted, and their molecular surface is shown except for Phe405 for clarity. The 5′-iodine group of T4 is represented by the green ball in the T4-bound TRα1 model. The hydrophobic contacts between Met256 and the phenolic outer ring are depicted as dashed lines. (c and d) Structural models of the TRα1-M256T mutant in complex with (c) T3 and (d) T4. (e and f) Structural models of the TRα1-M256A mutant in complex with (e) T3 and (f) T4. (g and h) Overlay of the structural orientation of the residue side-chains that face the (g) T3 and (h) T4 ligands at the 5′ position in WT (gray), M256T (blue), and M256A (red) mutant TRα1 models. All figures were created in YASARA Structure using PovRay imaging software.
We subsequently modeled the M256T (shortening of side-chain, hydrophilic moiety) mutant in both T3- and T4-bound TRα1 structures and analyzed the impact on the conformation of the ligand binding domain and direct substrate interactions [Fig. 2(c) and 2(d)]. The artificial M256A mutant was also modeled to reduce the side-chain length while maintaining the hydrophobic property of the residue [Fig. 2(e) and 2(f)]. Due to shortening of side-chain length in both mutants, direct hydrophobic interaction with the outer ring of T3 was lost [Fig. 2(c) and 2(e)]. Moreover, both mutants enlarged the niche surrounding the 5′ position of T3 due to reorientation of various residue side-chains in H11 and H12 and the subsequent outward shift of these helices. As a result, the niche adopts a structural configuration that resembles the WT receptor in the T4-bound state. These changes were more pronounced for the M256T than the M256A, exemplified by the degree of reorientation of His381, which was previously implicated to interact with the phenolhydroxyl group of T3 (18) [Fig. 2(g)]. In the case of T4, both mutations had little effect on structural organization [Fig. 2(d), 2(f), and 2(h)]. Based on these in silico predictions, we hypothesized that both substitutions would have a greater impact on T3 than on T4 binding and action.
Functional studies
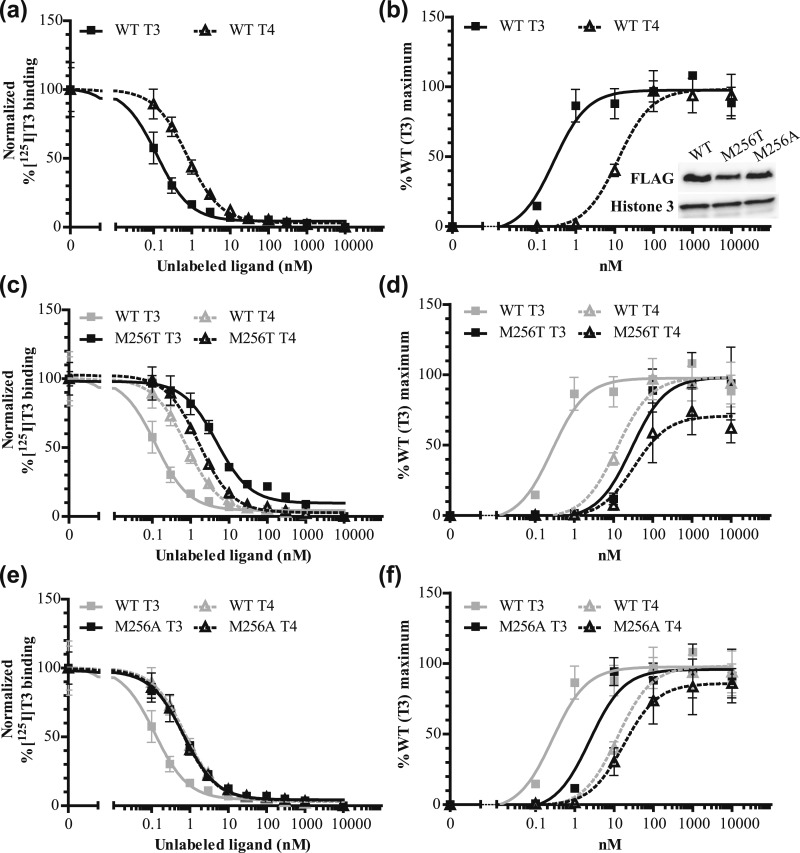
We performed in vitro studies to test this hypothesis. In line with previous literature (15–17), competitive binding assays showed that the affinity for T4 of WT TRα1 was approximately sevenfold lower than for T3, indicated by a higher Kd of T4 than T3 [Fig. 3(a); Table 1]. The TRα1-M256T mutant showed a ∼40-fold lower T3 binding affinity than WT, whereas T4 affinity was unchanged [Fig. 3(c); Table 1]. Also, the binding affinity of the TRα1-M256A mutant for T3 was selectively reduced (approximately sixfold) [Fig. 3(e); Table 1].
Figure 3.
(a, c, e) [125I]T3 dissociation curves showing that compared with (a) WT, (c) the TRα1-M256T mutation, and (e) TRα1-M256A mutation reduces the affinity for T3 (solid line) more than for T4 (dashed line) (mean ± SEM of three experiments for WT and M256T and two experiments for M256A performed in duplicate). (b, d, f) The TRα1-M256T and TRα1-M256A mutations also had a larger effect on T3- than on T4-dependent transcriptional activation (mean ±SEM of three experiments performed in triplicate). The effect of the Ala substitution on the ligand binding affinity and the transcriptional activity of TRα1 was less than the effect of the Thr substitution. The insert in (b) shows immunoblots confirming an equal expression of WT, M256T, and M256A FLAG-tagged TRα1 and Histone 3 as a loading control in the nuclear fraction of JEG-3 cells.
Table 1.
Summary of the Results of Competitive Binding, Transcriptional Activity, and Protein-Protein Interaction Assays of WT, TRα1-M256T, and TRα1-M256A Mutants
| T3 Stimulation | T4 Stimulation | |||||
|---|---|---|---|---|---|---|
| WT | M256T | M256A | WT | M256T | M256A | |
| LogKd | −0.91 ± 0.08 | 0.71 ± 0.10a | −0.16 ± 0.34b,c | −0.09 ± 0.10 | 0.22 ± 0.05 | −0.18 ± 0.02 |
| Kd(nM) | 0.12 | 5.14 | 0.69 | 0.81 | 1.67 | 0.66 |
| LogEC50-DR4 | −0.60 ± 0.10 | 1.51 ± 0.16a | 0.51 ± 0.08b,d | 1.16 ± 0.07 | 1.67 ± 0.11 | 1.44 ± 0.25 |
| EC50(nM) | 0.25 | 32.3 | 3.26 | 14.5 | 46.6 | 27.2 |
| LogIC50-NCoR1 | −1.26 ± 0.04 | 0.69 ± 0.18a | — | 0.02 ± 0.06 | 0.82 ± 0.14b | — |
| IC50(nM) | 0.06 | 4.87 | — | 1.05 | 6.64 | — |
| LogEC50-SRC1 | −0.76 ± 0.05 | 1.19 ± 0.07a | — | 0.42 ± 0.07 | 1.16 ± 0.08b | — |
| EC50(nM) | 0.17 | 15.5 | — | 2.65 | 14.6 | — |
Data are presented as mean ± SEM (one-way ANOVA with Tukey posttest).
P < 0.001 for WT vs mutant.
P < 0.01.
P < 0.001 for M256T vs M256A.
P < 0.01.
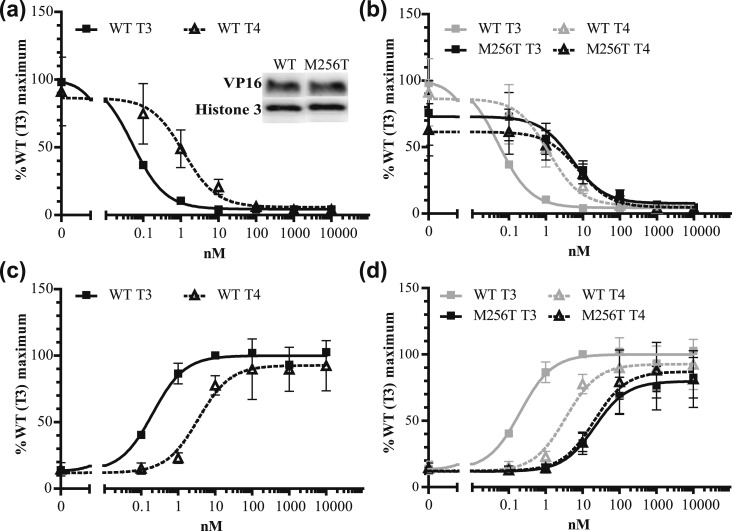
To evaluate the impact of both mutations on the transcriptional activity, WT and mutant receptors were cotransfected with a reporter construct in which luciferase expression is under control of a TH response element into JEG-3 cells with increasing concentrations of T3 or T4. Equal expression of WT and both mutants was confirmed by immunoblotting nuclear extracts with anti-FLAG antibodies [Fig. 3(b)]. In line with the binding assays and previous studies (16, 17), the transcriptional activation assay showed that the EC50 of WT TRα1 induced by T4 was ∼60-fold higher than that induced by T3 [Fig. 3(b); Table 1]. The EC50 of TRα1-M256T was 100-fold higher for T3 but was unchanged for T4 compared with WT [Fig. 3(d); Table 1]. The TRα1-M256A also selectively reduced transcriptional activity induced by T3 [Fig. 3(f); Table 1]. The transcriptional activity was also reduced when WT and TRα1-M256T were coexpressed compared with WT expressed alone, suggesting a dominant-negative effect of this mutant (data not shown). In mammalian two-hybrid assays compared with WT, the TRα1-M256T mutant also affected ligand-dependent interactions with the corepressor NCoR1 (fold increase IC50: ∼80-fold for T3 and approximately sixfold for T4) and the coactivator SRC1 (fold increase EC50: ∼90-fold for T3 and approximately sixfold for T4) [Fig. 4(a)–4(d); Table 1]. Together, our results indicate that the mutations located at the Met256 of TRα1 have a differential impact on the binding and activation of the receptor by T4 vs T3.
Figure 4.
The TRα1-M256T mutation had a larger effect on T3- than on T4-dependent (a and b) GAL4-NCoR1 dissociation and (c and d) GAL4-SRC1 association (mean ± SEM of at least three experiments performed in triplicate). The insert in (a) shows immunoblots confirming an equal expression of WT and M256T VP16 TRα1 fusion proteins and Histone 3 as loading control in the nuclear fraction of JEG-3 cells.
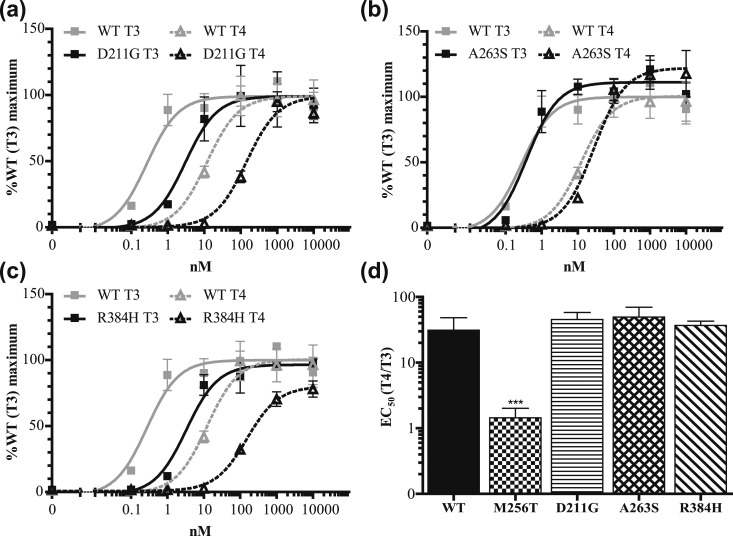
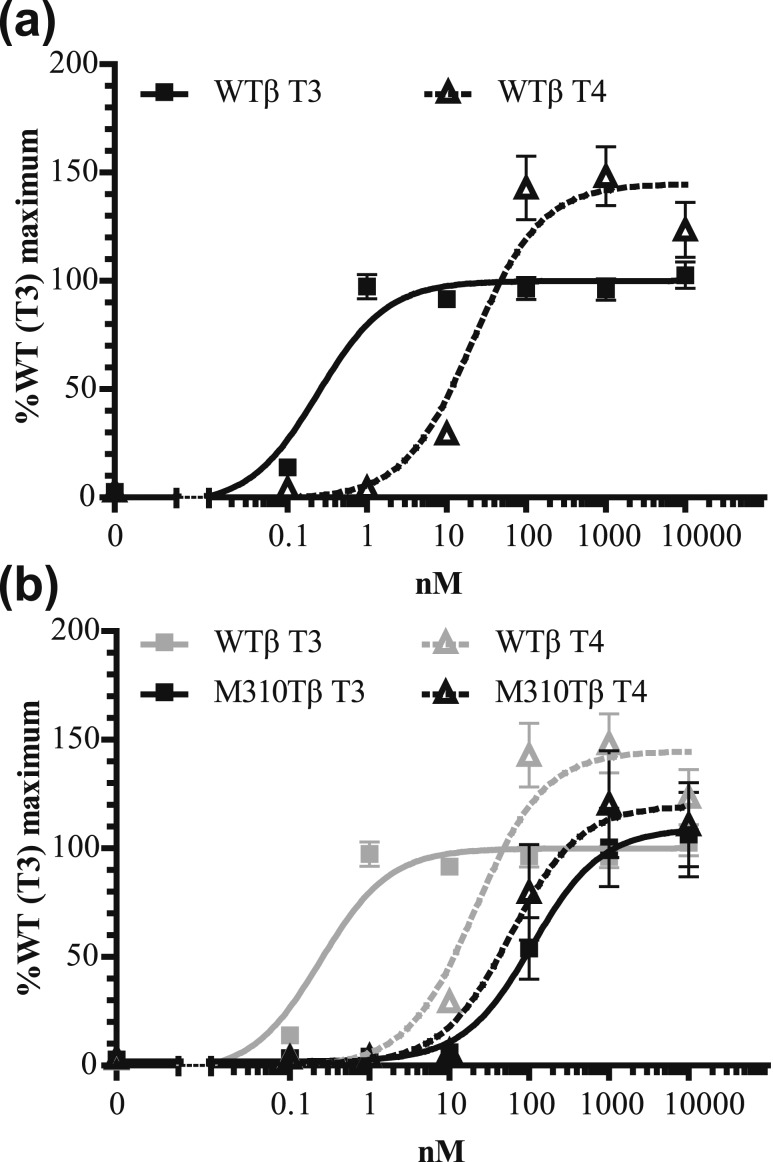
We next investigated if this T3 vs T4 difference is present in other TRα mutants located outside the niche surrounding the 5′-iodine position. However, these naturally occurring mutations (D211G, A263S, and R384H) had a similar impact on T3- and T4-induced transactivation, and, as for WT TRα, the EC50 values for T4 exceeded those for T3 by ∼30- to 50-fold [Fig. 5(a)–5(c)]. These transcriptional activation profiles were in contrast to the M256T mutant [Fig. 5(d)], strongly indicating that only this mutant has a predominant impact on T3 affinity. To extend our findings to TRβ, we studied the transcriptional activity of a corresponding mutation in TRβ1 (TRβ1-M310T). The EC50 of WT TRβ1 induced by T4 was ∼70-fold higher than that induced by T3 [Fig. 6(a)], which was similar to WT TRα1. The T3-induced transcriptional response of TRβ-M310T was greatly reduced, which contrasted with the T4-induced transcriptional activity (fold increase EC50: ∼350-fold for T3 and approximately threefold for T4) [Fig. 6(b)].
Figure 5.
(a–c) The T4-induced transcriptional activity of three TRα1 mutations identified in RTHα patients is lower than that is induced by T3, which is similar to WT [Fig. 2(d)] (mean ± SEM of three experiments performed in triplicate). (d) The EC50 of T4 is ∼30- to 50-fold higher than the EC50 of T3, except for TRα1-M256T. ***P < 0.001 (one-way ANOVA with Tukey post test).
Figure 6.
The T3- and T4-induced transcriptional activity of (a) WT and (b) TRβ1-M310T in JEG-3 cells shows that the TRβ1-M310T mutation affects T3- more than T4-dependent transcriptional activation (mean ± SEM of four experiments performed in triplicate), which is in line with the results of TRα1-M256T [Fig. 3(d)].
Discussion
Although the notion of T4 and T3 being the precursor and active hormones, respectively, is widely recognized in the clinical and scientific communities, the molecular and structural basis of this dogma has received little attention. In this study, we highlight the crucial role of residue Met256 of TRα1 and Met310 of TRβ1 in determining the differential bioactivity of T3 vs T4 using a novel mutant (TRα1-M256T) identified in an RTHα patient and a mutant at the corresponding position (TRβ1-M310T) identified in RTHβ patients (19–21). In contrast to WT TRα or TRβ and mutations involving other residues, mutations at these Met residues selectively affected binding and transactivation of TR by T3. These observations emphasize the key role of these residues in designating T4 as the prohormone and T3 as the major bioactive hormone.
In line with previous reports (15–17), our results showed that T3 has a higher binding affinity for WT TRα1 and stimulates receptor activity with a higher potency than T4. Previous structural studies in TRβ1 have suggested that the lower affinity for T4 is caused by decreased packing of the ligand binding domain in the presence of T4 vs T3, which particularly allows oscillation of H12 between liganded and unliganded states, resulting in a higher ligand dissociation rate (16). Here, we extend these observations by showing that the ligand binding domain of T3-liganded TR has a similar decrease in packing as observed in T4-liganded WT receptors upon substitution of Met256 in TRα1 or Met310 in TRβ1 by Thr. In contrast, these substitutions hardly changed the predicted structure of T4-liganded mutant receptors. Based on these models, we postulated that the extensive (hydrophobic) interactions of Met with surrounding residues are key in stabilizing interhelical interactions (e.g., between H6, H11, and H12), which facilitate the tight packing of the ligand binding domain as observed in T3-liganded receptors. Moreover, we observed a direct interaction between Met and the 5′ position of the outer ring of T3, which was not formed with T4. This suggests that Met256 in TRα1 and Met310 in TRβ1 have a critical role in achieving optimal folding and enthalpy in T3-liganded receptors, whereas their role in T4 binding is of less importance.
This in silico prediction was confirmed by in vitro studies indicating that TRα1-M256T selectively affected binding affinity for T3 as well as cofactor interactions and transcriptional activity of the T3-stimulated receptor. These properties seemed specific for the M256T mutant because the transactivation potency of T3 and T4 with TRα mutants identified in other RTHα patients [D211G (26), A263S, and R384H (28)] was affected equally. Additional testing of the naturally occurring mutation at the corresponding residue in the TRβ1 (M310T) (19–21) further substantiated the specificity of the findings.
Thr substitution at position 256 in TRα1 or 310 in TRβ1 not only alters the binding space but also affects the hydrophobicity of the ligand-binding pocket. Therefore, we tested the artificial TRα1-M256A mutant, which reduces the size of the side-chain but maintains the hydrophobic property of the ligand-binding pocket. Indeed, functional studies showed that TRα1-M256A also selectively impairs T3 binding affinity and T3-induced transcriptional activity, whereas T4 binding and activity are maintained. Although the effect of the TRα1-M256T mutation in our functional and structural models was slightly greater than that of TRα1-M256A, these findings support the notion that loss of the specific properties of Met, rather than the unfavorable impact of the hydrophilic moiety of Thr on the hydrophobic environment, are mainly responsible for the differential impact on T3 vs T4 signaling. Based on our studies and on a previous report (16), we propose that Met256 in TRα1 and Met310 in TRβ1 are crucial residues that determine specific affinity for T3 vs T4. Thr and Ala substitution at these Met positions significantly affected the hydrophobic interactions with T3 and altered the niche accommodating the outer ring of T3 to a “T4-bound” configuration, both resulting in a reduced binding affinity of the mutants for T3. In contrast, because the ligand binding domain of T4-liganded receptors already exhibit looser packing without direct interaction(s) between Met and the T4 molecule, mutations in the Met residue are better tolerated.
No unique phenotype was discernible in the newly identified M256T RTHα patient when compared with other cases of RTHα harboring missense mutations in the THRA gene (25, 26, 28–30) or in patients carrying TRβ-M310T (19–21) when compared with other RTHβ cases reported in the literature. These findings indicate that, although mutations at Met256-TRα1/Met310-TRβ1 residues preserve T4 binding to mutant receptor proteins, this property is not sufficient to prevent patients from developing features of RTH, implying that the phenotype of RTH is linked primarily to defective T3 rather than T4 binding by mutant TRs.
This study provides in vitro evidence for the importance of Met256 in TRα1 and Met310 in TRβ1 in ligand recognition. Our studies highlight the relevance of this Met residue in TRs for discrimination between T3 and T4, providing the molecular basis for the role of T4 as prohormone and T3 as bioactive hormone.
Acknowledgments
In the early stage of writing this manuscript, Professor Theo J. Visser suddenly and unexpectedly died. We highly value his contributions to the field, and we miss a great scientist, mentor, and friend. While deceased contributors are rightfully recognized and acknowledged, they cannot be added posthumously to an article’s byline.
Financial Support: This work is supported by Zon-MWTOP Grant 91212044, by an Erasmus MC Medical Research Advisory Committee grant (to R.P.P., M.E.M., and W.E.V.), by a grant from Chiang Mai University (to K.W.), and by Wellcome Trust Grant 210755/Z/18/Z and an NIHR Cambridge Biomedical Centre grant (to V.K.C.).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- FT3
free T3
- FT4
free T4
- RTH
resistance to thyroid hormone
- TH
thyroid hormone
- TR
thyroid hormone receptor
- WT
wild type
References
- 1. Gross J, Pitt-Rivers R. Physiological activity of 3:5:3′-L-triiodothyronine. Lancet. 1952;1(6708):593–594. [DOI] [PubMed] [Google Scholar]
- 2. Pitt-Rivers R. Metabolic effects of compounds structurally related to thyroxine in vivo: thyroxine derivatives. J Clin Endocrinol Metab. 1954;14(11):1444–1450. [DOI] [PubMed] [Google Scholar]
- 3. Mussett MV, Pitt-Rivers R. The thyroid-like activity of triiodothyronine analogues. Lancet. 1954;267(6850):1212–1213. [DOI] [PubMed] [Google Scholar]
- 4. Lerman J. The contribution of triiodothyronine to thyroid physiology. J Clin Endocrinol Metab. 1954;14(6):690–693. [DOI] [PubMed] [Google Scholar]
- 5. Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31(2):139–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh BK, Yen PM. A clinician’s guide to understanding resistance to thyroid hormone due to receptor mutations in the TRα and TRβ isoforms. Clin Diabetes Endocrinol. 2017;3(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lazar MA. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14(2):184–193. [DOI] [PubMed] [Google Scholar]
- 8. Refetoff S, DeWind LT, DeGroot LJ. Familial syndrome combining deaf-mutism, stuppled epiphyses, goiter and abnormally high PBI: possible target organ refractoriness to thyroid hormone. J Clin Endocrinol Metab. 1967;27(2):279–294. [DOI] [PubMed] [Google Scholar]
- 9. Sakurai A, Takeda K, Ain K, Ceccarelli P, Nakai A, Seino S, Bell GI, Refetoff S, DeGroot LJ. Generalized resistance to thyroid hormone associated with a mutation in the ligand-binding domain of the human thyroid hormone receptor beta. Proc Natl Acad Sci USA. 1989;86(22):8977–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dumitrescu AM, Refetoff S. The syndromes of reduced sensitivity to thyroid hormone. Biochim Biophys Acta. 2013;1830(7):3987–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bochukova E, Schoenmakers N, Agostini M, Schoenmakers E, Rajanayagam O, Keogh JM, Henning E, Reinemund J, Gevers E, Sarri M, Downes K, Offiah A, Albanese A, Halsall D, Schwabe JW, Bain M, Lindley K, Muntoni F, Vargha-Khadem F, Dattani M, Farooqi IS, Gurnell M, Chatterjee K. A mutation in the thyroid hormone receptor alpha gene [published correction appears in N Engl J Med 2012;367(15):1474]. N Engl J Med. 2012;366(3):243–249. [DOI] [PubMed] [Google Scholar]
- 12. van Mullem A, van Heerebeek R, Chrysis D, Visser E, Medici M, Andrikoula M, Tsatsoulis A, Peeters R, Visser TJ. Clinical phenotype and mutant TRα1. N Engl J Med. 2012;366(15):1451–1453. [DOI] [PubMed] [Google Scholar]
- 13. van Gucht ALM, Moran C, Meima ME, Visser WE, Chatterjee K, Visser TJ, Peeters RP. Resistance to thyroid hormone due to heterozygous mutations in thyroid hormone receptor alpha. Curr Top Dev Biol. 2017;125:337–355. [DOI] [PubMed] [Google Scholar]
- 14. Moran C, Chatterjee K. Resistance to thyroid hormone α-emerging definition of a disorder of thyroid hormone action. J Clin Endocrinol Metab. 2016;101(7):2636–2639. [DOI] [PubMed] [Google Scholar]
- 15. Apriletti JW, Eberhardt NL, Latham KR, Baxter JD. Affinity chromatography of thyroid hormone receptors: biospecific elution from support matrices, characterization of the partially purified receptor. J Biol Chem. 1981;256(23):12094–12101. [PubMed] [Google Scholar]
- 16. Sandler B, Webb P, Apriletti JW, Huber BR, Togashi M, Cunha Lima ST, Juric S, Nilsson S, Wagner R, Fletterick RJ, Baxter JD. Thyroxine-thyroid hormone receptor interactions. J Biol Chem. 2004;279(53):55801–55808. [DOI] [PubMed] [Google Scholar]
- 17. Schroeder A, Jimenez R, Young B, Privalsky ML. The ability of thyroid hormone receptors to sense t4 as an agonist depends on receptor isoform and on cellular cofactors. Mol Endocrinol. 2014;28(5):745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378(6558):690–697. [DOI] [PubMed] [Google Scholar]
- 19. Kim HK, Kim D, Yoo EH, Lee JI, Jang HW, Tan AH, Hur KY, Kim JH, Kim KW, Chung JH, Kim SW. A case of resistance to thyroid hormone with thyroid cancer. J Korean Med Sci. 2010;25(9):1368–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitchell CS, Savage DB, Dufour S, Schoenmakers N, Murgatroyd P, Befroy D, Halsall D, Northcott S, Raymond-Barker P, Curran S, Henning E, Keogh J, Owen P, Lazarus J, Rothman DL, Farooqi IS, Shulman GI, Chatterjee K, Petersen KF. Resistance to thyroid hormone is associated with raised energy expenditure, muscle mitochondrial uncoupling, and hyperphagia. J Clin Invest. 2010;120(4):1345–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takeda K, Weiss RE, Refetoff S. Rapid localization of mutations in the thyroid hormone receptor-beta gene by denaturing gradient gel electrophoresis in 18 families with thyroid hormone resistance. J Clin Endocrinol Metab. 1992;74(4):712–719. [DOI] [PubMed] [Google Scholar]
- 22. Nascimento AS, Dias SM, Nunes FM, Aparício R, Ambrosio AL, Bleicher L, Figueira AC, Santos MA, de Oliveira Neto M, Fischer H, Togashi M, Craievich AF, Garratt RC, Baxter JD, Webb P, Polikarpov I. Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function. J Mol Biol. 2006;360(3):586–598. [DOI] [PubMed] [Google Scholar]
- 23. Krieger E, Vriend G. YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics. 2014;30(20):2981–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wejaphikul K, Groeneweg S, Dejkhamron P, Unachak K, Visser WE, Chatterjee VK, Visser TJ, Meima ME, Peeters RP. Role of leucine 341 in thyroid hormone receptor beta revealed by a novel mutation causing thyroid hormone resistance. Thyroid. 2018;28(12):1723–1726. [DOI] [PubMed] [Google Scholar]
- 25. Moran C, Agostini M, McGowan A, Schoenmakers E, Fairall L, Lyons G, Rajanayagam O, Watson L, Offiah A, Barton J, Price S, Schwabe J, Chatterjee K. Contrasting phenotypes in resistance to thyroid hormone alpha correlate with divergent properties of thyroid hormone receptor α1 mutant proteins. Thyroid. 2017;27(7):973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Gucht AL, Meima ME, Zwaveling-Soonawala N, Visser WE, Fliers E, Wennink JM, Henny C, Visser TJ, Peeters RP, van Trotsenburg AS. Resistance to thyroid hormone alpha in an 18-month-old girl: clinical, therapeutic, and molecular characteristics. Thyroid. 2016;26(3):338–346. [DOI] [PubMed] [Google Scholar]
- 27. Kester MH, Kuiper GG, Versteeg R, Visser TJ. Regulation of type III iodothyronine deiodinase expression in human cell lines. Endocrinology. 2006;147(12):5845–5854. [DOI] [PubMed] [Google Scholar]
- 28. Demir K, van Gucht AL, Büyükinan M, Çatlı G, Ayhan Y, Baş VN, Dündar B, Özkan B, Meima ME, Visser WE, Peeters RP, Visser TJ. Diverse genotypes and phenotypes of three novel thyroid hormone receptor-α mutations. J Clin Endocrinol Metab. 2016;101(8):2945–2954. [DOI] [PubMed] [Google Scholar]
- 29. Tylki-Szymańska A, Acuna-Hidalgo R, Krajewska-Walasek M, Lecka-Ambroziak A, Steehouwer M, Gilissen C, Brunner HG, Jurecka A, Różdżyńska-Świątkowska A, Hoischen A, Chrzanowska KH. Thyroid hormone resistance syndrome due to mutations in the thyroid hormone receptor α gene (THRA). J Med Genet. 2015;52(5):312–316. [DOI] [PubMed] [Google Scholar]
- 30. Espiard S, Savagner F, Flamant F, Vlaeminck-Guillem V, Guyot R, Munier M, d’Herbomez M, Bourguet W, Pinto G, Rose C, Rodien P, Wémeau JL. A novel mutation in THRA gene associated with an atypical phenotype of resistance to thyroid hormone. J Clin Endocrinol Metab. 2015;100(8):2841–2848. [DOI] [PubMed] [Google Scholar]