Abstract
Camellia oleifera is one of the four largest woody edible oil plants in the world with high ecological and medicinal values. Due to frequent interspecific hybridization, it was difficult to study its genetics and evolutionary history. This study used C. oleifera that was collected on Hainan Island to conduct our research. The unique island environment makes the quality of tea oil higher than that of other species grown in the mainland. Moreover, a long-term geographic isolation might affect gene structure. In order to better understand the molecular biology of this species, protect excellent germplasm resources, and promote the population genetics and phylogenetic studies of Camellia plants, high-throughput sequencing technology was used to obtain the chloroplast genome sequence of Hainan C. oleifera. The results showed that the whole chloroplast genome of C. oleifera in Hainan was 156,995 bp in length, with a typical quadripartite structure of a large single copy (LSC) region of 86,648 bp, a small single copy (SSC) region of 18,297 bp, and a pair of inverted repeats (IRs) of 26,025 bp. The whole genome encoded a total of 141 genes (115 different genes), including 88 protein-coding genes, 45 tRNA genes, and eight rRNA genes. Among these genes, nine genes contained one intron, two genes contained two introns, and four overlapping genes were also detected. The total GC content of Hainan C. oleifera’s chloroplast genome was 37.29%. The chloroplast genome structure characteristics of Hainan C. oleifera were compared with mainland C. oleifera and those of the other eight closely related Theaceae species; it was found that the contractions and expansions of the IR/LSC and IR/SSC regions affected the length of chloroplast genome. The chloroplast genome sequences of these Theaceae species were highly similar. A comparative analysis indicated that the Theaceae species were conserved in structure and evolution. A total of 51 simple sequence repeat (SSR) loci were detected in the chloroplast genome of Hainan C. oleifera, and all Camellia plants did not have pentanucleotide repeats, which could be used as a good marker in phylogenetic studies. We also detected seven long repeats, the base composition of all repeats was biased toward A/T, which was consistent with the codon bias. It was found that Hainan C. oleifera had a similar evolutionary relationship with C. crapnelliana, through the use of codons and phylogenetic analysis. This study can provide an effective genomic resource for the evolutionary history of Theaceae family.
Keywords: Camellia oleifera, Island plant, Chloroplast genome, Repeat analysis, Codon usage, SSR, Evolution pressure
Introduction
The chloroplast genome, also known as chloroplast DNA, is often abbreviated as cpDNA. It shows the typical quadripartite structure generally consisting of four parts with a large single copy (LSC), a small single copy (SSC), and two inverted repeats (IRs) (Jansen et al., 2005; Palmer, 1991). The two IR regions are in the same sequence, but in the opposite directions. In general, 400–1,600 chloroplast genome copies are contained in plant cells (leaves) (Yang et al., 2010). The variation of chloroplast genome size is mainly affected by the length of the IR regions. For example, in the evolutionary process of the Geranium chloroplast genome, IR regions increased by 76 kb (Palmer, 1985). The IR regions of gymnosperms, such as Japanese black pine, which was only 495 bp in length (Tsudzuki et al., 1992), whereas the IR regions of legumes, such as Medicago, disappeared completely (Saski et al., 2005). This polymorphism of chloroplast genome has important research significance in phylogenetic and population genetics.
The chloroplast genome sequence has been successfully applied to the taxonomic and phylogenetic studies of many species. And the molecular evolution rate of the coding and non-coding regions are significantly different (Olmstead & Palmer, 1994). However, cpDNA is highly conserved (Wolfe, Li & Sharp, 1987), shows maternal inheritance (Corriveau & Coleman, 1988), and does not undergo genetic recombination. The control of chloroplast gene expression is affected by environmental factors and its developmental program, operating at several steps, including transcription, post-transcription, translation, and post-translation. The chloroplast gene expression is largely controlled at the post-transcriptional level (Sugita & Sugiura, 1996). Therefore, the plant chloroplast genome has significant advantages in revealing the relationship of species, and can provide a large amount of important data on chloroplast genetic transformation.
Camellia oleifera belongs to the genus Camellia of the family Theaceae. It is mainly distributed in the subtropical mountains of China (Zhou et al., 2013; Zhuang, 2008). A forest of C. oleifera has a good ecological value in maintaining soil and water and in regulating climate. In addition, its tea oil has a high medicinal value (He et al., 2007). In this study, we chose the Camellia species with a unique living environment (on Hainan Island), which is different from the other Camellia distribution provinces in China. The unique island climatic conditions have produced excellent and unique C. oleifera resources (Zheng et al., 2016). However, so far, the background in the genetics and genomics research of Camellia is rather weak. Due to frequent interspecific hybridization and polyploidization, the classification and phylogenetic studies of C. oleifera are quite difficult (Huang et al., 2014). Moreover, the C. oleifera on Hainan Island is an independent population, has a certain geographic isolation, with a relatively large genetic variation of its population. In the long run, this will gradually lead to the loss of the resources of germ-plasm. Therefore, this study will analyze the chloroplast genomic sequence of the Hainan C. oleifera to understand its similarities and differences with the other Camellia species in the process of natural selection, and identify its taxonomic groups, thereby providing valuable genomic resources for molecular breeding and phylogenetic research.
Materials and Methods
DNA extraction, sequencing, and annotation
Fresh leaves of C. oleifera were obtained by germinating the seeds that were collected from Chengmai, Hainan Province (110.00°E, 19.75°N) in China and the seeds were collected in areas that were not privately owned or protected in any way and no specific permits were required for this study. The treatment of plants and DNA extraction refer to the method of Zhang et al. (2017) and more experiment details could be got from our announcement.
The genome can be assembled directly by using overlap between sequenced reads; a total of 2.83G raw data was obtained, including 2.09G of clean data. The quality of the sequencing data of the samples was visually evaluated by the software Fastqc v 0.10.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). After the Illumina PCR adapter reads and low-quality reads were filtered from the paired-end and mate-pair library in the quality control step, all good-quality paired reads were assembled to contigs by using SOAPdenovo2 (Luo et al., 2012). The assembled contigs were joined into multiple scaffolds using SSPACE (Boetzer et al., 2011) to obtain the whole-genome sequence.
The programs DOGMA (Wyman, Jansen & Boore, 2004) and cpGAVAS (Liu et al., 2012) were used for the functional annotation and gene prediction of the whole chloroplast genome, respectively. Subsequently, the start and stop codons were adjusted manually by means of the Sequin tool to get the final open reading frame (ORF). The GeneMarkS (version 4.17) (http://topaz.gatech.edu/) program was used to retrieve the related coding genes. The predicted protein sequences were aligned using the National Center for Biotechnology Information (NCBI) Conserved Domain Database CDD (Marchlerbauer et al., 2015) to predict the Clusters of Orthologous Group of Proteins function of the genes and make the classification statistics (Yao et al., 2015). The circular chloroplast genome map of Hainan C. oleifera was generated with the OGDRAW (Lohse, Drechsel & Bock, 2007) software. The complete chloroplast genome sequence of Hainan C. oleifera has been submitted to the NCBI with GenBank accession number MF541730.
Comparative analysis of chloroplast genomes
The number of protein-coding genes, the GC content and the length of each region in the Hainan C. oleifera chloroplast genome and other nine Theaceae plants were compered by manually. A multiple sequence alignment of the 10 Theaceae chloroplast genomes was performed using mVISTA (Frazer et al., 2004; Mayor et al., 2000), with the annotated Hainan C. oleifera chloroplast genome sequence as a reference. The following nine species (with their GenBank accession numbers given in parentheses) are members of the genus Camellia: C. oleifera (JQ975031), C. luteoflora (KY626042), C. grandibracteata (NC_024659), C. sinensis (KC143082), C. leptophylla (NC_024660), C. pubicosta (NC_024662), C. crapnelliana (KF753632), C. huana (KY626040), and C. danzaiensis (NC_022460), which were downloaded from the GenBank database of NCBI (https://www.ncbi.nlm.nih.gov/pubmed). The visual comparisons of the IR/LSC and IR/SSC border regions of chloroplast genome from nine different species closely related to Hainan C. oleifera were conducted using the drawing software Visio 2013.
DNA barcode compared of Hainan C. oleifera and Mainland species
We selected the important DNA barcode sequences matK and trnH-psbA to compare Hainan C. oleifera and Mainland C. oleifera. The following objects: C. oleifera-matK (KP094074, KP094075, KR530501, KX216462) and C. oleifera-trnH-psbA (KX121760, KR533766, KP095297, GQ435325, GQ435326, GQ487355, KP095298) were involved. All registration numbers can be found in NCBI. The nucleotide sequence information of some C. oleifera was searched by NCBI database blast, and compared with the same nucleotide position of Hainan C. oleifera by the software MEGA 7.0.
SSRs characterization and long repeat sequences
The simple sequence repeats (SSRs) are also known as the short tandem repeats or microsatellites. Because of the presence of different nucleotides in the repeat units and different number of repeats, a high degree of variability in SSR length was caused. The microsatellite (di-, tri-, tetra-, penta-, and hexa-nucleotide repeats) assays were performed by using the software SSRHunter 1.3 (Li & Wan, 2005) with a minimum number of four repeat units for di- and tri-nucleotides and three for tetra-, penta-, and hexa-nucleotides.
The REPuter (Kurtz et al., 2001) software was used to predict the chloroplast genome long repeat sequences, which were divided into four categories, forward match, reverse match, palindromic match and complementary match, according to their comparison. In this paper, we examined the long repeat sequences with a minimum repeat length of 30 bp and a maximum of 60 bp.
Indices of codon usage
The relative synonymous codon usage (RSCU) (Sharp & Li, 1987) of Hainan C. oleifera chloroplast genome was calculated using DAMBE6 (Xia, 2017) based on the sequence of protein-coding genes in the chloroplast genome. The obtained RSCU values were statistically analyzed by SigmaPlot 10.0 and plotted into a histogram. The distribution of codon usage for the 10 Theaceae species is shown in the form of a heatmap, which was constructed using HemI 1.0 (Deng et al., 2014), according to the RSCU value. As an intuitive representation, it can graphically display the matrix data by describing each value in different color.
Sequence phylogenetic analysis
Evolutionary trees that indicate the evolutionary relationships among species which are thought to have a common ancestor are referred to as phylogenetic trees. The maximum likelihood method was used to determine the phylogenetic relationships of the Hainan C. oleifera clade with the clades of 21 other Theaceae species using MEGA 7.0 (Kumar, Stecher & Tamura, 2016) and an online software Interactive Tree of Life (iTOL) (Letunic & Bork, 2016). The GenBank databases was used to download the amino acid sequences 21 Camellia species, including C. oleifera (JQ975031), C. huana (KY626040), C. impressinervis (KF156835), C. pitardii (KF156837), C. danzaiensis (KF156834), C. grandibracteata (KJ806274), C. leptophylla (KJ806275), C. sinensis var. sinensis (KJ806281), C. sinensis var. dehungensis (KJ806279), C. crapnelliana (KF753632), C. taliensis voucher HKAS:S.X.Yang3157 (KF156839), C. taliensis voucher HKAS:S.X.Yang3158 (KF156836), C. reticulata (KJ806278), C. sinensis var. pubilimba (KJ806280), C. pubicosta (KJ806277), C. sinensis (KC143082), C. sinensis cultivar Longjing 43 (KF562708), C. cuspidata voucher HKAS: S.X.Yang3159 (KF156833), C. yunnanensis voucher HKAS:S.X.Yang1090 (KF156838), C. petelotii (KJ806276), C. luteoflora voucher CLUTE20161220 (KY626042). The model for phylogenetic assessment was based on the Newick tree file generated by MEGA 7.0. The tree was then imported to the iTOL online system. The bootstrap was quickly calculated with the iTOL’s in-built system, followed by the construction of the final circular phylogenetic tree. The same color-coded species in the picture represented similar phylogenetic relationships. We constructed phylogenetic trees using the following datasets due to the different molecular evolution rates of different cp genomic regions: (1) Complete chloroplast genome sequence; (2) Protein coding sequence; (3) LSC region; (4) SSC region. On the other hand, barcode labeling has been successfully applied to the study of phylogenetic relationships, and we have also compared the base sequences of DNA barcode (rbcL, matK, and trnH-psbA) of Hainan C. oleifera chloroplast genome and other 21 Theaceae plants. These three sites are commonly used research objects in DNA barcodes and are important molecular tools for identifying organisms (CBOL Plant Working Group, 2009; Yang et al., 2016).
Results and Analysis
Chloroplast genome characteristic of Hainan C. oleifera
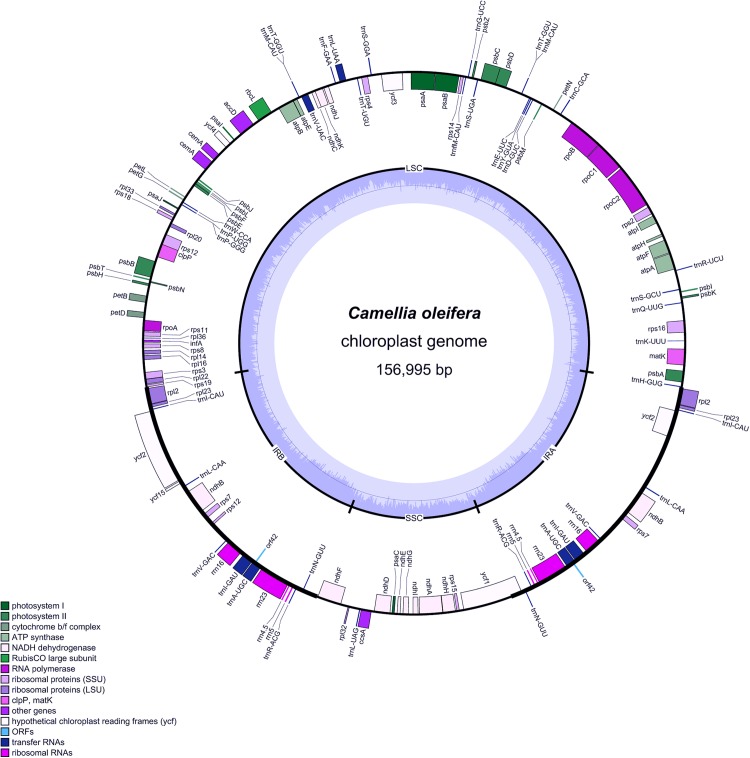
Like the majority of land plants, the whole chloroplast genome of Hainan C. oleifera showed a typical quadripartite genome organization with a size of 156,995 bp, including a LSC region of 86,648 bp and a SSC region of 18,297 bp, which were separated by two IR (IRa and IRb) regions of 26,025 bp (Fig. 1).
Figure 1. Gene map of the Hainan C. oleifera chloroplast genome.
The outermost colored blocks represent the physical location of different genes on the chloroplast genome, the inner circle is the physical location of the LSC, SSC, and IR regions on the genome and the different colors represent genes of different functional classes. Genes distributed outside the circle are transcribed counterclockwise, whereas genes inside are transcribed clockwise. The dark gray in the inner circle represents GC content, and the light gray represents AT content.
The chloroplast genome of Hainan C. oleifera was found to contain 141 predicted functional genes, including 88 protein-coding genes, 45 tRNA genes and eight rRNA genes, which were classified according to their function. Among the observed genes, 81 protein-coding genes, 30 tRNA genes, and four rRNA genes were unique. Among the protein-coding genes, 74 were single-copy genes and seven (ndhB, rps12, rps7, rpl2, rpl23, ycf2, and orf42) were duplicates. Among the tRNA genes, 19 were unique, and 11 (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnL-UAA, trnM-CAU, trnN-GUU, trnR-ACG, ttrnT-GGU, trnV-GAC, and trnV-UAC) were duplicates. The four rRNA genes were completely duplicated (Table 1). In addition, four overlapping genes (psbD-psbC, ndhK-ndhC, trnM-CAU-trnT-GGU, and trnP-GGG-trnP-UGG) were detected. Among these genes, nine genes (atpF, rpoC1, rpl2, ndhA, ndhB, ycf1, and trnI-GAU) contained a single intron, of which rpl2 and ndhB were identified as repetitive genes. Two genes (ycf3 and clpP) contained two introns. These two genes were similar to those in the Orpheus flower Haberlea rhodopensis and Broussonetia papyrifera (Ivanova et al., 2017; Xu et al., 2018). Unlike most angiosperm chloroplast genomes (Jin et al., 2014; Kong & Yang, 2015; Wang et al., 2016), among the 11 genes, four (atpF, rpoC1, ycf3, clpP) of which were located in the LSC region, two (ndhA and trnI-GAU) in the SSC region, other four (rpl2×2, ndhB×2) in the IR region, and one gene (ycf1) was a trans-spliced gene with its start codon located in the SSC region, stop codon in the IRb region, and possibly missing 3′ end. Additionally, the ndhA gene contained the largest intron (1,093 bp), and the ycf1 gene had the smallest one (27 bp) (Table S1).
Table 1. Genes present in the Hainan C. oleifera chloroplast genome.
| Group of genes | Gene names |
|---|---|
| Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome b/f complex | petA, petB, petD, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI |
| NADH dehydrogenase | ndhA*, ndhB(×2)*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| RubisCO large subunit | rbcL |
| RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 |
| Ribosomal proteins (SSU) | rps11, rps12(×2), rps14, rps15, rps16, rps18, rps19, rps2, rps3, rps4, rps7(×2), rps8 |
| Ribosomal proteins (LSU) | rpl14, rpl16, rpl2(×2)*, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 |
| Proteins of unknown function | ycf1*, ycf15, ycf2(×2), ycf3**, ycf4 |
| Transfer RNAs | trnA-UGC(×4), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC, trnH-GUG, trnI-CAU(×2), trnI-GAU(×4), trnK-UUU, trnL-CAA(×2), trnL-UAA(×2), trnL-UAG, trnM-CAU(×2), trnN-GUU(×2), trnP-GGG, trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU(×2), trnT-UGU, trnV-GAC(×2), trnV-UAC(×2), trnW-CCA, trnY-GUA |
| Ribosomal RNAs | rrn16(×2), rrn23(×2), rrn4.5(×2), rrn5(×2) |
| Other genes | infA, matK, clpP**, accD, ccsA, cemA, orf42(×2) |
Notes:
One or two asterisks after genes indicate that gene contains one or two introns, respectively.
The numbers in parentheses indicate the copy number of the gene.
Comparison with other Theaceae chloroplast genomes
The chloroplast genomic characteristics of Hainan C. oleifera were compared with nine other Theaceae species (Table 2). Similar to the reported results of the Theaceae chloroplast genome (Shi et al., 2013; Yang et al., 2013), the whole-genome length of Hainan C. oleifera was not much different from that of the other species. This change in sequence length depends mainly on the difference in the length of the LSC region (Li et al., 2015). The GC contents of the 10 species of Camellia were 37.29–37.34%, which were unevenly distributed in the whole genomes, with IRs exhibiting the highest GC content (42.94–43.01%), followed by the LSC (35.29–35.36%) and SSC regions (30.52–30.63%). Among them, the GC content in each area of Hainan C. oleifera and C. oleifera was the closest. As an important marker of molecular evolution, the IR region has a high GC content, which makes the sequence stable and highly conserved. In general, the protein-coding genes of two C. oleifera were the closest, although the total number of genes in Hainan C. oleifera was much larger than that of mainland C. oleifera, which has the largest number of 141 genes. This phenomenon may be caused by gene recombination and may affect its evolution.
Table 2. The basic composition of the Hainan C. oleifera chloroplast genome and other nine Theaceae plants.
| Total sequence length (bp)/GC content | LSC length/GC content | SSC length/GC content | IR length/GC content | Number of genes | Protein-coding genes | |
|---|---|---|---|---|---|---|
| C. oleifera Hainan | 156,995/37.29% | 86,648/35.29% | 18,297/30.55% | 26,025/42.98% | 141 | 88 |
| C. oleifera | 156,971/37.31% | 86,515/35.30% | 18,288/30.54% | 26,084/42.98% | 133 | 87 |
| C. luteoflora | 157,166/37.30% | 86,719/35.32% | 18,293/30.59% | 26,077/42.96% | 133 | 91 |
| C. grandibracteata | 157,127/37.29% | 86,656/35.32% | 18,285/30.52% | 26,093/42.94% | 127 | 91 |
| C. sinensis | 157,103/37.31% | 86,645/35.34% | 18,276/30.54% | 26,091/42.95% | 135 | 91 |
| C. leptophylla | 157,102/37.30% | 86,647/35.32% | 18,275/30.58% | 26,090/42.95% | 126 | 91 |
| C. pubicosta | 157,076/37.30% | 86,649/35.33% | 18,279/30.54% | 26,074/42.95% | 127 | 91 |
| C. crapnelliana | 156,997/37.30% | 86,655/35.30% | 18,406/30.60% | 25,968/43.01% | 136 | 91 |
| C. huana | 156,903/37.32% | 86,568/35.34% | 18,203/30.63% | 26,066/42.96% | 133 | 92 |
| C. danzaiensis | 156,576/37.34% | 86,204/35.36% | 18,259/30.59% | 26,056/42.98% | 135 | 92 |
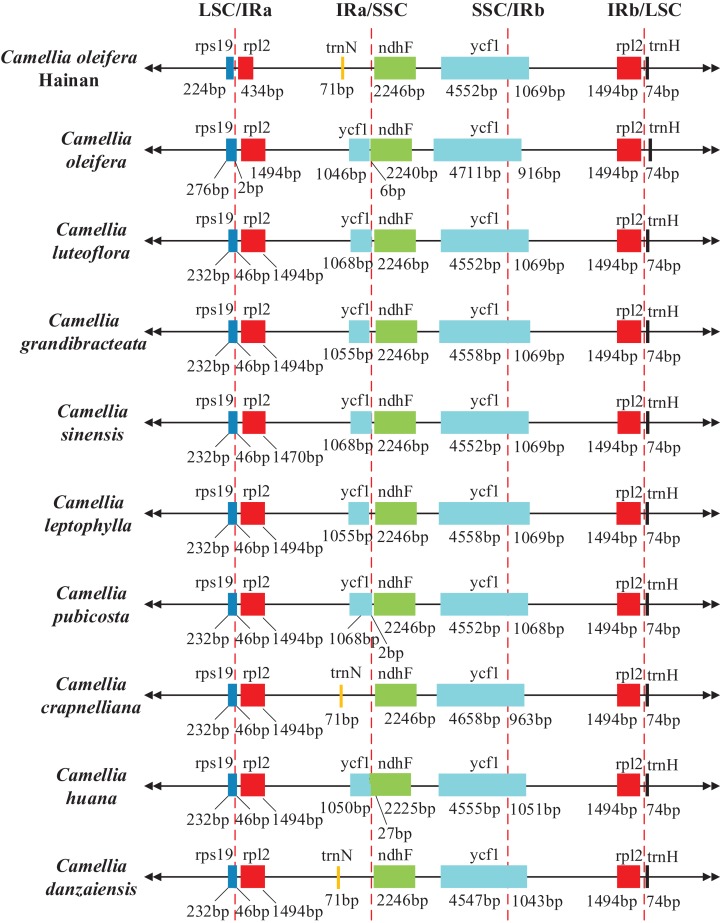
In the present paper, the IR/LSC and IR/SSC regions of the Hainan C. oleifera chloroplast genome were compared with the corresponding regions of the nine closely related Theaceae species (Fig. 2). As can be seen from the other Camellia chloroplast genome studies (Huang et al., 2014), the length of the chloroplast genome was variable, mainly due to the expansions and contractions of the border regions. Consistently, in all of the comparative chloroplast genomes, the IRb/SSC regions were located in the coding sequences of the ycf1 gene. The rpl2 (1,494 bp) and trnH (74 bp) genes were located on both sides of the IRb/LSC regions and were 106 and 2 bp away from the borders, respectively. However, the rps19 gene was located in the LSC region because of the expansion of the Hainan C. oleifera LSC region border, which was eight bp apart from the IRa/LSC region. Similarly, the rps19 gene from the mainland C. oleifera was also more biased towards the LSC region, whereas in the other eight Camellia species, with 232 bp occupying the LSC region and 46 bp occupying the IRa region. Like most plants, the ndhF gene involved in photosynthesis was located in the SSC region (Shen et al., 2017) and was 2,246 bp in length in all Camellia species, except in C. huana and C. oleifera. It should be emphasized that the ycf1 gene in the IRa regions was not observed in Hainan C. oleifera, C. crapnelliana, and C. danzaiensis because of the contraction of the IRa region border, which was replaced by the trnN gene. By comparing the border regions of the chloroplast genomes of 10 Theaceae species, it can be summarized as two different types and main feathered by gene structure of IRa/SSC. The trnN-ndhF genes located in above region of Hainan C. oleifera, C. crapnelliana and C. danzaniensis, while other species were ycf1-ndhF.
Figure 2. Comparison of the border positions of LSC, SSC, and IRs regions in chloroplast genome sequences of 10 Theaceae species.
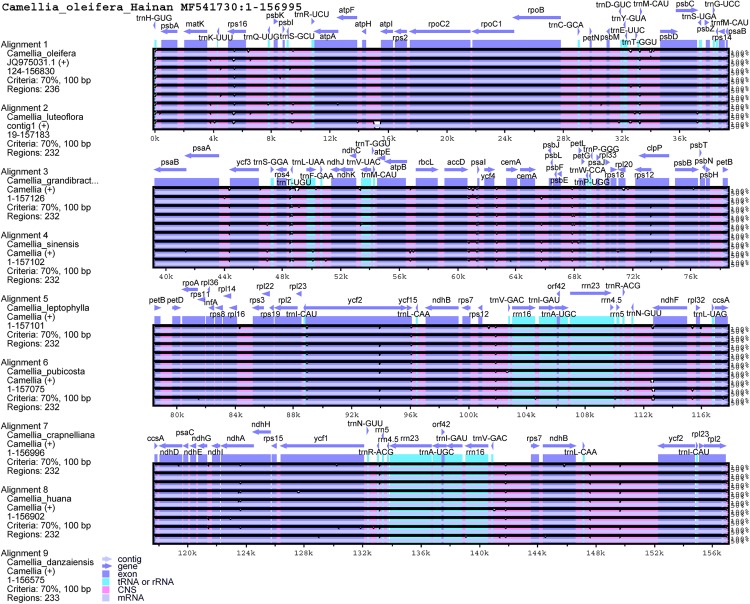
A multiple sequence alignment of the chloroplast genomes of the 10 Theaceae species was presented in Fig. 3. The result showed a high degree of similarity in the chloroplast genome sequences, suggesting that the chloroplast genome in the Theaceae species was evolutionarily conserved, despite of the discovery of several different regions in it, while the conservative of Theaceae plants is relatively rare in all angiosperms, which also makes the study of its evolution a certain pressure. Like other plants, the non-coding regions showed higher sequence diversity than the coding regions, whereas the IRs regions were more conserved than the single-copy regions (Huang et al., 2014; Ivanova et al., 2017; Yao et al., 2015). Among them, the rps16, clpP, ndhA, ycf2, and ycf1 genes showed high sequence divergence in the coding regions that makes such genes reliable markers for phylogenetic analysis. In addition, the non-coding regions showing a high degree of divergence included trnK-UUU-rps16, ycf3-trnS-GGA, trnL-UAA-trnF-GAA, accD-psaI, ycf4-cemA, psbE-petL, rps12-trnV-GAC, trnL-CAA-ycf2, and trnV-GAC-rps7.
Figure 3. Visualization alignments of chloroplast genome sequences among 10 Theaceae species with Hainan C. oleifera chloroplast genome as a reference.
The abscissa represents the position coordinates of the chloroplast genome of Hainan C. oleifera, and the ordinate represents the sequence similarity of the sample genome to the reference genome. Arrows indicate the annotated gene and its transcriptional direction, blue for the protein coding sequence (exons), green for tRNA or rRNA, and red for the conserved non-coding sequence (CNS).
Alignment of the matK gene and trnH-psbA IGR with other Camellia oleifera
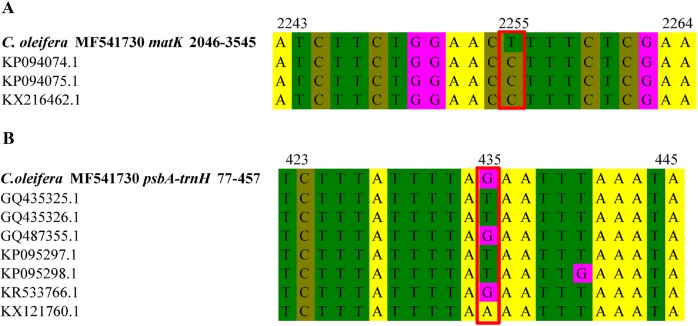
We compared Hainan C. oleifera with other mainland C. oleifera published in the NCBI database. The base sequences of the matK gene and trnH-psbA noncoding spacer were shown in Fig. 4. We selected some bases with differences, among which matK coding genes were compared. Although the protein base sequence is conservative, the results showed that there was a site change in Hainan C. oleifera, that is, at the position of 2,255 bp, all other three mainlands C. oleifera were base C, and object species was T. Similarly, in the comparison of trnH-psbA, it also changed at 435 bp, but it was the same as GQ487355 and KR533766, which were base G, and the other plants were A or T. This substitution of base pairs may cause changes in gene structure, leading to genetic mutations, which may be related to the living environment of Hainan C. oleifera.
Figure 4. Base sequence alignment of matK gene and trnH-psbA intergenic region between different C. oleifera.
(A) Partial nucleotide sequence of matK gene; (B) partial nucleotide sequence of trnH-psbA intergenic region.
SSR and long repeat sequences distribution
The SSRs are repetitive sequences of up to several dozen nucleotides consisting of one to six nucleotide repeat units, which are short in length and widely distributed in eukaryotic chloroplast genomes (Chen et al., 2006; Goldstein & Schlötterer, 1999). Owing to the presence of different nucleotides in the repeat units and different number of repeats, of which the most common is the dinucleotide repeat type, a high degree of variability is generated in SSR length. Therefore, we detected the SSRs distribution of the Hainan C. oleifera chloroplast genome by selecting the perfectly matched sequences, starting from the dinucleotide repeats. The statistical results were shown in Table 3. We detected a total of 51 SSR loci, all having a base composition biased toward A/T. Of these 51 SSRs, 28 were completely composed of A/T; this was consistent with previous studies (Cauzsantos et al., 2017; Shen et al., 2017; Yi & Kim, 2012). The distribution of SSRs in the chloroplast genome was heterogeneous, with up to 28 SSRs in the LSC region, five in the SSC region, and 18 in the IR region. Among all SSR loci, 23 were located in the 14 protein-coding genes (matK, atpA, rpoC2×2, rpoC1, ycf3, cemA, petA, rpoA, rol22, rpl2×2, ycf2×6, ndhB×2, ndhD×2, ndhA), and 26 in the intergenic and non-coding regions (trnS-UGA, rrn23×2). In terms of the SSR types, the most abundant repeat pattern was dinucleotide repeat unit (38 SSRs), with the largest type being AT/TA, accounting for 65.8% (25 SSRs). In addition, one tri- and 12 tetra-nucleotide repeats were found in the Hainan C. oleifera chloroplast genome. However, compared with other Theaceae species, Hainan C. oleifera and C. luteoflora were found to have no hexanucleotide repeats (Table S2). More specifically, all Camellia species have no pentanucleotide repeats (Huang et al., 2014). This is why these sequences were well-tagged in the studies of population phylogeny and taxonomy (Melotto-Passarin et al., 2011; Provan, Powell & Hollingsworth, 2001).
Table 3. Simple sequence repeats in Hainan C. oleifera chloroplast genome.
| ID | Repeat unit | Repeat number | Length (bp) | Start | End | Region | Annotation |
|---|---|---|---|---|---|---|---|
| 1 | TA | 4 | 8 | 2330 | 2337 | LSC | matK |
| 2 | AT | 4 | 8 | 4652 | 4659 | LSC | |
| 3 | AGAT | 3 | 12 | 6696 | 6707 | LSC | |
| 4 | TC | 4 | 8 | 9184 | 9191 | LSC | |
| 5 | GTCT | 3 | 12 | 11990 | 12001 | LSC | atpA |
| 6 | AT | 4 | 8 | 20077 | 20084 | LSC | rpoC2 |
| 7 | AT | 5 | 10 | 20842 | 20851 | LSC | rpoC2 |
| 8 | AT | 4 | 8 | 21871 | 21878 | LSC | rpoC1 |
| 9 | GA | 4 | 8 | 30227 | 30234 | LSC | |
| 10 | AG | 4 | 8 | 31910 | 31917 | LSC | |
| 11 | TCTT | 3 | 109 | 34002 | 34110 | LSC | |
| 12 | GA | 4 | 8 | 37363 | 37370 | LSC | trnS-UGA |
| 13 | AT | 4 | 8 | 38208 | 38215 | LSC | |
| 14 | TTTC | 3 | 12 | 45247 | 45258 | LSC | ycf3 |
| 15 | TA | 4 | 8 | 48399 | 48406 | LSC | |
| 16 | AT | 4 | 75 | 49339 | 49413 | LSC | |
| 17 | AT | 4 | 8 | 56936 | 56943 | LSC | |
| 18 | TA | 4 | 8 | 60906 | 60913 | LSC | |
| 19 | AAAT | 3 | 12 | 62713 | 62724 | LSC | |
| 20 | TC | 4 | 8 | 63432 | 63439 | LSC | cemA |
| 21 | AT | 4 | 8 | 64367 | 64374 | LSC | petA |
| 22 | AT | 4 | 8 | 65980 | 65987 | LSC | |
| 23 | TTC | 4 | 12 | 70097 | 70108 | LSC | |
| 24 | TA | 4 | 8 | 70419 | 70426 | LSC | |
| 25 | AT | 4 | 8 | 79616 | 79623 | LSC | |
| 26 | TA | 4 | 8 | 80572 | 80579 | LSC | rpoA |
| 27 | AT | 5 | 10 | 84341 | 84350 | LSC | |
| 28 | AT | 4 | 8 | 85931 | 85938 | LSC | rpl22 |
| 29 | TA | 5 | 10 | 87305 | 87314 | IRa | rpl2 |
| 30 | GA | 4 | 8 | 88915 | 88922 | IRa | ycf2 |
| 31 | GA | 4 | 8 | 89902 | 89909 | IRa | ycf2 |
| 32 | TCTA | 3 | 12 | 94648 | 94659 | IRa | ycf2 |
| 33 | TA | 4 | 8 | 95494 | 95501 | IRa | ycf2 |
| 34 | AG | 4 | 8 | 97417 | 97424 | IRa | ndhB |
| 35 | TA | 4 | 8 | 99589 | 99596 | IRa | |
| 36 | CT | 4 | 8 | 108654 | 108661 | IRa | rrn23 |
| 37 | CCCT | 3 | 12 | 110067 | 110078 | IRa | |
| 38 | AT | 4 | 8 | 116288 | 116295 | SSC | |
| 39 | GAAA | 3 | 12 | 118178 | 118189 | SSC | ndhD |
| 40 | AATA | 3 | 12 | 118328 | 118339 | SSC | ndhD |
| 41 | AAAT | 3 | 12 | 121323 | 121334 | SSC | |
| 42 | AT | 4 | 8 | 123859 | 123866 | SSC | ndhA |
| 43 | GAGG | 3 | 12 | 133565 | 133576 | IRb | |
| 44 | AG | 4 | 8 | 134983 | 134990 | IRb | rrn23 |
| 45 | TA | 4 | 8 | 144048 | 144055 | IRb | |
| 46 | CT | 4 | 8 | 146220 | 146227 | IRb | ndhB |
| 47 | TA | 4 | 8 | 148143 | 148150 | IRb | |
| 48 | ATAG | 3 | 12 | 148984 | 148995 | IRb | |
| 49 | TC | 4 | 8 | 153735 | 153742 | IRb | ycf2 |
| 50 | TC | 4 | 8 | 154722 | 154729 | IRb | ycf2 |
| 51 | AT | 5 | 10 | 156329 | 156338 | IRb | rpl2 |
To further explore the evolutionary characteristics of the Camellia species, we compared the long-repeat sequences in the chloroplast genome of Hainan C. oleifera with those of the other nine species (Table 4). We identified 28 forward, reverse and palindromic repeats with lengths ranging from 30 to 60 bp in 10 Theaceae species. Of these 28 repeat sequences, the ycf2 gene showed the presence of the largest number (60 bp in the chloroplast genome of C. pubicosta), and several repeats were also found in the intergenic regions. This phenomenon has also been reported in several angiosperms (Kong & Yang, 2017; Wang et al., 2016). Among them, seven long repeats ranging from 30 to 46 bp were found in the Hainan C. oleifera chloroplast genome. In total, six long repeats were located in the intergenic regions, and only one forward repeat was located in the protein-coding gene (ycf2). In the LSC regions, we observed only two pairs of palindromic repeats with lengths of 30 and 46 bp, respectively, and the remaining were located in the IR regions. Compared with the other nine species, the Hainan C. oleifera chloroplast genome had the same number of long-repeat sequences as C. crapnelliana, which had the least number of long-repeat sequences, while the most detected in C. oleifera (18 repeats) and the reverse match sequence was only detected in C. oleifera. Among the remaining seven species, eight long-repeat sequences were present in C. luteoflora, 12 in C. grandibracteata, 12 in C. sinensis, 10 in C. leptophylla, nine in C. pubicosta, 10 in C. huana, and 13 in C. danzaiensis.
Table 4. Long repeat sequences in the chloroplast genome of 10 Theaceae species.
| Type | Repeat sizes (bp) | Location | Region | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. oleifera Hainan | C. oleifera | C. luteoflora | C. grandibracteata | C. sinensis | C. leptophylla | C. pubicosta | C. crapnelliana | C. huana | C. danzaiensis | |||
| F | 56 | 56 | 56 | 56 | 56 | 48 | ycf2; ycf2 | IRa | ||||
| P | 56 | 56 | 56 | 56 | 56 | 48 | ycf2; ycf2 | IRa, IRb | ||||
| P | 48 | 48 | 60 | ycf2; ycf2 | IRa, IRb | |||||||
| F | 56 | 48 | 48 | 56 | 56 | 56 | 48 | ycf2; ycf2 | IRb | |||
| P | 48 | IGS (rpl2-trnH; rps19-rpl2) | LSC, IRa | |||||||||
| F | 48 | IGS (rpl2-trnH); rpl2 | LSC, IRb | |||||||||
| F | 47 | IGS (rpl2-trnH) | LSC, IRb | |||||||||
| P | 46 | IGS (petB-petD) | LSC | |||||||||
| P | 46 | 46 | 42 | 42 | 42 | 42 | 46 | 46 | 46 | petD; petD | LSC | |
| F | 42 | 42 | 41 | 42 | 42 | 42 | 42 | 42 | 42 | 42 | IGS (rps12-trnV); Intron (ndhA) | IRa, SSC |
| P | 42 | 42 | 41 | 42 | 42 | 42 | 42 | 42 | 42 | 42 | IGS (trnV-rps12); Intron (ndhA) | IRb, SSC |
| F | 38 | 38 | 38 | 38 | 38 | 38 | 38 | 38 | 38 | 30 | ycf2; ycf2 | IRa |
| P | 38 | 38 | 38 | 38 | 38 | 38 | 38 | ycf2; ycf2 | IRa, IRb | |||
| F | 38 | 38 | 30 | 30 | 38 | 38 | 38 | 38 | 30 | ycf2; ycf2 | IRb | |
| F | 38 | IGS (trnL-ycf2) | IRb | |||||||||
| P | 38 | ycf2; ycf2 | IRb | |||||||||
| P | 38 | ycf2; IGS (trnL-ycf2) | IRa, IRb | |||||||||
| F | 34 | IGS (rrn4.5-rrn5) | IRa | |||||||||
| P | 34 | IGS (rrn4.5-rrn5; rrn5-rrn4.5) | IRa, IRb | |||||||||
| F | 34 | IGS (rrn5-rrn4.5) | IRb | |||||||||
| R | 32 | IGS (atpB-rbcL) | LSC | |||||||||
| R | 31 | IGS (atpB-rbcL) | LSC | |||||||||
| F | 31 | IGS (atpB-rbcL) | LSC | |||||||||
| F | 34 | IGS (petA-psbJ) | LSC | |||||||||
| P | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | IGS (psbI-trnS; trnS-rps4) | LSC |
| R | 30 | IGS (atpB-rbcL) | LSC | |||||||||
| F | 30 | IGS (atpB-rbcL) | LSC | |||||||||
| P | 30 | 30 | 30 | ycf2; ycf2 | IRa, IRb | |||||||
Characteristic of codon usage
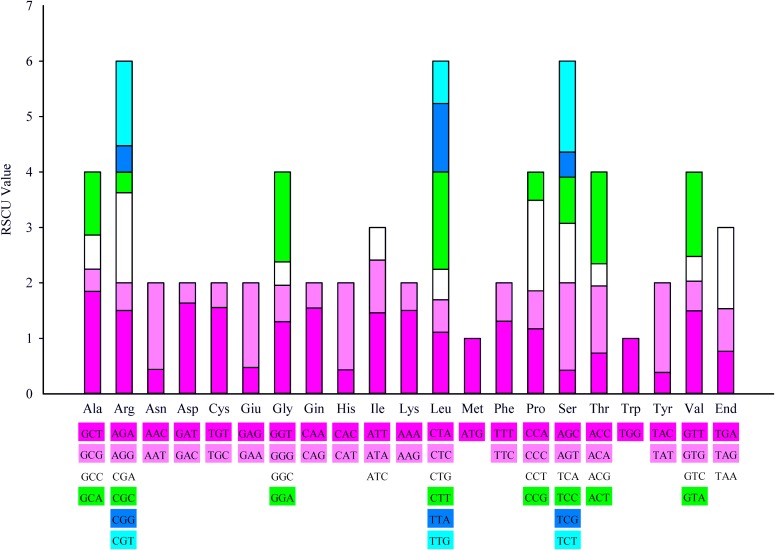
In the Hainan C. oleifera chloroplast DNA, the 88 protein-coding genes had 27,270 codons. More than 94% of the protein-coding genes used ATG as the start codon, and less than 6% of the remaining used ATT (petB and rps19), ATA (ndhD), and ATC (rpl16 and orf42) as start codons. Furthermore, ATA, ATC, ATT, and TTG were used as start codons in the Aquilaria sinensis chloroplast DNA (Wang et al., 2016), and GTG and ACT served as start codons in the rps19 and rps2 genes in Morus atropurpurea, respectively (Li et al., 2016). Among the stop codons, TAA was the most common, followed by TAG and TGA, with 73.86% of the stop codons ending in A, thus strongly reflecting the AT bias in codon usage. The RSCU value of the Hainan C. oleifera chloroplast genome was shown in Fig. 5; its value increased with the number of codons. Leucine accounted for the highest codon usage (10.84%), followed by serine (9.17%) and isoleucine (8.42%). Nearly one third of the total codons were represented by these three amino acids.
Figure 5. 20 amino acid codon and stop codon of the island plant Hainan C. oleifera chloroplast genome.
The color of the histogram corresponds to the color of the codon.
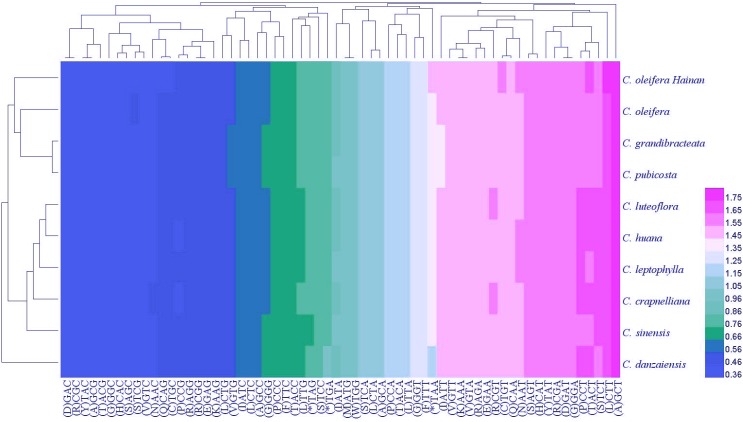
The distributions of codon usage in the form of heatmaps for 10 Theaceae species were shown in Fig. 6. It can be seen that about half of the codons with low RSCU values (shown in blue in the heatmaps) were infrequently used. All codons with RSCU >1 ended with A/T. A similar phenomenon has been found in other plant lineages, indicating that the A + T bias plays an important role in the plastid genome. In addition, we found that the codon usage of Hainan C. oleifera chloroplast genome was most similar to that of C. oleifera. As more chloroplast genome data are being continually explored, further validation of these findings will allow us to understand the relationship between different codon usage patterns and hosts, as well as the interactions among nuclear genomes (Liu & Xue, 2005).
Figure 6. The distributions of codon usage in the form of heat maps for 10 Theaceae species.
Color indication: red represents the larger RSCU values and blue represents the smaller RSCU values.
Phylogenetic relationship with near source species
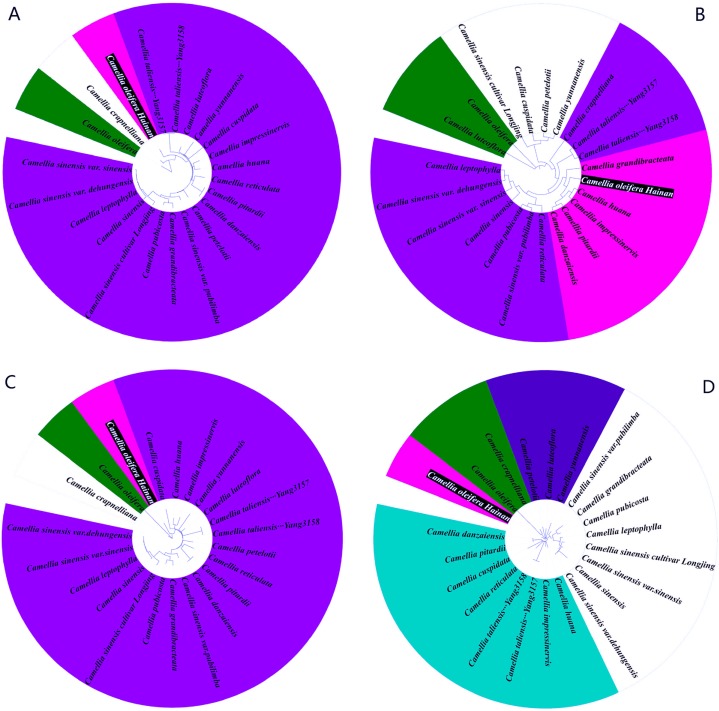
As an important part of plant cell organelles and photosynthetic organs, chloroplasts play an important role in the long history of biological evolution (Kim & Archibald, 2009). In this study, we not only constructed the phylogenetic tree of complete chloroplast genome of 22 Camellia species, but also extracted protein sequence information, LSC region sequences and SSC region sequences, and constructed the phylogenetic tree. The results showed a different evolutionary relationship, in which the whole chloroplast genome showed that Hainan C. oleifera, C. oleifera, and C. crapnelliana were independent branches and have similar evolutionary relationships (Fig. 7A). The protein coding sequence showed that Hainan C. oleifera was closely related to C. crapnelliana, C. danzaiensis, C. pitardii, C. huana, and C. impressinervis, which was closest to C. crapnelliana (Fig. 7B). The results of the LSC region (Fig. 7C) and the SSC region (Fig. 7D) were similar to those of the complete chloroplast genome sequence, showing that the three Camellia species of Hainan C. oleifera, C. oleifera, and C. crapnelliana were the same family. The evolutionary relationship was clear. There were similar parts in these results, especially C. oleifera and C. crapnelliana, but the protein coding sequence is more stable and not susceptible to the external environment, so its phylogenetic relationship is more informative.
Figure 7. Phylogenetic relationships of 22 species of Camellia plants inferred from different data partitions.
(A) Whole chloroplast genome; (B) protein coding region; (C) LSC region; (D) SSC region Species with similar evolutionary relationships are displayed in the same color.
The base sequence comparison of DNA barcodes (rbcL, matK, and trnH-psbA) revealed a high degree of conservation of the chloroplast genome sequence of the Camellia plants (Table S3). There were 10 locus in rbcL with differences (marked by red triangles below), but Hainan C. oleifera has the same base composition as most species. There were 22 locus differences in matK, similar to rbcL, the most occurred in a single base region of other species. Unlike the other species, Hainan C. oleifera was replaced by T at one base, and the rest were C. In the psbA-trnH comparison, there were nine positions in which the base changes slightly, while in the second change point, Hainan C. oleifera was C, and other species were A. Therefore, these single site changes can be used as good molecular markers.
Discussion
With the rapid development of molecular biology techniques, new molecular markers and detection methods are emerging in an endless stream. Several DNA analysis techniques suitable for chloroplast genome research on plant phylogeny and evolution will be available. Moreover, the research on population genetic changes will play an important role in the protection of biological resources and biodiversity (Xu & Wang, 2001). As an important evolutionary feature of the chloroplast genome, the codon usage bias has become an analytical tool widely used in several organisms (Nie et al., 2014; Sharp et al., 1988; Vicario, Moriyama & Powell, 2007). In this study, we systematically analyzed the RSCU value of the Hainan C. oleifera chloroplast genome. The results showed that nearly half of the values were higher than 1.00 and nearly half were lower than 1.00, and all high-valued codons ended with A or T, whereas the RSCU value was 1.00 without any codon usage bias (Gupta, Bhattacharyya & Ghosh, 2004). Therefore, we hypothesized that the codons in Hainan C. oleifera chloroplast genome preferably ended with A/T. This conjecture was also confirmed in this paper, and has also been found in other plant studies (Wang et al., 2016). It is generally accepted that codon usage bias reflects the balance between the mutational bias and natural selection for translation optimization (Bulmer, 1988; Sharp et al., 1993; Shin et al., 2015). In order to further clarify the role of the two in the evolution of the Asteraceae species, some researchers used the neutrality analysis, which reflected that the natural selection pressure played a major role in shaping codon usage (Nie et al., 2014). However, the C. oleifera population on Hainan Island has a certain geographical isolation; the genetic variation of population might occur. Whether the mutation or natural selection plays a leading role needs further confirmation.
Previous studies have shown that the chloroplast genome structure is highly conserved (Korpelainen, 2004). In this study, the genomic composition, gene sequence, GC content, and codon preference of the Hainan C. oleifera chloroplast genome were not much different from that of the other Camellia species, and were similar to the typical angiosperm chloroplast genome (Shinozaki et al., 1986; Wang et al., 2016; Yang et al., 2013), which revealed that the chloroplast genome of Hainan C. oleifera is highly conserved in structure and evolution. The conservation of these sequences makes Hainan C. oleifera have some pressure on evolutionary research, so that it difficult to determine the differential genes it contains. This also forced research on the evolution of C. oleifera on Hainan Island. In addition, we focused on the junctions between the IR/LSC and IR/SSC regions of the chloroplast genome, where more sequence data were available. In the long process of evolution, the structural order of the LSC, SSC, and IR regions remained unchanged. The differences among the chloroplast genomes of different species are mainly reflected in the length and orientation of the IRs regions. Among them, a similar situation was produced in Hainan C. oleifera, C. crapnelliana, and C. danzaiensis due to the change of IRa region, and the similarity at this level indicated that they shared a common ancestor. This can initially determine its source to reduce the pressure of its taxonomic research. As an important indicator of chloroplast genome evolution, the contractions, and expansions of the IR/LSC and IR/SSC boundaries often determine the chloroplast genome length variation (Goulding et al., 1996; Zhang et al., 2013). Moreover, the boundaries of the plastids of various species are often different even between members of the same family (Ravi et al., 2006). Therefore, it would be useful to compare the IR/LSC and IR/SSC junctions of chloroplast genome in different Camellia species for alleviating its evolutionary pressure.
It was difficult to study the classification and phylogeny of the genus Camellia because of various reasons, such as its frequent interspecific hybridization and polyploidization (Huang et al., 2014; Yang et al., 2013). However, the samples of C. oleifera used in this study were grown in a unique island environment, and their genetics and evolutionary history are still vague. Even if we used the complete chloroplast genome information for analysis, it was not enough to completely solve all phylogenetic relationships, as can be seen in previous studies (Petersen et al., 2011; Steane, 2005; Wortley et al., 2005). Besides, the comparison object we selected were no plant groups involved in the other genera of the family Theaceae, which might provide useful information for the evolutionary study of Hainan C. oleifera. In summary, our research can promote the exchange of information between the nuclear genomes of Camellia species and provide valuable genomic resources for phylogenetic studies.
Conclusions
In this study, we sequenced the complete chloroplast genome of C. oleifera on Hainan Island, China, using the Illumina high-throughput sequencing technology. The chloroplast genome of Hainan C. oleifera was found to have an intact quadripartite structure by genome sequencing and reassembly. The annotation and comparison with other Camellia species showed that the genomic composition, gene sequence, GC content, and codon preference of the Hainan C. oleifera chloroplast genome were not much different from that of the other Camellia species, and were similar to the typical angiosperm chloroplast genome. Therefore, we performed in-depth analysis of the border regions, long repeats, SSRs, and sequence diversity in the chloroplast genomes of 10 Camellia plants, indicated that their chloroplast genomes were highly conserved in structure and evolution. Long-sequence repeats, SSRs and DNA barcodes can be used as novel molecular markers. By studying the gene composition of the marginal regions, codon distribution and phylogeny, it was found that Hainan C. oleifera has similar evolutionary relationships with C. crapnelliana. Therefore, the data obtained in this study will be helpful to further study the evolutionary history of Hainan C. oleifera and to protect its excellent germplasm resources, and to provide valuable genome resources for phylogenetic and taxonomic studies and for the exchange of information between nuclear genomes.
Supplemental Information
*The ycf1 is a trans-spliced gene with the start located in the SSC region and the end in the IRb region, and possibly missing 3’ end.
Funding Statement
This research was supported by the Science and technology major project of Hunan province (2017NK1014), the Key Technology R&D Program of Hunan Province (2016TP2007, 2016NK2148), and the National Key Technology Research and Development Program of China (2014BAC09B03-02). There was no additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Wan Zhang analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper.
Yunlin Zhao conceived and designed the experiments.
Guiyan Yang conceived and designed the experiments, contributed reagents/materials/analysis tools.
Jiao Peng performed the experiments, contributed reagents/materials/analysis tools.
Shuwen Chen performed the experiments.
Zhenggang Xu analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
The complete chloroplast genome sequence of C. oleifera is available at GenBank: MF541730.
Data Availability
The following information was supplied regarding data availability:
Data access in NCBI: all complete chloroplast genome sequence are available with the following accessions Hainan C. oleifera (MF541730), C. oleifera (JQ975031), C. huana (KY626040), C. impressinervis (KF156835), C. pitardii (KF156837), C. danzaiensis (KF156834), C. grandibracteata (KJ806274), C. leptophylla (KJ806275), C. sinensis var. sinensis (KJ806281), C. sinensis var. dehungensis (KJ806279), C. crapnelliana (KF753632), C. taliensis voucher HKAS S.X.Yang3157 (KF156839), C. taliensis voucher HKAS S.X.Yang3158 (KF156836), C. reticulata (KJ806278), C. sinensis var. pubilimba (KJ806280), C. pubicosta (KJ806277), C. sinensis (KY626042), C. sinensis cultivar Longjing (KF562708), C. cuspidata (KF156833), C. yunnanensis (KF156838), C. petelotii (KJ806276), C. luteoflora (KY626042).
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
References
- Boetzer et al. (2011).Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Bulmer (1988).Bulmer M. Are codon usage patterns in unicellular organisms determined by selection-mutation balance? Journal of Evolutionary Biology. 1988;1(1):15–26. doi: 10.1046/j.1420-9101.1988.1010015.x. [DOI] [Google Scholar]
- Cauzsantos et al. (2017).Cauzsantos LA, Munhoz CF, Rodde N, Cauet S, Santos AA, Penha HA, Dornelas MC, Varani AM, Oliveira GC, Bergès H. The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: structural organization and phylogenomic studies in Malpighiales. Frontiers in Plant Science. 2017;8:334. doi: 10.3389/fpls.2017.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CBOL Plant Working Group (2009).CBOL Plant Working Group A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2006).Chen C, Zhou P, Choi YA, Huang S, Gmitter FG., Jr Mining and characterizing microsatellites from citrus ESTs. Theoretical and Applied Genetics. 2006;112(7):1248–1257. doi: 10.1007/s00122-006-0226-1. [DOI] [PubMed] [Google Scholar]
- Corriveau & Coleman (1988).Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 Angiosperm species. American Journal of Botany. 1988;75(10):1443–1458. doi: 10.1002/j.1537-2197.1988.tb11219.x. [DOI] [Google Scholar]
- Deng et al. (2014).Deng WK, Wang YB, Liu ZX, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLOS ONE. 2014;9(11):e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer et al. (2004).Frazer KA, Pachter LS, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Research. 2004;32(Web Server):W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein & Schlötterer (1999).Goldstein DB, Schlötterer C. Microsatellites: evolution and applications. Quarterly Review of Biology. 1999;83:633–634. [Google Scholar]
- Goulding et al. (1996).Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Molecular & General Genetics. 1996;252(1–2):195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- Gupta, Bhattacharyya & Ghosh (2004).Gupta SK, Bhattacharyya TK, Ghosh TC. Synonymous codon usage in Lactococcus lactis: mutational bias versus translational selection. Journal of Biomolecular Structure and Dynamics. 2004;21(4):527–535. doi: 10.1080/07391102.2004.10506946. [DOI] [PubMed] [Google Scholar]
- He et al. (2007).He GX, Li SL, Xu LC, Gao H. Growth characteristic and function value of Camellia oliefera. Jiangxi Forestry Science and Technology. 2007;4:39–42. [Google Scholar]
- Huang et al. (2014).Huang H, Shi C, Liu Y, Mao SY, Gao LZ. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evolutionary Biology. 2014;14(1):151. doi: 10.1186/1471-2148-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova et al. (2017).Ivanova Z, Sablok G, Daskalova E, Zahmanova G, Apostolova E, Yahubyan G, Baev V. Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Frontiers in Plant Science. 2017;8(402):204. doi: 10.3389/fpls.2017.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen et al. (2005).Jansen RK, Raubeson LA, Boore JL, DePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods in Enzymology. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- Jin et al. (2014).Jin GH, Chen SY, Yi TS, Zhang SD. Characterization of the complete chloroplast genome of Apple (Malus × domestica, Rosaceae) Plant Diversity and Resources. 2014;36:468–484. [Google Scholar]
- Kim & Archibald (2009).Kim E, Archibald JM. Diversity and evolution of plastids and their genomes. In: Sandelius AS, Aronsson H, editors. The Chloroplast: Interactions with the Environment. Berlin, Heidelberg: Springer Berlin Heidelberg; 2009. pp. 1–39. [Google Scholar]
- Kong & Yang (2015).Kong W, Yang J. The complete chloroplast genome sequence of Morus mongolica and a comparative analysis within the Fabidae clade. Current Genetics. 2015;62(1):165–172. doi: 10.1007/s00294-015-0507-9. [DOI] [PubMed] [Google Scholar]
- Kong & Yang (2017).Kong WQ, Yang JH. The complete chloroplast genome sequence of Morus cathayana and Morus multicaulis, and comparative analysis within genus Morus L. PeerJ. 2017;5(31):e3037. doi: 10.7717/peerj.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpelainen (2004).Korpelainen H. The evolutionary processes of mitochondrial and chloroplast genomes differ from those of nuclear genomes. Die Naturwissenschaften. 2004;91(11):505–518. doi: 10.1007/s00114-004-0571-3. [DOI] [PubMed] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz et al. (2001).Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic & Bork (2016).Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2016).Li QL, Guo JZ, Yan N, Li CC. Complete chloroplast genome sequence of cultivated Morus L. species. Genetics and Molecular Research. 2016;15:gmr15048906. doi: 10.4238/gmr15048906. [DOI] [PubMed] [Google Scholar]
- Li & Wan (2005).Li Q, Wan JM. SSRHunter: development of a local searching software for SSR sites. Hereditas. 2005;27:808–810. [PubMed] [Google Scholar]
- Li et al. (2015).Li XJ, Yang Z, Huang Y, Ji Y. Complete chloroplast genome of the medicinal plant paris polyphylla var. chinensis (Melanthiaceae) Journal of Tropical and Subtropical Botany. 2015;23:601–613. [Google Scholar]
- Liu et al. (2012).Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 2012;13(1):715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu & Xue (2005).Liu QP, Xue QZ. Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. Journal of Genetics. 2005;84(1):55–62. doi: 10.1007/BF02715890. [DOI] [PubMed] [Google Scholar]
- Lohse, Drechsel & Bock (2007).Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 2007;52(5–6):267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Luo et al. (2012).Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Qi P, Liu YJ. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchlerbauer et al. (2015).Marchlerbauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI. CDD: NCBI’s conserved domain database. Nucleic Acids Research. 2015;43(D1):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor et al. (2000).Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16(11):1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Melotto-Passarin et al. (2011).Melotto-Passarin DM, Tambarussi EV, Dressano K, De Martin VF, Carrer H. Characterization of chloroplast DNA microsatellites from Saccharum spp and related species. Genetics and Molecular Research. 2011;10(3):2024–2033. doi: 10.4238/vol10-3gmr1019. [DOI] [PubMed] [Google Scholar]
- Nie et al. (2014).Nie XJ, Deng PC, Feng KW, Liu PX, Du XH, You FM, Song WN. Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant Molecular Biology Reporter. 2014;32(4):828–840. doi: 10.1007/s11105-013-0691-z. [DOI] [Google Scholar]
- Olmstead & Palmer (1994).Olmstead RG, Palmer JD. Chloroplast DNA systematics: a review of methods and data analysis. American Journal of Botany. 1994;81(9):1205–1224. doi: 10.1002/j.1537-2197.1994.tb15615.x. [DOI] [Google Scholar]
- Palmer (1985).Palmer JD. Comparative organization of chloroplast genomes. Annual Review of Genetics. 1985;19(1):325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Palmer (1991).Palmer JD. CHAPTER 2 - Plastid chromosomes: structure and evolution. In: Bogorad L, Vasil IK, editors. The Molecular Biology of Plastids. Academic Press; 1991. pp. 5–53. [Google Scholar]
- Petersen et al. (2011).Petersen G, Aagesen L, Seberg O, Larsen IH. When is enough, enough in phylogenetics? A case in point from Hordeum (Poaceae) Cladistics. 2011;27(4):428–446. doi: 10.1111/j.1096-0031.2011.00347.x. [DOI] [PubMed] [Google Scholar]
- Provan, Powell & Hollingsworth (2001).Provan J, Powell W, Hollingsworth PM. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends in Ecology & Evolution. 2001;16(3):142–147. doi: 10.1016/S0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Ravi et al. (2006).Ravi V, Khurana JP, Tyagi AK, Khurana P. The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis. Tree Genetics & Genomes. 2006;3(1):49–59. doi: 10.1007/s11295-006-0051-3. [DOI] [Google Scholar]
- Saski et al. (2005).Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Molecular Biology. 2005;59(2):309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Sharp et al. (1988).Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Research. 1988;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp & Li (1987).Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp et al. (1993).Sharp PM, Stenico M, Peden JF, Lloyd AT. Codon usage: mutational bias, translational selection, or both? Biochemical Society Transactions. 1993;21(4):835–841. doi: 10.1042/bst0210835. [DOI] [PubMed] [Google Scholar]
- Shen et al. (2017).Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, Li X, Zhang B, Xu J, Chen S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 2017;22(8):1330–1343. doi: 10.3390/molecules22081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2013).Shi C, Liu Y, Huang H, Xia EH, Zhang HB, Gao LZ. Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: an exemplary study of ycf15 function and evolution in angiosperms. PLOS ONE. 2013;8(3):e59620. doi: 10.1371/journal.pone.0059620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin et al. (2015).Shin YC, Bischof GF, Lauer WA, Desrosiers RC. Importance of codon usage for the temporal regulation of viral gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(45):14030–14035. doi: 10.1073/pnas.1515387112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki et al. (1986).Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO Journal. 1986;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steane (2005).Steane DA. Complete nucleotide sequence of the chloroplast genome from the Tasmanian blue gum, Eucalyptus globulus (Myrtaceae) DNA Research. 2005;12(3):215–220. doi: 10.1093/dnares/dsi006. [DOI] [PubMed] [Google Scholar]
- Sugita & Sugiura (1996).Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Molecular Biology. 1996;32(1–2):315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- Tsudzuki et al. (1992).Tsudzuki J, Nakashima K, Tsudzuki T, Hiratsuka J, Shibata M, Wakasugi T, Sugiura M. Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Molecular & General Genetics. 1992;232:206–214. doi: 10.1007/BF00279998. [DOI] [PubMed] [Google Scholar]
- Vicario, Moriyama & Powell (2007).Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. BMC Evolutionary Biology. 2007;7(1):226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2016).Wang Y, Zhan DF, Jia X, Mei WL, Dai HF, Chen XT, Peng SQ. Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the Malvales order. Frontiers in Plant Science. 2016;7:1–13. doi: 10.3389/fpls.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, Li & Sharp (1987).Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortley et al. (2005).Wortley AH, Rudall PJ, Harris DJ, Scotland RW. How much data are needed to resolve a difficult phylogeny?: case study in Lamiales. Systematic Biology. 2005;54(5):697–709. doi: 10.1080/10635150500221028. [DOI] [PubMed] [Google Scholar]
- Wyman, Jansen & Boore (2004).Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Xia (2017).Xia XH. DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. Journal of Heredity. 2017;108(4):431–437. doi: 10.1093/jhered/esx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu & Wang (2001).Xu HF, Wang JB. Advances in molecular systematics. Chinese Journal of Ecology. 2001;20:41–46. [Google Scholar]
- Xu et al. (2018).Xu ZG, Yang GY, Dong M, Wu L, Zhang W, Zhao YL. The complete chloroplast genome of an economic and ecological plant, paper mulberry (Broussonetia kazinoki × Broussonetia papyifera) Mitochondrial DNA Part B. 2018;3(1):28–29. doi: 10.1080/23802359.2017.1419088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2016).Yang Y, Tao Z, Dong D, Jia Y, Li F, Zhao G. Comparative analysis of the complete chloroplast genomes of FiveQuercusSpecies. Frontiers in Plant Science. 2016;7:959. doi: 10.3389/fpls.2016.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2013).Yang JB, Yang SX, Li HT, Yang J, Li DZ. Comparative chloroplast genomes of camellia species. PLOS ONE. 2013;8(8):e73053. doi: 10.1371/journal.pone.0073053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2010).Yang M, Zhang XW, Liu GM, Yin YX, Chen KF, Yun QZ, Zhao DJ, Almssallem IS, Yu J. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.) PLOS ONE. 2010;5(9):e12762. doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao et al. (2015).Yao XH, Tang P, Li ZZ, Li DW, Liu YF, Huang HW. The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PLOS ONE. 2015;10(6):e0129347. doi: 10.1371/journal.pone.0129347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi & Kim (2012).Yi DK, Kim KJ. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLOS ONE. 2012;7(5):e35872. doi: 10.1371/journal.pone.0035872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2013).Zhang H, Li C, Miao H, Xiong S. Insights from the complete chloroplast genome into the evolution of Sesamum indicum L. PLOS ONE. 2013;8(11):e80508. doi: 10.1371/journal.pone.0080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2017).Zhang W, Zhao YL, Yang GY, Tang YC, Xu ZG. Characterization of the complete chloroplast genome sequence of Camellia oleifera in Hainan, China. Mitochondrial DNA Part B. 2017;2(2):843–844. doi: 10.1080/23802359.2017.1407687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2016).Zheng DJ, Pan XZ, Zhang DM, Xie LS, Zeng JH, Zhang ZL, Ye H. Survey and analysis on tea-oil Camellia resource in Hainan. Journal of Northwest Forestry University. 2016;31:130–135. [Google Scholar]
- Zhou et al. (2013).Zhou CF, Yao XH, Lin P, Wang KL, Chang J, Mo RH. Constituent changes associated with seeds development of Camellia oleifera Abel. Chinese Journal of Oil Crop Sciences. 2013;35:680–685. [Google Scholar]
- Zhuang (2008).Zhuang RL. Camellia oleifera of China. Beijing: China Forestry Press; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
*The ycf1 is a trans-spliced gene with the start located in the SSC region and the end in the IRb region, and possibly missing 3’ end.
Data Availability Statement
The following information was supplied regarding data availability:
Data access in NCBI: all complete chloroplast genome sequence are available with the following accessions Hainan C. oleifera (MF541730), C. oleifera (JQ975031), C. huana (KY626040), C. impressinervis (KF156835), C. pitardii (KF156837), C. danzaiensis (KF156834), C. grandibracteata (KJ806274), C. leptophylla (KJ806275), C. sinensis var. sinensis (KJ806281), C. sinensis var. dehungensis (KJ806279), C. crapnelliana (KF753632), C. taliensis voucher HKAS S.X.Yang3157 (KF156839), C. taliensis voucher HKAS S.X.Yang3158 (KF156836), C. reticulata (KJ806278), C. sinensis var. pubilimba (KJ806280), C. pubicosta (KJ806277), C. sinensis (KY626042), C. sinensis cultivar Longjing (KF562708), C. cuspidata (KF156833), C. yunnanensis (KF156838), C. petelotii (KJ806276), C. luteoflora (KY626042).
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.