Abstract
As a universal pathogen leading to neonatal defects and transplant failure, human cytomegalovirus (HCMV) has strict species specificity and this has prevented the development of a suitable animal model for the pathogenesis study. The mechanism of cross-species barrier remains elusive and there are so far no non-human cell culture models that support HCMV replication. The Chinese tree shrew (Tupaia belangeri chinensis) is a small laboratory animal and evolutionary closely related with primates. We investigated the susceptibility of primary tree shrew dermis fibroblasts (TSDF) to HCMV infection. Infection with a GFP-expressing HCMV virus resulted in green fluorescence in infected cells with the expression of IE1, UL44 and pp28. The titers of cell-free viruses reached 103 PFU/mL at 96 hpi, compared to titers of 104 PFU/mL observed in primary human foreskin fibroblasts. Our results suggested that TSDF was semi-permissive for HCMV infection. The TSDF model could be further used to investigate key factors influencing cross-species multiplication of HCMV.
Electronic supplementary material
The online version of this article (10.1007/s12250-019-00106-3) contains supplementary material, which is available to authorized users.
Keywords: Human cytomegalovirus (HCMV), Primary tree shrew dermis fibroblasts (TSDF), Cross-species infection, Semi-permissiveness
Introduction
Human cytomegalovirus (HCMV) is an opportunistic pathogen leading to life-threatening disease in immune-compromised patients and to significant congenital disease following primary infection of pregnant women. Various types of human cells are fully permissive for HCMV infection and support the expression of viral genes in temporally controlled cascade and the production of large amount of viral particles (Landolfo et al.2003). Immediate early (IE) genes (such as IE1) are expressed very early after the viral genome entry into the nucleus, followed by the expression of early (E) genes (such as UL44) which takes part in viral genome replication. Late gene expression (such as pp28) depends on expression of E genes and controls envelopment and egress of progeny viruses (Wathen et al.1981; Wathen and Stinski 1982). On the other hand, cell types in which viral protein synthesis and viral particle release are different from the situation in permissive cells were considered as semi-permissive or non-permissive. In semi-permissive cells, the kinetics of protein expression is usually delayed and little amount of progeny viruses are produced (Luo and Fortunato 2007). The viral protein synthesis and viral output were not measurable in non-permissive cells, and the abortive infection has been reported in cells from commonly used laboratory animals (Kim and Carp 1972; Fioretti et al.1973; Lafemina and Hayward 1988; Garcia-Ramirez et al.2001). This is the result of strict species specificity of HCMV infection and the lack of a suitable experimental animal model has hampered studies on HCMV pathogenesis. In order to overcome this limitation, it is important to investigate the molecular mechanism of the species specificity for HCMV infection.
In the past decades, both in vitro and in vivo infections of HCMV across species barrier have been tried by several groups. Rodents are the most popular experimental animals that the humanized-mouse engrafted with human tissues is so far the only animal model for direct in vivo study of HCMV, but viral infection is limited in implanted human cells (Crawford et al.2015). There were two reports of low level of HCMV replication in non-human cells, which were primary chimpanzee skin fibroblasts (CF) and porcine endothelial cells, respectively (Perot et al.1992; Millard et al.2010). Unfortunately, chimpanzees are not widely available for research mainly due to ethical concern and the endangered state of this species (Bennett and Panicker 2016). The viral growth kinetics in porcine endothelial cells was not well described. Despite the limitations of these studies, they have suggested that the species specificity of HCMV is stricter in vivo than in a non-human cell culture model and that an animal with close evolutionary relationship to humans is more likely to support productive infection of HCMV.
In recent years, the genome sequence of Chinese tree shrew (Tupaia belangeri chinensis) was annotated and found to be genetically close to primates, and its genes or pathways playing a role in infectious diseases have higher identity to human (Fan et al.2013). It is a small animal and provides advantages of a small body size and lower animal maintenance costs. These advantages enable its broad application in pathological research of many human viruses including Herpes Simplex Virus 1 (Li et al.2015, 2016; Xiao et al.2017; Yao 2017). This prompted us to investigate the susceptibility of primary cells of the Chinese tree shrew to HCMV infection.
Here, we used a bacterial artificial chromosome (BAC) clone of HCMV Towne strain expressing a gfp gene in the viral genome to monitor productive replication in primary tree shrew dermis fibroblasts (TSDF) (Marchini et al.2001). The detection of expression of IE1, UL44, and pp28 genes and small amount of viral progeny suggested that TSDF was semi-permissive for HCMV infection. The TSDF model would benefit the study of the molecular mechanisms of species specificity of HCMV.
Materials and Methods
Cells and Cell Culture
Primary TSDF and primary human foreskin fibroblasts (HFF) were cultured in minimal essential medium (MEM) medium (Hyclone, Waltham, USA) and supplemented with 10% fetal bovine serum (FBS), penicillin and streptomycin (100 U/mL and 100 µg/mL, respectively, Gibco, Waltham, USA). Low serum growth supplement (Gibco) was added for TSDF. The 3–5 months old first filial-generation Chinese tree shrews used for primary cells isolation were purchased from the Institute of Medical Biology, Chinese Academy of Medical Sciences. The sex of the animal did not influence the cell isolation. Primary TSDF was isolated by enzyme digestion method (Rittie and Fisher 2005). Briefly, the Chinese tree shrew was sacrificed and the lower part of the body was soaked in 75% ethanol for 10 min. The skin samples of the inner thigh were taken and cut into strips of 1-cm width and transferred into a 100 mm tissue culture dish. Subcutaneous tissue was removed by using a pair of forceps and rinsed with washing PBS containing 100 U/mL penicillin and 100 µg/mL streptomycin (Solarbio, Beijing, China). Excised samples were chopped into pieces approximately 1 mm3 in size and digested by 0.1% type I collagenase (Gibco) at 37 °C for 45 min. The cell suspension was filtered through a sterile steel mesh (150-µm mesh) and centrifuged at 200 ×g for 5 min at room temperature. The cell pellet was resuspended in culture medium and centrifuged again. The cell pellet was resuspended in culture medium and dispensed into T25 flasks. The medium was refreshed every 2–3 days and the cells were digested (0.25% trypsin–EDTA) when at 90% confluence and expanded in T75 flasks. Cells cultural conditions were maintained at 37 °C in a humidified atmosphere containing 5% CO2. The morphology of cells was observed with Leica Microsystems (Leica, Buffalo Grove, USA).
Virus and Viral Infection
A BAC clone of HCMV Towne strain expressing a gfp gene in the viral genome was propagated in HFF with low MOI (0.02) (Marchini et al.2001). The titers of virus stock, supernatant from infecting cells, and non-entried virus control were titrated by plaque assay on monolayer of HFF as described previously (Luo et al.2007). Cells were infected with HCMV at an MOI of 1 or 0.5 without removing the inoculum and harvest at indicated times for detection of viral replication. The viruses remaining in the supernatant of TSDF at 24 hpi was incubated in fresh culture medium and maintained under the same cultural condition and used as non-entried virus control. The GFP expression was observed under DMI3000B Manual Inverted Microscope (Leica). Cells emitted green fluorescence were considered GFP positive regardless of fluorescence intensity and the percentage of GFP-positive cells at indicated time was calculated by counting 1000 cells from randomly selected 3 fields.
DNA Isolation and Quantitative PCR Assay
DNA was isolated from cells with TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). Representative fragment of HCMV genome was amplified with 50 ng DNA and primers specific to ul83 (Forword -5′GCGAGACCGTGGAACTGC 3′, Reverse -5′ TTGCCCTGGATGCGATACTG 3′) by using ChamQ SYBR q-PCR Master Mix Kit (Vazyme, Nanjing, China) and followed by sequencing confirmation. The qPCR was performed as follow: 1 cycle of 5 min at 95 °C; 40 cycles of 10 s at 95 °C and 34 s at 60 °C; 1 cycle of 15 s at 95 °C, 60 s at 60 °C, and 15 s at 95 °C. The plasmid pcDNA3.0-UL83 was used to generate a standard curve, with which the copy number of HCMV genome in different cells was subsequently calculated.
Western Blot
Cells were collected and washed with PBS (Gibco). After centrifuge at 10,000 ×g, samples were added with 100 μL RIPA lysis buffer and incubated on ice for 1 h for total protein preparation. For each sample, 20 μL total protein quantified with BCA Protein Assay Kit (TaKaRa, Kusatsu, Japan) was used for protein separation with SDS-PAGE (Bio-Rad, Hercules, USA). After the protein was transferred to a polyvinylidene fluoride membrane (Milipore, Darmstadt, Germany), the membrane was blocked in PBS dissolved milk powder, followed by incubation with primary and secondary antibodies. Mouse monoclonal antibodies against IE1 (Thermo Fisher), UL44 (Abcam, Cambridge, USA), and pp28 (Abcam) were 1:1000 diluted, and mouse monoclonal antibodies against β-actin were 1:5000 diluted (Abcam). Horseradish peroxidase (HRP)-goat anti-mouse IgG (Abcam) was 1:5000 diluted. The membranes were treated with enhanced chemiluminescence reagents (GE, Fairfield, USA) and analyzed by using Imaging system (Bio-Rad).
Immunofluorescence Assay (IFA)
Cells were cultured in 10-mm dishes containing coverslips. At indicated times, the coverslips were rinsed with PBS, fixed with 4% formaldehyde, permeabilized with 1% Triton-100, blocked with corresponding antibodies, and counterstained with 1 μg/mL DAPI (Roche, Basel, Switzerland). For detection of expression of vimentin, TSDF-containing coverslips were first incubated with rabbit monoclonal antibody against human vimentin (1:1000 diluted, Abcam) and mouse monoclonal antibody against β-actin (1:1000 diluted, Abcam) for 2 h, then with FITC-conjugated goat polyclonal antibodies against rabbit IgG (1:500 diluted, Abcam) and rhodamine-conjugated goat polyclonal antibodies against mouse IgG (1:200, Thermo Fisher). The cells were visualized under DMI3000B Manual Inverted Microscope and image quantification was carried out using NIS-Elements F 4.00.00 software (Leica).
Statistical Statement
Statistical analyses were performed with GraphPad Prism 5 (GraphPad Software, San Diego, USA), using Student’s t test or Analysis of Variance (ANOVA) test. All data shown were as the representative results or the average from at least three independent experiments. The average results were shown as the mean ± standard deviation (SD). Significance was considered when P < 0.05.
Results
Isolation and Characterization of Primary Tree Shrew Dermis Fibroblasts
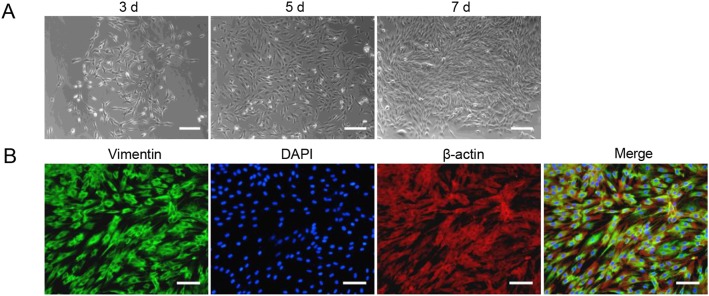
Given the reported permissiveness of primary skin fibroblasts from chimpanzees for HCMV infection and because of the close relationship to primates, primary TSDF were isolated from the groin of the Chinese tree shrews. After mechanical and enzymatic dissociation, the cells attached and spread on the plate, and gradually became confluent. Most cells had a uniform spindle shape and manifested a parallel arrangement when they were grown at higher density, the morphology of which was similar to that observed with fibroblasts (Fig. 1A). The expression of vimentin, the marker for fibroblasts (Franke et al.1978), was observed in most TSDF (green fluorescence) and co-localized with β-actin (red fluorescence) in cytoplasm (Fig. 1B), which was consistent with characteristics of fibroblasts. Therefore, we confirmed the characteristics of primary TSDF in culture.
Fig. 1.
Isolation and characterization of TSDF. A The morphology of TSDFs cultured for 3, 5 and 7 days, respectively. B Vimentin expressions in TSDF. The green fluorescence showed vimentin, β-actin marked cytoplasm (red fluorescence), and DAPI marked nuclear (blue fluorescence). Bar = 100 μm.
GFP Expression and CPE in TSDF Infected with HCMV
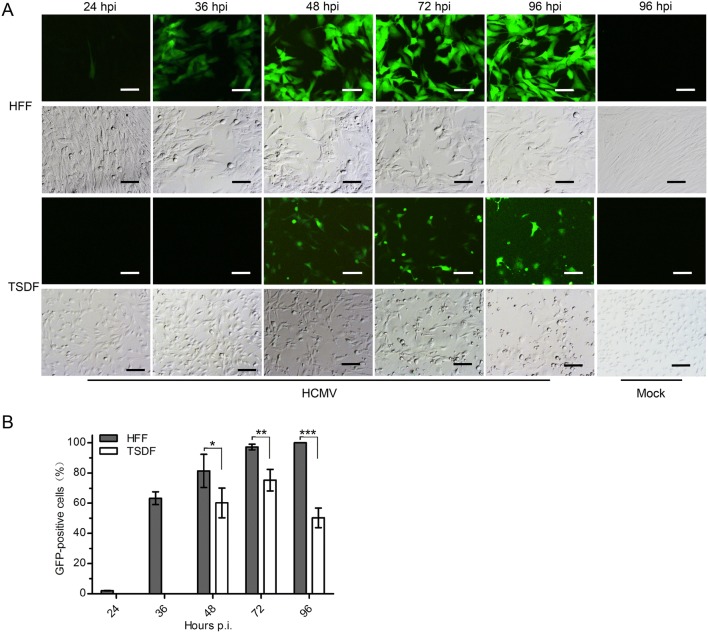
To initially screen for the susceptibility to HCMV infection, primary TSDF was infected at an MOI of 1 with GFP-expressing HCMV which is a convenient model for monitoring lytic replication (Marchini et al.2001; Cheng et al.2017). Both GFP expression and cytopathic effect (CPE) were observed at 24, 36, 48, 72, 96 h post infection (hpi) (Fig. 2). HFF was used as a positive control. In HFF, weak expression of green fluorescence was detected at 24 hpi and more than half cells were GFP positive from 36 hpi. The GFP-positive HFF was close to 100% at 96 hpi. In TSDF, GFP expression was delayed and weak green fluorescence appeared at 36 hpi. The maximum rate of GFP-positive TSDF was 70% at 72 hpi, and then the rate decreased to 50% at 96 hpi. The overall GFP intensity was much stronger in HFF than that in TSDF. The typical HCMV-induced CPE, including cell rounding and enlargement, were presented in HCMV-infected HFF, and infected TSDF showed cell rounding, enlargement as well as cell detachment at late infection period. However, in mock-infected cells, no visible CPE was observed. The expression of GFP indicated that HCMV entry and initiation of viral replication occurred in TSDF.
Fig. 2.
HCMV infection induced GFP expression and CPE in TSDF. A The expression of GFP and CPE were observed in HCMV-infected HFF and TSDF at indicated times. Mock-infected cells at 96 hpi were shown as negative controls. Bar = 100 μm. B The percentage of GFP-positive cells was calculated respectively for HFF and TSDF. * indicated P < 0.05, ** indicated P < 0.01, *** indicated P < 0.001.
TSDF Supported 3 Kinetic Classes of Genes Expression and Genome Replication of HCMV
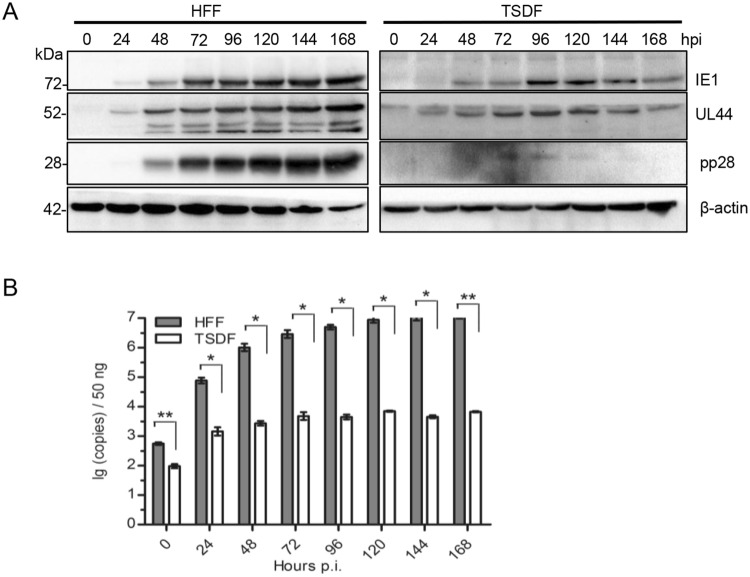
Previous studies showed that HCMV cross-infection failure was due to a blockage at post-entry stage that the expression of three kinetic classes of genes and DNA replication could not be achieved (DeMarchi 1983; Lafemina and Hayward 1988; Garcia-Ramirez et al.2001). In order to determine whether TSDF supported viral gene expression, TSDF and HFF were infected at an MOI of 0.5. The expressions of IE1, UL44, pp28, the representative proteins of IE, E and L phase, were detected by Western blot with β-actin as a reference control. HCMV genome copy number was analyzed by quantitative PCR (qPCR). As shown in Fig. 3A, these three viral genes expressed in HFF and the expression level increased over time. In TSDF, the expression of IE1 and UL44 was increased from 24 hpi onward and low level expression of pp28 was detected from 72 to 120 hpi. It should be noted that the late gene expression was barely detected in cross-species infection of cytomegaloviruses, even the expression of late protein gB of murine cytomegalovirus which has broader host range than HCMV was not detectable in human retinal pigment epithelial cells (Jurak and Brune 2006). The ability of TSDF supporting expression of the three viral genes, especially pp28, suggested that the susceptibility of this cell to HCMV infection is different from non-permissive cells. The genome replication level was quantified with ul83 primers, and the PCR fragments were confirmed by sequencing (Fig. 3B). In HFF, the viral DNA copy number increased from 102 to 107 per 50 ng total DNA from 0 to 168 hpi. In TSDF, the viral DNA copy number was 102 per 50 ng total DNA and reached 103 per 50 ng total DNA at 168 hpi. At 72 and 96 hpi, the viral genome copy number in TSDF was greater than at 48 hpi and the similar variation tendency was found in the expression intensity of the three viral proteins. Together, these results indicated a possibility of low level HCMV replication in TSDF.
Fig. 3.
TSDF supported the expression of 3 kinetic classes of genes and genome replication of HCMV. A The expression of IE1, UL44, pp28, and β-actin was detected by western blot. B The genome replication of HCMV was analyzed by qPCR with ul83 primers and the detection limit of the standard curve was 150 copies per 50 ng DNA.
TSDF Supported Production of Infectious Progeny Virus Which Can Infect HFF
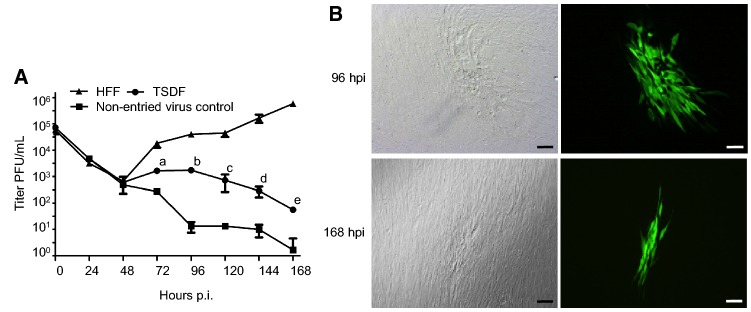
Besides viral protein synthesis, the production of cell-free viruses is of great importance to determine the permissiveness to HCMV infection. We next sought to investigate whether infectious progeny viruses could be produced in TSDF. The growth curve of HCMV in HFF and TSDF was determined by plaque forming assay of culture supernatants on fresh HFF monolayers (Fig. 4). Furthermore, in order to confirm whether progeny cell-free viruses were actually produced in TSDF, a non-entried virus control was conducted (Fig. 4A). The same amount of viruses remaining in the supernatant at 24 hpi was incubated to fresh medium and kept at the same culture condition. Since the virus could not replicate in cell culture medium, the newly produced viral particles would be identified by comparing the titer difference between in TSDF and in non-entried virus control. The GFP expression and plaques were detected in HFF inoculated with supernatant from infected TSDF (Fig. 4B), but it could not exclude the influence of residual input viruses. In non-entried virus control, the titer was detectable and kept decreasing from 24 hpi. However, it is interesting to find that both in HFF and TSDF the cell-free viruses decreased to similar amount from 24 to 48 hpi, but it began to increase from 72 hpi. The amount of cell-free viruses reached the highest level in TSDF at 96 hpi, which was 103 PFU/mL, and in HFF it reached 105 PFU/mL at 168 hpi. The titer in TSDF gradually decreased after 96 hpi, but it was significantly higher than in non-entried virus control at 72, 96, 120, 144, and 168 hpi (P < 0.05). At 168 hpi, there were almost no infectious viruses in the control while there were 5.59 ± 0.58 × 10 PFU/mL of cell-free viruses in TSDF. The decrease of amount of cell-free viruses in TSDF was consistent with cell detachment and decrease of pp28 expression level, which suggested the viral replication was somehow affected in late infection period. Based on the requirement of 48–72 h to complete one replication cycle of HCMV, the growth curve showed the achievement of at least one cycle of viral replication in TSDF. Taken together, our results confirmed productive replication of HCMV in TSDF and suggested the semi-permissiveness of this cell for HCMV infection.
Fig. 4.
TSDF supports production of infectious progeny virus which can infect HFF. A Growth kinetic of HCMV on HFF and TSDF was determined. A non-entried virus control was carried out to differentiate newly produced virus from residual input virus. The letters a, b, c, d, and e indicated the titer differences between TSDF and non-entried virus control at 72, 96, 120, 144, and 168 hpi, respectively, the P value of which were all less than 0.05. B The formation of plaques and expression of GFP were observed in HFF infected with supernatant from TSDF. Bar = 100 μm.
Discussion
The lack of an experimental animal model hampers studies on pathogenesis of HCMV infection, which is an important pathogen leading to neonatal defects and opportunistic infections in immune-compromised patents. A non-human cell model supporting replication of HCMV may be useful to study its strict species specificity. Previous reports implied that an animal with close evolutionary relationship to human such as the chimpanzee has a potential susceptibility to HCMV infection (Perot et al.1992). The Chinese tree shrew is genetically close to primates and has proved useful for pathological research of several human viruses (Yao 2017). Hence, we investigated the permissiveness of primary TSDF for HCMV infection. By detection of CPE, GFP expression, viral gene expression, viral genome replication, and cell-free viruses release, we showed that TSDF supported low level of HCMV replication. Based on synthesis of viral protein IE1, UL44 and pp28 and small amount of viral particle release, TSDF was suggested to be semi-permissive for HCMV infection.
Previous study of cross-species infection of CMVs gave us a hint that it is important to choose proper cell types and strains to establish a non-human cell model for this viral infection. In human, primary fibroblasts with skin or lung origin are mostly used to study in vitro HCMV infection because they are one of the main target cell types, ease culture, permissive for all strains, and support the production of high titer of cell-free virus (Fortunato 2014). Towne strain, one of the mostly used laboratory strain, has been reported to be permissive for HCMV infection of CF (Perot et al.1992). Therefore, TSDF was selected to be infected with a Towne strain variant expressing a gfp gene in the viral genome (Marchini et al.2001). The key factor to answer whether TSDF supported HCMV infection is the detection of the production of infectious progeny viruses. We found that the expression of pp28 which is dependent of viral DNA synthesis and essential for the production of infectious virus was detectable from 72 to 120 hpi. The amount of cell-free viruses kept increasing from 72 to 96 hpi and was significantly higher than in non-entried virus control from 72 to 168 hpi. Furthermore, we infected mouse embryonic fibroblasts (MEF) with HCMV Towne but could not detect UL44 expression by western blot (data not shown), which is consistent with reported non-permissiveness of MEF for this viral infection (Lafemina and Hayward 1988). We therefore demonstrated successful cross-species infection in TSDF. Meanwhile, compared with TSDF, we indeed observed different susceptibility of primary fibroblasts from lung (TSLF) and epithelial cells from kidney (TSKE) from the Chinese tree shrew to HCMV. The IE1 expression detected by IFA was less efficient in TSLF than in TSDF and no expression could be detected in TSKE (Supplementary Fig. S1). So far, TSDF is easy to be cultured and supports complete replication cycle of HCMV that this cell model would be useful for study cross-species infection of this virus.
The species specificity of HCMV is the result of co-evolution with human host that the interplay between the virus and the Chinese tree shrew would affect its cross-species infection. Although the mechanism of cross-species barrier of HCMV, the typical member of beta-herpesviruses, is still unknown, studies on other cytomegaloviruses (CMVs) have implied that multiple factors play a part in this process (Jurak and Brune 2006; Tang and Maul 2006; Schumacher et al.2010; Marsh et al.2015; Ostermann et al.2015; Burwitz et al.2016). UL128 and UL130 which are the subunit of pentameric complex in the viral envelope helped rhesus macaques CMV (RhCMV) infect cynomolgus macaques both in vitro and in vivo (Burwitz et al.2016). Since complete pentameric complex is necessary for infection of epithelial and endothelial cells, it suggested that cross-species infection would benefit from broadening the cell types a virus entering. There are various HCMV strains which could be classified as laboratory strains, such as Towne, and clinical strains. The Towne strains was highly adapted to fibroblasts and could produce high titer of cell-free viruses, while clinical strains have extended tropism and are able to infect epithelial and endothelial cells besides fibroblasts (Frascaroli and Sinzger 2014). Whether clinical strains of HCMV could productively infect TSDF and other cell types from the Chinese tree shrew should be investigated in future studies. Host factors are also involved in anti-viral response, including apoptosis and ND10 complex, and are reported to influence cross-species infection of CMVs (Jurak and Brune 2006; Tang and Maul 2006; Burwitz et al.2016). In this study, we observed cell detachment in HCMV-infected TSDF at late infection period. It is unclear what factors caused cell death and whether it has effects on virus multiplication. Fortunately, the annotated genome sequence of the Chinese tree shrew is available (Fan et al.2013), and this is helpful for further using TSDF model to exam factors that improve production of virus particle and relieve cell death and providing perspectives on mechanism of species specificity of HCMV.
In conclusion, the data of the present study indicated that TSDF was semi-permissive for HCMV infection. This TSDF model could be further used to investigate key factors influencing cross-species multiplication of HCMV and give us insights into the molecular mechanism of this process. Meanwhile, the characteristics of this viral infection of other types of cells from the Chinese tree shrew should be further studied to explore the utility of this animal for HCMV infection in vivo.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Key Research and Development Project of China (2017YFC1309302), the Applied Basic Research Projects of Yunnan Province (2018FD034), and the Yunnan Provincial Department of Education Science Research Fund Project (1405189906).
Author Contributions
SWD conceived the experiments. SWD and LSJ designed the experimental flow. LSJ, MY, YLD, and YBC performed the experiments. LSJ performed statistical analyses. SWD wrote the manuscript. FZ, AMZ, and LL reviewed the manuscript. MHL and XSX supervised the project. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare no conflict of interest.
Animal and Human Rights Statement
All experiments and protocols in this study were reviewed and approved by the Institutional Ethical Committee of Kunming University of Science and Technology.
Contributor Information
Min-Hua Luo, Phone: +86-27-87197600, Email: luomh@wh.iov.cn.
Xue-Shan Xia, Phone: +86-871-6592-0756, Email: xueshanxia@kmust.edu.cn.
References
- Bennett AJ, Panicker S. Broader impacts: international implications and integrative ethical consideration of policy decisions about US chimpanzee research. Am J Primatol. 2016;78:1282–1303. doi: 10.1002/ajp.22582. [DOI] [PubMed] [Google Scholar]
- Burwitz BJ, Malouli D, Bimber BN, Reed JS, Ventura AB, Hancock MH, Uebelhoer LS, Bhusari A, Hammond KB, Espinosa Trethewy RG, Klug A, Legasse AW, Axthelm MK, Nelson JA, Park BS, Streblow DN, Hansen SG, Picker LJ, Fruh K, Sacha JB. Cross-species rhesus cytomegalovirus infection of cynomolgus macaques. PLoS Pathog. 2016;12:e1006014. doi: 10.1371/journal.ppat.1006014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Jiang X, Yang B, Wen L, Zhao F, Zeng WB, Liu XJ, Dong X, Sun JY, Ming YZ, Zhu H, Rayner S, Tang Q, Fortunato E, Luo MH. Infected T98G glioblastoma cells support human cytomegalovirus reactivation from latency. Virology. 2017;510:205–215. doi: 10.1016/j.virol.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford LB, Streblow DN, Hakki M, Nelson JA, Caposio P. Humanized mouse models of human cytomegalovirus infection. Curr Opin Virol. 2015;13:86–92. doi: 10.1016/j.coviro.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarchi JM. Nature of the block in the expression of some early virus genes in cells abortively infected with human cytomegalovirus. Virology. 1983;129:287–297. doi: 10.1016/0042-6822(83)90168-X. [DOI] [PubMed] [Google Scholar]
- Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, He J, Hou HL, Hu L, Hu XT, Jiang XT, Lai R, Lang YS, Liang B, Liao SG, Mu D, Ma YY, Niu YY, Sun XQ, Xia JQ, Xiao J, Xiong ZQ, Xu L, Yang L, Zhang Y, Zhao W, Zhao XD, Zheng YT, Zhou JM, Zhu YB, Zhang GJ, Wang J, Yao YG. Genome of the Chinese tree shrew. Nat Commun. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- Fioretti A, Furukawa T, Santoli D, Plotkin SA. Nonproductive infection of guinea pig cells with human cytomegalovirus. J Virol. 1973;11:998–1003. doi: 10.1128/jvi.11.6.998-1003.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato EA. Use of diploid human fibroblasts as a model system to culture, grow, and study human cytomegalovirus infection. Methods Mol Biol. 2014;1119:47–57. doi: 10.1007/978-1-62703-788-4_4. [DOI] [PubMed] [Google Scholar]
- Franke WW, Schmid E, Osborn M, Weber K. Different intermediate-sized filaments distinguished by immunofluorescence microscopy. Proc Natl Acad Sci U S A. 1978;75:5034–5038. doi: 10.1073/pnas.75.10.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frascaroli G, Sinzger C. Distinct properties of human cytomegalovirus strains and the appropriate choice of strains for particular studies. Methods Mol Biol. 2014;1119:29–46. doi: 10.1007/978-1-62703-788-4_3. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez JJ, Ruchti F, Huang H, Simmen K, Angulo A, Ghazal P. Dominance of virus over host factors in cross-species activation of human cytomegalovirus early gene expression. J Virol. 2001;75:26–35. doi: 10.1128/JVI.75.1.26-35.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak I, Brune W. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 2006;25:2634–2642. doi: 10.1038/sj.emboj.7601133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Carp RI. Abortive infection of human diploid cells by murine cytomegalovirus. Infect Immun. 1972;6:793–797. doi: 10.1128/iai.6.5.793-797.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafemina RL, Hayward GS. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol. 1988;69(Pt 2):355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- Landolfo S, Gariglio M, Gribaudo G, Lembo D. The human cytomegalovirus. Pharmacol Ther. 2003;98:269–297. doi: 10.1016/S0163-7258(03)00034-2. [DOI] [PubMed] [Google Scholar]
- Li L, Li Z, Wang E, Yang R, Xiao Y, Han H, Lang F, Li X, Xia Y, Gao F, Li Q, Fraser NW, Zhou J. Herpes simplex virus 1 infection of tree shrews differs from that of mice in the severity of acute infection and viral transcription in the peripheral nervous system. J Virol. 2015;90:790–804. doi: 10.1128/JVI.02258-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li Z, Wang E, Yang R, Xiao Y, Han H, Lang F, Li X, Xia Y, Gao F, Li Q, Fraser NW, Zhou J. Herpes simplex virus 1 infection of tree shrews differs from that of mice in the severity of acute infection and viral transcription in the peripheral nervous system. J Virol. 2016;90:790–804. doi: 10.1128/JVI.02258-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MH, Fortunato EA. Long-term infection and shedding of human cytomegalovirus in T98G glioblastoma cells. J Virol. 2007;81:10424–10436. doi: 10.1128/JVI.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MH, Rosenke K, Czornak K, Fortunato EA. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J Virol. 2007;81:1934–1950. doi: 10.1128/JVI.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini A, Liu H, Zhu H. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol. 2001;75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh AK, Ambagala AP, Perciani CT, Russell JN, Chan JK, Janes M, Antony JM, Pilon R, Sandstrom P, Willer DO, MacDonald KS. Examining the species-specificity of rhesus macaque cytomegalovirus (RhCMV) in cynomolgus macaques. PLoS ONE. 2015;10:e0121339. doi: 10.1371/journal.pone.0121339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard AL, Haberli L, Sinzger C, Ghielmetti M, Schneider MK, Bossart W, Seebach JD, Mueller NJ. Efficiency of porcine endothelial cell infection with human cytomegalovirus depends on both virus tropism and endothelial cell vascular origin. Xenotransplantation. 2010;17:274–287. doi: 10.1111/j.1399-3089.2010.00594.x. [DOI] [PubMed] [Google Scholar]
- Ostermann E, Pawletko K, Indenbirken D, Schumacher U, Brune W. Stepwise adaptation of murine cytomegalovirus to cells of a foreign host for identification of host range determinants. Med Microbiol Immunol. 2015;204:461–469. doi: 10.1007/s00430-015-0400-7. [DOI] [PubMed] [Google Scholar]
- Perot K, Walker CM, Spaete RR. Primary chimpanzee skin fibroblast cells are fully permissive for human cytomegalovirus replication. J Gen Virol. 1992;73(Pt 12):3281–3284. doi: 10.1099/0022-1317-73-12-3281. [DOI] [PubMed] [Google Scholar]
- Rittie L, Fisher GJ. Isolation and culture of skin fibroblasts. Methods Mol Med. 2005;117:83–98. doi: 10.1385/1-59259-940-0:083. [DOI] [PubMed] [Google Scholar]
- Schumacher U, Handke W, Jurak I, Brune W. Mutations in the M112/M113-coding region facilitate murine cytomegalovirus replication in human cells. J Virol. 2010;84:7994–8006. doi: 10.1128/JVI.02624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Maul GG. Mouse cytomegalovirus crosses the species barrier with help from a few human cytomegalovirus proteins. J Virol. 2006;80:7510–7521. doi: 10.1128/JVI.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathen MW, Stinski MF. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathen MW, Thomsen DR, Stinski MF. Temporal regulation of human cytomegalovirus transcription at immediate early and early times after infection. J Virol. 1981;38:446–459. doi: 10.1128/jvi.38.2.446-459.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Liu R, Chen CS. Tree shrew (Tupaia belangeri) as a novel laboratory disease animal model. Zool Res. 2017;38:127–137. doi: 10.24272/j.issn.2095-8137.2017.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG. Creating animal models, why not use the Chinese tree shrew (Tupaia belangeri chinensis)? Zool Res. 2017;38:118–126. doi: 10.24272/j.issn.2095-8137.2017.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.