Abstract
Possible multidrug-resistant (MDR) mechanisms of four resistant strains of Escherichia coli to a model β-lactam, ampicillin, were investigated using contact angle measurements of wettability, crystal violet assays of permeability, biofilm formation, fluorescence imaging, and nanoscale analyses of dimensions, adherence, and roughness. Upon exposure to ampicillin, one of the resistant strains, E. coli A5, changed its phenotype from elliptical to spherical, maintained its roughness and biofilm formation abilities, decreased its length and surface area, maintained its cell wall integrity, increased its hydrophobicity, and decreased its nanoscale adhesion to a model surface of silicon nitride. Such modifications are suggested to allow these cells to conserve energy during metabolic dormancy. In comparison, resistant strains E. coli D4, A9, and H5 elongated their cells, increased their roughness, increased their nanoscale adhesion forces, became more hydrophilic, and increased their biofilm formation upon exposure to ampicillin. These results suggest that these strains resisted ampicillin through biofilm formation that possibly introduces diffusion limitations to antibiotics. Investigations of how MDR bacterial cells modify their surfaces in response to antibiotics can guide research efforts aimed at designing more effective antibiotics and new treatment strategies for MDR bacterial infections.
Keywords: adhesion force, AFM, antibiotics, β-lactam, biofilms, dimensions, E. coli, multidrug-resistance (MDR), permeability, roughness
Introduction
The world is intensely facing the growing health threat of infections caused by multidrug-resistant (MDR) microbes (Solomon & Oliver, 2014; Mohammad et al., 2015). Patients with such infections require more complicated treatment plans, because of the ability of bacterial cells to resist common antibiotics, longer hospital stays, and may die at a significantly higher rate compared with those who experience other infections (Wong et al., 2000; Livermore, 2004; Bonomo, 2018). During the last decade, global awareness of the emergence of MDR bacteria has risen but new MDR strains have increased as well (Solomon & Oliver, 2014; Rossolini et al., 2014). The latter is attributed largely to the increasing overuse and abuse of antibiotics in human and animal medicine (Vashishtha, 2010; Luyt et al., 2014; O’Brien, 2015). A report from the United States Centers for Disease Control and Prevention (CDC) in 2013 estimated that at least two million people get antibiotic-resistant infections yearly and more than 23,000 people die because of such infections (Mohammad et al., 2015).
β-Lactams represent a group of antibiotics that have been extensively used against most common bacterial infections (Kennedy et al, 1963; Coleman, 2011; Worthington & Melander, 2013; Zeng & Lin, 2013). Due to their widespread use, Gram-negative bacteria such as Escherichia coli developed complex means to resist them (Shaikh et al., 2015). Among these mechanisms are the modification of antibiotic target sites on bacterial surfaces, inactivation of antibiotic rings by β-lactamase, increase in cell wall impermeability to antibiotics, development of efflux pump at the bacterial cell membrane, and biofilm formation (Aeschlimann, 2003; Babic & Bonomo, 2006; Mohanty et al., 2012; Read & Woods, 2014). Because of the complex and interdependent mechanisms employed by bacterial cells to resist antibiotics, the treatment of MDR infections is still largely problematic and the emergence of new resistant bacterial strains to antibiotics is on the rise (Arias, 2016; Hawkey et al., 2018).
Peptidoglycans (PGs) in the inner cell membrane are the major target for β-lactams and their modification contributes to MDR (Ghuysen, 1994; Vollmer & Seligman, 2010). Upon binding to penicillin-binding proteins (PBPs), β-lactams inhibit the ability of PBPs to synthesize PGs (Jacoby & Medeiros, 1991; Dong et al., 2004). In addition, bacterial cells may utilize their negatively charged lipopolysaccharides (LPS), which decorate bacterial cells outer membranes, to provide a repulsion barrier to the diffusion of the hydrophilic β-lactams into the cell (Pagès & Winterhalter, 2008). Furthermore, hydrophobic membrane transporters, such as porins, present on the outer cellular membrane, regulate the traffic of hydrophilic molecules across the cell membrane (Pagès & Winterhalter, 2008; Delcour, 2009).
Many studies in the literature attempted to address the question of how Gram-negative bacterial cells such as E. coli resist antibiotics (Rology et al., 1993; Allen et al., 2010; Chroma & Kolar, 2010; Kumarasamy et al., 2010; Ali et al., 2014). The approach used in these studies was largely macroscopic. In some studies, genotypic methods were used to identify specific genes that confer antibiotic resistance in bacteria (Rology et al., 1993; Allen et al., 2010; Chroma & Kolar, 2010; Kumarasamy et al., 2010; Ali et al., 2014). Although interesting, genomic methods can be time and labor intensive and require prior knowledge of genes that confer MDR in bacteria (Shaikh et al., 2015). In other studies, physical examination of color changes in paper disks impregnated with substances that change color upon hydrolysis of antibiotics by β-lactamase enzymes produced by bacteria was used (Chroma & Kolar, 2010). An example of the latter is nitrocefin disks, which are composed of a chromogenic cephalosporin that changes color from yellow to red when the amide bond in the β-lactam ring is hydrolyzed by β-lactamase produced by bacteria (Brown et al., 2012). While the latter studies quantify the resistance of bacterial cells to antibiotics, they fail to address the fundamental means utilized by bacteria to resist antibiotics, such as changes that occur on the bacterial surface during treatment with antibiotics. To combat bacterial resistance to antibiotics, we need to improve our fundamental understanding of how bacterial cells develop such resistance and not just quantify the resistance.
In comparison to the macroscopic means described above, atomic force microscopy (AFM) offers an interesting means through which bacterial cellular properties and interactions with surfaces can be investigated in response to antibiotics. AFM is unique in its ability to probe individual cells for their nanoscale morphological features (Braga & Ricci 1998; Perry et al., 2009), mechanical properties (Perry et al., 2009; Longo et al., 2013), roughness (Alves et al., 2010), length and density of their surface biopolymers (Abu-Lail & Camesano 2003), and interactive forces to the variable surface under liquid media (Abu-Lail & Camesano 2003). AFM is also interesting in its ability to image bacterial cells and characterize their properties in parallel, in real-time, and under physiological conditions (Abu-Lail & Camesano 2003; Perry et al., 2009; Alves et al., 2010; Longo et al., 2013). This allows for structure-function relationships to be detailed under variable treatments. Furthermore, AFM requires minimal sample preparation, which is an advantage over multiple sample preparations used in electron microscopy (Golding et al., 2016) and bio-chemical analytical techniques (Chroma & Kolar, 2010a). However, AFM, as a technique, is limited to probing only surfaces and its analyses can be time-consuming. As such, it can only characterize the effects of given treatments on the bacterial surface properties and interactions. With that in mind, AFM studies can complement macroscopic studies in providing detailed mechanisms on how bacterial cells develop MDR. Here, we choose to utilize AFM to study the effects of β-lactams on bacterial surface properties and interactions. Not only that β-lactams are the most widely used class of antibiotics to treat bacterial infections (Shaikh et al., 2015), they interfere with cell metabolism leading to a decrease in the rate of cellular growth and impediment in cell wall synthesis of macromolecules (Braga & Ricci 1998; Longo et al., 2013). Such changes in the bacterial cell wall and physical properties can be measured in real-time using AFM techniques (Braga & Ricci 1998; Meincken et al., 2005; Chen et al., 2009; Perry et al., 2009; Longo et al., 2013).
Because of the above, the effects of β-lactams on bacterial cells have been extensively investigated by AFM (Braga & Ricci 1998; Yang et al., 2006; Perry et al., 2009; Longo et al., 2013). Most of these nanoscale studies though, focused mainly on the ability of AFM to quantify morphological changes induced by β-lactam antibiotics in susceptible bacteria via detailed imaging of the membranes of bacterial cells (Braga & Ricci 1998; Yang et al., 2006). Several forms of membrane damage of susceptible bacteria, ranging from the development of nanopores at the apex of the cell, the collapse of the bacterial cell wall and the loss of cellular content to actual cell death induced by β-lactams antibiotics, have been reported (Braga & Ricci 1998; Perry et al., 2009). Although these studies are interesting, they did not probe the changes in bacterial cell walls in a liquid environment as well as they only investigated susceptible cells and not resistant cells (Braga & Ricci 1998; Yang et al., 2006; Perry et al., 2009; Longo et al., 2013). Studies that probed nanoscale adhesion, cell elasticity, steric, hydrophobic interactions, membrane permeability, and biofilm formation of MDR bacteria as they get exposed to antibiotics or natural agents are limited in the literature.
Motivated by the limitations of current nanoscale and macroscale studies which investigated how bacterial cells respond to β-lactams, we focus on investigating how the MDR-E. coli resist ampicillin, a model β-lactam, using a multiscale approach. We hypothesize that upon exposure to ampicillin, cells will manipulate their roughness, morphology, hydrophobicity, membrane permeability, adhesion forces as well as their abilities to form biofilms to resist antibiotics. To validate our hypothesis, AFM was used to quantify the dimensions of the individual cells, the roughness of cellular surfaces and the nanoscale adhesion forces acting between the cell surface molecules of four-model MDR-E. coli strains and a model silicon nitride (Si3N4) cantilever in water for cells grown in the presence or absence of ampicillin. Furthermore, macroscopic assays that included membrane permeability, contact angle measurements of wettability, and biofilm formation were performed to provide insights into the role bacterial membranes play in how cells develop resistance to antibiotics.
Materials and Methods
Chemicals
Chemicals used in the present study were ampicillin (Sigma-Aldrich, St. Louis, MO, USA), Luria-Bertani (LB) broth/agar (RPICorp, Mount Prospect, IL), crystal violet (CV; Sigma-Aldrich, St. Louis, MO, USA), Syto 9 green fluorescent dye (Invitrogen Corp., Carlsbad, CA, USA), phenol red (Sigma-Aldrich, St. Louis, MO, USA), gelatin G2500–100G (Sigma-Aldrich, St. Louis, MO, USA), and Bacto agar (Difco, Detroit, Michigan, USA).
Bacterial Strains
Four domestic wild-type MDR Gram-negative E. coli strains were used here. These strains were obtained from Prof. Douglas R. Call of the Paul G. Allen School of Global Animal Health, Washington State University. All strains were collected from households in Tanzania under which they were exposed to water and were given the code names A5, D4, A9, and H5. These strains varied in their abilities to form biofilms and resist antibiotics including ampicillin.
Choice of β-Lactam and Model Surfaces
Ampicillin was chosen as the model β-lactam antibiotic to investigate because it is the most commonly used β-lactam against E. coli strains, the target bacteria investigated here (Kennedy & Murdoch, 1963; WHO, 2015). Furthermore, because AFM is a surface characterization technique, it can reveal the molecular dynamics of the bacterial cell surface undergoing a treatment with ampicillin (Braga & Ricci 1998; Yang et al., 2006; Perry et al., 2009; Longo et al., 2013). AFM measurements were carried out on cells using the most commonly available cantilevers, Si3N4. These cantilevers provide a base model to which bacterial adhesion can be quantified (Abu-Lail & Camesano 2003; 2012; Gordesli & Abu-Lail, 2012), as well as differences in physiochemical properties of bacterial cells such as roughness (Alves et al., 2010). Dimensions (Yang et al., 2006) can be compared across investigated strains in response to exposure to ampicillin. All measurements were done in water to maintain the electrostatic interactions between the positively charged surface and the negatively charged bacterial surface biopolymers (Lee et al., 2017) and to reflect the solvent to which bacterial cells were exposed to upon collection.
Determination of Bacterial Minimum Inhibitory Concentration
The MDR of the bacterial strains to standardized known concentrations of antibiotics was characterized by Prof. Call’s group using Mueller Hinton agar (Jenkins & Schuetz, 2012). The antibiotics used were ampicillin (32 μg/mL), amoxicillin (30 μg/mL), tetracycline (16 μg/mL), sulfonamide (512 μg/mL), and trimethoprim (8 μg/mL). The four E. coli strains selected for this experiment were all ampicillin resistant. Minimum inhibitory concentration (MICs) were determined in planktonic cultures using a broth dilution test as established in the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2017). Briefly, three colonies of each strain were taken from the LB plates, inoculated into 5 mL LB broth and incubated for 24 h at 37 °C with shaking at 150 rpm. Culture tubes containing ampicillin ranging between 0.2 and 400 μg/mL in LB media were prepared. The test was performed in 1:2 dilutions of ampicillin and each concentration was tested in a triplicate. The medium with ampicillin was then inoculated with a 100 μL of MDR-E. coli cells grown until the late exponential phase of growth as [optical density (OD)600 nm = 0.5]. After an overnight incubation, turbidity in the test tubes was used as an evidence of visible cell growth. The lowest concentration of the ampicillin needed to prevent cell growth was used as the MIC for subsequent experiments (CLSI, 2017). To verify the MIC obtained, a 100 μL of overnight cell suspension from all tested concentrations, including the identified MIC, were transferred into new tubes containing fresh LB medium without ampicillin (n = 3). All tubes that have ampicillin above MIC are expected not to regrow, while others containing concentrations below MIC will have some turbidity as evidence of growth. Indeed, the above was true. Strains A5 and H5 had a MIC of 50 μg/mL while strains D4 and A9 had a MIC of 45 μg/mL.
Determination of Minimum Biofilm Inhibitory Concentrations
The minimum biofilm inhibitory concentration (MBIC) is the concentration of an antimicrobial agent required to inhibit the growth of bacteria in a biofilm (Pickering et al., 2003). To determine MBIC, the protocol referenced in (Pantanella et al., 2008) and utilized the pH indicator phenol red was slightly modified. To prepare the LB-phenol red medium, a 25 mg of phenol red and a 20 g of LB broth were dissolved in a 1,000 mL of deionized (DI) water and the pH was adjusted to 7.2 (Pantanella et al., 2008). The final solution was sterilized at 121 °C, allowed to cool to room temperature and the final solution appeared clear red. The assay assumes that live cells are metabolically active and release acidic metabolites in the medium as they grow to form a biofilm. The buildup of acidic metabolites results in a drop in pH of the LB-phenol red medium, which is measured by a color change of the medium from red to yellow (Welch & Strømme, 2012). To prepare the biofilms to be tested for MBIC, MDR-E. coli strains were cultured in a polystyrene microtiter 96-well plate for 24 h at 37 °C without shaking (Pantanella et al., 2008; Welch & Strømme, 2012). Afterward, the colonized wells were washed three times with sterile 0.8% saline and refreshed with a new medium containing 2 mL of LB-phenol red medium supplemented with various concentrations of ampicillin that ranged from 15.63 to 1,000 μg/mL and each concentration was tested in a triplicate (n = 3). In addition to the wells tested with variable ampicillin concentrations, three wells were seeded with bacterial cells in the presence of LB-phenol red medium but without ampicillin (positive control). Similarly, three wells were tested with a phenol red medium without ampicillin or bacterial cells (negative control). After overnight incubation at 37 °C without shaking, the MBICs were estimated using the lowest concentration of ampicillin that inhibited a color change of the medium from red to yellow (Pantanella et al., 2008). To verify the obtained MBICs, the cultures from the 96-well plates were dumped on a sterile paper towel to remove loose cells and washed three times with 0.85% saline. Each well was filled with fresh LB-phenol red medium without ampicillin (n = 3). MBIC was confirmed as the concentration below which cells were able to develop biofilms and in wells with higher concentrations no color change was observed due to biofilm inhibition. E. coli strains A5 and H5 were characterized by MBICs of 500 μg/mL while strains D4 and A9 had MBICs of 350 μg/mL. Note that for the same strains, MBICs are eight totenfold higher than MICs.
Bacterial Growth
Three colonies of each strain taken from the LB plates were inoculated into 5 mL LB broth and incubated overnight at 37 °C with shaking at 150 rpm. The overnight cultures were then diluted in a 1:100 ratio in a fresh LB broth supplemented with or without ampicillin at MICs for the various strains (Perry et al., 2009). The ampicillin-treated samples were cultured for 3 h (180 min) or 8 h (480 min), respectively. The samples were cultured at 37 °C with shaking at 150 rpm until cells reached the late exponential phase of growth (OD600: 0.5–0.7). Cellular growth was measured with a UV/Vis spectrophotometer (Thermo Spectronic, USA) at a wavelength of 600 nm. The culture was performed in a triplicate using independent colonies. Cells cultured using this methodology were used later in our AFM and fluorescence microscopy experiments, biofilm formation assays, membrane permeability measurements, and contact angle studies with little to no modifications.
Preparation of Gelatin-Coated Mica Substrates for AFM Studies
E. coli has a negatively charged surface and most substrates used in AFM studies, such as mica, are negatively charged when unmodified (Doktycz et al., 2003; Allison et al., 2011). This will render the immobilization of E. coli on mica to be very challenging due to the expected electrostatic repulsion between the cell surface and the substrate (Doktycz et al., 2003; Allison et al., 2011). To solve this problem, the mica surface was coated with a positively charged gelatin layer capable of electrostatically attracting the negatively charged bacterial cells for improved adhesion. A 12 mm diameter mica disc (Ted Pella, Inc., Redding, CA, USA) was cleaved several times and hydrated in DI water and dried for 1 h to obtain a flat surface (Doktycz et al., 2003). A 0.5% (w/v) gelatin solution was prepared by dissolving 0.5 g of powder gelatin from porcine skin in a 100 mL of DI hot water and allowed to cool to 60–70 °C. Afterward, the freshly cleaved mica was gently submerged into the gelatin solution and withdrawn immediately. The gelatin-coated mica was supported on the edge of a paper towel to dry prior to use for bacterial immobilization (Allison et al., 2011).
Bacterial Cell Immobilization on Gelatin-Coated Mica
A 1,000 μL bacterial suspension of cells cultured until their late exponential phase of growth with or without ampicillin at MIC was pelleted by centrifugation at 5,000 g for 5 min. The supernatant was discarded, and the pellet was resuspended in a 1,000 μL DI water and centrifuged at 5,000 g for another 5 min. Finally, the pellet was resuspended in 500 μL DI water. A 100 μL of the bacterial cell suspension was applied to the freshly prepared gelatin-coated mica and incubated at room temperature for 10 min after which the surface was rinsed with DI water to remove nonadherent bacterial cells (Doktycz et al., 2003; Allison et al., 2011). Bacterial cells taken from three different cultures prepared with independent colonies for each strain were immobilized on three different gelatin-mica surfaces and used directly in the AFM measurements.
Fluorescence Microscopy
Prior to imaging with fluorescence microscopy (EVO FL AMG, Bothell, WA, USA), MDR-E. coli cells were grown in LB without ampicillin or in LB supplemented with ampicillin at the MIC quantified for each strain for 3 h with shaking at 150 rpm. The bacterial cells were allowed to colonize 12 mm diameter coverslips placed in a 24-well plate for 20 min. Unbounded cells were washed with phosphate-buffered saline (PBS) and coverslips were then incubated in 5 μM SYTO 9 green fluorescent dye for 15 min under dark conditions at room temperature (Yoon et al., 2011). We realize that washing with PBS may cover the effects of possible cell lysis as DNA from lysed cells can get solubilized in water and washed away. However, running these assays without washing may adversely affects the resolution of the images especially where the cellular debris and biopolymers are high. Three slides were made for each culture for a total of nine slides from three independent cultures per treatment. Five to seven fluorescent images were captured from each slide.
Measurement of Membrane Permeability with a Crystal Violet Assay
Briefly, a 5 mL of bacterial suspension grown as described above with or without ampicillin at MIC was centrifuged at 3,000 g for 10 min (Eriksson et al., 2002). The pellets were washed three times with PBS (pH 7.2) and the OD of the suspension at 600 nm was adjusted to 0.5. The suspension was then centrifuged at 3,000 g for 10 min, the supernatant was discarded, and the resulting pellet was suspended in 5 mL of PBS (pH 7.2) containing 0.1 g/mL (0.1% w/v) of CV. Afterward, the suspension was incubated at 37 °C for 10 min and subsequently centrifuged at 13,400 g for 15 min (Devi et al., 2010). The absorbances of the supernatants of treated and untreated samples were measured at 590 nm using an image reader (Cytation 5, BioTex, In, USA). The percentage of CV permeability of the cell wall was calculated as previously reported in the literature (Halder et al., 2015).
| (1) |
The CV assay measures the luminescence intensity. This dye acts like fluorochromes such as 1-N-phenylnaphthylamine (NPN) which are nonfluorescent in the nonlipid environment but become fluorescent when they meet lipids. When membranes of cells get compromised due to antibiotics’ exposure, cells release their internal content. Therefore, the higher the membrane damage, the higher the lipid content the CV dye will complex with, resulting in a higher fluorescence as predicted by equation (1) (Halder et al., 2015).
AFM Force Measurements
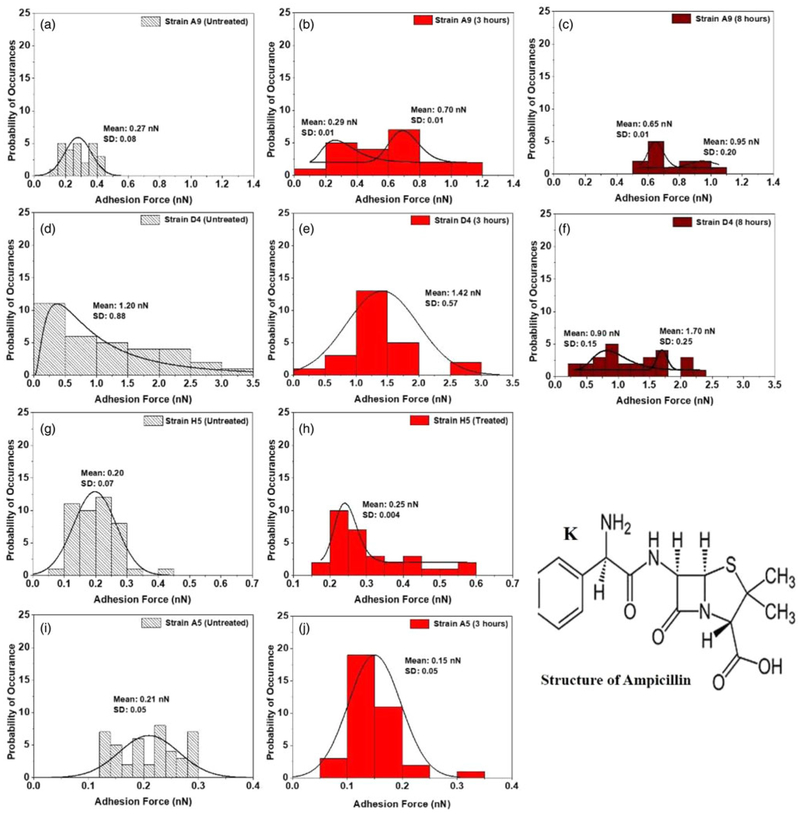
Forces between individual bacterial cells and model Si3N4 cantilevers (DNP-10, Bruker Inc., Camarillo, CA, USA) were measured in DI water with a Multimode AFM equipped with a Nanoscope IIIa controller and extender module (Bruker AXS Inc.). Here, we selected to perform the AFM force measurements under water because we collected these resistant bacteria from households in Tanzania in which the bacteria were exposed to water as the main solution. Approach and retraction force-distance curves were collected under tapping mode to avoid any bacterial surface damage caused by the lateral force exerted by the AFM cantilever (Dufrêne, 2002; Abu-Lail & Camesano, 2003). On each investigated cell and to have a statistical representation of the entire bacterial surface, the AFM point, and shoot function were employed to map 25 locations on the entire bacterium surface for force measurements (Supplementary Fig. S2a and inset) (Abu-Lail & Camesano, 2003). For each strain investigated, 13–45 individual cells taken from three independent cultures were probed. Note that a larger number of cells was easily identified on the substrates in the absence of antibiotics, while a lower number of intact cells was identified when cells were exposed to antibiotics. AFM retraction curves were analyzed using the AFM Nanoscope Analysis 1.5 software (Bruker) individually for the adhesion forces due to the expected heterogeneity in the acquired data (Supplementary Fig. S2b) (Park & Abu-Lail, 2011c). Due to such heterogeneity, histograms of all adhesion forces for each strain with and without ampicillin treatment were constructed using Microsoft Excel 2013 (Fig.5). Unimodal and multi-modal distributions in the histograms were fit to lognormal or Gaussian peak dynamic functions, respectively, using Origin 2018 v9.4, OriginLab Corp., Northampton, MA, USA (Arslan et al., 2018). Approach curves were not analyzed in this study.
Fig. 5.
Histograms showing the distributions of adhesion forces measured in water between Si3N4 and surface biopolymer of MDR-E. coli cells in the absence (a, d, g, and i) and presence of ampicillin (b, e, h, j, c, and f) at different time points (3 and 8 h). Log-normal and Gaussian dynamic peak functions were used to fit the unimodal and multi-modal distributions in the histograms (Origin v9.4, OriginLab Corp.) respectively. K is the chemical structure of the β-lactam (ampicillin) (Filho et al., 2010).
Prior to force measurements, the spring constant of each cantilever was determined from the power spectral density of the thermal noise fluctuations in DI water (Hutter & Bechhoefer, 1993; Park & Abu-Lail, 2011a). The average spring constant was found to be 0.07 ± 0.01 N/m (n = 18), close to the reported manufacturer value of 0.06 N/m. The deflection sensitivity was measured on a cleaned unmodified mica surface in water and found to be 45.26 ± 4.61 nm/V (n = 12) (Hutter & Bechhoefer, 1993). All bacterial cells images were captured at a scan rate of 0.41 ± 0.03 Hz (n = 12) and at a resolution of 256 samples per line. Furthermore, to confirm that the tip was not contaminated during measurements, force measurements were made on a bacteria-free area of the gelatin-coated mica disk before and after making the measurement on a bacterial cell (Park & Abu-Lail, 2011a; 2011b). Equality of the measurements ensured that the tip properties had not been altered by contact with the bacterial surface biopolymers (Park & Abu-Lail, 2011a; 2011b).
Biofilm Assays
The abilities of MDR-E. coli strains to form biofilms in the presence or absence of ampicillin were characterized using a microtiter plate technique as described previously (Stepanovic et al., 2007; O’Toole, 2011). The overnight grown culture was diluted in a ratio of a 1:100 in LB and cultured until the late exponential phase of growth. The OD was then adjusted to 0.5. A 100 μL of bacterial suspension was inoculated into wells of a sterile 96-well plate for 24 h at 37 °C without shaking. After incubation in LB without ampicillin, unattached cells and medium were gently dumped on an autoclaved stack of paper towels and wells were washed five times with sterile 0.85% (w/v) saline. Each well was refreshed with a new medium supplemented with or without ampicillin at the corresponding MBIC for a given strain and cultured for 3 h at 37 °C without shaking. The unattached cells and media were gently dumped on an autoclaved stack of paper towels and washed five times with sterile DI water by carefully pipetting 200 μL DI water into each well using a multichannel pipette. Afterward, each well was incubated at room temperature for 15 min with a 125 μL of a 0.1% (w/v) solution of crystal violate. The wells were rinsed again in 200 μL of sterilized DI water for four times and allowed to dry for 1 h. To solubilize the CV, each well was treated with a 125 μL of a 95% ethanol for 15 min. The absorbance of the supernatant was used to measure the degree of biofilm formation at OD600 nm values of the tested samples and the control using an image reader (Cytation 5, BioTex, In). The value of biofilm developed in each well was estimated using the formula shown in equation (2). Supplementary Table S1 below summarizes the criterion used to judge strains’ abilities to form biofilms.
| (2) |
Statistical Analysis
Variations between the ampicillin-treated samples and their corresponding controls were determined using one-way analysis of variance available in SigmaPlot 11.0 (Systat Software Inc., San Jose, CA, USA). Statistical significance was considered at 99.9% confidence interval (p < 0.001) unless otherwise stated.
Results
Effect of Ampicillin on Bacterial Growth
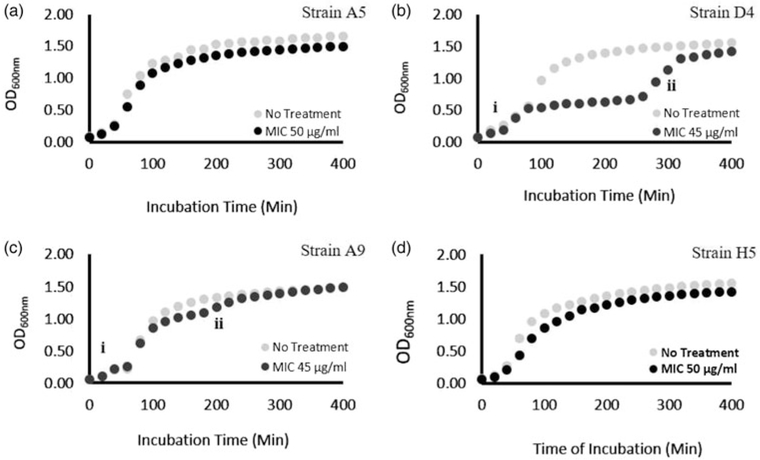
The growth curves of all investigated MDR E. coli bacterial strains are shown in Figure 1. For strains A5 and H5 and in the presence of ampicillin, the growth curves were typical with no indications of inhibition zones (Figs. 1a, 3d). Because of that, in further investigations, these strains were only tested after 3 h in their stationary phase of growth. However, the growth curves of strains D4 and A9 in the presence of ampicillin at MIC showed two exponential phases, followed by a stationary phase that coincided with data collected for the strains in the absence of ampicillin. The zone trapped between the two exponential phases is referred to as the inhibition zone and is a 280 min long for D4 and 180 min long for A9 (Figs. 1b, 1c). In a growth curve with inhibition, the first exponential phase corresponds to the growth of cells before the inhibition phase begins while the second exponential phase corresponds to the recovery stage under the inhibition of cellular growth to a regular pattern [labeled (i) and (ii) in Figs. 1b, 1c, respectively]. E. coli strain D4 was characterized by a lower specific growth constant (μ = 0.010/min) during the second exponential growth phase compared with that measured in the first exponential phase (μ = 0.024/min) (Fig. 1b). In the recovery second exponential growth phase (ii), the doubling time was found to be 71 min, 2.5-fold lower than that estimated in the first exponential phase (29 min). For E. coli strain A9, the specific growth rate constants were estimated to be (μ = 0.027 and 0.003/min) in the first and second exponential phases of growth (Fig. 1c). The doubling times were 26 and 267 min in (i) and (ii) exponential growth phases, respectively.
Fig. 1.
Growth curves of MDR-E. coli strains cultured in LB only (gray) or LB supplemented with ampicillin at MIC (black) at 37 °C with shaking at 150 rpm. E. coli strains D4 and A9 depict antimicrobial activity between ≈1–8 h, after which the growth curves for treated and untreated cells became similar. (i) The first exponential phase before the inhibition zone starts and (ii) the second exponential phase that represents the recovery stage.
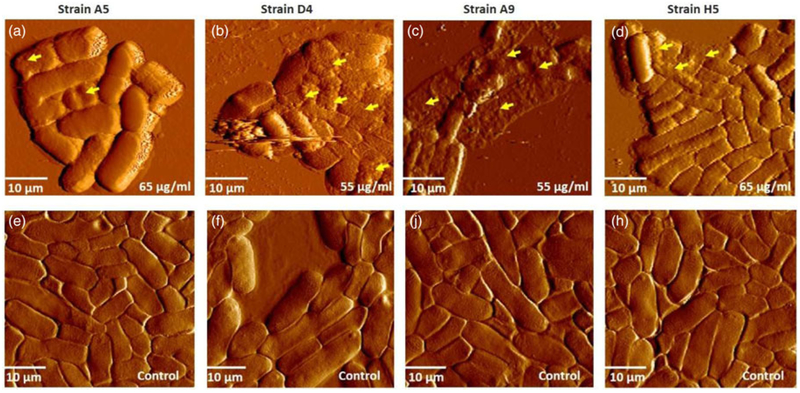
Fig. 3.
(a–d) Represents AFM deflection images of MDR-E. coli strains exhibiting different levels of membrane damage after exposure to ampicillin at concentrations slightly above the MIC (65 or 55 μg/mL). Yellow arrowheads indicate examples of membrane damage after exposure to ampicillin. (b, c) Show the collapse of cell membranes and cell lysis; (a and d) show increased roughness. (e–h) Represent deflection images of the control, untreated samples in air. All images were measured in air under contact mode, with the average scan rate of 0.93±0.01 Hz (n = 4) and spring constant of 0.070±0.02 N/m (n = 4).
Effect of Ampicillin on Bacterial Morphology
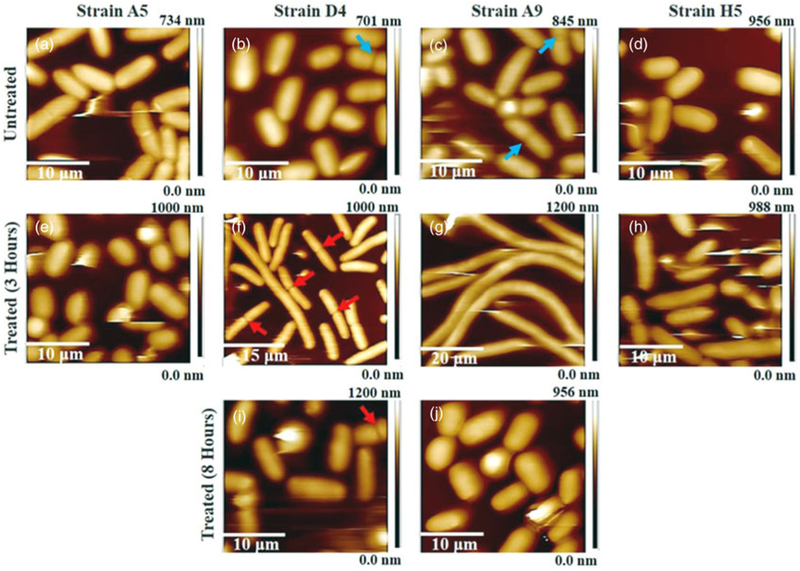
This section analyses captured AFM images (Fig. 2 and Supplementary Fig. S1) at the corresponding MIC which were used to quantify the width, length, and heights of cells (Supplementary Fig. S3). In all images, irrespective of the treatment condition, bacterial cells were close to each other and each bacterium was clearly distinguished from others in the same image (Fig. 2). Cells untreated with ampicillin appeared smooth and rod-like for all strains investigated with an average cell length of 2.98 ± 0.48 μm, width of 1.58 ± 0.18 μm, and a maximum height of 0.67 ± 0.08 μm for 36 cells, representative of the four strains investigated. Dimensions of cells representative of individual strains are reported in Supplementary Figure S3. Because of the presence of an inhibition zone in their growth curves when collected in the presence of ampicillin, E. coli strains D4 and A9 were imaged after 3 and 8 h of growth, respectively (Figs. 1b, 1c). The point was to check if the responses these strains had to ampicillin, as observed by the inhibition zone in the growth curve, were also associated with changes in their dimensions (Fig. 2, Supplementary Figs. S3, S4). Exposure to ampicillin at MIC for 3 h resulted in filamentous phenotypes in three of the strains investigated (D4, A9, and H5) (Fig. 2f-2h).
Fig. 2.
(a–j) Three-dimensional AFM height images. (a–d) Untreated MDR-E. coli cells (control), and (e–h) and (i, j) MDR-E. coli cells exposed to ampicillin at 3 and 8 h, respectively. The red arrowheads indicate elongated and dividing cells in presence of antibiotics and the blue arrowheads depict dividing cells in the absence of ampicillin.
After 3 h of ampicillin exposure, the average dimensions of cells representative of D4, A9, and H5 strains were 6.61 ± 0.213 μm in length, 1.81 ± 0.21 μm in width, and 0.86 ± 0.07 μm in maximum height for 28 cells (Supplementary Fig. S3). After 8 h of ampicillin exposure, the length, width, and height of the bacterial cells representative of D4 and A9 decreased compared with those values measured at 3 h and were found to be 3.41 ± 0.51, 1.60 ± 0.16, and 0.67 ± 0.08 μm, respectively (Supplementary Fig. S3). The recovery of bacterial dimensions toward the native shape at 8 h was statistically significant in strain D4 when compared with those estimated at 3 h treatment (Supplementary Fig. S3: p < 0.001). However, despite the reduction in the cell length at 8 h exposure to ampicillin, the dimensions of the cells in both strains (D4 and A9) were still longer than the untreated cells (Supplementary Fig. S3). Dimensions of cells representative of individual strains after 8 h are reported in Supplementary Figure S3. Note that elongation of cells upon exposure to ampicillin for up to 8 h did not reveal any sign of bacterial membrane damage within the resolution of the AFM tip (Fig. 2).
In addition to elongation as a resisting mechanism to ampicillin as discussed above, cells elongate as they divide. To discern the role of cellular division on elongation, the average lengths of dividing and nondividing cells in the presence and absence of ampicillin were quantified and compared. Our data indicated that upon division and compared with nondividing cells, A9 and D4 cells significantly elongated themselves by 1.3 and 1.4-fold compared with when no ampicillin was present. On the contrary and interestingly, A9 and D4 cells that were undergoing division in the presence of ampicillin were statistically similar in length to the nondividing cells exposed to ampicillin. Furthermore, comparing the lengths of A9 and D4 cells in the presence and absence of ampicillin shows that, upon exposure to ampicillin, nondividing cells were significantly elongated by 4.5 and 2.9-fold, respectively, and dividing A9 and D4 cells were significantly elongated by 2.9 and 1.7, respectively. Therefore, this analysis clearly points to that elongation observed in our study was due to ampicillin exposure and not due to cell division.
Contrary to the filamentous formation in strains D4, A9, and H5, exposure of E. coli A5 to ampicillin at MIC resulted in a reduction of their length and width compared with untreated cells (Fig. 2 and Supplementary Fig. S3). The average cell length and width of strain A5 decreased from 2.92 ± 0.64 to 2.57 ± 0.45 μm and from 1.91 ± 0.23 to 1.75 ± 0.19 μm, respectively (Supplementary Fig. S3: p < 0.001). Again, the integrity of the membrane was unaffected by exposure to antibiotics (Fig. 2). The AFM images are consistent with observations of the fluorescence images (Supplementary Fig. S5). According to the fluorescence images (Supplementary Fig. S5), no cell lysis was observed, and the elongated cells never showed DNA fragmentation.
Effect of Ampicillin on Modeled Cell Surface Area, Volume, and Surface Area to Volume Ratio
Using an ellipsoid as a prediction model for bacterial shape (Supplementary Fig. S1), we estimated the surface area (SA) and the surface area to volume ratio (SA/V) for all bacterial strains investigated under all culture conditions (Supplementary Fig. S4). The SA values of E. coli strains A5 and H5, after treatment with ampicillin, at MIC were not significantly different from their corresponding untreated controls (p = 0.122 and p = 0.668, respectively). The average SA values of E. coli strain D4 at 3 and 8 h were 3.5 and 1.4-fold greater than the untreated, respectively (Supplementary Fig. S4b: p < 0.001). For E. coli strain D4, the SA at 3 h of exposure to ampicillin was 2.5-fold greater than 8 h treatment (Supplementary Fig. S4b: p < 0.001). Differences between SA values of E. coli strain A9 treated at MIC at 3 and 8 h exposure to ampicillin were 6.3 and 4.5-fold greater than the untreated (Supplementary Fig. S4c: p < 0.001). The SA/V for treated cells was significantly different from the untreated for all conditions for the four strains (Supplementary Fig. S4c: p < 0.001).
Effect of Ampicillin on Membrane Permeability of MDR-E. coli
The CV uptake by MDR-E. coli in the presence or absence of ampicillin is presented in Supplementary Figure S6b. CV is a hydrophobic dye that poorly diffuses into an intact outer membrane but readily penetrates when the outer membrane is damaged (Devi et al., 2010; Halder et al., 2015). As such, the uptake of CV can be utilized to quantify disruptions in the outer membrane of bacteria (Devi et al., 2010; Halder et al., 2015). The uptake of CV into the cytoplasm of ampicillin-treated cells at MIC was not statistically significant from that of untreated cells for all investigated strains (Supplementary Fig. S6b). In contrast, the exposure of cells to concentrations slightly above MICs (65 or 55 μg/mL) showed significant uptake of CV and morphological damage to the cell membrane as seen through AFM imaging (Fig. 3). Morphological differences observed when cells were treated with higher ampicillin concentration than MIC demonstrated cell lysis, the collapse of the cell membrane, and rougher cellular surfaces compared with cells treated at MIC (Fig. 3).
Effect of Ampicillin on Bacterial Surface Roughness
The bacterial surface roughness values as a function of ampicillin treatment at various MICs (50 or 45 μg/mL) were quantified for all strains. Investigation of AFM captured height images indicated that exposure of MDR-E. coli cells to ampicillin at different MICs, irrespective of exposure time, increased the surface roughness values of cells without any sign of disruption of the bacterial cell envelope (Fig. 2 and Supplementary Fig. S6a). The average surface roughness values for treated strains A5 and H5 at their respective MICs for 3 h were 1.1 and 1.0-fold greater than their corresponding controls (Supplementary Fig. S6a, p > 0.001). Whereas, the average surface roughness values for E. coli D4 at 3 and 8 h were 2.5 and 1.8-fold greater than the control (Supplementary Fig. S6a, p < 0.001). When compared with the treatment group, E. coli strain D4, surface roughness at 3 h was 1.4-fold greater than 8 h treatment (Supplementary Fig. S6a, p < 0.001). The same trend was observed for strain A9 at 3 and 8 h exposure to ampicillin (45 μg/mL), where their roughness values were greater than that of the untreated by 1.6 and 1.8-fold, respectively (Supplementary Fig. S6a, p < 0.001). Interestingly, the cell surface roughness values of strains D4 and A9 increased after 3 h and was reduced after 8 h exposure to ampicillin but the roughness values at 8 h were still significantly larger than the control (Supplementary Fig. S6a, p < 0.001). This was the same trend observed with respect to their elongation. The responses to ampicillin apparently occurred at the shorter time periods. Note that for E. coli A5 and E. coli H5 with growth patterns with no inhibition (Figs. 1a, 1d), the surface roughness values were assessed only after 3 h exposure to ampicillin.
Effect of Ampicillin on the Nanoscale Adhesion Forces Measured between Bacterial Surface Biopolymers and Silicon Nitride
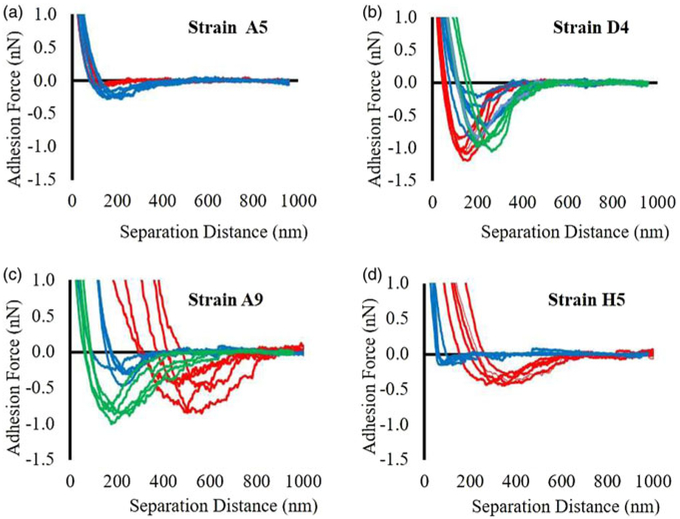
A summary of the measured adhesion forces measured between bacterial surface biopolymers and Si3N4 as a function of ampicillin exposure is shown in Figure. 5 and Supplementary Figure S6d, respectively. Adhesion forces were obtained from retraction curves as described in Supplementary Figure S2b. Examples of AFM retraction curves measured between the biopolymers of the four investigated strains and Si3N4 in the presence of ampicillin at MIC or in the absence of ampicillin are shown in Figure 4. When the retraction curves for stain A5 were compared in the presence (red lines) and absence (blue lines) of ampicillin, it was shown that treatment with ampicillin at MIC for 3 h significantly decreased the adhesion forces compared with the adhesion forces obtained from the untreated cells (Fig. 4a).
Fig. 4.
Representative adhesion force profiles in the absence of ampicillin (blue lines), 3 and 8 h after exposure to ampicillin at different MICs (red and green lines, respectively) for all four strains of MDR-E. coli, n = 5.
As can be seen from Figure 4, the retraction curves for strain D4 were overlapping in distances spanned for both treated and untreated cells, but forces for the treated cells at 3 h were noticeably higher than those measured for the untreated cells. The distances reported in these force profiles are directly proportional to the length of the bacterial surface biopolymers (Abu-Lail & Camesano, 2003). The real length of biopolymers can be longer or shorter than distances measured from retraction curves. This is contingent on the site of attachment of the chains to the AFM tip and whether the biopolymer chains are adequately elastic that the biomolecules stretch themselves past their contour lengths when an external force is applied on the cell surface (Abu-Lail & Camesano, 2003). When strain D4 was exposed to ampicillin for 8 h, the adhesion forces were comparable to the untreated cells and lower than magnitudes observed from the treatment for 3 h (Fig. 4b). When the retraction curves for strain A9 were compared for cells grown in the presence and absence of ampicillin, it was shown that shorter treatments with ampicillin at MIC for 3 h significantly increased the adhesion forces as well as the distances reflecting more extended bacterial surface biopolymers compared with untreated (Fig. 4c). In comparison, when strain A9 cells were exposed to ampicillin for 8 h, the distances deceased, while forces increased compared with the 3 h treatment as well as compared with untreated (Fig. 4c). Finally, when the retraction curves for H5 were compared for cells grown in the presence or absence of ampicillin, it was shown that ampicillin treatment at MIC for 3 h significantly increased the adhesion forces as well as the distances compared with untreated cells (Fig. 4d).
When quantified, ampicillin showed an anti-adhesive property in strain A5 at MIC for 3 h with its adhesion force being 1.3-fold, smaller than that of untreated cells (Supplementary Fig. S6d, p < 0.001). In contrast, the adhesion forces reported for strain D4 at MIC demonstrated 1.2 and 1.1-fold increases compared with untreated at 3 and 8 h of exposure, respectively (Supplementary Fig. S6d, p > 0.001). When compared with untreated cells, the average adhesion forces for strain A9 at MIC increased by 2.3 and 1.1-fold at 3 and 8 h, respectively (Supplementary Fig. S6d, p < 0.001). Strain H5 was also treated with ampicillin at MIC for 3 h and its average adhesion force was 1.6-fold higher than untreated cells (Supplementary Fig. S6d).
Effect of Ampicillin on Bacterial Surface Hydrophobicity
Contact angles on bacterial surfaces were measured using three probe liquids (water, formamide, and diiodomethane) (Supplementary Table S2a) and the results were used to calculate the surface energy components (mJ/m2) (Supplementary Table S2b) of the cells with and without exposure to ampicillin at respective MICs. Our results indicated that MDR-E. coli strains investigated are hydrophilic since their water contact angles were <45° (Absolom et al., 1983; van Loosdrecht et al., 1987; Daffonchio & Verstraete, 1995).
The MDR-E. coli cells grown in the presence of ampicillin appeared to be more hydrophilic than the untreated controls grown in LB, except for strain H5 (Supplementary Table S2a). Untreated cells were characterized by higher Lifshitz–van der Waals hydrophobic components compared with cells exposed to ampicillin (Supplementary Table S2b). On the contrary, the polar acid-based energy components were found to be higher for treated cells than for untreated cells with the electron acceptor energy component being dominant for all strains investigated (Supplementary Table S2b, p < 0.001).
In addition to utilizing the contact angles as means to assess the hydrophobicity of the bacterial cells upon exposure to ampicillin, the distributions of the adhesion histograms were also used to infer the hydrophobicity of the cells (Fig. 5). In order to do this, the famous chemistry rule “polar like polar” was utilized. Since Si3N4 is hydrophilic (Grant & Ducker, 1997), ampicillin is hydrophilic (Nikaido, 2003) and adhesion force measurements are done in hydrophilic water, the higher the adhesion forces measured between the bacterial surface biopolymers and Si3N4, the more hydrophilic the bacterial surface is. With the above in mind, peak forces obtained for the distributions of adhesion forces were compared (Fig. 5). Our results show that for all untreated cells, the adhesion peak value for the D4 strain was statistically higher compared with those obtained for all other strains indicating that it is the most hydrophilic strain. This was consistent with our contact angle measurements which also indicated that the water contact angle, as well as the polar acid–base energy component, were the highest for this strain (Supplementary Table S2). Upon exposure to ampicillin, cells of strain A5 statistically decreased their adhesion, cells of strain D4 kept a statistically similar adhesion and cells of strains H5 and A9 statistically increased their adhesion compared with untreated cells. This indicates that exposure to ampicillin made A5 slightly more hydrophobic, did not affect the hydrophobicity of D4 and increased the hydrophilicity of H5 and A9 cells. These results are more or less consistent with the contact angle measurements findings which also showed that A9 and D4 cells are hydrophilic. Our contact angle measurements though indicated that H5 cells became more hydrophobic, which was not what we observed from our adhesion force histograms. The peaks obtained for H5 cells prior to and after exposure to ampicillin were 0.20 and 0.25 nN, respectively (Fig. 5). Even though these values were statistically different, the differences are small and should be interpreted carefully as adhesion measurements are heterogeneous and dependent on numbers of cells analyzed (Camesano & Abu-Lail, 2002; Park & Abu-Lail, 2009; Park & Abu-Lail, 2011c).
Effect of Ampicillin on Biofilm Formation
Biofilms produced in the presence or absence of ampicillin were quantified for the four E. coli strains (Supplementary Fig. S6c). Biofilm formation was studied at 3 h exposure to ampicillin at different MBICs. The biofilm formation varied among the strains indicating that the amount of stainable extracellular polymeric substrates (EPS) attached to the walls of the 96-well plates varied. Quantification of biofilm strength in the four isolates before exposure to ampicillin indicated that all strains can form moderate biofilms, except for E. coli H5 which was only capable of forming a weak biofilm (Supplementary Table S1 and Fig. S6c). Strong biofilm formation was not identified for any of the four strains. All tested strains responded to ampicillin by significantly forming more biofilms compared with the untreated cells except for E. coli A5 (Supplementary Fig. S6c, p = 0.078). Biofilm formation was stimulated in E. coli D4 and A9 by 350 μg/mL ampicillin, for E. coli A5 and for E. coli H5 by 500 μg/mL ampicillin at 3 h exposure to ampicillin. Even though ampicillin showed anti-adhesive nanoscale forces in strain E. coli A5 compared with the control, it exhibited significant biofilm induction when compared with the control (Supplementary Fig. S6c, p < 0.001).
Discussion
Preservation of Membrane Integrity in MDR-E. coli During Exposure to Ampicillin
The integrity of the cell membranes of MDR-E. coli strains, when treated at MIC above the CLSI recommendation, was investigated using AFM and fluorescence microscopy imaging. If membrane integrity was compromised, a DNA leakage from the cells is to be expected (Yoon et al., 2011). Fluorescence microscopy of exposed cells to 3 h of ampicillin showed no cellular fragmentation compared with untreated cells (Supplementary Fig. S5). This was the case in both elongated cells, which contained uniform single long nucleoids and the DNA content was proportional to the length of the cell with no visible tendency for the long cell to separate (Supplementary Fig. S5), as well as in cells that adapted a spherical phenotype compared with an elliptical phenotype. AFM imaging confirmed the same findings (Fig. 2). DNA leakage from cells upon exposure to antibiotics was reported in the literature for susceptible bacterial cells (Bou et al., 2012). This was not the case here for resistant cells. Germán et al. studied the effect of oxacillin on A. baumannii cell wall integrity to determine the fragmentation of the DNA during antibiotics treatment (Bou et al., 2012). They observed nucleoid spreading of susceptible strains on the background indicated by scattered light from the emitted fluorescence, but the resistant strain did not show nucleoid spreading at any concentration (Bou et al., 2012).
To quantify the membrane integrity, the permeabilities of the bacterial outer membrane to CV, before and after exposure to ampicillin, were compared (Supplementary Fig. S6b) (Devi et al., 2010; Halder et al., 2015). MDR-E. coli strains tested in this study were not permeable to CV at their different MICs (Supplementary Fig. S6b). When treated with ampicillin above their MICs, MDR-E. coli strains A9 and D4 were permeable to CV (Supplementary Fig. S6b). The permeability was associated with clear evidence of compromised membranes as observed by AFM imaging (Figs. 3a-3d). In comparison, strains A5 and H5 contained a good number of undamaged cells with intact morphological structures like the control (Fig. 3). The lower frequency at which the membranes of strains A5 and H5 were damaged above the MIC exposure suggests that the time it may take for these cells to recover from the antibiotics may be less than that required for strains A9 and D4 (Fig. 3). As can be seen from our results, cells can vary in how much they allow ampicillin into their periplasmic space and, as such, how much disruption occurs in their membranes. Our results of CV permeability quantification as well as imaging of membrane integrity are consistent and support each other’s findings.
Studies which aimed at investigating the effects of antibiotics on bacterial membrane permeability are limited in the literature (Devi et al., 2010; Halder et al., 2015). Most of these studies were performed with cells susceptible to antibiotics. Findings from such studies showed that these susceptible cells, when exposed to antibiotics at a sub-lethal dose or at MIC, had a significant increase in their permeability and membrane damage compared with untreated cells (Devi et al., 2010; Halder et al., 2015). In comparison, our findings indicated that cells of MDR E. coli strains were intact when exposed to ampicillin at MIC. Preservation of membrane structural integrity at MIC, as revealed through the AFM and fluorescence microscopy imaging, suggest that MIC could only cause growth delay in MDR-E. coli, but not cell wall damage or cell lysis (Fig. 2 and Supplementary Fig. S5). Membrane damage occurred when MDR cells were exposed to a concentration above the MIC of ampicillin. Cells with compromised membrane integrity showed signs of the collapse of cell membranes, increase in roughness, and cell lysis compared with nonaffected cells (Fig. 3). It is not a surprise that ampicillin above MIC could cause a change in the cell membrane integrity of MDR-E. coli. As was discussed before, ampicillin inhibits PG synthesis by interfering with PBPs (Jacoby & Medeiros, 1991; Dong et al., 2004; Thomson & Bonomo, 2005). A higher concentration of antibiotics compared with MIC may result in arriving at the critical density of ampicillin needed to impede PG synthesis and disrupt cell wall integrity. In support of this claim, literature studies have demonstrated, for example, that Neisseria gonorrhoeae growing at or below MIC of β-lactam antibiotics showed a small increase in the PG synthesis, while a high concentration above MIC drastically reduced the quantifiable amount of PG synthesis (Brown & Harold, 1979; Fruci & Poole, 2016). Apart from the effects of β-lactam on PGs, the structure of LPS is presumed to play an important role in controlling bacterial outer membrane permeability (Wang et al., 2015). Susceptibility of bacterial cells to various antibiotics was related to the structure of LPS (Dasgupta et al., 1994). Studies on an LPS defect mutant of Pseudomonas aeruginosa (PAO1) showed a decreased resistance to gentamicin (Dasgupta et al., 1994). Here, the defected outer membranes that were associated with increased permeability to the hydrophobic CV suggest that cells were antibiotic sensitive above MIC. Furthermore, our results indicate that at the MIC, the membrane of MDR-E. coli strains are conserved, preventing ampicillin from diffusing inside the cell.
Effect of Ampicillin on Bacterial Dimensions and Roughness
Exposure to ampicillin increased the cell length and bacterial surface roughness compared with the untreated cells for three of the investigated E. coli strains (D4, A9, and H5) (Fig. 2, Supplementary Figs. S3-S5). This elongation in cells was associated with an increase in SA and volume (Supplementary Fig. S4). On the contrary, strain A5 modified its morphology from elliptical to spherical while its RMS values remained like the untreated cells after exposure to ampicillin (Supplementary Figs. S3, S5a). All cells maintained their cell wall integrity despite the morphological changes reported (Fig. 2 and Supplementary Fig. S5). The changes observed above can be justified by the mechanism of action employed by ampicillin to affect cell wall synthesis and, as such, bacterial surface properties and attachment (Jacoby & Medeiros, 1991; Yao et al., 2016). Upon binding to PBPs and mimicking the natural structure of D-Ala-D-Ala substrate that is principally bound to the enzyme that synthesizes PG, β-lactam antibiotics inhibit cell wall synthesis of PG (Jacoby & Medeiros, 1991; Yao et al., 2016). The inhibition of PG biogenesis has various effects on cell morphology such as delay or restriction of cell division, elongation and possible cell lysis that results in increased roughness (Jacoby & Medeiros, 1991; Aguayo et al., 2015).
Many rod-like microbes such as E. coli grow to employ two distinct, well-developed, PG machineries (Typas et al., 2012). In the first one, the actin-like MreB protein forms the rod system in E. coli and promotes the incorporation of new PG biomaterials along the lateral cell body to initiate elongation (Typas et al., 2012). This machinery requires PBP2 to function and cell elongation is an expected outcome. Elongation has been reported as means of cellular survival in response to stimuli such as temperature (Bhatti et al., 1976), antibiotics (Rolinson, 1980), and UV radiation (Ma et al., 2009). Cellular elongation allows cells to increase their SAs available for attachment to a model surface as well as promote their interactions with the host and neighboring cells (Young, 2006). Such interactions enable better nutrient uptake during their adaptation process to stress such as in the case of exposure to antibiotics (Young, 2006). Hence, we anticipate that elongation was employed by strains D4, A9, and H5 to resist ampicillin (Supplementary Figs. S3b-S3d).
In the second machinery, the tubulin-like FtsZ protein is responsible for the divisome and making PG in the new daughter cells (Bendezú & De Boer, 2008). This machinery requires PBP3 to catalyze the divisome (Typas et al., 2012) and a change in cell morphology is the expected outcome of antibiotics exposure. By decreasing the SA and conserving the volume, strains that adopt such a mechanism in response to antibiotics like strain A5 may be able to conserve energy by minimizing interactions with their extracellular environment. This mechanism will support ideal dormant state and decrease metabolic activities to reduce antibiotics targets (Shah et al., 2006).
The bacterial outer membrane can be regulated by changes in the surrounding environment. In a stress-free environment, cells use nutrients to divide, grow to mature, and produce toxins and products (Fruci & Poole, 2016). In the case of stress, like that imposed by ampicillin, they shift their use of nutrients to getting more SA, producing toxins and/or avoiding division (Cho & Bernhardt, 2014). Decisions on which mechanism to adapt with respect to the antibiotic stress are made by cells and evidence of these variable means to respond to antibiotics are reported in the literature (Satta & Canepari, 1979; Rolinson, 1980; Aeschlimann, 2003; Babic & Bonomo, 2006; Bendezú & De Boer, 2008; Mohanty et al., 2012; Read & Woods, 2014; Fruci & Poole, 2016). de Boer et al. demonstrated that inactivation of MreB protein causes cells to switch to a spherical shape that refuses to divide (Bendezú & De Boer, 2008). The MreB protein can be inactivated by β-lactam antibiotics (Satta, & Canepari, 1979). Satta et al. observed inhibition of cell wall elongation with K. pneumoniae and changes from rod to cocci structures after exposure to β-lactam mecillinam (Satta & Canepari, 1979). The authors concluded that the transition from rod to cocci phenotypes was because of a reduction in PG synthesis (Satta & Canepari, 1979). While the mechanism of changing the morphology of cells from elliptical to spherical is employed by bacterial strains in response to antibiotics, this mechanism has a problem. It has been reported in the literature that β-lactam antibiotics are more effective in dividing cells that are forming new septums than on the adult cells (Cho & Bernhardt, 2014). As such, cells like to mature into adult cells to avoid the effects of antibiotics on their membranes. One of the means to do that is through bacterial elongation and escaping division.
Furthermore, and besides morphological changes of cells, the increase in surface roughness due to the secretion of surface biopolymers can possibly enhance the attachment of bacterial cells to model surfaces (Young, 2006; Justice et al., 2007; Yoon et al., 2011). Having a heterogeneous population of surface biopolymers on the bacterial surface has been shown to correlate with higher adhesion energy (Park & Abu-Lail, 2011a). Bacterial surface biopolymers are essential for bacterial membrane integrity and have a key role in enhanced attachment and biofilm formation (Limoli et al., 2015). This implies that the rougher the surface of bacteria, the easier it is for cells to attach firmly to a surface and possibly lead to the formation of stronger biofilms (Supplementary Figs. S6a, S6c). A rougher bacterial surface has also been shown to promote attractive van der Waals forces which can be employed by bacterial cells to enhance their adhesion to surfaces (Abu-Lail & Camesano, 2006).
When it comes to nanoscale investigations of the role of antibiotics on surfaces and morphologies of bacterial cells, especially resistant cells, studies are very limited. Previously, AFM studies have revealed that sustained exposure to susceptible E. coli ATCC 25922 to cefodizime (cephalosporins) and ampicillin β-lactams result in the development of nanopores at the apex of the cell, the collapse of the cell wall, and loss of cellular content (Braga & Ricci 1998; Perry et al., 2009). In our study, resistant MDR E. coli strains were used, and no signs of membrane damage were observed at MIC within the AFM tip resolution (Fig. 2). Instead, we observed similar changes described in the literature when ampicillin was applied at higher concentrations compared with the MIC (Fig. 3) (Braga & Ricci 1998; Perry et al., 2009). Overall, the combined effects of bacterial surface roughness and changes in SAs of MDR-E. coli strains in response to antibiotic treatment have not been reported in the literature. Alves et al. measured the surface roughness of treated and untreated E. coli (ATCC 25922) cells susceptible to antimicrobial peptides (AMP) (Alves et al., 2010). They reported an increase in bacterial surface roughness and dramatic damage to the bacterial membrane after exposure to AMP at MIC (3 μM) and above MIC (5 μM) (Alves et al., 2010). The evident cell membrane destruction exerted by AMP was claimed by the authors to be responsible for the increase in surface roughness. Growing E. coli (ATCC 25922) in the presence of AMP below the MIC resulted in a decrease in the surface roughness and a minor surface damage (Alves et al., 2010).
Effect of Ampicillin on Bacterial Hydrophobicity and Contributions of the Latter to Adhesion
Because of its importance in governing bacterial interactions with surfaces (Stenstrom, 1989; Oliveira et al., 2001), the hydrophobicity of bacterial cells as a function of antibiotics treatment was investigated using contact angle measurements, as well as AFM adhesion measurements (Supplementary Table S2a). When the results of the two were considered, exposure to ampicillin made the cells of A9 and D4 more hydrophilic and the cells of H5 and A5 became slightly more hydrophobic. Since Si3N4 is a hydrophilic substrate in water (Eastman & Zhu, 1996), ampicillin as well is hydrophilic (Nikaido, 2003). Increased adhesion (Supplementary Fig. S6d), observed for cells exposed to antibiotics, requires increased hydrophilicity which was the case here for A9 and D4. The surface interactions of strains A5 and H5 to Si3N4 were dominated by electron acceptor and a very low electron donor surface energies in the presence and absence of ampicillin, respectively (Supplementary Table S2b). Overall, our data suggest that acid–base interactions are more critical to adhesion upon exposure to ampicillin compared with van der Waals interactions, which were dominant in controlling the adhesion of untreated cells to Si3N4. However, it is important to note that a high surface energy of the bacterial cell wall does not always guarantee a high cellular adhesion or biofilm formation to a hydrophilic surface (Renner & Weibel, 2011). Here, despite the same degree of hydrophilicity displayed by all the strains, nanoscale adhesion forces were quite variable among the strains tested (Supplementary Fig. S6d). This observation indicates that adhesion depends on more other factors than surface energies. Note that even when we utilized the adhesion forces to describe cellular hydrophobicity, these forces were heterogeneously distributed for each strain investigated, as is evident from widths of the histograms shown in Figure 5. Therefore, even though a peak value or a mean value can be used to represent the adhesion forces of each strain and thus the strain hydrophobicity, caution should be practiced when these results are interpreted as cells will have distributions of hydrophobic and hydrophilic locations on their surfaces and will not display a uniform hydrophobic or a hydrophilic surface. As such, and even though hydrophobic interactions are critical to adhesion forces, additional factors may still influence adhesion and biofilm formation. These include roughness, SA/V, the composition of surface molecules, length, conformation, and density of surface molecules, and biomechanics of cells (Durodie et al., 1995; Abu-Lail & Camesano, 2003; Yang et al., 2006; Yoon et al., 2011).
The change in the bacterial microenvironment forces bacterial cells to modify their surfaces to enhance their survival (Krasowska & Sigler, 2014). One of the mechanisms employed by cells to resist antibiotics involves protecting their hydrophilic outer membrane vehicles from damage which are rich in hydrophilic polysaccharides and/or LPS (Sundari & Balasubramanian, 1991). LPS is the major component of the outer membrane of the Gram-negative bacteria that provide the first line of defense against invading molecules such as ampicillin (Pagès & Winterhalter, 2008). LPS has the hydrophobic portion (lipid A) which anchors LPS to the cell membrane and the outer hydrophilic polysaccharide portion that extends from the cell surface (Pagès & Winterhalter, 2008). Having a hydrophilic bacterial surface facilitates the attachment of cells to Si3N4 in water, and thus enhances the ability of cells to form biofilms, the latter which can enhance bacterial resistance to antibiotics (Stewart, 2002). Two of the three strains that became more hydrophilic (D4 and A9) elongated themselves in response to ampicillin exposure. The higher SA is expected to be associated with a higher content of hydrophilic outer membrane LPS molecules that contribute to the hydrophilic nature of the cell. In support of the mechanism above, the loss of the phosphate group from the oligosaccharides that contribute to the makeup of the LPS outer membrane of E. coli increased the hydrophobicity of the bacterial cells and increased the outer membrane permeability to NPN (Wang et al., 2015). Even though NPN is an antioxidant and not an antibiotic, the study above confirms that the presence of LPS contributes to well-integrated hydrophilic cell membranes that can prevent diffusion of invader molecules such as ampicillin within the cell.
Having less hydrophobic molecules, produced by cells along with possible LPS conformations that expose the inner membrane, can lead to an overall more hydrophilic bacterial surface. For strains A5 and H5, the types of molecules expressed in response to ampicillin as well as the conformations of the LPS on the surface may be the reasons behind the observed wettabilities of cells. Future work should quantify the conformations of the LPS molecules on the surfaces of cells to provide insights on how these conformational properties affect hydrophobicity of cells.
Effect of Ampicillin on Nanoscale Bacterial Adhesion Forces and Biofilm Formation
Bacterial adhesion to surfaces is one of the main mechanisms employed by bacterial cells for variable functions including access to nutrients, building a biofilm community and having an improved antibiotics resistance (Donlan, 2002; Garrett & Zhang, 2008). Due to its vitality to cell function, many techniques, as well as studies, were devoted to quantifying cell adhesion (Abu-Lail & Camesano, 2003; Park & Abu-Lail, 2011a; Abu-Lail, 2012; Khalili & Ahmad, 2015). Some of these techniques quantify the adhesion of individual cells to surfaces and some quantify biofilm formation of cells (Abu-Lail & Camesano, 2003; Liu et al., 2006; Camesano et al., 2007; Wright et al., 2010; Abu-Lail, 2012; Huang et al., 2015). The adhesion of individual cells to surfaces in the colonization step of a surface is known as the first step of biofilm formation and is a critical step in establishing the bacterial community (Donlan, 2002; Garrett & Zhang, 2008; Huang et al., 2015; Wang et al., 2015a, 2015b). This adhesion can be easily quantified using AFM in the native cellular environment (Abu-Lail & Camesano, 2003; Park & Abu-Lail, 2011a; Abu- Lail, 2012). The following steps of bacterial adhesion continue through cell–cell adhesion and buildup of multiple layers of cells on surfaces. These cell layers relate to EPS and the entire community is referred to as a biofilm (Stepanovic et al., 2007; O’Toole, 2011; Gupta, 2015). Biofilms can be quantified through colorimetric assays or imaging (Huang et al., 2015; Wang et al., 2015). The two steps discussed above, although are often related, exceptions are sometimes reported when the two do not correlate nor scale up (Costerton et al., 1995; O’Toole, 2011; Yoon et al., 2011; Wang et al., 2015). As such, here and motivated by their importance as mechanisms that can contribute to MDR resistance, the nanoscale adhesion and biofilm formation of the four investigated strains to model surfaces were quantified as a function of ampicillin exposure dose and duration, and were related to each other (Supplementary Fig. S6c). We hypothesized that, in the strains that depicted increased roughness as well as elongation in response to ampicillin exposure, a higher adhesion forces to the Si3N4 model surface, as well as higher biofilm formation, will be observed. The opposite as well is true for the strain that reduced its SA and maintained its roughness in response to ampicillin exposure.
Our hypothesis was true for strains A9 and H5 which demonstrated elongation while adhesion forces were also increased compared with the untreated cells (Supplementary Figs. S3, S6d, p < 0.001). Strain D4 was similar to the control in adhesion even though it exhibited elongation and an increase in bacterial surface roughness upon exposure to ampicillin (Supplementary Fig. S6, p = 0.121). The morphological changes that led to a spherical shape for strain A5 upon exposure to ampicillin were associated with a reduction in adhesion compared with the control (Supplementary Fig. S6a, p < 0.001). These findings were consistent with our hypothesis. Strain D4 deviated from our hypothesis. When compared, the effects of SA on adhesion were more important than the effects of the roughness of bacterial cells on adhesion. Our results showed that roughness and nanoscale adhesion are proportional but not strongly correlated (data not shown) (Supplementary Figs. S6a, S6d). To our knowledge, there are no AFM studies in the literature which investigated the effects of antibiotics on nanoscale adhesion of MDR bacterial cells exposed to antibiotics, as well as how these forces correlate with bacterial surface roughness or dimensions and exposed area to interactions. Earlier work suggested that antibiotics can affect bacterial adhesion by promoting the expression or inhibition of synthesis of adhesins on the cell surface and/or by modifying bacterial morphology in a pattern that interferes with the ability of an antibiotic to approach receptors on host cell surface (Longo et al., 2013). Durodie et al. suggested that bacterial cellular protein content is stimulated after exposure to antibiotics leading to increased adhesion (Durodie et al., 1995). Hence, the type of molecular mechanisms that detail how cellular adhesion is affected by antibiotics treatment are not yet understood.
Our next step was to investigate biofilm formation upon exposure to ampicillin (Supplementary Fig. S6c). As we expected, ampicillin stimulated significant biofilm formation in three of the investigated strains (D4, A9, and H5, p < 0.001) compared with untreated cells (Supplementary Fig. S6c). Biofilm formation in strain A5 upon exposure to ampicillin was similar to that of the control (Supplementary Fig. S6c, p = 0.078). Our results for strains D4, A9, and H5 agree well with earlier studies in the literature which investigated the influence of antibiotics and elongation on biofilm formation in susceptible strains (Yoon et al., 2011). For example, Young et al. has previously shown that elongation is an important cellular factor for enhancement of biofilm formation in P. aeruginosa due to an increase in cell-to-cell contact (Yoon et al., 2011). It has also been reported that certain antibiotics such as ampicillin (Touhami et al., 2006), tobramycin (Hoffman et al., 2005), and methicillin (Kaplan, 2011) at their sub-MIC can enhance biofilm formation for different bacterial species. In our study, MDR-E. coli strains were treated with ampicillin at MIC and strains D4, A9, and H4 showed significant biofilm formation after 3 h exposure (Supplementary Fig. S6c, p < 0.001). Although strain A5 was not able to enhance its biofilm formation after exposure to ampicillin, its membrane is still expected to provide a similar level of protection for its cells upon exposure to antibiotics. Genetic evidence revealed that biofilm induction by β-lactam antibiotics has been associated with high expression of cps-lacZ which is identified in the colanic acid synthesis and further enhanced capsular polysaccharide matrix in E. coli (Sailer & Young, 2003; Kaplan, 2011). The development of such a thick layer of EPS leads to a potentially slower diffusion rate of antibiotics in biofilms (Costerton et al., 1995; O’Toole, 2011), new biofilm-specific phenotypes (Ito et al., 2009; O’Toole, 2011), and/or development of persister cells (Lewis, 2007). All these factors individually or combined lead to enhanced biofilm formation and enhanced antibiotic resistance by cells.
It is likely that the combination of elongation, increased surface roughness, and nanoscale adhesion of individual cells support the induction of biofilm formation after exposure to ampicillin. There are some studies which suggested that bacterial adhesion and biofilm formation depend, not only on the physical nature of the substratum, but on bacterial cell shape, size, production of surface proteins, and surface characteristics (Roosjen et al., 2006; Mei et al., 2011). The complexity associated with biofilm formation and its dependence on many factors clearly demonstrate why treatment of biofilm-related infections is a difficult task (O’Toole, 2011; Romling & Balsalobre, 2012). This complexity further clarifies, in part, factors behind why infections caused by biofilm-forming strains display complex resistance to a range of currently used antibiotics in comparison to nonbiofilm-related bacterial infections (O’Toole, 2011; Römling & Balsalobre, 2012).
Conclusions
Mechanisms of MDR E. coli Resistance to Ampicillin
Different bacterial strains can resist antibiotics differently (Russell, 1999). Here, four domestic E. coli strains known for their resistance of an array of commonly used antibiotics were explored for means they utilize to resist the cell-wall model β-lactam antibiotic, ampicillin. When strains investigated here were categorized for mechanisms they employ to resist antibiotics, two trends emerged. One of the strains, E. coli A5, in response to antibiotics, changed its phenotype from elliptical to spherical, maintained its roughness, decreased its length and SA, and maintained its integrity and biofilm formation in comparison to untreated cells (Fig. 2, Supplementary Figs. S5, S6c). With all the above, strain A5 resisted antibiotics by likely going into a dormancy state with decreased metabolic activity and division potential. This strain also decreased its adhesion to surfaces without requiring a further increase in biofilm formation. In our previous work, such reduction in adhesion was associated with collapsed bacterial biopolymers on the bacterial surface (Gordesli & Abu-Lail, 2012). If the same is assumed to be true for strain A5, such collapsed biopolymers will increase bacterial stiffness and may decrease the diffusion of antibiotics through the cell membrane. It also may hide the PBP necessary for ampicillin to attach E. coli cells and impede PG synthesis. The conformational properties of E. coli A5 in response to antibiotics are currently being tested in our laboratory. When extrapolated to strains similar to E. coli A5, our findings suggest that combating MDR infections may largely rely on developing antibiotics that make the cell membrane more hydrophobic by inhibiting the production of polysaccharides such that its interactions with hydrophilic surfaces, including mammalian cells, are decreased (Lambert, 2002) and its survival abilities that rely on forming biofilms decrease.
The second trend emerged for strains D4, A9, and H4. These strains responded to the antibiotic by elongating their cells, increasing their SA/V, increasing their roughness, nanoscale adhesion, and biofilm formation. These modifications are likely the result of increased expression of bacterial surface molecules such as LPS, proteins, and EPS (O’Toole, 2011; Römling & Balsalobre, 2012). With all the above modifications, these strains possibly resisted antibiotics through the creation of diffusion limitations to antibiotics within biofilms and enhancing abilities of some cells to become persisters (Lewis, 2007; Ito et al., 2009; O’Toole, 2011). Finally, for these strains, combating antibiotic resistance implies forming antibiotics that can diffuse easily within biofilms. Irrespective of the means they employed to resist antibiotics, all bacterial cells investigated kept their integrity and remained viable in the presence of antibiotics at their or below various MICs.
Supplementary Material
Acknowledgment.
The authors would like to acknowledge Prof. Douglas Call for providing the MDR-E. coli strains and for the training provided on how to perform the biofilm assays, Dr. Susmita Bose and Kevin Stenberg for helping us with the contact angle measurements, Dr. Viveka Vadyvaloo for helping us with the fluorescence imaging and National Institutes of Health (NIH) support for Samuel C. Uzoechi through the T32 GM008336.
Footnotes
Supplementary material. The supplementary material for this article can be found at https://doi.org/10.1017/S1431927618015696
Conflicts of interest. There are no conflicts to declare by authors.
References
- Absolom DR, Lamberti FV, Policova Z, Zingg W, Van Oss CJ, & Neumann WA (1983). Surface thermodynamics of bacterial adhesion. Appl Environ Microbiol 46(1), 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Lail L (2012). Using atomic force microscopy to measure anti-adhesion effects on uropathogenic bacteria, observed in urine after cranberry juice consumption. J Biomater Nanobiotechnol 03(04), 533–540. [Google Scholar]
- Abu-Lail NI & Camesano TA (2003). Role of ionic strength on the relationship of biopolymer conformation, DLVO contributions, and steric interactions to bioadhesion of pseudomonas putida KT2442. Biomacromolecules 4(4), 1000–1012. [DOI] [PubMed] [Google Scholar]
- Abu-Lail NI & Camesano TA (2006). The effect of solvent polarity on the molecular surface properties and adhesion of Escherichia coli. Colloids Surf B 51(1), 62–70. [DOI] [PubMed] [Google Scholar]
- Aeschlimann JR (2003). The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Pharmacotherapy 23(7), 916–924. [DOI] [PubMed] [Google Scholar]
- Aguayo S, Donos N, Spratt D & Bozec L (2015). Single-bacterium nanomechanics in biomedicine: Unravelling the dynamics of bacterial cells. Nanotechnology 26(6), 062001. [DOI] [PubMed] [Google Scholar]
- Ali R, Al-Achkar K, Al-Mariri A, & Safi M (2014). Role of polymerase chain reaction (PCR) in the detection of antibiotic-resistant Staphylococcus aureus. Egyptian Journal of Medical. [Google Scholar]
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J & Handelsman J (2010). Call of the wild: Antibiotic resistance genes in natural environments. Nature Reviews Microbiology 8(4), 251–259. [DOI] [PubMed] [Google Scholar]
- Allison DP, Sullivan CJ, Mortensen NP, Retterer ST & Doktycz M (2011). Bacterial immobilization for imaging by atomic force microscopy. j Visualized Exp (54), 5–7. doi: 10.3791/2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CS, Melo MN, Franquelim HG, Ferre R, Planas M, Lidia F, Bardají E, Kowalczyk W, Andreu D, Santos NC, Fernandes MX & Castanho MARB (2010). Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J Biol Chem 285(36), 27536–27544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan B, Colpan M, Gray KT, Abu-Lail NI & Kostyukova AS (2018). Characterizing interaction forces between actin and proteins of the tropomodulin family reveals the presence of the N-terminal actin-binding site in leiomodin Arch Biochem Biophys. Elsevier; 638(October 2017), 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic M & Bonomo RA (2006). What’s new in antibiotic resistance? Focus on beta-lactamases. Drug Resist Updates 9(3), 142–156. [DOI] [PubMed] [Google Scholar]
- Bendezú FO & De Boer PAJ (2008). Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190(5), 1792–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti AR, DeVoe IW & Ingram JM (1976). Cell division in Pseudomonas aeruginosa: Participation of alkaline phosphatase. J Bacteriol 126(1), 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo RA (2018). Multiple antibiotic-resistant bacteria in long-term-care facilities: An emerging problem in the practice of infectious diseases author (s): Bonomo Robert A. Published by: Oxford University Press Stable; 31(6), 1414–1422. Available at http://www.jstor.org/stable/4482385SPECIA. [DOI] [PubMed] [Google Scholar]
- Bou G, Otero MF, Santiso R, Tamayo M, Fernández MDC, Tomás M, Gosálvez J, & Fernández JL (2012). Fast assessment of resistance to carbapenems and ciprofloxacin of clinical strains of Acinetobacter baumannii. J Clin Microbiol 50(11), 3609–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga PC & Ricci D (1998). Atomic force microscopy: Application to investigation of Escherichia coli morphology before and after exposure to cefodizime. Dow. Antimicrob Agent Chemother 42(1), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AN, Smith K, Tova AS, Lu J, Obare SO & Scott ME (2012). Nanoparticles functionalized with ampicillin destroy multiple-antibiotic-resistant isolates of Pseudomonas aeruginosa and Enterobacter aerogenes and methicillin-resistant Staphylococcus aureus. Appl Environ Microbiol 78(8), 2768–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CA & Harold RP (1979). In Vitro Synthesis of Peptidoglycan by/β-Lactam-Sensitive and -Resistant Strains of Neisseria gonorrhoeae: Effects of/BLactam and Other Antibiotics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camesano TA & Abu-Lail NI (2002). Heterogeneity in bacterial surface polysaccharides, probed on a single-molecule basis. Biomacromolecules 3(4), 661–667. [DOI] [PubMed] [Google Scholar]
- Camesano TA, Liu Y & Pinzon-Arango PA (2007). Cranberry prevents the adhesion of bacteria: Overview of relevant health benefits. Agro Food Ind Hi Tech 18(1, S), 24–27. [Google Scholar]
- Chen YY, Wu CC, Hsu JL, Peng HL, Chang HY & Yew TR (2009). Surface rigidity change of Escherichia coli after filamentous bacteriophage infection. Langmuir 25(8), 4607–4614. [DOI] [PubMed] [Google Scholar]
- Cho H & Bernhardt TG (2014). Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159(6), 1310–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chroma M & Kolar M (2010). Genetic methods for detection of antibiotic resistance: Focus on extended-spectrum β-lactamases. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 154(4), 289–396. Available at http://www.ncbi.nlm.nih.gov/pubmed/21293539. [DOI] [PubMed] [Google Scholar]
- CLSI (2017). Performance standards for antimicrobial susceptibility testing 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing; 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Coleman K (2011). Diazabicyclooctanes (DBOs): A potent new class of non-β-lactam β-lactamase inhibitors. Curr Opin Microbiol 14(5), 550–555. [DOI] [PubMed] [Google Scholar]
- Costerton JW & Lewandowski Z (1995). Microbial biofilms. Annu Rev Microbiol 49, 711–745. [DOI] [PubMed] [Google Scholar]
- Daffonchio D & Verstraete W (1995). Contact angle measurement and cell hydrophobicity of granular sludge from upflow anaerobic sludge bed reactors. Appl Environ Microbiol 61(10), 3676–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, De Kievit TR, Masoud H, Altman E, Richards JC, Sadovskaya I, Speert DP & Lam JS (1994). Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infect Immun 62(3), 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour AH (2009). Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794(5), 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi KP, Nisha SA, Sakthivel R & Pandian SK (2010). Eugenol (an essential oil of clove) acts as an antibacterial agent against Salmonella typhi by disrupting the cellular membrane. J Ethnopharmacol 130(1), 107–115. [DOI] [PubMed] [Google Scholar]
- Doktycz MJ et al. (2003). AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy 97(1–4), 209–216. [DOI] [PubMed] [Google Scholar]
- Dong H, Chen M, Rahman S, Parekh HS, Cooper HM & Xu ZP (2003). AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy 97(1–4), 209–216. [DOI] [PubMed] [Google Scholar]
- Donlan RM (2002). Biofilms: Microbial life on surfaces. Emerg Infect Diseases. Available at http://www.cdc.gov/ncidod/EID/vol8no9/02-0063.htm, 8(9), 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF (2002). Atomic force microscopy, a powerful tool in microbiology. J Bacteriol 184(19), 5205–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durodie J, Coleman K, Simpson IN, Loughborough SH & Winstanley DW (1995). Rapid detection of antimicrobial activity using flow cytometry. Cytometry 21(4), 374–377. [DOI] [PubMed] [Google Scholar]
- Eastman T & Zhu D (1996). Adhesion forces between surface-modified AFM tips and a mica surface. Langmuir 12(11), 2859–2862. [Google Scholar]
- Eriksson M, Nielsen PE & Good L (2002). Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J Biol Chem 277(9), 7144–7147. [DOI] [PubMed] [Google Scholar]
- Filho RP, Polli MC, Filho SB, Garcia M & Ferreira EI (2010). Prodrugs available on the Brazilian pharmaceutical market and their corresponding bioactivation pathways. Brazilian J Pharmaceutical Sci 46(3), 393–420. [Google Scholar]
- Fruci M & Poole K (2016). Bacterial stress responses as determinants of antimicrobial resistance. Stress Environ Regul Gene Expression Adaptation Bacteria 1 (April), 115–136. [Google Scholar]
- Garrett TR & Zhang Z (2008). Bacterial adhesion and biofilms on surfaces. Prog Nat Sci 18(9), 1049–1056. [Google Scholar]
- Ghuysen J-M (1994). Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol 2(10), 372–80. [DOI] [PubMed] [Google Scholar]
- Golding CG, Lamboo LL, Beniac DR & Booth TF (2016). The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci Rep 6(February), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordesli FP & Abu-Lail NI (2012). The role of growth temperature in the adhesion and mechanics of pathogenic L. monocytogenes: An AFM study. Langmuir 28(2), 1360–1373. [DOI] [PubMed] [Google Scholar]
- Grant LM & Ducker WA (1997). Effect of substrate hydrophobicity on surface – aggregate geometry: Zwitterionic and nonionic surfactants. J Phys Chem B 101(27), 5337–5345. [Google Scholar]
- Gupta A (2015). Biofilm Quantification and Comparative Analysis of MIC (Minimum Inhibitory Concentration) & MBIC (Minimum Biofilm Inhibitory Concentration) Value for Different Antibiotics against E. coli. Int. J. Curr. Microbiol App. Sci 4(2), 198–224. [Google Scholar]
- Halder S, Yadav KK, Sarkar R, Sudipta M, Saha P, Haidar S, Karmakar S & Sen T (2015). Alteration of Zeta potential and membrane permeability in bacteria: A study with cationic agents. Springer Plus 4(1), 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey PM, Warren RE, Livermore DM, McNulty CAM, Enoch DA, Otter JA, Wilson A & Peter R (2018). Treatment of infections caused by multidrug-resistant gram-negative bacteria: Report of the British society for antimicrobial chemotherapy/healthcare infection society/British infection association joint working party. J Antimicrob Chemother 73(March 2015), iii2–iii78. [DOI] [PubMed] [Google Scholar]
- Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA & Miller SI (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436(7054), 1171–1175. [DOI] [PubMed] [Google Scholar]
- Huang Q, Wu H, Cai P, Fein JB & Chen W (2015). Atomic force microscopy measurements of bacterial adhesion and biofilm formation onto clay-sized particles. Sci Rep 5(November), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter JL & Bechhoefer J (1993). Calibration of atomic-force microscope tips. Rev Sci Instrum 64(7), 1868–1873. [Google Scholar]
- Ito A, Taniuchi A, May A, Kawata K & Okabe S (2009). Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl Environ Microbiol 75 (12), 4093–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA & Medeiros AA (1991). More extended-spectrum beta-lactamases. Antimicrob Agents Chemother 35(9), 1697–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SG & Schuetz AN (2012). Current concepts in laboratory testing to guide antimicrobial therapy. Mayo Clin Proc 87(3), 290–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Lynette C & Hultgren SJ (2007). Morphological plasticity as a bacterial survival strategy. Nat Rev Micro, advanced o. Available at file:///c:/pdf/rfd6662.pdf%5Cnfile:///home/ford/Documents/PDF/rfd6662.pdf. [DOI] [PubMed] [Google Scholar]
- Kaplan JB (2011). Antibiotic-induced biofilm formation. Int J Artif Organs 34(9), 737–751. [DOI] [PubMed] [Google Scholar]
- Kennedy WPU & Murdoch JMC (1963). Ampicillin in treatment of certain gram-negative bacterial infections. BMJ 2(5363), 962–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili AA & Ahmad MR (2015). A review of cell adhesion studies for biomedical and biological applications. Int J Mol Sci 16(8), 18149–18184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowska A & Sigler K (2014). How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol 4 (August), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis 10 (9), 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert PA (2002). Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J Appl Microbiol 92(s1), 46S–54S. [PubMed] [Google Scholar]
- Lee H, Myers C, Zaidel L, Nalam PC, Caporizzo MA, Daep Carlo A, Eckmann DM, Masters JG & Composto RJ (2017). Competitive adsorption of polyelectrolytes onto and into pellicle-coated hydroxyapatite investigated by QCM–D and force spectroscopy, doi: 10.1021/acsami.7b02774. [DOI] [PubMed] [Google Scholar]
- Lewis K (2007). Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5(1), 48–56. [DOI] [PubMed] [Google Scholar]
- Limoli DH, Jones CJ & Wozniak DJ (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr 3(3), 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Black MA, Caron L, & Camesano TA (2006). Role of cranberry juice on molecular-scale surface characteristics and adhesion behavior of Escherichia coli. Biotechnol Bioeng 93(2), 297–305. [DOI] [PubMed] [Google Scholar]
- Livermore DM (2004). The need for new antibiotics. Clin Microbiol Infect 10(Suppl 4), 1–9. [DOI] [PubMed] [Google Scholar]
- Longo G, Rio LM, Trampuz A, Dietler G, Bizzini A & Kasas S (2013). Antibiotic-induced modifications of the stiffness of bacterial membranes. J Microbiol Methods 93(2), 80–84. [DOI] [PubMed] [Google Scholar]
- Luyt CE, Bréchot N, Trouillet JL & Chastre J (2014). Antibiotic stewardship in the intensive care unit. Crit Care 18(5), 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Conover M, Lu H, Parsek MR, Bayles K & Wozniak DJ (2009). Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5(3). e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L et al. (2011). Influence of surface roughness on streptococcal adhesion forces to composite resins. Dent Mater 27(8), 770–778. [DOI] [PubMed] [Google Scholar]
- Meincken M, Holroyd DL & Rautenbach M (2005). Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli. Society 49(10), 4085–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad H, Mayhoub AS, Cushman M & Seleem MN (2015). Anti-biofilm activity and synergism of novel thiazole compounds with gly-copeptide antibiotics against multidrug-resistant Staphylococci. J Antibiot 68(4), 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S, Mishra S, Jena P, Jacob B, Sarkar B & Sonawane A (2012). An investigation on the antibacterial, cytotoxic, and antibiofilm efficacy of starch-stabilized silver nanoparticles. Nanomedicine 8(6), 916–924. [DOI] [PubMed] [Google Scholar]
- Munita JM & Arias CA (2016). Mechanisms of antibiotic resistance. Microbiol Spectr 4(2), 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev: MMBR 67(4), 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S (2015). Meeting the societal need for new antibiotics: The challenges for the pharmaceutical industry. Br J Clin Pharmacol 79(2), 168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA (2011). Microtiter dish biofilm formation assay. J Visualized Exp (47), 10–11. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R, Azeredo J, Teixeira P & Fonseca AP (2001). The role of hydrophobicity in bacterial adhesion. Bioline (October 2016), 11–22. Available at http://repositorium.sdum.uminho.pt/handle/1822/6706. [Google Scholar]
- Pagès JM & Winterhalter M (2008). The porin and the permeating antibiotic: A selective diffusion barrier in gram-negative bacteria. Nat Rev Microbiol 6 (12), 893–903. [DOI] [PubMed] [Google Scholar]
- Pantanella F, Valenti P, Frioni A, Natalizi T, Coltella L, Berlutti F (2008). Biotimer assay, a new method for counting Staphylococcus spp. in biofilm without sample manipulation applied to evaluate antibiotic susceptibility of biofilm. J Microbiol Methods 75(3), 478–484. [DOI] [PubMed] [Google Scholar]
- Park BJ & Abu-Lail NI (2009). A correlation between the virulence and the adhesion of Listeria monocytogenes to silicon nitride: An atomic force microscopy study. Colloids Surf B 73(2), 237–243. [DOI] [PubMed] [Google Scholar]
- Park BJ & Abu-Lail NI (2011a). The role of the pH conditions of growth on the bioadhesion of individual and lawns of pathogenic Listeria monocytogenes cells. J Colloid Interface Sci 358(2), 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ & Abu-Lail NI (2011b). Variations in the nanomechanical propertis of virulent and avirulent Listeria monocytogenes. Soft Matter 6(16), 3898–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B-J and Abu-Lail NI (2011c). Atomic force microscopy investigations of heterogeneities in the adhesion energies measured between pathogenic and non-pathogenic Listeria species and silicon nitride as they correlate to virulence and adherence. Biofouling 27(5), 543–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry CC, Weatherly M, Beale T & Randriamahefa A (2009). Atomic force microscopy study of the antimicrobial activity of aqueous garlic versus ampicillin against Escherichia coli and Staphylococcus aureus. J Sci Food Agric 89(6), 958–964. [Google Scholar]
- Pickering SAW, Bayston R & Scammell BE (2003). Electromagnetic augmentation of antibiotic efficacy in infection of orthopaedic implants. J Bone Joint Surg 85(4), 588–593. [DOI] [PubMed] [Google Scholar]
- Read AF & Woods RJ (2014). Antibiotic resistance management. Evol Med Public Health 2014(1), 147–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD & Weibel DB (2011). Physiochemical regulation of biofilm formation. MRS Bull 36(5), 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolinson GN (1980). Effect of lactam antibiotics on bacterial cell growth rate. Microbiology 120(2), 317–323. [DOI] [PubMed] [Google Scholar]
- Rology VI, Kitchin PA, Bootman JS & Lane B (1993). MEDICAL quality control of the polymerase chain reaction. Methods in Molecular Medicine 3, 107–114. [Google Scholar]
- Römling U & Balsalobre C (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med 272(6), 541–561. [DOI] [PubMed] [Google Scholar]
- Roosjen A, Busscher HJ, Norde W & Van der Mei HC (2006). Bacterial factors influencing adhesion of Pseudomonas aeruginosa strains to a poly(ethylene oxide) brush. Microbiology 152(9), 2673–2682. [DOI] [PubMed] [Google Scholar]
- Rossolini GM, Arena F, Pecile P & Pollini S (2014). Update on the antibiotic resistance crisis. Curr Opin Pharmacol 18, 56–60. [DOI] [PubMed] [Google Scholar]
- Russell AD (1999). Bacterial resistance to disinfectants: Present knowledge and future problems. J Hosp Infect 43(Suppl. 1), 57–68. [DOI] [PubMed] [Google Scholar]
- Sailer FC & Young KD (2003). β-lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol Lett 226(2), 245–249. [DOI] [PubMed] [Google Scholar]
- Satta G & Canepari P (1979). Peptidoglycan synthesis in cocci and rods of a morphologically conditional mutant of Klebsiella pneumoniae peptidoglycan in cocci and rods of a pH- dependent, morphologically conditional mutant of Klebsiella pneumoniae. Journal of Bacteriology 137(2), 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K (2006). Persisters: A distinct physiological state of E. coli. BMC Microbiol 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh S, Fatima J, Shakil S, Rizvi SM & Kamal MA (2015). Antibiotic resistance and extended spectrum betalactamases: Types, epidemiology and treatment. Saudi J Biol Sci 22(1), 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon SL & Oliver KB (2014). Antibiotic resistance threats in the United States: Stepping back from the brink. Am Fam Physician 89(12), 938–941. [PubMed] [Google Scholar]
- Stenstrom TA (1989). Bacterial hydrophobicity, an overall parameter for the measurement of adhesion potential to soil particles. Microbiology 55(1), 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovi S, Vukovi D, Hola V, Di Bonaventura G, Djuki S, Cirkovi I & Ruzicka F (2007). Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 115(3), 891–899. [DOI] [PubMed] [Google Scholar]
- Stewart PS (2002). Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 292(2), 107–113. [DOI] [PubMed] [Google Scholar]
- Sundari S & Balasubramanian D (1991). Are amphiphilic chains. Methods 1065, 35–41. [DOI] [PubMed] [Google Scholar]
- Thomson JM & Bonomo RA (2005). The threat of antibiotic resistance in gram-negative pathogenic bacteria: B-lactams in peril!. Curr Opin Microbiol 8(5), 518–524. [DOI] [PubMed] [Google Scholar]
- Touhami A, Jericho MH, Boyd JM & Beveridge TJ (2006). Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. Am Soc Microbiol 188(2), 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA & Vollmers W (2012). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10(2), 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdrecht MC, Johannes L, Norde W, Schraa G & Zhender AJB (1987). The role of bacterial cell wall hydrophobicity in adhesion. Appl Environ Microbiol 53(8), 1893–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha VM (2010). Growing antibiotics resistance and the need for new antibiotics. Indian Pediatr 47(6), 505–506. [DOI] [PubMed] [Google Scholar]
- Vollmer W & Seligman SJ (2010). Architecture of peptidoglycan: More data and more models. Trends Microbiol 18(2), 59–66. [DOI] [PubMed] [Google Scholar]
- Wang H, Wilksch JJ, Strugnell RA & Gee ML (2015a). Role of capsular polysaccharides in biofilm formation: An AFM nanomechanics study. ACS Appl Mater Interfaces 7(23), 13007–13013. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang J, Ren G, Li Y & Wang X (2015b). Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli. Mar Drugs 13(6), 3325–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch K & Strømme M (2012). A method for quantitative determination of biofilm viability. J Funct Biomater 3(4), 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2015). Model List of Essential Medicines--Explanatory notes—19th List. (April), p. 55 Available at http://www.who.int/medicines/publications/essentialmedicines/en/. [Google Scholar]
- Wong AHM, Wenzel RP & Edmond MB (2000). Epidemiology of bacteriuria caused by vancomycin-resistant enterococci--A retrospective study Am J Infect Control 28(4), 277–281. [DOI] [PubMed] [Google Scholar]
- Worthington RJ & Melander C (2013). Overcoming resistance to β-lactam antibiotics. J Org Chem 78(9), 4207–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Shah MK, Powell LC & Armstrong I (2010). Application of AFM from microbial cell to biofilm. Scanning 32(3), 134–149. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang K, Tan W, He X, Jin R, Li J & Li H (2006). Atomic force microscopy study of different effects of natural and semisynthetic ß-lactam on the cell envelope of Escherichia coli. Anal Chem 78(20), 7341–7345. [DOI] [PubMed] [Google Scholar]
- Yoon MY, Lee KM, Park YY & Sang S (2016). Combating antimicrobial resistance: Policy recommendations to save lives. Sci Rep. Nature Publishing Group, 22(2), 410–415. [Google Scholar]
- Yoon MY, Lee KM, Park YY & Sang S (2011). Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS ONE 6(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD (2006). The selective value of bacterial shape. Microbiol Mol Biol Rev 70(3), 660–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X & Lin J (2013). Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front Microbiol 4(May), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.