Abstract
Next-generation sequencing (NGS) at the HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1 and -DRB¾/5 loci was performed on 282 healthy unrelated individuals from different major regions of Spain. High-resolution HLA genotypes defined by full sequencing of class I loci and extended coverage of class II loci were obtained to determine allele frequencies and also to estimate extended haplotype frequencies. HLA alleles were typed at the highest resolution level (4-field level, 4FL); with exception of a minor deviation in HLA-DPA1, no statistically significant deviations from expected Hardy Weinberg Equilibrium (HWE) proportions were observed for all other HLA loci. This study provides new 4FL-allele and -haplotype frequencies estimated for the first time in the Spanish population. Furthermore, our results describe extended haplotypes (including the less frequently typed HLA-DPA1 and HLA-DQA1 loci) and show distinctive haplotype associations found at 4FL-allele definition in this Spanish population study. The distinctive allelic and haplotypic diversity found at the 4FL reveals the high level of heterozygosity and specific haplotypic associations displayed that were not apparent at 2-field level (2FL). Overall, these results may contribute as a useful reference source for future population studies, for HLA-disease association studies as a healthy control group dataset and for improving donor recruitment strategies of bone marrow registries. HLA genotyping data of this Spanish population cohort was also included in the 17th International Histocompatibility and Immunogenetics Workshop (IHIW) as part of the study of HLA diversity in unrelated worldwide populations using NGS.
Keywords: Human Leukocyte Antigen (HLA), Next-Generation Sequencing (NGS), Population study, Spain
1. Introduction
Mainland Spain is located on the Iberian Peninsula in Southwestern Europe and it is also very close to North Africa being just separated by the Strait of Gibraltar. The Spanish territory also includes the Balearic Islands in the Mediterranean Sea, the Canary Islands off the North African Atlantic coast and two cities, Ceuta and Melilla, located on the northern coast of Africa. As a consequence of its unique geographic location, Spain shows an extensive cultural diversity within its population (e.g. Spanish is the main language spoken but Catalan in the East, Galician in the Northwest and Euskera in the Western Pyrenees are also spoken languages). The Spanish population diversity resulting from migrations through its history is well documented (e.g. Christian Visigoths, North African Muslims and Sephardic Jews coexisted in the Iberian Peninsula for several centuries) [1]; recent migrations have further increased diversity (e.g. migrants who represent approximately 13% of the Spanish census, who are mainly coming from countries of Eastern Europe, South America and Northern Africa) [2].
In the clinical histocompatibility setting, assessment of HLA allelic and haplotypic diversity of each population is important, especially for hematopoietic stem cell transplantation (HSCT) [3] with unrelated donors. Previous studies have described allelic and haplotypic HLA frequency distribution in Spanish population [4–11]. However, most of these previous studies were only based on lower-resolution HLA typing data (allele resolution level at the 1-field or at the 2-field) generated by traditional HLA molecular typing techniques most commonly used in routine practice (sequence-specific primer (SSP) or sequence-specific oligonucleotide (SSO) probe technologies and sequence-based typing (SBT)). The majority of these earlier studies analyzed only sets of individuals coming from specific regions of Spain. At the same time, HLA typing was performed for certain loci but not for all major HLA loci. As a result, most of these previous studies did not define complete extended HLA haplotypes. Recent works have placed emphasis in the importance of elucidating HLA diversity for all major HLA genes and at a higher allele resolution level for all worldwide populations [12]; this is an unmet need for the Spanish population [4], that may improve current donor search criteria and therapeutic strategies in the HSCT field [3]. Application of next-generation sequencing (NGS) for high-resolution molecular HLA typing has enabled to obtain full-length and/or extended sequences and genotypes of all major HLA genes. This is based on the clonal sequencing nature and the increased read length, throughput, accuracy and resolution that NGS offers for describing the high polymorphism presented by HLA genes [13]. Therefore, NGS-based HLA typing methods permit to overcome many of the limitations of legacy techniques (SSP, SSO and SBT) and also facilitate detection of novel alleles [14]. At the same time, implementation of high-throughput platforms in this NGS technology allows cost-effective and large-scale population genetics studies [12].
The aim of the present study was to describe allelic and haplotypic frequency distribution by typing all major HLA class I and class II genes (HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1 and - DRB¾/5) at high-resolution (allele resolution level at the 4-field) and obtaining genotypes by using high-throughput NGS for a representative sample of the Spanish population including a cohort of 282 healthy unrelated individuals from different major regions of the country.
2. Materials and methods
2.1. Sample collection and testing methods
This population study includes 282 healthy unrelated individuals randomly selected from Spain in collaboration with the Spanish Working Group in Histocompatibility and Transplant Immunology (GETHIT) of the Spanish Society for Immunology (SEI). Collection of all genomic DNA samples consisted of 11 participant clinical laboratories that are situated in 10 different locations in Spain (Santander, Salamanca, Madrid (which included 2 different participant clinical laboratories), Barcelona, Valencia, Murcia, Córdoba, Sevilla, Málaga and Gran Canaria) which provided a set of 25–26 samples per institution (Figure 1). This HLA Spanish population study was approved by the Institutional Review Board (IRB) of the 17th International Histocompatibility and Immunogenetics Workshop (IHIW) as well as the respective local research and ethics committee of each Spanish participant institution and it was carried out in accordance with the principles of the Declaration of Helsinki. Samples were tested in parallel: i) All 282 samples were genotyped by using a commercial NGS-based HLA genotyping method [15] at the Stanford Histocompatibility, Immunogenetics and Disease Profiling Laboratory (HIDPL); ii) at the same time, the 11 Spanish participant clinical laboratories performed HLA typing tests (with a variable range of allele resolution level and number of HLA genes tested) of their respective sets of 25–26 samples by using other HLA molecular typing techniques (either using an in-house NGS platform or commercial/in-house SSO or SBT technologies).
Figure 1.
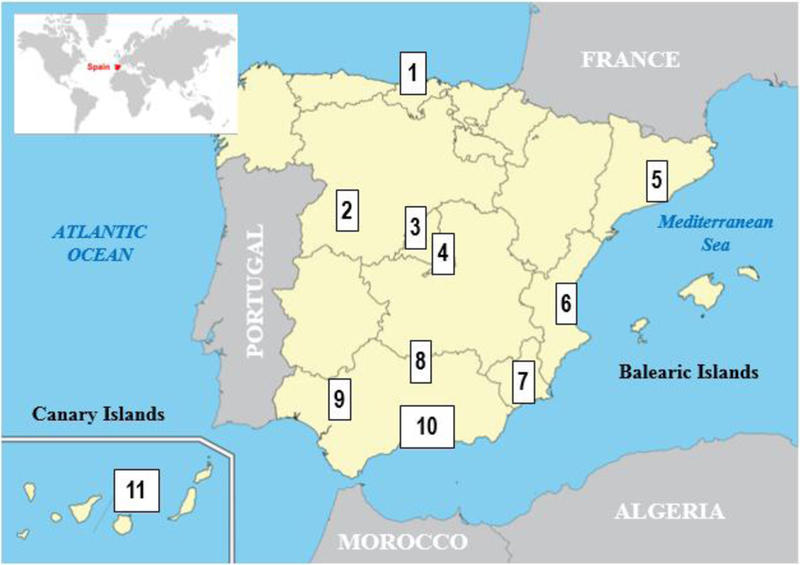
Map of the geographical location of Spain (Spanish territory colored in light yellow and Spanish provinces are delimited by grey borders) which shows the location of the 11 participant clinical laboratories (coded from 1 to 11) in the collection of samples (n=282 healthy unrelated individuals) for this study. In detail: [1] Immunology, Hospital Universitario Marqués de Valdecilla in Santander (n=25 samples); [2] Molecular Biology-Hematology, Hospital Clínico Universitario, in Salamanca (n=26 samples); [3] Immunogenetics and Histocompatibility, Instituto de Investigación Sanitaria Puerta de Hierro in Madrid (n=25 samples); [4] Histocompatibility, Centro de Transfusión de la Comunidad de Madrid in Madrid (n=26 samples); [5] Histocompatibility and Immunogenetics, Banc de Sang i Teixits in Barcelona (n=26 samples); [6] Histocompatibility, Centro de Transfusión de la Comunidad Valenciana in Valencia (n=26 samples); [7] Immunology, Hospital Clínico Universitario Virgen de la Arrixaca in Murcia (n=26 samples); [8] Immunology, Hospital Universitario Reina Sofía in Córdoba (n=26 samples); [9] Immunology, Hospital Universitario Virgen del Rocío in Sevilla (n=25 samples); [10] Histocompatibility, Centro de Transfusión de Málaga in Málaga (n=26 samples) and [11] Immunology, Hospital Universitario de Gran Canaria Dr Negrín in Las Palmas de Gran Canaria (n=25 samples).
*(Maps of this figure are a modified version from http://geology.com/world/map/map-of-spain.gif and from https://desfaziendoentuertos.prepress.es/mapas-de-espana-vectoriales-gratuitos/).
2.2. NGS-HLA sequencing and genotyping performed at Stanford HIDPL
All samples were genotyped for HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1 and - DRB¾/5 loci using the MIA FORA NGS FLEX HLA Typing 11 Kit 96 Tests (Immucor, Inc. Norcross, GA, USA), following manufacturer’s semi-automated protocol and as described previously [15]. Briefly, paired-end sequence reads were generated by using this aforementioned NGS-based HLA genotyping method, which specifically amplifies these 11 HLA genes with extensive coverage of the HLA genomic region by long-range polymerase chain reaction (PCR). All fragments of the final prepared DNA library were sequenced using sequencing by synthesis (SBS) chemistry in a massively parallel fashion on the Illumina® NGS sequencing platform (Illumina, Inc. San Diego, CA, USA). For assignment of HLA genotypes, NGS paired-end reads were analyzed using the MIA FORA FLEX version 3.0 software (Immucor, Inc. Norcross, GA, USA) and according to IPD-IMGT/HLA database version 3.25.0. All HLA genotyping calls automatically assigned by the software were manually reviewed (e.g for evaluation of ambiguities at the 4-field (see 2.3. Standardization of ambiguities at the 4-field)) and confirmed by the user.
2.3. Standardization of ambiguities at the 4-field
Some HLA assignments resulted ambiguous when trying to distinguish alleles at the 4-field allele resolution level (intronic and untranslated (UTR) sequence level). In these particular cases, called allele candidates present differences only in length of either homopolymer sequences or short tandem repeats (STRs); these were not sequenced with precision by the NGS method. In addition, few HLA-DPB1 ambiguities resulted from lack of coverage or phasing by the NGS sequencing methodology. Due to limitations for resolving this type of ambiguities, indistinguishable alleles at the 4-field level were merged to the lowest numbered allele according to IPD-IMGT/HLA database version 3.25.0. A complete list of indistinguishable alleles and their respective standardization criteria is shown in a separate report (Creary et al.; manuscript in preparation) and it is also available in the official 17th IHIW website (http://17ihiw.org/17th-ihiw-ngs-hla-data/).
2.4. HLA Spanish population data inclusion in the 17th IHIW database
Final standardized (according to aforementioned criteria) HLA typing data of this Spanish population cohort was validated and uploaded to the 17th International Histocompatibility and Immunogenetics Workshop database. Official version 3.25.0 of the IPD-IMGT/HLA database was used for this 17th IHIW database [16]. In addition, this HLA Spanish population dataset was also included as part of the study of HLA diversity in unrelated worldwide populations using NGS (Creary et al.; manuscript in preparation).
2.5. Statistical analyses
PyPop (Python for Population genomics) version 0.7.0 software was used to carry out allele frequencies determination, to estimate deviations from expected Hardy-Weinberg Equilibrium (HWE) proportions (based on exact test of Guo and Thompson), the Ewens-Watterson homozygosity (EWH) test of neutrality (tested by Slatkin’s implementation of the Monte-Carlo approximation of the Ewens-Watterson exact test, using a two-tailed test (p < 0.05) of the null hypothesis of neutrality) and all pairwise linkage disequilibrium (LD) estimates [17]. Hapl-o-Mat version 1.1 software was used to estimate extended haplotype frequencies from this current Spanish genotypic data using a maximum likelihood estimation via an expectation-maximization (EM) algorithm [18]. Finally, in order to compare allele frequencies (at the HLA-A, -B, -C, -DQB1 and -DRB1 loci) between the 3 different geographical Spanish regions established for this study (Northern-Central, Eastern and Southern Spain) as well as between the 10 Spanish locations studied in the present work, a population dendogram was constructed using POPTREEW (web version of POPTREE software) [19]. A total of 1000 dendrogram replicates based on the matrices of Nei genetic distances (DA) [20] were generated using the neighbor-joining (NJ) method [21].
3. Results
3.1. Evaluation of concordance of HLA typing results obtained from this study
HLA genotyping results obtained for all 282 samples by using this commercial NGS-based method [15] at Stanford HIDPL are 100% concordant with those available HLA typing results (e.g. HLA-DPA1 locus was not tested locally) obtained by using other HLA molecular typing techniques (either using an in-house NGS platform or commercial/in-house SSO or SBT technologies) respectively at the 11 local participant clinical laboratories from Spain (Supplementary Table 1). Therefore, we confirmed that all samples were tested correctly by all the participating laboratories without any sample-switching error, allele dropout (for the HLA loci tested respectively) and neither contamination.
3.2. Evaluation of deviations from expected Hardy-Weinberg equilibrium (HWE) proportions
At the 4-field allele resolution level, no overall deviations from expected HWE proportions are observed in any of the HLA loci analyzed with the exception of a minor but significant departure at the HLA-DPA1 locus (p-value = 0.0104) (Supplementary Table 2). To further investigate this HLA-DPA1 departure, collapsed 2-field and 3-field HLA genotyping datasets of this same Spanish population cohort (n=282) were evaluated (data not shown) and no HWE deviation was observed at any of the HLA loci. Furthermore, estimated homozygosity (Waterson’s homozygosity F statistic (F)) in HLA-DPA1 locus at the 4-field allele resolution level shows a much lower value (F=0.164) in comparison to collapsed 2-field (F=0.649) and 3-field (F=0.635) HLA genotyping datasets. Altogether, this can be interpreted as estimated deviations from HWE may not be corrected properly for multiple comparisons including low number counts of alleles or genotypes when they are present at the 4-field allele resolution level. Thus, this observed deviation may be explained by the fact that HLA alleles presenting low frequencies (e.g. HLA-DPA1 alleles) would not be considered properly when evaluating HWE proportions and their contribution to HWE deviation would be being estimated higher than it should be at this 4-field allele resolution level. Overall, the HLA dataset of this present study was considered valid for proceeding with the rest of statistical analyses.
3.3. HLA allelic frequencies in this Spanish population cohort
The frequency distribution of HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1, and -DRB¾/5 alleles at 4-field level of resolution are summarized in Supplementary Table 3. 36 HLA-A, 53 HLA-B, 40 HLA-C, 14 HLA-DPA1, 29 HLA-DPB1, 23 HLA-DQA1, 24 HLA-DQB1, 37 HLA-DRB1, 5 HLA-DRB3, 5 HLA-DRB4 and 3 HLA-DRB5 distinct alleles were identified. It can be observed how the most predominant HLA alleles (frequency higher than 5%) represent in the case of each locus: 6 HLA-A alleles (66 %), 6 HLA-B alleles (42%), 7 HLA-C alleles (55%), 7 HLA-DPA1 alleles (91%), 4 HLA-DPB1 alleles (66%), 9 HLA-DQA1 alleles (81%), 7 HLA-DQB1 alleles (73%), 6 HLA-DRB1 alleles (58%). In the case of HLA-DRB¾/5 alleles: HLA-DRB3*02:02:01:02 (15%), -DRB4*01:03:01:01 (16%) and -DRB5*01:01:01 (10%) are the most common.
3.4. Identification of two new HLA alleles in this Spanish population cohort
Two novel HLA alleles were identified during this Spanish population study using this aforementioned NGS-based HLA genotyping method [15] (Supplementary Figure 1). One individual (17th IHIW sample ID no. H00035F6, from Barcelona, Spain) presents a single base mismatch with HLA-B*38:20 allele reference sequence in exon 3 (codon 99), which leads to a synonymous substitution (Tyr (TAC) to Tyr (TAT)) (Supplementary Figure 1a-1c). Complete HLA genotyping result of this individual including the novel allele is:
HLA-A*29:02:01:01, HLA-A*25:01:01; HLA-C*12:03:01:01, HLA-C*03:03:01:01; HLA-B*38:20:02, HLA-B*15:01:01:01; HLA-DRB4*01:03:01:01, HLA-DRB3*02:02:01:02; HLA-DRB1*07:01:01:01, HLA-DRB1*13:01:01:01; HLA-DQA1*02:01:01:01, HLA-DQA1*01:03:01:02; HLA-DQB1*02:02:01:01, HLA-DQB1*06:03:01; HLA-DPA1*02:02:01, HLA-DPA1*01:03:01:01; HLA-DPB1*19:01, HLA-DPB1*02:01:02.
Also in another different subject (17th IHIW sample ID no. H00036D1, from Málaga, Spain), a single base mismatch with HLA-DRB3*02:02:01:01 allele reference sequence is detected in exon 3 (codon 166), which leads in this case to a non-synonymous substitution and, therefore, to an amino acid change (Arg (CGG) to Gln (CAG)) (Supplementary Figure 1d-1f). Complete HLA genotyping result of this other subject including the novel allele is:
HLA-A*11:01:01:01, HLA-A*11:01:01:01; HLA-C*05:01:01:02, HLA-C*15:02:01:01; HLA-B*44:02:01:01, HLA-B*51:01:01:01; HLA-DRB5*01:01:01, HLA-DRB3*02:71; HLA-DRB1*15:01:01:01, HLA-DRB1*03:01:01:01; HLA-DQA1*01:02:01:01, HLA-DQA1*05:01:01:01; HLA-DQB1*06:02:01, HLA-DQB1*02:01:01; HLA-DPA1*01:03:01:01, HLA-DPA1*01:03:01:02; HLA-DPB1*04:01:01:01, HLA-DPB1*02:01:02.
To confirm these findings, sequence-based typing (SBT) was performed using respective SBTexcellerator kits (GenDx, Utrecht, The Netherlands) on a 3130xL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and SBTengine HLA typing software version 3.14.0.2783 (GenDx, Utrecht, The Netherlands) at the corresponding local Spanish clinical laboratories of origin.
Reported sequences of both identified exon variants were submitted to GenBank and to the IPD-IMGT/HLA Database. These two new alleles have been officially assigned by the WHO HLA Nomenclature Committee for Factors of the HLA System [22]. In the case of the new HLA-B*38:20 allele, the official name given is HLA-B*38:20:02 (GenBank accession no. MG76848 and IPD-IMGT/HLA submission no. HWS10051845). Regarding the new HLA-DRB3*02:02:01:01 allele, the official name given is HLA-DRB3*02:71 (GenBank accession no. MG922498 and IPD-IMGT/HLA submission no. HWS10051607).
3.5. Ewens-Watterson homozygosity (EWH) test of neutrality
EWH test of neutrality was used for analysis of selective processes based on HLA allelic diversity at the 4-field allele resolution level of this Spanish population cohort. All HLA loci analyzed show levels of observed homozygosity (Fo) that are below the expected homozygosity under neutrality (Fe) with the exception of HLA-DPB1 locus (Table 1). Furthermore, HLA-B, -DQA1 and -DQB1 are the only loci that show statistically significant deviation from neutrality and, therefore, are consistent with a more pronounced balancing selection (Fnd << 0). As previously described across human populations [23][24], we also observed for this Spanish population cohort (in spite of presenting a relatively small sample size) an overall direction towards balancing selection for most of the classical HLA class I and II loci with the striking exception of HLA-DP genes. These latter (especially HLA-DPB1 locus, based on our results at the 4-field allele resolution level) seem to be more under directional selection, in which only a set of few alleles become selected (e.g. HLA-DPB1*04:01:01:01). These interpretations however need to be confirmed on a larger Spanish cohort, considering also the diverse nature of the regional subpopulations included in this study.
Table 1.
Ewens-Watterson Homozygosity (EWH) test of neutrality at the HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1 loci based on the 4-field allele resolution level HLA genotyping data (and according to IPD-IMGT/HLA database version 3.25.0) of this Spanish population study.
| Locus | Number of subjects typed (n) | k | Fo | Fe | Fnd | p-value of F |
|---|---|---|---|---|---|---|
| HLA-A | 276 | 36 | 0.0983 | 0.1048 | −0.1924 | 0.5208 |
| HLA-B | 276 | 53 | 0.0444 | 0.0661 | −1.2197 | 0.0331* |
| HLA-C | 276 | 40 | 0.0617 | 0.0934 | −1.0858 | 0.0577 |
| HLA-DPA1 | 275 | 14 | 0.1639 | 0.2766 | −1.0498 | 0.0806 |
| HLA-DPB1 | 274 | 29 | 0.1769 | 0.1336 | 0.9376 | 0.8572 |
| HLA-DQA1 | 272 | 23 | 0.0872 | 0.1708 | −1.3322 | 0.0094** |
| HLA-DQB1 | 269 | 24 | 0.0869 | 0.1630 | −1.2857 | 0.0120* |
| HLA-DRB1 | 272 | 37 | 0.0733 | 0.1015 | −0.8761 | 0.1443 |
The normalized deviate of the Ewens-Watterson homozygosity statistic (Fnd) was calculated based on the observed allele frequencies at each HLA locus and it is used to infer the action of balancing (Fnd << 0) or directional (Fnd >> 0) selection at each HLA locus. The results of the Ewens-Watterson Homozygosity Test are shown above. Number of unique alleles (k); Observed F (Fo); Expected F (Fe); Normalized deviate of F (Fnd).
p-value of F lower than 0.05 (p < 0.05) indicates a statistical significance at the 5% level.
p-value of F lower than 0.01 (p < 0.01) indicates a statistical significance at the 1% level.
Also when performing EWH test, HLA-DRB¾/5 loci were not included as they represent a particular virtual single “locus”. Where these HLA-DRB¾/5 genes characteristically behave as alleles of a single locus as the presence of one of these genes at the haplotype level excludes the presence of the other two genes. This is based on the linkage constraints that exist between the HLA-DRB¾/5 loci and the HLA-DRB1 locus, in which several HLA-DRB1 allele families can be differentiated [22].
3.6. 2-locus haplotype linkage disequilibrium (LD) analysis
Estimated 2-locus haplotype frequencies and measure of overall LD (Hedrickś D’ statistic) of pairs of neighboring genetic HLA loci (B~C, DPA1~DPB1, DQA1~DQB1 and DQB1~DRB1) at the 4-field allele resolution level are shown in Supplementary Table 4. Interestingly, it can be observed unique 2-locus haplotype associations in non-coding regions at the 4-field allele resolution level that are not apparent at the 2-field level. For instance, alleles of the HLA-B*35 allele group show very distinctive associations with HLA-C alleles at the the 4-field level. On one hand, at the 2-field level, HLA-B*35:01, HLA-B*35:02, HLA-B*35:03 and HLA-B*35:08 alleles show a strong and common association with HLA-C*04:01 allele. Nevertheless, at the 4-field level we observed that in the case of the intron variant HLA-B*35:01:01:01 it displays a specific association with HLA-C*04:01:01:01. Whereas, B*35:01:01:02 intron variant presents associations with not only HLA-C*04:01:01:01 allele but also with HLA-C*04:01:01:05 and HLA-C*04:01:01:06 alleles. In the case of HLA-B*35:02:01, we observed it seems to display a specific association with HLA-C*04:01:01:06 in Spanish population. As for HLA-B*35:03:01, it presents association with HLA-C*04:01:01:01. Finally, HLA-B*35:08:01 shows association with HLA-C*04:01:01:06. Furthermore, we also found distinctive haplotypic associations at the intronic level in several other HLA class I and class II loci pairs (e.g. HLA-DQA1*05:01:01 intron variants, HLA-B*18:01:01 intron variants, HLA-C*05:01:01 intron variants or HLA-C*06:02:01 intron variants). In contrast, HLA loci pairs as B*07:02:01~C*07:02:01:03, DQA1*01:01:01:02~DQB1*05:01:01:03 and DQB1*02:02:01:01~DRB1*07:01:01:01 are some examples of 4-field highly conserved associations found in this Spanish population cohort.
3.7. Global measures of pairwise linkage disequilibrium (LD) for HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1 and -DRB3/4/5 loci
To evaluate the overall linkage disequilibrium (LD) we considered (Table 2) two different locus-pair-level measures. The D’ (normalized Hedrick’s D’ statistic) parameter, expressed as the normalization of the product of allele frequencies at each locus, weights the LD contribution of specific allele pairs [17]. Whereas the second parameter, Wn (Cramer’s V statistic), calculates also a normalization in this case of the chi-square statistic for deviations between observed and expected haplotype frequencies [17]. The strongest associations are observed for the contiguous and/or physically close HLA loci pairs including DRB1~DRB5/4/3, DRB1~DQA1, DQA1~DQB1 and DRB1~DQB1 followed by B~C. HLA-DPA1~DPB1 pair appears associated with less strength. Interestingly, in spite of HLA-A~C pair being physically closer than HLA-A~B the strength of the LD between the latter is higher, suggesting that differences in diversity between HLA-B and -C loci may play a role in determining this measurement. Associations between HLA-A~B and HLA-B~DRB1 appear in similar ranges. HLA-DP loci show weaker LD associations than any of the other pairwise comparisons. As previously reported [25][26], LD patterns of HLA-DP loci seem to be driven primarily in a different manner compared to the other HLA loci (e.g. relatively higher rate of recombination and combined DPA1~DPB1 amino acid epitope have been suggested to contribute on this distinctive selection).
Table 2.
Global measures of pairwise linkage disequilibrium (LD) for HLA-A, -B, -C, -DPA1, -DPB1, - DQA1, -DQB1, -DRB1 and -DRB¾/5 loci at the 4-field resolution level (and according to IPD-IMGT/HLA database version 3.25.0) in this Spanish population study (n=282 subjects).
| Locus Pair HLA- | D’ | Wn |
|---|---|---|
| B~C | 0.93630 | 0.77226 |
| A~C | 0.59490 | 0.43926 |
| A~B | 0.64088 | 0.45530 |
| DPA1~DPB1 | 0.82896 | 0.71883 |
| DQA1~DQB1 | 0.97854 | 0.78901 |
| DQA1~DRB1 | 0.98990 | 0.86147 |
| DQB1~DRB1 | 0.97446 | 0.80953 |
| DPB1~DRB1 | 0.47923 | 0.37854 |
| DPB1~DQB1 | 0.43446 | 0.38512 |
| B~DRB1 | 0.70620 | 0.46705 |
| B~DQA1 | 0.66583 | 0.49954 |
| B~DQB1 | 0.65365 | 0.49127 |
| DRB1~DRB3 | 0.96724 | 0.86672 |
| DRB1~DRB4 | 0.97158 | 0.69772 |
| DRB1~DRB5 | 1 | 1 |
3.8. Estimation of extended HLA haplotype frequencies
Maximum likelihood estimation via an expectation-maximization (EM) algorithm is a statistical method commonly used for HLA haplotype inference and estimation of haplotype frequencies in unrelated individuals from a population-specific genotype data as in the present study. Moreover, this statistical method serves as an alternate approach when it is not possible to rely on family segregation studies [18]. Inferred extended HLA haplotypes (encompassing 6-locus, 7-locus and 9-locus respectively) were evaluated for the estimation of haplotype frequencies in this Spanish population cohort:
HLA~A~C~B~DRB¾/5~DRB1~DQB1 (Supplementary Table 5); HLA~A~C~B~DRB¾/5~DRB1~DQA1~DQB1 (Supplementary Table 6); and HLA~A~C~B~DRB¾/5~DRB1~DQA1~DQB1~DPA1~DPB1 (Supplementary Table 7).
Similarly to what we found in 2-locus haplotypes, it can be observed very distinctive extended haplotype associations in non-coding regions at the 4-field level that are not apparent at lower allele resolution level (2-field or 3-field) results that are obtained when using legacy methodologies (e.g. SSP or SSO) with important limitations in sequence coverage and phasing in comparison to NGS-based typing [12][13].
3.9. Different HLA allele distributions found between Spanish regions
Finally, we examined the disparity/similarity of allelic distributions within this Spanish population cohort based on the results (at the 4-field allele resolution level) of the current study. In this sense, we carried out a comparison of HLA distributions (based on allele frequencies found at HLA-A, -B, -C, - DQB1 and -DRB1 loci) between the 3 different geographical Spanish regions established (Northern-Central, Eastern and Southern Spain) as well as between the 10 Spanish locations studied in the present work (Supplementary Table 8).
Despite of limitations in the sample size shown by these different Spanish population sub-groups in the present study. At the HLA allele level, it can be observed that most frequent alleles at a national level (considering entire Spanish population, termed as “ESP”, n=282) are fairly evenly distributed and well represented among the different Spanish regions and locations evaluated here, with some minor exceptions (specifically found at the different 10 Spanish locations level presenting a limited and small sample size comparatively) that need to be further analyze by future larger-scale population studies (see Supplementary Table 8.b)). Taking into account genetic distances evaluated here (see Supplementary Tables 8.c) and 8.d)), the present entire Spanish population cohort shows a Mediterranean genetic substrate that seems to be represented more predominantly by Eastern and Central regions/locations situated within the Central Plateau as previously described [4–11]. Whereas the most Northern and Southern regions/locations (which are mountainous areas that are more isolated geographically unlike this Central Castilian Plateau region in mainland Spain; or even being very unique island areas such as Canary Islands) diverge from this aforementioned Mediterranean HLA distribution as reported in previous works [4–11]. For instance, although we considered Barcelona location as part of the Eastern region of Spain for this study, we clearly observed how this Catalan location seems to be more related to other Northern locations than to Mediterranean sites such as Valencia or Murcia. Interestingly, Salamanca location population group (situated very close to the frontier that separates Spain from Portugal) describes a pronounced distinctive HLA distribution in comparison to other Northern-Central locations in Spain as previously described and it also exemplifies the extensive HLA diversity of the Iberian Peninsula [4][8]. Furthermore, the striking divergence observed in Malaga and Gran Canaria locations (see Supplementary Table 8.d)) may be explained by the reported historic genetic contribution from North African Berber populations [1][5].
We also attempted to do this regional study at the extended HLA haplotype level (data not shown). However, due to these limited small sample sizes found at the different Spanish regions and locations it was not possible to estimate accurately haplotype frequencies via an expectation-maximization (EM) algorithm [18] to evaluate haplotype sharing between regions/locations.
Overall, in spite of presenting a relatively small sample size, the present Spanish population study has allowed us to see the great potential of NGS-based HLA population studies in order to identify 4-field HLA allele signatures at a regional level as a consequence of both differential regional historic events and the characteristic regional orography that favors more isolation of certain local populations. Nonetheless, future studies of larger population sample size at a wider geographic scale will be needed to assess more accurately the HLA diversity in Spanish population in order to confirm these observations and findings of our study as well as to reveal other unknown but significant polymorphism within the HLA system.
4. Discussion
In the present study, we characterized HLA allelic sequences of 11 major HLA genes with extensive coverage and phased-alleles with minimum heterozygous ambiguity per locus at the 4-field for a representative Spanish population cohort (n=282) by applying this novel high-throughput NGS-based HLA typing method [15]. We also examined allelic and haplotypic HLA frequency distributions at the 4-field allele resolution level in the Spanish population for the first time.
At the HLA allele level, regarding HLA class I loci we observed that HLA-B locus presents the highest allele diversity in comparison to HLA-A and -C loci in relation to the number of alleles (k) found in this population. Nevertheless, the 4-field allele resolution level has allowed us to see a significant diversity at the nucleotide level for HLA-A and -C loci in contrast to HLA-B locus. As an example, the only observed variant HLA-C*04:01 at the 2-field level shows a total of three different variants at the 4-field level (HLA-C*04:01:01:01, HLA-C*04:01:01:05 and HLA- C*04:01:01:06; representing 70.2%, 2.4% and 27.4% respectively inside this HLA-C*04:01 allele group). Similarly, HLA-DPA1 and HLA-DQA1 loci exemplify the higher level of heterozygosity found at the the 4-field level (molecular variation in non-coding regions as introns and UTR regions) in comparison to the 2-field level (specific HLA protein-coding alleles). For instance, the only observed variant HLA-DQA1*05:01 at the 2-field level shows a total of three different variants at the 4-field level (HLA-DQA1*05:01:01:01, HLA-DQA1*05:01:01:02 and HLA-DQA1*05:01:01:03; representing 39.7%, 53.4% and 6.8% respectively inside this HLA-DQA1*05:01 allele group). On the other hand, it can be observed how certain loci such as HLA-DPB1 (estimated homozygosity F=0.184 at 2-field in contrast to F=0.177 at 4-field), HLA-DQB1 (F=0.110 at 2-field in contrast to F=0.087 at 4-field) and HLA-DRB1 (F=0.074 at 2-field in contrast to F=0.073 at 4-field) loci show less differences regarding the allelic diversity found between the 2-field level and the 4-field level in this present study.
NGS technology also facilitates the identification of novel alleles [14], possible null or expression variant alleles [27] and also the detection of rare alleles [28]. In the present study, in addition to the aforementioned two identified and confirmed new alleles, we observed two distinct null alleles found with intermediate or relatively high frequency in this Spanish population cohort: HLA-C*04:09N allele (0.4%) and HLA-DRB4*01:03:01:02N allele (1.8%). In relation to previously considered rare alleles, HLA-C*12:166 (0.2%) and HLA-B*15:220 (0.4%) were detected in this Spanish population cohort in several instances. Regarding common and well-documented (CWD) HLA alleles [29][30], and as previously described in recent NGS HLA population studies [31–33], the 4-field allele resolution level reveals how in certain allele groups, an allele considered rare initially it actually presents a common occurrence while the lowest numbered allele is not the most frequent. For instance, in HLA-B locus (e.g. HLA-B*35:01:01:01 allele represents only 3.6% of this allele group whereas HLA-B*35:01:01:02 allele represents 96.4% of this allele group found in this Spanish population) and HLA-DRB1 locus (e.g. HLA-DRB1*12:01:01:03 allele represents 100% of this allele group whereas HLA-DRB1*12:01:01:01 allele is absent in this Spanish population).
At the HLA haplotype level, application of NGS-based methodology has allowed the assessment of distinctive 4-field haplotypic associations when evaluating non-coding region variation at both 2-locus and extended haplotype (encompassing 6-locus, 7-locus and 9-locus respectively) distributions in this Spanish population cohort. For instance, the HLA-DRB1*11:04:01 allele was found in two extended haplotypes that are similarly frequent in the Spanish population but differ strikingly at the 4-field at several HLA loci; these include:
HLA-A*24:02:01:01~C*04:01:01:06~B*35:02:01~DRB3*02:02:01:02~DRB1*11:04:01~DQA1*05:05:01:01~DQB1*03:01:01:02~DPA1*01:03:01:04~DPB1*04:01:01:01 (HF = 0.030), which is highly conserved and very common in Middle Eastern populations [34][35]; and HLA-A*01:01:01:01~C*07:01:01:01~B*18:01:01:02~DRB3*02:02:01:02~DRB1*11:04:01~DQA1*05:05:01:01~DQB1*03:01:01:03~DPA1*01:03:01:05~DPB1*04:02:01:01 (HF = 0.020). Therefore, intron variation (in the latter example according to HLA-DPA1 and -DQB1 loci) appears to be haplotype-specific.
Balas et al. study can be considered one of the most recent representative works evaluating the distribution of HLA alleles (HLA-A, -B, -C, -DQB1, -DRB1 and -DRB¾/5 loci) and extended haplotypes (defined by family segregation analysis) at a relatively high-resolution level in Spanish population [7]. Our present results at the HLA allele and haplotype level show very analogous common alleles and extended haplotypes in comparison to that previous study [7]. Moreover, when evaluating HLA-B~C haplotype distributions, we also found that HLA-B*51:01:01:01 allele displays a very broad distribution in relation to its association with HLA-C alleles (7 different associated HLA-C alleles were observed in this present study) (see Supplementary Table 4). Hence, HLA-B*51:01:01:01 allele could be considered as a negative predictive value to find a full-match unrelated donor (URD) for a Spanish patient [7]. In addition, we also found those same common haplotypes (e.g. HLA-A*30:02:01:01~C*05:01:01:01~B*18:01:01:01~DRB3*02:02:01:01~DRB1*03:01:01:01~DQB1*02:01:01; or HLA-A*25:01:01~C*12:03:01:01~B*18:01:01:02~DRB5*01:01:01~DRB1*15:01:01:01~DQB1*06:02:01) in Spanish population which are not as frequent as in other foreign unrelated bone marrow registries [36–38]. Therefore, this shows that development of local donor registries is also important for optimizing the URD search.
In comparison to other reported HLA populations datasets [36–38], it is noteworthy that the most common haplotype in North European and European American populations (extended haplotype at the 4-field level observed in this present study:
HLA-A*01:01:01:01~C*07:01:01:01~B*08:01:01:01~DRB3*01:01:02:01~DRB1*03:01:01:01~DQA1*05:01:01:02~DQB1*02:01:01) is not as frequent in Spanish population. In this sense, Spanish population seems to show an HLA haplotypic diversity with a distinctive and more spread haplotype frequency distribution in comparison to these other populations of European ancestry [36–38]. On the other hand, some of the most common HLA haplotypes described in the present Spanish population cohort (e.g. extended haplotype at the 4-field level observed in this Spanish population cohort: HLA-A*29:02:01:01~C*16:01:01:01~B*44:03:01:01~DRB4*01:01:01:01~DRB1*07:01:01:01~DQA1*02:01:01:01~DQB1*02:02:01:01) are also found with high frequencies in the registries of Argentina [33], exemplifying the significant Spanish genetic heritage that is still present in many populations from the entire American continent and more commonly within Hispanic ethnic groups [38–41].
Identification of null and rare alleles is important in determining donor-recipient matching especially for HSCT. Misidentification can have an impact on graft outcome and lead to complications such as acute graft-versus-host disease (GvHD) or graft failure [27][42]. NGS technology facilitates the characterization of null and rare alleles as well as defining respective null allele- and rare allele-carrying haplotypes. In this present study, those aforementioned null and rare alleles were found in certain inferred extended haplotypes of this Spanish population cohort (as some examples, tentative haplotypes that contain null alleles such as:
HLA-A*23:01:01~C*04:09N~B*44:03:01:01~DRB4*01:01:01:01~DRB1*07:01:01:01~DQA1*02:01:01:01~DQB1*02:02:01:01; or HLA-A*01:01:01:01~C*06:02:01:01~B*57:01:01~DRB4*01:03:01:02N~DRB1*07:01:01:01~DQA1*02:01:01:01~DQB1*03:03:02:01;s tentative haplotypes that contain rare alleles such as: HLA-A*02:01:01:01~C*12:166~B*52:01:01:02~DRB5*01:02~DRB1*15:02:01:02~DQA1*01:03:01:01~DQB1*06:01:01; or HLA-A*02:05:01~C*12:03:01:01~B*15:220~DRB4*01:03:01:01~DRB1*07:01:01:01~DQA1*02:01:01:01~DQB1*02:02:01:01). Nonetheless, future studies of larger population sample size at a wider geographic scale will be needed to obtain a more accurate determination of rare alleles and respective allele-carrying haplotypes [43].
In addition to other genetic markers (e.g. Y-chromosome and mitochondrial DNA), assessment of HLA haplotypes diversity within the worldwide populations and its geographical variation also contributes in the analysis of tracking migrations of modern populations as well as in anthropological studies [44][45]. Interestingly, we observed certain unique haplotypes that reflect both significant historic Sephardic Jewish and Arab genetic contributions (e.g. common haplotype: HLA-A*24:02:01:01~C*04:01:01:06~B*35:02:01~DRB3*02:02:01:02~DRB1*11:04:01~DQA1*05:05:01:01~DQB1*03:01:01:02~DPA1*01:03:01:04~DPB1*04:01:01:01) (HF = 0.030) [1][34][35][36] as well as a gene flow of relatively itinerant ethnic groups (e.g. common Spanish Gypsy haplotype:
HLA-A*01:01:01:01~C*15:02:01:01~B*40:06:01:02~DRB3*02:02:01:01~DRB1*14:04:01~DQA1*01:04:02~DQB1*05:03:01:01~DPA1*01:03:01:02~DPB1*02:01:02) (HF = 0.002) [46] in the present Spanish population cohort. In addition to this observed genetic substrate from various ethnic groups, it is well documented the relevant cultural heritage as well as the institutional recognition of all ethnic communities in Spain [47–49].
5. Conclusion
Results of the present Spanish population study show that NGS reveals distinctive allelic distributions and haplotypic associations. Moreover, HLA 4-field level data of worldwide populations may contribute to revise and to update CWD alleles list, knowledge of disease-associated alleles and/or haplotypes as well as current donor-recipient matching algorithms. Furthermore, our study shows that allelic and haplotypic HLA frequencies of Spanish population present a relative homogenous distribution that fits HWE proportions at all loci with the exception of HLA-DPA1 locus. At the same time, the distinctive HLA diversity found at both the allele and the haplotype levels is in concordance with the well-documented migrations presenting episodes of gene flow that have occurred through the course of history in Spain [1]. In this sense, application of NGS technology has allowed us to obtain a first glance of the HLA diversity at the 4-field level in the Spanish population. At the same time, future larger and wider geographic scale NGS studies will provide a more accurate description of this vast genetic diversity.
Overall, these results may contribute as a useful reference source for future population studies as well as a healthy control group dataset for evaluating HLA-disease associations. Furthermore, this HLA data may provide helpful information for improving donor recruitment strategies of bone marrow registries.
Supplementary Material
Acknowledgments
We are grateful to the reviewers of this manuscript for their helpful suggestions. We also thank all participant members of the Spanish Working Group in Histocompatibility and Transplant Immunology (GETHIT) of the Spanish Society for Immunology (SEI) for their significant contribution and collaboration to this study. This study was supported by Stanford Blood Center (as the official organizer and sponsor of the 17th International Histocompatibility and Immunogenetics Workshop-2017 (17th IHIW)) and by United States National Institutes of Health (NIH) grant U19NS095774.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have declared no conflicting interests.
References
- [1].Adams SM, Bosch E, Balaresque PL, Ballereau SJ, Lee AC, Arroyo E, López-Parra AM, Aler M, Grifo MSG, Brion M and Carracedo A The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. The American Journal of Human Genetics, 83(6) (2008) 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Anuario Estadístico de España 2017. http://www.ine.es/prodyser/pubweb/anuarios_mnu.htm, (last accessed November 2018).
- [3].Tiercy JM How to select the best available related or unrelated donor of hematopoietic stem cells?. Haematologica, 101(6) (2016) pp.680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Romòn I, Montes C, Ligeiro D, Trindade H, Sanchez-Mazas A, Nunes JM and Buhler S Mapping the HLA diversity of the Iberian Peninsula. Human Immunology, 77(10) (2016) 832–840. [DOI] [PubMed] [Google Scholar]
- [5].Martinez‐Laso J, Ramirez‐Puga A, Rivas‐García E, Fernández‐Tagarro E, Auyanet‐Saavedra I, Guerra‐ Rodríguez R, Díaz‐Novo N and García‐Cantón C North African‐Mediterranean HLA genetic contribution in a population of the kidney transplant waiting list patients of Canary origin (Gran Canaria). HLA (2018) (In press). [DOI] [PubMed]
- [6].Longás J, Martínez-Laso J, Rey D, Areces C, Casado EG, Parga-Lozano C, Luna F, de Salamanca ME, Moral P and Arnaiz-Villena A Las Alpujarras region (South East Spain) HLA genes study: evidence of a probable success of 17th century repopulation from North Spain. Molecular Biology Reports, 39(2) (2012) 1387–1394. [DOI] [PubMed] [Google Scholar]
- [7].Balas A, García‐Sánchez F and Vicario JL Allelic and haplotypic HLA frequency distribution in Spanish hematopoietic patients. Implications for unrelated donor searching. HLA, 77(1) (2011) 45–53. [DOI] [PubMed] [Google Scholar]
- [8].Alcoceba M, Marin L, Balanzategui A, Sarasquete ME, Chillon MC, Martín‐Jiménez P, Puig N, Santamaría C, Corral R, García‐Sanz R and San Miguel JF Frequency of HLA‐A,‐B and‐DRB1 specificities and haplotypic associations in the population of Castilla y León (northwest‐central Spain). HLA, 78(4) (2011) 249–255. [DOI] [PubMed] [Google Scholar]
- [9].Sanchez‐Velasco P, Gomez‐Casado E, Martinez‐Laso J, Moscoso J, Zamora J, Lowy E, Silvera C, Cemborain A, Leyva‐Cobián F and Arnaiz‐Villena A HLA alleles in isolated populations from North Spain: origin of the Basques and the ancient Iberians. HLA, 61(5) (2003) 384–392. [DOI] [PubMed] [Google Scholar]
- [10].Muro M, Marín L, Torío A, Moya-Quiles MR, Minguela A, Rosique-Roman J, Sanchis MJ, Garcia-Calatayud MC, Garcıá-Alonso AM and Álvarez-López MR HLA polymorphism in the Murcia population (Spain): in the cradle of the archaeologic Iberians. Human Immunology, 62(9) (2001) 910–921. [DOI] [PubMed] [Google Scholar]
- [11].Martinez‐Laso J, Juan D, Martinez‐Quiles N, Gomez‐Casado E, Cuadrado E and Arnaiz‐Villena A The contribution of the HLA‐A,‐B,‐C and‐DR,‐DQ DNA typing to the study of the origins of Spaniards and Basques. HLA, 45(4) (1995) 237–245. [DOI] [PubMed] [Google Scholar]
- [12].Sanchez-Mazas A and Meyer D The relevance of HLA sequencing in population genetics studies. Journal of Immunology Research, 2014 (2014) 971818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carapito R, Radosavljevic M and Bahram S Next-generation sequencing of the HLA locus: methods and impacts on HLA typing, population genetics and disease association studies. Human Immunology, 77(11) (2016) pp.1016–1023. [DOI] [PubMed] [Google Scholar]
- [14].Brown NK, Kheradmand T, Wang J and Marino SR Identification and characterization of novel HLA alleles: Utility of next-generation sequencing methods. Human Immunology, 77(4) (2016) pp.313–316. [DOI] [PubMed] [Google Scholar]
- [15].Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, Su LF, Levinson D, Fernandez-Viña MA, Davis RW, Davis MM and Mindrinos M High-throughput, high-fidelity HLA genotyping with deep sequencing. Proceedings of the National Academy of Sciences, 109(22) (2012) pp.8676–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chang CJ, Osoegawa K, Milius RP, Maiers M, Xiao W, Fernandez-Viña M and Mack SJ Collection and Storage of HLA NGS Genotyping Data for the 17 th International HLA and Immunogenetics Workshop. Human Immunology, 79(2) (2018) pp.77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lancaster AK, Single RM, Solberg OD, Nelson MP, and Thomson G PyPop update–a software pipeline for large-scale multilocus population genomics. HLA, 69(s1) (2007) 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schäfer C, Schmidt AH, and Sauter J Hapl-o-Mat: open-source software for HLA haplotype frequency estimation from ambiguous and heterogeneous data. BMC Bioinformatics, 18(1) (2017) 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Takezaki N, Nei M and Tamura K POPTREEW: Web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Molecular Biology and Evolution, 31(6) (2014) pp.1622–1624. [DOI] [PubMed] [Google Scholar]
- [20].Nei M, Tajima F and Tateno Y Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution, 19(2) (1983) pp.153–170. [DOI] [PubMed] [Google Scholar]
- [21].Saitou N and Nei M The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4) (1987) pp.406–425. [DOI] [PubMed] [Google Scholar]
- [22].Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK, Lau M, Lee KW, Mach B, Maiers M, Mayr WR, Müller CR, Parham P, Petersdorf EW, Sasazuki T, Strominger JL, Svejgaard, Terasaki PI, Tiercy JM and Trowsdale J Nomenclature for factors of the HLA system, 2010. Tissue Antigens, 75(4) (2010) p.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A and Thomson G Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Human Immunology, 69(7) (2008) pp.443–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brandt DY, César J, Goudet J and Meyer D The effect of balancing selection on population differentiation: a study with HLA genes. G3: Genes, Genomes, Genetics, 8(8) (2018) pp.2805–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cullen M, Perfetto SP, Klitz W, Nelson G and Carrington M High-resolution patterns of meiotic recombination across the human major histocompatibility complex. The American Journal of Human Genetics, 71(4) (2002) pp.759–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hollenbach JA, Madbouly A, Gragert L, Vierra-Green C, Flesch S, Spellman S, Begovich A, Noreen H, Trachtenberg E, Williams T and Yu N A combined DPA1~DPB1 amino acid epitope is the primary unit of selection on the HLA-DP heterodimer. Immunogenetics, 64(8) (2012) pp.559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Elsner HA and Blasczyk R Immunogenetics of HLA null alleles: implications for blood stem cell transplantation. HLA, 64(6) (2004) pp.687–695. [DOI] [PubMed] [Google Scholar]
- [28].Gonzalez‐Galarza FF, Mack SJ, Hollenbach J, Fernandez‐Vina M, Setterholm M, Kempenich J, Marsh SGE, Jones AR and Middleton D 16th IHIW: extending the number of resources and bioinformatics analysis for the investigation of HLA rare alleles. International Journal of Immunogenetics, 40(1) (2013) pp.60–65. [DOI] [PubMed] [Google Scholar]
- [29].Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D, Moraes ME, Pereira SE, Kempenich JH, Reed EF, Setterholm M, Smith AG, Tilanus MG, Torres M, Varney MD, Voorter CE, Fisher GF, Fleischhauer K, Goodridge D, Klitz W, Little AM, Maiers M, Marsh SG, Müller CR, Noreen H, Rozemuller EH, Sanchez-Mazas A, Senitzer D, Trachtenberg E and Fernández-Viña M Common and well‐documented HLA alleles: 2012 update to the CWD catalogue. HLA, 81(4) (2013) pp.194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sanchez‐Mazas A, Nunes JM, Middleton D, Sauter J, Buhler S, McCabe A, Hofmann J, Baier DM, Schmidt AH, Nicoloso G, Andreani M, Grubic Z, Tiercy JM and Fleischhauer K Common and well‐ documented HLA alleles over all of Europe and within European sub‐regions: A catalogue from the European Federation for Immunogenetics. HLA, 89(2) (2017) pp.104–113. [DOI] [PubMed] [Google Scholar]
- [31].Lind C, Ferriola D, Mackiewicz K, Papazoglou A, Sasson A and Monos D Filling the gaps–the generation of full genomic sequences for 15 common and well-documented HLA class I alleles using next-generation sequencing technology. Human Immunology, 74(3) (2013) pp.325–329. [DOI] [PubMed] [Google Scholar]
- [32].Fernandez-Vina MA, Wang C, Krishnakumar S, Levinson DF, Davis RW and Mindrinos M LBP19: extended coverage by next generation sequencing methods refines the characterization of the common and well documented HLA alleles. Human Immunology, 76(4) (2015) p.226. [Google Scholar]
- [33].Hurley CK, Hou L, Lazaro A, Gerfen J, Enriquez E, Galarza P, Cardozo M, Halagan M, Maiers M, Behm D and Ng J Next Generation Sequencing Characterizes the Extent of HLA Diversity in an Argentinian Registry Population. HLA, 91 (3) (2018) pp.175–186. [DOI] [PubMed] [Google Scholar]
- [34].Cano P, Testi M, Andreani M, Khoriaty E, Monsef JB, Galluccio T, Troiano M, Fernandez-Vina MA and Inati A (2012). HLA population genetics: a Lebanese population. HLA, 80(4) (2012) pp.341–355. [DOI] [PubMed] [Google Scholar]
- [35].Klitz W, Gragert L, Maiers M, Fernandez‐Viña M, Ben‐Naeh Y, Benedek G, Brautbar C and Israel S Genetic differentiation of Jewish populations. HLA, 76(6) (2010) pp.442–458. [DOI] [PubMed] [Google Scholar]
- [36].González-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MHT, Silva ALSD, Silva ALTE, Ghattaoraya GS, Alfirevic A, Jones AR and Middleton D Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Research, 43(D1) (2015) pp.D784–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pingel J, Solloch UV, Hofmann JA, Lange V, Ehninger G and Schmidt AH High-resolution HLA haplotype frequencies of stem cell donors in Germany with foreign parentage: how can they be used to improve unrelated donor searches?. Human Immunology, 74(3) (2013) pp.330–340. [DOI] [PubMed] [Google Scholar]
- [38].Gragert L, Madbouly A, Freeman J and Maiers M Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Human Immunology, 74(10) (2013) pp.1313–1320. [DOI] [PubMed] [Google Scholar]
- [39].Mack SJ, Tu B, Yang R, Masaberg C, Ng J and Hurley CK Human leukocyte antigen–A,-B,-C,-DRB1 allele and haplotype frequencies in Americans originating from southern Europe: Contrasting patterns of population differentiation between Italian and Spanish Americans. Human Immunology, 72(2) (2011) pp.144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zúñiga J, Yu N, Barquera R, Alosco S, Ohashi M, Lebedeva T, Acuña-Alonzo V, Yunis M, Granados-Montiel J, Cruz-Lagunas A, Vargas-Alarcón G, Rodriguez-Reyna TS, Fernandez-Vina M, Granados J and Yunis EJ HLA class I and class II conserved extended haplotypes and their fragments or blocks in Mexicans: implications for the study of genetic diversity in admixed populations. PLoS ONE, 8(9) (2013) p.e74442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Weiskopf D, Grifoni A, Arlehamn CSL, Angelo M, Leary S, Sidney J, Frazier A, Mack SJ, Phillips E, Mallal S and Cerpas C Sequence-based HLA-A, B, C, DP, DQ, and DR typing of 339 adults from Managua, Nicaragua. Human Immunology, 79(1) (2018) pp.1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Smith DM, Baker JE, Gardner WB, Martens GW and Agura ED HLA class I null alleles and new alleles affect unrelated bone marrow donor searches. HLA, 66(2) (2005) pp.93–98. [DOI] [PubMed] [Google Scholar]
- [43].Sanchez-Mazas A and Nunes JM Does NGS typing highlight our understanding of HLA population diversity? Some good reasons to say yes and a few to say be careful. Human Immunology (2018) (In press). [DOI] [PubMed]
- [44].Sanchez‐Mazas A, Fernandez‐Viña M, Middleton D, Hollenbach JA, Buhler S, Di D, Rajalingam R, Dugoujon JM, Mack SJ and Thorsby E Immunogenetics as a tool in anthropological studies. Immunology, 133(2) (2011) pp.143–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fernandez-Vina MA, Hollenbach JA, Lyke KE, Sztein MB, Maiers M, Klitz W, Cano P, Mack S, Single R, Brautbar C, Israel S, Raimondi E, Khoriaty E, Inati A, Andreani M, Testi M, Moraes ME, Thomson G, Stastny P and Cao K Tracking human migrations by the analysis of the distribution of HLA alleles, lineages and haplotypes in closed and open populations. Phil. Trans. R. Soc. B, 367(1590) (2012) pp.820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ramal LM, Pablo RD, Guadix MJ, Sánchez J, Garrido A, Garrido F, Jiménez‐Alonso J and López‐ Nevot MA HLA class II allele distribution in the Gypsy community of Andalusia, southern Spain. HLA, 57(2) (2001) pp.138–143. [DOI] [PubMed] [Google Scholar]
- [47].Boletin Oficial del Estado, Gobierno de España, 25 de Junio de 2015. http://www.boe.es/boe/dias/2015/06/25/pdfs/BOE-A-2015-7045.pdf, (last accessed November 2018).
- [48].Boletin Oficial del Estado, Gobierno de España, 12 de Noviembre de 1992. https://www.boe.es/boe/dias/1992/11/12/pdfs/A38214-38217.pdf, (last accessed November 2018).
- [49].Boletin Oficial de las Cortes Generales. Congreso de los Diputados, Gobierno de España, 22 de Junio de 2017. http://www.congreso.es/public_oficiales/L12/CONG/BOCG/D/BOCG-12-D-179.PDF, (last accessed November 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


