Graphical Abstract
Human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) exhibit a fetal phenotype that limits in vitro and therapeutic applications. Strategies to promote cardiomyocyte maturation have focused interventions on differentiated hPSC-CMs, but this study tests priming of early cardiac progenitor cells with polyinosinic-polycytidylic acid (pIC) to accelerate cardiomyocyte maturation. Cardiac progenitor cells were differentiated from hPSCs using a monolayer differentiation protocol with defined small molecule Wnt temporal modulation, and pIC was added during the formation of early cardiac progenitor cells. pIC priming did not alter the expression of cell surface markers for cardiac progenitor cells (>80% KDR+/PDGFRα+), expression of common cardiac transcription factors, or final purity of differentiated hPSC-CMs (~90%). However, cardiac progenitor cell differentiation in basal medium revealed that pIC priming resulted in hPSC-CMs with enhanced maturity manifested by increased cell size, greater contractility, faster electrical upstrokes, increased oxidative metabolism, and more mature sarcomeric structure and composition. To investigate the mechanisms of cardiac progenitor cell priming, RNAseq revealed that cardiac progenitor-stage pIC modulated early Notch signaling and cardiomyogenic transcriptional programs. Chromatin immunoprecipitation of cardiac progenitor cells showed that pIC treatment increased deposition of the H3K9ac activating epigenetic mark at core promoters of cardiac myofilament genes and the Notch ligand, JAG1. Inhibition of Notch signaling blocked the effects of pIC on differentiation and cardiomyocyte maturation. Furthermore, primed cardiac progenitor cells showed more robust formation of hPSC-CMs grafts when transplanted to the NSGW mouse kidney capsule. Overall, epigenetic modulation of CPCs with pIC accelerates cardiomyocyte maturation enabling basic research applications and potentially therapeutic uses.
Keywords: Cardiac progenitor cells, cardiomyocyte maturation, epigenetics, Notch signaling, human pluripotent stem cells
MeSH Terms: Stem Cells, Regenerative Medicine, Developmental Biology
Introduction
Human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) are powerful tools for disease modeling, drug development, basic research and possible therapies. However, hPSC-CMs exhibit developmental immaturity that limits these applications. Therefore, intense efforts to promote maturation of hPSC-CM have been undertaken including changing media to favor oxidative metabolism,1 manipulating action potentials by electrical pacing or Kir2.1 ion channel overexpression,2,3 creating three-dimensional tissue constructs,4 and extending cell culture over more than three months.5 Despite progress in enhancing maturity, these interventions are often time-consuming, cumbersome or promote only certain aspects of maturation (Table 1). Although environmental cues clearly modulate cardiomyocyte development, there is evidence that cell intrinsic processes also regulate the time course of maturation. For example, studies comparing mouse pluripotent stem cell (mPSC) and human pluripotent stem cell (hPSC) differentiation demonstrate species-specific differences in the kinetics of differentiation both in vitro and during in vivo teratoma formation suggesting a cell autonomous developmental clock.6 Consistent with an intrinsic developmental clock, distant cardiomyocytes found in pulmonary and azygous veins show maturational changes in troponin and myosin isoforms that are synchronous in time with cardiomyocytes of the heart proper, while spatially remote and subject to dramatically different hemodynamics and cell signaling.7 These observations suggest that both environmental and intrinsic processes control cardiac cellular maturation. Although the mechanisms controlling intrinsic pathways of maturation are largely unknown, epigenetic factors are likely important.8,9
Table 1.
Methods to Enhance hPSC-CM Maturation
 | ||||||
|---|---|---|---|---|---|---|
| Priming | Biochemical | Metabolic | Electrical | Mechanical | Temporal | |
| Stage | hPSC-CPCs | hPSC-CMs | hPSC-CMs | hPSC-CMs | hPSC-CMs | hPSC-CMs |
| Approach | Early epigenetic modulation | Activate maturation signals | Induce switch to FFA oxidation | Electrical pacing; hyperpolarization | Morphologic, force constraints | Prolonged cell culture |
| Reagents | pIC | Let-7 miRNA T3, IGF-1, dex. | IBMX, insulin, rosiglitazone, indomethacin, dex. | Pacing, co-culture, overexpress CSQ/Kir2.1 | Collagen, Hydrogel, PDMS, Matrigel, PEG/PCL | Basal medium |
| Equipment | Standard | Standard | Standard | Pacing device; 3D tissue; gene modification | Engineered materials | Standard |
| Speed | +++ | ++ | ++ | ++/+++ | ++ | + |
| Economy | +++ | ++ | ++ | +/++ | ++ | + |
| Scalable | +++ | +++ | +++ | +/++ | +/++ | +++ |
| References | This paper | 50–52 | 1 | 2,3,53–57 | 4,57–64 | 5,18,65,66 |
Dex. – dexamethasone; IBMX-isobutylmethylxanthine; FFA-free fatty acid; PDMS-polydimethylsiloxane; PEG-polyethylene glycol; PCL-polycaprolactone
We therefore hypothesized that appropriate priming of hPSC-derived cardiac progenitor cells (CPCs) can alter their epigenetic status and accelerate cardiomyocyte differentiation and maturation. For this purpose the double-stranded RNA and innate immunity activator, polyinosinic-polycytidylic acid (pIC), was investigated. pIC has previously been found to accelerate the reprogramming of skin fibroblasts to induced pluripotent stem cells by augmenting activating epigenetic marks at the transcription factor loci responsible for the observed lineage changes.10 Other agents that promote epigenetic changes have also been associated with accelerated cell type transitions in reprogramming,11 and pIC has also shown efficacy in enhancing reprogramming of fibroblasts to cells of the cardiovascular lineage such as cardiomyocytes and endothelial cells.12,13 Like reprogramming, the differentiation of hPSCs to the cardiomyocyte lineage involves a progression of epigenetic transitions;14 so we hypothesized that pIC could aid the transitions needed for hPSC-CM differentiation and maturation.
Here we demonstrate that pIC treatment of CPCs accelerates maturation of hPSC-CMs broadly assessed by structural, electrical, and metabolic assays. Primed CPCs have epigenetic changes in myofilament and Notch signaling-related genes, and inhibition of Notch signaling blocks the effect of pIC on maturation. Heterotopic transplantation of primed CPCs to the immunodeficient mouse leads to more robust cardiomyocyte differentiation. Thus, a simple biopolymer intervention at the CPC stage can substantially accelerate the differentiation and maturation of hPSC-CMs, which can improve in vitro applications and potentially therapeutic approaches.
Results
pIC priming of CPCs accelerates hPSC-CM maturation
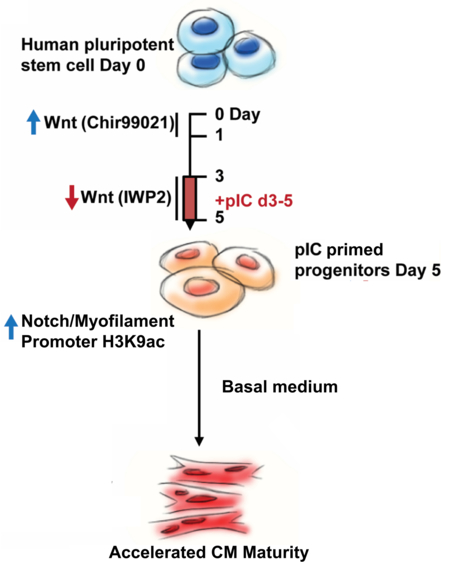
To test the effect of pIC on CPCs, we utilized a defined, small molecule hPSC cardiac differentiation protocol based on sequential activation of the Wnt pathway by GSK3β inhibition followed by Wnt inhibition (GiWi, Fig 1a).15 pIC was added on days 3–5 of the GiWi protocol during CPC formation, and we first assessed whether pIC impacted the formation of CPCs identified as the cell population co-labeled by KDR and PDGFRα.16 Approximately 80% of day 5 cells in the presence or absence of pIC express the KDR+/PDGFRα+ immunophenotype (Fig. S1a). pIC treatment does not alter the expression of key transcription factors present in CPCs including MESP1, GATA4, TBX5, ISL1, and NKX2–5 (Fig. S1a). Because prior studies utilized pIC treatment to promote lineage reprogramming, we also assessed if pIC treatment of CPCs altered the composition of the differentiated progeny. Flow cytometry for cardiac troponin T (cTnT) revealed that cells differentiated from pIC-treated and untreated CPCs using the GiWi protocol were primarily hPSC-CMs (80–95%) for both hES and hiPS cell lines (Fig. S1b). Thy1+ cells accounted for the majority of non-cardiomyocytes differentiated from the CPCs which were primarily a fibroblast population supported by co-labeling with other markers of cardiac fibroblasts such as WT1, MMP1, FSP1 (Fig. S1c).17 Endothelial and smooth muscle cells were rare based on CD31 and SM-MHC labeling (Fig. S1d). We detected no evidence for chamber-specific lineage changes in the mostly ventricular-fated GiWi cardiomyocytes as assessed by gene expression and immunostaining of atrial (HEY1, COUPTFII) and ventricular (IRX4, HAND1, HAND2) cardiomyocyte markers (Fig. S1e–f, action potential shape quantified by APD30/80 (Fig. S1g), or ventricular-specific myosin light chain 2v (Mlc2v) flow cytometry at day 80 (Fig. 1e). Thus, pIC treatment did not alter the formation of CPCs or their differentiation potential.
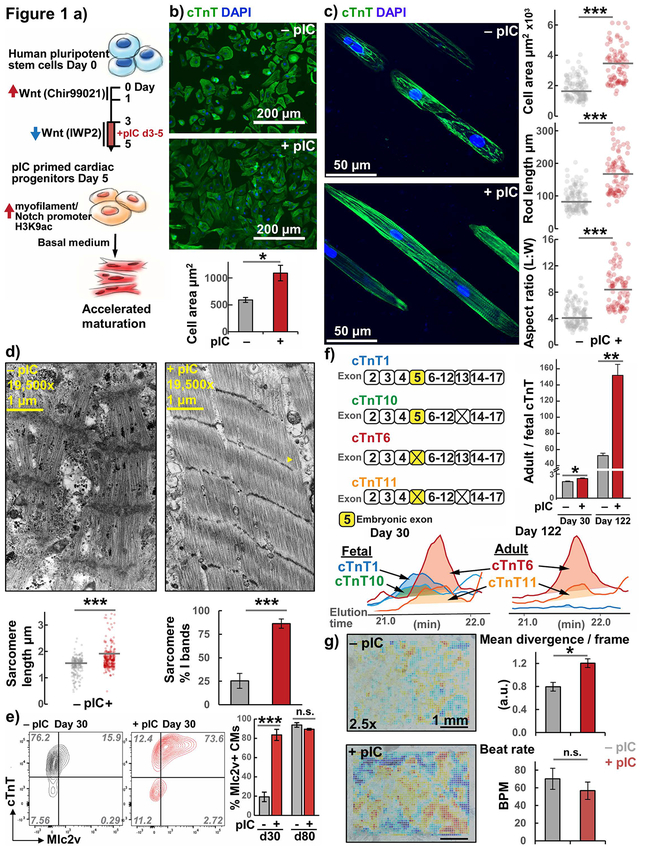
Figure 1. Increased structural maturation in hPSC-CMs from pIC primed CPCs.
a) Overview: Differentiation of hPSCs using a small molecule, biphasic Wnt modulation protocol with pIC treatment to generate primed CPCs that exhibit accelerated differentiation and maturation.
b) Cell size analysis of day 30 hiPSC-CMs quantified by the area of cTnT immunofluorescence in standard 2-D culture, n=4 passages ea. >50 cells.
c) Cell morphology analysis of cTnT immunolabeled hiPSC-CMs dissociated after day 30 and cultured for 1 week on 20 μm wide Matrigel patterned lanes on PDMS substrate.
d) Transmission electron micrographs of hESC-CMs showing I bands (yellow arrow). Individual sarcomere length measurements across all images and percentages of I banded sarcomeres in individual images quantified with means illustrated.
e) Flow cytometry plots for day 30 and day 80 hiPSC-CMs costained with cTnT and Mlc2v with average data, n=3.
f) Top-down mass spectrometry for cTnT isoforms area-under-the-curve (AUC) ratio of adult to fetal isoforms for day 30 and day 122 hESC-CMs, n=3.
g) Representative day 30 contractility heatmaps and average data calculated from divergence analysis of hiPSC-CM monolayers, n=7. Red indicates regions of higher divergence, and blue negative divergence (convergence).
t Test P *< 0.05, **< 0.01, ***< 0.001; hiPSC line DF19-9-11, hESC line H9-TNNT2-GFP.
We then assessed if pIC treatment of CPCs impacted the maturation of the differentiated hPSC-CMs. Compared to adult cardiomyocytes, hPSC-CMs are developmentally immature exhibiting smaller size, less organized sarcomeres, expression of fetal cardiac myofilament isoforms, decreased oxidative metabolism, smaller amplitude action potentials, and reduced contractility.7,15,18–22 We began by characterizing the morphological and structural properties of hPSC-CMs differentiated from pIC treated or untreated CPCs. Accordingly, we found pIC treatment of CPCs resulted in day 30 hPSC-CMs that were twice as large (Fig. 1b); however, the larger hPSC-CMs following pIC treatment still lacked adult rod-like morphology when cultured in this standard 2D tissue culture format. We therefore conducted cell size and morphology measurements on hPSC-CMs from treated and untreated CPCs that were dissociated and replated on 20 μm-wide lanes of Matrigel patterned on a soft PDMS substrate to mimic the stiffness found in the native heart (~10 kPa) to promote hPSC-CM structural organization.23,24 Rod-like hPSC-CMs emerged in this environment from pIC-treated CPCs that were again approximately twice as large and had nearly double the L:W aspect ratio (~8:1 vs 4:1) of hPSC-CMs from untreated CPCs (Fig. 1c, S2). We found that lane patterning was a necessary feature for rod-formation in this assay, wherein plating hPSC-CMs on unpatterned, soft PDMS did not result in rod formation (Fig. S2). Ultrastructural characterization by electron microscopy showed longer, more regular sarcomeres containing I-bands in hPSC-CMs from pIC-treated CPCs, which are features of sarcomeres found in more mature cardiomyocytes (Fig. 1d).18 The myofilament protein Mlc2v gradually increases in expression over 80 days as the ventricular-fated cardiomyocytes of the GiWi protocol mature, such that few early hPSC-CMs have detectable Mlc2v by flow cytometry.15,25 However, pIC treatment of CPCs markedly increased the percentage of cTnT+ hPSC-CMs that produced Mlc2v at day 30 compared to day 80 when more than 90% of hPSC-CMs were Mlc2v+ in both groups (Fig. 1e). This shows accelerated maturation of Mlc2v expression in hPSC-CMs from pIC-treated CPCs, and we next investigated markers which change at a later stage in hPSC-CM maturation. The myofilament protein cTnT is expressed throughout development, but it undergoes developmental changes in protein spliceoforms based on the presence or absence of embryonic exon 5,26 leading to initial expression of fetal isoforms 1 and 10 and subsequently adult isoforms 6 and 11 (Uniprot numbering). We performed top-down mass spectrometry to measure these isoforms of cTnT in hPSC-CMs and found that pIC treatment of CPCs increased the ratio of adult to fetal isoforms in day 30 hPSC-CMs. Notably, during prolonged culture to day 122, the adult to fetal ratio and the relative difference between hPSC-CMs from pIC treated and untreated CPCs increased dramatically (Fig. 1f). This suggests that while Mlc2v expression normalized at later-stage culture, myofilaments with slower maturation timelines were affected greater than 100 days after the day 3–5 pIC treatment and showcases the ability of top-down mass spectrometry for cTnT isoforms specifically to distinguish the maturation status of late-stage hPSC-CMs. Together these data indicate that pIC treatment of CPCs accelerates cardiomyocyte structural maturation based on hPSC-CM size, morphology, ultrastructure, and myofilament expression patterns.
To determine if the changes in maturation of structural properties translate into increased contractility of hPSC-CMs, we performed video analysis of contraction divergence patterns as previously described.27 Beat divergence is derived from a local cross-correlation (or “optical flow”) analysis of image sequences, and has been shown to distinguish active contraction from passive motion in mixed cultures.27 We found that hPSC-CMs from pIC-treated CPCs had increased average divergence (Fig. 1g, SV1), as well as increases in other parameters of sheet contractility (for extended data see Fig. S3).27,28 While some of these methods may be sensitive to the rate of beating, we found no significant difference between the spontaneous beat frequency in hPSC-CMs from pIC-treated and untreated CPCs (Fig. 1g). We then assessed for changes in other functional properties of hPSC-CMs. As cardiomyocytes mature, they exhibit faster action potential upstroke velocity and increased oxidative metabolism.20,29 We recorded spontaneous optical action potentials from day 30 hPSC-CMs using the voltage-sensitive dye RH237. The optical upstroke velocity in hPSC-CMs from pIC-treated CPCs was significantly faster (Fig. 2a, S4a), quantified as decreased rise time (the time required to change from 10 to 90% depolarization voltage amplitude).30 There was no difference in the spontaneous rate or other parameters measured for the spontaneous action potentials, except pIC treatment did decrease APD80 (Fig. S4a). To examine the metabolism of hPSC-CMs, we lactate-purified31 hPSC-CMs (>95% cTnT+) from both pIC-treated and untreated CPCs and used the Seahorse bioanalyzer to measure basal and maximal oxygen consumption in hPSC-CMs. hPSC-CMs from treated CPCs had approximately 2-fold greater basal and maximal oxygen consumption rates (Fig. 2b). These studies suggest an increase in maturation of functional properties of the hPSC-CMs differentiated from pIC-treated CPCs.
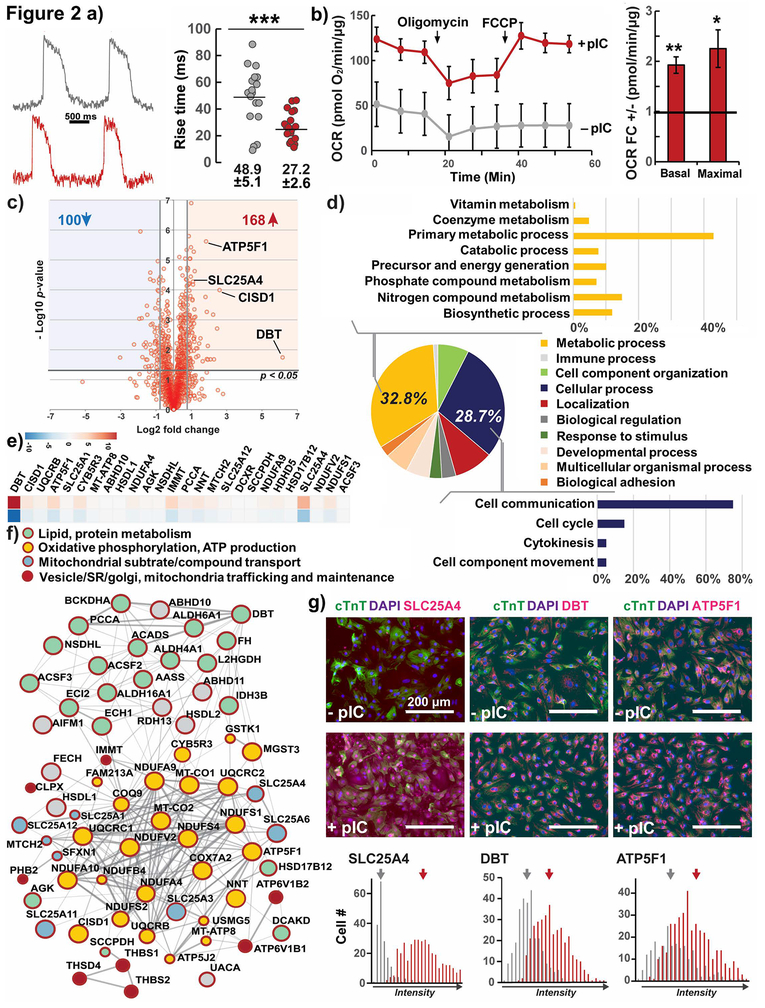
Figure 2. Functional and proteomic analysis of maturation in hPSC-CMs, effect of pIC treatment of CPCs.
a) Optical upstroke velocity with the voltage-sensitive dye RH237 (quantified by time required to traverse 10 to 90% action potential amplitude, rise time) for day 30 hESC-CMs, individual cells plotted from 4 cell preparations.
b) Seahorse quantification of Oxygen Consumption Rate (OCR) normalized to total protein fold change of hESC-CMs from primed to untreated CPCs, n=4
c) Global quantitative bottom up proteomic analysis of up- and downregulated proteins as volcano plot of hESC-CMs from primed CPCs compared to no treatment.
d) Ontology analysis revealed the majority of the up-regulated proteins were involved in primary metabolic processes and cell communication.
e) Heat-map of the top 34 upregulated proteins involved in metabolic processes and mitochondrial function.
f) Protein-protein interaction network of upregulated metabolic proteins, predominantly involved in oxidative phosphorylation and ATP production (yellow), as well as lipid/amino acid metabolism (green). Proteins important for mitochondrial substrate/compound transport (blue) and the crosstalk of mitochondrial and SR/Golgi compartments (red) were also upregulated. Full interactome see Fig. S4c.
g) Immunostaining and quantification of mean fluorescence of three top upregulated metabolic proteins. Means labeled. Unmerged images see Fig. S4d.
Test P *< 0.05, **<0.01, ***< 0.001; hESC line H9-TNNT2-GFP
To provide an unbiased evaluation of changes in protein expression in the hPSC-CMs generated by treated and untreated CPCs, we performed global label-free quantitative bottom-up proteomics on day 30 hPSC-CMs.32 We detected about 2,600 proteins (Fig. S4b) and found that overall, 168 proteins were significantly upregulated and 100 downregulated in hPSC-CMs from treated relative to untreated CPCs (p < 0.05 and > 50% change in expression, Fig. 2c). The upregulated proteins were mainly involved in metabolic and cell communication processes (Fig. 2d). Inspection of upregulated metabolic proteins suggested predominant roles in oxidative phosphorylation, mitochondrial transport, and the metabolism of amino acids and lipids (Fig. 2e–f, S4c, ST1). This was consistent with our functional data showing increased oxygen consumption rates. By quantitative immunostaining, we verified increased expression of four of the top upregulated proteins in the dataset involved in oxidative and lipid metabolism: branched chain amino acid dehydrogenase (DBT), NADP-dependent Steroid Dehydrogenase-Like (NSDHL), ATP synthase subunit ATP5F1, and the cytoplasmic mitochondrial DNA-stabilizing ATP transporter SLC25A4 (Fig. 2g, S4d). SLC25A4 protein in particular was almost absent from untreated CPC-derived cardiomyocytes and is a known mediator of mitochondrial function, where its loss in humans results in the mitochondrial myopathy MTDPS type 12.33 In contrast, down-regulated proteins were often involved in cytoskeletal organization (Myl6, Talin, Transgrelin, and collagens 18a1, 3a1, and 1a2, Fig. S4c). We found proteins involved in ribosomal biogenesis or mRNA synthesis and translation in both up and downregulated protein sets; however, we noticed mitochondrial ribosomal complex proteins MRPL1, MRPL44, and MRPS22 only among upregulated proteins in hPSC-CMs from pIC-treated CPCs.
Through the combination of this proteomics data with the functional and structural data above we conclude that there is increased maturation broadly assessed in structural, metabolic, and electrical properties of cardiomyocytes from pIC-treated CPCs, which we refer to hereafter as primed CPCs.
pIC epigenetically primes CPC Notch and cardiomyogenic transcriptional programs
We hypothesized that the effect of pIC on CPCs to promote hPSC-CM maturation involves modulation of cardiac developmental signaling networks and early cardiomyogenic transcriptional programs. To discover these networks, we performed RNA sequencing of day 5 pIC primed and untreated CPCs to evaluate gene expression.
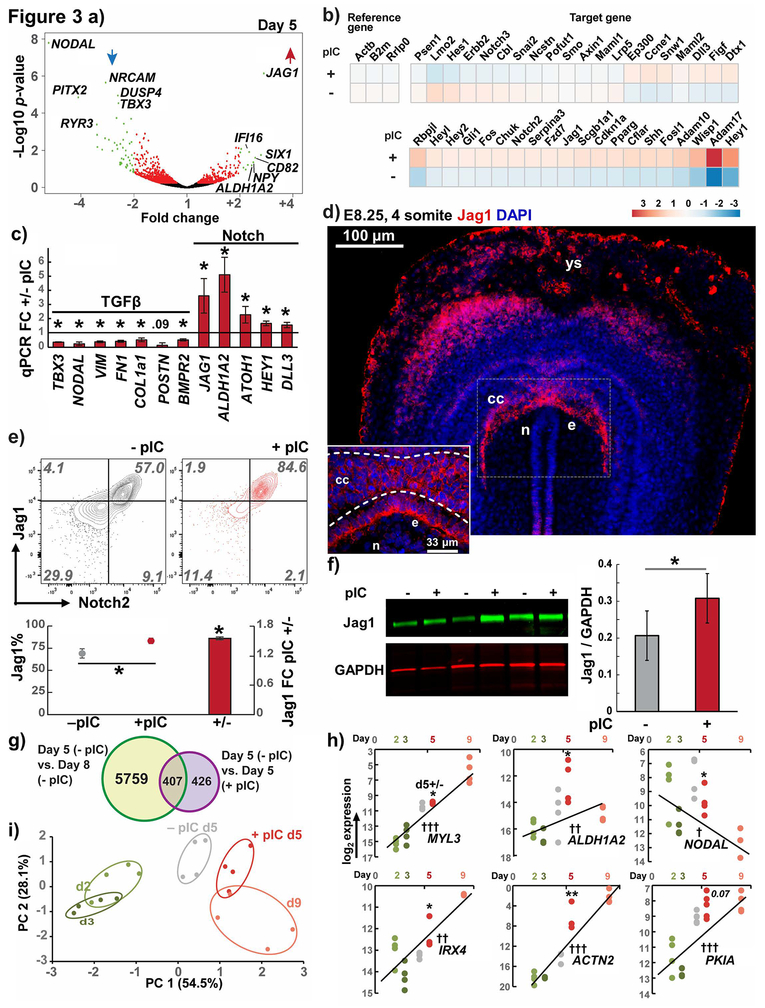
Among pIC-upregulated genes we found enrichment in GO terms associated with cardiac development, such as insulin signaling pathways, and we also noted upregulation of multiple genes in Notch signaling (ST2). We first validated this by targeted Notch pathway qPCR array, which demonstrated 48/65 Notch genes in this panel to be upregulated in primed CPCs (Fig. 3b). The top-enriched gene in RNA sequencing was the Notch pathway ligand JAG1 (Fig. 3a, ST2), and qPCR validation confirmed upregulation of JAG1 and other Notch-related genes such as HEY1, DLL3, ATOH1 and the retinoic acid signaling enzyme ALDH1A2 (RALDH2), (which requires Notch signaling for its expression in the heart34 and is also known to be necessary for early ventricular maturation during formation of the chambered heart in vivo).35–37 Jag1 protein expression has not been previously reported during heart development prior to the early heart tube myocardium.38 To determine if Jag1 is expressed in developing CPCs in vivo in analogy to our findings during in vitro differentiation, we performed immunostaining in cardiac crescent-stage E8.25/4-somite mouse embryos (Fig. 3d). Intense Jag1 immunolabeling was present in the CPCs of the cardiac crescent and the neighboring cells of the endoderm as well as in the yolk sac. JAG1 and NOTCH2 were the most abundant Notch ligand/receptor pair in the CPCs in RNAseq (Fig. S5a), so we investigated Jag1 and Notch2 protein surface localization on the hPSC-derived CPCs by flow cytometry. The majority of both primed and untreated CPCs co-expressed Jag1 and Notch2, but pIC treatment increased the percentage of Jag1+/Notch2+ cells (Fig. 3e). A pIC induced increase in Jag1 expression was also confirmed by quantitative, infrared western blotting (Fig. 3f). These data suggests that Jag1 and Notch2 function together in Notch signaling in CPCs, which is consistent with human genetic studies in which loss of function mutations in either JAG1 or NOTCH2 result in cardiomyopathy in patients with Alagille syndrome.39
Figure 3. Transcriptomics of primed progenitors reveals Notch and cardiomyogenic transcriptional program enrichment.
a) RNAseq volcano plot of day 5 pIC primed vs. untreated hiPSC-CPCs. Significantly altered genes with fold change > ±2 in green, > ±1 in red.
) Notch pathway RT-qPCR array showing upregulation in 48/65 Notch genes in pIC primed CPCs. Reference genes shown in left with no changes between cell groups.
c) RT-qPCR validation of RNAseq candidates in the TGFβ and Notch pathways, n = 4.
d) E8.25/4-somite stage mouse embryo transverse section immunolabeled for Jag1. ys - yolk sac, cc - cardiac crescent, n - notochord, e - endoderm.
e) Flow cytometry of day 5 CPCs for Jag1 and Notch 2 showing pIC treatment increased the number of Jag1 positive cells and median fluorescence signal-to-noise.
f) Licor near-IR western blot of CPC Jag1 with quantitation normalized to GAPDH, n = 3
g) Heatmap of top differentially expressed genes from RNAseq data comparing the earliest cardiomyocytes (day 8) and CPCs (day 5) in the GiWi protocol. Venn diagram illustrates overlap of a subset of these genes with those differentially expressed in RNAseq comparing day3–5 ±pIC.
h) RT-qPCR validation of overlap genes from (d) and comparison of expression levels between day 5 ±pIC with pre-progenitor (day 2,3) and post- (day9) time points. Regression computed from day 2, 3, and 9 with ANOVA test for slope P †< 0.05, ††< 0.01, †††< 0.001.
i) Principal component analysis of CPC priming genes and cell samples in (h).
t Test P *<0.05, **<0.01, *** <0.001; hiPSC line DF19-9-11
Gene ontology analysis of pIC downregulated genes showed pathways in cancer, pluripotency, and TGFβ signaling in the top four downregulated GO terms in KEGG pathways analysis (ST2), and we confirmed many of these genes by qPCR (Fig. 3a,c). We found that the downregulated TGFβ-related genes were largely classical epithelial-to-mesenchymal transition (EMT) markers such as FN1 and VIM, or found in the GO analysis among “networks involved in pluripotency signaling” (#4 downregulated GO term, ST2). Two of the top downregulated genes, TBX3 and NODAL, are established necessary factors for pluripotency.40,41 TGFβ also mediates the EMT of primitive streak-like mesendoderm formation in the initial phase of cardiac differentiation protocols.15,42 Thus, we interpreted pIC’s downregulation of TGFβ pathways as consistent with inhibiting residual expression of genes of pluripotency and early EMT/mesendoderm.
We hypothesized that pIC causes a change in gene expression in CPCs consistent with an acceleration of the differentiation process. We therefore sought to compare the transcriptional patterns of pIC primed and untreated CPCs to changes in gene expression occurring in early cardiac differentiation. We used a publicly available RNAseq dataset evaluating the time course of hPSC-CMs differentiation during the GiWi protocol (PCBC synapseID 25822579). We analyzed early days in this series (2, 3, 5, and 8) to detect leading indicator genes first differentially expressed in the earliest day 8 cardiomyocytes. Consistent with the initial formation of terminally differentiated cardiomyocytes, the day 8 reference dataset exhibited the greatest differences compared to days 2, 3 and 5 (Fig. S5b). 5,375 genes were differentially expressed between day 5 and day 8 in the reference RNA sequencing of the GiWi protocol (ST3). In our RNAseq of day 5 pIC primed and untreated CPCs, pIC affected a much smaller set of just 833 genes (out of ~10,500 genes detected, Fig. 3g). Nonetheless, there was significant overlaps in these datasets, with 49% of genes differentially expressed following pIC treatment overlapping the differentially expressed genes between day 5 and day 8 of the GiWi protocol. Gene ontology analysis shows that these overlap genes were predominately involved in cardiac processes and development (Fig. S5c, ST3). We then focused on validating several of these differentially expressed genes by qPCR on day 5 pIC primed and untreated CPCs, as well as time points before and after the progenitor stage (days 2, 3, and 9 of GiWi). We found increased expression of a subset of the genes discovered in our time course RNAseq analysis that have been previously discovered as early-upregulated cardiomyogenic factors in other large-scale transcriptomic and epigenetic studies – ACTN2, MYL3, ALDH1A2, IRX4, and PKIA.8,9 These genes increased in GiWi cultures between pre- (day 2 and 3) to post- (day 9) cardiac progenitor stages in a log-linear fashion and increased in pIC primed day 5 CPCs compared to untreated CPCs (Fig. 3h). Oppositely, we found that NODAL (as described above) decreased from day 2–3 to day 9, and was decreased in pIC primed CPCs in the direction of differentiation. When these markers were combined into principal component analysis (PCA), the statistically largest component of analysis, PC1 accounted for the majority of the variability in gene expression and convincingly separated earlier (day2, 3) and later (day 9) GiWi samples (Fig. 3i). PCA showed that pIC primed CPCs were most similar to the untreated day 5 CPCs, but had greater similarity to the later day 9 samples in this subset of early cardiomyocyte genes than did untreated CPCs.
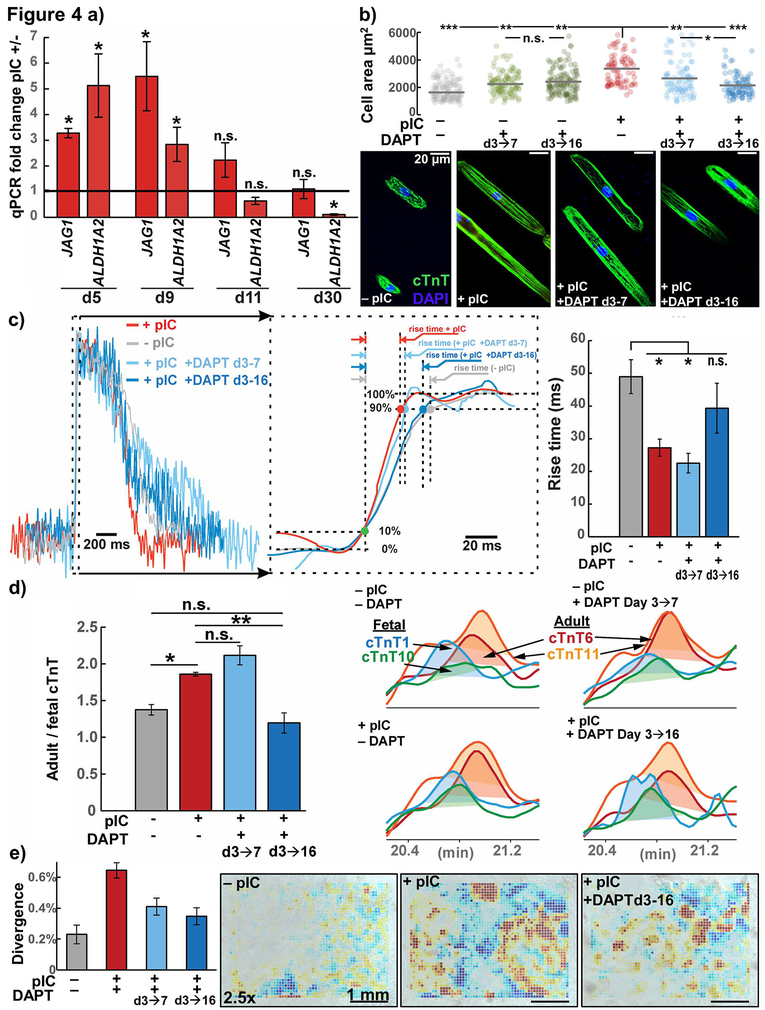
Given that the Notch pathway was upregulated in primed CPCs (Fig. 3b–c), we tested the effects of inhibiting Notch signaling with the gamma-secretase inhibitor DAPT. While JAG1 and ALDH1A2 were significantly enriched in day 5 CPCs, a time course experiment demonstrated that progenitor stage treatment with pIC from day 3–5 also had prolonged effects on the cells, with RNA levels of JAG1 and ALDH1A2 remaining upregulated until day 9–11 (Fig. 4a). We therefore assessed the maturation of hPSC-CMs from day 3–5 pIC-and untreated CPCs in the presence of DAPT inhibition. We first assessed the morphology of hPSC-CMs in the PDMS rod-formation assay and found that DAPT specifically blocked the increased cell area of hPSC-CMs from primed CPCs, but did not decrease the size of hPSC-CMs from untreated CPCs (Fig. 4b). This effect on primed CPCs was present with Notch inhibition with DAPT from day 3–7 and intensified when DAPT included the early cardiomyocyte window (day 3–16). Treatment from day 3–16 with DAPT also blocked other markers of maturation including the increase in optical upstroke velocity in cardiomyocytes from primed CPCs (decreased rise time) (Fig. 4c) and the protein isoform change toward adult cTnT 6 and 11 (Fig. 4d). On the other hand, we found that Notch inhibition from day 3–7 and day 3–16 blocked the increase in contractility of hPSC-CMs from primed CPCs to a similar extent (Fig. 4e). The role of Notch signaling in promoting maturation of hPSC-CMs is consistent with previous reports in vivo in which Jag1 deletion driven by the cardiomyocyte-specific Tnnt2 promoter inhibits ventricular maturation.43
Figure 4. Notch inhibition blocks acceleration of hPSC-CM maturation induced by pIC treatment of CPCs.
a) RT-qPCR expression of top Notch pathway associated genes from Fig. 3c, JAG1 and ALDH1A2, at the progenitor stage (day 5), earliest cardiomyocytes (day 9, 11), and later (day 30) using hiPSC line DF19-9-11, n = 3–4.
b) Confocal microscopy with morphology analysis cTnT-immunolabeled hiPSC-CMs from cells treated with indicated conditions, dissociated and cultured for 1 week on 30 μm wide Matrigel patterned lanes on PDMS substrate, n=6. ANOVA P < .001
c) Optical upstroke velocity measured using the voltage-sensitive dye RH237 (quantified by time required to traverse 10 to 90% action potential amplitude, rise time) from d30 hiPSC-CMs differentiated from primed and untreated CPCs, in the presence or absence of the Notch inhibitor DAPT at times indicated, n=17 individual cells from 4 passages. ANOVA P < .001
d) Top-down mass spectrometry for cTnT isoforms, AUC ratio of adult to fetal isoforms for day 30 hiPSC-CMs from primed and untreated CPCs, in the presence or absence of the Notch inhibitor DAPT, n=3. ANOVA P < .001
e) Video contractility analysis of hiPSC-CM monolayers from cells treated with indicated conditions.
t test or post-hoc Holm P * < .05, ** < .01, *** < .001; hiPSC line, DF19-9-11
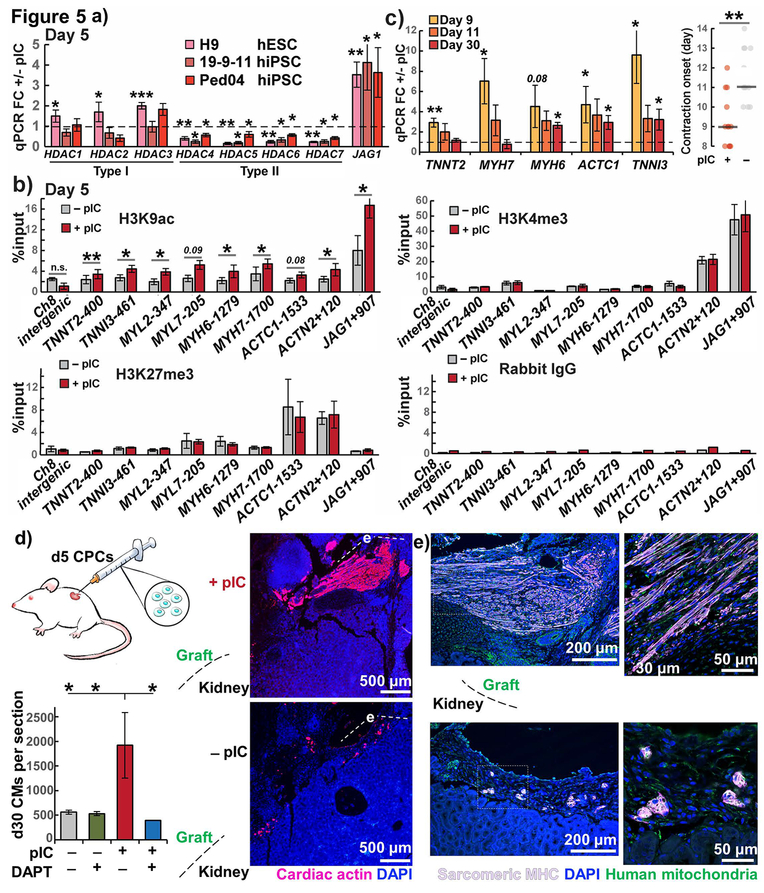
To uncover the upstream mechanisms for the transcriptional changes induced by pIC, we profiled the epigenetic status of both primed and untreated day 5 CPCs. Previous investigation demonstrated that pIC-mediated augmentation of fibroblast reprogramming is associated with decreased expression of repressive epigenetic modifiers, including type I and type II histone deacetylases, and this lead to increased activating marks at the loci of pluripotency factors.10 We examined the expression of type I and II histone deacetylases in CPCs differentiated from multiple hPSC lines. qPCR demonstrated that primed CPCs had universally lower levels of type II histone deacetylases (HDAC4–7), but unchanged to upregulated levels of type I HDAC1–3 (Fig. 5a). Type I HDACs are ubiquitous enzymes generally present in all cell types, but the heart is the only organ to express all type II HDACs 4–7, and changes in type II HDACs alone or in combination cause cardiac-specific phenotypes.44,45 We viewed predominant type II HDAC modulation in primed CPCs, compared to the mixed type I and II repression of pIC treatment in fibroblasts, as a distinct cardiac progenitor response suggesting that epigenetic activation in CPCs could involve the loci of the early cardiomyogenic and Notch transcriptional programs. In chIP-qPCR of day 5 primed and untreated CPCs, we found a significant pattern of increased deposition of the activating histone acetylation mark H3K9ac near the transcriptional start sites of both JAG1 and multiple cardiac myofilament genes: TNNT2, TNNI3, MYL2, MYL7, MYH6, MYH7, ACTC1, and ACTN2 (Fig. 5b). These changes with pIC treatment were specific for H3K9ac and not found in the H3K4me3 activating mark. There were also no changes in the repressive epigenetic modification H3K27me3 or H3K9ac deposition at a negative control intergenic region. Promoters with co-deposition of both H3K27me3 and H3K4me3 have previously been described in stem cells and progenitors as epigenetically “poised,”46 and we found greatest co-occupancy of these marks for ACTN2, which also saw the greatest relative increase in expression with pIC in day 5 CPC RT-qPCR (Fig. 3h, S1b). This suggests that poised genes may be expressed earlier in primed CPCs. These data and the pattern of increased H3K9ac across multiple cardiac myofilaments and JAG1 suggests greater epigenetic priming for differentiation along the cardiac lineage, and we hypothesized that it would lead to faster onset of myofilament transcription and cell contraction. A time course experiment of myofilament gene expression revealed that day 9 cells from primed CPCs had increased expression of all cardiac myofilaments studied compared to cells from untreated CPCs, and faster onset of contractions in the differentiated cells by day 9 (Fig. 5c). By day 30 the relative increases induced by pIC remained only for MYH6, TNNI3, and ACTC1, myofilament proteins with greater adult than fetal expression or force production. These results suggested that epigenetic priming causes earlier activation of the cardiomyogenic transcriptional program and accelerated differentiation.
Figure 5. Epigenetic and functional cardiac priming of CPCs by pIC.
a) Expression of type I and type II HDACs in pIC primed vs. untreated CPCs from three hPSC lines measured by RT-qPCR, n = 3–4. JAG1 was upregulated and type II HDACs downregulated in all three lines, confirming consistency of day 5 priming.
b) chIP-qPCR of cardiac myofilament genes, JAG1, and chromosome 8 intergenic negative control as raw percent input binding to activating marks H3K9ac and H3K4me3, repressive mark H3K27me3, or rabbit IgG negative control, in hiPSC line 19-9-11, n = 4.
c) RT-qPCR of myofilament genes for day 9, 11, and 30 cardiomyocytes, fold change +/− pIC treated CPCs and day of onset of contractions and in hiPSC line 19-9-11, n=3.
d) Cardiac actin (AC1–20.4.2) immunostaining of NSGW mouse kidney xenografts day 30-post transplantation of primed and untreated CPCs from hiPSC line 19-9-11, n=3–4. ANOVA P < .05.
e) Sarcomeric myosin heavy chain (MF20) immunostaining of areas in (d) showing overlap with anti-human mitochondria (113–1) in confocal microscopy.
t test or post-hoc Holm P * < .05, ** < .01, *** < .001
Given the impact of pIC on CPC epigenetics and acceleration of differentiation in the in vitro analyses, we sought to determine if pIC priming promoted the differentiation of CPCs and accelerated cardiomyocyte maturation following in vivo delivery. We therefore heterotopically grafted both primed and untreated day 5 CPCs into the kidney capsules of immunodeficient NSGW mice,47 and after 30 days the human grafts were identified by anti-human mitochondrial antibody. We observed approximately 4-fold more Cardiac actin and sarcomeric myosin heavy chain-expressing hPSC-CMs in grafts from primed CPCs, and treatment of the CPCs with the Notch inhibitor, DAPT, specifically reduced the number of differentiated hPSC-CMs (Fig. 5d–e). The grafts from primed CPCs exhibited hPSC-CMs with elongated morphology and aligned myofilaments in contrast to the more immature appearing circular clusters of hPSC-CMs from untreated CPCs. We cannot exclude that longer in vivo follow-up of animals would equalize certain of these differences, but the finding of accelerated differentiation of primed CPCs is consistent with the previously reported ability of later stage, committed CPCs to more readily differentiate into hPSC-CMs in the kidney capsule in contrast to early stage CPCs.48 In addition, these results from the heterotopic kidney capsule transplant model agree with the observed in vitro effects of pIC and suggest this strategy could have relevance for cell therapy applications.
Discussion
Here we describe the striking effects of pIC on hPSC-derived CPCs to accelerate differentiation and maturation of hPSC-CMs. Treatment of CPCs with pIC promotes H3K9ac deposition at the loci of Notch and cardiomyocyte genes, which leads to accelerated transcription of cardiac myofilaments and earlier onset of electromechanical contractions of the differentiated hPSC-CMs. Furthermore, pIC priming accelerates the overall maturation process of hPSC-CMs as broadly assessed by transcriptional, structural, electrical, and metabolic assays. We find that Notch signaling has an essential role in mediating this accelerated maturation, which is consistent with the observation that loss of function mutations in pIC’s top upregulated gene, JAG1, are a known cause of immature ventricular development and cardiomyopathy in humans.39 The effect of pIC priming of CPCs is also evident following heterotropic transplantation to the kidney capsule of NSG mice with formation of larger and more mature cardiac grafts.
Previous studies aiming to advance the maturation of hPSC-CMs have focused interventions on differentiated cardiomyocytes (Table 1),22 but in this study we demonstrate that early intervention limited to the CPC stage accelerates cardiac gene expression in CPCs and the subsequent time course of hPSC-CM maturation. These results suggest that pIC treatment of CPCs resets a developmental clock controlling the kinetics of formation of hPSC-CMs and subsequent maturation. Developmental clocks have been described in other mesodermal-derived progenitors to regulate the precisely timed formation of somites and segmentation of the vertebrate body. This segmentation clock results from cyclic activation of Notch, Wnt/β-catenin and FGF signaling leading to phasic expression of downstream genes.49 Species-specific differences in developmental kinetics also suggest developmental clock function in various progenitor populations.6 In cardiac development, a developmental clock regulating differentiation and maturation of cardiomyocytes has not been well defined, but our results suggest that subsets of early cardiomyogenic and Notch gene expression programs are involved in this function. This is consistent with a recent study demonstrating that loss of Jag1 disrupts ventricular cardiomyocyte development in the early embryonic heart leading to immature myocardium.43 Further studies will be needed to define upstream clock mechanisms in cardiac development and how pIC treatment impacts these mechanisms. Whether pIC has similar effects on other tissue-specific progenitors is also an intriguing question for future studies.
Because CPC priming affects cardiac lineage maturation by accelerating the early developmental clock, it is fundamentally different than other described methods for advancing maturation by manipulating differentiated hPSC-CMs (Table 1). Manipulations to hPSC-CMs that have been shown to advance maturation include applying exogenous microRNA or chemical signals to trigger developmental pathways;50–52 changing media to favor oxidative metabolism;1 electrical interventions either by pacing or Kir2.1 ion channel overexpression to hyperpolarize the cells;2,3,53–57 changing cardiomyocyte structural environment by mechanical loading, three-dimensional tissue construction, or constraining cell morphology into rod-like shapes;4,57–64 and extending cell culture over months.5,18,65,66 As these methods begin only after differentiation of hPSC-CMs, they entail costs of prolonged cell culture relative to CPC priming. As a simple single time-point additive, pIC is less laborious than other manipulations of hPSC-CMs. Early epigenetic CPC priming also provides the opportunity for synergy with later methods for further acceleration of maturation. Finally, certain of these methods, notably creation of structural or electrical micro-constructs for cells, may have significant limitations in scalability, which can constrain drug testing and represent a major barrier to clinical applications requiring large numbers of cells.
The most ambitious goal of exogenous cell therapy for the infarcted or failing heart is remuscularization of areas of scar and diseased myocardium with adult-like cardiac muscle, but the best cell type or preparation for this purpose remains unknown. Comparison of cardiomyocyte grafts from neonatal and fetal versus adult rat hearts found superiority of the less mature populations in repairing a rat infarct model.67 One explanation for such results is that immature cells have greater developmental or epigenetic plasticity. Furthermore, immature cardiomyocytes and CPCs have greater proliferation than mature cardiomyocytes,68,69 potentially leading to more successful grafts. These considerations suggest cells that are developmentally early (such as CPCs) yet primed for mature tissue formation (such as that shown in our heterotopic transplants) may represent a bridge between optimal final graft composition and optimal starting cell sources for exogenous cell therapy of the heart. Priming CPCs with pIC also has potential clinical relevance given that CPCs are the first human pluripotent stem cell derivative to be tested in a cardiovascular clinical trial.70 In this regard, pIC is a synthetic, chemically-defined polymer that has undergone safety evaluation as an FDA-approved agent for direct use in humans as an adjuvant in chemotherapy or vaccines. This makes priming CPCs with pIC practical to implement for clinical applications relative to other approaches to engineer cells such as genetic modification. Evaluation of the potency of primed CPCs to repair the injured heart is therefore an important future direction not addressed in the present study.
Methods
Refer to supplemental information.
Supplementary Material
Significance statement.
Here we have discovered that epigenetic priming of hPSC-CPCs with polyinosinic-polycytidylic acid accelerates differentiation and cardiomyocyte maturation in vitro and following heterotopic transplantation in vivo. Epigenetic and transcriptional profiling of primed cardiac progenitor cells reveals increased histone acetylation and earlier transcription of Notch and cardiomyogenic genes. This work unmasks pathways regulating the time course of cardiac progenitor cell differentiation and cardiomyocyte maturation, providing a novel strategy to rapidly induce cardiomyocyte maturation for research and clinical applications.
Acknowledgements
We wish to acknowledge the technical expertise of Joe Hardin and the UWCCC Experimental Pathology Laboratory, and Randall Massey at the Wisconsin Medical School EM Facility. In addition to authors AC and DGI, we also consulted Nathaniel Huebsch at Washington University in St. Louis for analysis of contractility. Access to RNAseq data housed in the Progenitor Cell Biology Consortium from multiple days of differentiation in the GiWi protocol was made generously available by the laboratory of Bruce Conklin at UCSF. Mass spectrometry was performed at the Wisconsin Human Proteomics Program, RNA sequencing at the Wisconsin Gene Expression Center, flow cytometry at the UWCCC flow lab, and Seahorse metabolic analysis at the Wisconsin Small Molecule Screening Facility. The project was supported by the University of Wisconsin ICTR TL1TR002375 and MSTP T32GM008692, AHA 15PRE2577004, and NIH F30 HL126452 fellowship grants (MB) as well as NIH grants R01 HL129798 (TJK), 1S10RR025644 (TJK) and 1U01HL134764 (TJK).
Footnotes
Accession Numbers
Day 5 CPC RNAseq GEO: GSE98941. Long time course GiWi protocol RNA from day 0–5, 8, 15, 30, and 60 PCBC synapseID: 25822579 (laboratory of Bruce Conklin, UCSF).
Conflicts of Interest
A.V.G., Y.G. declared employment at the University of Wisonsin-Madison. T.J.K. is a consultant for Cellular Dynamics International, a stem cell biotechnology company. All of the other authors declared no financial conflict of interest.
Data Availability Statement
Accession Numbers: Day 5 CPC RNAseq GEO: GSE98941. Long time course GiWi protocol RNA from day 0–5, 8, 15, 30, and 60 PCBC synapseID: 25822579 (laboratory of Bruce Conklin, UCSF).
References
- 1.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494(7435):105–110. doi: 10.1038/nature11799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan Y-C, Ting S, Lee Y-K, Ng K-M, Zhang J, Chen Z, Siu C-W, Oh SKW, Tse H-F. Electrical stimulation promotes maturation of cardiomyocytes derived from human embryonic stem cells. Journal of Cardiovascular Translational Research. 2013;6(6):989–999. doi: 10.1007/s12265-013-9510-z [DOI] [PubMed] [Google Scholar]
- 3.Vaidyanathan R, Markandeya YS, Kamp TJ, Makielski JC, January CT, Eckhardt LL. IK1-enhanced human-induced pluripotent stem cell-derived cardiomyocytes: an improved cardiomyocyte model to investigate inherited arrhythmia syndromes. American Journal of Physiology. Heart and Circulatory Physiology. 2016;310(11):H1611–1621. doi: 10.1152/ajpheart.00481.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Massé S, Gagliardi M, Hsieh A, et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nature Methods. 2013;10(8):781–787. doi: 10.1038/nmeth.2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lundy SD, Zhu W-Z, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells and Development. 2013;22(14):1991–2002. doi: 10.1089/scd.2012.0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry C, Schmitz MT, Jiang P, Schwartz MP, Duffin BM, Swanson S, Bacher R, Bolin JM, Elwell AL, McIntosh BE, et al. Species-specific developmental timing is maintained by pluripotent stem cells ex utero. Developmental Biology. 2017;423(2):101–110. doi: 10.1016/j.ydbio.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kracklauer MP, Feng H-Z, Jiang W, Lin JL-C, Lin JJ-C, Jin J-P. Discontinuous thoracic venous cardiomyocytes and heart exhibit synchronized developmental switch of troponin isoforms. FEBS Journal. 2013;280(3):880–891. doi: 10.1111/febs.12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piccini I, Araúzo-Bravo M, Seebohm G, Greber B. Functional high-resolution time-course expression analysis of human embryonic stem cells undergoing cardiac induction. Genomics Data. 2016;10:71–74. doi: 10.1016/j.gdata.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tompkins JD, Jung M, Chen C-Y, Lin Z, Ye J, Godatha S, Lizhar E, Wu X, Hsu D, Couture LA, et al. Mapping Human Pluripotent-to-Cardiomyocyte Differentiation: Methylomes, Transcriptomes, and Exon DNA Methylation “Memories.” EBioMedicine. 2016;4:74–85. doi: 10.1016/j.ebiom.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of Innate Immunity is Required for Efficient Nuclear Reprogramming. Cell. 2012;151(3):547–558. doi: 10.1016/j.cell.2012.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkinson CP, Pratt RE, Kirste I, Dal-Pra S, Cooke JP, Dzau VJ. Cardiomyocyte Maturation Requires TLR3 Activated Nuclear Factor Kappa B. Stem Cells (Dayton, Ohio). 2018;36(8):1198–1209. doi: 10.1002/stem.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayed N, Wong WT, Ospino F, Meng S, Lee J, Jha A, Dexheimer P, Aronow BJ, Cooke JP. Transdifferentiation of Human Fibroblasts to Endothelial Cells: Role of Innate Immunity. Circulation. 2015;131(3):300–309. doi: 10.1161/CIRCULATIONAHA.113.007394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151(1):206–220. doi: 10.1016/j.cell.2012.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(27):E1848–1857. doi: 10.1073/pnas.1200250109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 17.Witty AD, Mihic A, Tam RY, Fisher SA, Mikryukov A, Shoichet MS, Li R-K, Kattman SJ, Keller G. Generation of the epicardial lineage from human pluripotent stem cells. Nature Biotechnology. 2014;32(10):1026–1035. doi: 10.1038/nbt.3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, Hattori T, Ohno S, Kita T, Horie M, et al. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circulation Journal: Official Journal of the Japanese Circulation Society. 2013;77(5):1307–1314. [DOI] [PubMed] [Google Scholar]
- 19.O’Brien TX, Lee KJ, Chien KR. Positional specification of ventricular myosin light chain 2 expression in the primitive murine heart tube. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(11):5157–5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson C, Tran DD, George SC. Concise Review: Maturation Phases of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Stem cells (Dayton, Ohio). 2013. [accessed 2015 May 24];31(5). http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3749929/. doi: 10.1002/stem.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuchimochi H, Kuro-o M, Takaku F, Yoshida K, Kawana M, Kimata S, Yazaki Y. Expression of myosin isozymes during the developmental stage and their redistribution induced by pressure overload. Japanese Circulation Journal. 1986;50(10):1044–1052. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Pabon L, Murry CE. Engineering Adolescence: Maturation of Human Pluripotent Stem Cell-derived Cardiomyocytes. Circulation research. 2014;114(3):511–523. doi: 10.1161/CIRCRESAHA.114.300558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salick MR, Napiwocki BN, Sha J, Knight GT, Chindhy SA, Kamp TJ, Ashton RS, Crone WC. Micropattern width dependent sarcomere development in human ESC-derived cardiomyocytes. Biomaterials. 2014;35(15):4454–4464. doi: 10.1016/j.biomaterials.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herron TJ, Rocha AMD, Campbell K, Ponce-Balbuena D, Willis BC, Guerrero-Serna G, Liu Q, Klos M, Musa H, Zarzoso M, et al. Extracellular Matrix Mediated Maturation of Human Pluripotent Stem Cell Derived Cardiac Monolayer Structure and Electrophysiological Function. Circulation. Arrhythmia and electrophysiology. 2016. [accessed 2019 Mar 3];9(4). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4833010/. doi: 10.1161/CIRCEP.113.003638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional Cardiomyocytes Derived from Human Induced Pluripotent Stem Cells. Circulation research. 2009;104(4):e30–e41. doi: 10.1161/CIRCRESAHA.108.192237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei B, Jin J-P. Troponin T Isoforms and Posttranscriptional Modifications: Evolution, Regulation and Function. Archives of biochemistry and biophysics. 2011;505(2):144–154. doi: 10.1016/j.abb.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czirok A, Isai DG, Kosa E, Rajasingh S, Kinsey W, Neufeld Z, Rajasingh J. Optical-flow based non-invasive analysis of cardiomyocyte contractility. Scientific Reports. 2017. [accessed 2018 Oct 21];7 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5583397/. doi: 10.1038/s41598-017-10094-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, Mathur A, Nguyen TN, Yoo JC, Judge LM, et al. Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales. Tissue Engineering. Part C, Methods. 2015;21(5):467–479. doi: 10.1089/ten.tec.2014.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hom JR, Quintanilla RA, Hoffman DL, Karen L. de MB, Molkentin JD, Sheu S-S, Porter GA. The Permeability Transition Pore Controls Cardiac Mitochondrial Maturation and Myocyte Differentiation. Developmental cell. 2011;21(3):469–478. doi: 10.1016/j.devcel.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang D, Holzem K, Kang C, Xiao M, Hwang HJ, Ewald GA, Yamada KA, Efimov IR. Arrhythmogenic Remodeling of β2 versus β1 Adrenergic Signaling in the Human Failing Heart. Circulation: Arrhythmia and Electrophysiology. 2015. February 11:CIRCEP.114.002065. doi: 10.1161/CIRCEP.114.002065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12(1):127–137. doi: 10.1016/j.stem.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 32.Yang L, Gregorich ZR, Cai W, Zhang P, Young B, Gu Y, Zhang J, Ge Y. Quantitative Proteomics and Immunohistochemistry Reveal Insights into Cellular and Molecular Processes in the Infarct Border Zone One Month after Myocardial Infarction. Journal of Proteome Research. 2017;16(5):2101–2112. doi: 10.1021/acs.jproteome.7b00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Echaniz-Laguna A, Chassagne M, Ceresuela J, Rouvet I, Padet S, Acquaviva C, Nataf S, Vinzio S, Bozon D, Mousson de Camaret B. Complete loss of expression of the ANT1 gene causing cardiomyopathy and myopathy. Journal of Medical Genetics. 2012;49(2):146–150. doi: 10.1136/jmedgenet-2011-100504 [DOI] [PubMed] [Google Scholar]
- 34.Monte G del Casanova JC, Guadix JA, MacGrogan D, Burch JBE, Pérez-Pomares JM, Pompa JL de la. Differential Notch Signaling in the Epicardium Is Required for Cardiac Inflow Development and Coronary Vessel MorphogenesisNovelty and Significance. Circulation Research. 2011;108(7):824–836. doi: 10.1161/CIRCRESAHA.110.229062 [DOI] [PubMed] [Google Scholar]
- 35.Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dollé P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development (Cambridge, England). 2001;128(7):1019–1031. [DOI] [PubMed] [Google Scholar]
- 36.Lin S-C, Dollé P, Ryckebüsch L, Noseda M, Zaffran S, Schneider MD, Niederreither K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proceedings of the National Academy of Sciences. 2010;107(20):9234–9239. doi: 10.1073/pnas.0910430107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brade T, Kumar S, Cunningham TJ, Chatzi C, Zhao X, Cavallero S, Li P, Sucov HM, Ruiz-Lozano P, Duester G. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2. Development (Cambridge, England). 2011;138(1):139–148. doi: 10.1242/dev.054239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development (Cambridge, England). 2006;133(21):4381–4390. doi: 10.1242/dev.02607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Röpke A, Kujat A, Gräber M, Giannakudis J, Hansmann I. Identification of 36 novel Jagged1 (JAG1) mutations in patients with Alagille syndrome. Human Mutation. 2003;21(1):100. doi: 10.1002/humu.9102 [DOI] [PubMed] [Google Scholar]
- 40.Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463(7284):1096–1100. doi: 10.1038/nature08735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camus A, Perea-Gomez A, Moreau A, Collignon J. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Developmental Biology. 2006;295(2):743–755. doi: 10.1016/j.ydbio.2006.03.047 [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circulation research. 2012;111(9):1125–1136. doi: 10.1161/CIRCRESAHA.112.273144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Amato G, Luxán G, del Monte-Nieto G, Martínez-Poveda B, Torroja C, Walter W, Bochter MS, Benedito R, Cole S, Martinez F, et al. Sequential Notch activation regulates ventricular chamber development. Nature cell biology. 2016;18(1):7–20. doi: 10.1038/ncb3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125(3):453–466. doi: 10.1016/j.cell.2006.02.048 [DOI] [PubMed] [Google Scholar]
- 45.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews. Genetics 2009;10(1):32–42. doi: 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voigt P, Tee W-W, Reinberg D. A double take on bivalent promoters. Genes & Development. 2013;27(12):1318–1338. doi: 10.1101/gad.219626.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown ME, Zhou Y, McIntosh BE, Norman IG, Lou HE, Biermann M, Sullivan JA, Kamp TJ, Thomson JA, Anagnostopoulos PV, et al. A Humanized Mouse Model Generated Using Surplus Neonatal Tissue. Stem Cell Reports. 2018;10(4):1175–1183. doi: 10.1016/j.stemcr.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foo KS, Lehtinen ML, Leung CY, Lian X, Xu J, Keung W, Geng L, Kolstad TRS, Thams S, Wong AO, et al. Human ISL1+ Ventricular Progenitors Self-Assemble into an In Vivo Functional Heart Patch and Preserve Cardiac Function Post Infarction. Molecular Therapy. 2018;26(7):1644–1659. doi: 10.1016/j.ymthe.2018.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oates AC, Morelli LG, Ares S. Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development (Cambridge, England). 2012;139(4):625–639. doi: 10.1242/dev.063735 [DOI] [PubMed] [Google Scholar]
- 50.Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M, Sniadecki NJ, Ruohola-Baker H, Murry CE. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. Journal of Molecular and Cellular Cardiology. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu W-Z, Tulloch NL, Yang X, et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(21):E2785–E2794. doi: 10.1073/pnas.1424042112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V, Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, et al. Contractile Defect Caused by Mutation in MYBPC3 Revealed under Conditions Optimized for Human PSC-Cardiomyocyte Function. Cell Reports. 2015;13(4):733–745. doi: 10.1016/j.celrep.2015.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim C, Majdi M, Xia P, Wei KA, Talantova M, Spiering S, Nelson B, Mercola M, Chen HV. Non-Cardiomyocytes Influence the Electrophysiological Maturation of Human Embryonic Stem Cell-Derived Cardiomyocytes During Differentiation. Stem Cells and Development. 2010;19(6):783–795. doi: 10.1089/scd.2009.0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Fu JD, Siu CW, Li RA. Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: insights for driven maturation. Stem Cells (Dayton, Ohio). 2007;25(12):3038–3044. doi: 10.1634/stemcells.2007-0549 [DOI] [PubMed] [Google Scholar]
- 55.Lieu DK, Fu J-D, Chiamvimonvat N, Tung KC, McNerney GP, Huser T, Keller G, Kong C-W, Li RA. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circulation. Arrhythmia and Electrophysiology. 2013;6(1):191–201. doi: 10.1161/CIRCEP.111.973420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirt MN, Boeddinghaus J, Mitchell A, Schaaf S, Börnchen C, Müller C, Schulz H, Hubner N, Stenzig J, Stoehr A, et al. Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation. Journal of Molecular and Cellular Cardiology. 2014;74:151–161. doi: 10.1016/j.yjmcc.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 57.Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556(7700):239–243. doi: 10.1038/s41586-018-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feaster TK, Cadar AG, Wang L, Williams CH, Chun YW, Hempel JE, Bloodworth N, Merryman WD, Lim CC, Wu JC, et al. Matrigel Mattress: A Method for the Generation of Single Contracting Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circulation Research. 2015;117(12):995–1000. doi: 10.1161/CIRCRESAHA.115.307580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chun YW, Balikov DA, Feaster TK, Williams CH, Sheng CC, Lee J-B, Boire TC, Neely MD, Bellan LM, Ess KC, et al. Combinatorial Polymer Matrices Enhance In Vitro Maturation of Human Induced Pluripotent Cell Cell-Derived Cardiomyocytes. Biomaterials. 2015;67:52–64. doi: 10.1016/j.biomaterials.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circulation Research. 2011;109(1):47–59. doi: 10.1161/CIRCRESAHA.110.237206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D-H, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh K-Y, Tung L, Levchenko A. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proceedings of the National Academy of Sciences. 2010;107(2):565–570. doi: 10.1073/pnas.0906504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Liao M-LC, Levent E, Raad F, Zeidler S, Wingender E, et al. Defined Engineered Human Myocardium with Advanced Maturation for Applications in Heart Failure Modelling and Repair. Circulation. 2017;135(19):1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abilez OJ, Tzatzalos E, Yang H, Zhao M-T, Jung G, Zöllner AM, Tiburcy M, Riegler J, Matsa E, Shukla P, et al. Passive Stretch Induces Structural and Functional Maturation of Engineered Heart Muscle as Predicted by Computational Modeling. Stem Cells (Dayton, Ohio). 2018;36(2):265–277. doi: 10.1002/stem.2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shadrin IY, Allen BW, Qian Y, Jackman CP, Carlson AL, Juhas ME, Bursac N. Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nature Communications. 2017;8(1):1825. doi: 10.1038/s41467-017-01946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otsuji TG, Minami I, Kurose Y, Yamauchi K, Tada M, Nakatsuji N. Progressive maturation in contracting cardiomyocytes derived from human embryonic stem cells: Qualitative effects on electrophysiological responses to drugs. Stem Cell Research. 2010;4(3):201–213. doi: 10.1016/j.scr.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 66.Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E, Jaconi ME. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells (Dayton, Ohio). 2007;25(5):1136–1144. doi: 10.1634/stemcells.2006-0466 [DOI] [PubMed] [Google Scholar]
- 67.Hans Reinecke, Ming Zhang, Trudy Bartosek, Murry Charles E. Survival, Integration, and Differentiation of Cardiomyocyte Grafts. Circulation. 1999;100(2):193–202. doi: 10.1161/01.CIR.100.2.193 [DOI] [PubMed] [Google Scholar]
- 68.Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Developmental Cell. 2003;5(1):85–97. [DOI] [PubMed] [Google Scholar]
- 69.Takeuchi T Regulation of cardiomyocyte proliferation during development and regeneration. Development, Growth & Differentiation. 2014;56(5):402–409. doi: 10.1111/dgd.12134 [DOI] [PubMed] [Google Scholar]
- 70.Menasché P, Vanneaux V, Hagège A, Bel A, Cholley B, Cacciapuoti I, Parouchev A, Benhamouda N, Tachdjian G, Tosca L, et al. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: first clinical case report. European Heart Journal. 2015;36(30):2011–2017. doi: 10.1093/eurheartj/ehv189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.