Abstract
Members of the Hsp90 and Hsp70 families of molecular chaperones are important for the maintenance of protein homeostasis and cellular recovery following environmental stresses, such as heat and oxidative stress. Moreover, the two chaperones can collaborate in protein remodeling and activation. In higher eukaryotes, Hsp90 and Hsp70 form a functionally active complex with Hop (Hsp90-Hsp70 organizing protein) acting as a bridge between the two chaperones. In bacteria, which do not contain a Hop homolog, Hsp90 and Hsp70, DnaK, directly interact during protein remodeling. Although yeast possess a Hop-like protein, Sti1, Hsp90 and Hsp70 can directly interact in yeast in the absence of Sti1. Previous studies showed that residues in the middle domain of Escherichia coli Hsp90 are important for interaction with the J-protein binding region of DnaK. The results did not distinguish between the possibility that i) these sites were involved in direct interaction and ii) the residues in these sites participate in conformational changes which are transduced to other sites on Hsp90 and DnaK that are involved in the direct interaction. Here we show by crosslinking experiments that the direct interaction is between a site in the middle domain of Hsp90 and the J-protein binding site of Hsp70 in both E. coli and yeast. Moreover, J-protein promotes the Hsp70-Hsp90 interaction in the presence of ATP, likely by converting Hsp70 into the ADP-bound conformation. The identification of the protein-protein interaction site is anticipated to lead to a better understanding of the collaboration between the two chaperones in protein remodeling.
Keywords: Hsp82, HtpG, Ssa1, Hsp40, DnaJ
Introduction
Hsp90 and Hsp70 are evolutionarily conserved, ATP-dependent molecular chaperones that are important for the maintenance of protein homeostasis. Both chaperones are highly expressed proteins that are further induced during heat shock and other cellular stress conditions [1–3]. In eukaryotes, Hsp90 is essential and is involved in the stability, folding and activation of over 300 client (substrate) proteins, which include many protein kinases, steroid hormone receptors and transcription factors [3, 4]. In many bacteria including Escherichia coli, Hsp90 is not essential and phenotypes in strains lacking Hsp90 are minimal [1, 5–9]. Eukaryotic Hsp90 functions with numerous cochaperones that modulate its chaperone activity [3, 10]. In contrast, cochaperones for bacterial Hsp90 have not been observed.
Hsp90 is a homodimer with each monomer composed of three domains: an N-terminal ATP-binding domain (N-domain), a middle domain (M-domain) and a C-terminal domain (C-domain) that is involved in dimerization [3, 11–14]. While eukaryotic and bacterial Hsp90 have ~50% sequence similarity, there are two major differences between the Hsp90 sequences. First, eukaryotic Hsp90 usually has a flexible “charged linker” of variable length connecting the N-domain to the M-domain, which bacterial Hsp90 lacks [12, 15]. This linker has been proposed to act as a modulator of Hsp90 conformational changes and cochaperone binding [16–18]. Second, cytoplasmic eukaryotic Hsp90 has a C-terminal extension that is important for the binding of cochaperones containing tetratricopeptide repeat (TPR) domains [3, 10, 19].
Hsp90 is a dynamic molecule that undergoes conserved, large-scale conformational changes upon ATP binding, hydrolysis and ADP release [3, 12, 20, 21]. For example, in the absence of ATP, the Hsp90 dimer primarily populates an open, V-shaped conformation with the C-domains dimerized [3, 12, 15, 21, 22]. ATP binding causes the dimer to populate a closed conformation with the two N-domains interacting while maintaining the contacts in the C-domain [15]. When ATP is hydrolyzed, a more compact conformation has been observed [15, 19, 23] and upon ADP release, the open conformation is repopulated. The dynamics of Hsp90 and the length of time Hsp90 resides in the various conformations are modulated by the presence of nucleotide, client protein, and in eukaryotes, cochaperones and post-translational modifications [12, 20, 24–26].
In addition to the class of cochaperones that bind to the C-terminal extension, eukaryotic Hsp90 cochaperones have discrete binding sites on Hsp90, which can involve one or more of the individual domains [3, 4, 12, 14, 27]. Cochaperones interact with the various conformations that Hsp90 transitions through during its conformational cycle; for example, some cochaperones stabilize the closed or open conformation, which leads to the stimulation or inhibition of Hsp90 ATPase activity, respectively [3, 4, 10]. Other cochaperones can act as adaptor proteins for specific clients (substrates) or may have additional specialized activities [3, 4, 10].
Multiple client protein binding sites have been identified on Hsp90 and include regions of one or more Hsp90 domains [3, 12, 14, 28]. Many of these client binding sites are specific for individual clients as many clients are unrelated, both structurally and sequentially [3, 4, 12, 14, 27]. However, there are some commonalities in features such as surface area, charge and hydrophobicity [3, 4, 12, 28, 29]. Additionally, specific client proteins may require a cochaperone to target them to Hsp90 for activation or remodeling [3, 4, 12, 14].
In both eukaryotes and bacteria, Hsp90 collaborates with the ATP-dependent molecular chaperone Hsp70 and its cochaperones to remodel and activate proteins [3, 29–38]. Hsp70 and DnaK, bacterial Hsp70, are comprised of an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD) that are connected by a linker [39–41]. Hsp70/DnaK collaborates with two cochaperones, a J-domain cochaperone (Hsp40 in eukaryotes; DnaJ in bacteria) and a nucleotide exchange factor (NEF; GrpE in bacteria). The J-domain cochaperone stimulates ATP hydrolysis by Hsp70/DnaK and may be involved in presenting substrates to Hsp70/DnaK [42–47]. NEF promotes nucleotide exchange by Hsp70/DnaK, by triggering the release of ADP [39, 45, 48–50].
In recent work, we showed that E. coli Hsp90 and DnaK directly interact in vivo and in vitro. A region in the M-domain of Hsp90Ec and the J-protein binding site of DnaK were shown to be important for this interaction [30, 32, 38]. We further showed that yeast Hsp90, Hsp82, and yeast Hsp70, Ssa1, also directly interact in the absence of a Hop-like protein and require residues in Hsp82 homologous to those we identified in Hsp90Ec as important for interaction with DnaK [33]. However, there was a remaining question for both the bacterial and yeast systems; were the regions in Hsp90 and Hsp70 that we identified as important for the interaction, the regions of direct interaction or were they in regions of the chaperones involved in conformational changes important for the protein-protein interaction elsewhere on the chaperones?
In this study, we used chemical crosslinking to demonstrate the direct interaction between residues in the M-domain of Hsp90Ec and residues in the J-protein binding region of DnaK. We show that residues in the Ssa1 J-protein binding region, homologous to those on DnaK, are important for the interaction with Hsp82 as Ssa1 substitution mutants were defective in direct interaction with Hsp82. Additionally, residues in the J-protein binding region of Ssa1 can be specifically crosslinked to residues in the Hsp82 middle domain. Furthermore, we show that J-protein stimulates the interaction between Hsp90Ec and DnaK, likely by promoting the ADP-bound conformation of DnaK. Together, these results show that the sites of direct protein-protein interaction between Hsp90 and Hsp70 are conserved between bacteria and yeast.
Results
Residues in the J-protein binding region of DnaK interact directly with the Hsp90Ec middle-domain
Our previous mutagenesis studies, both in vivo and in vitro, as well as molecular modeling showed that residues in the middle domain of Hsp90Ec are important for interaction with DnaK [32, 38] and residues in the DnaJ binding region on DnaK are important for the interaction with Hsp90Ec [32]. However, both Hsp90Ec and DnaK undergo large conformational changes in response to ATP binding and hydrolysis and transduce conformational changes to distant regions of the chaperones [3, 12, 20, 21, 29, 51, 52]. Thus, there are two possible interpretations of these observations: i) a site in the middle domain of Hsp90Ec directly contacts a site in the DnaJ binding region of DnaK, and ii) the residues identified as important for the interaction may function indirectly by participating in conformational changes that are transduced to other sites on Hsp90Ec and DnaK which are involved in the direct interaction.
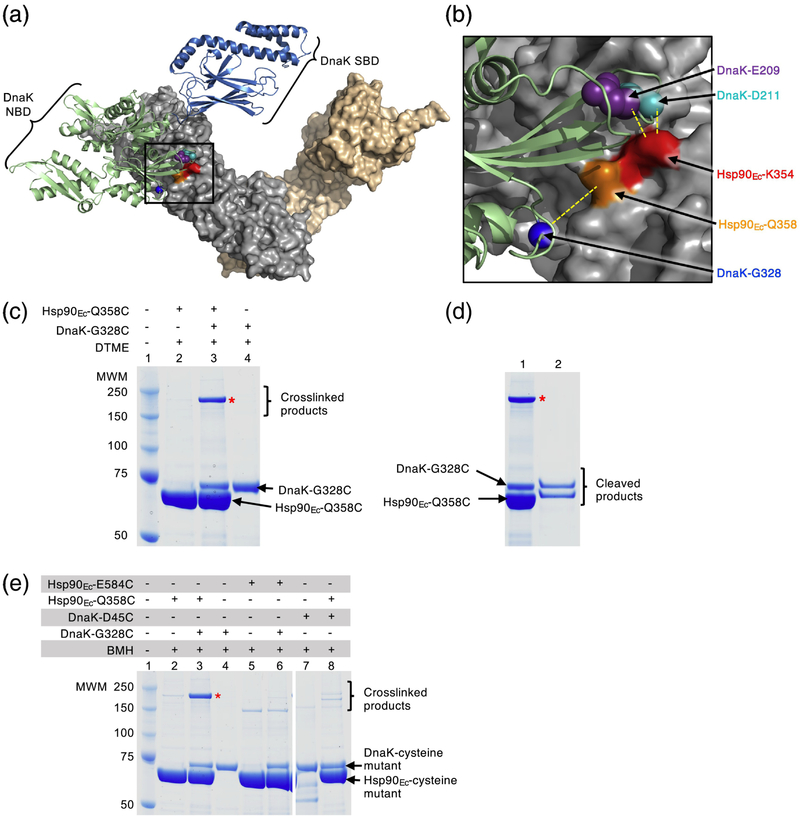
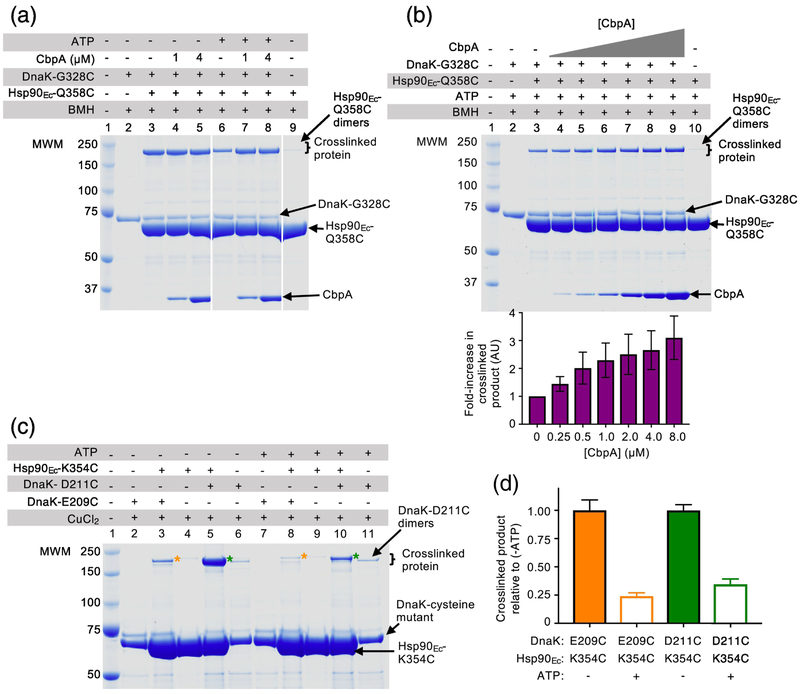
In the present study, we sought to unequivocally identify the site of direct interaction between DnaK and Hsp90Ec by performing chemical crosslinking experiments (Fig. 1a). To this end, we made cysteine substitutions in Hsp90Ec and DnaK residues predicted to be in close proximity to each other based on the docked model of the complex (Fig. 1b and Table 1 and 2) [32]. Both Hsp90Ec and DnaK are amenable to cysteine crosslinking studies because Hsp90Ec has no endogenous cysteines and DnaK has a single buried cysteine. An Hsp90Ec variant, Hsp90Ec-Q358C, was chosen to use in crosslinking experiments with DnaK because it was near, but not in the predicted interaction region (Fig. 1a and b; Table 1 and 2) [32]. In addition, we had previously observed that Hsp90Ec-Q358C was similar to wild-type in its ability to interact with DnaK and to collaborate in protein reactivation with DnaK (Supplementary Fig. S1a) [38]. We constructed DnaK-G328C since it was predicted to be close to Hsp90Ec-Q358C based on our docked model of the Hsp90Ec-DnaK complex (Fig. 1a and b; Table 1 and 2) [32]. In control experiments, DnaK-G328C exhibited properties indistinguishable from wild-type. Heat-denatured GFP was reactivated by DnaK-G328C in the presence of CbpA at a similar rate as wild-type DnaK and the reactivation reactions were similarly stimulated by Hsp90Ec wild-type or Q358C (Supplementary Fig. S1a).
Fig. 1.
Hsp90Ec and DnaK residues directly interact. (a) Docked model [32] of the apo structure of Hsp90Ec (PDB ID code 2IOQ) [15] and ADP-bound DnaK (PDB ID code 2KHO) [73]. The apo conformation of the Hsp90Ec dimer is shown as a surface rendering with one protomer in gray and one protomer in wheat. DnaK in the ADP-bound conformation [73] is shown as a ribbon model with the NBD in light green and the SBD in blue. Residues in Hsp90Ec, K354 and Q358, that were mutated in this study are shown in red and orange, respectively, while residues in DnaK, E209, D211 and G328, that were mutated in this study are shown as CPK models in purple, cyan and blue, respectively. The black square represents the field enlarged in (b). (b) Enlargement of the interaction region between Hsp90Ec and DnaK shown in (a). Hsp90Ec and DnaK are colored as in (a) and residues mutated in this study are also as in (a). Yellow dashed lines indicate the pairs of Hsp90Ec and DnaK residues that were used in crosslinking studies. (a) and (b) were both rendered using PyMOL (https://pymol.org/2/). (c) Hsp90Ec-Q358C and DnaK-G328C (4 μM each) were treated with the crosslinker DTME (13.3 Å linker) alone and in a mixture together and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. (d) The crosslinked product from (c) lane 3 was extracted and treated with a reducing agent and the products analyzed by SDS-PAGE followed by Coomassie blue staining. (e) Hsp90Ec mutants Hsp90Ec-Q358C and E584C (4 μM), and DnaK mutants DnaK-G328C and D45C (4 μM), were treated with BMH (13 Å linker) alone and in mixtures as indicated and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. Higher molecular weight bands in lanes 2, 5, 6 and 8 are likely crosslinked dimers of Hsp90Ec-Q358C (lane 2 and 8) and E584C (lane 5 and 6); the potential Hsp90Ec-E584C dimer appears to run at a lower molecular weight than other crosslinked products likely due to it attaining a more compact conformation following crosslinking. In (c-e), (*) indicates crosslinked product of interest and the gels shown are representative of at least three independent experiments.
Table 1.
Substitution mutants used in this study
Table 2.
Substitutions used for crosslinking and BTH assays
| Protein | Substitution | Residue tested for interaction | Distance (Å) between residues (CA)a |
|---|---|---|---|
| Hsp90Ec | K354C | DnaK E209C DnaK D211C |
10.6 7.8 |
| Hsp90Ec | Q358C | DnaK G328C DnaK D45C |
10.1 60.8 |
| Hsp90Ec | E584C | DnaK G328C | 73.0 |
| DnaK | E209C | Hsp90Ec K354C | 10.6 |
| DnaK | D211C | Hsp90Ec K354C Hsp90Ec Q358C |
7.8 13.4 |
| DnaK | G328C | Hsp90Ec Q358C Hsp90Ec E584C |
10.1 73.0 |
| DnaK | D45C | Hsp90Ec Q358C | 60.8 |
| Hsp82 | P281C | Ssa1 T219C | 9.1 |
| Hsp82 | E402C | Ssa1 K322C Ssa1 K323C |
10.7 11.6 |
| Hsp82 | E409C | Ssa1 K322C Ssa1 K323C Ssa1 T219C |
10.7 13.3 23.4 |
| Hsp82 | E635C | Ssa1 T219C | 86.0 |
| Ssa1 | T219C | Hsp82 P281C Hsp82 E409C Hsp82 E635C |
9.1 23.4 86.0 |
| Ssa1 | K322C | Hsp82 E402C Hsp82 E409C |
10.7 10.7 |
| Ssa1 | L323C | Hsp82 E402C Hsp82 E409C |
11.6 13.3 |
To monitor covalent interaction between Hsp90Ec-Q358C and DnaK-G328C, we incubated the two proteins together with a homobifunctional, cysteine reactive, reversible crosslinker, dithiobismaleimidoethane (DTME), that has a linker length of 13.3 Å. When the products were analyzed by SDS-PAGE, a prominent higher molecular weight species appeared, consistent with a Hsp90Ec-DnaK complex (Fig. 1c, lane 3). The higher molecular weight species was not present when either protein alone was treated with crosslinker (Fig. 1c, lanes 2 and 4). Following extraction from the gel, treatment with reducing agent and separation by SDS-PAGE, the higher molecular weight species was observed to be composed of approximately equal amounts of Hsp90Ec and DnaK (Fig. 1d, lane 2). Additionally, DnaK-G328C and Hsp90Ec-Q358C could be crosslinked using CuCl2, which promotes disulfide bond formation between cysteine sidechains that are within ~2 Å of one another (Supplementary Fig. S1b). However, less crosslinked product was observed than with the DTME or bismaleimidohexane (BMH) crosslinkers, which have longer linker arms (~13 Å) (Fig. 1c–1e and Supplementary Fig. S1b). These results demonstrate that the middle domain of Hsp90Ec directly interacts with the J-protein binding region of DnaK.
In control experiments, pairs of Hsp90Ec and DnaK residues that were predicted to be separated by > 60 Å in the docked model of Hsp90Ec and DnaK were also tested in the crosslinking assay (Fig. 1e and Supplementary Fig. S1c). No detectable crosslinked Hsp90Ec-DnaK complex was observed when either Hsp90Ec-E584C and DnaK-G328C or Hsp90Ec-Q358C and DnaK-D45C were treated with BMH and analyzed by SDS-PAGE (Table 1 and 2; Fig. 1e, lanes 5 and 6 and lanes 7 and 8).
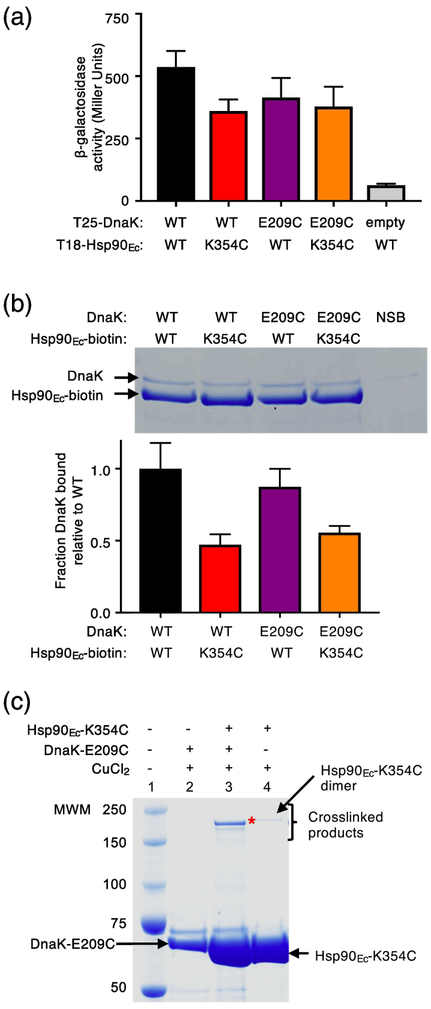
We constructed a second pair of DnaK-Hsp90Ec cysteine mutants in residues that were in close proximity based on the docked model, DnaK-E209C and Hsp90Ec-K354C (Fig. 1a and b; Table 1 and 2) [32], and tested the mutants in crosslinking experiments to substantiate the site of interaction. Both DnaK-E209C and Hsp90Ec-K354C had previously been shown to be partially defective for interaction with their wild-type partner using a bacterial two-hybrid assay and using purified proteins in a pull-down assay (Fig. 2a and 2b) [32, 38]. When we monitored the in vivo interaction between Hsp90Ec-K354C and DnaK-E209C we observed that they interacted with each other similarly to each mutant with their wild-type partner, although less well than the wild-type pair of chaperones, suggesting that the substitutions did not significantly disrupt the interaction surface (Fig. 2a). In vitro, less DnaK-E209C was retained by biotinylated Hsp90Ec-K354C compared to the amount of wild-type DnaK retained by biotinylated wild-type Hsp90Ec; however, the mutant-mutant interaction was consistent with the interaction observed for biotinylated Hsp90Ec-K354C and wild-type DnaK (Fig. 2b) [38]. We next treated Hsp90Ec-K354C and DnaK-E209C with CuCl2 and observed a slower migrating species by SDS-PAGE, consistent with the crosslinked Hsp90Ec-DnaK complex (Fig. 2c). DnaK-D211, another residue in the J-protein binding region of DnaK and near Hsp90Ec-K354, based on the interaction model (Fig. 1a and 1b) [32], was also substituted with a cysteine (Table 1 and 2). DnaK-D211C was found to form a covalent bond with Hsp90Ec-K354C in the presence of CuCl2 (Supplementary Fig. S2a and b). Similar to DnaK-E209C, DnaK-D211C interacts with Hsp90Ec-K354C both in vivo and in vitro (Supplementary Fig. S2c and d). These results provide additional support for the conclusion that Hsp90Ec and DnaK directly interact through residues in the J-protein binding region of DnaK and the Hsp90Ec M-domain.
Fig. 2.
Specific crosslinking of a pair of Hsp90Ec and DnaK residues important for interaction. (a) The in vivo interaction between Hsp90Ec wild-type or K354C and DnaK wild-type or E209C was monitored using a bacterial two-hybrid system that measured beta-galactosidase activity in liquid culture. β-galactosidase activity is shown as mean ± SEM (n=3). (b) The in vitro interaction between biotinylated Hsp90Ec wild-type or K354C and DnaK wild-type or E209C (4 μM each) was determined using a pull-down assay in the presence of L2, CbpA and ATP as described in Materials and Methods and analyzed by SDS-PAGE followed by Coomassie blue staining. DnaK wild-type or E209C associated with biotinylated Hsp90Ec wild-type or K354C was quantified using densitometry as described in Materials and Methods and is shown as a bar graph. For each lane of the gel the amount of DnaK was corrected for non-specific binding (NSB) and then was normalized to Hsp90Ec-biotin and the ratio of DnaK mutant to wild-type is plotted as the mean ± SEM (n=6). (c) Hsp90Ec-K354C and DnaK-E209C were treated with CuCl2 (~2 Å linker) alone and in a mixture together and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. (*) indicates crosslinked product of interest. A small amount of a slowly migrating species was likely the covalently linked Hsp90Ec-K354C dimer, since it was observed in reactions with and without Ssa1 (lanes 3 and 4). In (c), the gel shown is representative of at least three independent experiments.
From these results we conclude that residues, which are in the regions shown by mutagenesis studies and the docked model to be important for Hsp90Ec-DnaK binding, directly interact. The results eliminate the interpretation that the residues identified by mutagenesis studies are important for conformational changes required for interaction through other surfaces of the proteins. The results also validate our docked model of the Hsp90Ec-DnaK complex.
Substitution mutants in Ssa1 homologous to DnaK mutants defective in interaction with Hsp90Ec are defective in physical and functional interactions with Hsp82
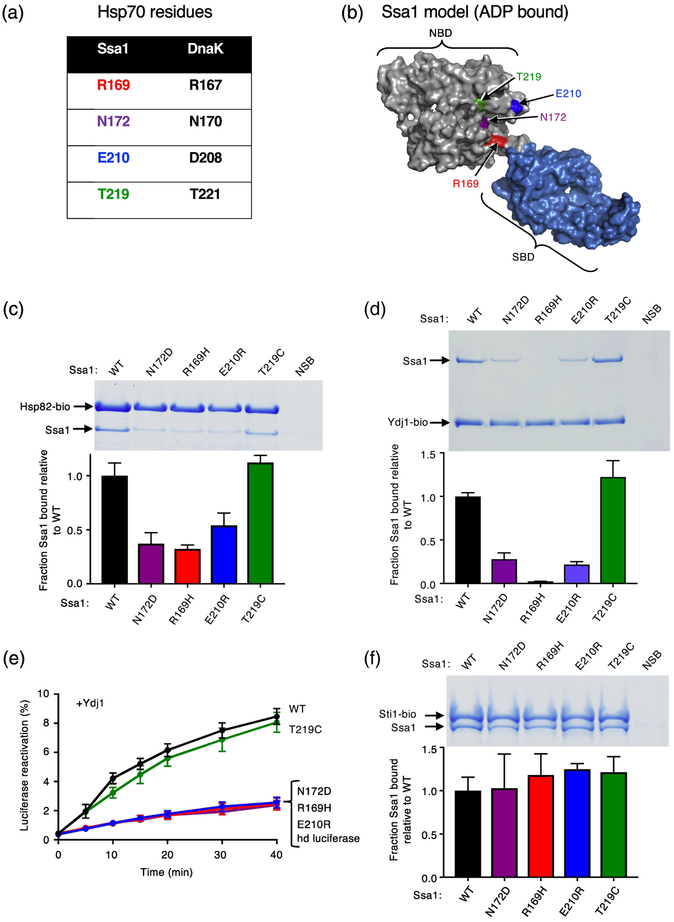
We wanted to determine if yeast Hsp70, Ssa1, and Hsp90, Hsp82, directly interact through regions homologous to those on DnaK and Hsp90Ec. The site on Ssa1 that is important for interaction with Hsp82 had not been previously determined, although our earlier work showed that a region in the Hsp82 middle domain, homologous to the region on Hsp90Ec that directly interacts with DnaK, is important for the interaction with Ssa1 [33]. Thus, before carrying out crosslinking experiments, we performed mutational analysis to identify residues on Ssa1 important for the interaction with Hsp82. Amino acid substitution in Ssa1 residues, which were predicted to be in the Hsp82 binding site homologous to DnaK residues in the Hsp90Ec (and J-protein) binding site [32], were constructed. The substituted residues included Ssa1-R169H, N172D, E210R and T219C (Fig. 3a and b; Table 1).
Fig. 3.
Ssa1 residues homologous to DnaK residues important for interacting with Hsp90Ec are important for Hsp82 and Ydj1 binding. (a) Chart showing residues in S. cerevisiae Hsp70, Ssa1, that align with four residues in E. coli DnaK, that were previously shown to be in the Hsp90Ec interaction region [32]. (b) Model of Ssa1 in the ADP-bound conformation (based on the ADP-bound conformation of DnaK (PDB ID code 2KHO) [33, 73] shown as a surface rendering with the NBD in gray and the SBD in blue and generated using PyMOL (https://pymol.org/2/). Residues mutated in this study and tested for interaction with Hsp82 are shown in color and labeled. (c) The in vitro interaction between biotinylated Hsp82 (2.5 μM) and Ssa1 wild-type or mutant (8 μM) was monitored using a pull-down assay as described in Materials and Methods. The results were analyzed by SDS-PAGE followed by Coomassie blue staining. Ssa1 wild-type or mutant associated with biotinylated Hsp82 was quantified using densitometry as described in Materials and Methods and is shown as a bar graph. For each lane of the gel, the amount of Ssa1 was corrected for non-specific binding (NSB) and then was normalized to Hsp82-biotin and the ratio of Ssa1 mutant to wild-type is plotted. (d) The in vitro interaction between biotinylated Ydj1 (1.2 μM) with Ssa1 wild-type or mutant (2 μM) was monitored using a pull-down assay as described in Materials and Methods. The results were analyzed by SDS-PAGE followed by Coomassie blue staining. Ssa1 wild-type or mutant associated with biotinylated Ydj1 was quantified using densitometry as described in Materials and Methods and is shown as a bar graph. For each lane of the gel, the amount of Ssa1 was corrected for non-specific binding (NSB) and then was normalized to Ydj1-biotin and the ratio of Ssa1 mutant to wild-type is plotted. (e) Heat-inactivated luciferase reactivation by Ssa1 wild-type or mutant and Ydj1 as indicated and described in Materials and Methods. Data from three or more replicates are presented as mean ± SEM. For some points, the symbols obscure the error bars. (f) The in vitro interaction between biotinylated Sti1 (2 μM) and Ssa1 wild-type or mutant (4 μM) was monitored using a pull-down assay as described in Materials and Methods. The results were analyzed by SDS-PAGE followed by Coomassie blue staining. Ssa1 wild-type or mutant associated with biotinylated Sti1 was quantified using densitometry as described in Materials and Methods and is shown as a bar graph. For each lane of the gel, the amount of Ssa1 was corrected for non-specific binding (NSB) and then was normalized to Sti1-biotin and the ratio of Ssa1 mutant to wild-type is plotted. In (c, d and f), the gels shown are representative of at least three independent experiments and quantification of the data from three or more replicate gels are presented as mean ± SEM.
The Ssa1 mutants were tested for the ability to interact with Hsp82 using an in vitro pull-down assay with Hsp82-biotin and Ssa1 wild-type or mutant. Three of the Ssa1 mutants, Ssa1-N172D, R169H and E210R, were retained ~50–60% less well than wild-type, suggesting they were partially defective in Hsp82 interaction (Fig. 3c). Ssa1-T219C was retained by Hsp82-biotin similarly to wild-type (Fig. 3c), suggesting that a cysteine substitution of this residue is compatible with the interaction or that this residue is not important for the interaction with Hsp82.
Since the three Ssa1 mutants defective in interacting with Hsp82 had substitutions in residues that in E. coli DnaK are also in the J-protein interacting region, we tested whether the Ssa1 mutant proteins were defective for interaction with Ydj1, a yeast J-protein. The interaction was monitored using an in vitro pull-down assay with Ydj1-biotin (Fig. 3d). Ssa1-R169H was defective in Ydj1 interaction and Ssa1-N172D and D210R were partially defective in Ydj1 interaction, with ~75% less mutant Ssa1 retained than wild-type (Fig. 3d). Ssa1-T219C interacted with Ydj1 similarly to wild-type (Fig. 3d). In a functional assay, Ssa1 wild-type and mutants were tested for the ability to function with Ydj1 in the reactivation of heat-denatured luciferase (Fig. 3e). The same three Ssa1 mutants, Ssa1-R169H, N172D and E210R, that were defective in Hsp82 and Ydj1 binding were defective in reactivating heat-denatured luciferase with Ydj1, consistent with the defects observed in Ydj1 interaction (Fig. 3d and 3e). Ssa1-T219C collaborated with Ydj1 in reactivation of heat-denatured luciferase, similar to Ssa1 wild-type (Fig. 3e) and consistent with its ability to bind Ydj1 like wild-type (Fig. 3d). In another functional assay using conditions in which reactivation of heat-denatured luciferase depends on Ssa1, Hsp82, Sis1 (a yeast J-protein) and Sti1 (an Hsp82 cochaperone that stabilizes the Hsp82-Ssa1 interaction [10, 33]), we found that Ssa1-R169H, N172D and E210R were defective in luciferase reactivation, while T219C was like wild-type (Supplementary Fig. S3a). With the mutant residues tested, we have not been able to differentiate between the binding site on Ssa1 for Hsp82 and Ydj1 (or Sis1). Despite the defects observed in luciferase reactivation and binding to Ydj1 and Hsp82, the Ssa1 mutants interacted with Sti1 similarly to wild-type (Fig. 3f), as expected since Sti1 interacts with the C-terminal VEEVD motif of Ssa1 [53, 54]. Additionally, all Ssa1 mutants could hydrolyze ATP (Supplementary Fig. S3b).
Together, the results show that a region on Ssa1 homologous to the Hsp90Ec binding site on DnaK [32] is important for the interaction between Ssa1 and both Hsp82 and J-protein (Ydj1/Sis1) [32, 44, 55].
Yeast Hsp90 and Hsp70 directly interact through regions homologous to those of E. coli Hsp90Ec and DnaK
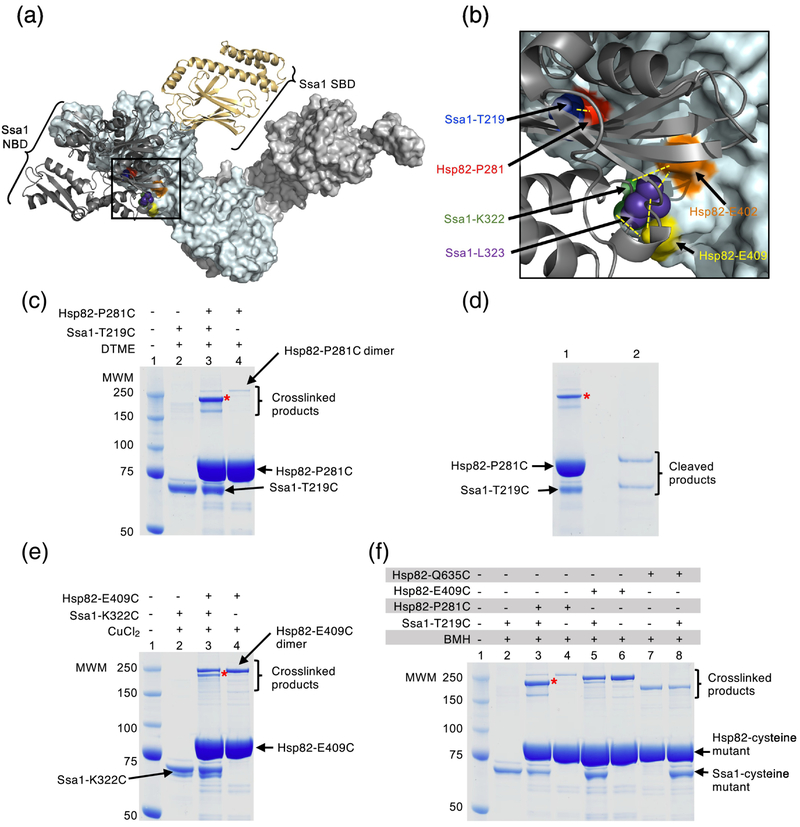
We next performed crosslinking experiments using Hsp82 and Ssa1 cysteine mutants to test if the regions important for the interaction between Hsp82 and Ssa1 were the sites of direct interaction, as they are for Hsp90Ec and DnaK. The experiments were feasible because Hsp82 has no endogenous cysteines and Ssa1 has three buried cysteines in the nucleotide-binding domain. We used Hsp82-P281C since it was predicted by the docked model of the Hsp82-Ssa1 complex to be in the interaction region [33] and because it is homologous to Hsp90Ec-K238, a residue shown to be important for interaction with DnaK (Fig. 4a and b and Table 1 and 2) [38]. However, Hsp82-P281C was able to interact with Ssa1 similar to Hsp82 wild-type both in vivo and in vitro [33], suggesting that the cysteine substitution did not alter the interaction or that it was near, but not in the immediate site of interaction. We selected Ssa1-T219C for crosslinking with Hsp82-P281C because it was predicted to be in close proximity to Hsp82-P281C based on the Hsp82-Ssa1 docked model and it functioned similarly to wild-type in in vitro assays (Fig. 3e, 4a and b; Table 1 and 2) [33]. Following treatment of the Hsp82-P281C and Ssa1-T219C mixture with DTME (13.3 Å), a prominent higher molecular weight product was observed by SDS-PAGE that was consistent with the molecular weight of a covalently linked Hsp82-Ssa1 complex (Fig. 4c, lane 3). It was not visible when each chaperone was treated separately with DTME (Fig. 4c, lanes 2 and 4). A small amount of a slowly migrating species was likely the covalently linked Hsp82-P281C dimer, since it was observed in reactions with and without Ssa1 (Fig. 4c, lanes 3 and 4). The minor species that migrated slightly faster than the covalently linked Hsp82-Ssa1 complex was not characterized (Fig. 4c, lane 3). Following extraction of the prominent crosslinked species from the gel, treatment with reducing agent and analysis by SDS-PAGE, about equimolar amounts of Hsp82 and Ssa1 were observed (Fig. 4d, lane 2), showing a direct interaction between the two chaperones in the predicted regions of interaction.
Fig. 4.
Hsp82 and Ssa1 residues directly interact. (a) Docked homology models of apo yeast Hsp82 and ADP-bound Ssa1 [33]. The apo conformation of the Hsp82 dimer is shown as a surface rendered model in light cyan and light gray and the ADP-bound conformation of Ssa1 is shown as a ribbon model with the NBD in dark gray and the SBD in wheat. Residues in Hsp82, P281, E402 and E409, that were mutated in this study are shown in red, orange and yellow, respectively, while residues in Ssa1, T219, K322 and L323, that were mutated in this study are shown as CPK models in blue, green and purple, respectively. The black square represents the field that is enlarged in (b). (b) Enlargement of the interaction region between Hsp82 and Ssa1 shown in (a). Hsp82 and Ssa1 are colored as in (a) and residues mutated in this study are also as described in (a). Yellow dashed lines indicate the pairs of Hsp82 and Ssa1 residues that were used for crosslinking experiments. (a) and (b) were both rendered using PyMOL (https://pymol.org/2/). (c) Hsp82-P281C and Ssa1-T219C (4 μM each) were treated with the crosslinker DTME (13.3 Å linker) alone and in a mixture together and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. The major crosslinked species (*) is consistent with crosslinked Hsp82-P281C and Ssa1-T219C (lane 3). A small amount of a slowly migrating species was likely the covalently linked Hsp82-P281C dimer, since it was observed in reactions with and without Ssa1 (lanes 3 and 4). The minor species seen in crosslinking reactions with Hsp82-P281C and Ssa1-T219C that migrated slightly faster than the predominant species was not characterized (lane 3). (d) The predominant crosslinked species (*) from (c) lane 3 was extracted and treated with a reducing agent and the products analyzed by SDS-PAGE followed by Coomassie blue staining. (e) Hsp82-E409C and Ssa1-K322C (4 μM each) were treated with CuCl2 (~2 Å linker) alone and in a mixture together and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. The Hsp82-P281C and Ssa1-T219C crosslinked species is indicated (*, lane 3) and a second species consistent with the covalently linked Hsp82-P281C dimer, since it was observed in reactions with and without Ssa1, was also observed. (f) Hsp82 mutants including Hsp82-P281C, E409C and Q635C (4 μM), and Ssa1-T219C (4 μM) are treated with BMH (13 Å linker) alone and in mixtures together as indicated and covalent bond formation is monitored by SDS-PAGE followed by Coomassie blue staining. Crosslinked dimers of Hsp82-P281C (lane 3 and 4), E409C (lane 5 and 6) and Q635C (lane 7 and 8) can be observed; Hsp82-Q635C runs at a lower molecular weight than other crosslinked products likely due to it attaining a more compact conformation following crosslinking. In (c-f), (*) indicates crosslinked product of interest and the gels shown are representative of at least three independent experiments.
Crosslinking was also observed between another pair of residues predicted to be close to one another in the complex, Hsp82-E409C and Ssa1-K322C (Fig. 4b), following treatment with CuCl2 (Fig. 4e, lane 3 and Table 2). For Hsp82-E409C, a second higher molecular weight crosslinked product was observed in the reactions with and without Ssa1, suggesting that the species is a crosslinked Hsp82 dimer (Fig. 4e, lanes 3 and 4). Crosslinked complexes were also observed between (i) Hsp82-E402C and Ssa1-K322C, (ii) Hsp82-E402C and Ssa1-L323C, (iii) Hsp82-E409C and Ssa1-K322C, and (iv) Hsp82-E409C and Ssa1-L323C when using the crosslinker BMH (13 Å) (Table 1 and 2 and Supplementary Fig. S4a). In control experiments (Fig. 4f and Supplementary Fig. S4b), a crosslinked species was not detected between Hsp82-Q635C and Ssa1-T219C (Fig. 4f, lane 8), which were predicted to be ~86 Å apart (Table 2). In addition, when Hsp82-E409C and Ssa1-T219C, which were predicted to be ~23 Å apart (Table 2), were treated with BMH, only a small amount of crosslinked complex was observed (Fig. 4f, lane 5). Altogether, the results show that the site of direct interaction between the Hsp90 M-domain and the J-protein binding region of Hsp70 is conserved in bacteria and yeast.
J-protein promotes the interaction between Hsp90 and Hsp70 in the presence of ATP
In this study and in previous work we observed that the binding sites for Hsp90 and the J-protein on Hsp70 overlap (Fig. 3d and 3e) [32, 33, 38], suggesting that J-proteins may affect the interaction between Hsp90 and Hsp70. To further explore the potential role of a J-protein in the Hsp70-Hsp90 interaction, we performed BMH crosslinking experiments using DnaK-G328C and Hsp90Ec-Q358C with and without CbpA, which contains no cysteines. We analyzed crosslinked products using SDS-PAGE and observed that in the absence of added ATP (a condition under which DnaK is in the closed conformation [52] and Hsp90Ec is in the open conformation [3, 12]) crosslinking between the chaperones was high and the amount of crosslinked complex did not increase or decrease when CbpA was present (Fig. 5a, lanes 3–5). However, in the presence of ATP, ~60% less crosslinked DnaK-Hsp90Ec complex was observed (Fig. 5a, lane 6) compared to the amount of complex formed without ATP (Fig. 5a, lane 3). We further observed that in the presence of ATP, the amount of crosslinked product increased significantly when CbpA was added to the reaction mixtures (Fig. 5a, lanes 7–8). By titrating CbpA in crosslinking reactions with DnaK-G328C, Hsp90Ec-Q358C and ATP, we observed that the amount of crosslinked Hsp90Ec-DnaK species increased with increasing CbpA concentration (Fig. 5b; see quantification beneath). In control experiments CbpA did not promote the formation of crosslinked Hsp90Ec dimers or DnaK dimers (Supplementary Fig. S5a, lanes 11 and 12). These results suggest that one role of J-proteins may be to stimulate the conversion of the ATP-bound conformation of Hsp70 to the ADP-bound conformation, which is then able to interact with Hsp90.
Fig. 5.
J-protein promotes the interaction between DnaK and Hsp90Ec in the presence of ATP. (a) Hsp90Ec-Q358C (4 μM) and DnaK-G328C (4 μM) alone and in mixtures with CbpA (1 or 4 μM) (no endogenous cysteines) in the absence or presence of 4 mM ATP as indicated were treated with BMH (13 Å linker) and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. (b) In the presence of ATP (4 mM), Hsp90Ec-Q358C (4 μM) and DnaK-G328C (4 μM) alone and in mixtures with increasing concentrations of CbpA as indicated were treated with BMH (13 Å linker) and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. Crosslinked products were quantified using ImageJ (http://imagej.nih.gov/ij) and standardized to crosslinked products in the absence of CbpA (lane 3) and plotted as the mean ± SEM (n=3). (c) Hsp90Ec-K354C (4 μM) and DnaK-E209C or DnaKD211C (4 μM) in the presence or absence of 4 mM ATP as indicated, were treated with CuCl2 and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. (*) indicates crosslinked product of interest for DnaK-E209C and Hsp90Ec-K354C (lanes 3 and 8) and (*) indicates crosslinked product of interest for DnaK-D211C and Hsp90Ec-K354C (lanes 5 and 10). (d) Crosslinked products from (c) were quantified using ImageJ (http://imagej.nih.gov/ij) and standardized to crosslinked products in the absence of ATP (-ATP) and plotted as the mean ± SEM (n=3). In (a-c), the gels shown are representative of at least three independent experiments.
Crosslinking experiments using Hsp90Ec-K354C and either DnaK-E209C or DnaK-D211C also showed that not as much crosslinked complex, about 75% less, was formed in the presence of ATP than in the absence of ATP (Fig. 5c, compare lane 3 with 8 and lane 5 with 10; Fig. 5d, quantification of 5c). Also consistent with these data are previous experimental results [31–33, 38, 56, 57], and molecular modeling results showing that the ADP form and not the ATP form of DnaK interacts with Hsp90Ec [32].
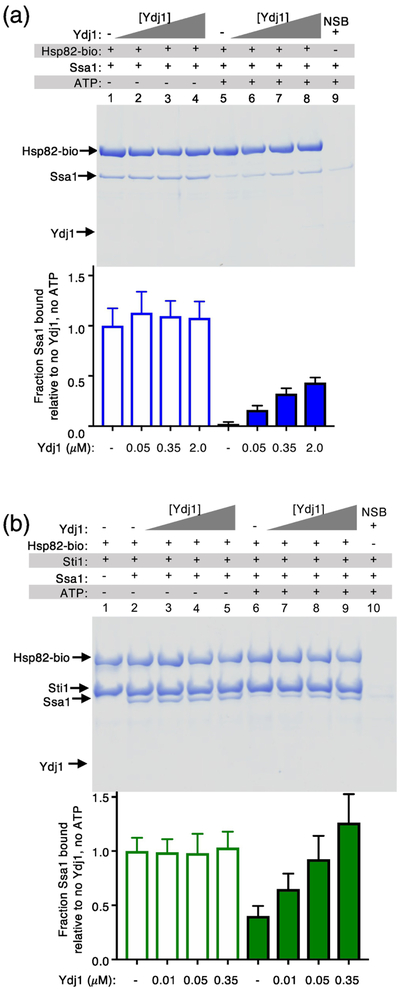
We next tested if a yeast J-protein similarly increases the amount of Ssa1-Hsp82 complex by performing pull-down experiments using biotin-labeled Hsp82 and Ssa1. Ssa1 readily associated with Hsp82-biotin in the absence of ATP but could not be detected in complexes with Hsp82 in the presence of ATP (Fig. 6a, compare lanes 1 and 5; see quantification beneath). As observed with the bacterial chaperones, there was no detectable effect of Ydj1 on the amount of Ssa1 associated with Hsp82-biotin in the absence of ATP (Fig. 6a, lanes 1–4). However, when Ydj1 was titrated into mixtures of biotin-labeled Hsp82 and Ssa1 in the presence of ATP, Ssa1 was observed in association with Hsp82 and the amount of Ssa1 associated with Hsp82 increased as the concentration of Ydj1 increased (Fig. 6a, lanes 5–8; quantification beneath). Similar results were obtained when Sti1 was included in the reaction, although the amount of Ssa1 associated with Hsp82-biotin was significantly more (Fig. 6b; quantification beneath), as previously observed [33]. These results show that in the presence of ATP the interaction of yeast Hsp90 and Hsp70 is also stimulated by a J-protein and suggest that J-proteins in both bacteria and yeast have a role in facilitating the interaction between Hsp70 and Hsp90, possibly by promoting conversion of the ATP-bound form of Hsp70 to the ADP-bound form, which is then able to interact with Hsp90.
Fig. 6.
J-protein promotes the interaction between Ssa1 and Hsp82 in the presence of ATP. (a) The in vitro interaction between biotinylated Hsp82 (2.6 μM) and Ssa1 (11 μM) alone and in mixtures with increasing concentrations of Ydj1 in the absence or presence of 2 mM ATP was monitored using a pull-down assay as described in Materials and Methods. (b) The in vitro interaction between biotinylated Hsp82 (1.6 μM) and Ssa1 (8 μM) with Sti1 (1 μM) and in mixtures with increasing concentrations of Ydj1 in the absence or presence of 2 mM ATP was monitored using a pull-down assay as described in Materials and Methods. In both (a) and (b) the results were analyzed by SDS-PAGE followed by Coomassie blue staining. Ssa1 associated with biotinylated Hsp82 was quantified using densitometry as described in Materials and Methods and is shown as a bar graph. For each lane of the gel, the amount of Ssa1 was corrected for non-specific binding (NSB) and then was normalized to Hsp82-biotin and the ratio of Ssa1 bound relative the amount bound in the absence of Ydj1 and ATP is plotted. The gels shown are representative of in (a) five and in (b) three independent experiments and quantification of the data from the replicate gels are presented as mean ± SEM.
Working model for the collaboration between the Hsp70 system and Hsp90
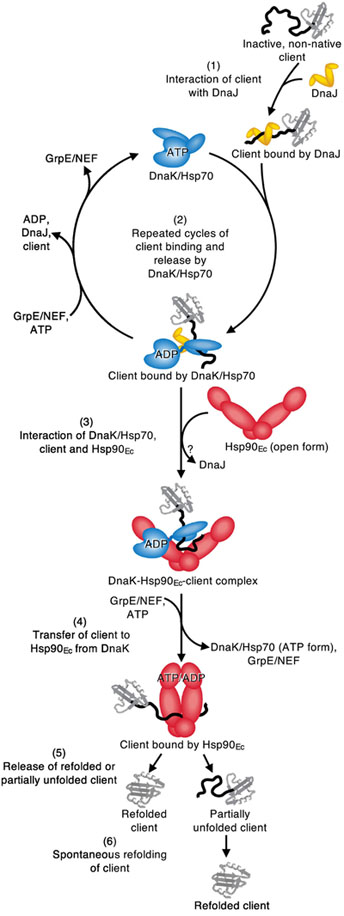
Our working model for the collaboration between bacterial DnaK and Hsp90, although speculative, seeks to incorporate the current knowledge of the Hsp90 and Hsp70 chaperone systems (Fig. 7). It is well established by the work of many groups that the DnaK/Hsp70 system works prior to Hsp90 [3, 12, 29, 51]. This begins when the J-protein first interacts with client and recruits DnaK/Hsp70 to the client (Fig. 7, step 1) [42, 44, 52]. In this reaction, the J-protein interacts with the ATP-bound conformation of DnaK/Hsp70 and promotes ATP-hydrolysis and client transfer to DnaK/Hsp70 [44, 46, 52, 55]. Repeated cycles of client binding and release by DnaK/Hsp70 can prevent client misfolding and/or promote partial client remodeling (Fig. 7, step 2) [41, 44, 46, 52]. Alternatively, DnaK/Hsp70 in the ADP-bound conformation can interact with Hsp90 (Fig. 5a and 5b and Fig. 7, step 3) [31–33, 36, 38, 56, 57]. Specifically, the interaction occurs between the J-protein binding region of Hsp70 and the M-domain of Hsp90 [32, 33, 38]. The J-protein may be displaced at this step in the cycle. Hsp90 is likely in the open conformation upon DnaK/Hsp70 binding to accommodate interactions with clients that bind to surfaces buried when Hsp90 is in the closed conformation. However, the DnaK/Hsp70 interacting residues in the Hsp90 M-domain are also surface exposed in the closed conformation [32, 33], and it may not be essential for Hsp90 to be in the open conformation for DnaK/Hsp70 to bind. Concomitant client release by DnaK/Hsp70 and transfer to Hsp90 is triggered by i) ADP release and ATP binding by DnaK/Hsp70, promoted by GrpE/NEF [39, 45, 48–50], ii) Hsp90 promoting substrate release from DnaK/Hsp70 [31, 37], and iii) DnaK/Hsp70 stimulation of Hsp90 ATPase activity to transfer client to Hsp90 [28, 29, 31, 34, 36, 38, 51, 57]. Simultaneous with or following client transfer, DnaK/Hsp70 likely dissociates from Hsp90 (Fig. 7, step 4) [3, 28, 31, 37]. Lastly, Hsp90 releases the client, which may be in a native or near native conformation (Fig. 7, step 5) [28, 29, 31] or in a partially folded conformation that spontaneously refolds or is rebound by other chaperones (Fig. 7, step 6).
Fig. 7.
Working model for the collaboration of bacterial Hsp70 and Hsp90 in protein remodeling and activation. (1) The J-protein, DnaJ in E. coli, engages inactive, non-native client. (2) DnaK/Hsp70 is recruited to the client by DnaJ/J-protein and through rounds of client binding and release prevents client misfolding or initiates protein remodeling. (3) DnaK/Hsp70 in the ADP-bound conformation stably interacts with substrate and Hsp90 is recruited via the interaction between the DnaK/Hsp70 J-protein binding site and the M-domain of Hsp90. (4) Nucleotide induced conformational changes likely in both chaperones promote client transfer from DnaK/Hsp70 to Hsp90. (5) Client is released from Hsp90 in either a partially remodeled state or in an active conformation. (6) Spontaneous refolding of partially remodeled client can occur. See Results for details of the model.
The basic working model for the mechanism of the eukaryotic Hsp70-Hsp90 client remodeling system is similar to that suggested in Fig. 7 for the simpler bacterial system [3, 13, 28, 29, 33, 34, 51]. As with bacterial chaperones, the Hsp70 system acts first to hold non-native regions of clients and/or to partially remodel clients in iterative cycles of binding and release [29, 30, 34, 51]. In the next step, clients are transferred to Hsp90. However, in eukaryotes, Hsp90 co-chaperones modulate the interaction between Hsp70 and Hsp90 and can further regulate Hsp90 during client remodeling [3, 10, 12, 14]. For example, Hop/Sti1 stabilizes the Hsp70-Hsp90 interaction and promotes substrate reactivation [3, 37, 56]. Nevertheless, even in the presence of Hop/Sti1, the ADP-bound conformation of Hsp70 preferentially interacts with Hsp90 [33, 56], and a J-protein is important for Hsp70-Hop/Sti1-Hsp90 complex formation and activity [33, 37, 56]. Hop/Sti1 binding to Hsp90 inhibits ATP hydrolysis by shifting the conformational equilibrium of Hsp90 towards the apo conformation [31], while Hsp70 binding to the Hsp90-Hop complex promotes ATP binding by Hsp90, leading to ATP hydrolysis and client transfer [31, 37, 56]. Taken together, a model for the collaboration of Hsp70 and Hsp90, from bacteria to eukaryotes, in protein remodeling is becoming clearer.
Discussion
Previous studies using site-directed mutagenesis and molecular modeling identified regions important for the interaction between Hsp90 and Hsp70 of E. coli and yeast. While they suggested the identified regions were the sites of direct interaction, they left open the interpretation that the identified regions may be important for conformational changes that alter the direct binding at a distant site on the proteins. In this work we showed by crosslinking experiments that Hsp90 and Hsp70 from both E. coli and yeast directly interact through residues on the M-domain of Hsp90 and the J-protein binding region of Hsp70. Further, our results suggest that the J-protein promotes the direct interaction between Hsp70 and Hsp90 by converting the ATP-bound conformation of Hsp70 into the ADP-bound conformation.
The region on yeast Hsp82 where Ssa1 interacts may be a more general protein interaction surface. For example, residues in the Ssa1 interaction region of the Hsp82 M-domain have also been identified as interacting with cochaperones Aha1 and Sba1 [3, 23, 58, 59]. In addition, human Hsp90β residues homologous to those in Hsp82 shown to interact with Ssa1 were shown to interact with the Hsp90 cochaperone, Cdc37 [60]. Moreover, several client proteins, including Cdk4, Tau and p53, interact with the Hsp82 M-domain in regions that include some of the homologous Ssa1 binding residues or are adjacent to homologous Ssa1 binding residues [3, 4, 12, 14, 28, 61]. Together these results suggest that the Hsp90 M-domain may act as a multiprotein binding surface and that the collaboration between Hsp90 and Hsp70 in protein remodeling may be modulated by Hsp70 and Hsp90 cochaperones and clients competing for the same binding surfaces.
Similar to the overlapping protein binding sites on the yeast Hsp82 M-domain, the region of the DnaK and Ssa1 NBD where Hsp90 interacts is also involved in binding additional proteins, namely, the J-domain cochaperones (Fig. 3c and d) [40, 43, 44, 55, 62–64]. Furthermore, Hsp70 undergoes ATP-dependent conformational changes where a portion of the Hsp70 substrate-binding domain interacts with the NBD through some of the residues involved in Hsp90 binding. The ability of J-protein to bind to the ATP-bound conformation of Hsp70 and promote the ADP-bound conformation, which is competent for binding to Hsp90, is additional evidence of the interplay of proteins at this site on Hsp70. Again, a shared protein interaction surface appears to be important for the modulation of Hsp70 activity and collaboration with Hsp90.
Previous research has investigated larger complexes including various combinations of eukaryotic Hsp90, Hsp70, Hop/Sti1 and clients. These complexes were studied using different structural methods including electron microscopy, cryo-electron microscopy and chemical crosslinking followed by mass spectrometry [3, 31, 56, 59, 60, 65, 66]. In these studies, different conformations of the chaperones and alternative interactions between Hsp90 and Hsp70 were observed compared with those directly identified in this study by crosslinking. Moreover, the stoichiometry of some of the larger complexes was different depending on the study; for example, in the Hsp90:Hsp70:Hop:GR complex Hsp70 was observed to be either a monomer [31, 65] or a dimer [66]. Perhaps some of these differences could be attributed to different steps in the protein remodeling pathway being captured by the various methods utilized.
In summary, this work establishes how Hsp90 and Hsp70 of bacteria and yeast directly interact during the process of protein remodeling. A better understanding of the interaction between the two chaperones provides insight into the collaboration in protein remodeling. Further, this interaction is conserved between bacteria and yeast and possibly higher eukaryotes and would provide additional insight into the interplay between the two ATP-dependent molecular chaperones.
Materials and Methods
Plasmids and Strains-
Single substitution mutations of Hsp90Ec, DnaK, Hsp82 and Ssa1 were made with the QuikChange Lightning mutagenesis system (Agilent) using pET-HtpG [30], pET-DnaK [67], pT18-Hsp90Ec [30], pT25-DnaK [30], pET-Hsp82 [33] and pET-Ssa1 [33]. All mutations were verified by DNA sequencing.
Proteins-
Hsp90Ec wild-type and mutants [30], DnaK wild-type and mutants [68], CbpA [69], Hsp82 wild-type and mutants [27, 33], Ssa1 wild-type and mutants [67, 70], Ydj1 [67, 70], Sis1 [33], Sti1 [27] and His-tagged L2 [71] were isolated as described. All proteins were >95% pure as determined by SDS-PAGE. Luciferase and luciferin were from Promega. Concentrations given are for Hsp90Ec, Hsp82, CbpA, Sis1 and Ydj1 dimers and DnaK, Ssa1, Sti1, GFP, L2 and luciferase monomers. Previously unpublished mutant proteins including DnaK-G328C and D211C, Ssa1-R169H, D172N, E210R, T219C, K322C and L323C, and Hsp82-E402C and E409C were shown to have trypsin digestion patterns and functional activity similar to the wild-type protein (Supplementary Fig. S1a; Supplementary Fig. S2e and f; Supplementary Fig. S3a and b; Supplementary Fig. S6a–d). Hsp90Ec, Hsp82, Ydj1 and Sti1 were labeled with biotin using a 1.5-fold excess of NHS-PEG4-Biotin (Thermo, Life Technologies). Excess biotin reagent was removed using 7K MWCO Zeba Spin Desalting Columns.
Crosslinking-
DTME (dithiobismaleimidoethane, Thermo Fisher) crosslinking reactions (25 μL) contained 20 mM Tris-HCl, pH 7.5, 75 mM KCl, 10 mM MgCl2, 25 mM EDTA (TKME Buffer) and 4 μM Hsp90Ec, DnaK, Hsp82 and Ssa1 cysteine mutants as indicated. Proteins were crosslinked in the presence of 0.8 mM DTME at 23 °C for 1 hr and analyzed by Coomassie staining following SDS-PAGE. Crosslinked species were excised and treated with LDS Sample Buffer containing DTT (50 mM) and re-analyzed by Coomassie staining following SDS-PAGE. CuCl2 crosslinking reactions (25 μL) contained TKME Buffer, 4 mM ATP, 4 μM Hsp90Ec, DnaK, Hsp82 and Ssa1 cysteine mutants as indicated. Proteins were crosslinked in the presence of 1 mM (DnaKHsp90Ec) or 0.2 mM (Ssa1-Hsp82) CuCl2 at 23 °C for 1 hr and analyzed by Coomassie staining following SDS-PAGE. BMH (bismalimidohexane, Thermo Fisher) or BMOE (bismaleimidoethane, Thermo Fisher) crosslinking reactions (25 μL) contained TKME Buffer, 4 mM ATP, 4 μM Hsp90Ec, DnaK, Hsp82 and Ssa1 cysteine mutants and CbpA as indicated. Proteins were crosslinked in the presence of 0.8 mM BMH or BMOE at 23 °C for 1 hr and analyzed by Coomassie staining following SDS-PAGE.
Bacterial two-hybrid assay-
Bacterial two-hybrid assays were performed as previously described [30, 72].
In vitro protein-protein interaction assay–
Association of DnaK with Hsp90Ec-biotin was measured using a pull-down assay. Hsp90Ec wild-type or K354C-biotin (2 or 4 μM) was incubated for 5 min at 23 °C in reaction mixtures (50 μL) containing PD buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 5% glycerol (vol/vol), 0.01% Triton X-100 (vol/vol), 2 mM DTT, 10 mM MgCl2, 2 mM ATP) with DnaK wild-type or mutant (4 μM), L2 (2.3 μM) and CbpA (1 μM). Neutravidin agarose (40 μL 1:1 slurry) (Thermo, Pierce) was then added and incubated 10 min at 30 °C with mixing. The reactions were centrifuged 1 min at 1000 × g and the recovered agarose beads were washed two times with 0.5 mL PD buffer and centrifuged 1 min at 1000 × g. Bound proteins were eluted with buffer containing 2 M NaCl and analyzed by Coomassie blue staining following SDS-PAGE. Interaction of Hsp82 with Ssa1 was measured using a pull-down assay. Hsp82-biotin wild-type (2.5 μM) was incubated for 5 min at 23 °C in reaction mixtures (50 μL) containing PD buffer with Ssa1 wild-type or mutant (8 μM). Neutravidin agarose (40 μL 1:1 slurry) (Thermo, Pierce) was then added and incubated 10 min at 30 °C with mixing. The reactions were diluted with 0.5 mL PD buffer, centrifuged 1 min at 1000 × g and the recovered agarose beads were washed once with 0.5 mL PD buffer. Bound proteins were eluted with buffer containing 2 M NaCl and analyzed by Coomassie blue staining following SDS-PAGE. Interaction of Hsp82 and Ssa1 in the absence and presence of Sti1, Ydj1 and ATP was measured using a pull-down assay as previous described [33]. Protein concentrations utilized are indicated in the figure legends.
Interaction of Ssa1 with Sti1 or Ydj1 was measured using a pull-down assay. Sti1-biotin (2 μM) and Ssa1 wild-type or mutant (4 μM) or Ydj1-biotin (1.2 μM) with Ssa1 wild-type or mutant (2 μM) were incubated for 5 min at 23 °C in reaction mixtures (50 μL) containing PD buffer as indicated. Ydj1-Ssa1 reactions also contained 2 mM ATP and 10 mM MgCl2. Reactions were processed as for DnaK with Hsp90Ec-biotin reactions above.
Where indicated, protein band intensities from replicate gels were quantified using ImageJ (http://imagej.nih.gov/ij). For each lane the biotinylated protein was used for normalization after any non-specific binding protein was subtracted. DnaK mutants and Ssa1 mutants were plotted relative to the corresponding wild-type protein. Ssa1 in the presence of Ydj1 with or without ATP was plotted relative to Ssa1 in the absence of ATP and Ydj1.
Heat-denatured luciferase reactivation –
Luciferase reactivation was performed as previously described [27, 33]. 20 nM heat-denatured luciferase, prepared as described [30], was incubated at 24 °C in reaction mixtures (75 μL) containing 25 mM Hepes, pH 7.5, 50 mM KCl, 0.1 mM EDTA, 2 mM DTT, 10 mM MgCl2, 50 μg/ml bovine serum albumin (BSA), 1 mM ATP, an ATP regenerating system (25 mM creatine phosphate, 6 μg creatine kinase), 1 μM Ssa1 wild-type or mutant and 0.025 μM Ydj1 as indicated. Aliquots were removed at the indicated times and light output was measured using a Tecan Infinite M200Pro in luminescence mode with an integration time of 1000 ms following the injection of luciferin (50 μg/ml). Reactivation was determined compared to a non-denatured luciferase control. Luciferase and luciferin were from Promega.
Supplementary Material
Hsp90 interacts with Hsp70 in bacteria and yeast
Specific crosslinking is used to define the sites of direct Hsp90-Hsp70 interaction
Residues in the Hsp90 M-domain directly contact the Hsp70 J-protein binding site
The J-protein promotes the Hsp90-Hsp70 interaction in the presence of ATP
The Hsp90-Hsp70 regions of direct interaction are conserved in E. coli and yeast
Acknowledgements:
We thank the reviewers for helpful comments and suggestions that significantly improved this manuscript. DNA sequencing was conducted at the CCR Genomics Core at the NIH, National Cancer Institute. This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Abbreviations used:
- Hsp90
90-kDa heat shock protein
- Hsp70
70-kDa heat shock protein
- Hop
Hsp90-Hsp70 organizing protein
- NBD
nucleotide-binding domain
- SBD
substrate-binding domain
- NEF
nucleotide exchange factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests: The authors declare no competing financial interests.
References
- [1].Bardwell JC, Craig EA. Ancient heat shock gene is dispensable. Journal of Bacteriology. 1988;170:2977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lindquist S The heat-shock response. Annu Rev Biochem. 1986;55:1151–91. [DOI] [PubMed] [Google Scholar]
- [3].Schopf FH, Biebl MM, Buchner J. The Hsp90 chaperone machinery. Nature Reviews Molecular Cell Biology. 2017;18:345–60. [DOI] [PubMed] [Google Scholar]
- [4].Radli M, Rudiger SGD. Dancing with the Diva: Hsp90-Client Interactions. J Mol Biol. 2018. [DOI] [PubMed] [Google Scholar]
- [5].Grudniak AM, Pawlak K, Bartosik K, Wolska KI. Physiological consequences of mutations in the htpG heat shock gene of Escherichia coli. Mutat Res. 2013;745–746:1–5. [DOI] [PubMed] [Google Scholar]
- [6].Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol. 2007;189:950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Press MO, Li H, Creanza N, Kramer G, Queitsch C, Sourjik V, et al. Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90. PLoS Genetics. 2013;9:e1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Thomas JG, Baneyx F. Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG In vivo. J Bacteriol. 1998;180:5165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proceedings of the National Academy of Sciences. 2011;108:20136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Johnson JL. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochimica et Biophysica Acta - Molecular Cell Research. 2012;1823:607–13. [DOI] [PubMed] [Google Scholar]
- [11].Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure. 2004;12:1087–97. [DOI] [PubMed] [Google Scholar]
- [12].Mayer MP, Le Breton L. Hsp90: breaking the symmetry. Molecular cell. 2015;58:8–20. [DOI] [PubMed] [Google Scholar]
- [13].Prodromou C Mechanisms of Hsp90 regulation. Biochem J. 2016;473:2439–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Röhl A, Rohrberg J, Buchner J. The chaperone Hsp90: Changing partners for demanding clients. Trends in Biochemical Sciences. 2013;38:253–62. [DOI] [PubMed] [Google Scholar]
- [15].Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell. 2006;127:329–40. [DOI] [PubMed] [Google Scholar]
- [16].Hainzl O, Lapina MC, Buchner J, Richter K. The charged linker region is an important regulator of Hsp90 function. Journal of Biological Chemistry. 2009;284:22559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jahn M, Rehn A, Pelz B, Hellenkamp B, Richter K, Rief M, et al. The charged linker of the molecular chaperone Hsp90 modulates domain contacts and biological function. Proc Natl Acad Sci U S A. 2014;111:17881–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tsutsumi S, Mollapour M, Prodromou C, Lee CT, Panaretou B, Yoshida S, et al. Charged linker sequence modulates eukaryotic heat shock protein 90 (Hsp90) chaperone activity. Proc Natl Acad Sci U S A. 2012;109:2937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huck JD, Que NL, Hong F, Li Z, Gewirth DT. Structural and Functional Analysis of GRP94 in the Closed State Reveals an Essential Role for the Pre-N Domain and a Potential Client-Binding Site. Cell Rep. 2017;20:2800–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Graf C, Lee CT, Eva Meier-Andrejszki L, Nguyen MT, Mayer MP. Differences in conformational dynamics within the Hsp90 chaperone family reveal mechanistic insights. Front Mol Biosci. 2014;1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Southworth DR, Agard DA. Species-Dependent Ensembles of Conserved Conformational States Define the Hsp90 Chaperone ATPase Cycle. Molecular Cell. 2008;32:631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple Conformations of E. coli Hsp90 in Solution: Insights into the Conformational Dynamics of Hsp90. Structure. 2008;16:755–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ali MMU, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Flynn JM, Mishra P, Bolon DN. Mechanistic Asymmetry in Hsp90 Dimers. J Mol Biol. 2015;427:2904–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Retzlaff M, Hagn F, Mitschke L, Hessling M, Gugel F, Kessler H, et al. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol Cell. 2010;37:344–54. [DOI] [PubMed] [Google Scholar]
- [26].Soroka J, Wandinger SK, Mausbacher N, Schreiber T, Richter K, Daub H, et al. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell. 2012;45:517–28. [DOI] [PubMed] [Google Scholar]
- [27].Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, et al. Uncovering a Region of Heat Shock Protein 90 Important for Client Binding in E. coli and Chaperone Function in Yeast. Molecular Cell. 2013;49:464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Karagöz GE, Rüdiger SGD. Hsp90 interaction with clients. Trends in Biochemical Sciences. 2015;40:117–25. [DOI] [PubMed] [Google Scholar]
- [29].Moran Luengo T, Mayer MP, Rudiger SGD. The Hsp70-Hsp90 Chaperone Cascade in Protein Folding. Trends Cell Biol. 2019;29:164–77. [DOI] [PubMed] [Google Scholar]
- [30].Genest O, Hoskins JR, Camberg JL, Doyle SM, Wickner S. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kravats AN, Doyle SM, Hoskins JR, Genest O, Doody E, Wickner S. Interaction of E. coli Hsp90 with DnaK Involves the DnaJ Binding Region of DnaK. Journal of Molecular Biology. 2017;429:858–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kravats AN, Hoskins JR, Reidy M, Johnson JL, Doyle SM, Genest O, et al. Functional and physical interaction between yeast Hsp90 and Hsp70. Proc Natl Acad Sci U S A. 2018;115:E2210–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Moran Luengo T, Kityk R, Mayer MP, Rudiger SGD. Hsp90 Breaks the Deadlock of the Hsp70 Chaperone System. Mol Cell. 2018;70:545–52 e9. [DOI] [PubMed] [Google Scholar]
- [35].Morishima Y, Murphy PJM, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor??hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. Journal of Biological Chemistry. 2000;275:18054–60. [DOI] [PubMed] [Google Scholar]
- [36].Nakamoto, Fujita K, Ohtaki A, Watanabe S, Narumi S, Maruyama T, et al. Physical interaction between bacterial heat shock protein (Hsp) 90 and Hsp70 chaperones mediates their cooperative action to refold denatured proteins. Journal of Biological Chemistry. 2014;289:6110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. Journal of Molecular Biology. 2006;356:802–11. [DOI] [PubMed] [Google Scholar]
- [38].Genest O, Hoskins JR, Kravats AN, Doyle SM, Wickner S. Hsp70 and Hsp90 of E. coli Directly Interact for Collaboration in Protein Remodeling. Journal of Molecular Biology. 2015;427:3877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–51. [DOI] [PubMed] [Google Scholar]
- [40].Mayer MP. Intra-molecular pathways of allosteric control in Hsp70s. Philos Trans R Soc Lond B Biol Sci. 2018;373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mayer MP, Kityk R. Insights into the molecular mechanism of allostery in Hsp70s. Front Mol Biosci. 2015;2:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Acebron SP, Fernandez-Saiz V, Taneva SG, Moro F, Muga A. DnaJ recruits DnaK to protein aggregates. J Biol Chem. 2008;283:1381–90. [DOI] [PubMed] [Google Scholar]
- [43].Ahmad a, Bhattacharya a, McDonald Ra, Cordes M, Ellington B, Bertelsen EB, et al. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proceedings of the National Academy of Sciences. 2011;108:18966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Craig EA, Marszalek J. How Do J-Proteins Get Hsp70 to Do So Many Different Things? Trends Biochem Sci. 2017;42:355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–57. [DOI] [PubMed] [Google Scholar]
- [46].Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature reviews Molecular cell biology. 2010;11:579–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, et al. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A. 1999;96:5452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bracher A, Verghese J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front Mol Biosci. 2015;2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bracher A, Verghese J. GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones. Subcell Biochem. 2015;78:1–33. [DOI] [PubMed] [Google Scholar]
- [50].Harrison C GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones. 2003;8:218–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Genest O, Wickner S, Doyle SM. Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J Biol Chem. 2019;294:2109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mayer MP, Gierasch LM. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J Biol Chem. 2019;294:2085–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. The Biochemical journal. 2007;404:159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. [DOI] [PubMed] [Google Scholar]
- [55].Kityk R, Kopp J, Mayer MP. Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones. Mol Cell. 2018;69:227–37 e4. [DOI] [PubMed] [Google Scholar]
- [56].Hernandez MP, Sullivan WP, Toft DO. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperonecomplex. J Biol Chem. 2002;277:38294–304. [DOI] [PubMed] [Google Scholar]
- [57].Sun M, Kotler JLM, Liu S, Street TO. The ER chaperones BiP and Grp94 selectively associate when BiP is in the ADP conformation. J Biol Chem. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Meyer P, Prodromou C, Liao C, Hu B, Mark Roe S, Vaughan CK, et al. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:511–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pearl LH. Review: The HSP90 molecular chaperone-an enigmatic ATPase. Biopolymers. 2016;105:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Verba KA, Wang RY-R, Arakawa A, Liu Y, Shirouzu M, Yokoyama S, et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 2016;352:1542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Verba KA, Agard DA. How Hsp90 and Cdc37 Lubricate Kinase Molecular Switches. Trends Biochem Sci. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gassler CS, Buchberger A, Laufen T, Mayer MP, Schrder H, Valencia A, et al. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone (Hsp70hheat shock proteinshprotein folding). Biochemistry. 1998;95:15229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. Journal of molecular biology. 1999;289:1131–44. [DOI] [PubMed] [Google Scholar]
- [64].Suh W-C, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. 1998;95:15223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Alvira S, Cuéllar J, Röhl A, Yamamoto S, Itoh H, Alfonso C, et al. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nature communications. 2014;5:5484. [DOI] [PubMed] [Google Scholar]
- [66].Morgner N, Schmidt C, Beilsten-Edmands V, Ebong IO, Patel NA, Clerico EM, et al. Hsp70 Forms Antiparallel Dimers Stabilized by Post-translational Modifications to Position Clients for Transfer to Hsp90. Cell Reports. 2015;11:759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, et al. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci U S A. 2011;108:6915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Skowyra D, Wickner S. The interplay of the GrpE heat shock protein and Mg2+ in RepA monomerization by DnaJ and DnaK. J Biol Chem. 1993;268:25296–301. [PubMed] [Google Scholar]
- [69].Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:1054–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Reidy M, Sharma R, Roberts BL, Masison DC. Human J-protein DnaJB6b Cures a Subset of Saccharomyces cerevisiae Prions and Selectively Blocks Assembly of Structurally Related Amyloids. J Biol Chem. 2016;291:4035–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Motojima-Miyazaki Y, Yoshida M, Motojima F. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochemical and Biophysical Research Communications. 2010;400:241–5. [DOI] [PubMed] [Google Scholar]
- [72].Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.