SUMMARY
Gentsch et al. (2018) recently reported that a common side effect of translation-blocking morpholino antisense oligonucleotides is the induction of a set of innate immune response genes in Xenopus embryos, and that splicing-blocking morpholinos lead to unexpected off-target mis-splicing events. Here we present an analysis of all publicly available Xenopus RNA-seq data in a reexamination of effects of translation-blocking morpholinos on the innate immune response. Our analysis does not support the authors’ general conclusion, which was based on a limited number of RNA-seq datasets. Moreover, the strong induction of an immune response appears to be specific to the tbxt/tbxt2 morpholinos. The more comprehensive study presented here indicates that using morpholinos for targeted gene knockdowns remains of considerable value for the rapid identification of gene function.
Keywords: translational blocking morpholino, innate immune response, tbxt, brachyury, loss of function, reverse genetics, gene knockdown, Xenopus, zebrafish
eTOC BLURB
Gene expression interference by morpholinos has been questioned due to unwanted side-effects, including immune response induction by a limited set of morpholinos. By performing a metaanalysis of available transcriptomic datasets, Paraiso et al. show that induction of an immune response is not a general side-effect of morpholinos during early embryogenesis.
INTRODUCTION
Morpholino antisense oligonucleotides (MO) have been used widely for nearly two decades in both the Xenopus and zebrafish research communities to transiently knockdown the function of targeted genes (Heasman et al., 2000; Nasevicius et al., 2000). The method is relatively inexpensive and quite rapid, as the analysis of morphants can be directly performed in injected F0 embryos. However, use of MOs in zebrafish was suggested to induce unwanted side effects including the induction of cell death in the nervous system and expression of tp53 transcripts derived from an alternative promoter (Robu et al., 2007), whereas no evidence of such issues has been reported in Xenopus or other species. In addition, the appropriateness of MOs as a loss-of-function (LOF) tool has been questioned, because the majority of phenotypes resulting from a subset of MO knockdown experiments in zebrafish were not seen in corresponding genetic LOF mutants (Kok et al., 2015). Others have suggested that these differences could be explained by genetic compensation in LOF mutants (Rossi et al., 2015). The utility of MOs as a genetic tool has been met by opposing views in both Xenopus and zebrafish (Blum et al., 2015; Stainier et al., 2015).
In the January 2018 issue of Developmental Cell, a report using RNA-seq analysis suggested that a side effect of the use of translation-blocking MOs targeting tbxt/brachyury paralogs in Xenopus tropicalis embryos caused induction of a significant number of genes involved in the innate immune response, and that injection of splice-blocking MOs led to off-target splicing defects (Gentsch et al., 2018). This study examined a limited set of published RNA-seq datasets from MO-mediated LOF and concluded that the induction of an innate immune response by translation-blocking MOs is a common side effect. The earliest time point whereby embryonic cells can induce an innate immune response is unclear. Induction of innate immune response related genes tp53, tp53inp1 and c3ar1 by MOs in RNA-seq datasets generated as early neurula stage/stage 14 suggest that relevant immune cells might not be required as migrating myeloid progenitor and hemangioblast progenitor cells only appear at stage 14 and stage 18, respectively (Briggs et al., 2018). This study suggested that this immune response is cell intrinsic and can be activated in all embryonic cells; and that this initiates at least as early as neurula stage. We are interested in the function of maternal effect gene products in early stage embryos and these proteins are synthesized from spliced mRNAs deposited in the egg during oogenesis. Therefore we re-examined whether translation blocking MOs cause induction of innate immune genes, and not whether spliceblocking MOs result in off-target mis-splicing. Because the published genome-wide analysis was based only on a limited number of MO knockdown experiments, we wished to address whether the effects of MOs on an innate immunity response are indeed a common occurrence. Since the previous analysis was restricted to embryos of mid-neurula and later stages, we also wanted to determine whether induction of an innate immune response occurs in the period between the onset of zygotic transcription and neurula stages. The question is fundamentally important, as both the Xenopus and zebrafish research communities have used MOs to uncover the function of many genes. Contrary to Gentsch et al., our analysis of 54 publicly available Xenopus MO knockdown datasets with their corresponding control datasets demonstrates that cohorts of Xenopus innate immune response genes are not commonly activated by translation-blocking MOs, but we did find infrequent activation of a few genes reported. Based on currently available transcriptomic datasets, we suggest that the strong effects observed by Gentsch et al. are confined to the use of tbxt/tbxt2 (formerly known as t/brachyury and t2/brachyury2), and that the use of translation-blocking MOs remains a useful approach to uncovering the biological function of genes during early Xenopus embryogenesis.
RESULTS
Strong induction of tp53, tp53inp1, and c3ar1 genes is confined to the injection of tbxt/tbxt2 MO oligonucleotides
To validate the results by Gentsch et al. (2018) we first searched for all current publicly available X. tropicalis and X. laevis RNA-seq datasets in the NCBI Sequence Read Archive (SRA), the European Nucleotide Archive (ENA), and the DNA Databank of Japan Sequence Read Archive (DRA) for data involving MO knockdown experiments. We found 16 projects comprised of 48 X. laevis and 91 X. tropicalis RNA-seq datasets (Table S1). All 16 projects used MOs except for the Gazdag et al. (2016) datasets, which used alternative stabilized antisense oligonucleotides. Among the 16, 10 projects (Kwon et al., 2014; Chiu et al., 2014; Yasuoka et al., 2014; Marlétaz et al., 2015; Dichmann et al., 2015; Nakamura et al., 2016; Campbell et al., 2016; Noiret et al., 2016; Ding et al., 2017; Gentsch et al., 2018) contained experiments where the GeneTools standard control or experimental MO injected sample could be compared with a non-MO injected control (i.e., uninjected or water injected). We used only the datasets that had these controls for our analysis. The extent of our analysis includes morphant sequencing datasets ranging from stage 9 through stage 36, encompassing those experiments analyzed by Gentsch et al. from stage 14 through stage 36. A majority of the sequencing datasets we analyzed overlapped with stages analyzed by Gentsch et al. during neurula (N = 18), early tailbud (N = 14) and late tailbud (N = 6). We extended the analysis to early embryogenesis by including datasets from blastula (N = 2) and gastrula (N = 14) stages.
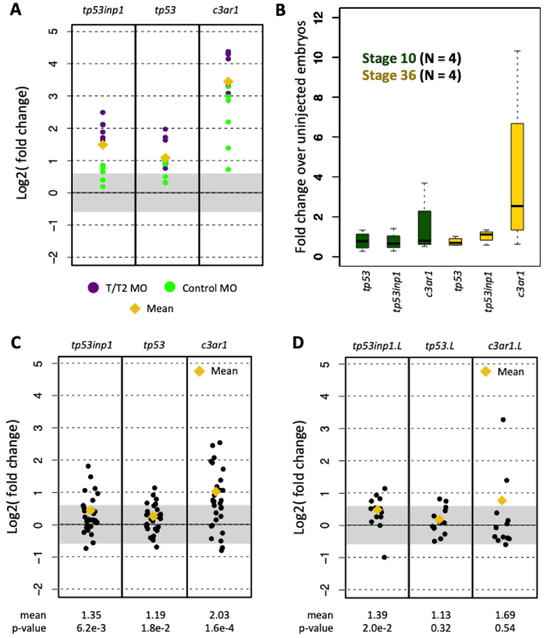
Among the innate immune response genes induced by MOs, the expression of tp53inp1, tp53, and c3ar1 were those most extensively studied in Gentsch et al., and therefore we sought to reproduce their results in our initial analyses by examining the expression of each of these genes in the newly collected MO knockdown data (Figure S1). Gentsch et al. reported that these three genes were induced not only following an injection of a tbxt/tbxt2 quadruple MO cocktail (Figure 1A), but also after control MO injection, although the inductions were weaker than in the tbxt/tbxt2 MO injections. To further validate these results, we performed our own microinjections of the standard control MO into X. tropicalis embryos in biological replicates. Contrary to their findings, RT-qPCR shows that tp53 and tp53inp1 are generally not induced across all biological replicates, regardless of developmental stage (Figure 1B). c3ar1 induction, on the other hand appears consistent with the published findings.
Figure 1. Expression of innate immune response genes in X. tropicalis and X. laevis RNA-seq datasets.
(A) Fold change in induction caused by the tbxt/tbxt2 MOs and the control MOs. (B) Fold change caused by control MO in biological replicates at stages 10 and 36 using RT-qPCR. (C,D) Fold change induction of innate immune response genes in X. tropicalis (C) and X. laevis (D) datasets.
We then examined the expression of these genes among all other available X. tropicalis and X. laevis datasets and found that the inductions of tp53inp1 and tp53 were clearly weaker than in the expression data reported by Gentsch et al., i.e., mean inductions < 1.5-fold. For c3ar1, the mean induction was < 2-fold (Figure 1C, D). Because we aligned the reads to the version 9 X. tropicalis genome assembly using Bowtie2 and RSEM while the published study aligned to the version 7 assembly using STAR, we also examined the possibility that discrepancies between conclusions might have arisen based on the use of different bioinformatics analysis protocols. The fold changes reported in Gentsch et al. were comparable to those in our experiments, with the exception of X. laevis c3ar1.L (the c3ar1 homeologous gene copy found on the long chromosome subset of the allotetraploid X. laevis genome). This difference with c3ar1.L was likely due to its low expression levels in the exosc9 MO experiment, resulting in high variance in fold change quantitation (Table S2). Overall, our experiments and meta-analysis of public RNA-seq datasets suggest that there is no induction of tp53 and tp53inp1, while the induction of c3ar1 is variable.
A cohort of innate immune response genes are not commonly activated by morpholino oligonucleotides
As we find little evidence of an innate immune response by assaying for the expression of tp53inp1, tp53, and c3ar1, we wondered whether we could detect this biological process from the transcriptomic datasets by looking at a larger cohort of genes. Because a list of the innate immune response genes in Xenopus was not available in the Gentsch et al. paper, we performed differential expression analysis using the same software and parameters as used in their study. We compared available RNA-seq datasets from tbxt/tbxt2 morphants and tbxt−/−;tbxt2−/− mutant embryos and identified 1,154 genes that were specifically activated in the morphants compared to their respective controls. Among these genes, Gene Ontology (GO) analysis identified three innate immune response-related terms: ‘innate immune response’ (GO:0045087), ‘regulation of innate immune response’ (GO:0045088), and ‘positive regulation of innate immune response’ (GO:0045089). We combined the genes corresponding to these three GO terms and generated two gene lists (one for X. tropicalis and the other for X. laevis, Table S3), and used these lists for subsequent analyses. The 77 X. tropicalis and 120 X. laevis lists are comprised of genes involved in various subsystems of the innate immune response including complement system genes such as c1r, c1s, c3, c4a, and c9; the signaling molecule nfkb1 (which regulates cytokine production); and interferon regulatory transcription factor genes such as irf1, irf7, and irf9.
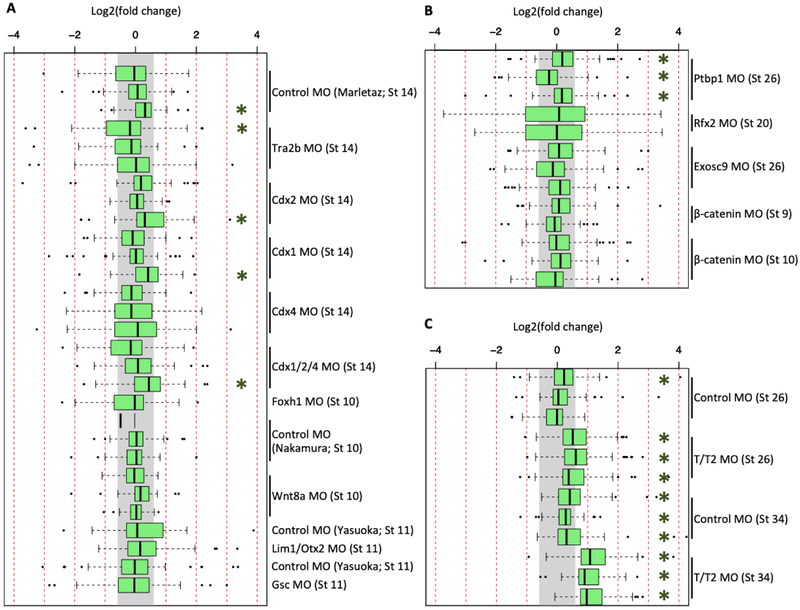
We then determined whether any of the other publicly available RNA-seq datasets involving MO experiments showed activation of the genes from our combined list for Xenopus innate immune response discussed above. These innate immunity genes are generally not activated in either the X. tropicalis or X. laevis datasets when a 1.5-fold expression level difference is used as a cutoff value (Figure 2A,B). While a few datasets showed statistically significant activation, that was not a consistent occurrence among biological replicates. On the contrary, and as expected, the majority of the tbxt/tbxt2 datasets did show an up-regulation of the cohort of innate immunity genes in stage 26 and stage 34 embryos (Figure 2C). The standard control MO injection at stage 34 displayed a weak up-regulation that, while the median was < 1.5 fold, was nevertheless statistically significant (Figure 2C). We conclude that gene cohort analysis using the GO-identified genes does not detect statistically significant induction of innate immune response genes resulting from the injection of MOs.
Figure 2. Expression of GO-identified innate immune response genes in X. tropicalis and X. laevis RNA-seq datasets.
Fold change expression of innate immune response genes across 29 datasets in X. tropicalis (A), 13 datasets in X. laevis (B), and in 12 the tbxt/t2 MO datasets (C). Gray region indicates fold change of < 1.5x. Green asterisk (*) indicates a T-test p-value of < 0.01.
The analysis we performed thus far might not provide a complete view of the induction of innate immune response genes. Large cohort analysis can carry a risk of minimizing the contributions of specific genes in the analysis pipeline. Additionally, the innate immune genes induced in tbxt/tbxt2 morphants might be inductions specific to this MO cocktail, but might not reveal a set of innate immune genes that are induced by other MOs. Therefore, we employed two additional analyses. First, because Robert and Ohta (2009) had provided an annotated list of innate immune response genes conserved between mammals and Xenopus, we worked from that list to identify corresponding gene models in the X. tropicalis v9.0 and X. laevis v9.2 genome assemblies by means of both gene name matching and BLAST alignments. That analysis identified a set of 53 X. tropicalis gene models and 81 X. laevis gene models. The lists included categories such as leukocyte receptors, signaling molecules, cytokines, cytotoxic killing genes, antibacterial peptides, and the complement system (Table S3). When these lists were compared with the set of genes from the previous GO-identified cohort, only 13/53 of X. tropicalis and 15/81 X. laevis gene models overlapped. Therefore, using the Robert and Ohta gene collection expands our analysis beyond the list derived from GO annotations. We then determined whether any of the innate immunity genes from the Robert and Ohta were induced in the available MO-injected datasets. We did not detect significant activation (p-value of < 0.01) of innate immunity genes with the exception of four samples (Figure 3A,B). The literature-identified cohort of innate immune response genes was again seen to be most activated by the tbxt/tbxt2 MO cocktail at stage 34, and less strongly at stage 26 (Figure 3C).
Figure 3. Expression of literature-identified innate immune response genes in X. tropicalis and X. laevis RNA-seq datasets.
Fold change expression of innate immune response genes across 29 datasets in X. tropicalis (A), 13 datasets in X. laevis (B), and in 12 the tbxt/t2 MO datasets (C). Gray region indicates fold change of < 1.5x. Green asterisk (*) indicates a T-test p-value of < 0.01.
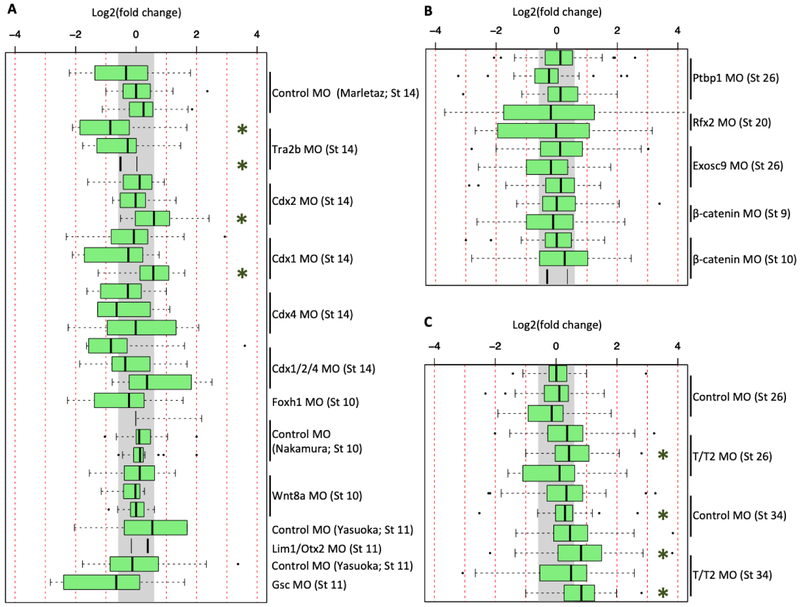
In a second analysis, we examined the list of differentially expressed genes from each of the available datasets to determine whether different subsets of innate immunity genes were significantly induced by different MOs. Analysis was performed on all datasets containing at least two replicates to obtain lists of genes that are differentially expressed in individual MO-injected samples relative to uninjected (or water injected) sibling embryos. We applied the same cutoff criteria described by Gentsch et al. (2018) of 1.5-fold change with an adjusted p-value of < 0.1, to create these gene lists. GO enrichment analysis was then performed on each gene list. GO terms related to innate immune response are significantly enriched in the tbxt/tbxt2 MO dataset at stage 34, but less significantly at stage 26 (Figure S2). The control MO from Gentsch et al. showed some enrichment of GO terms related to innate immune response at stage 34, but not at stage 26. When we performed similar differential expression analyses with all the other available datasets we were unable to detect any enrichment of GO terms related to innate immune response (Figures S3, S4). Taking these observations together with other analyses, we conclude that innate immune response induction is not a common feature of a MO-injected transcriptome. The robust induction of the innate immune seems to be specific to the tbxt/tbxt2 MO. We did not observe the excessive induction of an immune response by control MO prior to stage 34 (Figure 2A, 2C, 3A, 3C).
tbxt/tbxt2 MOs are unusual in inducing a subset of innate immune response genes
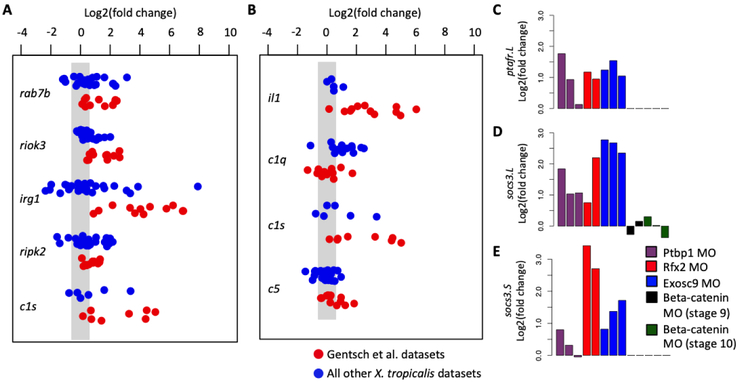
GO term enrichment analysis did not reveal innate immune induction, with the exception of the tbxt/tbxt2 MOs and control MOs (but only at stage 34). Therefore, we next examined whether individual genes other than tp53inp1, tp53, and c3ar1 were consistently activated by MO injection. If MO injections generally induce innate immune responses, then a key set of innate immunity genes should be up-regulated across embryos injected with different MOs. We combined the GO-identified innate immune genes with those identified by Robert and Ohta (Table S3) and searched for genes meeting the following two criteria: a t-test p-value < 0.01, and a fold change up-regulation > 1.5. Among all the X. tropicalis datasets, five genes (rab7b, riok3, irg1, ripk2, and c1s) in the GO-identified cohort and four genes (il1, c1q, c1s, and c5) in the Robert and Ohta cohort were up-regulated (Figure 4A,B). However, if the tbxt/tbxt2 MO datasets are removed from the analysis, none of these genes are upregulated in a statistically significant manner. These results indicate a strong contribution from the tbxt/tbxt2 datasets to the outcome. As this effect is not seen in other MO injection experiments, we suggest that the strong upregulation of select innate immune response genes is not a general phenomenon related to MO injection, but rather is a peculiarity associated with tbxt/tbxt2 datasets (Figure 1).
Figure 4. Specific induction of innate immune response genes.
Fold change expression of genes which were identified to be significantly activated in the X. tropicalis datasets in both the GO-identified (A) and the literature-identified (B) cohort of genes. Fold change expression of X. laevis genes ptafr.L/gene13059 (C), socs3.L/gene3766 (D) and socs3.S/gene50103, which were identified to be significantly activated. We used the criteria p-value < 0.01 and fold change > 1.5 to define significant.
A similar analysis was performed for the five X. laevis MO datasets using the combined gene lists from the GO-identified genes and those identified by Robert and Ohta (Tables S3). Among these, activation of only two genes, ptafr.L (platelet activating factor receptor), socs3.L and socs3.S (suppressor of cytokine signaling 3), were statistically significant (Figure 4C,D). Neither of these were found in the analysis of the X. tropicalis datasets above. At present, the role of Xenopus ptafr.L in innate immunity is not well understood. Much of what is known about the socs3 gene concerns its role during regeneration following wounding wherein socs3 is induced after epithelial (Kuliyev et al., 2005) and retinal ganglion optic nerve (Whitworth et al., 2017) wounding, as well as in spinal cord (Lee-Liu et al., 2014) and limb (Grow et al., 2006) regeneration models. Thus, a common set of genes does not appear to be upregulated between X. tropicalis and X. laevis as part of an immune response.
DISCUSSION
The combined use of MOs and RNA-seq has become a powerful tool in assaying genome-wide functions of developmental genes. Particularly, as we are interested in establishing gene regulatory networks in the early embryo using these technologies, the findings by Gentsch et al. (2018) whereby MOs can induce innate immune response from early embryos, has been a cause of concern in the analysis of transcriptomic datasets. However, when we examined publically available RNA-seq datasets generated from multiple labs including our own, we find no compelling evidence that MOs cause an innate immune response prior to late tailbud stages/stage 34. The only strong effects we identified appear to be particular to the tbxt/tbxt2 quadruple MO experiments. Interestingly, Gentsch et al. have suggested that the induction of an immune response could be dependent on the GC content of MOs as stronger induction of innate immune response genes was detected with MOs having higher GC content. However, we note that our analysis using foxh1 and gsc MOs having relatively high GC content, 60% and 56%, respectively, did not induce immune response genes during gastrula stages (compared to the standard control MO GC content of 32%). Therefore, we believe that MOs still remain a powerful knockdown tool with proper controls in assaying for gene function, especially combined with the use of RNA sequencing methods.
Stage dependence of an immune response
Because later stage samples from the standard control MO and tbxt/tbxt2 MOs by Gentsch et al. showed induction (Figure 2A,C), this finding suggests that there is stage dependence in eliciting an immune response. Consistent with this finding, the standard control MO experiments by Marlétaz et al. (2015), Nakamura et al. (2016), and Yasuoka et al. (2015) which were performed at stage 14 or earlier, did not show a strong induction of innate immune response genes. Most of the available Xenopus datasets we analyzed had been generated on or prior to stage 14 except for three from X. laevis. But when we analyzed these three later-stage X. laevis datasets, one from stage 20 (rfx2 MO) and two from stage 26 (ptbp1 and exosc9 MOs), none of the three showed any statistically significant induction of innate immune response genes (Figure 2C).
The lack of innate immune response in early embryonic stages is consistent with the biology of the early immune system in Xenopus embryos. Functional primitive myeloid cells are reported to be first detected during early tailbud stages (stage 26) (Costa et al., 2008). In addition, recent single cell RNA-seq datasets (Briggs et al., 2018) have shown that the initial appearance of migrating myeloid progenitor and hemangioblast progenitor cells occurs during neurulation, at stages 14 and 18, respectively. At present, it is unclear as to whether these progenitor cells are competent to perform immune-related functions during these stages. We note that our analysis largely considers the effects of MOs on embryos at neurula or earlier stages due to the scarcity of transcriptomic datasets in later stages. Therefore, the induction of an innate immune response and the mechanism thereof during these later stages is still unknown.
Explaining the discrepancy
How then can we explain the discrepancy between our conclusions and that of Gentsch et al.? As shown here, the tbxt/tbxt2 MO cocktail’s effects are an outlier when compared to other MOs. While some MOs can up-regulate a small number of genes related to an innate immune response, a larger scale genomewide effect seen with the tbxt/tbxt2 MOs is not seen in these other experiments.
Why are c3ar1, socs3 and ptafr, genes associated with innate immune responses, up-regulated in some MO experiments? Of these three genes, when examining X. tropicalis RNA-seq datasets, only c3ar1 is found to be up-regulated (after tbxt/tbxt2 datasets are excluded from analysis), albeit in inconsistent manner. Interestingly, c3ar1 expression is upregulated by MOs targeting mesodermally-active transcription factors during gastrula stages (e.g., Cdx1, Cdx2, Cdx4, Gsc), but not regulators of epidermal development during tailbud stages (e.g., Ptbp1, Rfx2, or Exosc9). Perhaps perturbation of TFs in the mesoderm leads to up-regulation of c3ar1, which is broadly expressed in this tissue during gastrula stages (McLin et al., 2008). C3ar1 is a chemotactic receptor that, along with its ligand, C3, plays a role in numerous developmental events where morphogenetic movements require chemotaxis. c3ar1.L is required for radial intercalation during epiboly and cohesive migration of neural crest cells (Carmona- Fontaine et al., 2011; Szabo et al., 2016). c3ar1 (and c3) is also expressed in the developing eye, otic placodes, and in the presumptive liver of the tailbud embryo (McLin et al., 2008)., Thus, disruption of various processes during early development might result in induction of c3ar1, independent of this gene’s role in innate immunity.
When analyzing X. laevis (Figure 4C-E), but not X. tropicalis, datasets, injection of ptbp1, rfx2, or exosc9 MOs leads to up-regulation of both homeologs of socs3, and ptafr.L, but again only inconsistently. During normal development, socs3 is expressed at tailbud stages in neural tube, neural crest cells, the dorsal epidermis, and somites, suggesting a developmental role for this factor in these ecto- and mesodermal derivatives (Yan et al., 2015). This finding is interesting in that ptbpl, rfx2, and exosc9 are all involved in normal epidermal development. Phenotypically, ptbp1 and exosc9 morphants exhibit blister formation underneath the dorsal fin of tailbud embryos and display disruptions of epidermal layer formation (Noiret et al., 2016). The rfx2 gene encodes a critical transcription factor involved in regulation of ciliogenesis in the epidermis (Chung et. al, 2014; Kwon et al., 2014). Thus, induction of socs3.L after ptbpl, rfx2, or exosc9 MO injections is likely the result of perturbations to normal epidermal development. A role for ptafr in early Xenopus development has not been reported. Thus, while we see infrequent activation of a handful of genes by MO injection, these could be due to developmental regulation, rather than an immune response.
How can the differences in expression of numerous genes (including c3ar1)between tbxt/tbxt2 MO knockdowns and mutants in the Gentsch et al. study be explained? Currently, it is difficult to answer this question decisively, however there are a number of possible explanations. First, it is tempting to speculate that a compensation mechanism, as has been proposed in zebrafish to explain reported discrepancies between some morphants and their mutant counterparts (Rossi et al., 2015), might be operational here. Other alternatives might include the efficacy of the tbxt/tbxt2 MOs by the tailbud stages where RNA-seq was performed, or how different genetic backgrounds of the mutant and morphant embryos contributes to the observations reported.
Like Gentsch et al., a recent MO experiment in zebrafish (doi: https://doi.org/10.1101/479188) has noted increased expression of a selected group of interferon-stimulated genes, particularly during segmentation stage (equivalent to Xenopus tailbud stage). Hence, it remains possible that MOs may induce an immune response during later development, and should be used with proper controls. In addition, as available transcriptomic datasets are largely generated for early embryonic development, neither ours nor Gentsch et al.’s findings are conclusive to determine whether an immune response is induced by MOs specifically during later embryonic development. Based on our extensive analysis, we conclude that MOs do not elicit an innate immune response during early Xenopus embryogenesis
STAR METHODS
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ken W.Y. Cho (kwcho@uci.edu).
Experimental model and subject details
Xenopus tropicalis adults were obtained either from NASCO (University of Virginia stock) or raised in the laboratory; and were maintained in agreement with the University of California, Irvine Institutional Animal Care Use Committee (IACUC). X. tropicalis females were injected with 10 units of Chorulon HCG (Merck and Co.) 1-3 nights prior to embryo collection, and 100 units of HCG on the day of embryo collection. Eggs were collected in dishes coated with 0.1% BSA in 1/9× MMR. Sperm suspension in 0.1% BSA in 1/9× MMR was obtained from sacrificed adult X. tropicalis males and the eggs were in vitro fertilized with sperm suspension (Ogino et al., 2006). The embryos were dejellied with 3% cysteine in 1/9x MMR pH 7.8 for 10 minutes after fertilization, and were then ready for manipulation. Embryos were staged using the Nieukwoop-Faber developmental table (Nieuwkoop and Faber, 1958; Khokha et al., 2002).
Method Details
Standard control MO microinjection
The standard control MO (5'-CCTCTTACCTCAGTTACAATTTATA-3') was obtained from GeneTools, LLC. X. tropicalis embryos were injected with 20 ng of the standard control MO at 1-2 cells stage. RNA is harvested from whole embryos at either stage 10 or stage 36 based on the NF developmental table using previously described methods (Chomczynski et al., 1987). RNA samples were reverse transcribed, and gene expression was assayed with qPCR using the Roche Lightcycler 480 II and the Roche SYBR green I master with the default SYBR green protocol. Fold change in gene expression between uninjected and control MO injected was calculated using the ΔΔCp approach.
Identification of cohorts of innate immune response genes using Gene Ontology
The RNA-seq datasets from Gentsch et al. (2018) were obtained from NCBI GEO using the accession number GSE96655. The reads were aligned to the X. tropicalis genome v9.0 (Hellsten et al., 2010; Karimi et al., 2018) using Bowtie2 v2.2.7 (Langmead and Salzberg, 2012) and RSEM v1.2.12 (Li and Dewey, 2011). Differential expression was performed using DEseq2 (Love et al., 2014) using the cutoffs of > 1.5 fold change and < 10% FDR. The control MO and tbxt/tbxt2 MO RNA-seq experiments were compared to their respective sibling uninjected controls; while the tbxt−/−;tbxt2−/− mutant RNA-seq experiments were compared to their respective wild type controls. From this analysis, we identified the list of genes that are upregulated in the control MO or the tbxt/tbxt2 MOs, that are not upregulated in the tbxt−/−tbxt2−/− mutants. Gene Ontology analysis was performed using the Gene Ontology Consortium online tool (Ashburner et al., 2000; The Gene Ontology Consortium, 2017) and obtained three GO terms related to innate immune response. From these three terms, we obtained a list of genes in our differential expression analysis that are associated either one of the three GO terms.
Identification of cohorts of innate immune response genes from Robert and Ohta
Xenopus genes that are associated with innate immunity were identified from Robert and Ohta (2009). We then searched for their corresponding gene models in the X.tropicalis genome v9.0 (Hellsten et al., 2010; Karimi et al., 2018) and the X. laevis genome v9.2 (Session et al., 2016; Karimi et al., 2018).
Meta-analysis of published RNA-seq datasets using MOs
We searched for RNA-seq datasets that involved the use of knockdown technologies in X. tropicalis and X. laevis in the NCBI Sequence Read Archive (SRA), the European Nucleotide Archive (ENA), and the DNA Databank of Japan Sequence Read Archive (DRA). We obtained datasets from 16 projects (Table S1) (Tandon et al., 2013; Gentsch et al., 2013; Kwon et al., 2014; Chiu et al., 2014; Yasuoka et al., 2014; Marlétaz et al., 2015; Dichmann et al., 2015; Wills et al., 2015; Nakamura et al., 2016; Campbell et al., 2016; Gazdag et al., 2016; Gao et al., 2016; Noiret et al., 2016; Ding et al., 2017; Gentsch et al., 2018; Skariah et al., 2018). We aligned the reads to the appropriate the X. tropicalis genome v9.0 (Hellsten et al., 2010; Karimi et al., 2018) or the X. laevis genome v9.2 (Session et al., 2016; Karimi et al., 2018) using Bowtie2 v2.2.7 (Langmead and Salzberg, 2012) and RSEM v1.2.12 (Li and Dewey, 2011) to obtain the expression pattern in transcripts per million (TPM) or normalized read counts. Data figures were generated using the functions boxplot, plot and barplot; and statistical significance of fold changes was tested using the function t.test in R v3.1.0, all using the expression in TPM (R Core Team, 2014). For Gene Ontology analysis, we first performed differential expression using DEseq2 (Love et al., 2014) using the cutoffs of > 1.5 fold change and < 10% FDR. Metascape (Tripathi et al, 2015) was used to perform Gene Ontology analysis and visualize enrichment results, with default parameters whereby significant GO terms were identified with a minimum overlap of 3, p-value > 0.01, and a minimum enrichment of 1.5. Datasets that did not yield any GO terms due to low number of differentially expressed genes were not reported.
Quantification and statistical analysis
Data quantification and statistical analysis are described in the method details.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical Commercial Assays | ||
| mMessage mMachine Sp6 Transcription Kit | Thermo Fisher Scientific | Cat#AM1340 |
| Deposited Data | ||
| Wnt8a morpholino and control RNA-seq | Nakamura et al., 2016 | GEO: GSE72657 |
| Foxh1 morpholino and control RNA-seq | Chiu et al., 2014 | GEO: GSE53654 |
| E2a morpholino and control RNA-seq | Wills et al., 2015 | GEO: GSE56169 |
| Lim/Otx2 morpholino, Gsc morpholinos and control RNA-seq | Yasuoka et al., 2014 | DRA: DRA000516, DRA000517, DRA000518, DRA001093, DRA001094, DRA001095 |
| Mov10 morpholino and control RNA-seq | Skariah et al., 2018 | GEO: GSE86382 |
| Beta-catenin morpholino and control RNA-seq | Ding et al., 2017 | GEO: GSE93195 |
| Tbp/Tlf/Tbp2 morpholino, Gcn5 antisense DNA and control RNA-seq | Gazdag et al., 2015 | GEO: GSE76995 |
| Ascl1 morpholino and control RNA-seq | Gao et al., 2016 | GEO: GSE76915 |
| Rfx2 morpholino and control RNA-seq | Kwon et al., 2014 | GEO: GSE50593 |
| Tcf21 morpholino and control RNA-seq | Tandon et al., 2013 | GEO: GSE45786 |
| Cdx1, Cdx2, Cdx4 and Cdx1/2/4 morpholinos, and control RNA-seq | Marlétaz et al., 2015 | GEO: GSE71006 |
| Tbxt/Tbxt2 morpholino and control RNA-seq | Gentsch et al., 2013 | GEO: GSE48663 |
| Foxn4 morpholino and control RNA-seq | Campbell et al., 2016 | GEO: GSE89271 |
| Ptbp1 morpholino, Exosc9 morpholino and control RNA-seq | Noiret et al., 2016 | GEO: PRJEB8711 |
| Tra2b morpholino and control RNA-seq | Dichmann et al., 2015 | GEO: PRJNA266550 |
| Tbxt/Tbxt2 morpholino and control RNA-seq | Gentsch et al., 2018 | GEO: GSE96655 |
| X. tropicalis genome version 9.0 | Hellsten et al., 2010; Karimi et al., 2018 | RRID:SCR_003280; URL:http://www.xenbase.org/ |
| X. laevis genome v9.2 | Session et al., 2016; Karimi et al., 2018 | RRID:SCR_003280; URL:http://www.xenbase.org/ |
| Experimental Models: Organisms/Strains | ||
| X. tropicalis, out-bred Nigerian | University of Virginia, NASCO | URL:https://www.enasco.com/ |
| Oligonucleotides | ||
|
X. tropicalis smn2 RT primer forward: AAATTCCCAGGACCAAAAGG |
Integrated DNA Technologies | N/A |
|
X. tropicalis smn2 RT primer reverse: ACACGTGTCGCCTACTCTCC |
Integrated DNA Technologies | N/A |
|
X. tropicalis tp53 RT primer forward: CCCTCAACTGAGGATTACGC |
Integrated DNA Technologies | N/A |
|
X. tropicalis tp53 RT primer reverse: CTTGTTGAGGTCGGTGGAGT |
Integrated DNA Technologies | N/A |
|
X. tropicalis tp53inp1 RT primer forward: CCCAGCCCTGATAGAACAGA |
Integrated DNA Technologies | N/A |
|
X. tropicalis tp53inp1 RT primer reverse: TTTCATTCGAGCAGCAAGAG |
Integrated DNA Technologies | N/A |
|
X. tropicalis c3ar1 RT primer forward: CAATATCAGGAATGGGACGAA |
Integrated DNA Technologies | N/A |
|
X. tropicalis c3ar1 RT primer reverse: TTCACTTCCGGTAACGTGCT |
Integrated DNA Technologies | N/A |
| Standard control morpholino: 5’-CCTCTTACCTCAGTTACAATTTATA-3’ |
GeneTools | N/A |
| Software and Algorithms | ||
| RSEM v.1.2.12 | Li and Dewey, 2011 | RRID:SCR_013027; URL:http://deweylab.biostat.wisc.edu/rsem/ |
| Bowtie 2 v2.2.7 | Langmead and Salzberg, 2012 | RRID:SCR_016368; URL:http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| DEseq2 | Love et al., 2014 | RRID:SCR_016533;URL:https://github.com/PF2-pasteur-fr/SARTools |
| R v3.1.0 | R Core Team, 2014 | RRID:SCR_001905; URL:http://www.r-project.org/ |
| Metascape | Tripathi et al., 2015 | RRID:SCR_016620;URL:http://metascape.org/gp/index.html#/main/step1 |
| Gene Ontology | Ashburner et al., 2000; The Gene Ontology Consortium, 2017 | RRID:SCR_002143;URL:http://www.geneontology.org/ |
HIGHLIGHTS.
Analyzed publicly available Xenopus morphant RNA-seq datasets
Innate immune response gene induction is not a general effect related to morpholinos
Strong induction of an immune response is specific to the tbxt/tbxt2 morpholinos
ACKNOWLEDGEMENTS
We thank Xenbase for genomic and community resources (http://www.xenbase.org/, RRID: SCR_003280) in addition to their bioinformatic assistance; and the UC Irvine High Performance Computing Cluster (https://hpc.oit.uci.edu/) for their valuable resources and helpful staff. This research was funded by the following grants awarded K.W.Y.C.: NIH R01GM126395 and NSF 1755214. K.D.P. was a recipient of T32-HD60555.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, De Robertis EM, Wallingford JB, Niehrs C, 2015. Morpholinos: Antisense and Sensibility. Dev Cell 35, 145–9. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM, 2018. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EP, Quigley IK, Kintner C, 2016. Foxn4 promotes gene expression required for the formation of multiple motile cilia. Development 143, 4654–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Theveneau E, Tzekou A, Tada M, Woods M, Page KM, Parsons M, Lambris JD, Mayor R, 2011. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev Cell 21, 1026–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WT, Charney LR, Blitz IL, Fish MB, Li Y, Biesinger J, Xie X, Cho KW, 2014. Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program. Development 141, 4537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N, 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162, 156–9. [DOI] [PubMed] [Google Scholar]
- Chung MI, Kwon T, Tu F, Brooks ER, Gupta R, Meyer M, Baker JC, Marcotte EM, Wallingford JB, 2014. Coordinated genomic control of ciliogenesis and cell movement by RFX2. Elife 3, e01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Soto X, Chen Y, Zorn AM, Amaya E, 2008. spib is required for primitive myeloid development in Xenopus. Blood 112, 2287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichmann DS, Walentek P, Harland RM, 2015. The alternative splicing regulator Tra2b is required for somitogenesis and regulates splicing of an inhibitory Wnt11b isoform. Cell Rep 10, 527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Ploper D, Sosa EA, Colozza G, Moriyama Y, Benitez MD, Zhang K, Merkurjev D, De Robertis EM, 2017. Spemann organizer transcriptome induction by early beta-catenin, Wnt, Nodal, and Siamois signals in Xenopus laevis. Proc Natl Acad Sci U S A 114, E3081–E3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Zhu X, Chen G, Ma X, Zhang Y, Khand AA, Shi H, Gu F, Lin H, Chen Y, Zhang H, He L, Tao Q, 2016. A novel role for Ascl1 in the regulation of mesendoderm formation via HDAC-dependent antagonism of VegT. Development 143, 492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdag E, Jacobi UG, van KI, Weeks DL, Veenstra GJ, 2016. Activation of a T-box-Otx2-Gsc gene network independent of TBP and TBP-related factors. Development 143, 1340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch GE, Owens ND, Martin SR, Piccinelli P, Faial T, Trotter MW, Gilchrist MJ, Smith JC, 2013. In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep 4, 1185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch GE, Spruce T, Monteiro RS, Owens NDL, Martin SR, Smith JC, 2018. Innate Immune Response and Off-Target Mis-splicing Are Common Morpholino-Induced Side Effects in Xenopus. Dev Cell 44, 597–610.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow M, Neff AW, Mescher AL, King MW, 2006. Global analysis of gene expression in Xenopus hindlimbs during stage-dependent complete and incomplete regeneration. Dev Dyn 235, 2667–85. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C, 2000. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol 222, 124–34. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. , 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328, 633–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi K, Fortriede JD, Lotay VS, Burns KA, Wang DZ, Fisher ME, Pells TJ, James-Zorn C, Wang Y, Ponferrada VG, et al. , 2018. Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res 46, D861–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. , 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn 225, 499–510. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van IA, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, et al. , 2015. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 32, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliyev E, Doherty JR, Mead PE, 2005. Expression of Xenopus suppressor of cytokine signaling 3 (xsOcS3) is induced by epithelial wounding. Dev Dyn 233, 1123–30. [DOI] [PubMed] [Google Scholar]
- Kwon T, Chung MI, Gupta R, Baker JC, Wallingford JB, Marcotte EM, 2014. Identifying direct targets of transcription factor Rfx2 that coordinate ciliogenesis and cell movement. Genom Data 2, 192–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Liu D, Moreno M, Almonacid LI, Tapia VS, Munoz R, von MJ, Gaete M, Melo F, Larrain J, 2014. Genome-wide expression profile of the response to spinal cord injury in Xenopus laevis reveals extensive differences between regenerative and non-regenerative stages. Neural Dev 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN, 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. doi: 10.1101/002832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marletaz F, Maeso I, Faas L, Isaacs HV, Holland PW, 2015. Cdx ParaHox genes acquired distinct developmental roles after gene duplication in vertebrate evolution. BMC Biol 13, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLin VA, Hu CH, Shah R, Jamrich M, 2008. Expression of complement components coincides with early patterning and organogenesis in Xenopus laevis. Int J Dev Biol 52, 1123–33. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, de PAE, Veenstra GJ, Hoppler S, 2016. Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment to cis-regulatory modules. Development 143, 1914–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC, 2000. Effective targeted gene ’knockdown’ in zebrafish. Nat Genet 26, 216–20. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J, 1958. Normal Table of Xenopus Laevis (Daudin). Copeia 1958, 65. doi: 10.2307/1439568 [DOI] [Google Scholar]
- Noiret M, Mottier S, Angrand G, Gautier-Courteille C, Lerivray H, Viet J, Paillard L, Mereau A, Hardy S, Audic Y, 2016. Ptbp1 and Exosc9 knockdowns trigger skin stability defects through different pathways. Dev Biol 409, 489–501. [DOI] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM, 2006. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat Protoc 1, 1703–10. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2014. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/ [Google Scholar]
- Robert J, Ohta Y, 2009. Comparative and developmental study of the immune system in Xenopus. Dev Dyn 238, 1249–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC, 2007. p53 activation by knockdown technologies. PLoS Genet 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DY, 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–3. [DOI] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. , 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skariah G, Perry KJ, Drnevich J, Henry JJ, Ceman S, 2018. RNA helicase Mov10 is essential for gastrulation and central nervous system development. Dev Dyn 247, 660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Kontarakis Z, Rossi A, 2015. Making sense of anti-sense data. Dev Cell 32, 7–8. [DOI] [PubMed] [Google Scholar]
- Szabo A, Cobo I, Omara S, McLachlan S, Keller R, Mayor R, 2016. The Molecular Basis of Radial Intercalation during Tissue Spreading in Early Development. Dev Cell 37, 213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM, Conlon FL, 2013. Tcf21 regulates the specification and maturation of proepicardial cells. Development 140, 2409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium, 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. , 2015. Meta- and Orthogonal Integration of Influenza OMICs Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 18, 723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth GB, Misaghi BC, Rosenthal DM, Mills EA, Heinen DJ, Watson AH, Ives CW, Ali SH, Bezold K, Marsh-Armstrong N, Watson FL, 2017. Translational profiling of retinal ganglion cell optic nerve regeneration in Xenopus laevis. Dev Biol 426, 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AE, Baker JC, 2015. E2a is necessary for Smad2/3-dependent transcription and the direct repression of lefty during gastrulation. Dev Cell 32, 345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Ranganathan R, Maynard T, Streit A, Moody SA, 2015. Microarray identification of novel genes downstream of Six1, a critical factor in cranial placode, somite, and kidney development. Dev Dyn 244, 181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuoka Y, Suzuki Y, Takahashi S, Someya H, Sudou N, Haramoto Y, Cho KW, Asashima M, Sugano S, Taira M, 2014. Occupancy of tissue-specific cis-regulatory modules by Otx2 and TLE/Groucho for embryonic head specification. Nat Commun 5, 4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.