Abstract
Precision medicine focuses on DNA abnormalities, but not all tumors have tractable genomic alterations. The WINTHER trial ( NCT01856296) navigated patients to therapy on the basis of fresh biopsy-derived DNA sequencing (arm A; 236 gene panel) or RNA expression (arm B; comparing tumor to normal). The clinical management committee (investigators from five countries) recommended therapies, prioritizing genomic matches; physicians determined the therapy given. Matching scores were calculated post-hoc for each patient, according to drugs received: for DNA, the number of alterations matched divided by the total alteration number; for RNA, expression-matched drug ranks. Overall, 303 patients consented; 107 (35%; 69 in arm A and 38 in arm B) were evaluable for therapy. The median number of previous therapies was three. The most common diagnoses were colon, head and neck, and lung cancers. Among the 107 patients, the rate of stable disease ≥6 months and partial or complete response was 26.2% (arm A: 23.2%; arm B: 31.6% (P=0.37)). The patient proportion with WINTHER versus previous therapy progression-free survival ratio of >1.5 was 22.4%, which did not meet the pre-specified primary end point. Fewer previous therapies, better performance status and higher matching score correlated with longer progression-free survival (all P<0.05, multivariate). Our study shows that genomic and transcriptomic profiling are both useful for improving therapy recommendations and patient outcome, and expands personalized cancer treatment.
There are now numerous approved drugs affecting the molecular pathways frequently aberrant in tumors, and hundreds of novel targeted drugs, including immune-checkpoint modulators, are in clinical development. Not unexpectedly, meta-analyses demonstrate that biomarker-driven trials have better outcomes than trials lacking biomarkers1–3. Some of the most rapid advances have been achieved with investigation of DNA structural abnormalities. Today, there are several models for choosing genome-targeted or immune-targeted therapies according to molecular abnormalities (for example, individual mutations or microsatellite instability/high mutational burden)4. Successes have been reported across diverse malignancies, with a few examples as follows: EGFR (encoding epidermal growth factor receptor) mutation with erlotinib, KIT mutation with imatinib, BRAF mutation with vemurafenib and ALK translocation with crizotinib5–8.
Unfortunately, not all patients’ tumors have pharmacologically tractable DNA alterations. Thus, extending the application of precision medicine requires a deeper understanding of cancer biology. There is a need to explore oncogenic mechanisms beyond the identification of genomic driver aberrations and to incorporate new methodologies, such as those interrogating gene expression. Hence, we initiated an international trial— WINTHER—that prospectively navigated patients to therapy according to either DNA-guided next-generation sequencing (NGS) or transcriptional analysis that specifically compared tumor to matched normal tissue9. Grounded in the precision medicine knowledge at the trial start, the protocol prioritized genomic (DNA) matches, and RNA-guided therapy was exploratory. The design and primary end point centered on the Von Hoff model that uses the patient as their own control—comparing progression-free survival (PFS) on the trial (PFS2) to the PFS recorded on the therapy administered immediately prior to enrollment (PFS1)10. As integration of transcriptomic investigation in the clinical setting is new, other trial objectives included evaluating trial data to generate important methodological evolution and enhance predictive performance for future investigations, as well as determining clinical benefit (response, PFS and survival) and examining these factors in each arm, as guided by DNA and RNA information, respectively.
The WINTHER protocol was conducted under the auspices of the Worldwide Innovative Network (WIN) for personalized cancer medicine (WIN Consortium) and included investigators from five countries in North America, Europe and the Middle East.
From April 2013 to December 2015, 303 patients consented; evaluable patients included 107 participants (35%) who received treatment consistent with one or more of the recommendations identified by the clinical management committee (CMC) (69 patients (22.7% of consented patients; 64.5% of treated patients) on arm A (DNA guided) and 38 patients (12.5% of consented patients; 35.5% of treated patients) on arm B (RNA guided)) (Extended Data Fig. 1). The most common reasons for patient attrition included poor-quality biopsies and health deterioration or death prior to treatment initiation.
Median age was 57 yr; this did not differ between arm A and arm B. The most common diagnoses were colon cancers (N = 34 patients; 32% of the 107 treated patients) followed by head and neck cancers (N = 21; 20%) and lung cancers (N = 21; 20%). Sixty-two patients were men (58%). The median number of previous therapies was 3 (range = 1–12); 28 patients (26.2%) had ≥5 previous therapies. Eastern Cooperative Oncology Group (ECOG) performance status was good (0 or 1) for all 107 patients at the time of consent. The patients were accrued at four centers located in four countries: Spain, Israel, France and Canada (Supplementary Table 1); the two US sites did not accrue patients, mainly due to regulatory delays9.
Overall, 253 patients (83.4% of consented patients) had dual tumor and normal biopsies. The main sites of tumor biopsy were the liver, lung, lymph nodes, gastrointestinal track and head and neck. Supplementary Table 2 describes the organ-matched normal tissues for each type of tumor.
The most common reasons that patients were not biopsied included difficulty accessing the lesion, patient weakened or succumbed and consent withdrawal. Of these 253 patients, 158 (52% of consented patients; 62.5% of patients with dual biopsies) had adequate biopsy quality for both genomic and transcriptomic analyses (Extended Data Fig. 1).
Complications following biopsy that qualified as serious adverse events occurred in only 1.2% of patients and were related only to tumor biopsies; two participants had a pneumothorax that required chest tubes and 24 h of hospitalization for observation, and one individual had a convulsion (unrelated to biopsy) for which the patient was also hospitalized for 24 h.
The efficacy analysis was based on 107 patients who received treatment consistent with CMC recommendations (Extended Data Fig. 1). Overall, 63 patients received single agents and 44 patients received ≥2 drugs. Of the 159 drugs given, 115 were approved off-label, whereas 22 were approved on-label and 22 were investigational. Gene product-targeted agents were often given, but the drug classes administered included immunotherapy, chemotherapy and hormonal agents (Supplementary Tables 3 and 4).
The overall rate (95% confidence interval (CI)) of stable disease (SD) ≥ 6 months/partial or complete responses (SD ≥ 6 months/PR/CR) was 26.2% (18.1–35.6%) (PR/CR: 11.2% (5.9–18.8%) (arm A: 23.2% (13.9–34.9%) (PR/CR: 13%); arm B: 31.6% (17.5–48.7%) (PR/CR: 7.9%)) (Table 1). There were no significant differences between the two arms with regard to the PR/CR or SD ≥6 months/PR/CR rates (P = 0.37).
Table 1 |.
Outcome data including SD ≥6 months and PR/CR as well as PFS2/PFS1 in the WINTHER trial
| Arm A (DNA)a | Arm B (RNA)a | All patients | |
|---|---|---|---|
| SD ≥6 months/PR/CRb | 16 (23.2%) | 12 (31.6%) | 28 (26.2%) |
| Response: CR/PR | 9 (13.0%) | 3 (7.9%) | 12 (11.2%) |
| SD ≥6 months | 7 (10.1%) | 9 (23.7%) | 16 (15.0%) |
| PD or SD <6 monthsa | 53 (76.8%) | 26 (68.4%) | 79 (73.8%) |
| Total | 69 (100%) | 38 (100%) | 107 (100%) |
| Frequency (N (%))c | |||
| PFS2/PFS1 of >1.5 | |||
| Yesd | 14 (20.3%) | 10 (26.3%) | 24 (22.4%) |
| No | 55 (79.7%) | 28 (73.7%) | 83 (77.6%) |
| Total | 69 (100%) | 38 (100%) | 107 (100%) |
Patients who died or whose tumors clinically progressed before the first evaluation were considered as having progressive disease (PD).
The percentages of patients with SD ≥6 months/PR/CR were not significantly different between the two arms (two-sided Fisher’s exact test, P=0.37).
Of the 17 enrolled patients who were not evaluable for WINTHER therapy because the CMC recommendations were not followed (see Fig. 1), the PFS2/PFS1 ratio of >1.5 was 18%; the PFS2/PFS1 ratio of >1.3 was also 18%.
The percentages of patients with a PFS2/PFS1 ratio of >1.5 were not significantly different between the two arms (two-sided Fisher’s exact test, P = 0.48).
Using the patient as their own control, as previously described10, we calculated the patient proportion for whom duration of PFS on WINTHER (designated PFS2) versus duration of PFS on the patients’ last previous therapy (PFS1) was greater than 1.5 (PFS2/PFS1 >1.5) (Table 1). For all treated patients, patient proportion with PFS2/PFS1 >1.5 was 22.4% (14.9–31.5%) (arm A: 20.3% (11.6–31.7%); arm B: 26.3% (13.4–43.1%)). This was the primary study end point and it was not achieved (P = 1 and P=0.81 (arms A and B, respectively)). There were no statistically significant differences between the two arms in the patient proportion with PFS2/PFS1 >1.5 (Fisher’s exact test, P=0.48). The patient proportion with a PFS2/PFS1 ratio of >1.3 was 25%.
Beyond the 107 evaluable patients, there were an additional 17 enrolled patients who were not evaluable because CMC recommendations were not followed (Extended Data Fig. 1); the patient proportion with a PFS2/PFS1 ratio of >1.5 was 18%, and the PFS2/PFS1 ratio of >1.3 was also 18%.
An exploratory matching score (based on drugs administered) was assigned for all 107 evaluable patients (see Methods for details). For DNA (arm A), the matching score was derived from the number of alterations matched to a drug (or drugs) received divided by the total number of alterations for any given patient11–14; for RNA, the matching score was assigned by adding the reciprocal of the ranks of each matched drug received by the patient according to the WINTHER algorithm (or simply the reciprocal of the rank if only one drug was matched)15.
A higher matching score reflected a greater degree of matching between the administered treatment and the alterations. (Matching scores were determined post-hoc by an investigator (R.K.) and the statistician (J.J.L.) who were both blinded to the treatment outcome at the time of matching score determination.) The recursive partitioning and regression trees (RPART) method identified that the optimal cut-off points for arm A and arm B were matching scores of 0.25 and 0.30, respectively, for predicting PFS. As DNA and RNA matching scores are derived differently, RPART was performed separately for arm A and arm B to identify the patients with high and low score. Once the status was determined separately, a post-hoc combined analysis of low and high matching score was performed for all 107 patients.
Factors associated with PFS were examined by the univariate and multivariate Cox model (Table 2). The median PFS was 2.01 months in all patients, and 1.94 and 2.43 months in arm A and arm B, respectively.
Table 2 |.
Multivariate analysis for arm A (DNA), arm B (RNA) and all patients (N = 107) with factors associated with PFS and OSa
| Covariate | Univariate | Multivariateb | ||
|---|---|---|---|---|
| HR (PFS) (95% CI) | P valuec | HR (PFS) (95% CI)e | P value | |
| Arm A (DNA): factors associated with PFS | ||||
| Age 60yr (N = 33) versus ≤60 yr (N = 36) | 1.089 (0.669–1.772) | 0.73 | - | NS |
| Sex (women (N = 33) versus men (N = 36)) | 0.925 (0.569–1.502) | 0.75 | - | NS |
| Diagnosis: lung (N = 17) versus other (N = 52) | 0.515 (0.29–0.913) | 0.023 | - | NS |
| ECOG PS = 0 (N = 21) versus PS > 0(N = 48) | 0.358 (0.206–0.625) | 0.0003 | 0.367 (0.21–0.642) | 0.0004 |
| Previous treatments ≤ 2 (N = 23) versus >2 (N = 46) | 0.628 (0.373–1.056) | 0.0795 | - | NS |
| Matching score high versus lowe of ≥0.25 (N = 50) versus <0.25 (N = 19) | 0.482 (0.277–0.836) | 0.0095 | 0.508 (0.291–0.884) | 0.0167 |
| Arm B (RNA): factors associated with PFS | ||||
| Age >60 yr (N = 17) versus ≤60 yr (N = 21) | 0.492 (0.249–0.971) | 0.04 | - | NS |
| Sex (women (N = 12) versus men (N = 26)) | 0.429 (0.2–0.92) | 0.03 | - | NS |
| Diagnosis: lung (N = 4) versus other (N = 34) | 1.216 (0.419–3.525) | 0.7 | - | NS |
| ECOG PS = 0 (N = 15) versus PS > 0 (N = 23) | 0.747 (0.374–1.495) | 0.4 | - | NS |
| Previous treatments ≤ 2 (N = 11) versus >2 (N = 27) | 0.322 (0.136–0.76) | 0.0097 | 0.322 (0.136–0.76) | 0.0097 |
| Matching score high (≥0.3) (N = 30) versus low (<0.3) (N = 8)d | 0.561 (0.245–1.283) | 0.17 | - | NS |
| Combined arms A and B (PFS) | ||||
| Age >60 yr (N = 50) versus ≤60 yr (N = 57) | 0.824 (0.556–1.221) | 0.3 | - | NS |
| Sex (women (N = 45) versus men (N = 62)) | 0.725 (0.487–1.081) | 0.1 | - | NS |
| Diagnosis: lung (N = 21) versus other (N = 86) | 0.647 (0.395–1.059) | 0.08 | - | NS |
| ECOG PS = 0 (N = 36) versus PS > 0 (N = 71) | 0.485 (0.316–0.745) | 0.009 | 0.585 (0.373–0.917) | 0.02 |
| Previous treatments ≤ 2 (N = 34) versus >2 (N = 73) | 0.517 (0.334–0.801) | 0.003 | 0.629 (0.398–0.996) | 0.048 |
| Matching score high (N = 80) versus low (N = 27)d | 0.487 (0.308–0.771) | 0.002 | 0.519 (0.328–0.822) | 0.005 |
| Combined arms A and B (OS) | ||||
| Age >60 yr (N = 50) versus ≤60 yr (N = 57) | 0.981 (0.637–1.510) | - | - | NS |
| Sex (women (N = 45) versus men (N = 62)) | 0.865 (0.559–1.336) | 0.5125 | - | NS |
| Diagnosis: lung (N = 21) versus other (N = 86) | 0.585 (0.330–1.037) | 0.0665 | - | NS |
| ECOG PS = 0 (N = 36) versus PS > 0 (N = 71) | 0.297 (0.177–0.496) | <0.0001 | 0.31 (0.182–0.511) | <0.0001 |
| Previous treatments ≤ 2 (N = 34) versus >2 (N = 73) | 0.436 (0.264–0.719) | 0.0012 | - | NS |
| Matching score high (N = 80) versus low (N = 27)d | 0.536 (0.330–0.870) | 0.0116 | 0.581 (0.357–0.944) | 0.0285 |
Median (mean ± s.d.) in days between scans was 52.0 (60.6 ± 5.4) (excluding patients whose PFS was determined because of clinical progression prior to the first scan): arm A, 52.5 (63.4 ± 7.9); arm B, 51.0 (56.2 ±5.9); two-sided Wilcoxon test for comparing the timing of scans between arm A (N=69) and arm B (N = 38), P=0.63, no difference.
Univariate variables with P<0.05 were chosen for multivariate analysis.
P values were determined by a two-sided Wald test (note that for the Kaplan-Meier curves, see Fig. 1, P values were determined by a log-rank test)).
Cut-off points for matching scores were chosen by the recursive partition program rpart.
HR, hazard ratio provided for variables significant in multivariate analysis; NS, not significant.
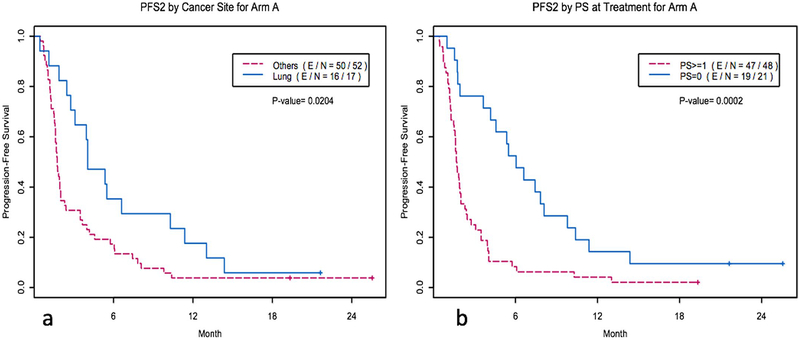
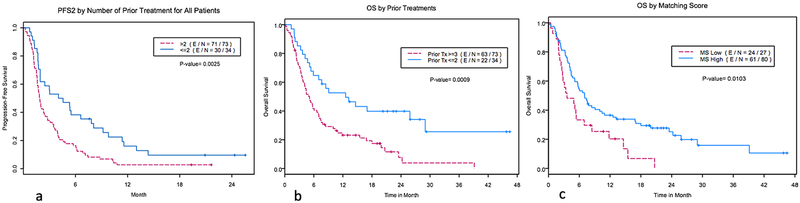
In the univariate analysis, for arm A, the disease site of the lung (versus others), an ECOG performance status of 0 (versus >0) and a higher matching score (versus lower matching score) correlated with longer PFS (Cox model, P = 0.023, 0.0003 and 0.0095, respectively) (Table 2, Fig. 1a and Extended Data Fig. 2). The number of previous treatments of ≤2 (versus >2) was marginally significant (P = 0.08).
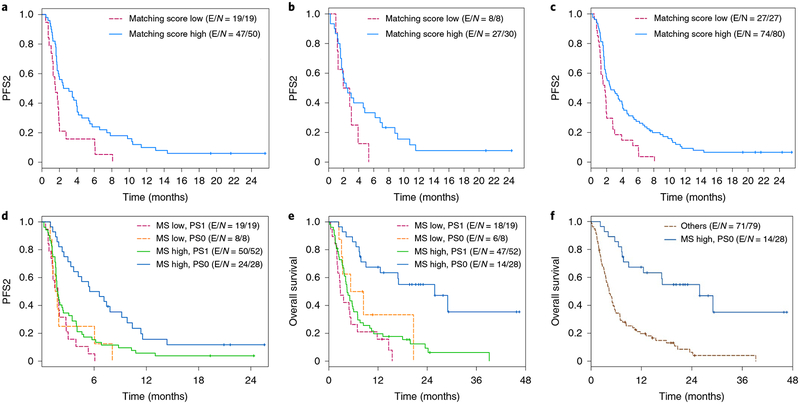
Fig. 1 |. Kaplan-Meier curves of PFS and OS by matching score and performance status.
a, Kaplan-Meier curves of PFS for arm A by matching score (high: matching score > 0.25 chosen from the recursive partitioning program rpart) for high (N = 50) versus low (N = 19) groups. HR (95% CI) = 0.482 (0.277–0.836); P = 0.008 by two-sided log-rank test. E, event; N, number. b, Kaplan-Meier curves of PFS for arm B by matching score (high: matching score >0.3 chosen from the recursive partitioning program rpart) for high (N = 30) versus low (N = 8) groups. HR (95% CI) = 0.561 (0.245–1.283); P = 0.16 by two-sided log-rank test. c, Kaplan-Meier curves of PFS for all patients by matching score for high (N = 80) versus low (N = 27) groups. HR (95%CI) = 0.487 (0.308–0.771); P = 0.002 by two-sided log-rank test. d, Kaplan-Meier curves of PFS for all patients, showing the top predictive factors issued from the multivariate analysis: the performance status (PS) at consent and the matching score (MS). The four groups are MS low, PS1 (N = 19); MS low, PS0 (N = 8); MS high, PS1 (N = 52); MS high, PS0 (N = 28) (P = 0.0002 by two-sided log-rank test). e, Kaplan-Meier curves of OS for all patients, showing the top predictive factors issued from the multivariate analysis: the performance status at consent and the matching score. The four groups are the same as in panel d; P < 0.0001 by two-sided log-rank test. f, Kaplan-Meier curves of OS for all patients combining the two predictive factors issued from the multivariate analysis: the performance status at consent and the matching score. The two groups are MS high and PS0 (N = 28) and others (MS low and PS> 0) (N = 79); P < 0.0001 by two-sided log-rank test.
For arm B, age >60 yr (versus ≤60 yr), female (versus male) and ≤2 previous treatments were associated with longer PFS (Cox model, P = 0.04, 0.03 and 0.0097, respectively). A higher matching score was not associated with longer PFS (P = 0.17) (Table 2, Fig. 1b and Extended Data Fig. 3).
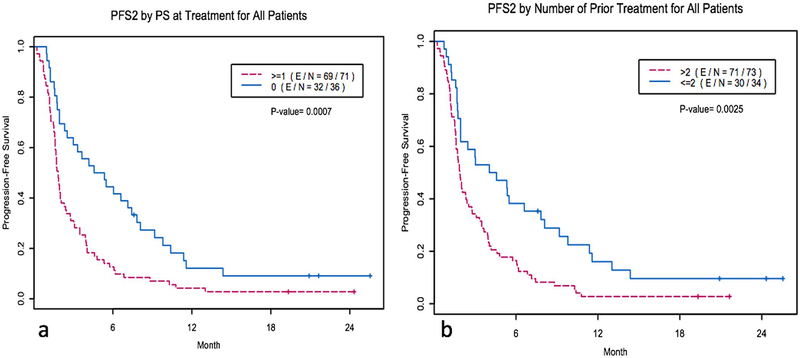
For all patients, an ECOG performance status of 0, the number of previous treatments ≤2 and a higher matching score correlated with longer PFS (Cox model, P = 0.009, 0.003 and 0.002, respectively) (Table 2, Fig. 1c and Extended Data Fig. 4). Trends of wider separation between the high versus low matching score groups were observed as time progressed. At the PFS = 0.25, the PFS times were 5.75 versus 2.01 months for arm A, 7.13 versus 3.47 months for arm B and 6.72 versus 2.79 months for all patients in high versus low matching scores groups, respectively (Fig. 1).
In the multivariate analysis, for arm A, an ECOG performance status of 0 and a higher matching score correlated with a longer PFS (Cox model, P=0.0004 and 0.02, respectively). For arm B, only previous treatment ≤2 was associated with a longer PFS (Cox model, P = 0.01). For all patients, an ECOG performance status of 0, ≤2 previous treatments and a higher matching score correlated with a longer PFS (Cox model, P=0.02, 0.048 and 0.005, respectively) (Table 2).
By combining ECOG performance status and matching score, patients with a performance status of 0 and a high matching score had the longest PFS compared with all other subgroups. Patients with a performance status of >0 and/or a low matching score had a substantially shorter PFS (Fig. 1d). The median PFS for patients with a high matching score and a performance status of 0 was 6.1 months; the median PFS for patients with a low matching score or a performance status of >0 was 1.8 months (P = 0.0002).
The median overall survival (OS) was 5.9 months in 107 patients; 5.1 and 7.4 months in arm A and arm B, respectively. In the univariate analysis, an ECOG performance status of 0 (versus >0), the number of previous treatments ≤2 (versus >2) and a higher matching score (versus a lower matching score) were associated with a longer OS (Cox model, P < 0.0001, P=0.001 and P=0.012, respectively) (Table 2 and Extended Data Fig. 5). From the figures (Extended Data Fig. 5c), significant separation between the high versus low matching score groups were observed (P = 0.01); the median OS times were 7.3 versus 3.6 months in all patients for high versus low matching score groups, respectively. With time, the OS separation increased; at the OS = 0.25, the OS times were 24.2 versus 11.8 months for all patients (high versus low matching score) (Extended Data Fig. 5c).
In the multivariate analysis, an ECOG performance status of 0 (P < 0.0001) and a higher matching score (P = 0.029) correlated with a longer OS (Table 2). Combining the ECOG performance status and matching score, patients with a performance status of 0 and a high matching score had the longest OS compared with all other subgroups. The OS in patients with either a performance status of >0 and/or a low matching score was substantially shorter (Fig. 1e).The median OS for patients with a high matching score and a performance status of 0 was 25.8 versus 4.5 months for a low matching score and a performance status of >0 (P < 0.0001) (Fig. 1f).
We also examined the use of tumor versus normal tissue as a comparator for transcriptomics. Extended Data Fig. 6 shows that the level of basal gene expression is highly variable between individuals. Hence, to eliminate host gene expression variability as a confounder, the transcriptomic expression level was derived by comparing the tumor tissue to its histological normal counterpart.
High-grade toxicity data were captured and curated for 84 of the 107 evaluable patients; the French Ethics Committee for Gustave Roussy considered this trial a navigation trial and hence toxicities could not be captured. Overall, 0 of 21 patients (0%) with low versus 11 of 63 patients (17%) with high matching scores (P = 0.06) had grade 3 toxicity or higher, at least possibly related to a drug (total = 11 of 84 patients (13%)), during the first month of therapy. Most of the patients received one or two drugs. The most common toxicities were diarrhea, rash and fatigue or weakness. Toxicity was reversible when the drug was held or the dose was reduced.
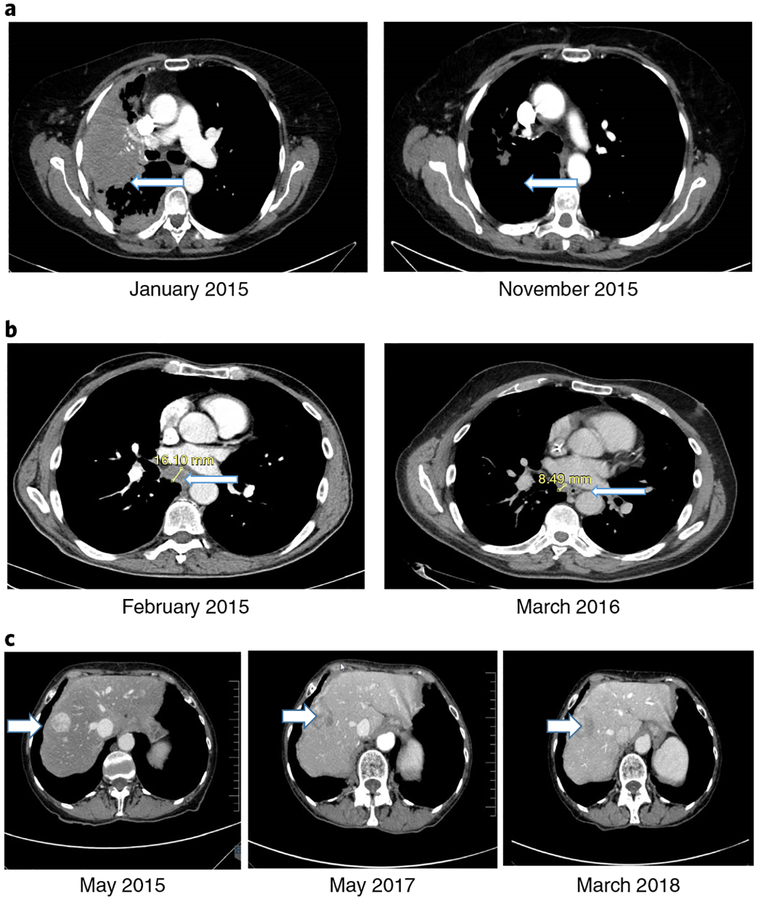
Figure 2 shows examples of patients with exceptional responses to gene-targeted therapy16 or immunotherapy17 according to either DNA NGS matches (Fig. 2a,b) (CR (PFS of 13 months) and PR (PFS ongoing at >36 months), respectively) or to gene-targeted therapy according to an RNA match (Fig. 2c) (SD ongoing at >34 months). None of these patients experienced significant side-effects.
Fig. 2 |. Examples of exceptional responses with CT scans.
a, A 72-year-old woman with non-small-cell lung adenocarcinoma progressing after erlotinib and pemetrexed enrolled on the WINTHER trial (November 2014). NGS matches found a EGFR T790M mutation (which leads to a resistance to the drugs available at the time). The WINTHER CMC recommended the EGFR small-molecule inhibitor afatinib with the antibody cetuximab, according to data showing responses with this combination16. (Osimertinib (an EGFR inhibitor targeting EGFR T790M) was not yet approved.) Outcome refers to a CR (PFS of 13 months). The left panel is pre-treatment and shows a large lung mass (arrow). The right panel shows tumor resolution (arrow). b, A 68-year-old man with progressive metastatic colorectal cancer after Xelox (capecitabine and oxaliplatin) and FOLFIRI (folinic acid, 5-fluoruracil, irinotecan)-cetuximab. NGS matches found a MSH6 mutation (a mismatch repair gene alteration causing microsatellite instability). The WINTHER CMC recommended pembrolizumab, according to data newly emerging at the time (and later validated17) regarding checkpoint inhibitor efficacy in a mismatch repair gene defect setting. Pembrolizumab was initiated in March 2015. Outcome refers to a PR (PFS of >36 months). The left panel shows baseline mediastinal adenopathy (arrow). The right panel shows mediastinal adenopathy regression (arrow). c, A 69-year-old woman with a well-differentiated, neuroendocrine, small gut tumor and peritoneal metastasis underwent debulking surgery and received lanreotide (a long-acting somatostatin analog) for residual disease (April 2011 to June 2012). She then developed bowel obstruction (due to peritoneal progression), which required surgery. The somatostatin analog continued until April 2014, when progression to the liver occurred. Axitinib (on a clinical trial) was given for 10 months before progression. In April 2015, she enrolled on the WINTHER trial. NGS found no DNA alterations; RNA matches revealed AKT2 and AKT3 overexpression. The WINTHER CMC recommended an mTOR inhibitor. Everolimus was started (May 2015). Outcome refers to prolonged disease stabilization (PFS of >34 months). (Everolimus was later approved for this indication in 2016.) The left panel shows baseline liver metastases (arrow). The middle panel shows stable liver metastases (arrow) at 1 yr. The right panel shows ongoing stable liver metastases (arrow) at >34 months. The examples of the exceptional responses shown in a-c can be found in Supplementary Table 4 (ID156, ID183 and ID203, respectively).
The underlying concept for the WINTHER study consisted of considering patient therapy options at an individual level, depending on the characteristics of each person’s tumor, and not at the aggregate level on global results obtained from large groups. Although the trial did not meet its primary end point related to a PFS2/PFS1 ratio of >1.5, there were several novel paradigm shifts in the WINTHER trial that merit highlighting: (1) deployment of a transcriptomic arm (arm B) to navigate patients with various solid tumors prospectively to a large panel of therapies, in addition to a genomic arm (arm A); (2) using a 236 NGS gene panel, rather than less comprehensive testing, for the genomic arm; (3) timely CMC teleconference discussions that included investigators from centers in France, Spain, Israel, Canada and the United States; and (4) navigating patients to a clinical trial or to on-label-approved or off-label-approved drugs. Importantly, the WINTHER trial incorporated transcriptomics to expand personalized treatment to a larger fraction of patients. Given the knowledge at the time of trial planning, the protocol gave priority to the genomic matches. RNA-guided therapies were selected only when there were no actionable DNA alterations or when the DNA-selected drugs were unavailable.
To translate RNA investigations into therapeutic decisions for the physicians, a novel strategy and algorithm were developed, based on three pillars: (1) dual biopsies, enabling differential gene expression and microRNA profile assessment in tumor versus matched normal organ tissues so as to discard most genetic variability between indi-viduals15–18 (Extended Data Fig. 6); (2) a knowledge database that included standard-of-care and investigational drug information; and (3) an innovative algorithm that enabled linking the patients’ RNA information to the knowledge database (see Methods) and provided a ranked drug list for each patient.
The primary study objective was to attain a PFS2/PFS1 ratio of >1.5 in 50% of patients in arm A and in 40% of patients in arm B (normally, the PFS would be expected to decrease with each therapy line). At the time of study development, it was felt that this objective required 200 treated patients (60 in arm A and 140 in arm B) to achieve appropriate statistical power. Secondary objectives included improving algorithm performance and increasing knowledge in handling biopsies and DNA or RNA preparation, as well as examining response, PFS and survival.
Overall, 107 of the 303 enrolled patients (35%) received treatment consistent with CMC recommendations; 69 of the 107 patients (64.5%) on arm A and 38 out the 107 (35.5%) on arm B. During trial planning (around 2012), it was anticipated that for every 60 patients enrolled in arm A, 140 would enroll in arm B, according to the limited number of druggable DNA alterations known at the time. However, the reverse occurred, with more patients enrolled in arm A as the number of actionable DNA alterations expanded rapidly during the following years. Even so, keeping in mind that the protocol was designed to preferentially steer patients to therapy on the basis of genomics if a tractable alteration was found, it seems that transcriptomics added substantially to the number of patients treated. Indeed, without transcriptomics, about 23% (rather than 35%) of consented patients would have been treated. Most previous studies of genome-guided therapy have been hindered by low matching rates, often in the range of ~10–25% (refs.11,12,14,19–22). Our study suggests that incorporating transcriptomic analysis could attenuate this problem to some extent. Of interest in this regard, the benefit rate (SD ≥ 6 months/PR/CR) was 26.2% and was numerically (but not statistically) higher in arm B than in arm A (31.6% versus 23.2%) (Table 1).
There were several challenges that accounted for patient attrition (Extended Data Fig. 1): biopsy access or quality difficulties and early deterioration or death. Patients’ acceptance of the dual biopsies was high and safe procurement of normal tissues was possible in most cases. Much of the attrition was related to histological or RNA quality obtained from fresh-frozen biopsies, a lot of which were resolved as individual centers gained experience. However, the use of formalin-fixed paraffin-embedded tissue in future trials will need to be evaluated. Attrition due to patient health decline was also an important factor and may have been due to the fact that these patients were very heavily pretreated; the median number of previous therapies was three, and over one-quarter of patients had five or more previous therapy lines. Thus, future studies might consider enrolling participants earlier in the disease course.
For all treated patients, the patient proportion with the WINTHER versus previous therapy PFS ratio (PFS2/PFS1) of >1.5 was 22.4% (20.3% in arm A; 26.3% in arm B (P = 0.48)). The PFS2/PFS1 ratio of >1.3 was 25%. Thus, the primary end point was not met and we proceeded with further investigations.
In the WINTHER study, the physicians chose the therapy given. Consequently, we performed a post-hoc calculation (while blinded to the outcome) of a matching score, which indicated the degree to which the actual drugs administered matched the complex DNA or RNA alterations in each patient’s cancer (see Methods)11,12,23–28. Higher matching scores reflected a greater degree of matching. Certain factors were independently associated with both longer PFS and OS: a better ECOG performance status (ECOG of 0), the number of previous treatments ≤2 and a higher matching score. In the multivariate analysis, patients with a performance status of 0 and a high matching score had the longest PFS and the best OS compared to all other subgroups. Indeed, the median PFS for patients with a high matching score and a performance status of 0 was 6.1 versus 1.8 months (compared to a low matching score and a performance status of >0) (P=0.0002). The median OS for patients with a high matching score and a performance status of 0 was 25.8 versus 4.5 months (compared to a low matching score and a performance status of >0) (P < 0.0001) (Fig. 1f). Furthermore, for both PFS and OS, a wider separation between the high versus low matching score groups occurred with time, suggesting a protracted correlation with efficacy.
Several cases of exceptional responses are presented. For instance, a patient with lung adenocarcinoma and an EGFR T790M mutation—known to be resistant to the EGFR inhibitor monotherapy available at the time—was successfully treated (starting in January 2015) with afatinib and cetuximab, achieving a CR (Fig. 2a); afatinib and cetuximab were used based on literature showing responses in patients with this resistant mutation (afatinib response rate: <10%; afatinibcetuximab response rate: 32%)16. Since that time, osimertinib, a drug with potent activity against EGFR T790M alterations has been approved by the US Food and Drug Administration (FDA) (November 2015) and the European Medicines Agency (EMA) (February 2016). A second patient with colorectal cancer and an MSH6 (mismatch repair) mutation received the checkpoint inhibitor pembrolizumab and achieved a partial remission ongoing at >36 months (Fig. 2b). Treatment was started in March 2015, just as data about the efficacy of this type of match were emerging and predating FDA approval (in May 2017) of pembrolizumab for microsatellite unstable solid tumors by >2 yr; this indication is not yet approved in Europe by the EMA (as of 31 December 2018). Finally, a third patient with a refractory gastrointestinal neuroendocrine tumor showed no DNA alterations; however, transcriptomics revealed elevated expression of AKT2 and AKT3, and she was successfully treated with the mechanistic target of rapamycin (mTOR) inhibitor everolimus, resulting in disease stability ongoing at almost 3 yr. This response is of interest for several reasons: (1) effective therapy was based on transcriptomics; and (2) mTOR inhibitors as single agents (in contrast to their use in combinations) are rarely effective when matched to DNA alterations, presumably because of frequent genomic co-alterations14,29–37. This patient started everolimus treatment in May 2015; in 2016, both the FDA and the EMA approved everolimus for this indication according to studies showing improved outcome with a median PFS of ~11 months (ref.38). The PFS is considerably longer in our patient, perhaps because she had transcriptomic indicators of phosphatidylinositol-3-OH kinase (PI3K) pathway component overexpression. As noted above, at the date of start of some of these treatments, they were exploratory and used off-label, but their approval at a later stage pointed to the pertinence of the WINTHER predictive tools and therapeutic choices.
There were limitations to the study that might have affected trial results. First, patients had heavily pretreated disease, which may have limited their capacity for response. Second, the number of patients in each arm was small and the trial was not designed to compare arms. Third, the fact that participating centers were located over a wide geographical territory meant that each center had to separately learn the methodology for specimen processing and access to the CMC’s recommended treatment was unequal. Fourth, the frequency of imaging follow-up was not prescribed or documented for PFS1 (the standard-of-care therapy that preceded the WINTHER treatment (PFS2)); thus, the timing of scans might influence (either negatively or positively) the PFS2/PFS1 ratio. Importantly, however, OS (a parameter that would not be confounded by these considerations) correlated independently with the degree of matching. Fifth, as this was not a randomized trial, it is not possible to eliminate the possibility that prognostic or other factors confounded the results. Sixth, recursive partitioning analysis of the DNA-recommendation patients identified a statistically significant cut-off point that was optimized on these 69 patients; one cannot know whether this cut-off point would retain statistical significance in a validation cohort. It is plausible that a different cutoff point may be optimal in another population or that the degree of matching correlates linearly with outcome. Understanding this issue should be an objective of future studies. Seventh, adjustments for multiple comparisons were not made; thus, these data need to be validated by future studies. Last, therapy recommendations were not locked down, but based on CMC suggestions. However, the latter also afforded flexibility to incorporate dynamic knowledge changes in the rapidly evolving omic fields.
Despite these limitations and other challenges related to implementation of a precision medicine trial across multiple institutions, as previously described9, the WINTHER trial made several interesting contributions. The WIN consortium trial was an international precision medicine study that involved investigators from centers in five countries with differing drug and clinical trial availability, yet successfully navigated 35% of patients with refractory cancers to therapy using either genomic or transcriptomic analysis. The WINTHER trial integrated a new generation of genomic and transcriptomic tools (a large panel NGS-based test and WINTHER RNA-based algorithms) in the decision-making process, but their interpretation by the physicians, cognizant of rapidly evolving knowledge, remained essential. One of the assets of the trial was the successful weekly CMC teleconferences in which principal investigators from participating centers in North America, Europe and the Middle East discussed the biological results and therapeutic options available on a web portal (analogous to a global ‘Molecular Tumor Board’). Transcriptomics (comparing tumor to histological normal counterpart) substantially increased the percentage of patients who could be administered matched therapy. Of interest in this regard, we hypothesized that normal variability of gene expression may have limited previous utility of transcriptomics in the clinical setting15,18,39,40 and we showed that accounting for this variability was important for accurate assessment of the gene expression level (Extended Data Fig. 6). The most common reasons for patient drop off included difficulty with biopsy access or quality and early deterioration or death. For the former challenge, WINTHER’s regular teleconferences improved knowledge in handling biopsies of tumor and normal tissues and optimized histological preparation for DNA and RNA extractions, identified critical failure points in the process and found appropriate solutions to implement transcriptomics in clinical research. The latter challenge—patient deterioration—is not unprecedented; reports from other precision medicine trials show that many patients go to hospice or die before treatment, and considering that these patients were very heavily pretreated, the data indicate that precision oncology trials are being offered too late in the disease course11. On the basis of the findings of the current WINTHER study, future investigations should consider integrating genomic and transcriptomic data, as is being performed in our recently activated SPRING trial (ClinicalTrials.gov identifier: NCT03386929), as well as enrolling patients earlier in the disease course.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-019-0424-4.
Methods
Patients and sites.
Patients had advanced cancers that had progressed on standard treatment. The protocol represented a prospective navigational trial in which genomic or transcriptomic data were used to direct patients to therapy that might include specific clinical trials of investigational drugs, or on-label or off-label use of approved drugs. Approved off-label refers to a drug that is approved by the local medicine regulatory agency (such as the FDA in the United States and the EMA in Europe) for one indication (that is, it is no longer an experimental drug), but it was used for a different indication. Patients had a fresh biopsy of tumor and corresponding normal tissue after they enrolled in the study, and these biopsies were used for the genomic and transcriptomic assays. Treatment was initiated after progression on the previous therapy.
A CMC made the treatment recommendations, which at times entailed differentially ranked possibilities, but the physician chose the therapy. Participating principal investigators were located at Gustave Roussy (Villejuif, France), Centre Leon Berard (Lyon, France), Vall d’Hebron Institute of Oncology (VHIO) (Barcelona, Spain), the Chaim Sheba Medical Center (CSM) (Tel Hashomer, Israel), Segal Cancer Centre, Jewish General Hospital, QCROC-Quebec Cancer Consortium and Rossy Cancer Network, McGill University (Montreal, Quebec, Canada), The University of Texas MD Anderson Cancer Center (Houston, TX, USA) and the University of California San Diego, Moores Cancer Center (San Diego, CA, USA). All patients signed an institutional-review-board-approved informed consent form for the WINTHER protocol ( NCT01856296). The protocol was approved at all sites that recruited patients. If the patient was navigated to an investigational clinical trial, the patient signed a consent form for that trial as well.
A study coordinator who traveled to the sites monitored all study data, confirming them with the source documents, and resolved all queries.
CMC.
Per the protocol, the CMC recommended a drug (or drugs) prospectively. The CMC convened via weekly teleconferences throughout the time that the protocol was active. The site lead investigators from the five participating countries (France, Spain, Israel, Canada and the United States) (seven participating institutes) attended the call via webex (unless they were unavailable that week), as well as any physician designees and a study manager. The calls were conducted in the English language. The local investigator presented the patient (without identifiers such as names) and reviewed online WINTHER portal information handled by Ben-Gurion University of the Negev (Beersheva, Israel) that included all of the patients’ clinical, genomic and transcriptomic reports. The CMC suggested therapies according to factors such as drug or clinical trial availability in the country or institute and patient comorbidities. Per protocol, first it was determined whether there was a potentially actionable DNA alteration according to arm A genomic data. If there was no potential match, the patient was assigned a drug or drugs according to the arm B transcriptomic data. Guiding principles for the CMC included providing the best possible drug matches for the patient, prioritizing genomics over transcriptomics and preferably providing at least three ranked choices (ranking based on the best match for the DNA or RNA alteration) for the physician to consider.
Biopsies.
The WINTHER study was based on a new concept for selecting the most appropriate therapy for each patient, using a wide spectrum of biological information and including both genomic and transcriptomic technologies to navigate patients with advanced refractory cancer to a matched targeted drug. Consented patients underwent a dual biopsy from the metastasis and from the histologically matched normal tissue.
The interventional radiologist selected for biopsy one of the non-necrotic and >2-cm metastatic lesions. The image-guided biopsies were performed under standard operating rules and guidelines using adequate devices, such as trucut 18-G needles placed at the periphery of the lesion. For each patient, the normal tissue of origin of the tumor, as defined in Supplementary Table 2, was biopsied. For example, in the case of a tumor tissue biopsy of a liver metastasis from a rectal adenocarcinoma, the matched normal tissue was the rectal mucosa obtained separately by rectal endoscopy.
All biopsies obtained were stored in cryogenic tubes containing RNAlater, a stabilizing reagent that prevents the degradation of nucleic acids (without the need for freezing in the imaging facilities) and preserves the structural morphology to enable subsequent pathology review.
Histology preparation and quality control.
Biopsies were embedded in optimal cutting temperature (OCT) wax. Slices of 0.5 μm were cut and stained with H&E to assess the percentage of tumor cells and to identify any contamination with adjacent tissues, which was attenuated by microdissection that enriched for tumor content (≥50% tumor content was needed).
DNA and RNA preparations.
The tissue biopsies that had passed quality control were then lysed with a Polytron, by homogenization in the lysis buffer RLT Plus provided in the kit, and DNA was obtained with a specific affinity silica matrix column, specifically retaining the DNA, whereas RNAs and proteins were collected from the through flow. DNA was washed and eluted. The through flow containing RNAs was mixed with tri-reagent and, subsequently, RNA was obtained by isopropanol precipitation. This procedure enables the collection of all types of RNAs, including mRNAs and small microRNA species. RNAs pelleted through centrifugation were washed with ethanol 75% and solved in nuclease-free water. Quality control for DNAs and RNAs was performed using spectrophotometry absorbance (Nanodrop) and through electrophoresis using lab-on-a-chip technology from Agilent Technologies.
NGS (arm A (DNA)).
Tumor specimens collected for tissue NGS analysis were sent directly to Foundation Medicine. The FoundationOne genomic assay (http://www.foundationone.com/) (Clinical Laboratory Improvement Assessment-approved standards) assessed genetic alterations from the entire coding sequence of 236 cancer-related genes, plus rearrangements found in introns of an additional 28 cancer-related genes. Equivocal amplification in this assay refers to copy number 6 or 7, whereas amplification means copy number ≥8 (except for ERBB2, for which equivocal amplification means copy number 5 or 6 and amplification means copy number ≥7). To be reported by the FoundationOne assay, specific alterations must have been identifiable in at least 10% of tumor DNA and a sequencing coverage of 500 × (ref.13).
Transcriptomics (matched tumor and normal (arm B (RNA)).
All patients included in the study, whatever the status of their DNA analysis, had an analysis of their transcriptome attempted. But, per protocol, the transcriptomic information was only considered for navigation to a therapy in cases that had no match found or therapy available per arm A (DNA).
The drug selection in arm B is based on the biological characteristics of the tumor of the individual to be treated in comparison to a normal sample from the same individual. Based on a score calculated (examining deregulated target genes), the match between genes deregulated to a literature-derived knowledge base of gene-drug-directed connections, the relative efficacy of the drugs was predicted for the individual. The method used is based on the following pillars:
Characterizing molecular anomalies of the tumor tissue (metastasis) in comparison to the normal from the same patient, thereby determining the deregulated genes in the tumor according to tumor/normal differences;
Providing a knowledge database comprising the target genes for each drug or the plurality of drugs;
Determining a score for each drug of the plurality of drugs essentially according to the percentage of deregulated genes among the target genes implicated in the efficacy of each drug in the tumor sample from the patient.
Characterizing molecular anomalies of the tumor tissue (metastasis) in comparison to the normal using a gene expression method.
Gene expression direct comparison of tumor tissue and normal tissue RNAs was performed using Agilent inkjet printing 8 × 60k oligo-arrays and dual color technology using standard operating procedures and reagents supplied by Agilent Technologies. Of each tumor and normal tissue, 100 ng total RNA was used to generate double-stranded complementary DNA using MMLV reverse transcriptase and oligo DT primers coupled with the promoter sequence of T7 RNA. Probe labeling and linear amplification were generated using Agilent Technologies reagents and T7 RNA polymerase that generated labeled complementary labeled RNAs (tumor labeled with Cy5 and normal tissue with complementary RNAs with Cy5). After fragmentation and purification of labeled complementary RNA, hybridizations were performed as dual-dye-swapped (direct and inversed labeling) experiments with direct co-hybridization of equal amounts of labeled tumor and normal probes. After washing and drying, microarrays were read using the Agilent 2000 scanner version C. After processing with Agilent Feature extractions software, data were used for direct comparison of intensities and editing of a report containing a detailed data quality report, identifying mRNAs differentially expressed (overexpressed or underexpressed in tumor versus normal), and providing for each gene fold changes and intensities together with P values for each measure, type of structural abnormalities (amplifications or deletions), threshold, fold changes and intensities.
WINTHER knowledge database.
The drugs considered in the database included both registered drugs and drugs in development (for example, contemporary authorization of use or clinical trials). The knowledge database developed consisted of publications linking gene expression with the efficacy of a drug for a plurality of drugs. The WINTHER database had been generated by extracting information from the Cytotoxicity Database, which was in turn generated by literature review (http://ctdbase.org/) or by reviewing the literature for those drugs for which no information could be extracted from the Cytotoxicity Database. This knowledge base was periodically updated and improved during the trial.
The therapies included in the database were:
All compounds available in the consolidated clinical trial portfolio of the cancer centers participating in the WINTHER trial.
Registered molecular targeted and standard chemotherapy drugs.
The WINTHER algorithm.
The WINTHER algorithm scores each drug by matching between the knowledge base and the gene expression detected in the tumor to be treated in an individual. The algorithm can be intuitively described as the fold-change-adjusted fraction of target genes (that is, genes known from the literature to be associated with drug efficacy) that are differentially expressed in the tumor, with the contribution of every gene adding or subtracting from the score according to its concordance (positive) or discordance (negative) with the knowledge base. Formally, the score of drug x in individual i (Si) can be defined as follows.
Let the concordance-adjusted fold change, be defined for gene g and drug x in individual i as
where fg,i represents the log fold change of gene g in individual i between the tumor and normal tissue (in base 2), and dg,x is a direction coefficient for gene g and drug x extracted from the knowledge base (positive = +1, negative = −1).
The WINTHER score for drug x in individual i (Si) thus becomes
Where Mi reflects the subset of genes for which Fg,i ≠ 0 and |Mi| is the size of the Mi set, and Ig is the intensity of gene g. The results were arbitrarily multiplied by 100 to make the values more readable. As the score depends on the knowledge database (through dg,x), this database was updated three times during the trial (on July 2013, August 2013 and September 2014).
Initial analysis of published tumor-normal matched sets (the CHEMORES data set15) suggested that drugs associated with only one gene tended to rank higher than drugs with multiple genes. As a result, we separated the ranked lists into two tables, one containing information about monogenic drugs and one containing multigenic drugs. Both tables were presented to the CMC for consideration.
Matched therapy and matching score.
Because physicians could choose the therapy, all of the outcome analysis was evaluated based on the actual drug (or drugs) given to the patient (or patients). Physicians might elect to choose a drug or drugs lower ranked by the CMC owing to availability or other considerations. Physicians could also give unmatched drugs as part of the regimen; these drugs were not scored. Hence, a matching score11,12 was calculated as part of a post-hoc blinded analysis to ascertain the correlation between the degree of DNA or RNA matching and outcome. The matching score was calculated by one investigator (R.K.) who was blinded to response, PFS and OS data at the time of calculation; the score was cross-checked by the study statistician (J.J.L.) in face-to-face meetings and teleconferences, still under blinding to outcomes. This matching score is distinct from the WINTHER score resulting from the RNA algorithm.
Arm A (genomics).
For DNA, the pathogenicity of variants was derived from the Foundation Medicine genomic reports. Alterations were considered matched if the gene product was differentially expressed in tumor versus normal cells and could be targeted by a drug, or if a drug inhibited or modified the oncogenic activity of the gene product (at low IC50 (half-maximum inhibitory concentrations) for small-molecule inhibitors); or if the gene product was the primary target recognized by an antibody. Anti-programmed cell death 1 (PD1)-PD1 ligand 1 (PDL1) immunotherapy was considered a match if there was a mismatch repair gene alteration or if there were more than eight characterized alterations (including mutations, deletions, amplifications, insertions, and fusions or rearrangements) (which may reflect a high tumor mutational burden); the matching score was 1.0 (100%) in these cases. The tumor mutational burden was not available as a clinical-grade assay at the time that patients accrued onto the trial; hence, we counted characterized alterations as a surrogate.
The matching score was calculated as the number of characterized DNA alterations affected by the drug (or drugs) administered divided by the total number of characterized alterations. For example, if a patient had the DNA alterations BRAF V600E and ERBB2 amplification and received the BRAF inhibitor vemurafenib, the matching score would be 1/2 = 0.5. Conversely, if the patient had the DNA alterations BRAF V600E, ERBB2 amplification, TP53 mutation, MET amplification and PIK3CA H1047R mutation, and received the ERBB2 antibody trastuzumab as well as vemurafenib, the matching score would be 2/5 = 0.4
If two drugs were given and they matched the same alterations, they were scored as two hits. If a drug was given that affected the downstream mediators of an alteration, it was scored as one, as long as the molecule that was affected was no more than two effectors downstream of the alteration. If several mutations occurred in the same gene, but they had a similar effect, they were counted once; however, if they were known to have differing effects, they were counted individually. Everolimus or other PI3K-AKT-mTOR inhibitors alone in patients with PI3K pathway and MEK co-mutations or two or more co-alterations received a score of zero because MEK and multiple other co-alterations are a resistance pathway for PI3K signaling, and targeting the PI3K pathway alone in this situation is known to not be an effective strategy14,32,36.
Arm B (transcriptomics (matched tumor versus normal)).
For arm B, the matching score was calculated as the reciprocal of the rank of the WINTHER score rank.If there was more than one drug used, the reciprocals of the ranks of each drug were added together to obtain the matching score. For example, if the physician investigator selected the drug ranked 1, the matching score was 1; if the physician investigator selected the drug ranked 2, the matching score was 0.5; if the physician investigator selected the drug ranked 3, the matching score was 0.3, and so on. If the physician investigator selected 2 drugs and they were ranked as 4 and 5, the matching score was 0.25 + 0.2 = 0.45.
In some cases, a translation was done between equally targeted drugs according to availability. In arm B, if the algorithm offered an anti-T cell drug, it was interpreted as a sign of infiltrating T cells and converted to a recommendation to use a checkpoint inhibitor.
Response assessment.
Responses were assessed by local physicians at the participating centers using the RECIST 1.1 criteria. PFS and OS were analyzed from the date of starting the therapy to the time of progression or death, respectively, using the method of Kaplan and Meier. Patients still without progression (for PFS) or alive (for OS) at the time of analysis were censored on that date. PFS2 refers to the PFS on the WINTHER therapy; PFS1 refers to the PFS on the therapy immediately preceding the WINTHER therapy. Patients were considered evaluable for PFS2 if they had their baseline scan within 30 d of the start of therapy and follow-up scans about every 6–10 weeks thereafter.
Statistical methods and outcome end points.
All statistical analyses were performed by our statistician (J.J.L.). Efficacy end points include response rate, rate of SD ≥ 6 months, PFS and OS. (When referring to SD ≥ 6 months/PR/CR, the 6-month requirement pertains only to SD). The ratio of the PFS of the current treatment (PFS2) over PFS of the previous treatment (PFS1) was targeted to be as follows: PFS2/PFS1 > 1.5 in 50% of patients in arm A and in 40% of patients in arm B, respectively.
Descriptive statistics were applied to summarize the data. Point estimates and the associated 95% CIs were provided. Exact binomial CIs were provided for binary end points. Standard statistical tests including the chi-squared test and Fisher’s exact test for categorical data and the t-test for continuous data were applied. Time-to-event end points, such as PFS and OS, were analyzed by the Kaplan-Meier estimate. The log-rank test and Cox proportional hazards model (Wald test) were applied to test the effect of covariates on PFS and OS. The RPART method was applied to find the optimal cut-off point for dichotomizing a continuous covariate (for example, matching score) in predicting an outcome. As DNA and RNA matching scores are derived differently, RPART was performed separately for arm A (DNA) and arm B (RNA) to identify the patients with high and low score. Once the status was determined separately, a post-hoc combined analysis of low and high matching scores was performed for all 107 patients. All statistical tests were performed as two-sided tests. Statistical packages including SAS and R were used for statistical analysis.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The availability of the data is described in the Nature Research Reporting Summary. Detailed clinical and biological information for each patient is available in Supplementary Table 4; further biological data are available at http://www.winconsortium.org", containing: (1) tumor mutations data in XML format; and (2) expression data in a table format (providing information about tumor versus normal fold change and tumor intensity alone for all of the cases for which mRNA was analyzed). The BAM and XML files for normal tissue are deposited in dbGaP with a controlled access mechanism for private information.
Code availability
No custom code or mathematical algorithm was used. Statistical analysis was performed using standard software including SAS and R.
Extended Data
Extended Data Fig. 1 |.
Consort diagram.
Extended Data Fig. 2 |. Kaplan-Meier curves of various factors influencing PFS and OS.
a, Kaplan-Meier curves of PFS for arm A by cancer site (lung site, N = 17 versus other site, N = 52). PFS2 denotes the PFS of the WINTHER treatment. P = 0.0204 by two-sided log-rank test. b, Kaplan-Meier curves of PFS for arm A by ECOG performance status at treatment (PS = 0, N= 21 versus PS = 1, N = 48). PFS2 denotes the PFS of the WINTHER treatment. P= 0.0002 by two-sided log-rank test.
Extended Data Fig. 3 |. Kaplan-Meier curves of various factors influencing PFS and OS.
a, Kaplan-Meier curves of PFS for arm B by age group. Age >60yr, N = 17 versus age ≤60yr, N = 21. PFS2 denotes the PFS of the WINTHER treatment. P = 0.0361 by two-sided log-rank test. b, Kaplan-Meier curves of PFS for arm B by sex. Sex = female, N = 12 versus sex = male, N= 26. PFS2 denotes the PFS of the WINTHER treatment. P = 0.0252 by two-sided log-rank test. c, Kaplan-Meier curves of PFS for arm B by the number of previous treatments. For the number of previous treatments of ≤2, N = 11 versus >2, N = 27. PFS2 denotes the PFS of the WINTHER treatment. P = 0.0066 by two-sided log-rank test.
Extended Data Fig. 4 |. Kaplan-Meier curves of various factors influencing PFS and OS.
a, Kaplan-Meier curves of PFS all patients by ECOG PS at treatment. PS = 0, N = 36 versus PS = 1, N = 71. PFS2 denotes the PFS of the WINTHER treatment. P = 0.0007 by two-sided log-rank test. b, Kaplan-Meier curves of PFS of all of the patients by the number of previous treatments. The number of previous treatments of ≤2, N = 34 versus >2, N = 73. PFS2 denotes the PFS of the WINTHER treatment. P= 0.0025 by two-sided log-rank test.
Extended Data Fig. 5 |. Kaplan-Meier curves of various factors influencing PFS and OS.
a, Kaplan-Meier curves of OS of all patients by ECOG PS at treatment. PS = 0, N = 36 versus PS = 1, N = 71 (P < 0.0001 by two-sided log-rank test). b, Kaplan-Meier curves of OS all patients by the number of previous treatments (Tx). The number of prior Tx of ≤ 2, N = 34 versus >2, N = 73 (P= 0.0009 by two-sided log-rank test). c, Kaplan-Meier curves of OS of all patients by matching score. MS high, N=80 versus MS low, N = 27 (P = 0.0103 by two-sided log-rank test).
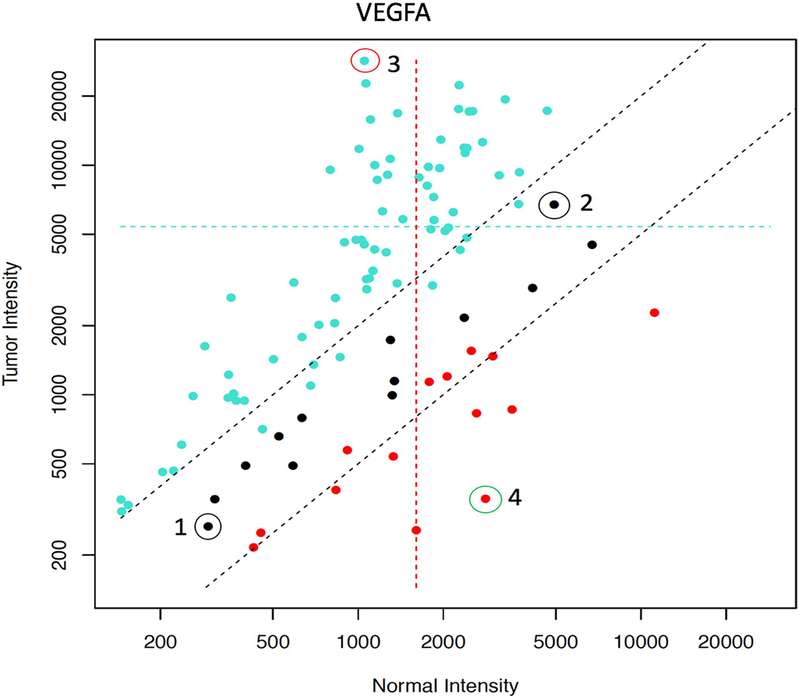
Extended Data Fig. 6 |. Effect of individual variability of normal VEGFA RNA expression on the assessment of VEGFA levels in tumors.
On the y axis, the transcript intensity in tumors is shown, and on the x axis the transcript intensity in matched normal biopsies is shown. Intensities are measured as a relative fluorescence unit (RFU) signal as assessed with Agilent microarray technology. Overexpression in the tumor is denoted in turquoise points, underexpression is denoted in red and no change is denoted in black. The twofold threshold (both high and low) is indicated by two dotted black lines. All 101 patients of the WINTHER study with evaluable RNA data were considered. Example 1 shows a patient with a low level of basal expression (300 RFU) in the tumor and 300 RFU in the normal biopsies, with no differential expression between the normal and tumor biopsies. Example 2 shows a patient with a high level of basal expression of 6,000 RFU in the tumor versus 6,000 RFU in the normal biopsies, but again no differential expression between the tumor and normal counterpart. Example 3 marked in turquoise shows the pattern of a higher expression in tumor versus normal tissue. Example 4 marked in red shows the pattern of a lower expression in tumor versus normal tissue. This current study hypothesizes that simultaneously investigating the matched phenotypically normal tissue can help to optimize transcriptomic data. With this approach, each patient serves as his or her own control, hence avoiding the use of pooled tumor or normal tissues. Our data demonstrate that the level of basal gene expression is highly variable between individuals. All patients presented with black points had no differential expression between tumor and normal tissue, but others show a large variability between individuals in the basal level of normal expression of VEGFA.
Supplementary Material
Acknowledgements
Dr John Mendelsohn, President Emeritus of The University of Texas, MD Anderson Cancer Center, Director of Sheikh Khalifa Bin Zayad Al Nahyan Institute for Personalized Cancer Therapy (IPCT), Houston, TX, USA, Chairman of the WIN Association—WIN Consortium, Villejuif, France, and co-author of the Letter died on 7 January 2019. Dr Mendelsohn was a brilliant scientist and visionary, an optimist and a truly inspirational leader. The WINTHER study was one of his very last projects.
The research leading to these results has received funding from the European Union Seventh Framework Program (FP7/2007–2013 under grant agreement no. 306125). This work was funded in part by the ARC Foundation for Cancer Research (France), Pfizer Oncology, Lilly France SAS and Novartis Pharmaceuticals Corporation. This work was also funded in part by The Fero/J.P. Morgan Private Bank Clinical Oncology Research Grant, the National Cancer Institute grant P30 P30-CA023100 (R.K.), the Israeli Science Foundation grant 1188/16 (E.R.), Instituto Salud Carlos III—Programa Rio Hortega Contract grant CM15/00255 (I.B.), the Canadian Institutes for Health Research (grant MOP-142281 to W.H.M.) and the Canadian Cancer Society (grant 703811 to W.H.M.).
Footnotes
Competing interests
J.Rodon reports non-financial support and reasonable reimbursement for travel from the European Journal of Cancer, Vall d’Hebron Institut of Oncology, the Chinese University of Hong Kong, SOLTI, Elsevier, GlaxoSmithKline; receives consulting and travel fees from Novartis, Eli Lilly, Orion Pharmaceuticals, Servier Pharmaceuticals, Peptomyc, Merck Sharp & Dohme, Kelun Pharmaceutical/Klus Pharma, Spectrum Pharmaceuticals, Pfizer, Roche Pharmaceuticals and Ellipses Pharma (including serving on the scientific advisory board from 2015 to present); receives research funding from Bayer and Novartis; and serves as investigator in clinical trials with Spectrum Pharmaceuticals, Tocagen, Symphogen, BioAtla, Pfizer, GenMab, CytomX, Kelun Pharmaceutical/Klus Pharma, Takeda-Millenium, GlaxoSmithKline and Ipsen. J.-C.S. received consultancy fees from AstraZeneca, Roche, Sanofi, Servier, Pierre Fabre and is a full-time employee of Medimmune/AstraZeneca since September 2017. W.H.M. receives speaking and/or consulting fees from BMS, Merck, Roche, Novartis and Amgen. E.R. received consultant fees from Teva, Carmentix and Hinoman, is receiving consultant fees from Equinom and has ownership interest in Carmentix. A.O. receives consulting fees from Roche Israel, MSD Israel, Boehringer Ingelheim and AstraZeneca. A.T. received consultant fees from Roche and receives research funding from EMD Serono, Baxter, Foundation Medicine, ONYX and Bayer. PS. collaborates in research with Roche, AstraZeneca, BMS and Novartis. I.B. receives consultant fees from Orion Pharma, receives speaking fees from BMS, AstraZeneca and Merck Serono, and is principal investigator and receives funding for clinical trials from AstraZeneca, BMS, Celgene, Gliknik, GSK, Janssen, KURA, MSD, Novartis, Orion Pharma and Pfizer. Y.L. collaborates in research with Merck, Roche, AstraZeneca, Sanofi, Pfizer, Janssen, Astellas and BMS. M.A. is an employee and shareholder of Ariana Pharmaceuticals. VM. is an employee (salary and equity) of Foundation Medicine. J.-F.M. is a full-time employee and stockholder of Pfizer. G.B. collaborates in formal clinical trial contracts, investigator initiated trials (IITs) and in joint grants funded by the Canadian and Quebec governments with Roche, Merck, Novartis, AstraZeneca, Bayer, Esperas, Aurka, Caprion and MRM-P. J.T. declares a scientific consultancy role for Array Biopharma, AstraZeneca, Bayer, BeiGene, Boehringer Ingelheim, Chugai, Genentech, Genmab A/S, Halozyme, Imugene, Inflection Biosciences, Ipsen, Kura Oncology, Lilly, MSD, Menarini, Merck Serono, Merrimack, Merus, Molecular Partners, Novartis, Peptomyc, Pfizer, Pharmacyclics, ProteoDesign SL, Rafael Pharmaceuticals, F. Hoffmann-La Roche, Sanofi, SeaGen, Seattle Genetics, Servier, Symphogen, Taiho, VCN Biosciences, Biocartis, Foundation Medicine, HalioDX SAS and Roche Diagnostics. R.K. has research funding from Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, Guardant Health, Grifols, Konica Minolta and OmniSeq, as well as consultant fees from LOXO, X-Biotech, Actuate Therapeutics, Roche and NeoMed. She serves as an advisor to Soluventis. She receives speaker fees from Roche and also has equity in IDbyDNA, CureMatch and Soluventis. F.W., C.B. and V.L. are full-time employees of WIN Association-WIN Consortium. WIN Association-WIN Consortium is the owner of the patent family entitled ‘Method for predicting efficacy of drugs in a patient’ (WINTHER). The inventors are V.L., J.-C.S., Michel Ducreux and Thomas Tursz.
Extended data is available for this paper at https://doi.org/10.1038/s41591-019-0424-4.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-019-0424-4.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schwaederle M et al. Association of biomarker-based treatment strategies with response rates and progression-free survival in refractory malignant neoplasms: a meta-analysis. JAMA Oncol. 2, 1452–1459 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Jardim DL et al. Impact of a biomarker-based strategy on oncology drug development: a meta-analysis of clinical trials leading to FDA approval. J. Natl. Cancer Inst. 107, djv253(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwaederle M et al. Impact of precision medicine in diverse cancers: a meta-analysis of phase II clinical trials. J. Clin. Oncol 33, 3817–3825 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman AM et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol.Cancer Ther 16, 2598–2608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsao MS et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N. Engl. J. Med 353, 133–144 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Demetri GD et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med 347, 472–480 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Chapman PB et al. BRIM-3 study group: improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med 364, 2507–2516 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solomon BJ et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med 371, 2167–2177 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Rodon J et al. Challenges in initiating and conducting personalized cancer therapy trials: perspectives from WINTHER, a Worldwide Innovative Network (WIN) Consortium trial. Ann. Oncol 26, 1791–1798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Von Hoff DD et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J. Clin. Oncol 28, 4877–4883 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Wheler JJ et al. Cancer therapy directed by comprehensive genomic profiling: a single center study. Cancer Res. 76, 3690–3701 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Schwaederle M et al. Precision oncology: the UC San Diego Moores Cancer Center PREDICT experience. Mol. Cancer Ther 15, 743–752 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Frampton GM et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol 31, 1023–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsimberidou AM et al. Initiative for molecular profiling and advanced cancer therapy (IMPACT): an MD Anderson precision medicine study. JCO Precis. Oncol 2017, 2017(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazar V et al. A simplified interventional mapping system (SIMS) for the selection of combinations of targeted treatments in non-small cell lung cancer. Oncotarget 6, 14139–14152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janjigian YY et al. Dual inhibition of EGFR with afatinib and cetuximab in kinase inhibitor-resistant EGFR-mutant lung cancer with and without T790M mutations. Cancer Discov. 4, 1036–1045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koscielny S Why most gene expression signatures of tumors have not been useful in the clinic. Sci. Transl. Med 2, 14ps2(2010). [DOI] [PubMed] [Google Scholar]
- 19.Tsimberidou AM et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin. Cancer Res 18, 6373–6383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massard C et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 7, 586–595 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Le Tourneau C et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 16, 1324–1334 (2015). [DOI] [PubMed] [Google Scholar]
- 22.ECOG-ACRIN Cancer Research Group & National Cancer Institute Executive Summary: Interim Analysis of the NCI-MATCH Trial (ECOG-ACRIN Cancer Research Group & National Cancer Institute, 2016); http://ecog-acrin.org/nci-match-eay131/interim-analysis [Google Scholar]
- 23.Gerlinger M et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciriello G et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet 45, 1127–1133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bieg-Bourne CC et al. Next-generation sequencing in the clinical setting clarifies patient characteristics and potential actionability. Cancer Res. 77, 6313–6320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kandoth C et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martincorena I & Campbell PJ Somatic mutations in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Kato SM et al. Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary. Cancer Res. 77, 4238–4246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janku F et al. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep. 6, 377–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesan P et al. Target-based therapeutic matching in early-phase clinical trials in patients with advanced colorectal cancer and PIK3CA mutations. Mol. Cancer Ther 12, 2857–2863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holsinger FC et al. Biomarker-directed therapy of squamous carcinomas of the head and neck: targeting PI3K/PTEN/mTOR pathway. J. Clin. Oncol 31, e137–e140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janku F et al. PIK3CA mutations in advanced cancers: characteristics and outcomes. Oncotarget 3, 1566–1575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janku F et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 73, 276–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrido-Laguna I et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS ONE 7, e38033(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janku F et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS ONE 6, e22769(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janku F et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J. Clin. Oncol 30, 777–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodon J, Dienstmann R, Serra V & Tabernero J Development of PI3K inhibitors: lessons learned from early clinical trials. Nat. Rev. Clin. Oncol 10, 143–153 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Yao JC et al. RAD001 in advanced neuroendocrine tumours, fourth trial (RADIANT-4) study group. Everolimus for the treatment of advanced, non-functional neuroendocrine tumours of the lung or gastrointestinal tract (RADIANT-4): a randomised, placebo-controlled, phase 3 study. Lancet 387, 968–977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc 57, 289–300 (1995). [Google Scholar]
- 40.Ioannidis JP Why most published research findings are false. PLoS Med. 2, e124(2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The availability of the data is described in the Nature Research Reporting Summary. Detailed clinical and biological information for each patient is available in Supplementary Table 4; further biological data are available at http://www.winconsortium.org", containing: (1) tumor mutations data in XML format; and (2) expression data in a table format (providing information about tumor versus normal fold change and tumor intensity alone for all of the cases for which mRNA was analyzed). The BAM and XML files for normal tissue are deposited in dbGaP with a controlled access mechanism for private information.