Abstract
Stimuli-responsive release of drugs from a nanocarrier in spatial-, temporal-, and dosage-controlled fashions is of great interest in the pharmaceutical industry. Paclitaxel is one of the most effective and popular chemotherapeutic drugs against a number of cancers such as metastatic or nonmetastatic breast cancer, non–small cell lung cancer, refractory ovarian cancer, AIDS-related Kaposi’s sarcoma, and head and neck cancers. Here, by taking the advantage of RNA nanotechnology in biomedical and material science, we developed a three-dimensional pyramid-shaped RNA nanocage for a photocontrolled release of cargo, using paclitaxel as a model drug. The light-triggered release of paclitaxel or fluorophore Cy5 was achieved by incorporation of photocleavable spacers into the RNA nanoparticles. Upon irradiation with ultraviolet light, cargos were rapidly released (within 5 min). In vitro treatment of breast cancer cells with the RNA nanoparticles harboring photocleavable paclitaxel showed higher cytotoxicity as compared to RNA nanoparticles without the photocleavable spacer. The methodology provides proof of concept for the application of the light-triggered controlled release of drugs from RNA nanocages.
Keywords: RNA nanotechnology, controlled release, paclitaxel, drug delivery, RNA nanoparticles, Phi29 three-way junction
1. Introduction
Smart nanomaterials have opened new horizons for the development of ever more efficient and high-performance nanocarriers via which selective delivery of cargos is achievable [1]; such materials can be exploited for biomedical applications, particularly in the design of stimuli-responsive delivery approaches [2]. Implementation of these advanced nanodevices requires easy manipulation of biocompatible materials at a nanoscale to make them susceptible to a specific stimulus [3–6]. Although nanoparticle-based therapeutics have shown advantages in specific cancer targeting, high delivery efficiency remains a challenge as a result of nonspecific Fickian diffusion of therapeutics from nanocarriers [7, 8]. To achieve a controlled release of cargo on nanodelivery platforms, various stimuli including variations in temperature [9] and in ultrasound intensity [10], pH changes [11], redox gradients [12], or light [13, 14] have been employed in responsive nanoparticulate systems. Among the external stimuli, light has attracted special attention due to ease of production, noninvasive nature, controllable intensity, and spatially confined application for finely controlled durations [15–19]. Thus, light has been utilized in an abundance of applications for intelligent targeted delivery systems that can provide control of treatment processes and allow for transport and release of drugs at disease sites [20–22].
The development of DNA nanotechnology has enabled custom predesigned nanostructures with high spatial addressability. A plethora of DNA-based assemblies have been explored to serve as drug and gene delivery platforms [23, 24]. With the advancement of RNA nanotechnology in the past decade, there was an explosion of interest in developing RNA-based therapeutics, which have been envisioned as the third milestone in the drug development [25–27]. In particular, the packaging RNA (pRNA) from the phi29 DNA packaging motor has been extensively explored as a building unit for the construction of structurally ordered nanostructures [28, 29]. Of note, the three-way junction (3WJ) motif of the pRNA recently manifested unparalleled advantages as a nanodelivery system owing to its high thermodynamic stability, programmability, and favorable in vivo pharmacokinetic and pharmacodynamic profiles [30–33]. These characteristics are well exemplified by 3WJ-based two-dimensional (2D) therapeutic delivery platforms [34–36] as well as recently developed three-dimensional (3D) RNA nanoparticles [37, 38]. 3D RNA tetrahedron nanoparticles show specific in vivo cancer targeting with low accumulation in healthy organs [38]; RNA nanoprisms have been proven to effectively protect the encapsulated payloads such as small RNA from RNase degradation [37]. To fulfill the promises of RNA nanocage–based controlled drug delivery for disease treatment, the precise control of the size, shape, and cargo encapsulation has been achieved, while the cargo release profile is critical yet remains uncontrollable [39].
Paclitaxel is a chemical isolated from the bark of Pacific Yew (Taxus brevifolia) [40]. As one of the most effective and popular chemotherapeutic drugs against a number of cancers [41, 42], it induces cell death by promoting and stabilizing microtubules and then stalling G2 or M phases of the cell cycle [43]. Nonetheless, the formulation with Cremophor EL or dehydrated ethanol [44] causes severe side effects [45] and has unpredictable pharmacokinetics. Therefore, alternative paclitaxel formulations such as nanoparticle-based delivery systems [46, 47] have been devised. For example, Abraxane, a nanoparticle-based delivery system composed of paclitaxel and albumin was approved by the US FDA in 2005 for the treatment of metastatic breast cancer and non–small cell lung cancer. Nevertheless, improving therapeutic efficacy of nanoparticle-based paclitaxel delivery systems remains challenging due to the lack of control over their release profile at a tumor site. With several types of paclitaxel nanodelivery platforms under development—including polymer–drug conjugates [48], inorganic nanoparticles [49], lipid-based nanoparticles [50], polymer-based nanoparticles [51], carbon nanotubes [52], and nanocrystals [53]—an RNA nanotechnology-based platform for paclitaxel delivery with a tailored release profile has never been reported.
In this proof-of-concept study, we developed 3D RNA pyramidal nanocages as a photoresponsive system for on-demand release of a model drug with ease of fabrication and high programmability. In this study, the RNA pyramids were constructed by modular design, where multiple RNA motifs were incorporated to enhance the structural rigidity and stability, leading to uniform size and structurally ordered nanostructures. A photocleavable (PC) spacer was inserted into the RNA nanoparticle to enable highly efficient photoinduced cleavage. Specifically, fluorophore Cy5 or paclitaxel were tethered to RNA nanocages via chemical conjugation through the PC spacer. Upon irradiation with 365 nm ultraviolet (UV) light, the conjugated fluorophore and drug were rapidly released (within 5 min). In vitro cancer cell treatment with the RNA nanoparticles harboring PC paclitaxel revealed higher cytotoxicity as compared to RNA nano-particles without PC spacers. For the first time, we employed a controllable cargo release strategy on 3D RNA nanocages, which in principle can be generalized to the delivery and release of diverse biomolecules, thus contributing to the development of RNA-based therapeutics and possibly leading to successful translation into the clinic.
2. Results
2.1. Construction of RNA pyramid nanoparticles
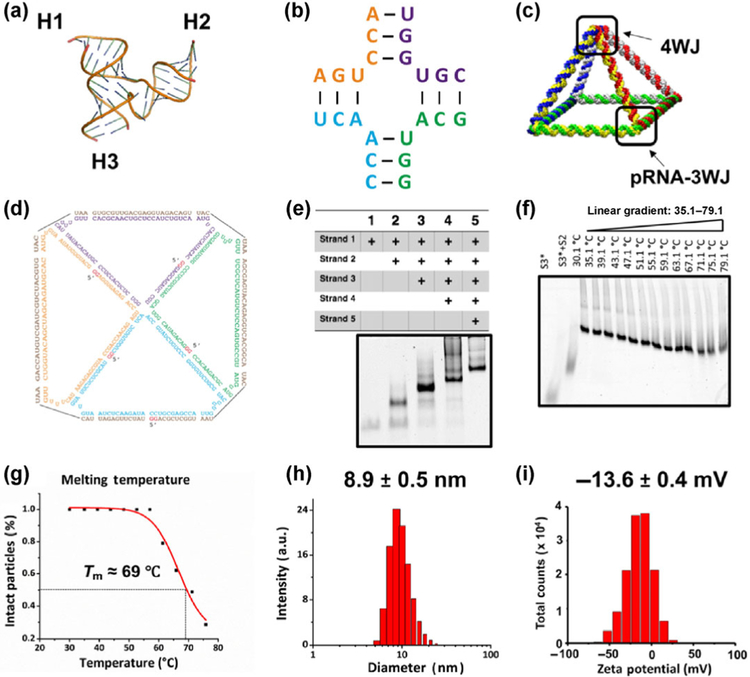
The in silico design of the RNA pyramid was primarily based on the 3WJ motif (Protein Data Bank [PDB] entry: 4KZ2, Fig. 1(a)) of pRNA and the four-way junction (4WJ) motif of a hairpin ribozyme molecule (Fig. 1(b)) [29, 54]. The pRNA 3WJ composed of three short fragments has been reported to be unusually thermodynamically stable allowing for its use as a building unit to drive the formation of more complex nanostructures [30, 33, 55]. Each corner of the RNA pyramid was composed of a 3WJ module [30], which was extended creating a 4WJ at the vertex. Five RNA oligo strands 1 to 5 (Table S1 in the Electronic Supplementary Material (ESM)) were allowed to self-assemble in vitro in a one-pot reaction into a pyramid-shaped nanocage (Fig. 1(c)) with four triangular side faces and one square bottom face composed of eight double-stranded edges (top view with the sequence: Fig. 1(d)). Swiss PDB viewer and PyMOL Molecular Graphics System were used to align four corners and vertices with RNA helix linkage. One-pot bottom-up assembly of such a nano-particle was achieved by thermal denaturation of equimolar amounts of RNA oligonucleotides at 85 °C and slow cooling down to 4 °C (during 45 min). To obtain intact RNA pyramid nanoparticles, all five strands were synthesized by in vitro transcription and then mixed in a stoichiometric ratio and annealed in 1× Tris buffer in a one-pot manner to form the RNA complex with high efficiency.
Figure 1.
Design and characterization of the RNA pyramid. (a) Crystal structure of the 3WJ of pRNA composed of three branches: H1, H2, and H3. (b) The 4WJ core derived from a hairpin ribozyme [54]. (c) A schematic of the RNA pyramid composed of four 3WJs at the bottom and one 4WJ at the top vertex. (d) Sequence and 2D structure of the RNA pyramid. (e) Native PAGE in a 6% gel showing the stepwise assembly of RNA pyramid. “+” indicates the presence of strands. (f) TGGE showing the thermostability and melting temperature (Tm) of the RNA pyramid. (g) Measurement of Tm of RNA pyramid. (h) DLS analysis showing the size of the RNA pyramid. (i) Zeta potential of the RNA pyramid.
To verify the successful assembly of the RNA pyramids, a native 6% polyacrylamide gel electrophoresis (PAGE) was carried out (Fig. 1(e)). Strands 1 to 5 were added step by step in 1× Tris buffer for assembly followed by gel electrophoresis to verify the participation of each strand in the assembled complex. As shown in Fig. 1(e), electrophoretic mobility in lanes 1–5 indicates a stepwise upshift due to the increased strand proportion in the complex. Fluorophore Cy5–labeled strands also indicated the incorporation of each strand into the RNA pyramid nanoparticle (Fig. S1 in the ESM).
2.2. Characterization of the RNA pyramid nanoparticles
To examine stability of the assembled pyramids, thermostability of the nanoparticles was investigated by temperature gradient gel electrophoresis (TGGE) [56]. TGGE is commonly conducted for measuring melting temperature (Tm) of large and complex nucleic acid structures. A gradient temperature (30–80 °C) was applied perpendicular to the electrical current on the 4% native acrylamide gel, with an increasing temperature that gradually led to the dissociation of RNA nanostructures (Fig. 1(f)). As suggested in Fig. 1(g), half of the particles remained intact at 69 °C (Tm), indicating the high thermostability of the RNA pyramid nanostructures. The average hydrodynamic diameter of the RNA pyramid was measured in a dynamic light scattering (DLS) assay. As revealed in Fig. 1(h), the apparent hydrodynamic size of the RNA pyramid turned out to be 8.9 ± 0.5 nm. Even though the RNA pyramid nanoparticles did not have a globular shape, we assumed that the measured size was in good agreement with the predicted size, considering the rapid tumbling of RNA nanoparticles in solution. The surface charge of the RNA pyramid was also determined as previously described [57]. The surface charge typically influences the aggregation propensity of nanoparticles in solution. Figure 1(i) illustrates the highly negative charge of the RNA pyramid with a single peak at −13.6 ± 0.4 mV, effectively preventing aggregation of the nanoparticles. The anionic nature of the RNA nanocage is also desirable for in vivo applications because nonspecific cell binding and unfavorable interactions with the reticuloendothelial system could be minimized [25, 27, 58]. Taken together, the results from PAGE, TGGE, and DLS strongly indicated the formation of a compact and intact RNA complex composed of five strands.
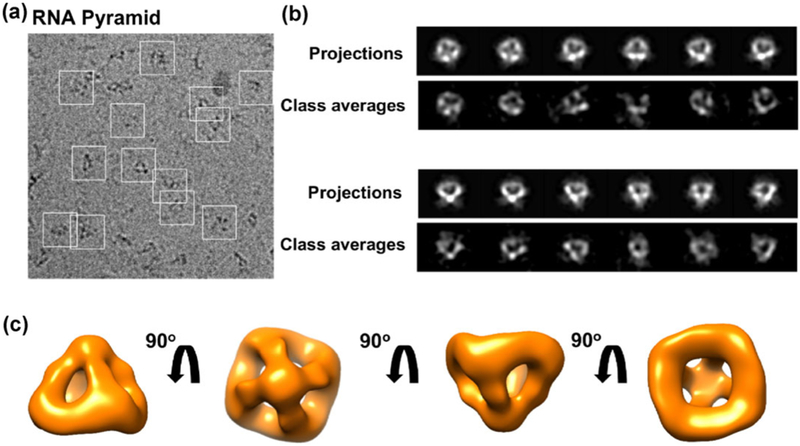
To visualize and validate the pyramid-shaped structure, we performed single-particle analysis by cryoelectron microscopy (cryo-EM). The pyramid nanoparticles were well distributed, as shown in a raw image (Fig. 2(a)). Besides, 2D computer-generated projections of the reconstructed 3D RNA pyramid nanoparticle matched well with 2D class averages (Fig. 2(b)). The final 3D map of the RNA pyramid at ~ 25 Å resolution was constructed from 1,103 particles in EMAN2 (see also the Experimental section). Four different views of the RNA pyramid nanoparticle (Fig. 2(c)) with a 90° rotation showed the structural features consistent with the predicted 3D model displayed in Fig. 1(c), further confirming the successful assembly of the pyramid-shaped RNA nanocage.
Figure 2.
Cryo-EM analysis of the RNA pyramid. (a) A representative raw cryo-EM micrograph with individual RNA pyramid nanoparticles in white square boxes. (b) Representative projections and corresponding class averages of the 3D model. (c) The final 3D cryo-EM map in four views.
2.3. Conjugation of RNA pyramids with RNA moieties having original functionalities
To develop a potential drug delivery platform, the programmability and versatility of nanocarriers are critical for enabling the incorporation of multiple targeting ligands and therapeutic modules without compromising their original structural and functional properties [59]. For the incorporation of five RNA-based functional modules including aptamers (such as malachite green [60], spinach [61], ATP [62], and streptavidin aptamers [63]) and hepatitis B virus (HBV) ribozyme [64], the 5′ end of each of the five strands was extended with a functional RNA motif (Table S2 in the ESM) to construct a multifunctional RNA pyramid, thereby placing the functional group on the center of the faces of the pyramid (Fig. S2(a) in the ESM). After that, we performed functional assays to verify the authentic functionalities of different modules (Figs. S2 and S3 in the ESM). The data indicated that the aptamers and HBV ribozyme that were fused with the pyramid retained their authentic folding and functionalities without interference with the folding of the pyramid core structure. This finding points to the good potential of these pyramid nanoparticles as a platform for multifunctional-drug delivery.
2.4. The design and characterization of conjugates of PC drugs with RNA pyramid nanocages
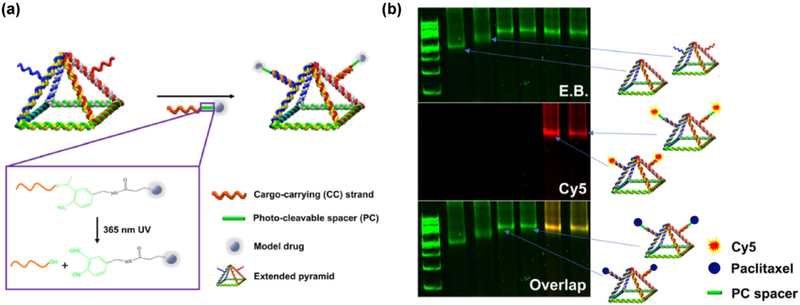
By virtue of the noninvasiveness of light compared to other exogenous stimuli, we chose a light source as a trigger for implementing the controlled release of drugs. Prior to anchoring chemical drugs on RNA nanocages, the Cy5 fluorophore was evaluated as a model drug because of the ease of its release characterization. As illustrated in Fig. 3(a), an extended pyramid was designed based on the aforementioned well-characterized nanocage by extending strands 1 and 3 with the same dangling ends (Table S3 in the ESM). The two sticky ends displayed on the pyramid nanoparticle were hybridized with two short cargo-carrying (CC) strands. Poly-uracil (poly-U) was embedded to provide structural flexibility of the dangling ends. A PC spacer with an o-nitrobenzyl group (highlighted in green in the box in Fig. 3(a)) was incorporated into the short cargo-carrying strand (CC strand and CC-PC strand; Table S3 and Fig. S4(a) in the ESM). Model drugs were conjugated to cargo-carrying strands. Upon irradiation with 365 nm UV light, an n-transition occurs within the o-nitrobenzyl group. The excited singlet state is transformed into a triplet state. An oxygen of the nitro group abstracts a proton from the methylene carbon at the γ-H position, and an aci-nitro intermediate is formed [65]. Subsequently, rearrangement and cleavage of the resonance-stabilized five-membered ring rapidly regenerates a hydroxyl group at the 5′ end of RNA and produces nitroso aldehyde as an additional moiety on the model drug, thus releasing it from the RNA pyramid.
Figure 3.
Design and construction of photoresponsive RNA pyramid nanoparticles. (a) A schematic of the RNA pyramid conjugated with photo-cleavable cargo. (b) Native PAGE imaged with E.B. and Cy5 channels to demonstrate coassembly of Cy5- or paclitaxel-conjugated RNA oligomers into RNA pyramid nanoparticles.
After the synthesis of CC and CC-PC strands with reactive groups at the 5′ end, the model drug was covalently conjugated to the RNA. Two different commonly used bioconjugation chemistries were applied to model drug conjugation. For fluorophore Cy5, an amine modifier was attached at the 5′ end of RNA for labeling via an NHS ester reaction as previously described (Figs. S4(b) and S4(d) in the ESM) [66]. For paclitaxel, an alkyne group was attached to RNA for a click reaction with azido-modified paclitaxel (Figs. S4(c) and S4(e) in the ESM). Urea PAGE characterization corroborated successful model drug conjugation to CC and CC-PC strands by different bioconjugation methods. We confirmed the self-assembly of RNA nanocages harboring paclitaxel with or without the PC spacer by native PAGE in a 6% gel (Fig. 3(b)). The naked pyramid without extension strands (lane 1) showed faster mobility, whereas the extended pyramidal nanocages with dangling strands (lane 2) Manifested slower mobility in gel. Anchoring of chemical drugs (lanes 3 and 4, without or with the PC spacer, respectively) or fluorophores (lanes 5 and 6, without or with the PC spacer, respectively) resulted in an upshift due to increased molecular weight of the nanocages.
2.5. Characterization of the model drug release
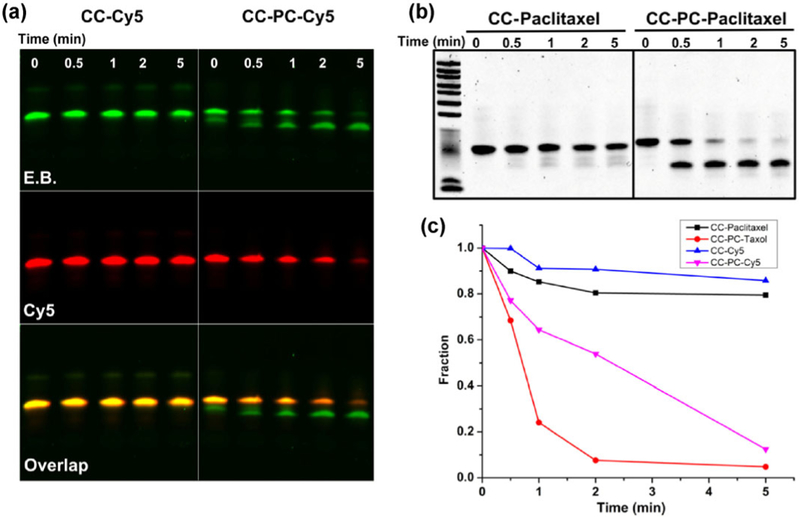
After confirming the successful conjugation, we characterized the release of Cy5 and paclitaxel. Upon irradiation with 365 nm UV light, Cy5 on the RNA strand with the PC spacer was rapidly released within 5 min, whereas its counterpart without the PC spacer did not show a clear release of the fluorophore as characterized by denaturing PAGE (Fig. 4(a)). In the CC-PC-Cy5 moiety, a lower band in the ethidium bromide (E.B.) channel indicated RNA strands with the release of the Cy5 fluorophore that was conjugated to the RNA strands. The Cy5 fluorescence in CC-PC-Cy5 conjugates also decreased due to the cleavage and release. To demonstrate the light-induced release of chemical drugs, paclitaxel was chosen as an example. As in the fluorophore release profile, more than 95% of the conjugated paclitaxel was cleaved from short CC-PC oligos within 5 min of UV irradiation. Due to the loss of the conjugated chemical drug, RNA showed faster electrophoretic mobility during denaturing urea PAGE (Fig. 4(b)). In contrast, RNA–paclitaxel without the PC spacer did not undergo noticeable release of the conjugated drug molecules. Quantification of the fluorescence intensity of Cy5 in the ImageJ software confirmed the on-demand release of cargo from the CC-PC strand upon UV irradiation (Fig. 4(c)).
Figure 4.
The photocontrolled release of Cy5 and paclitaxel from RNA strands. (a) Urea PAGE imaged with E.B. and Cy5 channels showing the release of Cy5 from an RNA strand under UV irradiation. Left panel: An RNA strand conjugated with Cy5 without a PC spacer. Right panel: The release of Cy5 within 5 min from the complex with the PC spacer. (b) Urea PAGE showing the light-triggered release of paclitaxel from CC and CC-PC RNA strands. (c) Cy5 and paclitaxel release profiles of RNA strands with and without PC spacer.
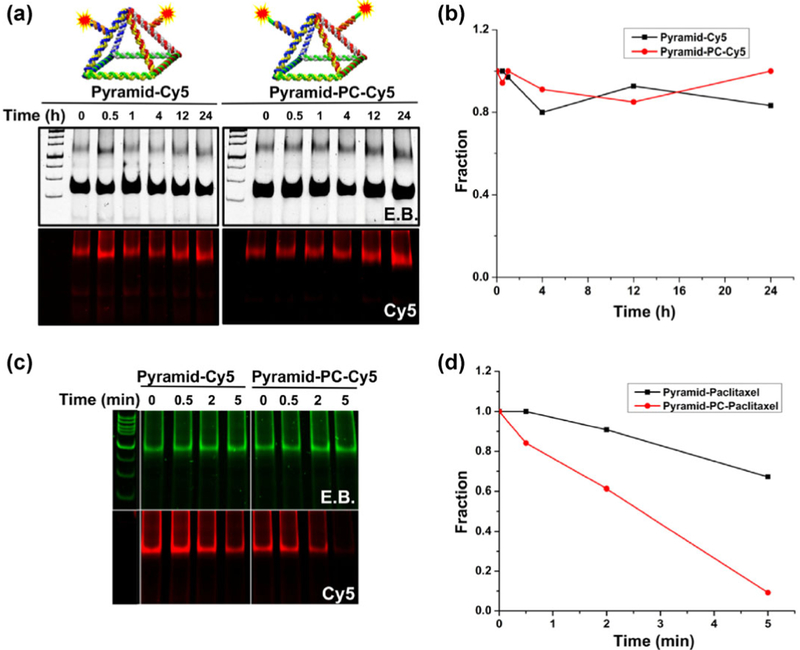
To test the profile of a Cy5 release from the RNA nanocage under physiological conditions without any trigger, we implemented one-pot self-assembly of extended RNA pyramid strands with short RNA oligos; this procedure generated RNA nanocages displaying two Cy5-tethered dangling ends (Fig. 3(b)). We then incubated the RNA nanocages harboring Cy5 with 10% fetal bovine serum (FBS) at 37 °C for different periods (0–24 h). As presented in the native PAGE gel (Fig. 5(a)), an image of the E.B. channel indicated an intact RNA pyramid during 24 h incubation in 10% FBS. No significant degradation was observed for both types of pyramid nanoparticles. In the Cy5 channel, the fluorescent signal from both types of pyramids did not obviously decrease, indicating the stable caging of fluorophores under physiological conditions, as confirmed by the quantification data (Fig. 5(b)). This finding raises a concern that most of the nanocarriers may not be able to release the cargo under physiological conditions especially when the on-demand release is not inducible. To demonstrate feasibility of the controlled release of fluorophores from pyramidal RNA nanocages, UV irradiation was applied for different durations. The two types of RNA nanocages had distinct release profiles. Again, the E.B. channel for native PAGE in the 6% gel did not show an obvious decrease in the signal, indicating the intact RNA pyramid under UV irradiation. In the Cy5 channel, RNA pyramids with the PC spacer (right panel, Pyramid–PC–Cy5) yielded a quick release of fluorophores as suggested by a decreased fluorescent signal (Fig. 5(c)). More than 90% of Cy5 was released within 5 min from Pyramid–PC–Cy5 as quantified in the ImageJ software (Fig. 5(d)). Because the gel mobility shift caused by the release of paclitaxel from pyramid nanoparticles was unnoticeable, the data on the drug release from pyramid nanoparticles are not presented here.
Figure 5.
Comparison of Pyramid–Cy5 and Pyramid–PC–Cy5 in FBS and under UV irradiation. (a) Native PAGE of RNA pyramids harboring the Cy5 fluorophore with (right panel) or without the PC spacer (left panel) after incubation with 10% FBS for 0–24 h. (b) Normalized fluorescence intensity of RNA pyramids indicating the stability of the PC spacer in FBS. (c) Native PAGE imaged with E.B. and Cy5 channels showing RNA pyramids harboring Cy5 with (left panel) or without (right panel) the PC spacer after UV irradiation for 0–5 min. (d) The release profile of Cy5 from RNA pyramids.
2.6. A light-triggered release of paclitaxel for in vitro treatment of breast cancer cells
Covalent attachment of chemotherapeutic drugs provides stable caging and prevents nonspecific diffusion from the carrier; however, there are concerns as to whether the drugs conjugated via a cross-linker retain their biological activity. To verify the cytotoxicity of paclitaxel conjugated to RNA pyramid nanoparticles, an MTT assay was performed on breast cancer cells to assess the cell viability after treatment. The RNA pyramid nanoparticles harboring paclitaxel with or without the PC spacer caused a certain degree of inhibition of tumor cell growth without UV irradiation. We expected that the cytotoxicity would be caused by paclitaxel slowly being released from the RNA nanoparticles owing to intracellular RNA degradation. The cytotoxicity of pyramid nanoparticles harboring paclitaxel increased in a concentration-dependent manner. RNA pyramid nanoparticles themselves without paclitaxel did not have obvious cytotoxicity, considering the fact that paclitaxel-conjugated RNA pyramids with or without the PC spacer had similar inhibitory effects on cancer cell growth (Fig. S5(a) in the ESM). These results suggested that as a prodrug, paclitaxel retains its activity after conjugation to the RNA and RNA pyramid.
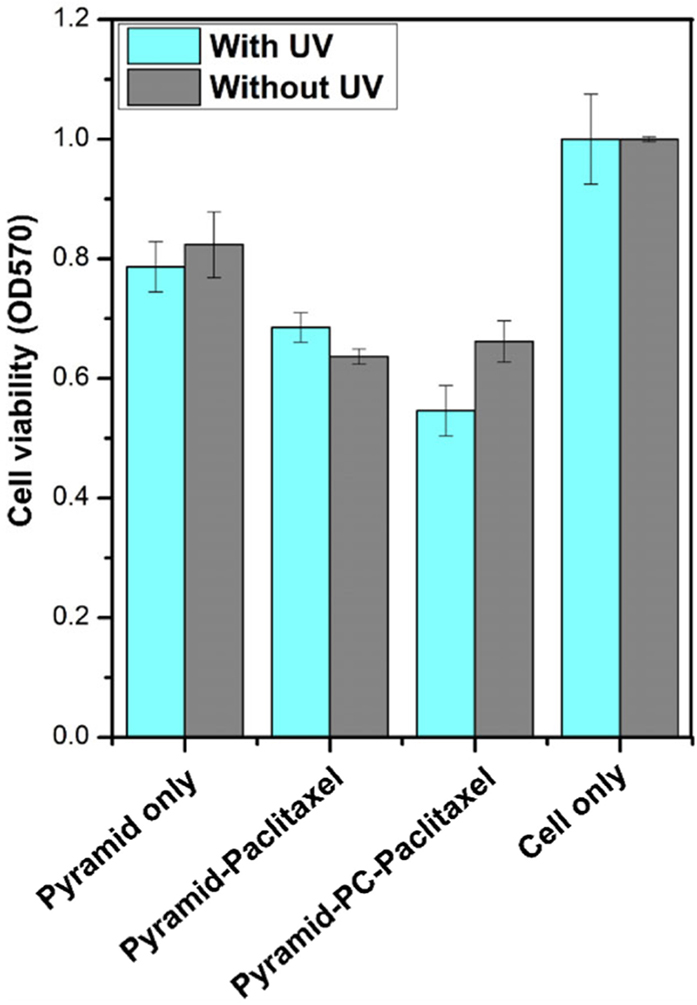
To further compare the paclitaxel release from pyramid nano-particles with or without the PC spacer in the presence of UV light, the MTT assay was performed to verify the release of paclitaxel (Fig. 6). Both pyramid–paclitaxel and pyramid–PC–paclitaxel at 400 nM exerted cytotoxicity after 72 h incubation with MDA-MB-231 cells because of the presence of drug molecules. Treatment with pyramid–paclitaxel without the PC spacer showed no obvious difference in cell viability with or without UV irradiation. In contrast, pyramid–PC–paclitaxel induced higher cytotoxicity with the UV trigger, thus pointing to the UV-light–facilitated release of paclitaxel from the pyramid nanoparticles.
Figure 6.
The cytotoxic effect of the RNA pyramid nanoparticles. Pyramid–PC–paclitaxel exerted higher cytotoxicity after UV irradiation showing the controlled release, whereas pyramid–paclitaxel without PC showed no enhancement. Error bars represent standard deviation.
3. Discussion
RNA, as a unique polymer, has emerged as an attractive material for nanobiotechnology [25, 26, 67, 68]. Despite the success of various 2D RNA-based nanodelivery systems in animal studies, development of 3D RNA nanocages for efficacious controlled release of cargo is still intriguing [37, 38]. The stochastic ligand–receptor interaction and an incontrollable drug release in most delivery systems make it difficult to achieve specific delivery with precise spatiotemporal and dosage control. Herein, we report fabrication of 3D pyramid-shaped RNA nanocages as a platform for the controllable drug release. The newly developed RNA pyramids were demonstrated to form well-defined and stable structures. DLS and cryo-EM data show that the nanocages are homogeneous in size and have the predicted shape. Furthermore, thermostability assays revealed that the pyramids are stable well beyond the 37 °C requirement for in vivo applications. The light-controlled release of paclitaxel was found to effectively increase cytotoxicity toward breast cancer cells.
Some technologies in cancer therapeutics that do not allow for the on-demand delivery of chemotherapeutic agents can result in severe side effects and toxicities caused by nonspecific diffusion and release. This phenomenon is evidenced in FDA-approved paclitaxel formulations, owing to a lack of a controlled release. For the first time, we engineered RNA nanocages with a stimuli-responsive property that allows for a controlled release of their cargo, including paclitaxel. Via strategic positioning of PC o-NB groups in RNA strands, the light-induced liberation of attached cargos was demonstrated. Upon exposure to UV light (λ = 365 nm), the covalently attached cargos were efficiently released from RNA nanocages on demand and were completely liberated within a relatively short period. Although UV light may not be amenable to in vivo applications because of its poor tissue penetration, the UV light source can be replaced by a less attenuated light source such as near-infrared light, along with utilization of a two-photon photoactivation molecule.
This novel RNA nanocage further extends the field of RNA nanotechnology, via an alternative delivery mechanism as compared to that previously seen in RNA nanoparticles [35, 36]. Although future studies are necessary to examine the suitability of paclitaxel-labeled RNA nanoparticles for treatment of cancers, here we provide proof of concept by creating a new therapeutic platform involving RNA nanoparticles as a photostimulated material. Adjustments to the platform may be required for in vivo applications for a longer-wavelength light source; however, similar principles of the particle design and construction remain valid.
In summary, we fabricated a novel RNA nanocage capable of releasing compounds in response to UV irradiation. The nanocages were found to have a high degree of programmability, which could accommodate a larger variety of biomolecules, thereby providing a promising platform for stimuli-responsive drug delivery. We believe that in the coming years, many innovative breakthroughs and discoveries will continue to happen in the emerging field of RNA nanotechnology–based smart nanoplatforms for improved cancer therapy.
4. Experimental
4.1. RNA pyramid design, preparation, and self-assembly
Synthetic DNA molecules were purchased from Integrated DNA Technologies and amplified with primers containing the T7 RNA polymerase promoter sequence. All RNA strands were prepared by in vitro transcription of amplified DNA templates using Y693F mutant T7 polymerase and 2′-F modified cytosine (C) and uracil (U) nucleotides followed by purification via 8 M Urea 8% denaturing PAGE. Equimolar concentrations of five strands (1 μM) were mixed in 1× Tris buffer (100 mM NaCl, 50 mM Tris pH 8.0) for bottom-up self-assembly of RNA pyramids. The one-pot assembly was achieved by heating the strands at 85 °C for 5 min and slowly cooling them down (during 45 min) to 4 °C on an Eppendorf Mastercycle thermocycler. Then, the assembled nanoparticles were analyzed by native PAGE.
4.2. Cryo-EM imaging
Two microliters of an RNA pyramid nanoparticle solution (2 μM) was applied onto a glow-discharged 200-mesh R1.2/1.3 Quantifoil grid. The grids were blotted for 1.5 s and rapidly frozen in liquid ethane using a Vitrobot Mark IV (FEI). Next, the girds were transferred to a JEM2200FS cryo-electron microscope (JEOL) operated at 200 kV with an in-column energy filter with a slit of 20 eV for screening. Micrographs of the RNA pyramid nanoparticles were captured by a 4K × 4K CCD camera (Gatan) at 80,000× magnification (corresponding to a calibrated sampling of 1.36 Å per pixel) and a dose rate of ~ 30 electrons per second per Å2 with a total exposure time of 1.5 s. A total of 27 images were acquired with a defocus range of 2–4 μm.
4.3. Synthesis of short RNA oligos with the PC spacer and functional groups
RNA synthesis was performed at a 200 nmol scale on an automated oligo synthesizer. Universal 1,000 Å controlled pore glass solid support and standard reagents including 2′-F modified RNA phosphoramidites, PC spacer phosphoramidite (Cat. # 10-4913-90), 5′-hexynyl phosphoramidite (Cat. # 10-1908-90), and 5′-amino-modifier C6-PDA (Cat. # 10-1947-90) were acquired from Glen Research. Sequences of the short RNA oligomers are listed in the 5′-to-3′ orientation (“r” denotes a 2′-OH base, “f” indicates a 2′-F-modified base, and “PC” represents the photocleavable spacer). For fluorophore conjugation, 5′-amino modifier C6-PDA was used to modify the strand with a primary amine group for a reaction with Cyanine 5 NHS ester. For paclitaxel conjugation, 5′-hexynyl phosphoramidite was employed to modify the strand with an alkyne group for a click reaction with paclitaxel-N3. After the synthesis, the oligomers were deprotected and desalted by conventional methods. Each strand was synthesized using 2′-fluorinated cytidine and uracil. Urea PAGE in a 16% gel was conducted to characterize the synthesized strands.
4.4. UV irradiation–induced cargo release
A hand-held low-power UV lamp served as a light source (4 W, 0.5 mW·cm−2, irradiation at 365 nm), and 10 μL of RNA oligomers (3 μM) and self-assembled RNA nanocages (1 μM) with or without PC spacers were irradiated for different durations. The samples after UV irradiation were kept on ice in the dark. RNA oligomer samples were analyzed by urea PAGE in a 16% gel. All RNA nanocage samples were analyzed by native PAGE in a 6% gel. The Cy5-labeled RNA and RNA nanocages were analyzed on the Typhoon fluorescent imaging system in the Cy5 and E.B. channels. Quantitative analysis was performed in the ImageJ software. Equal-sized boxes were drawn around the lanes corresponding to the square complexes, and corresponding quantified values for each hybrid square were divided by the sum of the values presented in the corresponding lane. Plots were generated in Origin 8.0 software.
4.5. MTT assay
To assay the toxicity of pyramid–paclitaxel and pyramid–PC–paclitaxel nanoparticles toward breast cancer MDA-MB-231 cells, the CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega) was carried out to evaluate cell viability changes. Briefly, 5 × 103 MDA-MB-231 breast cancer cells were seeded in 96-well plates a day prior to the assay. On the second day, pyramid–paclitaxel or pyramid–PC–paclitaxel nanoparticles were added into the wells at a final concentration of 50, 100, 200, or 400 nM in triplicate. Paclitaxel concentration was calculated as the double RNA nanoparticle concentration based on the stoichiometric ratio. Unconjugated pyramid nanoparticles served as a control at the same testing concentrations. The plate was then incubated at 37 °C for 48 h in a humidified atmosphere containing 5% of CO2. After incubation, 15 μL of the Dye Solution was added into each well, and the plate was incubated at 37 °C for up to 4 h in the humidified atmosphere containing 5% of CO2. Next, 100 μL of the Solubilization Solution/Stop Mix was added into each well and incubated for 2 h. Finally, the contents of the wells were mixed to get a uniformly colored solution and their absorbance at 570 nm was recorded on a Synergy 4 microplate reader (Bio-Tek).
To further verify the facilitated release of paclitaxel from pyramid–PC–paclitaxel nanoparticles upon UV irradiation, 400 nM concentration of each RNA sample was chosen for the assay after 72 h incubation with the cells. In the UV light treatment plate, each well was irradiated with 365 nm UV light from a handheld UV lamp for 15 min.
Supplementary Material
Acknowledgements
The research in P. G.’s lab was supported by NIH grants R01EB019036, R01CA186100 and U01CA207946 to Peixuan Guo. The cryo-EM work was supported by NIH grants P41GM103832 (W. C.) and P50 GM103297 (W. C.). P. G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. We would like to thank Dr. Farzin Haque and Dr. Daniel Jasinski for helpful discussions. We also would like to thank Dr. Yi Shu for synthesis of paclitaxel-N3.
Footnotes
Conflict of interests
P. G. is the cofounder of ExonanoRNA LLC. He is also the consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd, as well as the cofounder of Shenzhen P&Z Bio-medical Co. Ltd and its subsidiary US P&Z Biological Technology LLC. His inventions at the University of Kentucky have been licensed to Matt Holding and Nanobio Delivery Pharmaceutical Co., Ltd. His inventions at the Ohio State University have been licensed to ExonanoRNA LLC.
Electronic Supplementary Material: Supplementary material (RNA sequences, RNA nanoparticle characterization and conjugation characterization) is available in the online version of this article at https://doi.org/10.1007/s12274-018-2174-x.
References
- [1].Karimi M; Ghasemi A; Sahandi ZP; Rahighi R; Moosavi Basri SM; Mirshekari H; Amiri M; Shafaei PZ; Aslani A; Bozorgomid M et al. Smart micro/nanoparticles in stimulus-responsive drug/gene delivery systems. Chem. Soc. Rev 2016, 45, 1457–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hoffman AS Stimuli-responsive polymers: Biomedical applications and challenges for clinical translation. Adv. Drug Deliv. Rev 2013, 65, 10–16. [DOI] [PubMed] [Google Scholar]
- [3].Blum AP; Kammeyer JK; Rush AM; Callmann CE; Hahn ME; Gianneschi NC Stimuli-responsive nanomaterials for biomedical applications. J. Am. Chem. Soc 2015, 137, 2140–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rahoui N; Jiang B; Taloub N; Huang YD Spatio-temporal control strategy of drug delivery systems based nano structures. J. Control. Release 2017, 255, 176–201. [DOI] [PubMed] [Google Scholar]
- [5].Liu D; Yang F; Xiong F; Gu N The smart drug delivery system and its clinical potential. Theranostics 2016, 6, 1306–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kahn JS; Hu YW; Willner I Stimuli-responsive DNA-based hydrogels: From basic principles to applications. Acc. Chem. Res 2017, 50, 680–690. [DOI] [PubMed] [Google Scholar]
- [7].Mura S; Nicolas J; Couvreur P Stimuli-responsive nanocarriers for drug delivery. Nat. Mater 2013, 12, 991–1003. [DOI] [PubMed] [Google Scholar]
- [8].Torchilin VP Multifunctional, stimuli-sensitive nanoparticulate systems for drug delivery. Nat. Rev. Drug Discov 2014, 13, 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lin ZQ; Gao W; Hu HX; Ma K; He B; Dai WB; Wang XQ; Wang JC; Zhang X; Zhang Q Novel thermo-sensitive hydrogel system with paclitaxel nanocrystals: High drug-loading, sustained drug release and extended local retention guaranteeing better efficacy and lower toxicity. J. Control. Release 2014, 174, 161–170. [DOI] [PubMed] [Google Scholar]
- [10].Rapoport NY; Kennedy AM; Shea JE; Scaife CL; Nam KH Controlled and targeted tumor chemotherapy by ultrasound-activated nanoemulsions/microbubbles. J. Control. Release 2009, 138, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Alam MM; Han HS; Sung S; Kang JH; Sa KH; Al Faruque H; Hong J; Nam EJ; Kim IS; Park JH et al. Endogenous inspired biomineral-installed hyaluronan nanoparticles as pH-responsive carrier of methotrexate for rheumatoid arthritis. J. Control. Release 2017, 252, 62–72. [DOI] [PubMed] [Google Scholar]
- [12].Li J; Huo MR; Wang J; Zhou JP; Mohammad JM; Zhang YL; Zhu QN; Waddad AY; Zhang Q Redox-sensitive micelles self-assembled from amphiphilic hyaluronic acid-deoxycholic acid conjugates for targeted intracellular delivery of paclitaxel. Biomaterials 2012, 33, 2310–2320. [DOI] [PubMed] [Google Scholar]
- [13].Veetil AT; Chakraborty K; Xiao K; Minter MR; Sisodia SS; Krishnan Y Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat. Nanotechnol 2017, 12, 1183–1189. [DOI] [PubMed] [Google Scholar]
- [14].Spring BQ; Bryan SR; Zheng LZ; Mai ZM; Watanabe R; Sherwood ME; Schoenfeld DA; Pogue BW; Pereira SP; Villa E et al. A photoactivable multi-inhibitor nanoliposome for tumour control and simultaneous inhibition of treatment escape pathways. Nat. Nanotechnol 2016, 11, 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Karimi M; Sahandi ZP; Baghaee-Ravari S; Ghazadeh M; Mirshekari H; Hamblin MR Smart nanostructures for cargo delivery: Uncaging and activating by light. J. Am. Chem. Soc 2017, 139, 4584–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bansal A; Zhang Y Photocontrolled nanoparticle delivery systems for biomedical applications. Acc. Chem. Res 2014, 47, 3052–3060. [DOI] [PubMed] [Google Scholar]
- [17].Wang YY; Deng YB; Luo HH; Zhu AJ; Ke HT; Yang H; Chen HB Light-responsive nanoparticles for highly efficient cytoplasmic delivery of anticancer agents. ACS Nano 2017, 11, 12134–12144. [DOI] [PubMed] [Google Scholar]
- [18].Shim G; Ko S; Kim D; Le QV; Park GT; Lee J; Kwon T; Choi HG; Kim YB; Oh YK Light-switchable systems for remotely controlled drug delivery. J. Control. Release 2017, 267, 67–79. [DOI] [PubMed] [Google Scholar]
- [19].Kohman RE; Cha SS; Man HY; Han X Light-triggered release of bioactive molecules from DNA nanostructures. Nano Lett. 2016, 16, 2781–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Geng SY; Wang YZ; Wang LP; Kouyama T; Gotoh T; Wada S; Wang JY A light-responsive self-assembly formed by a cationic azobenzene derivative and SDS as a drug delivery system. Sci. Rep 2017, 7, 39202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Basuki JS; Qie FX; Mulet X; Suryadinata R; Vashi AV; Peng YY; Li LL; Hao XJ; Tan TW; Hughes TC Photo-modulated therapeutic protein release from a hydrogel depot using visible light. Angew. Chem., Int. Ed 2017, 56, 966–971. [DOI] [PubMed] [Google Scholar]
- [22].Lajunen T; Nurmi R; Kontturi L; Viitala L; Yliperttula M; Murtomaki L; Urtti A Light activated liposomes: Functionality and prospects in ocular drug delivery. J. Control. Release 2016, 244, 157–166. [DOI] [PubMed] [Google Scholar]
- [23].Sun WJ; Jiang TY; Lu Y; Reiff M; Mo R; Gu Z Cocoon-like self-degradable DNA nanoclew for anticancer drug delivery. J. Am. Chem. Soc 2014, 136, 14722–14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun WJ; Ji WY; Hall JM; Hu QY; Wang C; Beisel CL; Gu Z Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem., Int. Ed 2015, 54, 12029–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shu Y; Pi FM; Sharma A; Rajabi M; Haque F; Shu D; Leggas M; Evers BM; Guo PX Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv. Drug Deliv. Rev 2014, 66, 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jasinski D; Haque F; Binzel DW; Guo PX Advancement of the emerging field of RNA nanotechnology. ACS Nano 2017, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li H; Lee T; Dziubla T; Pi FM; Guo SJ; Xu J; Li C; Haque F; Liang XJ; Guo PX RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today 2015, 10, 631–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shu Y; Haque F; Shu D; Li W; Zhu Z; Kotb M; Lyubchenko Y; Guo P Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA 2013, 19, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 2012,7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shu D; Shu Y; Haque F; Abdelmawla S; Guo PX Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol 2011, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Piao XJ; Wang HZ; Binzel DW; Guo PX Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2’-F RNA, and LNA in the context of Phi29 pRNA 3WJ. RNA 2018, 24, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Binzel D; Shu Y; Li H; Sun MY; Zhang QS; Shu D; Guo B; Guo PX Specific delivery of MiRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol. Ther 2016, 24, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Binzel DW; Khisamutdinov EF; Guo PX Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry 2014, 53, 2221–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jasinski DL; Khisamutdinov EF; Lyubchenko YL; Guo PX Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano 2014, 8, 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shu D; Li H; Shu Y; Xiong GF; Carson WE; Haque F; Xu R; Guo PX Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano 2015, 9, 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cui DX; Zhang CL; Liu B; Shu Y; Du T; Shu D; Wang K; Dai FP; Liu YL; Li C et al. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Sci. Rep 2015, 5, 10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Khisamutdinov EF; Jasinski DL; Li H; Zhang KM; Chiu W; Guo PX Fabrication of RNA 3D nanoprisms for loading and protection of small RNAs and model drugs. Adv. Mater 2016, 28, 10079–10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li H; Zhang KM; Pi FM; Guo SJ; Shlyakhtenko L; Chiu W; Shu D; Guo PX Controllable self-assembly of RNA tetrahedrons with precise shape and size for cancer targeting. Adv. Mater 2016, 28, 7501–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xu CC; Haque F; Jasinski DL; Binzel DW; Shu D; Guo PX Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy. Cancer Lett. 2018, 414, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wani MC; Taylor HL; Wall ME; Coggon P; McPhail AT Plant antitumor agents. VI. Isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc 1971, 93, 2325–2327. [DOI] [PubMed] [Google Scholar]
- [41].Spencer CM; Faulds D Paclitaxel. Drugs 1994, 48, 794–847. [DOI] [PubMed] [Google Scholar]
- [42].Rowinsky EK; Donehower RC Paclitaxel (Taxol). N. Engl. J. Med 1995, 332, 1004–1014. [DOI] [PubMed] [Google Scholar]
- [43].Horwitz SB Mechanism of action of Taxol. Trends Pharmacol. Sci 1992, 13, 134–136. [DOI] [PubMed] [Google Scholar]
- [44].Singla AK; Garg A; Aggarwal D Paclitaxel and its formulations. Int. J. Pharm 2002, 235, 179–192. [DOI] [PubMed] [Google Scholar]
- [45].Gelderblom H; Verweij J; Nooter K; Sparreboom A Cremophor EL: The drawbacks and advantages of vehicle selection for drug formulation. Eur. J. Cancer 2001, 37, 1590–1598. [DOI] [PubMed] [Google Scholar]
- [46].Shu Y, Yin H, Rajabi M, Li H, Vieweger M, Guo S, Shu D, Guo P. RNA-based micelles: A novel platform for paclitaxel loading and delivery. J. Control. Release, 2018, 14, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim SC; Kim DW; Shim YH; Bang JS; Oh HS; Kim SW; Seo MH In vivo evaluation of polymeric micellar paclitaxel formulation: Toxicity and efficacy. J. Control. Release 2001, 72, 191–202. [DOI] [PubMed] [Google Scholar]
- [48].Bedikian AY; Plager C; Papadopoulos N; Eton O; Ellerhorst J; Smith T Phase II evaluation of paclitaxel by short intravenous infusion in metastatic melanoma. Melanoma Res. 2004, 14, 63–66. [DOI] [PubMed] [Google Scholar]
- [49].Hwu JR; Lin YS; Josephrajan T; Hsu MH; Cheng FY; Yeh CS; Su WC; Shieh DB Targeted paclitaxel by conjugation to iron oxide and gold nanoparticles. J. Am. Chem. Soc 2009, 131, 66–68. [DOI] [PubMed] [Google Scholar]
- [50].Yoshizawa Y; Kono Y; Ogawara K; Kimura T; Higaki K PEG liposomalization of paclitaxel improved its in vivo disposition and anti-tumor efficacy. Int. J. Pharm 2011, 412, 132–141. [DOI] [PubMed] [Google Scholar]
- [51].Hamaguchi T; Kato K; Yasui H; Morizane C; Ikeda M; Ueno H; Muro K; Yamada Y; Okusaka T; Shirao K et al. A phase I and pharmacokinetic study of NK105, a paclitaxel-incorporating micellar nanoparticle formulation. Br. J. Cancer 2007, 97, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lay CL; Liu HQ; Tan HR; Liu Y Delivery of paclitaxel by physically loading onto poly(ethylene glycol) (PEG)-graftcarbon nanotubes for potent cancer therapeutics. Nanotechnology 2010, 21, 065101. [DOI] [PubMed] [Google Scholar]
- [53].Deng JX; Huang L; Liu F Understanding the structure and stability of paclitaxel nanocrystals. Int. J. Pharm 2010, 390, 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Walter F; Murchie AI; Lilley DMJ Folding of the four-way RNA junction of the hairpin ribozyme. Biochemistry 1998, 37, 17629–17636. [DOI] [PubMed] [Google Scholar]
- [55].Binzel DW; Khisamutdinov E; Vieweger M; Ortega J; Li JY; Guo PX Mechanism of three-component collision to produce ultrastable pRNA three-way junction of Phi29 DNA-packaging motor by kinetic assessment. RNA 2016, 22, 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Benkato K; O’Brien B; Bui MN; Jasinski DL; Guo PX; Khisamutdinov EF Evaluation of thermal stability of RNA nanoparticles by temperature gradient gel electrophoresis (TGGE) in native condition In RNA Nanostructures. Methods in Molecular Biology, vol 1632 Bindewald E; Shapiro B, Eds.; Humana Press: New York, NY, 2017; pp 123–133. [DOI] [PubMed] [Google Scholar]
- [57].Lee TJ; Haque F; Shu D; Yoo JY; Li H; Yokel RA; Horbinski C; Kim TH; Kim SH; Kwon CH et al. RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget 2015, 6, 14766–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tiemann K; Rossi JJ RNAi-based therapeutics–current status, challenges and prospects. EMBO Mol. Med 2009, 1, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shu D; Khisamutdinov EF; Zhang L; Guo PX Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor. Nucleic Acids Res. 2014, 42, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kolpashchikov DM Binary malachite green aptamer for fluorescent detection of nucleic acids. J. Am. Chem. Soc 2005, 127, 12442–12443. [DOI] [PubMed] [Google Scholar]
- [61].Pothoulakis G; Ceroni F; Reeve B; Ellis T The spinach RNA aptamer as a characterization tool for synthetic biology. ACS Synth. Biol 2014, 3, 182–187. [DOI] [PubMed] [Google Scholar]
- [62].Sassanfar M; Szostak JW An RNA motif that binds ATP. Nature 1993, 364, 550–553. [DOI] [PubMed] [Google Scholar]
- [63].Srisawat C; Engelke DR Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA 2001, 7, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hoeprich S; Zhou Q; Guo S; Qi G; Wang Y; Guo P Bacterial virus Phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus. Gene Ther. 2003, 10, 1258–1267. [DOI] [PubMed] [Google Scholar]
- [65].Gaplovsky M; Il’ichev YV; Kamdzhilov Y; Kombarova SV; Mac M; Schwörer MA; Wirz J Photochemical reaction mechanisms of 2-nitrobenzyl compounds: 2-Nitrobenzyl alcohols form 2-nitroso hydrates by dual proton transfer. Photochem. Photobiol. Sci 2005, 4, 33–42. [DOI] [PubMed] [Google Scholar]
- [66].Jasinski DL; Yin HR; Li ZF; Guo PX The hydrophobic effect from conjugated chemicals or drugs on in vivo biodistribution of RNA nanoparticles. Hum. Gene Ther, in press, DOI: 10.1089/hum.2017.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pi F; Binzel D; Lee TJ;Li Z; Sun M; Rychahou P; Li H; Haque F; Wang S; Croce CM et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol 2018. , 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.