Abstract
RNA is a versatile biomaterial that can be used to engineer nanoassemblies for personalized treatment of various diseases. Despite promising advancements, the design of RNA nanoassemblies with minimal recognition by the immune system remains a major challenge. Here, an approach is reported to engineer RNA fibrous structures to operate as a customizable platform for efficient coordination of siRNAs and for maintaining low immunostimulation. Functional RNA fibers are studied in silico and their formation is confirmed by various experimental techniques and visualized by atomic force microscopy (AFM). It is demonstrated that the RNA fibers offer multiple advantages among which are: i) programmability and modular design that allow for simultaneous controlled delivery of multiple siRNAs and fluorophores, ii) reduced immunostimulation when compared to other programmable RNA nanoassemblies, and iii) simple production protocol for endotoxin-free fibers with the option of their cotranscriptional assembly. Furthermore, it is shown that functional RNA fibers can be efficiently delivered with various organic and inorganic carriers while retaining their structural integrity in cells. Specific gene silencing triggered by RNA fibers is assessed in human breast cancer and melanoma cell lines, with the confirmed ability of functional fibers to selectively target single nucleotide mutations.
Keywords: endotoxin-free fibers, immunology, RNA interference, RNA nanotechnology, therapeutic nucleic acids
1. Introduction
RNA regulates a myriad of biological processes at different levels. RNA interference (RNAi),[1] for instance, is one of the therapeutically relevant pathways that allows the regulation of gene expression using exogenous RNAs. Notably, the very first therapy based on RNAi has just been approved by FDA.[2] Aside from synthetic RNAi inducers, several other promising classes of therapeutic nucleic acids (TNAs) have been developed, such as antisense oligos, aptamers, ribozymes, and mRNAs.[3,4] TNAs are being increasingly considered for the treatment of a wide variety of conditions, including cancers, metabolic disorders, viral infections, cardiovascular disorders, and inflammatory diseases, especially where traditional small molecule drugs fail.[5]
The simultaneous use of multiple TNAs is anticipated to have significant synergistic effects. One example is combinatorial RNAi, used for the simultaneous suppression of multiple genes, which in the case of HIV prevents the possibility of its mutation-assisted escape from treatment.[6] If a combination of TNAs is chosen to be used, its optimal delivery formulation could be achieved via programmable nucleic acid assemblies[7–10] that support the localization of the therapeutic elements with precise composition and stoichiometry.[11–16] Based on crystal and NMR structures of natural RNAs, as well as general knowledge about RNAs’ folding and interacting principles, different RNA motifs with specific tertiary structures can be combined to form RNA assemblies of nanometer size.[17,18] Programmable RNA nanoassemblies can be further functionalized with aptamers, fluorescent dyes, proteins, and siRNAs[10,11,13,14,19–24] resulting in a new class of TNAs, called “nano-TNAs.” These nano-TNAs can then utilize intracellular biochemical processes, such as RNAi, to down-regulate expression of specific genes, as was confirmed by numerous animal studies.[11,12,15,21,25] However, the clinical translation of TNAs in general and nano-TNAs in particular, is commonly complicated by dose-limiting toxicities related to nucleic acids recognition by the immune system.[26–29]
It has become apparent that the immunomodulatory effects of nano-TNAs are largely unknown and the designs of new RNA nanoassemblies must be characterized, in terms of their immunotoxicity, to further facilitate their successful development. A common roadblock for traditional TNAs is the induction of proinflammatory responses with fever and fever-like reactions.[26,29] Many host cells, both immune and nonimmune, have a variety of receptors that can sense nucleic acids and initiate an inflammatory response.[28] Various strategies have been developed to decrease the immunological recognition of traditional TNAs, including backbone and 2′-OMe modification of nucleotides, as well as the removal of 5′-triphosphates.[30–34] However, the applicability of these strategies to nano-TNAs has never been fully investigated.
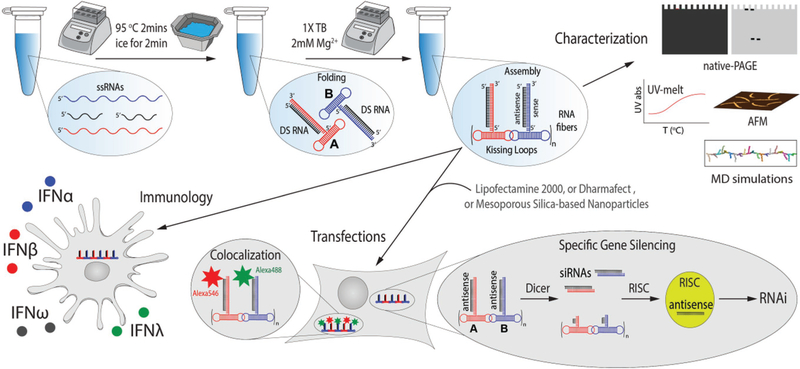
In recent studies, we found that, in general, fibrous RNA nanoassemblies are less proinflammatory than planar or globular RNA nanostructures.[35] For this reason, we set out to explore the use of RNA fibers[36] for the coordinated delivery of siRNAs (Figure 1). In addition, we investigate how the functionalization of RNA fibers with siRNAs affect their immune recognition and compare it to previously characterized functional RNA nanoparticles (globular cubes[22] and planar rings[11]). The overall goal of this study is to determine the conditions for efficacious siRNA delivery without inducing overwhelming proinflammatory responses.
Figure 1.
Schematics of assembly, physicochemical characterization, and further assessment of biological activity of functional RNA fibers.
In our earlier work, we demonstrated that type I interferon (IFN) response can be used as a consistent marker of nano-TNA immunorecognition.[22,37,38] As result, this analysis is used as a foundation to assess proinflammatory response of RNA fibers by measuring the induction of IFNs in normal human peripheral blood mononuclear cells (PBMC), freshly collected from healthy donors. Additionally, we show that programmable RNA fibers can be optimized to achieve efficient delivery of siRNAs without amplifying the undesirable proinflammatory response.
2. Results and Discussion
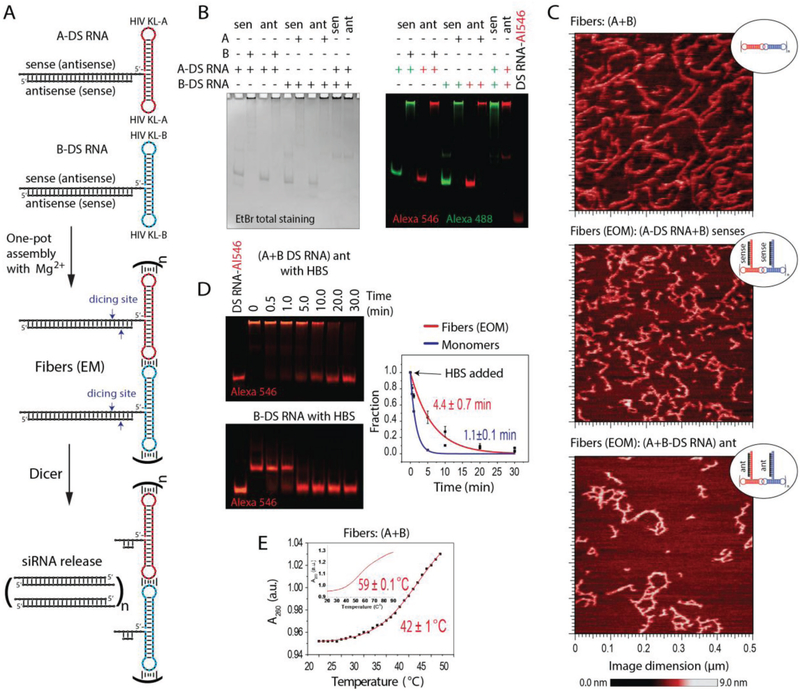
The assembly of RNA fibers is promoted by HIV-like (≈180°) kissing loop interactions.[39,40] We have developed an experimental scheme[36] that allows each kissing loop, positioned at the side of the dumbbell-shaped hairpins (here named A and B), to be programmed to interact with its cognate partner and form RNA fibers with an A/B repeating unit. The RNA fibers can be easily formed by mixing the individual strands prior to a simple one-pot assembly protocol.[41] The A/B self-assembling system provides programmable building blocks to which different functionalities can be added. As illustrated in Figure 1, fiber monomer can be decorated, for example, with a therapeutic Dicer substrate (DS) RNAs.[42] This functionalization is achieved through the extension of the 3′-ends of individual monomers with either DS RNA sense or antisense strands bound to their complementary strands. Once inside the cells, DS RNAs are recognized and cleaved by Dicer, an endoribonuclease involved in the RNAi pathway, producing functional siRNAs that further trigger the silencing of specific genes.[8] The functionalized monomers (A or B) will be later referred to as (A (or B)-DS RNA) sense (or antisense). To study the physical impact of the functional groups, we compare RNA fibers with and without attached DS RNAs. We also designed RNA fibers functionalized with scrambled DS RNA sequences, which are to be used as negative controls in all gene silencing experiments.
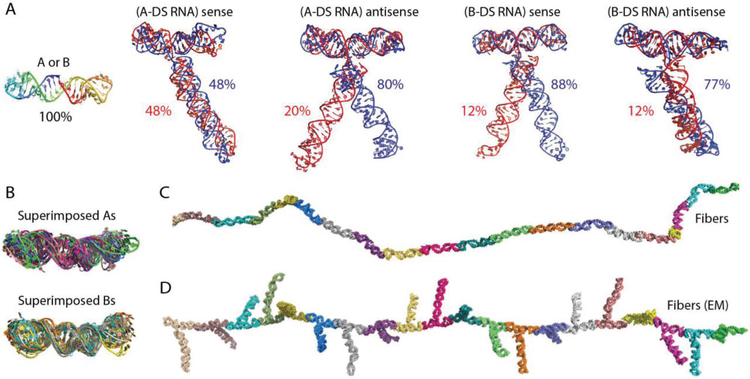
We used discrete molecular dynamics (DMD) simulations to model the structure and dynamics of RNA monomers and RNA fibers.[43–47] This analysis confirmed that A and B monomers form dumbbell structures with two arms at a 180° orientation (Figure 2A,B). This orientation is also preserved in the monomers with functional groups (Figure 2A). Clustering analysis of selected minimal energy structures revealed that in most of the structures the functional groups rest on the N-terminal part of the dumbbell. We observed only two minor clusters for (A-DS RNA) sense and for (B-DS RNA) antisense with functional groups resting on the opposite side of the dumbbell, representing 20% and 12% of all structures respectively. For (A-DS RNA) sense and (B-DS RNA) antisense, we observed two clusters with functional groups twisted around their helical axes (Figure 2A). By running equilibrium DMD simulations, we assessed the dynamics of functionalized and nonfunctionalized RNA fibers. We observed that the bending of fibers predominantly occurs due to the flexibility of hairpin loops at the ends of the dumbbell structures, and not due to the opening of the dumbbell structure in the middle (Figure 2C,D and Videos S1 and S2, Supporting Information). This observation was further confirmed by superimposition of selected minimal energy structures that align well in the middle of the dumbbell but not at the ends (Figure 2B).
Figure 2.
In silico characterization of RNA fibers. Molecular dynamic (MD) simulations of A) nonfunctionalized and functionalized with Dicer substrate RNAs (DS RNAs) monomers, with the most stable conformation shown. B) Superimposed monomers. Single snapshot of trajectory of C) nonfunctionalized RNA fiber and D) RNA fiber functionalized with Dicer substrate RNAs at every monomer (EM).
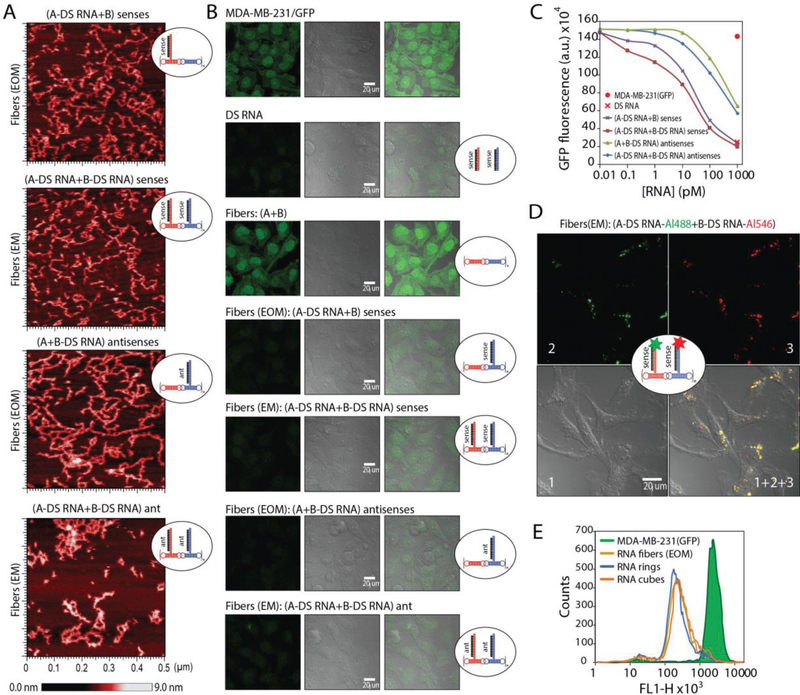
All assemblies were confirmed by nondenaturing polyacrylamide gel electrophoresis (native-PAGE) and visualized by atomic force microscopy (AFM) (Figure 3 and Figure S1, Supporting Information). Due to the possibility of alternative stacking, the DS RNA-functionalized fibers become prone to structural kinks, yet still maintain the fibrous structure. The removal of three nucleotide single-stranded linkers separating DS RNAs from the fibers promotes the formation of ring-like structures observed by AFM (Figure S2, Supporting Information); however, only the fibers with linkers are the focus of this study.
Figure 3.
In vitro characterization of RNA fibers. EM and EOM denote functionalization at every monomer and every other monomer of A/B fibers, respectively. A) Schematic of functionalized RNA fibers assembly and intracellular siRNA release through dicing. B) Native-PAGE, confirming the successful formation of fibers. “green +” – Alexa 488 labeled; “red +” – Alexa 546 labeled. C) AFM images showing fibers with and without DS RNA designed against GFP. D) Relative blood stabilities of functionalized fibers and individual monomers analyzed by native-PAGE gel. Error bars are ±SEM (N = 3). E) Melting temperatures measured for RNA fibers via UV-melt.
We found that RNA fibers are more resistant to nuclease degradation in human blood serum than individual monomers used to prepare these fibers (Figure 3D). The half-lives, as measured by blood stability assay, were t1/2 = 4.4 ± 0.7 min and t1/2 = 1.1 ± 0.15 min for fibers and monomers, respectively. Even with the use of a delivery carrier, the stability in blood is important for TNAs efficacy in biological systems because it allows these therapeutics to reach the target cell. Our data suggest that the long double-stranded structure of the functional RNA fibers would allow them to circulate longer and reach the cells of choice. Melting temperatures (Tm), measured for RNA fibers with and without functionalization, revealed several melting steps present in their melting profile (Figure 3E). The measured Tms of 42 ± 1 and 59 ± 0.1 °C correspond to the disruption of intermolecular (kissing loop interactions) and intramolecular (denaturation of the dumbbell) hydrogen bonds, respectively. The results confirm thermodynamic stability of RNA fibers at physiologically relevant temperatures and conditions.
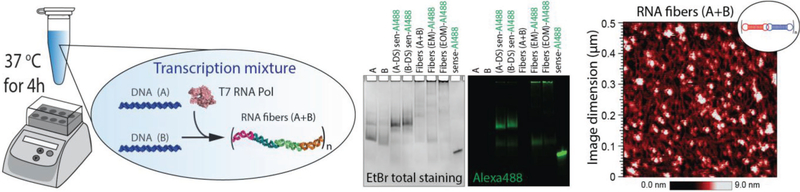
Cotranscriptional assembly of RNA nanoparticles[48–50] allows for their potential scaled-up production in mammalian cells. This represents an important feature that makes nano-TNAs amenable to assembly in patients’ cells and their further use as personalized medicine. We confirm the possibility for RNA fibers to self-assemble at isothermal conditions, during the T7 RNA polymerase-driven in vitro transcription (Figure 4).
Figure 4.
In vitro cotranscriptional assembly of RNA fibers confirmed by native-PAGE and AFM. EM and EOM denote functionalization at every monomer and every other monomer of A/B fibers, respectively. Note that for, native-PAGE, when fibers are formed, the monomer bands disappear. The blobs in AFM can be attributed to the composition of the transcription mixture.
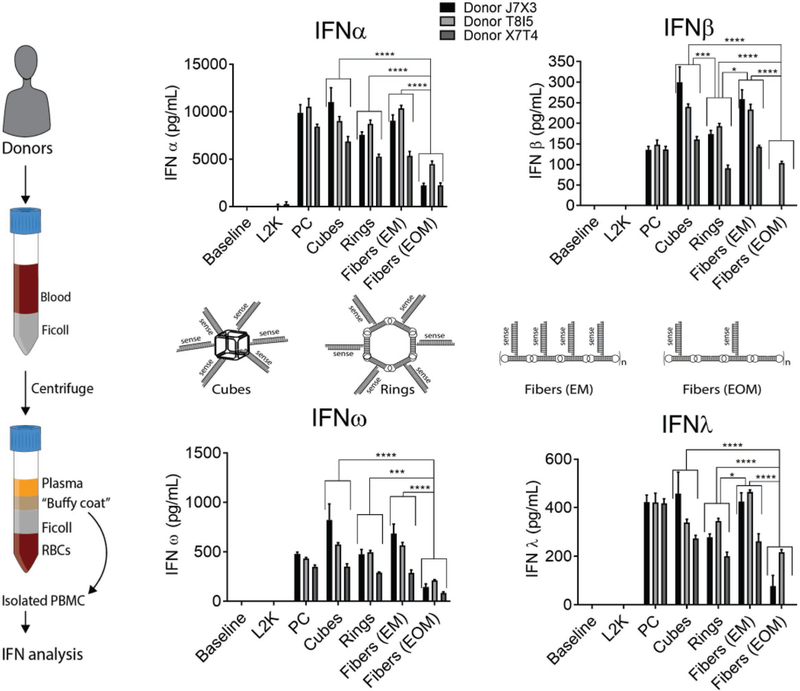
Next, we analyze the response of the human immune cells by exposing RNA fibers to PBMC cultures derived from the freshly collected blood of three healthy donors. When tested at equimolar concentration, RNA cubes,[22] RNA rings,[11] and RNA fibers with every monomer (EM) functionalized with DS RNAs induce high levels of type I (IFNα, IFNβ, and IFNω) and type III (IFNλ) interferon response, comparable to that of the assay positive control (PC) ODN2216, a CpG oligonucleotide known to be a potent IFN inducer (Figure 5, cubes vs rings vs fibers (EM) vs PC). However, when DS RNA moieties were attached to every other monomer (EOM) of the RNA fibers, the induction of IFN response significantly decreased (Figure 5, fibers (EM) vs fibers (EOM)). The data suggests that reducing the number of DS RNAs attached to the RNA fibers aids in the decrease of the undesirable proinflammatory responses while retaining the delivery of all functional siRNA.
Figure 5.
Interferon response in human PBMC. Isolation of PBMC from freshly collected blood samples is schematically shown in the left panel. RNA cubes, rings and fibers functionalized with siRNA (schematically shown in the middle panel) were tested for the ability to induce type I (IFNα, IFNβ, and IFNω) and type III (IFNλ) interferons. PBMCs from three healthy donors were used. All RNA assemblies were complexed with Lipofectamine 2000 (L2K) before addition to cells. L2K alone was included as control. Untreated cells were used as the baseline. Concentration of all tested particles was 10 × 10−9 m. Fibers with every monomer (EM) and every other monomer (EOM) functionalized were compared. Culture supernatants collected 24 h after the treatment were analyzed in duplicate by the multiplex ELISA. Percent coefficient of variation between individual replicates was less than 25. Error bars are ±SD (N = 3). Statistical analysis was performed by one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
To confirm the desired functionality of RNA fibers, we transfected human breast cancer cells expressing green fluorescent protein (GFP) with RNA fibers decorated with DS RNAs designed against GFP[42] (Figure 6A–C). GFP is used to visually confirm Dicer-assisted release of siRNAs from functionalized RNA fibers. The relative extent of GFP downregulation can be visually assessed by fluorescent microscopy and statistically analyzed by flow cytometry. The results confirm the efficient gene silencing triggered by DS RNA functionalized RNA fibers at sub-nanomolar concentrations. The fibers (EM) and (EOM) were compared, and although all functionalized fibers showed silencing, fibers with the 3′-end extension for DS RNA anti-sense (with DS RNA sense added) showed the best GFP knock-down. The less immunotoxic fibers (EOM), extended with antisense showed an increased silencing over the fibers (EM) functionalized with the sense extension. This may be due to the dicing position of the DS RNA and changes in thermodynamic asymmetry of the resulting siRNAs.[51,52] We also show that the presence of a cationic lipid-like carrier (Lipofectamine 2000 or L2K) is required for successful gene silencing (Figure S4, Supporting Information) and that nonfunctional RNA fibers or RNA fibers functionalized with scramble DS RNAs have no effect on GFP expression (Figure S4, Supporting Information), thus confirming that the GFP silencing can be solely attributed to the presence of correct DS RNAs in the fiber structure. Collectively, the data support our original objective of designing the RNA fibers with both effective functional capabilities of specific gene silencing and reduced immunostimulation. Colocalization confocal microscopy of fluorescently functionalized RNA fibers (labeled with both Alexa 488 and Alexa 546) in human breast cancer cells confirm that the RNA fibers maintain their structural integrity during cellular uptake (Figure 6D and Figure S3, Supporting Information). Once the RNA fibers enter the cytoplasm, Dicer cleaves off the siRNAs and separates the fluorophores, thus diluting the signal that can no longer be observed by microscopy (data not shown).
Figure 6.
Delivery of RNA fibers to human breast cancer cells triggers RNA interference. A) AFM images of different fibers functionalized at every monomer (EM) or every other monomer (EOM) with DS RNAs designed to silence GFP. B) Microscope images of human breast cancer cells expressing GFP before and after transfections with RNA fibers. C) Dose-dependent silencing efficiencies of GFP for different fibers introduced in picomolar range. D) Intracellular structural integrity of RNA fibers simultaneously labeled with two different fluorophores is confirmed by confocal microscopy colocalization experiment. E) Relative silencing efficiencies of functionalized RNA fibers (EOM) compared to functionalized RNA cubes and rings functionalized with six DS RNAs each. All assemblies in E were tested at 10 × 10−9 m concentrations as in Figure 4.
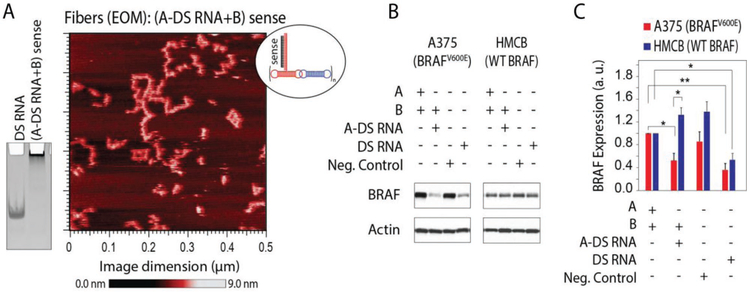
To demonstrate therapeutic potential, we study RNA fibers functionalized with DS RNAs designed to target the mutant BRAFV600E oncogene (Figure 7 and Figure S5, Supporting Information). BRAF encodes a serine-threonine kinase that is a component of the MAP kinase signaling pathway. BRAF mutations are detected in ≈50% of cutaneous melanomas and represent the most common genetic event in this disease.[53,54] The clear majority (90%) of BRAF mutations result in substitutions for valine at residue 600, most commonly with glutamic acid (V600E), and substantially increases BRAF kinase activity that leads to constitutive activation of the MAP kinase pathway promoting uncontrolled proliferation. The clinical development of selective small-molecule inhibitors of BRAFV600E proteins (e.g., vemurafenib) demonstrate significant improvements in clinical response and increased free survival rates compared to chemotherapy. We designed RNA fibers to specifically target only the mutated BRAFV600E by first converting the verified mutant specific siRNA[55] into the DS RNAs and then using these sequences for functionalization of RNA fibers. Interestingly, BRAFV600E DS RNAs target both mutant and WT BRAF, whereas BRAFV600E RNA fibers are efficient in targeting mutant BRAFV600E rather than WT BRAF (Figure 7B,C). This can be explained by the possible dicing of both sides of free DS RNAs resulting in some siRNAs not specific to the mutations. However, DS RNAs attached to the RNA fibers limit the possibility of dicing just to one end of DS RNA, thus producing the BRAFV600E mutant sensitive siRNAs.
Figure 7.
RNA fibers designed to specifically downregulate expression of mutated BRAFV600E but not the wild type (WT) BRAF. A) AFM image of RNA fibers with DS RNAs at every other monomer (EOM) designed against mutant BRAFV600E and a native-PAGE showing RNA fibers assembly. B,C) Specific gene silencing triggered by RNA fibers (0.5 × 10−9 M) in human melanoma cells compared to equimolar DS RNA. Relative BRAF silencing is normalized to a house keeping gene, actin. Shown are the results for RNA fibers transfected into cell line A375 with BRAFV600E and HMCB cells with the WT BRAF gene. Error bars are ±SEM (N = 3). Statistical analysis was performed by one-way ANOVA (*p < 0.05, **p < 0.01).
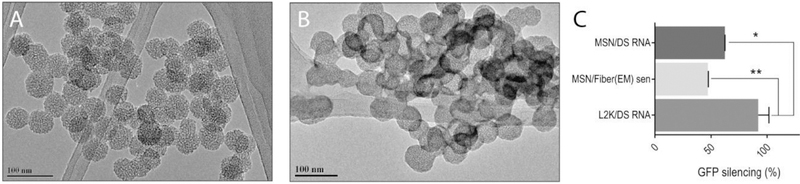
It is well known that neither L2K nor DharmaFECT 1 is suitable for biomedical application due to their in vivo toxicities. Therefore, in an attempt to make a clinically relevant formulation and to test if the delivery agent influences the functionality of RNA fibers, we chose mesoporous silica-based nanoparticles (MSNs) as an alternative non-lipid carrier. MSNs have demonstrated tremendous potential for delivering active pharmaceuticals[56–59] and TNAs[58,60] targeting numerous diseases. Versatility of this delivery platform relies on MSNs’ high surface area, tunable surface chemistry, biocompatibility, and well-defined structure of pores. To promote the delivery of RNA fibers, we develop a multistep synthetic approach, modifying the surface of MSNs with polyethylene imine (PEI) and polyethylene glycol (PEG) polymer chains.[61,62] The amine groups in the PEI polymer are protonated under physiological conditions rendering an overall positive charge on the surface of the carrier (Table S2, Supporting Information) and enabling the loading of negatively charged RNA fibers via electrostatic interactions. Moreover, it has been shown that PEI promotes the endo-lysosomal escape (“proton-sponge” effect) critical for the RNAi therapeutic outcome.[62,63] Finally, the modification of PEI-MSNs with PEG polymer imparts “stealth-like” properties to the RNA fiber loaded MSNs protecting the platform from immune recognition and nucleases while increasing the blood circulation time in vivo.[64] The newly synthesized PEG-MSNs are characterized by transmission electron microscopy (TEM), dynamic light scattering (DLS), and measured ζ-potential. The average diameter of the PEG-MSNs is determined to be 40 ± 5 nm (average of 40 particles) based on TEM results (Figure 8A). The loading of RNA fibers does not alter the size of the PEG-MSNs, with average diameter of 41 ± 3 nm (Figure 8B). Additionally, the higher contrast observed on the edges of the loaded PEG-MSNs can be associated with RNA fibers.
Figure 8.
Delivery of RNA fibers with MSN. A) TEM image of PEG-MSN. B) TEM image of PEG-MSN with RNA fibers. C) GFP-silencing efficiencies for functionalized RNA fibers and for DS RNA, with either PEG-MSN or L2K (control) used as the transfection vector. Statistical analysis was performed by one-way ANOVA (*p < 0.05, **p < 0.01).
The analysis of the hydrodynamic diameter of the PEG-MSNs (157 ± 15 nm) show a slight aggregation under physiological conditions due to the reduced electrostatic repulsion. The changes on the ζ-potential values through the different functionalization steps for the synthesis of the PEG-MSN confirm the successful modifications of the material (Table S2, Supporting Information). The electrostatically driven binding between the negatively charged phosphate backbone of nucleic acids and positive amine groups on the PEG-MSN is evaluated at different N/P ratios (N = number of moles of free amines on MSN as determined by a ninhydrin assay;[65] P = number of moles of phosphates) by electrophoretic mobility shift assay (Figure S6, Supporting Information). The optimal N/P ratio is determined to be 10 and then used for all subsequent studies. The efficient internalization of RNA fibers (labeled with Alexa 546) by PEG-MSN (labeled with FITC) is confirmed by flow cytometry (Figure S7, Supporting Information) as well as subsequent GFP silencing (Figure 8C). While the silencing efficiency is higher for L2K, the overall data suggest that the PEG-MSNs could be used as an alternative carrier for biomedical applications and further in vivo studies[66] need to be carried out.
In summary, programmable RNA fibers present modular scaffolds that are easy to functionalize for coordinating siRNAs, fluorophores, and other small molecules and their controlled delivery to human cells. The success of the very first RNA interference therapeutic agent (Patisiran[2]), approved by FDA on August 10th 2018, makes all further developments within this innovative technology timely and of a great importance. The immunostimulatory properties of DS RNA functionalized RNA fibers can be optimized by adding DS RNAs to the nanoscaffold in the alternating mode. This modular design and the unique shape of the scaffold allows the researchers to finetune the immunorecognition of RNA fibers to optimize their immunological safety and therapeutic efficacy. Our results suggest the possibility for RNA fibers to become a pharmaceutical option for personalized medicine as we demonstrate that the BRAFV600E silencing with RNA fibers has the potential to be used topically to treat melanoma in situ. Treatment using RNA fibers presents potentially lower toxicity than DS RNA alone since fibers target only the mutant BRAF in tumor cells and not WT BRAF in normal cells. The use of functional RNA fibers that stay intact in cells will provide a higher concentration and desired stoichiometry of therapeutic siRNAs locally and improve the loading of RNAi machinery present only in specific cytoplasmic locations.[67] Most importantly, the precisely controlled therapeutic composition of the functional RNA fibers can be easily altered by changing the functionalized monomers.[41] We further suggest that the ability to control the amounts of type I IFNs that the cells produce in response to RNA fibers, as demonstrated in the present study, could also be used as additional therapeutic modality, thereby contributing to the gene-silencing function of these constructs. This hypothesis stems from the known role of the type I IFN response in mobilizing the body’s natural immunity against tumors. Recombinant IFNs were shown to improve cancer therapy due to their ability to make the tumors “hot,” i.e., more responsive to immunotherapies.[68] The RNA fibers with controlled immunostimulation may serve as a source of natural IFN response produced by the body’s own cells, as opposed to the recombinant IFNs delivered from external sources and associated with systemic toxicity.[69] Moreover, the unique shape of RNA fibers somehow mimics double-stranded DNA plasmids and therefore the delivery agents that are efficiently used for plasmids in vivo may become suitable for RNA fibers. This further expands the possibilities of RNA fibers as nanoscaffolds since they can be functionalized and size-modified to work in conjunction with other delivery methods. These hypotheses warrant further studies of RNA fibers in conjunction with different delivery carriers as well as towards future immunotherapies.
3. Experimental Section
Molecular Dynamics Simulations
The replica exchange simulations[70] were used to model the structure of A and B monomers both alone and functionalized with DS sense and antisense RNA. Eight replicas were run at the following temperatures 0.200, 0.208, 0.214, 0.220, 0.225, 0.230, 0.235, and 0.240, where temperatures are expressed in kcal (mol kB)−1 units. Each simulation was run for one million steps. The trajectory snapshots were saved every 1000 steps. Following the simulations, 100 of the snapshots with the least energy from each trajectory were selected and clustered by root mean square deviation (RMSD). The centroids of clusters were chosen as representative structures. To model functionalized and non-functionalized RNA fibers, DMD simulations were performed at constant temperature T = 0.25 kcal (mol kB)−1 run for one millions steps. Each fiber in our simulations consists of 20 monomers.
Fibers Preparation and Assembly
All RNA sequences used in this project are listed in the Supporting Information. Individual DNAs coding RNA fibers, cubes, and rings, short DS RNA sense, and antisense strands and all fluorescently labeled oligos were purchased from Integrated DNA Technologies Inc (IDT). DNA oligos containing T7 RNA Polymerase promoter sequences and coding for RNA strands were amplified by the polymerase chain reaction using MyTaq Mix (Bioline), purified using Zymo Research spin columns, transcribed in vitro with T7 polymerase (80 × 10−3 m HEPES-KOH, pH 7.5; 2.5 × 10−3 m spermidine; 50 × 10−3 m DTT; 25 × 10−3 m MgCl2; 5 × 10−3 m NTPs), and purified by 8 M urea-PAGE (15% polyacrylamide gel electrophoresis). RNA bands were visualized with a UV lamp (short wavelength), cut, eluted overnight in 300 × 10−3 m NaCl, 89 × 10−3 m Tris-borate (pH 8.2), 2 × 10−3 m EDTA, precipitated in 2.5 volumes of 100% ethanol, rinsed with 90% ethanol, vacuum dried, and dissolved in ultrapure water (17.8 MΩ cm). For cotranscriptional assembly, DNA oligos containing T7 RNA Polymerase promoter sequences for individual monomers were mixed and added to the transcription mixture. Transcriptions were stopped after 4 h of incubation at 37 °C by adding DNase and additional incubation for 30 min. Cotranscriptional assemblies were visualized by native-PAGE.
RNA fibers and rings were assembled by mixing monomers at an equimolar ratio, heating to 95 °C for 2 min, snap cooling on ice for 2 min, adding 20% volume of 5× assembly buffer (final concentration: 89 × 10−3 m tris-borate (pH 8.2), 50 × 10−3 M KCl, 2 × 10−3 M MgCl2), and then incubated at room temperature for 20 min.
RNA cubes were assembled by mixing all constituent strands at an equimolar ratio and heated to 95 °C for 2 min. The samples were then snap cooled to 45 °C and incubated for 2 min. Finally, 5× assembly buffer was added to reach the desired concentration and the samples were incubated at 45 °C for an additional 30 min.
Following all assembly protocols, assemblies mixed with gel loading buffer (assembly buffer with 50% glycerol) were confirmed via nondenaturing polyacrylamide gel electrophoresis, native-PAGE (8% acrylamide (37.5:1), 89 × 10−3 m tris-borate (pH 8.2), 2 × 10−3 m MgCl2) visualized with a Bio-Rad ChemiDoc MP System using total staining with ethidium bromide or fluorescently labeled oligonucleotides. Gels were run in a cold room (4 °C) at 300 V, 150 mA for 30 min.
Blood Stability Assays
Alexa-546 labeled fibers (0.5 × 10−3 m final) were incubated with 10% (v/v) human blood serum at 37 °C. At each time point, 5 μL of the mixture was aliquoted out, mixed with the gel loading buffer, and placed on dry ice to stop the reaction. All samples were loaded in reverse order and analyzed on native-PAGE. The data obtained was plot using OriginPro2016 software. The half-life (t1/2) was obtained by using the first-order rate equation: γ = A0 e −kt1/2, where A0 = 1, k is the rate constant, and t1/2 is the half-life. OriginPro2016 provided the uncertainty of k, which was used in error propagation to determine the uncertainty of t1/2.
Atomic Force Microscopy
All samples (1 × 10−6 m) were diluted either 5× or 10× in assembly buffer, then 6 μL of diluted sample was added to freshly cleaved mica and allowed to adsorb for ≈30 s. Next 25 μL of assembly buffer was added onto the sample on the mica, then 25 μL of 60 × 10−3 M NiCl2 was added onto the mixture. Finally, 25 μL of assembly buffer was added to the tip and the sample was imaged. Images were obtained with a Digital Instruments Multimode AFM, equipped with a Nanoscope III controller. Sharp Nitride Lever (SNL) tips from Bruker with a nominal spring constant of 0.24 N m−1 were used for imaging, with a drive frequency of 9–10 kHz.
UV-Melting Experiments
To measure the melting temperatures (Tm) of RNA fibers (at 0.5 × 10−6 m), temperature-dependent absorption measurements were recorded at 260 nm on an Agilent 8453 spectrophotometer coupled with the Agilent 89 090 Peltier Temperature Controller.
In Vitro Immunology
The study was performed using peripheral blood mononuclear cells isolated from the whole blood of healthy donor volunteers using the Ficoll Paque gradient density method described earlier.[71] Donor blood was collected into vacutainers containing Li-heparin as anticoagulant under the NCI protocol OH-9-C-N046. Cells were cultured in RPMI supplemented with 10% of heat inactivated fetal bovine serum (Hyclone Labs, GE Healthcare Life Sciences, Logan, UT). Prior to treatment, all particles were complexed with lipofectamine 2000 as follows. First, 20 μL of RNA fiber, RNA ring, or RNA cube stocks with the final concentration of 1 × 10−6 m were mixed with 4 μL of the commercial stock of Lipofectamine 2000 (L2K) purchased from ThermoFisher. The mixture was incubated at room temperature for 30 min. Following this, 376 μL of the serum-free Opti-MEM (ThermoFisher) medium was added to the complexes and 40 μL of this working solution was used for the treatment of PBMCs in complete culture medium containing 10% heat-inactivated fetal bovine serum. The final concentration of RNA constructs in cell cultures was 10 × 10−9 m. For the samples assessing L2K alone, buffer used to store the stocks of RNA constructs (89 × 10−3 m tris-borate (pH 8.2), 50 × 10−3 m KCl, 2 × 10−3 M MgCl2) was used in place of the fiber constructs, and the procedure described above was performed to allow the equivalent manipulation and volume ratios as in the samples treated with RNA fibers. The positive control oligonucleotide was added directly to the cells treated with the negative control mixture. The samples where cells were treated with a negative control only were considered as L2K alone. The untreated cells were denoted as baseline. Incubation with particles and controls continued for 24 h, then the supernatants were collected and analyzed for the presence of type I interferons by multiplex ELISA (Quansys BioSciences Inc., Logan, UT). Since endotoxin may confound the results of cytokine secretion studies, all particles were screened using kinetic turbidity LAL assay for potential contamination. Endotoxin was undetectable (<0.5 EU mL−1) in all tested particles (Table S1, Supporting Information). Reagents and PyrosFlex instrument from Associates of Cape Code Inc. (East Falmouth, MA) were used for the LAL assays.
Transfection of Human Cell Lines and Gene Silencing Experiments
For GFP silencing experiments, human breast cancer cells (MDA-MB-231 expressing GFP) were grown in D-MEM media supplemented with 10% FBS at 37 °C in a 5% CO2 incubator at 2 × 104 or 3 × 104 cells per well in either 12- or 24-well plates, respectively. The media were supplemented with 10% FBS and penicillin–streptomycin. All transfection experiments were carried out using Lipofectamine 2000 (L2K). The RNA ring, RNA cube, RNA fiber (EM), RNA fiber (EOM), and DS RNA were incubated with L2K at RT for 30 min, then Opti-MEM was added, for a final concentration of 10 × 10−9 m. The cells were incubated for 4 h followed by a media change with D-MEM. Dilutions were made prior to incubation with L2K for other tested concentrations. Silencing of GFP was assessed 72 h later using the BD Accuri C6 flow cytometer. Nontreated cells were used as a positive control. At least 10 000 events were collected for each sample and analyzed. CellQuest or the CFlow Sampler software were used to retrieve the geometric mean fluorescence intensity (gMFI) and the standard error of the mean.
BRAFV600E homozygous A375 melanoma cells were transfected with BRAFV600E DS RNAs or functionalized RNA fibers. HMCB melanoma cells, which carry the wild-type (WT) BRAF, were used as a nontarget controls. Briefly, A375 and HMCB were plated at 6 × 104 cells per well (12-well plate). On the next day, DS RNAs or (A-DS RNA) sense fibers were incubated with Opti-MEM for 5 min at 25 °C (final volume of 50 μL) and 2 μL DharmaFECT 1 (ThermoFisher) with Opti-MEM for 5 min at 25 °C (final volume of 50 μL). After incubation, RNA and DharmaFECT 1 were mixed and incubated for 20 min at 25 °C (final volume of 100 μL). After incubation, 100 μL per well of RNA/DharmaFECT solution was added for 48 h, after which cells were harvested with RIPA Buffer (10 × 10−3 m Tris pH 7.4, 150 × 10−3 m NaCl, 1 × 10−3 m EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton-X). Whole cell lysates were analyzed by immunoblotting with antibodies to BRAF (Santa Cruz) and beta-actin (Santa Cruz).
Nonfunctionalized RNA fibers were used as a negative control in all gene silencing experiments. Additionally, RNA fibers carrying Mission Universal Negative Control siRNAs (Sigma-Aldrich, SIC001) were designed as a negative control. These fibers were designed to be physically identical to the therapeutic fibers tested, but carry scramble siRNAs that are known to have no homology to any known human, mouse, or rat gene.
Fluorescence Microscopy
To assess the intracellular integrity of RNA fibers, colocalization experiments were carried out using an LSM 710 confocal microscope (Carl Zeiss) with a 63×, 1.4 NA magnification lens. MDA-MB-231 (no GFP) cancer cells plated in glass bottom dishes (Ibidi, Madison) were transfected the next day with 100 × 10−9 m of the fluorescently labeled RNA fibers as indicated above. Upon the 4 h incubation at 37 °C, the cells were washed three times with PBS and incubated at room temperature for 20 min with a 4% paraformaldehyde solution to fix the samples. The samples were washed again in PBS and stored at 4 °C prior to visualization. For Alexa 488 imaging, a 488 nm line of an argon laser was used as excitation and the emission was collected between 493 and 557 nm. For Alexa 546 imaging, a DPSS 561 laser was used for excitation and emission was collected between 566 and 680 nm. To ensure that there is no bleed through with the conditions used, cells transfected with RNA fibers containing only Alexa 488 or only Alexa 546 are used as controls.
Preparation of PEG-MSNs
Tetraethylorthosilicate (TEOS, 98%), cetyltrimethylammonium bromide (CTAB, 95%), fluorescein isothiocyanate (FITC, 90%), 3-trihydroxysilylpropyl methylphosphonate (THPMP)s, and aminopropyltriethoxysilane (APTES) were from Sigma (St. Louis, MO). Polyethylenimine (PEI, MW 10 kD) was purchased from Alfa Aesar. monomethoxyPEG-NHS (mPEG-NHS) was procured from Creative PEGworks. MSNs were synthesized by a surfactant-templated approach, following our previous work.[72] Briefly, 0.78 g of CTAB was dissolved in a solution of 21.6 mL water, and 3.32 mL ethanol. To this, 0.1 mL diethylamine (0.4 × 10−3 m) was added and, the solution was heated to 60 °C. TEOS (2.19 mL) was added dropwise into the aqueous solution of CTAB, and the reaction was allowed to run for 18–21 h at 60 °C to obtain as-made MSNs. Later, the surface of MSNs was modified by grafting methylphoshonate groups to increase the negative charge. For phosphonate modification, THPMP (0.3 mL diluted in 1.0 mL water) was added dropwise to the as-synthesized MSNs and stirred for 6–8 h at 60 °C. The CTAB surfactant were then removed from the pores using an acidic wash approach. They were redispersed in acidic methanol and heated to 60 °C, to afford negatively charged MSNs. To incorporate fluorescein molecules in the silicate framework, fluorescein-modified silane was first synthesized and added to negatively charged MSN dispersion. To synthesize fluorescein-modified silane, 7.3 μL of APTES was mixed with 3.3 mg of FITC in 0.6 mL of dry DMF and stirred for 2 h under inert atmosphere. The fluorescein-modified silane was then mixed with the heated dispersion of PgMSN (150 mg), and stirred overnight. The FITC labeled MSNs were then centrifuged and washed with methanol.
After removal of the surfactant, MSNs were coated with PEI polymer to switch the surface charge to highly positive values (PEI-PgMSNs). To perform PEI coating, 10 mg of negatively charged MSNs was dispersed in a solution containing 5 mg of PEI in ethanol. After stirring for 2 h at room temperature, PEI-coated particles (PEI-MSN) were washed with PBS. The amount of polymer coated onto the surface of MSNs was 20 wt% as determined by the ninhydrin test. Finally, the surface of PEI-MSNs was functionalized with PEG (MW = 2 K) chains through a conjugation reaction with mPEG-NHS (Scheme S1, Supporting Information). To carry out the PEGylation, 10 mg of PEI-MSN was dispersed in absolute ethanol. To this dispersion, a solution of mPEG-NHS (3 mg mL−1) in ethanol was added. After stirring for 24 h, the PEGylated PEI-PgMSN (PEG-MSN) were washed with PBS, and used for further studies.
Optimization of PEG-MSNs for Gene Delivery
In order to optimize the binding affinity of the PEG-MSNs with gene/DNA/RNA material, the ds-DNA (bp = 38) was complexed with PEG- MSN at different N/P ratios (N/P = 0.5, 1, 2, 5, 10, 20, and 40). The mixture of PEG-MSNs and ds-DNA was mixed in assembly buffer for 30 min at room temperature. The agarose gel retardation assay was performed following a procedure reported in the literature with slight modifications.[61]
Transmission Electron Microscopy
The nanoparticles were dispersed in water/buffer, and the suspension (5 μL) was deposited on a carboncoated copper grid and dried for at least 24 h. For PEG-MSN-RNA fiber sample, second step of negative staining was performed, using phosphotungstic acid,[73] wherein a drop of negative staining agent was deposited, once the sample was dried onto the copper grid. Images were taken with a JEOL JEM 2100 LaB6 TEM. Particle size distributions were calculated by ImageJ, from different images taken over different quartiles.
Dynamic Light Scattering
The hydrodynamic diameter, and the surface zeta potential of the nanoparticles was measured by DLS and ζ-potential measurements were carried out using a Malvern Instrument Zetasizer Nano (red laser 633 nm) (Malvern Instrument Ltd., Malvern, UK). The samples were prepared by suspending the nanoparticles in phosphate buffer solution (0.1 m).
Cellular Uptake of PEG-MSN-RNA Fiber
Cellular uptake of PEGMSN-RNA fiber was examined by flow cytometry. Briefly, MDA-MB-231 cells were seeded in 24-well plates with a density of 2.5 × 104 cells per well, and incubated with fluorescein-labeled PEG–MSN, Alexa 546 RNA fiber, and dilabeled (fluorescein/Alex 546) PEG-MSN-RNA fiber with 30 μg mL−1, and 30 × 10−9 m as final concentration of the MSNs and RNA fiber, respectively. During the whole process, the cells were maintained in D-MEM media. After 24 h of incubation time period, the cells were collected and tested for fluorescein and Alexa546 using the flow cytometer.
Gene Silencing In Vitro with PEG-PEI-MSN as Transfection Agent
For evaluation of silencing efficiency, MDA MB 231 expressing GFP were cultured following the same protocol as mentioned above. Briefly, MDA-MB-231-GFP cells were seeded in 24-well plates with a density of 2.5 × 104 cells per well and incubated with the dilabeled PEG-MSN-RNA fiber, at two different concentration (30 × 10−9, and 60 × 10−9 m), and with PEG-MSN as a control. The cells were incubated for 72 h before they were harvested. Throughout the process the cells were consistently maintained in D-MEM media. The cells were collected, and tested for GFP using the flow cytometer.
Statistics
Statistical analysis was done by one-way analysis of variance (ANOVA) using GraphPad Prism software. All column means were compared by Tukey’s multiple comparison test. A p value of less than 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM120487 (to K.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. L.R. was supported in part by funding provided by NSF-REU and DOD-ASSURE under NSF Grant No. CHE 1460867. This project has been funded in part with federal funds from the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (J.S. and P.F.J.) and the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E (M.V., A.S., and M.A.D.) The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work was also supported by the U.S. National Science Foundation under CAREER grant DMR-1450747 (to E.F.). N.V.D. also acknowledges support from NIH grants R01-GM114015, R01-GM064803, and R01-GM123247.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Lauren Rackley, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
Jaimie Marie Stewart, Department of Bioengineering, University of California, Riverside, CA 92521, USA.
Jacqueline Salotti, Mouse Cancer Genetics Program, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Andrey Krokhotin, Department of Biochemistry and Biophysics, University of North Carolina Chapel Hill, NC 27514, USA.
Ankit Shah, Nanotechnology Characterization Lab, Cancer Research Technology Program, Frederick National Laboratory for Cancer Research, Sponsored by the National Cancer Institute, Frederick, MD 21702, USA.
Justin R. Halman, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA
Ridhima Juneja, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
Jaclyn Smollett, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
Lauren Lee, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
Kyle Roark, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
Mathias Viard, Cancer and Inflammation Program, Leidos Biomedical Research Inc., Frederick National Laboratory for Cancer Research, Frederick, MD 21702, USA.
Mubin Tarannum, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
Juan Vivero-Escoto, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
Peter F. Johnson, Mouse Cancer Genetics Program, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA
Marina A. Dobrovolskaia, Nanotechnology Characterization Lab, Cancer Research Technology Program, Frederick National Laboratory for Cancer Research, Sponsored by the National Cancer Institute, Frederick, MD 21702, USA
Nikolay V. Dokholyan, Department of Biochemistry and Biophysics, University of North Carolina Chapel Hill, NC 27514, USA Department of Pharmacology, Department of Biochemistry & Molecular Biology, Penn State College of Medicine Hershey, PA 17033, USA.
Elisa Franco, Department of Mechanical Engineering, University of California, Riverside, CA 92521, USA.
Kirill A. Afonin, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA The Center for Biomedical Engineering and Science, University of North Carolina at Charlotte, Charlotte, NC 28223, USA.
References
- [1].Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC, Nature 1998, 391, 806. [DOI] [PubMed] [Google Scholar]
- [2].Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, Lin KP, Vita G, Attarian S, Plante-Bordeneuve V, Mezei MM, Campistol JM, Buades J, Brannagan TH 3rd, Kim BJ, Oh J, Parman Y, Sekijima Y, Hawkins PN, Solomon SD, Polydefkis M, Dyck PJ, Gandhi PJ, Goyal S, Chen J, Strahs AL, Nochur SV, Sweetser MT, Garg PP, Vaishnaw AK, Gollob JA, Suhr OB, Engl N J. Med 2018, 379, 11. [Google Scholar]
- [3].Burnett JC, Rossi JJ, Chem. Biol 2012, 19, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sullenger BA, Nair S, Science 2016, 352, 1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alvarez-Salas LM, Curr. Top. Med. Chem 2008, 8, 1379. [DOI] [PubMed] [Google Scholar]
- [6].Grimm D, Kay MA, Mol. Ther 2007, 15, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, Li C, Haque F, Liang XJ, Guo P, Nano Today 2015, 10, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M, Shapiro BA, Acc. Chem. Res 2014, 47, 1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jasinski D, Haque F, Binzel DW, Guo P, ACS Nano 2017, 11, 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guo P, Nat. Nanotechnol 2010, 5, 833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, Heldman E, Reiser J, Chiu W, Freed EO, Shapiro BA, Nano Lett. 2014, 14, 5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, Mustain W, Valentino J, Guo P, Evers BM, ACS Nano 2015, 9, 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, Xu R, Guo P, ACS Nano 2015, 9, 9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Binzel DW, Shu Y, Li H, Sun M, Zhang Q, Shu D, Guo B, Guo P, Mol. Ther 2016, 24, 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG, Nat. Nanotechnol 2012, 7, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shibata T, Fujita Y, Ohno H, Suzuki Y, Hayashi K, Komatsu KR, Kawasaki S, Hidaka K, Yonehara S, Sugiyama H, Endo M, Saito H, Nat. Commun 2017, 8, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Grabow WW, Jaeger L, Acc. Chem. Res 2014, 47, 1871. [DOI] [PubMed] [Google Scholar]
- [18].Geary C, Chworos A, Verzemnieks E, Voss NR, Jaeger L, Nano Lett. 2017, 17, 7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shu D, Shu Y, Haque F, Abdelmawla S, Guo P, Nat. Nanotechnol 2011, 6, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee TJ, Haque F, Vieweger M, Yoo JY, Kaur B, Guo P, Croce CM, Methods Mol. Biol 2015, 1297, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Feng L, Li SK, Liu H, Liu CY, LaSance K, Haque F, Shu D, Guo P, Pharm. Res. 2014, 31, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Afonin KA, Viard M, Kagiampakis I, Case CL, Dobrovolskaia MA, Hofmann J, Vrzak A, Kireeva M, Kasprzak WK, KewalRamani VN, Shapiro BA, ACS Nano 2015, 9, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stewart JM, Viard M, Subramanian HK, Roark BK, Afonin KA, Franco E, Nanoscale 2016, 8, 17542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bui MN, Brittany Johnson M, Viard M, Satterwhite E, Martins AN, Li Z, Marriott I, Afonin KA, Khisamutdinov EF, Nanomed.: Nanotechnol. Biol. Med 2017, 13, 1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Afonin KA, Viard M, Martins AN, Lockett SJ, Maciag AE, Freed EO, Heldman E, Jaeger L, Blumenthal R, Shapiro BA, Nat. Nanotechnol 2013, 8, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dobrovolskaia MA, DNA RNA Nanotechnol. 2016, 3, 1. [Google Scholar]
- [27].Dobrovolskaia MA, J. Controlled Release 2015, 220, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dobrovolskaia MA, McNeil SE, Expert Opin. Drug Delivery 2015, 12, 1163. [DOI] [PubMed] [Google Scholar]
- [29].Dobrovolskaia MA, McNeil SE, Expert Opin. Biol. Ther 2015, 15, 1023. [DOI] [PubMed] [Google Scholar]
- [30].Kornbrust D, Cavagnaro J, Levin A, Foy J, Pavco P, Gamba-Vitalo C, Guimond A, Nucleic Acid Ther. 2013, 23, 21. [DOI] [PubMed] [Google Scholar]
- [31].Levin AA, Biochim. Biophys. Acta, Gene Struct. Expression 1999, 1489, 69. [DOI] [PubMed] [Google Scholar]
- [32].Levin AA, Engl N J. Med 2017, 376, 86. [DOI] [PubMed] [Google Scholar]
- [33].Lindow M, Vornlocher HP, Riley D, Kornbrust DJ, Burchard J, Whiteley LO, Kamens J, Thompson JD, Nochur S, Younis H, Bartz S, Parry J, Ferrari N, Henry SP, Levin AA, Nat. Biotechnol 2012, 30, 920. [DOI] [PubMed] [Google Scholar]
- [34].Schubert D, Levin AA, Kornbrust D, Berman CL, Cavagnaro J, Henry S, Seguin R, Ferrari N, Shrewsbury SB, Nucleic Acid Ther. 2012, 22, 211. [DOI] [PubMed] [Google Scholar]
- [35].Hong E, Halman JR, Shah AB, Khisamutdinov EF, Dobrovolskaia MA, Afonin KA, Nano Lett. 2018, 18, 4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L, Nano Lett. 2011, 11, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Halman JR, Satterwhite E, Roark B, Chandler M, Viard M, Ivanina A, Bindewald E, Kasprzak WK, Panigaj M, Bui MN, Lu JS, Miller J, Khisamutdinov EF, Shapiro BA, Dobrovolskaia MA, Afonin KA, Nucleic Acids Res. 2017, 45, 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Johnson MB, Halman JR, Satterwhite E, Zakharov AV, Bui MN, Benkato K, Goldsworthy V, Kim T, Hong E, Dobrovolskaia MA, Khisamutdinov EF, Marriott I, Afonin KA, Small 2017, 13, 1701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39] [39].Laughrea M, Jette L, Biochemistry 1996, 35, 1589. [DOI] [PubMed] [Google Scholar]
- [40].Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P, Nat. Struct. Biol 2001, 8, 1064. [DOI] [PubMed] [Google Scholar]
- [41].Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L, Nat. Protoc 2011, 6, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA, Nucleic Acids Res. 2005, 33, 4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV, RNA 2008, 14, 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Krokhotin A, Houlihan K, Dokholyan NV, Bioinformatics 2015, 31, 2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Proctor EA, Ding F, Dokholyan NV, J. Mol. Biol 2011, 408, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Proctor EA, Dokholyan NV, Curr. Opin. Struct. Biol 2016, 37, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shirvanyants D, Ding F, Tsao D, Ramachandran S, Dokholyan NV, J. Phys. Chem. B 2012, 116, 8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L, Shapiro BA, Nano Lett. 2012, 12, 5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L, Nat. Nanotechnol 2010, 5, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Geary C, Rothemund PWK, Andersen ES, Science 2014, 345, 799. [DOI] [PubMed] [Google Scholar]
- [51].Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD, Cell 2003, 115, 199. [DOI] [PubMed] [Google Scholar]
- [52].Khvorova A, Reynolds A, Jayasena SD, Cell 2003, 115, 209. [DOI] [PubMed] [Google Scholar]
- [53].Bucheit AD, Davies MA, Biochem. Pharmacol 2014, 87, 381. [DOI] [PubMed] [Google Scholar]
- [54].Poulikakos PI, Rosen N, Cancer Cell 2011, 19, 11. [DOI] [PubMed] [Google Scholar]
- [55].Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA, Cancer Res. 2003, 63, 5198. [PubMed] [Google Scholar]
- [56].Slowing II, Vivero-Escoto JL, Wu CW, Lin VS, Adv. Drug Delivery Rev. 2008, 60, 1278. [DOI] [PubMed] [Google Scholar]
- [57].Alvarez-Berrios MP, Sosa-Cintron N, Rodriguez-Lugo M, Juneja R, Vivero-Escoto JL, Chem J 2016, 2672740/1–2672740/15, https://www.hindawi.com/journals/jchem/2016/2672740/.
- [58].Darvishi B, Farahmand L, Majidzadeh AK, Mol. Ther. – Nucleic Acids 2017, 7, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tang F, Li L, Chen D, Adv. Mater 2012, 24, 1504. [DOI] [PubMed] [Google Scholar]
- [60].Cha W, Fan R, Miao Y, Zhou Y, Qin C, Shan X, Wan X, Li J, Molecules 2017, 22, E782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Meng H, Liong M, Xia T, Li Z, Ji Z, Zink JI, Nel AE, ACS Nano 2010, 4, 4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xia T, Kovochich M, Liong M, Meng H, Kabehie S, George S, Zink JI, Nel AE, ACS Nano 2009, 3, 3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tu J, Wang T, Shi W, Wu G, Tian X, Wang Y, Ge D, Ren L, Biomaterials 2012, 33, 7903. [DOI] [PubMed] [Google Scholar]
- [64].Petros RA, De Simone JM, Nat. Rev. Drug Discovery 2010, 9, 615. [DOI] [PubMed] [Google Scholar]
- [65].Xu Z, Xu T, Cheng Y, Ma M, Xu P, Qu H, Wen L, Anal. Lett 2008, 41, 444. [Google Scholar]
- [66].Dreau D, Moore LJ, Alvarez-Berrios MP, Tarannum M, Mukherjee P, Vivero-Escoto JL, J. Biomed. Nanotechnol 2016, 12, 2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, Nakahara K, Preall JB, Bellare P, Sontheimer EJ, Carthew RW, Nat. Cell Biol 2009, 11, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Di Franco S, Turdo A, Todaro M, Stassi G, Front. Immunol 2017, 8, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Foa P, Massaro P, Caldiera S, LaTargia ML, Iurlo A, Clerici C, Fornier M, Bertoni F, Maiolo AT, Eur. J. Haematol 1998, 60, 273. [DOI] [PubMed] [Google Scholar]
- [70].Ding F, Tsao D, Nie H, Dokholyan NV, Structure 2008, 16, 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ilinskaya AN, Clogston JD, McNeil SE, Dobrovolskaia MA, Nanomed.: Nanotechnol., Biol., Med 2015, 11, 1925. [Google Scholar]
- [72].Walker WA, Tarannum M, Vivero-Escoto JL, J. Mater. Chem. B 2016, 4, 1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Smith KO, Melnick JL, Virology 1962, 17, 480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.